Abstract
BACKGROUND
T-cell large granular lymphocytic leukemia is a rare lymphoproliferative disorder characterized by the expansion of clonal CD3+CD8+ cytotoxic T lymphocytes (CTLs) and often associated with autoimmune disorders and immune-mediated cytopenias.
METHODS
We used next-generation exome sequencing to identify somatic mutations in CTLs from an index patient with large granular lymphocytic leukemia. Targeted resequencing was performed in a well-characterized cohort of 76 patients with this disorder, characterized by clonal T-cell–receptor rearrangements and increased numbers of large granular lymphocytes.
RESULTS
Mutations in the signal transducer and activator of transcription 3 gene (STAT3) were found in 31 of 77 patients (40%) with large granular lymphocytic leukemia. Among these 31 patients, recurrent mutational hot spots included Y640F in 13 (17%), D661V in 7 (9%), D661Y in 7 (9%), and N647I in 3 (4%). All mutations were located in exon 21, encoding the Src homology 2 (SH2) domain, which mediates the dimerization and activation of STAT protein. The amino acid changes resulted in a more hydrophobic protein surface and were associated with phosphorylation of STAT3 and its localization in the nucleus. In vitro functional studies showed that the Y640F and D661V mutations increased the transcriptional activity of STAT3. In the affected patients, downstream target genes of the STAT3 pathway (IFNGR2, BCL2L1, and JAK2) were up-regulated. Patients with STAT3 mutations presented more often with neutropenia and rheumatoid arthritis than did patients without these mutations.
CONCLUSIONS
The SH2 dimerization and activation domain of STAT3 is frequently mutated in patients with large granular lymphocytic leukemia; these findings suggest that aberrant STAT3 signaling underlies the pathogenesis of this disease. (Funded by the Academy of Finland and others.)
T-cell large granular lymphocytic leukemia was initially described as a clonal disorder of large granular lymphocytes involving blood, bone marrow, spleen, and liver.1 This disorder is characterized by the presence of abnormal CD3+CD8+CD57+ lymphocytes corresponding to activated effector cytotoxic T lymphocytes (CTLs).2,3 Large granular lymphocytic leukemia is frequently accompanied by autoimmune processes such as rheumatoid arthritis (often manifested as Felty’s syndrome) and immune-mediated cytopenias.4 Many cases are indolent, and distinguishing large granular lymphocytic leukemia from reactive processes involving large granular lymphocytosis may be difficult, since both conditions can be associated with a skewed CTL antigen-receptor repertoire (i.e., oligoclonal expansion rather than the normal polyclonal distribution) and can be linked to viral infections and autoimmune conditions.5–8 Furthermore, biclonal subtypes of large granular lymphocytic leukemia have been described, as well as clonal switching — both of which are suggestive of a pervasive antigenic drive.9 Thus, large granular lymphocytic leukemia may represent an extreme pole in the spectrum of CTL responses to antigen.
No recurrent chromosomal aberrations or mutations have been found to be associated with large granular lymphocytic leukemia. On the intracellular level, gene-expression studies have suggested dysregulation of apoptotic pathways (e.g., the FAS–FAS ligand and sphingolipid pathways) and activation of survival signaling pathways (e.g., the RAS and phosphatidylinositol 3-kinase [PI3K]–AKT pathways).10–14
Massively parallel second-generation sequencing technology has been used successfully to uncover the genetic background of some malignant hematologic disorders, such as acute myeloid leukemia,15,16 hairy-cell leukemia,17 the myelodysplastic syndrome,18 and chronic lymphocytic leukemia.19 The underlying genetic defects in large granular lymphocytic leukemia have not been elucidated. In hopes of illuminating the molecular pathogenesis of this disorder and providing insights into the aberrant regulation of T-cell activation in relation to the associated autoimmune disorders, we performed whole-exome and RNA sequencing of leukemic CTLs and matched healthy control cells obtained from a patient with large granular lymphocytic leukemia to identify somatic genetic changes.
METHODS
STUDY PATIENTS
The study population consisted of 77 patients with large granular lymphocytic leukemia. All patients had had CD3+CD8+ large granular lymphocytes in the peripheral blood for more than 6 months, and at the time of diagnosis, the clonality of T cells was confirmed by a polymerase-chain-reaction (PCR) assay for T-cell–receptor rearrangement. (The main characteristics of the patients are summarized in Tables S1 and S2 in the Supplementary Appendix, available with the full text of this article at NEJM.org.)
WHOLE-EXOME AND RNA SEQUENCING AND BIOINFORMATIC ANALYSES
A commercial platform (Agilent) was used to prepare sample libraries of genomic DNA from the leukemic (CD8+) cells and nonleukemic (CD4+) cells obtained from the index patient (Patient 1 in Table S1 in the Supplementary Appendix), followed by in-solution exome capture. Massively parallel sequencing was performed with the Genome Analyzer II (Illumina). Candidate somatic mutations were identified according to a bioinformatic protocol (Fig. S1 in the Supplementary Appendix). Reads from RNA sequencing were aligned and analyzed to detect fusion genes. Gene expression was evaluated visually from RNA-sequence read alignments with the Integrative Genomics Viewer (Broad). (Whole-exome sequencing, bioinformatics, and other methods are fully described in the Supplementary Appendix.)
VALIDATION OF CANDIDATE SOMATIC MUTATIONS AND STAT3 SCREENING
After exome sequencing, the candidate mutations were validated with the use of Sanger sequencing. For the STAT3 mutation screening, five primer pairs were designed to cover the six exons encoding the Src homology 2 (SH2) domain of STAT3 (exons 19 to 24). (The primers are described in Table S3 in the Supplementary Appendix.)
STATISTICAL ANALYSIS
All statistical analyses are described in detail in the Supplementary Appendix.
RESULTS
SOMATIC MUTATIONS IDENTIFIED BY EXOME SEQUENCING IN THE INDEX PATIENT
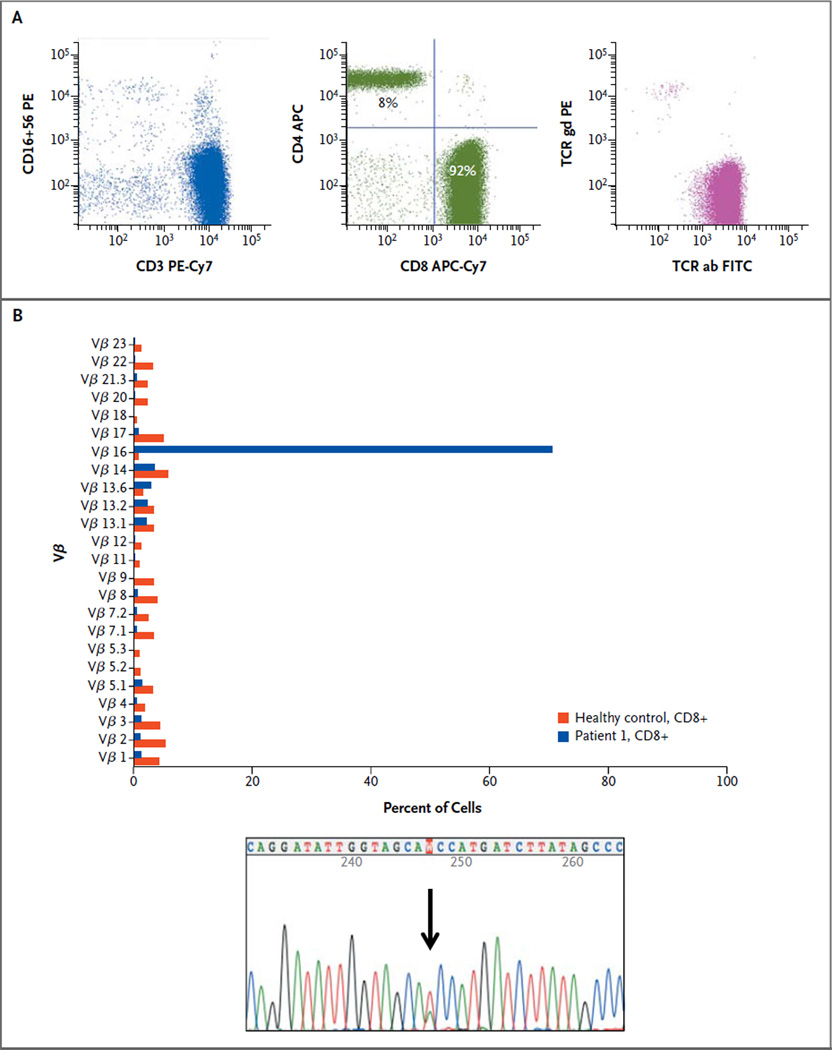
The index patient was a 70-year-old man with untreated typical T-cell large granular lymphocytic leukemia. Figure 1A shows the results on flow cytometry. Whole-exome sequencing of purified CD8+ leukemic cells and CD4+ control cells from the index patient produced approximately 61.1 million and 60.2 million reads, respectively, that could be mapped to the reference genome (Fig. S1 in the Supplementary Appendix). The pairedend read length was 82 nucleotides. After somatic mutation calling and validation by capillary sequencing, the genes for signal transducer and activator of transcription 3 (STAT3), macrophage scavenger receptor 1 (MSR1), and homeobox C9 (HOXC9) were found to harbor somatic heterozygous mutations specific to the CD8+ cell population. The mutations in STAT3 and MSR1 introduced an amino acid substitution in the encoded proteins, whereas the mutation in HOXC9 was a deletion of an amino acid (Table S4 in the Supplementary Appendix). (The STAT3 mutation is shown in Fig. 1B, and in Fig. S2 in the Supplementary Appendix.) No fusion genes were identified by RNA sequencing.
Figure 1. Results of Flow Cytometry and Sequencing in the Index Patient.
The index patient was a 70-year-old man with untreated large granular lymphocytic leukemia and grade 3 neutropenia. Panel A shows the results of flow cytometry. CD3 and CD16/56 staining of the lymphocyte population indicated that the majority of cells were CD3-positive and CD16/56-negative (left plot); 92% of CD3+ gated cells were CD4-negative and CD8-positive (middle plot). CD8+ T cells expressed T-cell receptor alpha and beta (right plot). APC denotes allophycocyanin, Cy7 cyanine 7, FITC fluorescein isothiocyanate, PE phycoerythrin, TCR ab T-cell receptor alpha and beta, and TCR gd gamma and delta. Panel B shows the results of Vβ and validation sequencing of the CD8 sorted cells. When the sample was obtained for exome-sequencing analysis, the patient had one large, predominantly T-cell clone: 94% of CD8+ cells consisted of a single Vβ16 clone (upper graph). A heterozygous STAT3 SH2 mutation T→A (protein D661V) was detected in CD8+ T cells (lower graph).
STAT3 D661V MUTATION IN THE INDEX PATIENT
The STAT3 mutation in chromosome 17 in the index patient was located in exon 21, encoding a portion of the STAT3 SH2 domain (Fig. 2A and 2B). A heterozygous missense mutation (T→A) resulted in a D661V variant protein. In the germline CD4+ control sample, only normal reads (63) were detected, whereas in the CD8+ tumor sample, 29 mutant reads and 39 germline reads were detected, resulting in a mutant allele frequency of 43% (Fig. S2 in the Supplementary Appendix). Also, sequencing of RNA derived from the CD8+ leukemic cells identified the D661V mutation as a prominent clone (Fig. S2A in the Supplementary Appendix), whereas it was absent from the CD8+ RNA from healthy control cells.
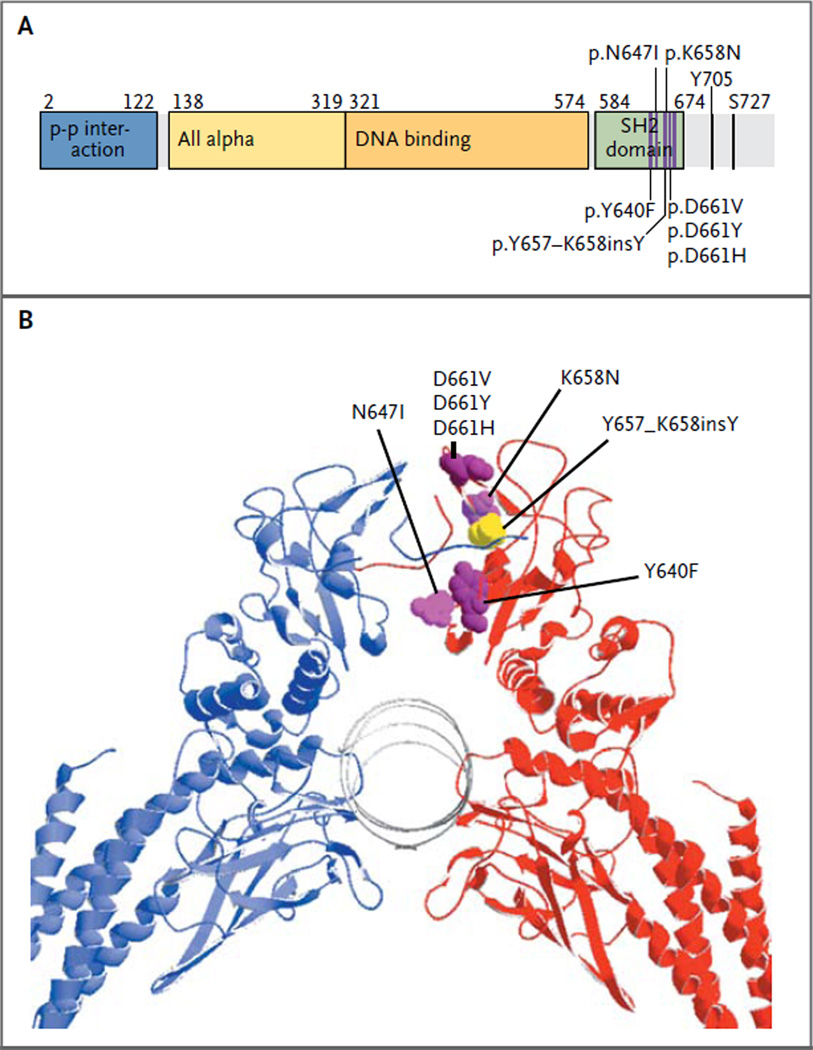
Figure 2. Locations of STAT3 Mutations and Crystal Structure of STAT3 Homodimer.
All seven STAT3 mutations (D661V, D661Y, D661H, N647I, K658 N, Y640F, and Y657_K658insY) are found in the SH2 domain of the STAT3 protein and are only a couple of amino acids apart, as shown in a linear representation of the STAT3 primary sequence (Panel A). The structure of the STAT3 domain is represented according to the definition in the protein families database (Sanger Institute, http://pfam.sanger.ac.uk). A three-dimensional model of the STAT3 dimer (Panel B) shows that the mutated residues are located at the dimerization interface of the SH2 domain. Six STAT3 mutations are shown in one of the two subunits (magenta), as is the in-frame insertion mutation (yellow).
STAT3 SCREENING IN THE PATIENT COHORT
STAT3 is an oncogene, and its activation plays a key role in cell signaling in many types of cancer.20 Since the D661V mutation was located in the SH2 domain (Fig. 2), which mediates dimerization and activation of the STAT3 protein, we next designed primers for the secondary screening of the six exons encoding the STAT3 SH2 region in the remaining patients. Five other recurrent somatic missense mutations and one inframe insertion were identified in 30 additional patients (Fig. 2, and Fig. S3 and Tables S1 and S5 in the Supplementary Appendix), with a STAT3 mutation frequency of 40% (31 of all 77 patients in the cohort). The most common mutation hot spot was located at amino acid D661, resulting in three different variant proteins: D661V (mutation frequency, 9% [7 patients]), D661Y (mutation frequency, 9% [7 patients]), and D661H (mutation frequency, 1% [1 patient]). Y640F mutations were detected in 17% of patients (13 of 77), N647I mutations in 4% (3 of 77), and a K658N mutation in 1 patient, as well as an in-frame insertion mutation (Y657_K658insY) in an additional patient at the same site.
In the other 46 patients with large granular lymphocytic leukemia who had more than 20% clonal cells in the DNA sample (on the basis of CD8+ T-cell counts and Vβ expression), no mutations were detected in the STAT3 SH2 domain (Table S2 in the Supplementary Appendix). Also, no HOXC9 or MSR1 mutations were found in any other patient in the study cohort. In addition, two T-cell acute lymphoblastic leukemia cell lines (Jurkat and MOLT-4) and samples from 9 patients with acute myeloid leukemia and from 6 patients with acute lymphocytic leukemia were screened, and no STAT3 mutations were found. Similarly, CD8+ cells from 6 patients with reactive large granular lymphocytosis during tyrosine kinase inhibitor therapy,8 as well as from 2 patients with cytomegalovirus infection and from 1 patient with graft-versus-host disease, were studied and found to be negative for STAT3 mutations. Moreover, none of the identified mutations were present in two population-variation data sets: 1000 Genomes (www.1000genomes.org/data) and dbSNP (www.ncbi.nlm.nih.gov/projects/SNP).
Further sequencing analysis of sorted B-cell, T-cell, and natural killer (NK)–cell fractions was performed for Patients 1, 19, and 30 in the group of patients with STAT3 mutations. STAT3 mutations were found only in the T-cell (CD8+) fraction, whereas NK-cell and B-cell fractions showed wildtype STAT3 (Fig. S4 in the Supplementary Appendix).
All identified STAT3 mutations were located only a few codons apart in exon 21 (Fig. 2A). Furthermore, they all induced an amino acid change that made the protein surface more hydrophobic. The protein structure shows that they are positioned very close to each other in the dimerization interface (Fig. 2B). Only one of the six STAT3 mutations (Y640F) has been described previously, in inflammatory hepatocellular adenoma.21 This mutation alters a conserved tyrosine residue, leading to cytokine-independent tyrosine phosphorylation and STAT activation, as well as to cytokine-dependent hyperactivation.21,22
CLINICAL CHARACTERISTICS AND STAT3 MUTATION STATUS
A comparison of the clinical characteristics of patients with large granular lymphocytic leukemia showed that neutropenia was more common among patients with a STAT3 mutation (24 of 31 patients [77%]) than among those without a STAT3 mutation (23 of 46 patients [50%]) (P=0.02) (Tables S1 and S2, respectively, in the Supplementary Appendix). Similarly, patients with STAT3 mutations had rheumatoid arthritis more often than did those without STAT3 mutations (26% vs. 6%, P=0.02). No differences were found between the groups with and those without STAT3 mutations in terms of age, sex, hemoglobin level, and leukocyte or lymphocyte values, and the frequency of signs of B-cell dyscrasia, anemia, or thrombocytopenia was similar in the two groups. Basic laboratory values and the presence or absence of related autoimmune disorders did not differ significantly according to the specific STAT3 mutation (D661V, D661Y, or Y640F), but the number of patients with each mutation was small (Fig. S5 in the Supplementary Appendix).
In one patient with the Y640F mutation (Patient 31), SNP-A karyotyping identified unisomal parental disomy in 17q, where STAT3 is located, resulting in a homozygous mutation (Fig. S6 in the Supplementary Appendix). This patient had both anemia and neutropenia and had been treated with cyclosporine and alemtuzumab.
PHOSPHORYLATION AND TRANSLOCATION INTO NUCLEUS OF STAT3
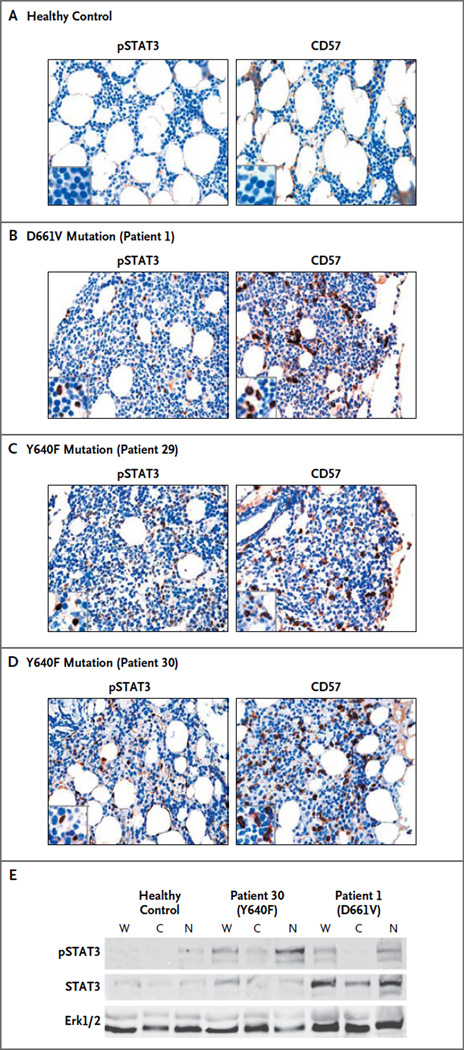
Bone marrow–biopsy samples were obtained from one healthy control and from three patients with large granular lymphocytic leukemia, one with the D661V mutation and two with the Y640F mutation (Fig. 3). Immunohistochemical staining showed that the lymphocyte infiltration in the patients with leukemia expressed CD57, which is typical of large granular lymphocytes (Fig. 3B, 3C, and 3D). Unlike the healthy control (Fig. 3A), the patients with leukemia had positive staining of the nuclei of infiltrated lymphocytes, with phosphorylated STAT3 antibody indicating STAT3 activation. These findings were confirmed by Western blot analysis of nuclear extract samples from two of the patients (one with the Y640F mutation and one with the D661V mutation) and from the healthy control (Fig. 3E).
Figure 3. Immunohistochemical Staining of Bone Marrow–Biopsy Samples from a Healthy Control and from Three Patients with Large Granular Lymphocytic Leukemia.
Panels A through D show paraffin sections from bone marrow–biopsy samples stained with monoclonal antibodies against phosphorylated STAT3 (pSTAT) (left) and CD57 (right). No staining is observed in the samples obtained from healthy controls (a representative example of which is shown in Panel A). The samples obtained from three patients who had large granular lymphocytic leukemia with STAT3 mutations (Panels B, C, and D) show infiltration by lymphocytes, which stain for pSTAT3 (in the nucleus) and for CD57 (in the cytoplasm). Panel E shows fractionated mononuclear cells from a healthy donor and from Patient 30 and Patient 1, who had Y640F and D661V STAT3 mutations, respectively. The mutant forms are hyperphos-phorylated and localized predominantly in the nucleus. Normalized aliquots of whole-cell (W), cytosolic (C), and nuclear (N) fractions were resolved on sodium dodecyl sulfate–polyacrylamide-gel electrophoresis and transferred to polyvinylidene fluoride membrane, and Western blot analysis was performed with the use of anti-pSTAT3, anti-STAT3, and anti-Erk1 antibodies. Erk1/2 denotes extracellular regulated kinases 1 and 2.
UP-REGULATION OF STAT3 TRANSCRIPTIONAL TARGETS IN PATIENTS WITH STAT3 MUTATIONS
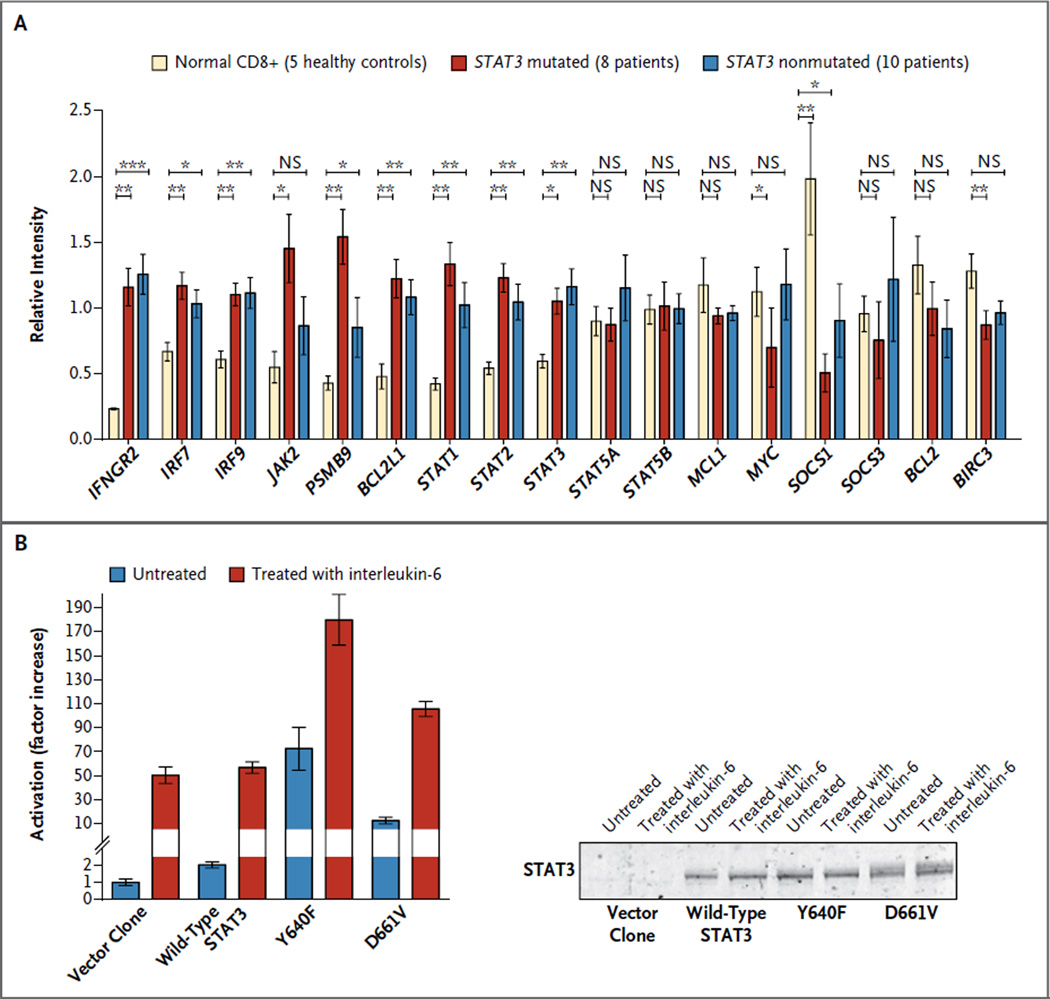
With the use of the HumanHT-12 v4 Expression BeadChip array, RNA expression analysis was performed on CD8+ sorted cells from 8 patients with STAT3 mutations (D661H, D661Y, D661V, and Y640F) and from 10 patients without STAT3 mutations. CD8+ cells from 5 healthy volunteers were obtained for comparison. INFGR2, IRF7, IRF9, PSMB9, BCL2L1, STAT1, STAT2, and STAT3 were significantly up-regulated and SOCS1 was down-regulated in the samples from the patients with and those without STAT3 mutations (Fig. 4A). The expression of JAK2 was significantly higher only in the patients with a STAT3 mutation, whereas expression of interleukin-6, one of the main up-stream regulators of STAT3, was not significantly changed in these patients, although the values ranged widely. No significant differences were found in BCL2 or MCL1 expression in between-group comparisons (Fig. 4A). Analysis of the RNA sequencing data from the index patient with the D661V mutation showed similar up-regulation of STAT3 target genes, including the FAS ligand (Fig. S7 in the Supplementary Appendix).
Figure 4. Quantification of STAT3-Responsive Genes in Patients with STAT3 Mutations, and Activation of STAT3-Dependent Transcription by Wild-Type and Mutant Protein Variants.
Panel A shows the mean levels of RNA expression of STAT3-responsive genes from CD8 sorted cells obtained from 5 healthy controls, 8 patients with STAT3 mutations, and 10 patients without STAT3 mutations, with the use of the HumanHT-12 v4 Expression BeadChip Array (Illumina). I bars represent standard errors. One asterisk denotes P<0.05, two asterisks P<0.01, and three asterisks P<0.001. Panel B shows human embryonic kidney cells that were left untreated or treated with interleukin-6 to induce STAT3 phosphorylation and activation. These cells, which harbored a luciferase reporter gene under the control of a STAT3-responsive sis-inducible element (SIE), were transfected with vector alone, wild-type STAT3, or the two mutants most frequently identified (Y640F and D661V), and each condition was tested in triplicate.
Y640F AND D661V MUTATIONS AND INCREASED TRANSCRIPTIONAL ACTIVITY OF STAT3
To explore the functional implications of the Y640F and D661V mutations, we generated expression constructs for wild-type, Y640F, and D661V variants of the STAT3 protein. When expressed in HEK293 human embryonic kidney cells carrying a luciferase reporter gene under the control of a STAT3-responsive sis-inducible element (SIE), the Y640F mutant exhibited strikingly increased activity under both basal (unstimulated) and interleukin-6–stimulated conditions (Fig. 4B). The D661V mutant caused more modest (but still significant) basal and interleukin-6–stimulated transcriptional activation (Fig. 4B).
DISCUSSION
By means of whole-exome and targeted sequencing, we found that 40% of patients with large granular lymphocytic leukemia had activating mutations in STAT3. The STAT3 missense mutations (D661V, D661Y, D661H, Y640F, N647I, and K658N), as well as the insertion mutation (Y657_K658insY), were located in the SH2 domain on the dimerization interface that mediates STAT3 activation.23 Structural modeling indicated that all the observed mutations increased the hydrophobicity of the STAT3 SH2 dimerization surface. The mutational hot spot in large granular lymphocytic leukemia resides close to the transcriptional activation domain, and constitutive activation may result from increased stabilization of STAT3 homodimers or heterodimers through enhanced hydrophobic attraction between STAT monomers.23 Leukemic large granular lymphocytes have been shown to express high levels of activated STAT3,24 supporting the functional role of the STAT3 SH2 domain mutations. In accordance with this finding, an in vitro assay showed up-regulation of STAT3 target gene expression in patients with large granular lymphocytic leukemia who had the mutation and increased transcriptional activity of the mutated STAT3 isoforms. Furthermore, STAT3 was phosphorylated in the patients with the mutation.
Studies done in T-cell–specific Stat3-deficient mice show that Stat3 activation is responsible for interleukin-6–dependent T-cell proliferation by preventing apoptosis independently of Bcl-2.25 Furthermore, in a liver-specific mouse model, constitutively active Stat3 was shown to provide protection against Fas-mediated liver injury, whereas liver-specific Stat3-knockout mice failed to survive a typically nonlethal dose of FAS agonist.26 Similarly, leukemic large granular lymphocytes appear to be resistant to FAS-induced apoptosis, despite high levels of FAS and FAS ligand expression27; when STAT3 expression is reduced with an antisense oligonucleotide approach, large granular lymphocytes appear to regain their FAS sensitivity.24,28 Also, other antiapoptotic and survival-promoting signaling pathways, including some regulated by STAT3, have been shown to be upregulated in leukemic large granular lymphocytes.10,13,24 These findings support the hypothesis that constitutive STAT3 activation in large granular lymphocytic leukemia cells enhances antiapoptotic pathways. New, specific STAT3 inhibitors (e.g., OPB-3112)29 have recently been evaluated in early clinical trials for the treatment of other hematologic cancers, and our findings suggest that preclinical and clinical studies of these drugs are also warranted in patients with large granular lymphocytic leukemia.
The diagnosis of large granular lymphocytic leukemia can be difficult to establish, since benign large granular lymphocytosis with clonal T cells may occur in conjunction with viral infections (e.g., cytomegalovirus infection) or autoimmune disorders. Somatic STAT3 mutations may provide a diagnostic tool for classifying some cases of large granular lymphocytosis as true T-cell leukemias that are distinct from clonally skewed reactive processes and from hyperreactive or persistent T-cell responses. We included in our study only patients with one major Vβ clone in order to ensure that the study cohort would be clinically and genetically uniform. More sensitive techniques (e.g., Vβ clone sorting, mutation-specific real-time quantitative PCR assay) would allow for the detection of mutations in patients with smaller or multiple clones as well; thus, the true mutational frequency in large granular lymphocytic leukemia may be higher or lower. Since expression analysis showed that patients with and those without STAT3 mutations had many common overexpressed STAT3 target genes, further studies are warranted to determine whether STAT3 mutation–negative patients have mutations in other STAT-related pathways.
Patients with STAT3 mutations had neutropenia and rheumatoid disorders more often than did patients with wild-type STAT3. Large granular lymphocytes can secrete several cytokines that may play a role in immune-mediated cytopenias and autoimmune disorders.30,31 The associated clinical findings may also be due to antihost activity of expanded CTLs, since it has been shown that leukemic large granular lymphocytes can directly lyse pulmonary-artery endothelial cells and human synovial cells.32 Constitutive activation of STAT3 has been observed in patients with autoimmune disorders such as rheumatoid arthritis and Crohn’s disease.33–35 The strong link between large granular lymphocytic leukemia and these disorders warrants screening for STAT3 mutations in T lymphocytes from patients who have autoimmune disease in the absence of overt large granular lymphocytic leukemia.
In conclusion, STAT3 is frequently mutated in large granular lymphocytic leukemia, highlighting key steps in the molecular pathogenesis of this rare disease. Structural and functional data suggest that the observed mutations result in STAT3 hyperactivation and may confer ligand-independent signaling.
Supplementary Material
Acknowledgments
Supported by a special subsidy for health sciences, research, and training provided by the Finnish government and by grants from the Academy of Finland, the Finnish Cancer Societies, the Sigrid Juselius Foundation, the Finnish Association of Hematology, the National Clinical Graduate School, the Finnish Funding Agency for Technology and Innovation, the European Regional Development Fund, the Jane and Aatos Erkko Foundation, the K.A. Johansson Foundation, and the Signe and Ane Gyllenberg Foundation, and by grants from the National Institutes of Health (R01 CA127264A, to Mr. Clemente; R01AI085578, to Ms. Guinta; CA98472, to Dr. Loughran; and 2K24HL077522, to Dr. Maciejewski).
We thank the personnel at the Hematology Research Unit Helsinki and Institute for Molecular Medicine Finland for their expert clinical and technical assistance.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Loughran TP, Jr., Kadin ME, Starkebaum G, et al. Leukemia of large granular lymphocytes: association with clonal chromosomal abnormalities and autoimmune neutropenia, thrombocytopenia, and hemolytic anemia. Ann Intern Med. 1985;102:169–175. doi: 10.7326/0003-4819-102-2-169. [DOI] [PubMed] [Google Scholar]
- 2.Loughran TP., Jr. Clonal diseases of large granular lymphocytes. Blood. 1993;82:1–14. [PubMed] [Google Scholar]
- 3.Sokol L, Loughran TP., Jr. Large granular lymphocyte leukemia. Oncologist. 2006;11:263–273. doi: 10.1634/theoncologist.11-3-263. [DOI] [PubMed] [Google Scholar]
- 4.Burks EJ, Loughran TP., Jr. Pathogenesis of neutropenia in large granular lymphocyte leukemia and Felty syndrome. Blood Rev. 2006;20:245–266. doi: 10.1016/j.blre.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Rossi D, Franceschetti S, Capello D, et al. Transient monoclonal expansion of CD8+/CD57+ T-cell large granular lymphocytes after primary cytomegalovirus infection. Am J Hematol. 2007;82:1103–1105. doi: 10.1002/ajh.20981. [DOI] [PubMed] [Google Scholar]
- 6.Gentile TC, Hadlock KG, Uner AH, et al. Large granular lymphocyte leukaemia occurring after renal transplantation. Br J Haematol. 1998;101:507–512. doi: 10.1046/j.1365-2141.1998.00712.x. [DOI] [PubMed] [Google Scholar]
- 7.Mohty M, Faucher C, Vey N, et al. Features of large granular lymphocytes (LGL) expansion following allogeneic stem cell transplantation: a long-term analysis. Leukemia. 2002;16:2129–2133. doi: 10.1038/sj.leu.2402645. [DOI] [PubMed] [Google Scholar]
- 8.Mustjoki S, Ekblom M, Arstila TP, et al. Clonal expansion of T/NK-cells during tyrosine kinase inhibitor dasatinib therapy. Leukemia. 2009;23:1398–1405. doi: 10.1038/leu.2009.46. [DOI] [PubMed] [Google Scholar]
- 9.Clemente MJ, Wlodarski MW, Makishima H, et al. Clonal drift demonstrates unexpected dynamics of the T-cell repertoire in T-large granular lymphocyte leukemia. Blood. 2011;118:4384–4393. doi: 10.1182/blood-2011-02-338517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang R, Shah MV, Yang J, et al. Network model of survival signaling in large granular lymphocyte leukemia. Proc Natl Acad Sci U S A. 2008;105:16308–16313. doi: 10.1073/pnas.0806447105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah MV, Zhang R, Irby R, et al. Molecular profiling of LGL leukemia reveals role of sphingolipid signaling in survival of cytotoxic lymphocytes. Blood. 2008;112:770–781. doi: 10.1182/blood-2007-11-121871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epling-Burnette PK, Bai F, Wei S, et al. ERK couples chronic survival of NK cells to constitutively activated Ras in lymphoproliferative disease of granular lymphocytes (LDGL) Oncogene. 2004;23:9220–9229. doi: 10.1038/sj.onc.1208122. [DOI] [PubMed] [Google Scholar]
- 13.Schade AE, Powers JJ, Wlodarski MW, Maciejewski JP. Phosphatidylinositol-3-phosphate kinase pathway activation protects leukemic large granular lymphocytes from undergoing homeostatic apoptosis. Blood. 2006;107:4834–4840. doi: 10.1182/blood-2005-08-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schade AE, Wlodarski MW, Maciejewski JP. Pathophysiology defined by altered signal transduction pathways: the role of JAK-STAT and PI3K signaling in leukemic large granular lymphocytes. Cell Cycle. 2006;5:2571–2574. doi: 10.4161/cc.5.22.3449. [DOI] [PubMed] [Google Scholar]
- 15.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiacci E, Trifonov V, Schiavoni G, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papaemmanuil E, Cazzola M, Boult-wood J, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Lawrence MS, Wan Y, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [ Erratum, Cell 1999;99:239.] [DOI] [PubMed] [Google Scholar]
- 21.Pilati C, Amessou M, Bihl MP, et al. Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. J Exp Med. 2011;208:1359–1366. doi: 10.1084/jem.20110283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scarzello AJ, Romero-Weaver AL, Maher SG, et al. A mutation in the SH2 domain of STAT2 prolongs tyrosine phosphorylation of STAT1 and promotes type I IFN-induced apoptosis. Mol Biol Cell. 2007;18:2455–2462. doi: 10.1091/mbc.E06-09-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker S, Groner B, Müller CW. Three-dimensional structure of the Stat-3beta homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 24.Epling-Burnette PK, Liu JH, Catlett-Falcone R, et al. Inhibition of STAT3 signaling leads to apoptosis of leukemic large granular lymphocytes and decreased Mcl-1 expression. J Clin Invest. 2001;107:351–362. doi: 10.1172/JCI9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J Immunol. 1998;161:4652–4660. [PubMed] [Google Scholar]
- 26.Haga S, Terui K, Zhang HQ, et al. Stat3 protects against Fas-induced liver injury by redox-dependent and -independent mechanisms. J Clin Invest. 2003;112:989–998. doi: 10.1172/JCI17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamy T, Liu JH, Landowski TH, Dalton WS, Loughran TP., Jr Dysregulation of CD95/CD95 ligand-apoptotic pathway in CD3(+) large granular lymphocyte leukemia. Blood. 1998;92:4771–4777. [PubMed] [Google Scholar]
- 28.Yang J, Epling-Burnette PK, Painter JS, et al. Antigen activation and impaired Fasinduced death-inducing signaling complex formation in T-large-granular lymphocyte leukemia. Blood. 2008;111:1610–1616. doi: 10.1182/blood-2007-06-093823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayakawa F, Sugimoto K, Kurahashi S, Sumida T, Naoe T. A novel STAT3 inhibitor OPB-31121 induces tumor-specific growth inhibition in a wide range of hematopoietic malignancies without growth suppression of normal hematopoietic cells. Program and abstracts of the 53rd American Society of Hematology annual meeting and exposition; December 10–13, 2011; San Diego, CA. abstract. [Google Scholar]
- 30.Liu JH, Wei S, Lamy T, et al. Chronic neutropenia mediated by Fas ligand. Blood. 2000;95:3219–3222. [PubMed] [Google Scholar]
- 31.Kothapalli R, Nyland SB, Kusmartseva I, Bailey RD, McKeown TM, Loughran TP., Jr Constitutive production of proinflammatory cytokines RANTES, MIP-1beta and IL-18 characterizes LGL leukemia. Int J Oncol. 2005;26:529–535. [PubMed] [Google Scholar]
- 32.Chen X, Bai F, Sokol L, et al. A critical role for DAP10 and DAP12 in CD8+ T cell-mediated tissue damage in large granular lymphocyte leukemia. Blood. 2009;113:3226–3234. doi: 10.1182/blood-2008-07-168245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Sengupta TK, Zhong Z, Ivashkiv LB. Regulation of the balance of cytokine production and the signal transducer and activator of transcription (STAT) transcription factor activity by cytokines and inflammatory synovial fluids. J Exp Med. 1995;182:1825–1831. doi: 10.1084/jem.182.6.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramos HL, Valencia-Pacheco G, Alcocer-Varela J. Constitutive STAT3 activation in peripheral CD3(+) cells from patients with primary Sjögren’s syndrome. Scand J Rheumatol. 2008;37:35–39. doi: 10.1080/03009740701606010. [DOI] [PubMed] [Google Scholar]
- 35.Lovato P, Brender C, Agnholt J, et al. Constitutive STAT3 activation in intestinal T cells from patients with Crohn’s disease. J Biol Chem. 2003;278:16777–16781. doi: 10.1074/jbc.M207999200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.