Abstract
Unlike other Salmonellae, the intracellular bacterial human pathogen Salmonella Typhi exhibits strict host specificity. The molecular bases for this restriction are unknown. Here we show that the expression of a single type III secretion system effector protein from broad-host Salmonella Typhimurium allows Salmonella Typhi to survive and replicate within macrophages and tissues from mice, a non-permissive host. We found that this effector proteolytically targets Rab32, which controls traffic to lysosome-related organelles in conjunction with components of the biogenesis of lysosome-related organelle complexes (BLOCs). RNAi-mediated depletion of Rab32 or of an essential component of a BLOC complex was sufficient to allow S. Typhi to survive within mouse macrophages. Furthermore, Salmonella Typhi was able to survive in macrophages from mice defective in BLOC components. These findings provide insight into the molecular bases of S. Typhi host restriction and uncover a previously unknown mechanism of pathogen control in macrophages.
Keywords: typhoid fever, Rab GTPases, Rab32, membrane traffic, bacterial pathogenesis, type III secretion, macrophages, innate immunity, lysosomes, lysosome-related organelles
The bacterial pathogen Salmonella enterica comprises thousands of serovars (i. e. variants that can be distinguished by their surface antigen composition) that as a whole can infect a large number of vertebrate species ranging from reptiles to humans (1, 2). While some serovars can infect a broad range of hosts, others are extremely host specific. This is the case of Salmonella enterica serovar Typhi (S. Typhi), which naturally can only infect humans although experimentally it can also infect higher primates (3). S. Typhi causes typhoid fever, a life-threatening systemic disease that every year kills 200,000 people worldwide (4-6). Although the genome sequences of several host-specific and broad-host Salmonellae are available, the molecular bases for this remarkable host adaptation are unknown and likely to be multifactorial (7, 8). It is believed that genome reduction most likely played a central role in the narrowing of S. Typhi’s host range (9). This host restriction is also manifested at the cellular level since, in contrast to human macrophages, S. Typhi is unable to survive within macrophages of mice, a non-permissive species (10, 11). The interaction of Salmonella enterica with host cells is largely determined by the activities of two type III secretion systems (T3SS), which deliver bacterial effector proteins into host cells to modulate a variety of cellular processes (12-15). We recently discovered that differences in the composition of the membrane compartment that harbors S. Typhi and S. Typhimurium is due to differences in the assortment of the T3SS effector proteins that they encode (16). While vacuoles containing S. Typhi efficiently recruit Rab29, those containing the broad-host serovar S. Typhimurium do not because its T3SS effector GtgE, absent from S. Typhi, proteolytically degrades it (16).
We hypothesized that the presence or absence of a Rab GTPase in the vacuoles harboring S. Typhi or S. Typhimurium must translate into fundamental differences in the intracellular biology of these pathogens, potentially influencing host-cell restriction. We expressed the S. Typhimurium T3SS effector GtgE in S. Typhi and examined its ability to survive within primary bone-marrow-derived macrophages (BMDM) obtained from mice, a non-permissive species (17). Surprisingly, we found that expression of gtgE significantly increased the ability of S. Typhi to survive within mouse macrophages (Fig. 1A and S1). While very few colony-forming units (c. f. u.) were recovered from mouse macrophages 48 hs after infection with wild-type S. Typhi, large numbers of c. f. u. were recovered from macrophages infected with S. Typhi expressing gtgE. In fact, the number of c. f. u. recovered from macrophages infected with S. Typhi expressing gtgE were equivalent to those recovered from macrophages infected with the broad-host serovar S. Typhimurium (Fig. 1A and S1). These results indicate that expression of a single effector protein from broad host Salmonellae allows S. Typhi to overcome host-cell restriction and survive in a non-permissive host cell.
Fig. 1. GtgE expression limits host-cell restriction.
(A) Survival of S. Typhi expressing GtgE in bone-marrow-derived mouse macrophages (BMDM). Macrophages were infected with S. Typhi (Ty), S. Typhi expressing GtgE (Ty GtgE), or S. Typhimurium (Tm). Cells were lysed at the indicated time points and c. f. u. enumerated. Values are means ± SEM of three independent measurements. p values (for the difference relative to values obtained from cells infected with S. Typhi) were determined by the Student’s t test: *<0.05; **<0.01; ***<0.001. (B) and (C) C. f. u. recovered from the spleens of C57BL/6 Nramp+/+ mice infected with S. Typhi (wt) or S. Typhi expressing GtgE (GtgE) 4 days after intraperitoneal (B) or oral (C) inoculation. Horizontal bars indicate the means. The p value was determined by the Wilcoxon rank sum test.
To investigate the consequences of overcoming host-cell restriction in the ability of S. Typhi to replicate within a non-permissive host, we infected mice with the S. Typhi strain expressing gtgE. We found a significantly higher number of c. f. u. in the tissues of mice infected with S. Typhi expressing gtgE compared to those infected with wild-type (Fig. 1B, 1C, and S2). These results indicate that expression of gtgE allows S. Typhi to overcome some of the host restriction barrier. Furthermore, these results suggest that the inability of S. Typhi to replicate within non-human hosts is at least in part due to its inability to survive within macrophages of the non-permissive species. However, additional factors must contribute to host restriction since the virulence of S. Typhi expressing GtgE did not match that of S. Typhimurium (18), which is highly virulent for mice.
To gain insight into the mechanisms by which GtgE allows S. Typhi to overcome the host-cell restriction barrier, we examined the effect of depleting Rab29, a target of its protease activity (16), on the ability of S. Typhi to survive within primary mouse BMDM. Surprisingly, we found that siRNA-mediated depletion of Rab29 had no effect on the ability of S. Typhi to survive in non-permissive macrophages (Fig. S3), which indicated that GtgE might have additional cellular target(s) for its protease activity responsible for its phenotype. We therefore examined the ability of GtgE to target other Rab GTPases. We found that, despite the broad conservation of residues surrounding the GtgE cleavage site in several GTPases (Fig. S4), GtgE was only able to target Rab32 and Rab38 (Fig. 2A, 2B, 2C, and S5). Consistent with these findings, phylogenetic analysis of all Rab-family GTPases indicated that Rab32 and Rab38 are the GTPases most highly related to Rab29 (Fig. 2D and S6). However, GtgE did not cleave Rab23, the next most highly related GTPase, nor did it cleave other GTPases phylogenetically close to Rab29, Rab32 or Rab38 (Fig. 2A, 2C, 2D, S4, S5, and S6). Consistent with these observations, Rab32 and Rab38 (but not Rab23) were cleaved in cells infected with S. Typhimurium, which expresses GtgE, but not in cells infected with S. Typhi, which does not (Fig. 2C). These results indicate that GtgE is a specific protease that targets a very restricted sub-group of highly related Rab GTPases.
Fig. 2. Rab32 and Rab38 are targets of the GtgE protease activity.
(A) GtgE cleaves Rab32 and Rab38 in cultured cells. COS-1 cells were co-transfected with plasmids expressing the indicated GFP or RFP-tagged Rab GTPases and wild-type GtgE (wt) or a GtgE catalytic mutant (GtgEH151A) (cat). Twenty-four hours after transfection, cells were lysed and samples were analyzed by Western blotting using a rabbit anti-GFP antibody or a rabbit anti-RFP antibody. (B) GtgE cleaves Rab32 and Rab38 in vitro. Purified MBP-tagged Rab32 or MBP-tagged Rab38 were incubated with MBP-tagged GtgE, separated by SDS-PAGE and stained with Coomassie. (C) GtgE targets Rab32 and Rab38 during Salmonella infection. COS-1 cells expressing GFP-Rab29, YFP-Rab32, YFP-Rab38 or RFP-Rab23 were infected with S. Typhi (Ty), or S. Typhimurium (Tm). Two and a half hour after infection cells were lysed and samples were analyzed by Western blotting using a rabbit anti-GFP antibody or a rabbit anti-RFP antibody. (D) Phylogenetic tree of the human Rab and Rab-like GTPases. The locations of Rab29, Rab32 and Rab38 within the tree are indicated in red.
We have previously shown that shortly after infection, Rab29 is readily recruited to the S. Typhi-containing vacuoles and remains associated with these vacuoles for several hours after infection (16). We therefore investigated whether Rab32 and Rab38 were also recruited to the S. Typhi-containing vacuole. We found that, like Rab29, Rab32 and Rab38 were recruited to the S. Typhi-containing vacuole with similar kinetics (Fig. 3A). Similar observations were made in cells infected with the related human-restricted serovar Salmonella Paratyphi, which, like S. Typhi, does not encode gtgE (Fig. 3B). In contrast, Rab32 and Rab38 were not recruited to the S. Typhimurium vacuole (Fig. 3B), which is consistent with the degradation of these GTPases in infected cells by GtgE (Fig. 2C). Similar results were observed in mouse primary BMDM (Fig. 3C). These results indicate that there are very significant differences between the composition of the intracellular compartments that harbor the human-restricted S. Typhi and the broad-host Salmonellae such as S. Typhimurium, which may contribute to their differences in host specificity.
Fig. 3. Rab32 and Rab38 are recruited to the Salmonella Typhi-containing vacuole.
Henle-407 cells (A and B) or mouse primary BMDM (C) expressing YFP- or CFP-tagged Rab32 or Rab38 (green) were infected with S. Typhi (A and C), S. Paratyphi (B and C), or S. Typhimurium (B and C) expressing mCherry (red) and imaged at the indicated times (A) or 2 hours (B and C) after infection. The images shown represent maximum intensity projections of Z-stacks. Bars, 10 μm.
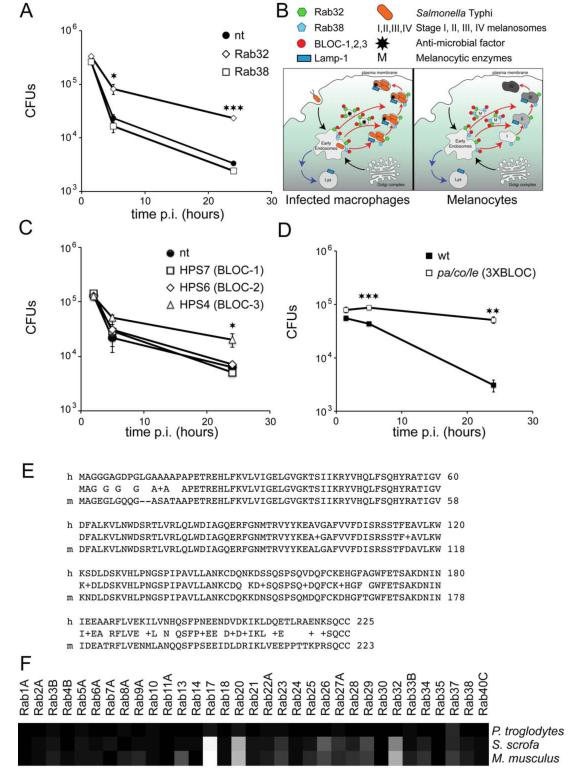
To ascertain which of the GtgE targets restricts the survival of S. Typhi within non-permissive macrophages, we investigated the effect of siRNA-mediated depletion of Rab32 or Rab38 on the ability of S. Typhi to survive within primary mouse BMDM. We found that depletion of Rab38 had no effect on the ability of S. Typhi to survive in non-permissive macrophages (Fig. 4A and S7). However, depletion of Rab32 significantly increased the ability of S. Typhi to survive within mouse macrophages, essentially phenocopying S. Typhi expressing gtgE (Fig. 4A and S7). These results indicate that GtgE confers to S. Typhi the capacity to survive within mouse macrophages by degrading Rab32.
Fig. 4. Rab32 and BLOC-3 are required for S. Typhi host-cell restriction.
(A) Intracellular survival of S. Typhi in BMDM depleted of Rab32 and Rab38. BMDM from mice were transfected with a non-targeting siRNA smart pool (nt) or siRNA smart pools targeting Rab32 or Rab38. Three days after transfection cells were infected with S. Typhi, lysed at the indicated time points after infection and c. f. u. were enumerated. Values are means ± SEM of three independent measurements. (B) Schematic representation of the comparison between the maturation of the Salmonella-containing vacuole and melanosomes (an example of a lysosome-related organelle). (C) Intracellular survival of S. Typhi in BMDM depleted of BLOCs. BMDM from mice were nucleoporated with a non-targeting siRNA smart pool (nt) or specific siRNA smart pools targeting HPS7 (subunit of BLOC-1), HPS6 (subunit of BLOC-2) or HPS4 (subunit of BLOC-3). Three days after transfection, cells were infected with S. Typhi, lysed at the indicated time points and c. f. u. were enumerated. Values are means ± SEM of three independent experiments. (D) Intracellular survival of S. Typhi in BMDM defective for BLOC-1, -2, and -3. BMDM cells from mice simultaneously defective in BLOC-1, 2, and -3 (pa/co/le) were infected with S. Typhi, lysed at the indicated time points after infection and c. f. u. were enumerated. Values are means ± SEM of three independent measurements. (E) Alignment of human and mouse Rab32 shows sequence variation in the N-terminal and C-terminal regions. (F) Heat-map of the degrees of identity of the human Rab GTPases with the orthologs in three mammalian species: Pan troglodytes, Sus scrofa, Mus musculus. Black represents 100% identity, white represents 75% identity. p values were determined by the Student’s t test: *<0.05; **<0.01; ***<0.001.
Rab32 and Rab38 have been implicated in the biogenesis of specialized compartments distinct from lysosomes that are collectively known as “lysosome-related-organelles” (LRO) such as melanosomes or specialized granules in platelet and T-cells (19-23)(24). Rab32 and Rab38, in conjunction with BLOC 1, 2, and 3, coordinate the delivery of specific cargo to LROs including enzymes required for melanine synthesis or a variety of antimicrobial proteins (20, 23, 25, 26). Intriguingly, the Salmonella- containing vacuole (SCV) exhibits features of LROs such as the presence of the lysosomal glycoprotein 1 (Lamp-1), the absence of lysosomal degradative enzymes (27, 28) and, as shown here, the presence of Rab32 and Rab38 (Fig. 4B). We therefore hypothesized that Rab32 may restrict S. Typhi growth in mouse macrophages by delivering an antimicrobial activity to the SCV. If this hypothesis were correct, interfering with components of the BLOC complexes should allow S. Typhi to survive in mouse macrophages. We found that depletion of HSP-7 or HSP-6, essential components of BLOC-1 and BLOC-2, respectively, did not have an effect on the ability of S. Typhi to survive within mouse macrophages (Fig. 4C and S8). In contrast, depletion of HSP-4, an essential component of BLOC-3, allowed S. Typhi to survive within mouse macrophages (Fig. 3C and S8). Furthermore, we found that S. Typhi survived in BMDM obtained from a mouse (pa/co/le) simultaneously defective in BLOC-1, -2, and -3 (29-31) (Fig. 4D). Together, these results indicate that S. Typhi restriction in cells from a non-permissive host is due to an antimicrobial activity delivered to its intracellular vacuole by a machinery akin to that utilized in the genesis of specialized compartments such as melanosomes and T-cell granules. The rapid kinetics of S. Typhi loss of c. f. u. after infection of non-permissive cells (Fig. 1) suggests that this activity must kill S. Typhi rather than restrict its growth. It is therefore possible that differences in the antimicrobial activity of human macrophages may have relieved S. Typhi from the need to acquire (or retain) gtgE thus contributing to its host specialization.
Rab GTPases are highly conserved across vertebrate species exhibiting very little amino acid sequence variation (32). However, Rab32 exhibits relatively more amino acid sequence variation across mammalian species than most other members of this family (Fig. 4E and 4F). We hypothesize that this variation may have been driven by the action of virulence factors that, like GtgE, may target this host-defense pathway. Mutations in the components of the machinery involved in melanosome formation such as the BLOCs or a geranylgeranyl transferase that modifies Rab32, lead to a variety of pathologies such as Hermansky Pudlak syndrome (33, 34). These deficiencies not only lead to albinism but to other clinical manifestations including increased susceptibility to infections (35). It is possible that deficiencies in macrophage microbial killing functions may account at least in part for the observed increased susceptibility to bacterial infections. In this context, it is also intriguing that a recent genome-wide association study has uncovered a genetic polymorphism in Rab32 that is linked to increased susceptibility to Mycobacterium leprae infection (36). Furthermore, Rab32 has been reported to be present in the Mycobacterium tuberculosis-containing vacuole (37). Therefore, it is likely that the mechanism described here may be important in the control of other important intracellular bacterial pathogens that, like Salmonella, reside in this specialized membrane compartment.
We have described here a novel mechanism that restricts the growth of the human restricted Salmonella Typhi within mouse macrophages, a mechanism that could be overcome by the expression of a single effector protein from broad host Salmonellae. These findings provide major insight into the mechanisms of pathogen host-adaptation and describe a novel mechanism of host defense to target pathogens located at a unique intracellular compartment.
Supplementary Material
Fig. S1. GtgE expression overcomes host-cell restriction. Bone-marrow-derived mouse macrophages were infected with S. Typhi (Ty), S. Typhi expressing GtgE (Ty GtgE), or S. Typhimurium (Tm) using an m. o .i. of 5 for all strains. Cells were lysed at the indicated time points and c. f. u. enumerated. Data are standardized relative to the c. f. u. values 1.5 h after infection (considered 1).
Fig. S2. GtgE expression limits host restriction. Colony forming units recovered from the spleens of Nramp −/− mice infected with S. Typhi (wt) or S. Typhi expressing GtgE (GtgE) 4 days after intraperitoneal inoculation. Horizontal bars indicate the means. The p value was determined by the Wilcoxon rank sum test.
Fig. S3: Depletion of Rab29 does not rescue S. Typhi survival in macrophages. BMDM cells were transfected with a non-targeting siRNA smart pool (nt) or a siRNA smart pool targeting Rab29. Three days after transfection cells were infected with S. Typhi, lysed at the indicated time points and c. f. u. were enumerated. Values are means ± SEM of three independent measurements.
Fig. S4. Rab GTPase sequence alignment around the GtgE cleavage site. Sequences of the human Rab GTPases corresponding to GtgE cleavage site (marked with a vertical dashed line) were aligned using ClustalW. Sequences immediately surrounding the cleavage site that correspond exactly as those of Rab29, a GtgE substrate, are shown in red. Different degrees of substitutions in the sequences of the other Rab GTPases are shown in a color gradient from red to blue. Representative Rab GTPases for the groups with the highest similarity (red, orange and yellow) were selected for testing their susceptibility to GtgE. Results of those experiments are indicated in the right column. n. t. : not tested
Fig. S5. COS-1 cells were co-transfected with plasmids expressing the indicated Rab GTPases (fused to GFP or RFP) and wild type GtgE (wt), or GtgE catalytic mutant GtgEH151A (cat). Twenty-four hours after transfection cells were lysed and samples were analyzed by Western blotting using a rabbit anti-GFP antibody or a rabbit anti-RFP antibody. * Indicate the position of the Rab GTPase cleavage products.
Fig. S6. Multiple alignment of the closely related Rab GTPases Rab29, Rab32, Rab38 and Rab23. The arrow indicates the GtgE cleavage site. Black shading indicates identical residues and gray shading indicates conserved residues.
Fig. S7. Transcript levels of Rab32 and Rab38 in nucleoporated BMDM cells. BMDM cells were nucleoporated with non-targeting siRNA (nt) or siRNAs targeting Rab32 or Rab38. Three days later cells were analyzed for transcript content by RT-PCR.
Fig. S8. Transcript levels of BLOC components in nucleoporated BMDM cells. BMDM cells from caspase 1 knock-out mice were nucleoporated with a non-targeting siRNA smart pool (nt) or specific siRNA smart pools targeting either HPS7 (subunit of BLOC-1), HPS6 (subunit of BLOC-2) or HPS4 (subunit of BLOC-3). Three days after transfection cells were infected with S. Typhi, and 24 hs after infection they were analyzed for transcript content by RT-PCR. Values are means ± SEM of three independent experiments.
Table S1. List of plasmids used in this study
Acknowledgments
We thank Walther Mothes for generously providing primary bone-marrow-derived macrophages from pa/co/le mice, which had been kindly provided to him by Richard Swank. We also thank Craig Roy’s laboratory for providing different Rab GTPase constructs and members of the Galán laboratory for critical reading of the manuscript. This work was supported by NIAID Grants AI079022 and AI055472 to J. E. G.
References and Notes
- 1.Ohl ME, Miller SI. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 2001;52:259. doi: 10.1146/annurev.med.52.1.259. [DOI] [PubMed] [Google Scholar]
- 2.Grassl G, Finlay B. Pathogenesis of enteric Salmonella infections. Curr Opin Gastroenterol. 2008;24:22. doi: 10.1097/MOG.0b013e3282f21388. [DOI] [PubMed] [Google Scholar]
- 3.Edsall G, et al. Studies on infection and immunity in experimental typhoid fever. I. Typhoid fever in chimpanzees orally infected with Salmonella typhosa. J. Exp. Med. 1960;112:143. doi: 10.1084/jem.112.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parry C, Hien TT, Dougan G, White N. J. Farrar, Typhoid fever. N Engl J Med. 2002;347:1770. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 5.Crump J, Mintz E. Global trends in typhoid and paratyphoid Fever. Clin Infect Dis. 2010;50:241. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raffatellu M, Wilson R, Winter S, Bäumler A. Clinical pathogenesis of typhoid fever. J Infect Dev Ctries. 2008;2:260. doi: 10.3855/jidc.219. [DOI] [PubMed] [Google Scholar]
- 7.Jacobsen A, Hendriksen R, Aaresturp F, Ussery D, Friis C. The Salmonella enterica pan-genome. Microb Ecol. 2011;62:487. doi: 10.1007/s00248-011-9880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabbagh S, Forest C, Lepage C, Leclerc J, Daigle F. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett. 2010;305:1. doi: 10.1111/j.1574-6968.2010.01904.x. [DOI] [PubMed] [Google Scholar]
- 9.Parkhill J, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001 Oct 25;413:848. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 10.Schwan WR, Huang XZ, Hu L, Kopecko DJ. Differential bacterial survival, replication, and apoptosis-inducing ability of Salmonella serovars within human and murine macrophages. Infect Immun. 2000 Mar;68:1005. doi: 10.1128/iai.68.3.1005-1013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vladoianu I, Chang H, Pechère J. Expression of host resistance to Salmonella typhi and Salmonella typhimurium: bacterial survival within macrophages of murine and human origin. Microb. Pathog. 1990;8:83. doi: 10.1016/0882-4010(90)90072-x. [DOI] [PubMed] [Google Scholar]
- 12.Galan JE. Salmonella interactions with host cells: type III secretion at work. Annual review of cell and developmental biology. 2001;17:53. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- 13.Waterman SR, Holden DW. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. 2003 Aug;5:501. doi: 10.1046/j.1462-5822.2003.00294.x. [DOI] [PubMed] [Google Scholar]
- 14.Ibarra J, Steele-Mortimer O. Salmonella--the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol. 2009;11:1579. doi: 10.1111/j.1462-5822.2009.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srikanth C, Mercado-Lubo R, Hallstrom K, McCormick B. Salmonella effector proteins and host-cell responses. Cell Mol Life Sci. 2011;68:3687. doi: 10.1007/s00018-011-0841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spano S, Liu X, Galan JE. Proteolytic targeting of Rab29 by an effector protein distinguishes the intracellular compartments of human-adapted and broad-host Salmonella. Proc Natl Acad Sci U S A. 2011 Nov 8;108:18418. doi: 10.1073/pnas.1111959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.See Supplementary Material for experimental details.
- 18.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 19.Wasmeier C, et al. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J Cell Biol. 2006;175:271. doi: 10.1083/jcb.200606050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bultema J, Ambrosio A, Burek C, Di Pietro S. BLOC-2, AP-3, and AP-1 Proteins Function in Concert with Rab38 and Rab32 Proteins to Mediate Protein Trafficking to Lysosome-related Organelles. J. Biol. Chem. 2012;287:19550. doi: 10.1074/jbc.M112.351908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raposo G, Marks M. Melanosomes--dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8:786. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holt O, Gallo F, Griffiths G. Regulating secretory lysosomes. J Biochem. 2006;140:7. doi: 10.1093/jb/mvj126. [DOI] [PubMed] [Google Scholar]
- 23.Benado A, Nasagi-Atiya Y, Sagi-Eisenberg R. Protein trafficking in immune cells. Immunobiology. 2009;214:507. doi: 10.1016/j.imbio.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 24.Raposo G, Tenza D, Murphy D, Berson J, Marks M. Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J. Cell Biol. 2001;152:809. doi: 10.1083/jcb.152.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dell’Angelica E. The building BLOC(k)s of lysosomes and related organelles. Curr Opin Cell Biol. 2004;16:458. doi: 10.1016/j.ceb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Aspengren S, D H, HN S, M W. New insights into melanosome transport in vertebrate pigment cells. Int Rev Cell Mol Biol. 2009;272:245. doi: 10.1016/S1937-6448(08)01606-7. [DOI] [PubMed] [Google Scholar]
- 27.Bakowski M, Braun V, Brumell J. Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic. 2008;9:2022. doi: 10.1111/j.1600-0854.2008.00827.x. [DOI] [PubMed] [Google Scholar]
- 28.Steele-Mortimer O. The Salmonella-containing vacuole: moving with the times. Curr Opin Microbiol. 2008;11:38. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, et al. Murine Hermansky-Pudlak syndrome genes: regulators of lysosome-related organelles. Bioessays. 2004;26:616. doi: 10.1002/bies.20042. [DOI] [PubMed] [Google Scholar]
- 30.Gautam R, et al. Interaction of Hermansky-Pudlak Syndrome genes in the regulation of lysosome-related organelles. Traffic. 2006;7:779. doi: 10.1111/j.1600-0854.2006.00431.x. [DOI] [PubMed] [Google Scholar]
- 31.Chan W, et al. Murine leukemia virus spreading in mice impaired in the biogenesis of secretory lysosomes and Ca2+-regulated exocytosis. PloS one. 2008;3:e2713. doi: 10.1371/journal.pone.0002713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brighouse A, Dacks J, Field M. Rab protein evolution and the history of the eukaryotic endomembrane system. Cell Mol Life Sci. 2010;67:3449. doi: 10.1007/s00018-010-0436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dell’Angelica E, Shotelersuk V, Aguilar R, Gahl W, Bonifacino J. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Molecular cell. 1999;3:11. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- 34.Di Pietro S, Dell’Angelica E. The cell biology of Hermansky-Pudlak syndrome: recent advances. Traffic. 2005;525-33:525. doi: 10.1111/j.1600-0854.2005.00299.x. [DOI] [PubMed] [Google Scholar]
- 35.Stinchcombe J, Bossi G, Griffiths G. Linking albinism and immunity: the secrets of secretory lysosomes. Science. 2004;305:55. doi: 10.1126/science.1095291. [DOI] [PubMed] [Google Scholar]
- 36.Zhang F, et al. Identification of two new loci at IL23R and RAB32 that influence susceptibility to leprosy. Nature genetics. 2011 Dec;43:1247. doi: 10.1038/ng.973. [DOI] [PubMed] [Google Scholar]
- 37.Seto S, Tsujimura K, Koide Y. Rab GTPases regulating phagosome maturation are differentially recruited to mycobacterial phagosomes. Traffic. 2011;12:407. doi: 10.1111/j.1600-0854.2011.01165.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. GtgE expression overcomes host-cell restriction. Bone-marrow-derived mouse macrophages were infected with S. Typhi (Ty), S. Typhi expressing GtgE (Ty GtgE), or S. Typhimurium (Tm) using an m. o .i. of 5 for all strains. Cells were lysed at the indicated time points and c. f. u. enumerated. Data are standardized relative to the c. f. u. values 1.5 h after infection (considered 1).
Fig. S2. GtgE expression limits host restriction. Colony forming units recovered from the spleens of Nramp −/− mice infected with S. Typhi (wt) or S. Typhi expressing GtgE (GtgE) 4 days after intraperitoneal inoculation. Horizontal bars indicate the means. The p value was determined by the Wilcoxon rank sum test.
Fig. S3: Depletion of Rab29 does not rescue S. Typhi survival in macrophages. BMDM cells were transfected with a non-targeting siRNA smart pool (nt) or a siRNA smart pool targeting Rab29. Three days after transfection cells were infected with S. Typhi, lysed at the indicated time points and c. f. u. were enumerated. Values are means ± SEM of three independent measurements.
Fig. S4. Rab GTPase sequence alignment around the GtgE cleavage site. Sequences of the human Rab GTPases corresponding to GtgE cleavage site (marked with a vertical dashed line) were aligned using ClustalW. Sequences immediately surrounding the cleavage site that correspond exactly as those of Rab29, a GtgE substrate, are shown in red. Different degrees of substitutions in the sequences of the other Rab GTPases are shown in a color gradient from red to blue. Representative Rab GTPases for the groups with the highest similarity (red, orange and yellow) were selected for testing their susceptibility to GtgE. Results of those experiments are indicated in the right column. n. t. : not tested
Fig. S5. COS-1 cells were co-transfected with plasmids expressing the indicated Rab GTPases (fused to GFP or RFP) and wild type GtgE (wt), or GtgE catalytic mutant GtgEH151A (cat). Twenty-four hours after transfection cells were lysed and samples were analyzed by Western blotting using a rabbit anti-GFP antibody or a rabbit anti-RFP antibody. * Indicate the position of the Rab GTPase cleavage products.
Fig. S6. Multiple alignment of the closely related Rab GTPases Rab29, Rab32, Rab38 and Rab23. The arrow indicates the GtgE cleavage site. Black shading indicates identical residues and gray shading indicates conserved residues.
Fig. S7. Transcript levels of Rab32 and Rab38 in nucleoporated BMDM cells. BMDM cells were nucleoporated with non-targeting siRNA (nt) or siRNAs targeting Rab32 or Rab38. Three days later cells were analyzed for transcript content by RT-PCR.
Fig. S8. Transcript levels of BLOC components in nucleoporated BMDM cells. BMDM cells from caspase 1 knock-out mice were nucleoporated with a non-targeting siRNA smart pool (nt) or specific siRNA smart pools targeting either HPS7 (subunit of BLOC-1), HPS6 (subunit of BLOC-2) or HPS4 (subunit of BLOC-3). Three days after transfection cells were infected with S. Typhi, and 24 hs after infection they were analyzed for transcript content by RT-PCR. Values are means ± SEM of three independent experiments.
Table S1. List of plasmids used in this study