Abstract
The STAT3 transcription factor is an important regulator of stem cell self-renewal, cancer cell survival, and inflammation. In the pancreas, STAT3 is dispensable for normal development whereas the majority of pancreatic ductal adenocarcinomas (PDAC) show constitutive activation of STAT3, suggesting its potential as a therapeutic target in this cancer. Here, we sought to define the mechanisms of STAT3 activation and its functional importance in PDAC pathogenesis. Large-scale screening of cancer cell lines with a JAK2 inhibitor that blocks STAT3 function revealed a >30-fold range in sensitivity in PDAC, and showed a close correlation of sensitivity with levels of tyrosine-phosphorylated STAT3 and of the gp130 receptor, an upstream signaling component. Correspondingly, upregulation of the IL6/LIF-gp130 pathway accounted for the strong STAT3 activation in PDAC subsets. To define functions of STAT3 in vivo, we developed mouse models that test the impact of conditional inactivation of STAT3 in KRAS-driven PDAC. We showed that STAT3 is required for the development of the earliest pre-malignant pancreatic lesions, acinar-to-ductal metaplasia (ADM) and pancreatic intraepithelial neoplasia (PanIN). Moreover, acute STAT3 inactivation blocked PDAC initiation in a second in vivo model. Our results demonstrate that STAT3 has critical roles throughout the course of PDAC pathogenesis, supporting the development of therapeutic approaches targeting this pathway. Moreover, our work suggests that gp130 and phospho-STAT3 expression may be effective biomarkers for predicting response to JAK2 inhibitors.
Keywords: Pancreatic cancer, STAT3, Pancreatic intraepithelial neoplasia, mouse models, gp130
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer death in the United States and carries a dismal 5-year survival rate of <5% (1). Activating mutations in the KRAS oncogene are the defining lesion in this malignancy, present in 70–95% of cases (2–4). PDAC is believed to arise from precursor lesions called pancreatic intraepithelial neoplasia (PanIN). Molecular pathology analysis of human specimens and the development of genetically engineered mouse models support a model in which PanINs proceed through multiple stages of increasingly severe cellular atypia culminating in the development of invasive carcinoma. This histologic progression is associated with KRAS activation as an early event and the subsequent step-wise accumulation of inactivating mutations in the tumor suppressors, Ink4a/Arf, p53, and SMAD4 (5,6). As this genetic information has not yet led to the development of effective targeted therapeutic strategies in PDAC, there is considerable focus on defining additional molecular pathways driving the progression and maintenance of this disease.
The Signal transducer and activator of transcription (STAT) family transcription factors are constitutively activated in a wide range of human malignancies (7). STAT proteins are present in the cytoplasm under basal conditions and are activated by phosphorylation on a single tyrosine residue, which triggers dimerization and nuclear localization (8,9). Classically, STAT tyrosine phosphorylation is mediated by the Janus (JAK) family of tyrosine kinases, which themselves are activated by cytokine and growth factor receptors (10,11). Other tyrosine kinases, such as src, have also been reported to mediate tyrosine phosphorylation of STAT proteins (12). The STAT proteins were originally identified as factors required for downstream signaling in response to interferon and other inflammatory cytokines (8). Subsequent studies identified key functions for STAT proteins in the maintenance of self-renewal of embryonic stem cells and in the activation of proliferative, anti-apoptotic and inflammatory pathways to initiate and maintain growth of a number of tumor types (7,13,14).
STAT3 has been identified as a key oncogenic factor in a number of epithelial malignancies and is required for oncogenesis in mouse models of skin and gastric cancers (15,16). In PDAC, constitutive activation of STAT3 by phosphorylation of Tyr705 has been reported in 30–100% of human tumor specimens, as well as in many PDAC cell lines (17,18). By contrast, this pathway is inactive in normal pancreas, and correspondingly STAT3 is not required for pancreatic development or homeostasis, as demonstrated by conditional knockout studies in mice (19). Several lines of evidence suggest that aberrant activation of STAT3 in PDAC is functionally important. Firstly, STAT3 is required for the process of acinar-to-ductal metaplasia (ADM)—thought to be an early event in PDAC pathogenesis—upon ectopic expression of the Pdx1 transcription factor, a key regulator of early pancreatic development (20). In addition to this potential role in early PDAC, STAT3 has been suggested as a therapeutic target in established PDAC since examination of a limited number of cell lines for the impact of chemical STAT3 pathway inhibitors and dominant-negative STAT3 constructs has shown that the pathway may contribute to the proliferation of some PDAC cell lines in vitro and the tumorigenicity of some PDAC xenografts (17,18,21,22). These data support the need for more detailed studies to define the basis for STAT3 activation in PDAC and to rigorously establish specific roles for STAT3 in the initiation and progression of PDAC in vivo.
In this study, we examined the sensitivity of a large series of PDAC cells lines to pharmacologic STAT3 inhibition and defined biomarkers of sensitivity as well as key upstream activators of the pathway in this cancer. We also employed genetically engineered mouse models to determine the impact of genetic inactivation of STAT3 on the progression of PDAC. Collectively our results demonstrate that upregulation of the gp130 receptor and strong STAT3 phosphorylation point to a subset of PDAC that are highly sensitive to pharmacologic inhibition of the JAK2/STAT3 pathway, and that STAT3 plays an important role in driving PDAC progression at multiple stages of pancreatic tumorigenesis in vivo, thereby supporting STAT3 as a potential therapeutic target in PDAC.
MATERIALS AND METHODS
Cell Lines
PDAC cell lines were grown in DMEM/F12 (GIBCO) with 10% FBS and assayed in DMEM/F12 with 5% FBS and were obtained from the MGH Center for Molecular Therapeutics (CMT), which performs routine cell line authentication testing by SNP and STR analysis. For drug sensitivity studies, data from over 500 solid tumor cell lines were obtained from the CMT drug screen database (30). Viable cell titer relative to untreated cells was determined using Cell Titer Glo assay (Promega). For apoptosis assays, cells were stained with propidium iodide and Annexin V Cy5 (Biovision) according to the manufacturer’s protocol and assayed on a LSRII flow cytometer (BD Biosciences).
Mouse Strains and Histologic Analysis
The Pdx1-Cre transgenic mouse strain and the LSL-KRASG12D knock-in mouse strain have been previously described (23,24). The STAT3lox/lox mouse strain was kindly provided by David Levy (25). These strains were intercrossed to produce the experimental cohorts. Pancreata isolated from 12 week-old mice were analyzed in blinded fashion by a single pathologist (V.D.) to determine the percent of each pancreas occupied by ADM or PanIN lesions.
Pancreatic ductal cells
Pancreatic ductal cells where isolated from 9 week-old Pdx1-Cre; LSL-KRASG12D mice as previously described (26) and propagated in laminin (BD Biosciences). Mouse shp53 retroviral construct (MLS sh-p53.1224, 27) was transfected in Ecopack 293T cells and media collected at 48 and 72 hours. One week after overnight incubation with retrovirus in the presence of 8μg/mL polybrene, GFP-positive infected cells were FACS-sorted and propagated.
Molecular Analyses
Western blotting was performed using standard methods. Immunostaining was performed using standard methods on formalin-fixed, paraffin embedded tissues. After deparaffinization slides are washed with 9.83% NaCl for 3 min followed by a PBS wash and a wash in distilled water for 5min. Antigen retrieval was performed with pressure cooker (2100 Retriever, PickCell Laboratories) and R-Buffer A (pH6.0, Electron Microscopy Sciences).
Orthotopic tumor model
SCID mice (C3SnSmn.CB17-Prkdcscid/J, Jackson Labs) were subjected to general anesthesia according to MGH SRAC policies. Orthotopic injections of the pancreas were performed as previously described (28), using 2×104 PanIN cells suspended in 50μl of Duct Media (26), mixed with 50μl of Matrigel (BD Biosciences).
More detailed Materials & Methods are presented in the Supplementary files, online.
RESULTS
Phospho-STAT3 levels predict the sensitivity of PDAC cell lines to JAK2 inhibition
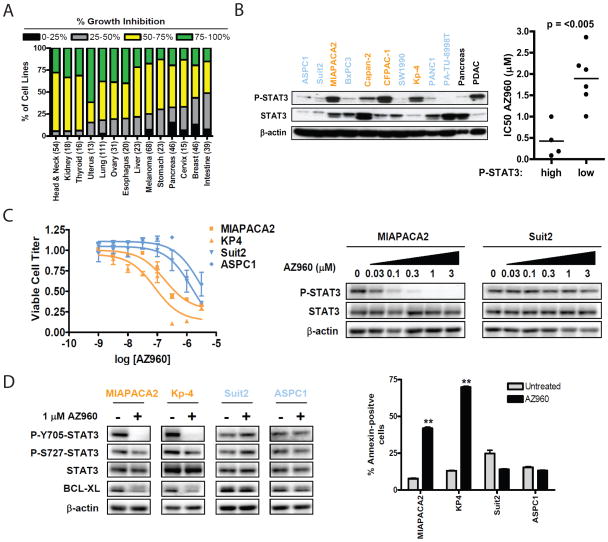
Previous studies of a limited number of human PDAC cell lines have shown that subsets are sensitive to pharmacologic or genetic inactivation of the JAK2/STAT3 pathway (17,18,21). To more broadly define the role of this pathway in PDAC tumor maintenance, we evaluated the drug sensitivity profile for the JAK2-selective inhibitor AZ960 across a large-scale cell line repository containing over 500 solid tumor cell lines, including 46 PDAC cell lines (29,30). These cell lines are characterized at the molecular level for regional changes in chromosomal copy number (SNP Chip analysis), global mRNA expression, and common cancer gene mutations, enabling correlations of sensitivity in relation to specific molecular features. PDAC cell lines as a group showed intermediate sensitivity to JAK2 inhibition (Fig. 1A). However, we found that a subset (20%) of PDAC cell lines displayed high sensitivity (>75% inhibition of viable cell number relative to untreated control), suggesting that JAK2 inhibitors could be useful in selected PDAC patients. To explore this possibility further, we evaluated P-STAT3 levels in a panel of 10 randomly selected PDAC cell lines (Fig. 1B). While P-STAT3 could be detected in all but one cell line, high levels of P-STAT3 in comparison with normal pancreatic tissue were detected in 4 of the 10 cell lines; these high levels in cell lines were comparable to those observed in PDAC tissue specimens.
Fig. 1. P-STAT3 levels predict sensitivity of PDAC cell lines to JAK2 inhibition.
A, Analysis of the drug sensitivity profile of AZ960 (3μM) over a panel of >500 solid tumor cell lines based on tumor type (total number of cell lines in parenthesis). Bar color indicates percent growth inhibition relative to control. Bar height represents the percentage of cell lines of each tumor type showing the indicated degree of growth inhibition. B, left, western blot of P-STAT3 and total STAT3 levels in 10 PDAC cell lines, compared to normal pancreas and PDAC tissue. Right, comparison of the IC50 of AZ960 in PDAC cell lines with high vs. low P-STAT3 levels. Bar represents the mean IC50 value for each group. P-value is shown. C, left, PDAC cell lines with high (orange) or low (blue) P-STAT3 levels were treated in triplicate with the indicated concentrations of AZ960 for 72h. Viable cell titer was determined, and average values are shown relative to untreated controls for each cell line. Error bars represent SD for each measurement. Right, western blot of PDAC cell lines treated with the indicated concentrations of AZ960 for 24h. D, left, PDAC cell lines were treated in the presence or absence of 1μM AZ960 for 24h. Lysates were probed with the indicated antibodies. Right, PDAC cell lines were treated in the presence or absence of 1μM AZ960 for 72h. Percent apoptotic cells was determined by Annexin V staining (**p<0.01).
Notably, there was a strong correlation between high levels of P-STAT3 and sensitivity to AZ960 (Fig. 1B, and Supplementary Fig. S1). In cell lines with elevated P-STAT3, AZ960 inhibited cell viability in a dose-dependent manner that correlated with inhibition of STAT3 Tyr705-phosphorylation (Fig. 1C). This was associated with down-regulation of BclXL (Fig. 1D), an anti-apoptotic protein and an established STAT3 target (29), as well as a corresponding pronounced induction of apoptosis. By contrast, cell lines with low P-STAT3 showed no changes in P-STAT3 or BclXL in response to AZ960, and did not undergo apoptosis, suggesting that low basal levels of STAT3 Tyr705-phosphorylation in these cell lines are not due to JAK2 (Figs. 1C, D). As expected, AZ960 did not affect STAT3 Ser727-phosphorylation (Fig. 1D), a modification known to be JAK2-independent (31,32). These data indicate that strong STAT3 activation seen in a subset PDAC cell lines is mediated by JAK2 and may predict sensitivity to JAK2 inhibitors.
IL-6 family cytokine signaling regulates P-STAT3 levels in PDAC cell lines
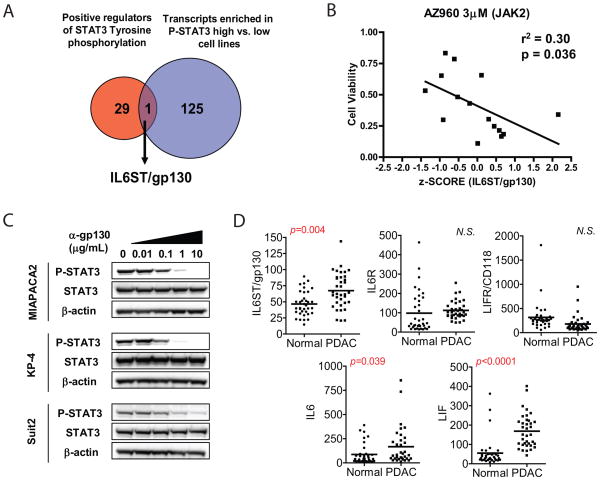
Next, we sought to establish the upstream signaling pathways responsible for STAT3 activation in PDAC. To this end, we took advantage of the genome-wide mRNA expression profiles available for our cell line repository to identify transcripts enriched in PDAC cell lines with elevated P-STAT3 levels. This analysis identified 126 transcripts associated with strong STAT3 activation. We cross-referenced this gene set with a list of 30 genes identified by a gene ontology search as positive regulators of STAT3 tyrosine phosphorylation (Figs. 2A, Supplementary Fig. S2). Notably, the only gene found to be present in both gene sets was the interleukin 6 signal transducer (IL6ST), which encodes membrane glycoprotein 130 (gp130). This protein is the common signal transducing component of the IL6 cytokine receptor family, which forms complexes with the ligand-binding receptor subunits of multiple IL6 cytokine family members (including the IL6, leukemia inhibitory factor (LIF), oncostatin M, and ciliary neurotrophic factor receptors), thereby serving as an important activator of JAK-STAT signaling (33,34). Significantly, the level of IL6ST transcript in PDAC cell lines was strongly correlated with the degree of sensitivity to the JAK2 inhibitor AZ960 in our large-scale cell line screen (Fig. 2B). This effect was specific to JAK-STAT pathway inhibition, as IL6ST transcript levels showed no correlation with sensitivity to inhibitors of EGFR, MEK1/2, and IGF1R (using erlotinib, AZD6244, and AEW541, respectively; Supplementary Fig. S3), pathways that are under active investigation as PDAC drug targets. While the mechanism of IL6ST upregulation in PDAC cell lines is not clear—no focal gene amplifications were detected, and Stat3 knockdown (Fig. S4), these data show that IL6ST transcript levels specifically predict sensitivity to JAK2 inhibition in PDAC cell lines, and suggest that IL6ST may be a key upstream mediator of JAK-STAT3 activity in this cancer.
Fig. 2. P-STAT3 is regulated by IL6 cytokine family signaling in PDAC cell lines.
A, Gene expression micorarray data from P-STAT3 high and low PDAC lines were analyzed to identify transcripts differentially enriched in P-STAT3 high cell lines. 126 transcripts found to be enriched in P-STAT3 high cell lines were cross-referenced with 30 positive regulators of STAT3 tyrosine phosphorylation identified by gene ontology search. Only one gene, IL6ST (encoding gp130) was present in both data sets. B, IL6ST transcript level z-score for 15 PDAC cell lines was correlated with drug sensitivity data to 3μM AZ960 from a large cell line repository drug screen. P value and r2 value are shown. C, PDAC cell lines were treated for 24h with increasing concentrations of gp130-neutralizing antibody. Cell lysates were probed with the indicated antibodies. D, Gene expression microarray data from 36 human PDACs and matched normal pancreas controls (normal) were analyzed for expression of IL6 cytokine family members. Bars represent mean of each group. P values are shown (N.S., not significant).
To test directly whether IL6ST signaling regulates P-STAT3 in PDAC cell lines, cells were treated with a monoclonal neutralizing antibody to gp130, known to block signaling by multiple IL6 cytokine family members (35). This antibody inhibited P-STAT3 in a dose-dependent manner in PDAC cell lines with high P-STAT3 levels, while only modestly affecting basal levels in low P-STAT3 lines (Fig. 2C). Since these findings suggest that IL6 cytokine family signaling through IL6ST regulates STAT3 activation in PDAC cell lines, we evaluated the expression of IL6 cytokine family members in human PDAC, in comparison to normal pancreas. Consistent with our cell line data, IL6ST transcript levels were significantly increased in PDAC, relative to normal pancreas, whereas levels of IL6R and LIFR/CD118 were not up-regulated in PDAC tissue. Transcript levels of IL6 cytokine family ligands IL6 and LIF were also significantly increased in PDAC, relative to normal pancreas. Thus, upregulation of gp130 and IL6/LIF are likely to contribute to STAT3 activation in human PDACs.
STAT3 phosphorylation is observed at multiple stages of KRAS-induced pancreatic tumorigenesis
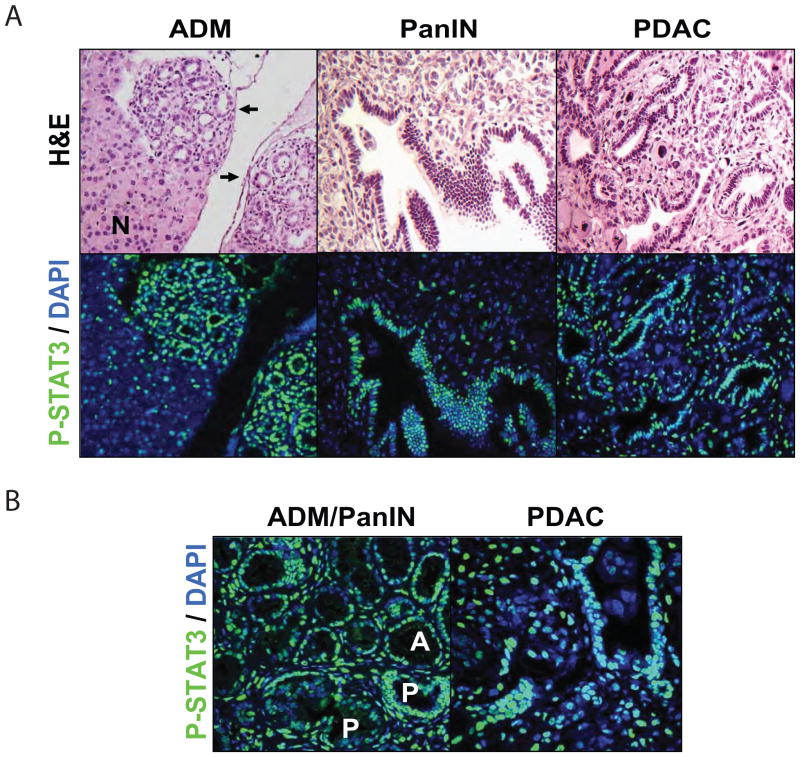
Since some PDAC cell lines appear to be highly dependent on STAT3 activity, we chose to explore the contributions of STAT3 to different stages PDAC evolution in vivo. First, we sought to establish the tyrosine-phosphorylation status of STAT3 at different stages of PDAC pathogenesis. We employed a genetically engineered model of multi-stage tumor progression (Pdx1-Cre; LSL-KRASG12D mice) in which activation of an oncogenic KRASG12D allele in the pancreas results in gradual formation of ADM and PanIN lesions. PanINs progress to PDAC with long latency (~1 year), which is greatly accelerated by genetic inactivation of the p53 or Ink4a/Arf tumor suppressor loci (24,36–38). Immunofluorescence analysis showed that P-STAT3 staining was undetectable in normal pancreas. By contrast, nuclear P-STAT3 expression was present at all stages of PDAC progression, with robust staining observed in both ADM and PanINs (Figs. 3A, B) as well as in fully developed invasive PDAC, consistent with elevated P-STAT3 levels in tumor tissue seen by western blot analysis (Fig. 1B). At each stage of tumorigenesis, there was also evidence of sporadic P-STAT3 staining in the stromal tissue. Thus, STAT3 activation occurs at the earliest stages of pancreatic tumorigenesis and is maintained in invasive cancers, implying possible roles for STAT3 in both tumor initiation and in continued propagation of advanced PDAC. Additionally, the staining pattern may indicate functions of STAT3 in both the tumor epithelium and stroma.
Fig. 3. STAT3 is phosphorylated at multiple stages of pancreatic tumorigenesis.
A, ADM and late-stage PanIN tissue from Pdx1-Cre; LSL-KRASG12D mice and PDAC tissue from Pdx1-Cre; LSL-KRASG12D; p53+/− mice were analyzed for the presence of P-STAT3 (green) by immunofluoresence with DAPI nuclear counterstain (blue). After image capture, slides were stained with hematoxylin and eosin (H&E). Arrows indicate regions of ADM. Normal pancreas is indicated by (N). B, higher magnification images of P-STAT3 staining (green) in ADM (upper half of image, A) or early-stage PanIN (lower half of image, P) lesions or in PDAC are shown.
Loss of STAT3 reduces ADM and PanIN formation induced by oncogenic KRAS
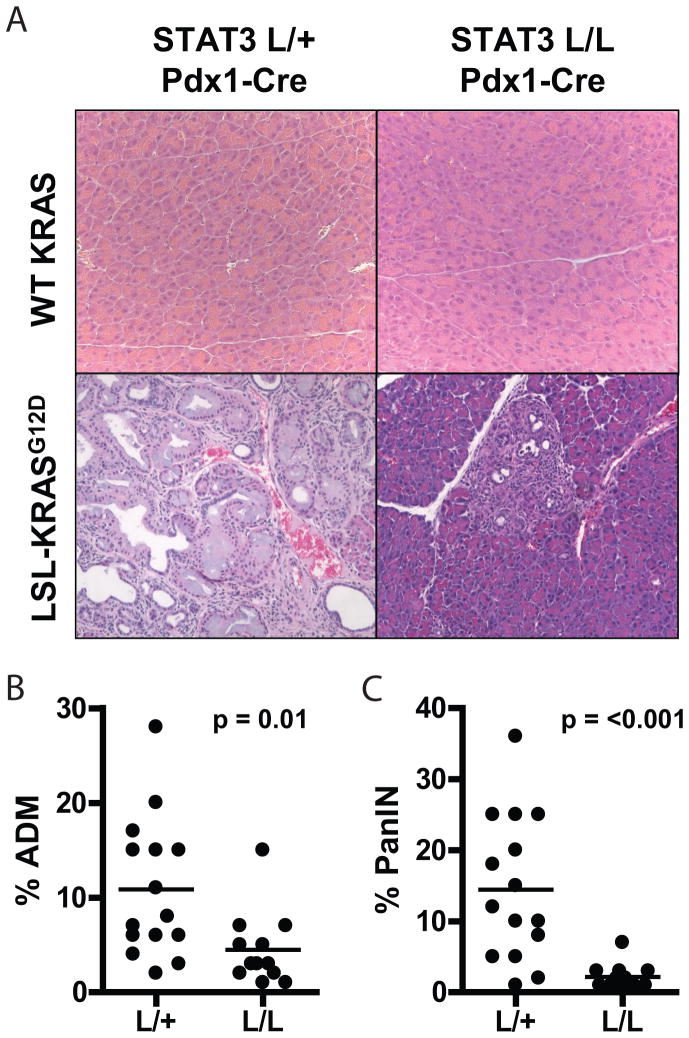
ADM is the earliest change observed in models of KRAS-induced pancreatic oncogenesis and is a potential precursor to PanIN (39,40). Based on the expression pattern of P-STAT3 in these lesions, we examined the requirement of STAT3 in early pancreatic tumorigenesis by crossing Pdx1-Cre; LSL-KRASG12D mice with mice containing a conditional STAT3-knockout allele (STAT3lox). The gradual accumulation of ADM and PanIN and long latency for formation of invasive PDAC in the Pdx1-Cre; LSL-KRASG12D model makes it ideal for evaluating early stages of tumorigenesis. Pdx1-Cre; LSL-KRASG12D; STAT3lox/lox mice exhibited complete loss of P-STAT3 and total STAT3 in the pancreas (Supplementary Fig. S5A, B), indicating effective Cre-mediated recombination. Consistent with previous reports, the pancreas of Pdx1-Cre; STAT3lox/lox mice developed normally, and these mice did not show any evident physiologic alterations (Fig. 4A). We isolated pancreata from Pdx1-Cre; LSL-KRASG12D; STAT3lox/lox mice or from Pdx1-Cre; LSL-KRASG12D; STAT3lox/+ controls at 12 weeks of age and quantified the extent of pancreatic lesions (see Methods). At this time point, control mice showed extensive ADM and PanIN formation (Fig. 4A), resulting in largely distorted pancreatic architecture. By contrast, in mice with STAT3 ablation, the majority of the pancreas had normal structure with only sporadic foci of ADM and PanIN. Correspondingly, the formation of ADM and PanIN was reduced 2.4-fold (p=0.01) and 6.6-fold (p<0.001), respectively, in Pdx1-Cre; KRASG12D; STAT3lox/lox mice, relative to controls (Figs. 4B, C). Thus, STAT3 plays a critical role in the robust formation of ADM and PanIN induced by oncogenic KRAS. While STAT3 ablation did not completely eliminate formation of these lesions, it led to a greatly attenuated ADM/PanIN phenotype.
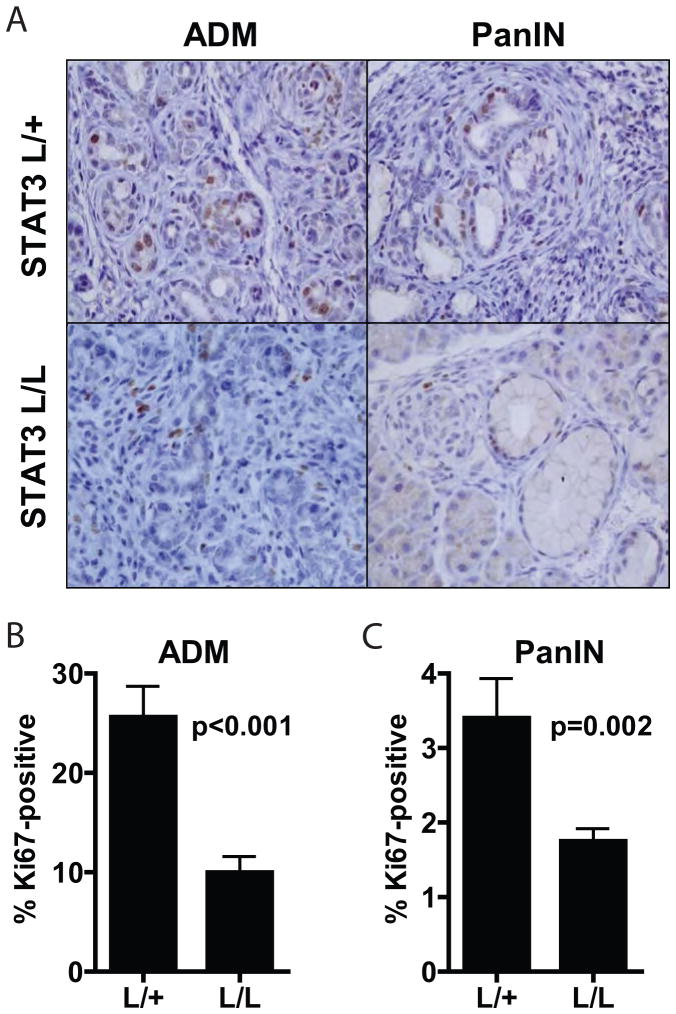
Fig. 4. Loss of STAT3 decreases KRAS-induced ADM and PanIN formation.
A, pancreatic tissue from 12 week-old Pdx1-Cre; STAT3lox/+ (L/+) or Pdx1-Cre; STAT3lox/lox (L/L) mice homozygous for wild-type KRAS alleles (upper panels) or heterozygous for the LSL-KRASG12D allele (lower panels) were harvested and stained with hematoxylin and eosin. B–C, the percent of each pancreas occupied by ADM (B) or PanIN (C) was calculated for each mouse. Each point represents a single mouse, and horizontal bars represent mean percentage for each group. P-values are shown.
Previous work has suggested a link between STAT3 and ADM. In a transgenic mouse model of ADM driven by persistent pancreatic expression of Pdx1, genetic inactivation of STAT3 can block ADM (20). Our data suggest that loss of STAT3 can also inhibit ADM in the more physiologically relevant setting of KRAS activation. The loss of polarized epithelium and reduced cell contacts associated with ADM may create a permissive environment for further processes of cellular transformation. Therefore, it is possible that the involvement of STAT3 in KRAS-induced pancreatic tumorigenesis could be restricted to its role in the formation of these very early lesions. The associated reduction in PanINs could thus simply reflect decreased frequency of ADM precursors, whereas STAT3 maybe dispensable for continued pancreatic tumorigenesis once ADM has occurred. To begin to answer this question, we examined proliferation rates of ADM and PanIN lesions that formed in STAT3-null and control pancreata. Notably, STAT3 ablation resulted in significantly reduced levels of proliferation, as assessed by Ki67 staining, in both ADM and PanIN lesions (Figs. 5A–C), indicating a continued role for STAT3 in the proliferation and progression of early pancreatic lesions, even after ADM has occurred. An ongoing role for STAT3 in pancreatic tumorigenesis is consistent with the persistent expression of P-STAT3 observed at multiple stages of tumorigenesis, including PanINs and fully developed PDAC (Figs. 3A, B).
Fig. 5. ADM and PanIN lesions that form in the absence of STAT3 show decreased proliferation.
A, pancreatic tissue from 12 week-old Pdx1-Cre; LSL-KRASG12D; STAT3lox/+ (L/+) or Pdx1-Cre; LSL-KRASG12D; STAT3lox/lox (L/L) mice was harvested and stained for Ki67. B-C, the percentage of Ki67-positive nuclei in ADM (B) or PanIN (C) lesions was calculated for each genotype, and mean percentage is shown. Error bars represent SD. P-values are shown.
STAT3 is required for the progression to invasive PDAC
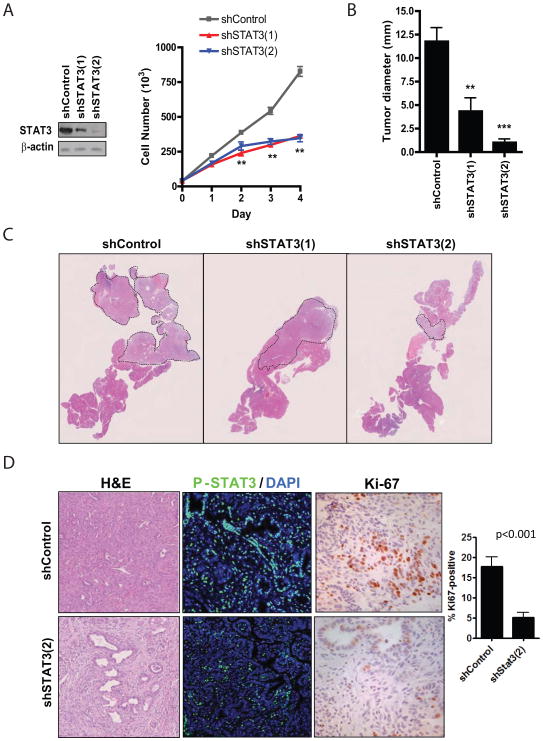
We next sought to determine the role of STAT3 in the progression to advanced PDAC. To this end, we isolated pancreatic ductal cells from 9 week-old Pdx1-Cre; LSL-KRASG12D mice. These cells are not tumorigenic upon orthotopic injection into recipient mice, whereas shRNA-mediated inactivation of p53 enables these cells to rapidly progress to form invasive PDAC. To examine the effect of STAT3 loss on the tumorigenic potential of KRAS-shp53 ductal cells, we used two shRNA constructs targeting STAT3, which led to varying degrees of STAT3 knockdown and caused a marked decrease in the proliferation of KRAS-shp53 ductal cells, compared to control shRNA (Fig. 6A). When these cells were injected orthotopically, expression of shSTAT3 dramatically reduced PDAC formation compared to control shRNA, reducing the tumor volume from 2.9-fold (shSTAT3-1) to 11.5-fold (shSTAT3-2), consistent with the degree of STAT3 knockdown produced (Figs. 6B, C). The tumors that formed from shControl cells showed mainly features moderately-differentiated and poorly-differentiated PDAC (Fig. 6D, upper left panel), with only focal areas of PanINs. By contrast, the small tumors that form upon STAT3 knockdown showed a higher proportion of PanIN in addition to regions of moderately-differentiated PDAC (Fig. 6D, lower left panel). Immunofluorescence analysis confirmed that the shSTAT3 tumors lacked detectable STAT3 expression (Fig. 6D, middle panels). Notably, Ki-67 staining analysis of areas of invasive cancer showed that the shSTAT3 PDAC had a >3-fold reduction in proliferation rates (p<0.001) (Fig 6D, right panels and chart). Thus, reduction in STAT3 expression attenuates progression to invasive PDAC, and impairs the growth of the tumors that ultimately form. Collectively our studies demonstrate that STAT3 is an important component of the molecular program driving PDAC progression and identify critical roles of this transcription factor at multiple stage of disease pathogenesis.
Fig. 6. STAT3 knockdown prevents PDAC initiation in vivo.
A, KRAS-shp53 ductal cells were infected with the indicated shRNAs and analyzed by western blot (left panel) and by cell counting. Measurements were performed in triplicate, and average values are shown (**p<0.001). Error bars represent SD. B, Equal numbers of KRAS-shp53 ductal cells infected with the indicated shRNAs were injected orthotopically into the pancreata of recipient mice. After 4 weeks, pancreatic tissue was harvested and the maximum tumor diameter was determined. Values represent the average tumor diameter for each group, and error bars represent SD (**p<0.01, ***p<0.001). C, upper, low magnification view of sectioned pancreas from representative mice harvested as in C. Dashed lines outline tumor tissue. D, Representative images of orthotopic tumors expressing shControl or shSTAT3(2) analyzed by H&E staining (Left, 200x), immunofluorescence for P-STAT3 staining (Middle; green, P-STAT3, blue, DAPI nuclear counterstain), and immunohistochemical analysis of Ki-67 staining (Right, 400x). The percentage of Ki-67-positive nuclei represented in the graph; the great majority of Ki-67 staining was in the tumor epithelial cells, whereas only occasional stromal cells were Ki-67+.
DISCUSSION
PDAC carry extremely poor prognosis, and, in contrast to recent advances in several other common epithelial cancers, studies to date have not defined molecular features in PDAC patients that predict sensitivity to specific targeted therapies. Here, by systematic screening of >500 cancer cell lines—including 46 derived from PDAC—we identified a subset of PDAC cell lines with high sensitivity to JAK2 inhibition, and showed that this subset is characterized by activation of the gp130-STAT3 pathway. Importantly, we validated the functional role of STAT3 in PDAC pathogenesis in vivo. In keeping with the pronounced activation of STAT3 seen in ADM and PanIN in tissue specimens, genetic inactivation of STAT3 dramatically reduced both ADM and PanIN formation driven by oncogenic KRAS (Figs. 4A–C). Moreover, STAT3 inactivation blocked malignant progression to invasive PDAC despite concurrent knockdown of p53 in these cells (Figs. 6C, D). Thus, our data support a critical requirement for aberrant activation of STAT3 at multiple stages of PDAC initiation, progression, and maintenance. Importantly, several JAK2 inhibitors are in advanced clinical development (41), and our studies suggest the potential of using levels of P-STAT3 and gp130 as biomarkers for patient selection in future clinical trials for PDAC using these compounds.
P-STAT3 is first detected in ADM (Fig. 3A), the earliest pre-neoplastic lesions arising in KRAS-driven PDAC models (40). Correspondingly, while STAT3 is completely dispensable for normal pancreatic development and function, its loss dramatically reduces formation of ADM induced by oncogenic KRAS (Fig. 4A, B). This requirement for STAT3 in KRAS-induced ADM is consistent with previous data demonstrating the importance of STAT3 for ADM induced by aberrant pancreatic expression of Pdx1, a homeobox transcription factor that controls the specification and expansion of early pancreatic progenitors in the embryo (20). In addition to facilitating ADM formation and ensuing development of PanIN, activated STAT3 has an ongoing role in sustaining PanIN proliferation and progression to PDAC, and in the viability of a subset of PDAC cell lines. The broad requirement for STAT3 at early stages of PDAC tumorigenesis and its more restricted role in established PDAC cell lines suggest that STAT3 may have temporally specific functions during tumor evolution. STAT3 regulates several processes that potentially contribute to tumorigenesis, including subverting cellular differentiation programs, controlling energy metabolism, regulating an inflammatory transcriptional program, and promoting cellular survival (7,42–45). The early role of STAT3 in ADM driven either by oncogenic KRAS or by the aberrant expression of Pdx1, may reflect a requirement for STAT3 in developmental reprogramming as is observed in glioma (45,46), whereas, alternate processes may be operative in more advanced lesions that harbor additional gene mutations. Although additional investigation will be required to define functions of STAT3 in evolving PDAC, the marked inhibition of cell proliferation and survival upon STAT3 knockdown in KRAS-shp53 ductal cells and treatment of PDAC cells with the JAK2 inhibitor demonstrate a key cell autonomous role for JAK-STAT3 signaling.
STAT3 is activated by numerous growth factor and cytokine signaling pathways as well as by oncogenic RAS (7,34,43). Despite the prevalence of oncogenic KRAS mutations in human PDAC, they do not appear to contribute to STAT3 tyrosine phosphorylation in this setting since shRNAs targeting KRAS did not reduce P-STAT3 levels in PDAC cell lines (data not shown). Rather, our data demonstrate that high levels of P-STAT3 seen in ~40% of PDAC cell lines are due to differential expression of the gp130 receptor. In particular, P-STAT3 and gp130 levels showed a close correlation, and gp130 blocking antibodies specifically extinguished STAT3 tyrosine phosphorylation in the subset of PDAC cell lines showing strong activation of the pathway. We also observed increased gp130 expression in human PDAC relative to normal pancreas. Gp130 is a component of the IL6 receptor complex, and consistent with the importance of this pathway, its ligands, LIF and IL6 were also prominently elevated in human PDAC tissues. The data in cell lines indicate an important role for autocrine signaling for STAT3 activation in established PDAC, however, our findings also suggest a potential mechanism linking inflammation with ADM and pancreatic tumor initiation. ADM is observed under conditions of chronic inflammation, such as chronic pancreatitis, and chronic pancreatitis is a risk factor for PDAC (47,48). It appears likely that release of cytokines, particularly members of the IL6 cytokine family, during inflammatory conditions may lead to activation of STAT3 and may cooperate with mutated KRAS to promote ADM and PanIN formation.
In summary, our findings in mouse models and human cell lines support the therapeutic targeting of STAT3 signaling in PDAC and indicate that JAK2 inhibitors may have utility in this cancer. Recent clinical successes with targeted therapies directed at subsets of solid tumors harboring specific genetic or protein biomarkers, such as mutations in EGFR, ALK, or BRAF or amplification/overexpression of HER2 have created a paradigm for personalized approaches to cancer therapy (49). It is possible that P-STAT3 or IL6 cytokine family signaling could serve as biomarkers to guide similar approaches to applying therapies targeted against the STAT3 pathway in PDAC.
Supplementary Material
Acknowledgments
GRANT SUPPORT
R.B.C was supported by NIH training grant T32 CA071345. G.C. was supported by Fondazione Umberto Veronesi and Associazione Italiana per la ricerca sul Cancro. This work was support by grants to N.B. from the AACR-Pancreatic Cancer Action Network, Waxman Foundation for Cancer Research, NIH (NCI 2P01CA117969-06), and Dana-Farber/Harvard Cancer Center Gastrointestinal Cancer SPORE grant P50 CA127003 (to N.B. and J.A.E.)
References
- 1.Warshaw AL, Fernández-del Castillo C. Pancreatic carcinoma. N Engl J Med. 1992;326:455–65. doi: 10.1056/NEJM199202133260706. [DOI] [PubMed] [Google Scholar]
- 2.Hruban RH, van Mansfeld AD, Offerhaus GJ, van Weering DH, Allison DC, Goodman SN, et al. K-ras oncogene activation in adenocarcinoma of the human pancreas. A study of 82 carcinomas using a combination of mutant-enriched polymerase chain reaction analysis and allele-specific oligonucleotide hybridization. Am J Pathol. 1993;143:545–54. [PMC free article] [PubMed] [Google Scholar]
- 3.Klimstra DS, Longnecker DS. K-ras mutations in pancreatic ductal proliferative lesions. Am J Pathol. 1994;145:1547–50. [PMC free article] [PubMed] [Google Scholar]
- 4.Moskaluk CA, Hruban RH, Kern SE. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57:2140–3. [PubMed] [Google Scholar]
- 5.Rozenblum E, Schutte M, Goggins M, Hahn SA, Panzer S, Zahurak M, et al. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 1997 May 1;57(9):1731–4. [PubMed] [Google Scholar]
- 6.Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6:2969–72. [PubMed] [Google Scholar]
- 7.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Shuai K, Stark GR, Kerr IM, Darnell JE., Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science. 1993;261:1744–6. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 9.Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE., Jr Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–8. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 10.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–21. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 11.Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–8. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 12.Cao X, Tay A, Guy GR, Tan YH. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol Cell Biol. 1996;16:1595–603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–73. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 14.Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–5. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan KS, Sano S, Kiguchi K, Anders J, Komazawa N, Takeda J, DiGiovanni J. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J Clin Invest. 2004;114:720–8. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins BJ, Grail D, Nheu T, Najdovska M, Wang B, Waring P, et al. Hyperactivation of Stat3 in gp130 mutant mice promotes gastric hyperproliferation and desensitizes TGF-beta signaling. Nat Med. 2005;11:845–52. doi: 10.1038/nm1282. [DOI] [PubMed] [Google Scholar]
- 17.Scholz A, Heinze S, Detjen KM, Peters M, Welzel M, Hauff P, et al. Activated signal transducer and activator of transcription 3 (STAT3) supports the malignant phenotype of human pancreatic cancer. Gastroenterology. 2003;125:891–905. doi: 10.1016/s0016-5085(03)01064-3. [DOI] [PubMed] [Google Scholar]
- 18.Toyonaga T, Nakano K, Nagano M, Zhao G, Yamaguchi K, Kuroki S, et al. Blockade of constitutively activated Janus kinase/signal transducer and activator of transcription-3 pathway inhibits growth of human pancreatic cancer. Cancer Lett. 2003;201:107–16. doi: 10.1016/s0304-3835(03)00482-8. [DOI] [PubMed] [Google Scholar]
- 19.Lee JY, Hennighausen L. The transcription factor Stat3 is dispensable for pancreatic beta-cell development and function. Biochem Biophys Res Commun. 2005;334:764–8. doi: 10.1016/j.bbrc.2005.06.162. [DOI] [PubMed] [Google Scholar]
- 20.Miyatsuka T, Kaneto H, Shiraiwa T, Matsuoka TA, Yamamoto K, Kato K, et al. Persistent expression of PDX-1 in the pancreas causes acinar-to-ductal metaplasia through Stat3 activation. Genes Dev. 2006;20:1435–40. doi: 10.1101/gad.1412806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei D, Le X, Zheng L, Wang L, Frey JA, Gao AC, Peng Z, Huang S, Xiong HQ, Abbruzzese JL, Xie K. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22:319–29. doi: 10.1038/sj.onc.1206122. [DOI] [PubMed] [Google Scholar]
- 22.Jaganathan S, Yue P, Turkson J. Enhanced sensitivity of pancreatic cancer cells to concurrent inhibition of aberrant signal transducer and activator of transcription 3 and epidermal growth factor receptor or Src. J Pharmacol Exp Ther. 2010;333:373–81. doi: 10.1124/jpet.109.162669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–57. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 24.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003 Dec;4(6):437–50. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 25.Lee CK, Raz R, Gimeno R, Gertner R, Wistinghausen B, Takeshita K, et al. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002;17:63–72. doi: 10.1016/s1074-7613(02)00336-9. [DOI] [PubMed] [Google Scholar]
- 26.Agbunag C, Lee KE, Buontempo S, Bar-Sagi D. Pancreatic duct epithelial cell isolation and cultivation in two-dimensional and three-dimensional culture systems. Methods Enzymol. 2006;407:703–10. doi: 10.1016/S0076-6879(05)07055-2. [DOI] [PubMed] [Google Scholar]
- 27.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–95. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 28.Mohammad RM, Al-Katib A, Pettit GR, Vaitkevicius VK, Joshi U, Adsay V, et al. An orthotopic model of human pancreatic cancer in severe combined immunodeficient mice: potential application for preclinical studies. Clin Cancer Res. 1998;4:887–94. [PubMed] [Google Scholar]
- 29.Gozgit JM, Bebernitz G, Patil P, Ye M, Parmentier J, Wu J, et al. Effects of the JAK2 inhibitor, AZ960, on Pim/BAD/BCL-xL survival signaling in the human JAK2 V617F cell line SET-2. J Biol Chem. 2008;283:32334–43. doi: 10.1074/jbc.M803813200. [DOI] [PubMed] [Google Scholar]
- 30.McDermott U, Sharma SV, Dowell L, Greninger P, Montagut C, Lamb J, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–41. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen Z, Darnell JE., Jr Mapping of Stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic Acids Res. 1997;25:2062–7. doi: 10.1093/nar/25.11.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Blenis J, Li HC, Schindler C, Chen-Kiang S. Requirement of serine phosphorylation for formation of STAT-promoter complexes. Science. 1995;267:1990–4. doi: 10.1126/science.7701321. [DOI] [PubMed] [Google Scholar]
- 33.Taga T, Narazaki M, Yasukawa K, Saito T, Miki D, Hamaguchi M, et al. Functional inhibition of hematopoietic and neurotrophic cytokines by blocking the interleukin 6 signal transducer gp130. Proc Natl Acad Sci U S A. 1992;89:10998–1001. doi: 10.1073/pnas.89.22.10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kishimoto T, Akira S, Narazaki M, Taga T. Interleukin-6 family of cytokines and gp130. Blood. 1995;86:1243–54. [PubMed] [Google Scholar]
- 35.Kitamura T, Tange T, Terasawa T, Chiba S, Kuwaki T, Miyagawa K, et al. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol. 1989;140:323–34. doi: 10.1002/jcp.1041400219. [DOI] [PubMed] [Google Scholar]
- 36.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–26. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005 May;7(5):469–83. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 38.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006;103:5947–52. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi C, Hong SM, Lim P, Kamiyama H, Khan M, Anders RA, et al. KRAS2 mutations in human pancreatic acinar-ductal metaplastic lesions are limited to those with PanIN: implications for the human pancreatic cancer cell of origin. Mol Cancer Res. 2009;7:230–6. doi: 10.1158/1541-7786.MCR-08-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu L, Shi G, Schmidt CM, Hruban RH, Konieczny SF. Acinar cells contribute to the molecular heterogeneity of pancreatic intraepithelial neoplasia. Am J Pathol. 2007;171:263–73. doi: 10.2353/ajpath.2007.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–97. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–6. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–7. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–92. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–25. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowenfels AB, Maisonneuve P, DiMagno EP, Elitsur Y, Gates LK, Jr, Perrault J, et al. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J Natl Cancer Inst. 1997;89:442–6. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]
- 48.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–49. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 49.McDermott U, Settleman J. Personalized cancer therapy with selective kinase inhibitors: an emerging paradigm in medical oncology. J Clin Oncol. 2009;27:5650–9. doi: 10.1200/JCO.2009.22.9054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.