Summary
Age-associated changes in immune response increase the risk of infection and promote inflammation and auto-immunity in older adults. The newly discovered cytokine IL-23 contributes to the maintenance and expansion of Th-17 cells, which promote proinflammatory responses. Our preliminary findings suggested that Th-17 responses are increased in aged mice. IL-23 consists of p40 and p19 subunits. Expression of the p19 subunit is regulated at the transcriptional level by NF-κB p65 and c-Rel transcription factors. Using bone-marrow-derived dendritic cells (DCs) from C57BL/6 mice, we show that IL-23 protein production and p19 subunit mRNA levels are significantly increased in DCs from aged mice after activation with TLR ligands (LPS + R848) when compared with DCs of young adult mice. We found that the increase in p19 expression in aged cells is associated with chromatin remodeling characterized by di- and tri-methylation of histone H3K4 and binding of mainly c-Rel at the p19 promoter. In young DCs, the promoter is tri-methylated only at H3K4 and bound by both p65 and c-Rel. C-Rel knockdown restores p65 binding in aged cells but does not activate p19 expression, suggesting that c-Rel is critical for p19 expression. In addition, p65 knockdown significantly increases c-Rel binding and p19 expression in young DCs to levels close to those detected in old cells. Furthermore, the decrease in p65 binding at the p19 promoter in old DCs was specific to the p19 gene since p65 binding to the IL-12p40 promoter was not significantly different between old and young DCs. Our results demonstrate that selective changes in H3K4 methylation, and c-Rel and p65 binding at the p19 promoter occur in DCs and contribute to the upregulation of the p19 subunit expression and IL-23 protein production observed in aged mice. This suggests epigenetic and transcriptional mechanisms contribute to dysregulated inflammatory and autoimmune responses associated with aging.
Keywords: aging, epigenetics, IL-23, inflammation
Introduction
Aging is associated with immunosenescence, characterized by increases in inflammatory and autoimmune responses and declines in protective immunity (Trebilcock & Ponnappan, 1996; Gangemi et al., 2003; Wikby et al., 2005; Dinarello, 2006; Yung & Julius, 2008). Previous studies have reported decline in cell-mediated immunity, including T and B cell function, in aging humans with increased frequency of autoantibody production and organ-specific autoimmune diseases, such as Hashimoto’s thyroiditis and rheumatoid arthritis (Boren & Gershwin, 2004; Yung & Julius, 2008). In addition, some studies have reported impaired function of innate immune cells in elderly humans and in genetically deficient mice (Liu et al., 2007; Van & Shaw, 2007). These studies suggest that abnormalities in the aging immune system are because of various deficiencies in T cell-dependent adaptive responses and concomitant increases in the production of proinflammatory cytokines. The loss of protective immunity is accompanied by chronic inflammation in aging because of changes in the expression of inflammatory mediators (Yung & Julius, 2008). For example, the levels of circulating inflammation-associated cytokines, such as IL-6, TNFα, sIL-2 receptor (Gangemi et al., 2003; Wikby et al., 2005; Dinarello, 2006; Yung & Julius, 2008) and chemokines, such as IL-8, MCP-1, MIP-1α, and RANTES (Swift et al., 2001; Chen et al., 2003) are higher in aged than in young humans. Of note, are recent studies showing that age-related changes in cytokine and immune responses depend largely on the T helper (Th)17 inflammatory subset of CD4 T cells, which has recently been implicated in mediating host defense against infection and promoting autoimmunity and inflammation by producing IL-17 and other cytokines (Bettelli et al., 2008; Dong, 2008; Huang et al., 2008). It has become increasingly evident that Th-17 immunity plays a significant role in host defense and autoimmune responses, and older adult T cells appears to be predisposed to a Th-17 response (Huang et al., 2008).
IL-23 is a member of the IL-12 cytokine family, which includes IL-12, IL-23, IL-35, and IL-27. IL-23 consists of two subunits: IL-12p40 subunit (which is shared with IL-12) and IL-23p19 subunit (which shares sequence homology with IL-12p35 subunit but is distinct and leads to unique biologic effects). Like IL-12, IL-23 is predominantly produced by macrophages and dendritic cells (DCs) activated by bacterial products, such as Gram-negative bacterial lipopolysaccharides (LPS), Sendai virus, and proinflammatory cytokines, such as TNFα and IL-1β (Fiorentino et al., 1991; Hunter, 2005; Dong, 2008). Recently, the role of IL-23 in inflammatory responses has been demonstrated in animal models (Cua et al., 2003; Murphy et al., 2003). IL-23 selectively promotes IL-17 cytokine production by maintaining and expanding Th17 cell expansion/maintenance (Cua et al., 2003) and promoting the production of other proinflammatory mediators, including IL-8, IL-6, MCP-1, and G-CSF (Libermann & Baltimore, 1990; Dong, 2008). Synthesis of biologically active IL-23 heterodimer requires the expression of both p40 and p19 subunits (Dong, 2008). Mice lacking expression of the IL-23p19 subunit are protected against experimentally induced arthritis (Kontgen et al., 1995; Cua et al., 2003; Murphy et al., 2003). This protection is associated with defects in IL-17 production (Langrish et al., 2005). In addition, mice overexpressing the p19 subunit gene suffer systemic inflammation (Gerondakis et al., 1996). Recent studies have demonstrated that the p19 subunit gene is mainly regulated at the transcriptional level by NF-κB transcription factors p65 and c-Rel that bind to p19 promoter and are necessary for TLR-induced activation of the p19 subunit expression in murine macrophages and DCs (Carmody et al., 2007; Mise-Omata et al., 2007).
Chromatin constitutes a platform for the assembly of transcription factors and cofactors, and can be broadly divided into heterochromatin, which is closed and transcriptionally silent, and euchromatin, which is open and transcriptionally active (Grunstein et al., 1995; Sedivy et al., 2008). Post-translational modifications on histones (the basic building units of chromatin) affects chromatin configuration and transcription factor binding. Methylation of histone (H)3 at lysine 4 (H3K4) has been linked to gene activation whereas methylation at lysine 9 (H3K9) and lysine 27 (H3K27) are associated with gene repression (Sims et al., 2003; Kouzarides, 2007). Distinct methylation states are associated with different transcriptional outcomes. H3K4 can be mono-, di-, and tri-methylated (Strahl et al., 1999). Both di-methylated H3K4 (H3K4me2) and tri-methylated H3K4 (H3K4me3) are detected on active genes (Noma et al., 2001; Bernstein et al., 2002). In particular, tri-methylation is preferentially associated with promoters of active genes (Schneider et al., 2004). In addition, mammalian aging is associated with remodeling of chromatin structure (Sedivy et al., 2008). In particular, there is an overall decrease in heterochromatic structures, and many senescent cells show dramatic changes in chromatin structure.
Chromatin remodeling because of histone modifications greatly affect the binding and activity of chromatin-associated cofactors and transcription factors, including NF-κB, to their cognate DNAs thereby controlling gene expression (Sheppard et al., 1999; Cheung et al., 2000b; Chen et al., 2002; Kiernan et al., 2003). The NF-κB/Rel family of transcription factors plays a crucial role in immune and inflammatory responses by activating a wide variety of genes involved in host defense (Baeuerle & Henkel, 1994; Verma et al., 1995; Baldwin, 1996). Five NF-κB family members have been identified in mammals. These include p65, p50, p52, c-Rel, and RelB (Baldwin, 1996). These proteins share a common sequence known as Rel homology domain and exist as homo- and heterodimer complexes that are maintained in an inactive form in the cytoplasm through association with inhibitory IκB proteins (Verma et al., 1995; Ghosh et al., 1998). Cellular activation by various signals leads to the phosphorylation and degradation of IκB, resulting in nuclear translocation of NF-κB complexes where they bind to NF-κB binding sites present in the promoters and enhancers of many genes, including inflammatory cytokine genes, such as IL-2, IL-12, IL-6, IL-1β, TNFα, and IL-23 (Bull et al., 1989; Collart et al., 1990; Libermann & Baltimore, 1990; Hiscott et al., 1993; Verma et al., 1995; Carmody et al., 2007).
Our preliminary studies suggested that IL-23 expression might be dysregulated during aging, because of the increased expression of Th-17 immunity in aged mice (High et al., 2007). In this study, we investigated transcriptional regulation of the IL23p19 subunit gene, which is critical for IL-23 protein production, in murine (C57BL/6) bone marrow-derived DCs. Our results show increased production of IL-23 in TLR-activated DCs derived from aged mice compared with young mice. We also show that changes in chromatin configuration affect the NF-κB transcription factor p65 and c-Rel binding to p19 proximal promoter. The promoter is di- and tri-methylated at H3K4 and bound almost exclusively by c-Rel in aged DCs. In young cells, however, the promoter is only tri-methylated and bound by both p65 and c-Rel proteins. Our studies demonstrate that differential epigenetic changes and transcriptional factor binding underlie the increase in p19 expression and IL-23 production that is associated with aging, and suggest that epigenetic and transcriptional mechanisms in DCs likely play a role in the dysregulated inflammatory and autoimmune responses in the aged.
Results
IL-23 protein production and p19 gene subunit expression are significantly increased in aged DCs
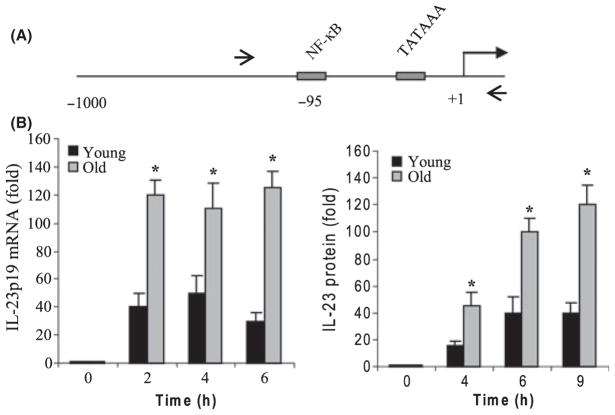
As mentioned earlier, IL-23 maintains and expands Th17 cells that promote inflammatory and autoimmune responses through increased production of IL-17. Our preliminary studies (High et al., 2007) suggested that IL-17 production is elevated in aged mice and IL-23 production enhanced from aged murine BM-derived DCs after TLR stimulation (R. Myer & K. High, unpublished data). We reasoned that IL-23 expression may be dysregulated during aging. We first measured IL-23 secretion by activated DCs. Bone marrow-derived DCs from young and aged C57BL/6 mice were stimulated for 4, 6, and 9 h with LPS (TLR4 agonist) plus R848 (TLR7 and 8 agonist). It should be noted that our preliminary experiments indicated that the p19 expression was significantly higher upon combined stimulation with LPS plus R848 compared with either ligand alone. Therefore, we aimed to investigate the p19 transcription under these strong inducing conditions to clearly identify any age-related changes. We observed a marked increase in IL-23 secretion by DCs from both young and aged mice, starting as early as 2 h poststimulation (Fig. 1B). However, DCs from aged mice produced significantly more IL-23 as compared with young mice. Because p19 subunit expression is critical for forming biologically active IL-23 protein, we next determined p19 subunit gene expression by real-time PCR. mRNA level in unstimulated cells was assigned a value of 1 and all other values were calculated relative to 1. As shown in Fig. 1B, p19 mRNA expression was induced in both young and aged cells as early as 2 h poststimulation and remained elevated thereafter. In addition, mRNA level in aged DCs was significantly higher than that seen in young cells. These data suggest that p19 subunit expression and IL-23 protein production are dysregulated during aging.
Fig. 1.
(A) Diagram of the 5′ proximal promoter region of the murine IL-23p19 gene. The NF-κB binding site at −95 bp relative to the transcription start site is shown. (B) IL-23p19 mRNA and IL-23 protein are significantly upregulated in aged DCs. Bone marrow-drived DCs were left unstimulated or stimulated with 100 ng mL−1 LPS plus 3 μM R848 for the indicated times. Total RNA was isolated and analyzed for IL-23p19 subunit gene expression by real-time PCR. Culture supernatants were collected and assayed by ELISA for IL-23 protein production. Data are the mean ± SEM of three experiments. Values from unstimulated cells (0 h) were assigned the value 1 and all other values were calculated relative to 1. *Statistically (P ≤ 0.05) significant (young vs. old).
Selective changes in p19 promoter binding by p65 and c-Rel transcription factor during aging
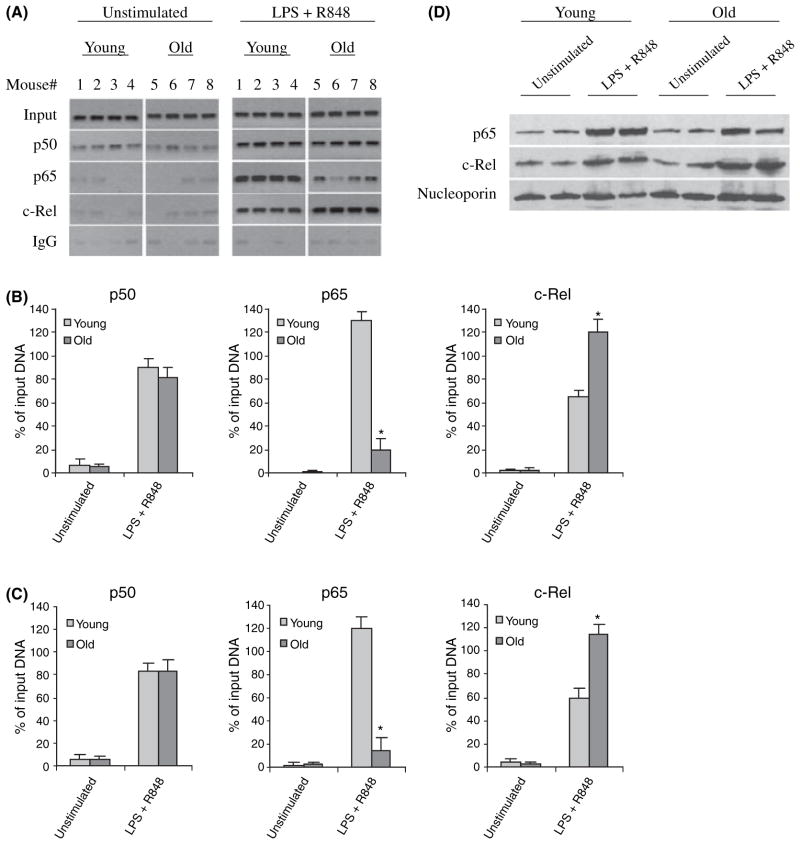
The p19 subunit gene expression is regulated at the transcriptional level by the NF-κB p65 and c-Rel proteins that bind to and activate the promoter in murine macrophages and DCs upon dual agonist TLR stimulation. The promoter contains two NF-κB/Rel binding sites at −95 and −935 bp (Carmody et al., 2007) and the most proximal site (at −95 bp) is indispensible for the TLR-mediated activation of p19 promoter (Mise-Omata et al., 2007). To investigate whether the increase in p19 mRNA seen in aged DCs is because of changes in the transcription factor binding and/or activity, we analyzed the 300-bp promoter sequences flanking the transcription start site and containing the proximal NF-κB site at −95 bp (see Fig. 1A) for p50, p65, and c-Rel binding after the stimulation of DCs for 4 h. Semiquantitative PCR on ChIP DNA (Fig. 2A,B) showed low levels of p50, but not p65 or c-Rel, bound to the promoter in unstimulated cells. After stimulation with LPS plus R848, the levels of p50 protein (which forms active heterodimers with p65 and c-Rel) binding to the promoter was markedly increased to almost similar levels in both young and aged DCs. We also detected marked increases in p65 and c-Rel binding in young DCs. In aged cells, however, we observed a marked decrease in p65 binding concomitant to an increase in c-Rel binding compared with younger DCs. Quantitative real-time PCR confirmed there was a significant decrease in p65 and increase in c-Rel binding, respectively, in aged cells (Fig. 2C). To determine whether the differential changes in p65 and c-Rel binding were because of increase or decrease in their protein level, we performed western blot analysis using nuclear extract from DCs stimulated for 4 h. As shown in Fig. 2D, p65 and c-Rel protein levels were markedly increased in the nuclei of both young and aged cells after stimulation. Densitometric analysis of the protein bands revealed no significant difference in the nuclear levels of p65 or c-Rel in aged DCs compared with young DCs (Fig. S1). These results demonstrate significant differential changes in p65 and c-Rel transcription factor binding to the p19 promoter during aging in mice despite a normal nuclear translocation of both proteins.
Fig. 2.
c-Rel and p65 binding to the IL-23p19 promoter in bone marrow-derived DCs from young and old mice. (A) DCs were left unstimulated or stimulated for 4 h with 100 ng mL−1 LPS plus 3 μM R848. DNA-protein complexes were cross-linked with 1% formaldehyde. Cross-linked chromatin was isolated and immunoprecipitated with c-Rel, p50, or p65 Ab. One tenth of each chromatin sample was reserved before immunoprecipitation and used as an internal control (input). The rest of the sample was used for the immunoprecipitation. DNA was recovered from the immunoprecipitation and input (10× dilution) samples and then analyzed for the presence of IL-23p19 promoter sequences by PCR. Data from four mice in each group are shown. (B) Densitometric analysis of the bands shown in (A). Band intensities were analyzed using Quantity One Imager. IgG control values were subtracted and the sample values were normalized to input DNA and are presented as percentage of input (diluted 10×). Data are the mean ± SEM and are representative of three experiments. *Statistically (P ≤ 0.05) significant (young vs. old). (C) Real-time PCR analysis of p50, p65, and c-Rel binding. Data represent the mean ± SEM of three experiments. (D) c-Rel and p65 protein expression in the nucleus of DCs. Cells were left unstimulated or stimulated for 4 h and nuclear proteins were extracted and analyzed for the expression of c-Rel and p65 by western blotting. Membranes were first probed with c-Rel Ab, stripped and re-probed with p65 and then nucleoporin Ab. Results from two mice in each group are shown.
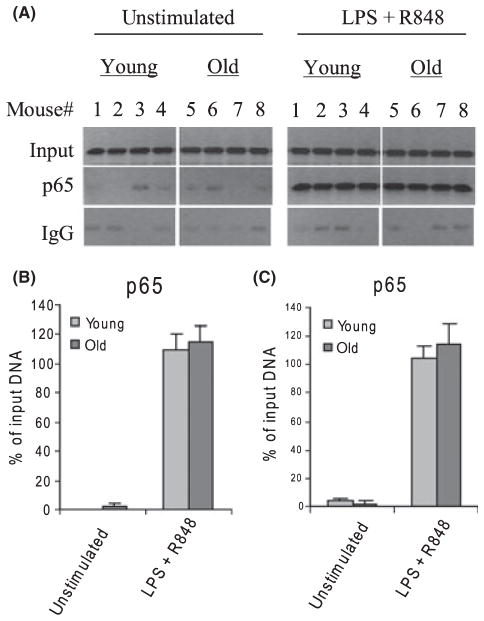
To test whether the change in p65 binding was specific to the p19 promoter, we measured its binding to the IL-12p40 subunit gene, whose expression is also regulated by p65 and c-Rel (Sanjabi et al., 2000; Wen et al., 2008). Figure 3 shows that p65 binds to the IL-12p40 promoter at almost equal levels in young and old DCs after TLR stimulation, as determined by standard (B) and real-time (C) PCR.
Fig. 3.
p65 binding to the IL-12p40 promoter. (A) Chromatin was isolated and immunoprecipitated with p65 Ab and then analyzed by PCR for the enrichment of IL-12p40 promoter sequences in the immunoprecipitated DNA. (B) Band intensities were analyzed as described in Fig. 2. Data are the mean ± SEM and are representative of three experiments. (C) Real-time PCR analysis. Data represent the mean ± SEM of three experiments.
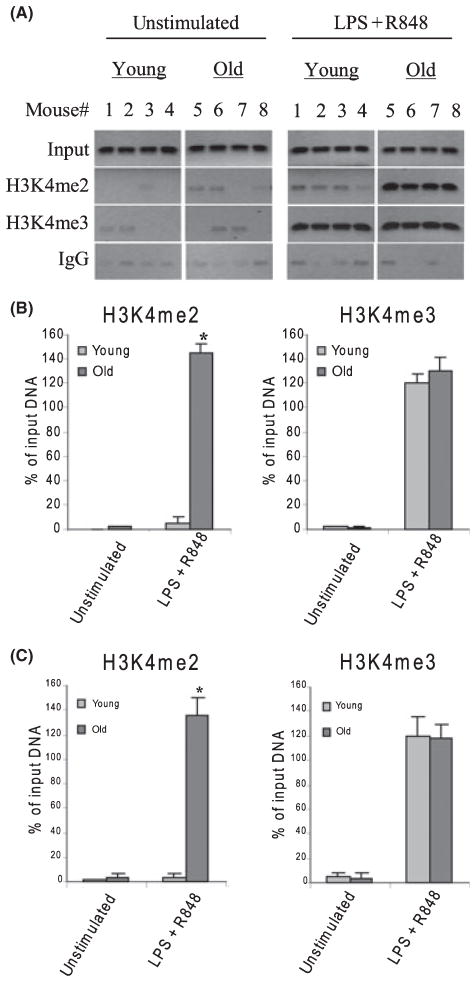
Histone H3K4 is differentially methylated at the p19 promoter during aging
To investigate whether the changes in the p19 promoter binding by p65 and c-Rel are because of changes in chromatin structure, we measured H3K4me2 and H3K4me3, which have been associated with chromatin remodeling and activation of gene transcription (Wysocka et al., 2005), using PCR with ChIP DNA. As shown in Fig. 4A,B, there was background binding before stimulation of both H3K4me2 and H3K4me3 in young and aged DCs. This background was also seen with DNA immunoprecipitated with IgG control antibody. After stimulation, we observed a robust increase in H3K4me2 in aged cells only. In addition, we detected a very significant increase in the H3K4me3 level both in young and aged cells. However, the difference in the level of H3K4me3 was not statistically different by age. Real-time PCR analysis confirmed similar methylation patterns (Fig. 4C). These results demonstrate the differential induction of H3K4 methylation at the p19 promoter nucleosome during its activation in DCs and further suggest that aging is associated with changes in H3K4 methylation pattern.
Fig. 4.
IL-23p19 promoter is bound by di- and tri-methylated H3K4 in aged DCs. Cells were stimulated and treated as described in Fig. 2. Chromatin was isolated and immunoprecipitated with H3K4me2 or H3K4me3 antibody. (A) DNA was recovered from the immunoprecipitated and input samples and then analyzed for the presence of IL-23p19 promoter sequences by PCR. Data from four mice each group are shown. (B) Band intensities were analyzed as described in Fig. 2. Data are the mean ± SEM and are representative of three experiments. *Statistically (P ≤ 0.05) significant (young vs. old). (C) Real-time PCR analysis. Data represent the mean ± SEM of three experiments.
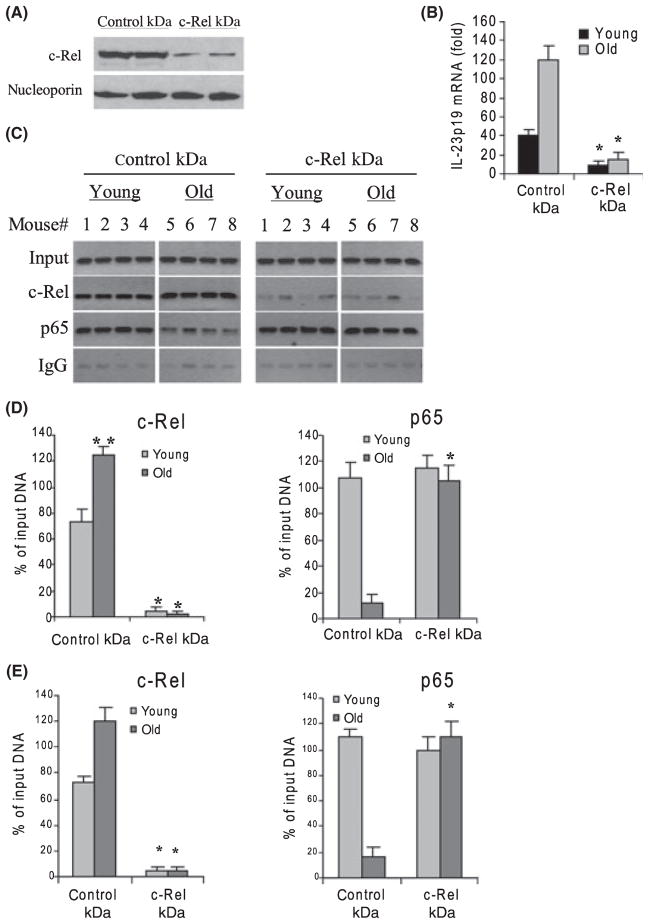
c-Rel, but not p65, is critical for the upregulation of p19 expression in aged DCs
The results presented in Fig. 2 show a significant increase in c-Rel binding to the p19 promoter in aged DCs. To determine the relative contribution of c-Rel binding to the upregulation of p19 expression in aged DCs, we analyzed c-Rel binding and p19 mRNA levels after c-Rel knockdown. Cells were transfected with a pool of c-Rel-specific siRNA for 24 h and then stimulated for 4 h with LPS plus R848. As shown in Fig. 5A, siRNA transfection resulted in dramatic decrease of c-Rel protein expression in aged DCs. Next, we measured the p19 mRNA levels in young and aged DCs after c-Rel knockdown. We observed a nearly complete loss of the p19 mRNA in both young and aged cells (Fig. 5B). These results indicated that c-Rel was critical for the activation of p19 transcription. In addition, standard PCR showed a complete loss of c-Rel binding to the promoter after c-Rel knockdown (Fig. 5C,D). While there were no obvious changes in p65 binding in young DCs, we observed a significant increase in p65 binding in aged cells (compare Fig. 5C with Fig. 2A). Real-time PCR showed similar results (Fig. 5E). These results demonstrate that c-Rel is critical for the induction of p19 transcription in young and aged DCs and that the increase in p65 binding seen in aged cells (after c-Rel knockdown) does not compensate for c-Rel in activating p19 mRNA transcription.
Fig. 5.
c-Rel knockdown decreases IL-23p19 expression in aged DCs. Cells were transfected with nonspecific or c-Rel-specific siRNA. After 24 h, cells were washed and stimulated for 4 h with 100 ng mL−1 LPS plus 3 μM R848. (A) Western blot analysis of c-Rel expression in stimulated cells after c-Rel knockdown. Results from two old mice are shown. (B) c-Rel knockdown significantly decreases IL-23p19 mRNA level. RNA was isolated and analyzed by real-time PCR. Data are the mean ± SEM from four mice and represent the average of three experiments. KD, knockdown; *P ≤ 0.05 (control KD vs. c-Rel KD). (C) p65 binds IL-23p19 promoter in aged DCs in the absence of c-Rel. Cells were transfected and treated as described above. Chromatin was isolated and immunoprecipitated with c-Rel or p65 Ab and then analyzed by PCR. (D) Band intensities were analyzed as described in Fig. 2. Data are the mean ± SEM and are representative of three experiments. *Statistically (P ≤ 0.05) significant (control KD vs. c-Rel KD). **Statistically (P ≤ 0.05) significant (young vs. old). (E) Real-time PCR analysis. Data represent the mean ± SEM of three experiments.
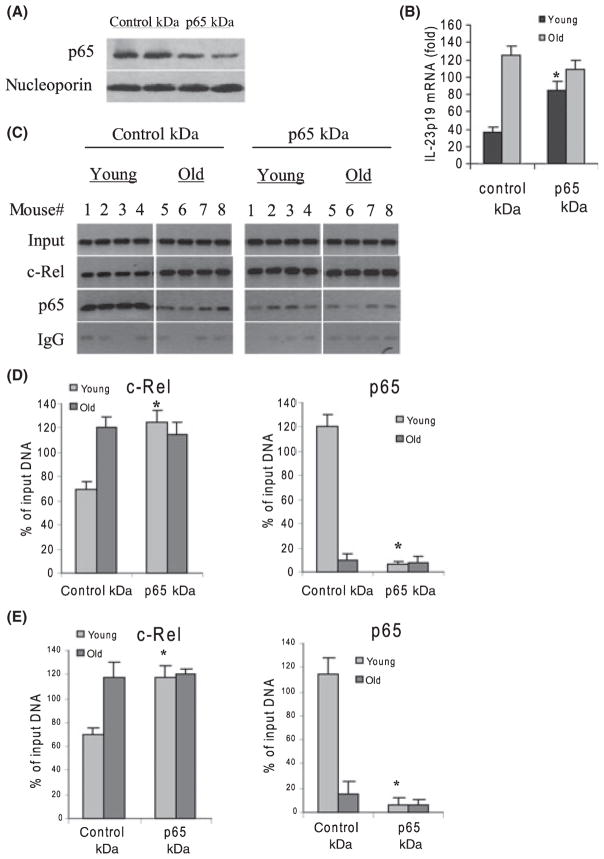
Loss of p65 expression increases c-Rel binding and upregulates p19 expression in young DCs
To determine whether the increase in p65 binding in young DCs (Fig. 2) interferes with the transcription activation by c-Rel, we measured p65 and c-Rel binding and p19 mRNA levels after p65 knockdown. Results (Fig. 6) show a concurrent increase in c-Rel binding at the p19 promoter in young cells (Fig. 6C). The amount of p65 binding reached levels similar to those detected in old cells without p65 knockdown (compare Figs 6C and 2A). Meantime, we detected a significant increase in the p19 mRNA levels after p65 knockdown (Fig. 6B). Moreover, we detected similar levels of c-Rel and p65 binding by real-time PCR (Fig. 6E). In addition, western blotting showed a marked decrease in p65 protein expression after p65 siRNA transfection (Fig. 6A). These results suggest that p65, although dispensible for p19 induction in both young and old DCs (Fig. 5), can compete for binding by c-Rel and subsequent p19 upregulation in young DCs.
Fig. 6.
p65 knockdown increases c-Rel binding and p19 expression in young DCs but has no effects in old DCs. Cells were transfected with nonspecific or p65-specific siRNA. After 24 h, cells were washed and stimulated for 4 h with 100 ng mL−1 LPS plus 3 μM R848. (A) Western blot analysis of p65 expression in stimulated cells after p65 knockdown. Results from two old mice are shown. (B) p65 knockdown significantly increases p19 mRNA level in young DCs. RNA was isolated and analyzed by real-time PCR. Data are the mean ± SEM from four mice and represent the average of three experiments. KD, knockdown; *P ≤ 0.05 (control KD vs. p65 KD). The left panel shows western blotting of p65 protein in stimulated cells after p65 knockdown. Results from two mice are shown. (C) p65 binding to the p19 promoter is increased in young DCs after p65 KD. Cells were transfected and treated as described above. Chromatin was isolated and immunoprecipitated with c-Rel or p65 Ab and then analyzed by PCR. (D) Band intensities were analyzed as described in Fig. 2 and are the mean ± SEM and represent three independent experiments *Statistically (P ≤ 0.05) significant (control KD vs. p65 KD). (E) Real-time PCR analysis. Data represent the mean ± SEM of three experiments.
Discussion
Our results demonstrated that selective epigenetic changes in chromatin configuration and transcription factor binding in DCs play a role in the dramatic upregulation of the IL-23p19 subunit gene expression and IL-23 protein production associated with aging. The p19 subunit gene promoter is di- and tri-methylated on H3K4 and binds NF-κB c-Rel transcription factor at significantly higher levels in aged DCs after activation with LPS plus R848. By contrast, activated DCs from young mice show increase in the level of H3K4me3 only and binding of NF-κB p65 and c-Rel transcription factors at the p19 promoter, suggesting that age-associated changes in chromatin structure and transcription factor assembly represent interdependent mechanisms that contribute to dysregulated expression of the p19 subunit and IL-23 production during aging.
Recent studies have shown that various inflammatory genes, such as IL-4, IL-5, IL-12, TNFα, and MCP-1 (Weinmann et al., 1999; Fields et al., 2002; Barthel & Goldfeld, 2003; Boekhoudt et al., 2003; Goriely et al., 2003, 2003; Wen et al., 2008) undergo epigenetic changes characterized by histone modifications affecting chromatin structure and transcription factor binding (Lee et al., 2006). In addition, studies investigating IL-12 (which shares the p40 subunit with IL-23) transcription have reported that chromatin remodeling played a role in the induction of IL-12 p40 and p35 subunit expression in LPS-activated macrophages (Weinmann et al., 1999; Goriely et al., 2003) and that p300-mediated histone acetylation reinforced c-Rel-mediated transcriptional activation of p40 subunit (Sun et al., 2004). Our results showed a significant increase and differential changes in H3K4me2 and H3K4me3 after activation of DCs. While we detected H3K4me3 at the p19 promoter in both young and aged DCs, H3K4me2 was exclusively detected in activated DCs from aged mice.
Chromatin remodeling because of histone modifications greatly affect the binding and activity of chromatin-associated cofactors and transcription factors, including NF-κB, to their cognate DNAs thereby controlling gene expression (Sheppard et al., 1999; Cheung et al., 2000b; Chen et al., 2002; Kiernan et al., 2003). Both H3K4me2 and H3K4me3 can be detected in the promoter of many genes (Bernstein et al., 2002). H3K4me2 is detected in active euchromatic regions but not in silent heterochromatic sites (Noma et al., 2001). In addition, it has been demonstrated that, at least in yeast, H3K4me2 occurs at both active and inactive genes, whereas H3K4me3 is present exclusively at active genes (Santos-Rosa et al., 2002). Interestingly, we found that p19 gene mRNA was induced in activated DCs from both young and old mice, but was dramatically higher in aged mice. However, in contrast to young DCs, we detected H3K4me2 in aged DCs only. It has recently been proposed in yeast that when genes are active, H3K4me3 invariably appears and H3K4me2 often persists (Santos-Rosa et al., 2002). A recent study has reported that the chronic regulation of IL-12 gene expression in postseptic mice is associated with an epigenetic mechanism of gene regulation by the presence of H3K4me3 at the IL-12 p35 and p40 subunit promoters, leading to transcription activation (Wen et al., 2008). The role of di-methylated H3K4 may be to determine a transcriptionally permissive state, whereas the tri-methylated state may allow for an active chromatin conformation (Santos-Rosa et al., 2002). This supports the concept of methylation state as a major determinant of the level of gene activity. Thus, the presence of H3K4me2 at the p19 promoter in aged, but not young, DCs may be to ensure persistently active transcription of the p19 subunit.
The mechanism by which H3K4 methylation mediates transcription activation is presently unknown. Two possible models are that; methylated H3K4 marks a gene for the recruitment of complexes involved in transcription activation and/or displaces factors involved in transcription repression, such as histone de-acetylases (Nishioka et al., 2002; Sims et al., 2003; Wysocka et al., 2005; Li et al., 2007). In this regard, it has been shown that histone H3 acetylation activates the IL-17 promoter in T cells (Akimzhanov et al., 2007) and reinforces the c-Rel-mediated activation of the IL-12 p40 subunit gene (Sun et al., 2004). In addition, a molecular effector that specifically recognizes methylated H3K4 has been identified in human cells (Wysocka et al., 2005). WDR5 binds to H3K4me2 and H3K4me3 and forms large protein complexes that include histone acetyltransferase and methyltransferase activities, which in turn can generate di- and tri-methylated H3K4 (Wysocka et al., 2005). Recently, Wysocka et al., (2005) have demonstrated that WDR5 knockdown in mammalian cells leads to decreases in tri-methylation but not di-methylation and that WDR5, although not required for the association of methyltransferase complexes with chromatin or for H3K4me2 formation, is essential for conversion of H3K4me2 to H3K4me3 by the H3K4 methyltransferase complex. Based on these findings, those authors proposed a model where WDR5 binds to H3K4me2, which can be further tri-methylated by WDR5-containing methyltransferase complexes, such as Set1 and MLL1. Thus, the presence of H3K4me2 at the p19 promoter in aged DCs could provide an unlimited substrate for forming H3K4me3 by WDR5, resulting in a persistently high level of trimethylated H3K4 and the p19 transcription upregulation seen in aged DCs. Epigenetic-mediated changes in chromatin structure may enhance accessibility to binding sites in promoter sequences and increase recruitment of general transcription co-factors, leading to transcription induction (Cheung et al., 2000a; Saccani et al., 2002). Recent reports suggest that mammalian aging is associated with epigenetic remodeling of chromatin structures. For example, there is an age-associated changes in the redistribution of histone modifying enzymes and chromatin binding proteins as well as decline in total genomic DNA methylation, i.e. an overall decrease in heterochromatin formation [reviewed in (Sedivy et al., 2008)].
Our current results do not explain the increase in H3K4me2 in aged DCs. Since H3K4 was increased after cell activation, it is likely that protein co-factors may interact with c-Rel to mediate this effect. The chromain remodeling protein Chd1 associates with histone acetyltransferase activity and preferentially acetylates histone H3 (Pray-Grant et al., 2005). It is possible that c-Rel may interact with Chd1 to mediate histone acetylation. Interstingly, a recent study has shown that inhibition of histone acetylation abrogated p65 binding to and transcriptional activation of IL-12p40 promoter (Bode et al., 2007), which is largely regulated in a manner similar to the p19 gene. Thus, in aged DCs, c-Rel may interact with a chromatin effector that induces histone acetylation which in turn attenuates p65 binding at the p19 promoter while maintaining H3K4me2 as an unlimited source for H3K4me3. This may suggest interdependency of transcription factor binding and histone modification in regulation p19 in aged DCs. In addition, c-Rel has been shown to synergize with the histone acetyltransferase p300 to enhance IL-12p40 transcription (Goriely et al., 2003). In addition, a recent study has shown that while c-Rel is required for IL-12p40 efficient transcription, it is not required for chromatin remodeling across the promoter (Weinmann et al., 2001). By contrast, Rao et al. (2003) reported that c-Rel was essential for global changes in chromatin structure across the IL-12 promoter but the underlying mechanism has not been identified.
Another interesting finding by this study is the selective binding of p19 promoter by p65 and c-Rel. Both proteins play a role in the activation of p19 expression in macrophages and DCs (Carmody et al., 2007; Mise-Omata et al., 2007), with c-Rel being indispensible for transcriptional activation. Our data indicated that, in contrast to young DCs, there was a marked decrease in p19 promoter binding by p65 in aged DCs concomitant to an increase in c-Rel binding and p19 transcriptional upregulation. The decrease in p65 binding was not because of decrease in p65 protein expression in aged DCs, because densitometric analysis of the western blots did not reveal any significant difference in the nuclear levels of p65 between young and aged DCs. In addition, the decrease in p65 binding in old DCs was specific to the p19 subunit gene because we did not detect any significant differences in p65 binding level to the IL-12p40 subunit gene promoter in both young and old DCs. Interestingly, IL-12 protein expression level was not significantly different in young compared with old DCs (our unpublished observations) Furthermore, we did not detect significant changes in the nuclear level of c-Rel protein in aged DCs compared with young. Thus, a certain chromatin configuration or chromatin-associated factor could favor the binding of c-Rel, thus precluding p65 binding.
Our finding that p65 knockdown by siRNA in young DCs increases p19 mRNA contradicts those by Mise-Omata et al. (2007) and Utsugi et al. (2006). While both studies showed that c-Rel was essential (as suggested by our study) for TLR-mediated induction of p19 expression, they reported a decrease in p19 mRNA levels in p65-deficient macrophages and monocytederived DCs. Given that c-Rel is essential for the induction of p19, one would expect that in the absence of p65, the p19 expression will be maintained or increased, or at least would not change, because of binding of c-Rel. Our data (Fig. 6D) showed an increase in c-Rel binding at the p19 promoter in young DCs after p65 knockdown. This is in line with our conclusion that p65 may compete with c-Rel for promoter binding, and differences between our data vs. those of Mise-Omata et al. (2007) or Utsugi et al. (2006) may be because of cell type (macrophages or monocyte-derived DCs vs. bone marrow-derived DCs), species (human vs. mouse) and/or differences in knockdown vs. knockout techniques. Overall, however, our data suggest that in the absence of p65, the increase in c-Rel binding activity may account for the increase in p19 expression in murine DCs. P65 may play a role in transcription activation of p19 but may not be required if c-Rel is present. In support of our argument that p65 may not be required for transcriptional activation, Carmody et al. (2007) have recently shown that c-Rel−/− murine DCs fail to transcribe p19 (despite the normal expression of p65) and concluded that other members of the NF-κB, including p65, cannot compensate for the role of c-Rel. In addition, a recent study (Sanjabi et al., 2000) has suggested that the minor reduction in IL-12p40 (which is regulated in the same manner as p19) expression in p65-deficient macrophages may be because of apoptotic cell death and that c-Rel is essential for IL-12p40 gene activation.
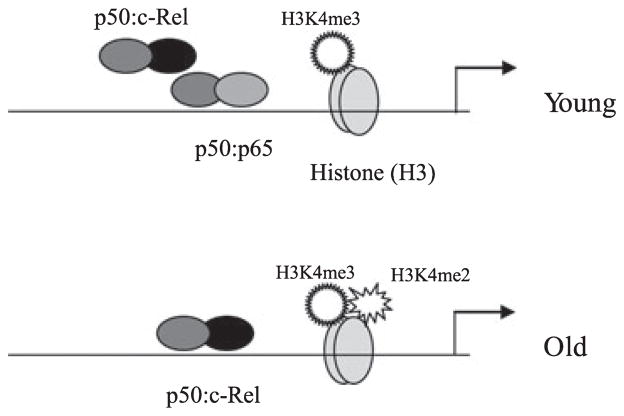
It is unclear how the increase in c-Rel expression and binding upregulate p19 expression in aged DCs. A recent study has shown that c-Rel is essential for changes in chromatin structure and transcription induction of the IL-2 promoter in T cells (Rao et al., 2003). In addition, p50:c-Rel dimers are suggested to be stronger transcriptional activators than p50:p65 dimers (Carmody et al., 2007; Mise-Omata et al., 2007). Thus, it is possible that the increase in c-Rel binding at the p19 promoter in aged DCs provides an opportunity for enhancing transcription activation. C-Rel may recruit transcriptional coactivators to the p19 promoter. Transcription factors may also recruit chromatin co-activators (Cheung et al., 2000a; Fischle et al., 2003). Our c-Rel knockdown experiments showed that c-Rel is essential for p19 gene expression in both young and aged DCs, as demonstrated by the significant decrease in p19 mRNA. The data also showed that p65 was not critical for p19 activation in aged DCs, because c-Rel knockdown resulted in a significant increase in p65 binding to the p19 promoter, yet we could not detect p19 mRNA. Together, these results suggest that c-Rel, while required for p19 transcription activation in young DCs, is necessary for the upregulated expression in aged DCs and that this selective requirement depends on the chromatin configuration imposed by the H3K4 methylation state. Our results also suggest that p65, although dispensible for p19 induction in both young and old DCs, may compete with c-Rel binding in young cells and, therefore, may be responsible for the lower expression levels of p19 in young DCs. We propose a model (Fig. 7) for transcription activation of the p19 subunit gene in DCs. Upon activation with LPS plus R848, the p19 promoter is tri-methylated on H3K4 (me3) and binds p50:p65 and p50:c-Rel heterodimers in young DCs. In aged cells, H3K4 is di- (me2) and tri-methylated (me3), which may result in a chromatin configuration that precludes p65, leading to increases in p50:c-Rel binding and transcription upregulation.
Fig. 7.
A model depicting age-associated differences in transcription factor binding and chromatin structure during p19 subunit gene activation in murine DCs. Upon activation with LPS plus R848, the p19 promoter is tri-methylated on H3K4 and bind p50:p65 and p50:c-Rel heterodimers in young DCs. In aged DCs, H3K4 is di- (H3K4me2) and tri-methylated (H3K4me3), which may result in a chromatin configuration that precludes p65, leading to increases in p50:c-Rel binding and transcription upregulation.
In conclusion, IL-23 is crucial for the development of inflammatory and autoimmune diseases. It maintains and expands Th17 inflammatory cells and IL-17 production that may be responsible, at least in part, for major age-related changes in inflammatory cytokines (Bettelli et al., 2008; Dong, 2008; Huang et al., 2008), and is associated with inflammatory diseases, including lupus erythmatosus, chronic intestinal inflammation, rheumatoid arthritis, and asthma (Bettelli et al., 2008; Dong, 2008). Because deficiencies in IL-23p19 subunit gene expression result in a decrease in Th17 cell numbers and IL-17 cytokine production, our results showing that upregulation of p19 expression and IL-23 production in aged mice is associated with changes in NF-κB p65 and c-Rel transcription factor assembly because of epigenetic changes in chromatin structure suggest that epigenetic targeting of the c-Rel-p19 axis could provide an opportunity for controlling dysregulated cytokine and inflammatory responses associated with aging.
Experimental procedures
Mice and culture of bone marrow cells
Young (3 months) and old (22 months) male C57BL/6 mice were purchased from Harlan Sprague (Indianapolis, Indiana) and housed under pathogen-free conditions at the animal research facility and treated in accordance with approved protocols of the Animal Care and Use Committee of Wake Forest University School of Medicine. Bone marrow cells were collected from femur bones and washed in RPMI-1640 supplemented with 10% FBS (Hyclone, Logan, UT, USA). Red Blood Cells were lysed with ACK lysing buffer (BioWhittaker, Walkersville, MD, USA) for 2 min. Cells were washed once with HBSS with 3% FBS and once with RPMI-1640. Cells were suspended in RPMI-1640 supplemented with 10% FBS, 2 mM L-glutamine, 20 ng mL−1 gentamicin, and 20 ng mL−1 recombinant mGM-CSF (Invitrogen, Carlsbad, CA, USA), seeded at 0.5 × 106 cells mL−1 in 24-well plates and incubated at 37 °C under 5% CO2. On day 3 and day 5, two-thirds of the media was replaced with fresh medium (containing mGM-CSF). On day 6, DCs were harvested, washed with complete medium, and then stimulated with 100 ng mL−1 Gram-negative bacterial LPS (Escherichia coli 0111:B4; Sigma, St. Louis, MO, USA) plus 3 μM R848 (3 M, Saint Paul, MN, USA) (GL Synthesis, Worcester, MA, USA) for the indicated times. Cells were harvested at 0, 2, 4, and 6 h, for mRNA assay. Supernatants were collected at 0, 4, 6, and 9 h poststimulation, for IL-23 protein assay.
ELISA
IL-23 protein concentration in the culture supernatants was measured using an ELISA kit according to the manufacturer’s instructions (eBioscience, San Diego, CA, USA).
Western blot analysis
Nuclear extracts were prepared from DCs by incubation in lysis buffer (10 mM Tris–HCl (pH 7.5), 1.5 mM MgCl2, 10 mM KCl, 0.2 mM EDTA, 0.2 mM DTT, 0.5 mM PMSF, 0.1 mM NaV04, 0.1% Titon X-100, and protease inhibitor cocktail. Supernatants (cytoplasmic fraction) were removed by centrifugation at 3400 g for 10 min at 4 °C. The nuclear pellets were resuspended in lysis buffer containing 20 mM Tris–HCl (pH 7.5), 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 0.2 mM DTT, 0.5 mM PMSF, 5% glycerol, 1% NP-40, 0.1% SDS, and protease inhibitor cocktail. After incubation on ice for 30 min, lysates were centrifuged at 11000 g for 15 min at 4 °C. The supernatants were aliqoted and kept at −70 °C, after measuring the total protein content. Equal amounts of proteins (30 μg) were resolved on SDS–PAGE and electroblotted onto polyvinylidene fluoride (PVDF) membranes. The blots were blocked and probed overnight at 4 °C with primary antibodies against c-Rel or p65. Membranes were washed and incubated with appropriate HRP-conjugated secondary antibodies. Proteins were detected using enhanced chemiluminescence (ECL) reagent (Pierce Biotechnology, Rockford, IL, USA). Membranes were stripped and re-probed with nucleoporin antibody (Pendergrass et al., 2006).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation assays were performed according to the protocol provided by the manufacturer (Active Motif, Carlsbad, CA, USA), with some modifications. Cells grown to 80% confluence were harvested and washed twice with cold PBS. Cells were treated with formaldehyde at a final concentration of 1% for 10 min at room temperature, to cross-link protein- DNA complexes. Cells were then incubated with glycine for 5 min at room temperature and washed with PBS. Cells (5 × 106) were lysed in 1% SDS for 30 min at 4 °C and centrifuged. The chromatin (supernatant) was sheared by sonication three times for 15 s each at 40% power in an ice bath (Branson, Model 250). These shearing conditions generate DNA fragments in the range of 200–500 bp. Ten microliters of the cleared chromatin were reserved as ‘input’ DNA sample. Chromatin was then immunoprecipitated overnight at 4 °C with antibodies specific to p50, p65, c-Rel, H3K4me2, H3K4me3, or IgG Ab. Immunoprecipitates were washed sequentially and then eluted from the beads. Protein-DNA complexes cross-links, along with the ‘input’ sample were reversed at 95 °C for 3 h, and then treated with proteinase K for 1 h at 37 °C. The resulting DNA was kept at −20 °C until analyzed by PCR.
PCR
Semiquantitative PCR was performed to measure the relative abundance of the p19 promoter fragments in the immunoprecipitated (ChIP) DNA. PCR reaction contained 1 μM of each primer, 2 mM MgCl2, 0.2 μM dNTPs, 5 μl of the ChIP DNA and 0.04 U μL−1 AmpliTag Gold DNA polymerase (Applied Biosystems, Foster City, CA, USA). The PCR conditions were as follows: 1 cycle at 94 °C for 5 min, 30 cycles at 94 °C, 58 °C, and 72 °C for 30 s each, and a final cycle at 72 °C for 5 min. Our preliminary experiment indicated that the PCR amplification was in the linear range at 30 cycle, while sequences were poorly enriched at 25 cycles and highly enriched at 35 cycles. Therefore, 30 cycles were chosen for our analysis. Equal amounts of PCR products were run on 1.5% ethidium bromide-stained agarose gel. The images were captured and the band intensities were quantitated using Quantity One Imager (Bio-Rad, Hercules, CA, USA). Sample data were normalized to input DNA and are presented as percentage of input. The primers used in PCR were designed to amplify a sequence in the murine IL-23p19 promoter containing the NF-κB site at −95 bp relative to the transcription start site (Carmody et al., 2007) and were as follows: p19F (5′-CACTCATTTCCCCTGGAACT- 3′) and p19R (5′-GAGTCTAACTCTAGTCCG- 3′). The primers used for amplifying the IL-12p40 promoter sequences (−150 to +3) were: p40F (5′-GGGGGAGGGAGGAACTTCTT- 3′) and p40R (5′-TCTGCTGCCTTGGCTGCTCCT- 3′) (Wen et al., 2008).
Real-time PCR
Real-time PCR was performed to measure IL-23p19 mRNA expression and to quantify the IL23p19 DNA sequences in the immunoprecipitated (ChIP) DNA.
For ChIP DNA, the following oligonucleotides were used: forward primer (5′-CACTCATTTCCCCTGGAACT-3′), reverse primer (5′-GAGTCTAACTCTAGTCCG-3′), and internal fluorescence probe (5′-FAM-GAAGCGGCATACCTGGGCTC-3′) (Applied Biosystems) The PCR reaction (25 μL) contained 5 μL ChIP DNA, 12.5 μL of 2 × TaqMan Universal Master Mix containing DNA polymerase and dNTPs, 300 nM of each primer, and 100 nM internal probe. Reactions were run in triplicates at 50 °C for 2 min, 95 °C for 10 min followed by 30 cycles at 95 °C for 15 s and 60 °C for 1 min (combined annealing and extension), using ABI Prism 7000 Sequence Detection System (Applied Biosystems). Sample data were normalized to the input DNA and calculated as percentage of input.
For mRNA expression, total RNA was isolated using STAT-60 extraction kit, according to the manufacturer’s protocol (Tel- Test, Friendswood, TX, USA). Two micrograms RNA were reverse-transcribed to cDNA in a 25-μL volume containing 0.2 μM dNTPs, 2.5 μM oligo d(T), 5 mM MgCl2, and 0.25 U μL−1 of murine leukemia reverse transcriptase (Applied Biosystems). The RT reaction was incubated for 1 h at 42 °C and 5 min at 99 °C. The PCR was performed using 5 μL cDNA and IL-23p19 and GAPDH predesigned TaqMan primer/probe sets (Applied Biosystems). The PCR conditions were as described above. Sample data were normalized to GAPDH mRNA values and are presented as fold change relative to mRNA from unstimulated cells (set as 1-fold).
siRNA transfection
Cells were transfected with nonspecific, c-Rel-specific, or p65-specific small interfering RNAs (siRNA) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) using siRNA transfection kit according to the manufacturer’s instructions (AMAXA, Gaithersburg, MD, USA). After 24 h, cells were washed with medium and stimulated for 4 h with 100 ng mL−1 LPS plus 3 μM R848.
Statistical analysis
Data are presented as the mean ± SEM and are representative of three or more experiments. Significant differences between groups were determined by Student’s t-test. P-values ≤ 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Mark Chappell (Center for Vascular Research) for providing the nucleoporin antibody. This work was supported in part by NIAID grant AI057952 (KPH).
Footnotes
Additional Supporting Information may be found in the online version of this article:
Fig. S1 Densitometric analysis of western blot shown in Fig. 2D. Data were normalized to nucleoporin and plotted as fold change relative to unstimulated cells (set as 1-fold). These results suggest that there is no significant increase in the nuclear level of c-Rel in aged cells compared with young cells.
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- Baeuerle PA, Henkel T. Function and activation of NF-kappa B in the immune system. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Barthel R, Goldfeld AE. T cell-specific expression of the human TNF-alpha gene involves a functional and highly conserved chromatin signature in intron 3. J Immunol. 2003;171:3612–3619. doi: 10.4049/jimmunol.171.7.3612. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA. 2002;99:8695–8700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Korn T, Oukka M, Kuchroo VK. Induction and effector functions of T(H) 17 cells. Nature. 2008;453:1051–1057. doi: 10.1038/nature07036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode KA, Schroder K, Hume DA, Ravasi T, Heeg K, Sweet MJ, Dalpke AH. Histone deacetylase inhibitors decrease Toll-like receptor-mediated activation of proinflammatory gene expression by impairing transcription factor recruitment. Immunology. 2007;122:596–606. doi: 10.1111/j.1365-2567.2007.02678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhoudt GH, Guo Z, Beresford GW, Boss JM. Communication between NF-kappa B and Sp1 controls histone acetylation within the proximal promoter of the monocyte chemoattractant protein 1 gene. J Immunol. 2003;170:4139–4147. doi: 10.4049/jimmunol.170.8.4139. [DOI] [PubMed] [Google Scholar]
- Boren E, Gershwin ME. Inflamm-aging: autoimmunity, and the immune-risk phenotype. Autoimmun Rev. 2004;3:401–406. doi: 10.1016/j.autrev.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Bull P, Hunter T, Verma IM. Transcriptional induction of the murine c-rel gene with serum and phorbol-12-myristate-13-acetate in fibroblasts. Mol Cell Biol. 1989;9:5239–5243. doi: 10.1128/mcb.9.11.5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RJ, Ruan Q, Liou HC, Chen YH. Essential roles of c-Rel in TLR-induced IL-23 p19 gene expression in dendritic cells. J Immunol. 2007;178:186–191. doi: 10.4049/jimmunol.178.1.186. [DOI] [PubMed] [Google Scholar]
- Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Mo R, Lescure PA, Misek DE, Hanash S, Rochford R, Hobbs M, Yung RL. Aging is associated with increased T-cell chemokine expression in C57BL/6 mice. J Gerontol A Biol Sci Med Sci. 2003;58:975–983. doi: 10.1093/gerona/58.11.b975. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000a;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Cheung WL, Briggs SD, Allis CD. Acetylation and chromosomal functions. Curr Opin Cell Biol. 2000b;12:326–333. doi: 10.1016/s0955-0674(00)00096-x. [DOI] [PubMed] [Google Scholar]
- Collart MA, Baeuerle P, Vassalli P. Regulation of tumor necrosis factor alpha transcription in macrophages: involvement of four kappa B-like motifs and of constitutive and inducible forms of NF-kappa B. Mol Cell Biol. 1990;10:1498–1506. doi: 10.1128/mcb.10.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, Zurawski S, Wiekowski M, Lira S, Gorman D, Kastelein RA, Sedgwick JD. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- Dinarello CA. Interleukin 1 and interleukin 18 as mediators of inflammation and the aging process. Am J Clin Nutr. 2006;83:447S–455S. doi: 10.1093/ajcn/83.2.447S. [DOI] [PubMed] [Google Scholar]
- Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O’Garra A. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147:3815–3822. [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- Gangemi S, Basile G, Merendino RA, Minciullo PL, Novick D, Rubinstein M, Dinarello CA, Lo BC, Franceschi C, Basili S, D’ UE, Davi G, Nicita-Mauro V, Romano M. Increased circulating Interleukin-18 levels in centenarians with no signs of vascular disease: another paradox of longevity? Exp Gerontol. 2003;38:669–672. doi: 10.1016/s0531-5565(03)00061-5. [DOI] [PubMed] [Google Scholar]
- Gerondakis S, Strasser A, Metcalf D, Grigoriadis G, Scheerlinck JY, Grumont RJ. Rel-deficient T cells exhibit defects in production of interleukin 3 and granulocyte-macrophage colony-stimulating factor. Proc Natl Acad Sci USA. 1996;93:3405–3409. doi: 10.1073/pnas.93.8.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Goriely S, Demonte D, Nizet S, De WD, Willems F, Goldman M, Van LC. Human IL-12(p35) gene activation involves selective remodeling of a single nucleosome within a region of the promoter containing critical Sp1-binding sites. Blood. 2003;101:4894–4902. doi: 10.1182/blood-2002-09-2851. [DOI] [PubMed] [Google Scholar]
- Grunstein M, Hecht A, Fisher-Adams G, Wan J, Mann RK, Strahl-Bolsinger S, Laroche T, Gasser S. The regulation of euchromatin and heterochromatin by histones in yeast. J Cell Sci Suppl. 1995;19:29–36. doi: 10.1242/jcs.1995.supplement_19.4. [DOI] [PubMed] [Google Scholar]
- High KP, Prasad R, Marion CR, Schurig GG, Boyle SM, Sriranganathan N. Outcome and immune responses after Brucella abortus infection in young adult and aged mice. Biogerontology. 2007;8:583–593. doi: 10.1007/s10522-007-9106-6. [DOI] [PubMed] [Google Scholar]
- Hiscott J, Marois J, Garoufalis J, D’Addario M, Roulston A, Kwan I, Pepin N, Lacoste J, Nguyen H, Bensi G. Characterization of a functional NF-kappa B site in the human interleukin 1 beta promoter: evidence for a positive autoregulatory loop. Mol Cell Biol. 1993;13:6231–6240. doi: 10.1128/mcb.13.10.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MC, LiaoJ J, Bonasera S, Longo DL, Goetzl EJ. Nuclear factor-kappaB-dependent reversal of aging-induced alterations in T cell cytokines. FASEB J. 2008;22:2142–2150. doi: 10.1096/fj.07-103721. [DOI] [PubMed] [Google Scholar]
- Hunter CA. New IL-12-family members: IL-23 and IL-27, cytokines with divergent functions. Nat Rev Immunol. 2005;5:521–531. doi: 10.1038/nri1648. [DOI] [PubMed] [Google Scholar]
- Kiernan R, Bres V, NgR W, Coudart MP, El MS, Sardet C, Jin DY, Emiliani S, Benkirane M. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem. 2003;278:2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- Kontgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, Gerondakis S. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin- 2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Libermann TA, Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol Cell Biol. 1990;10:2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Fergusson MM, Castilho RM, Liu J, Cao L, Chen J, Malide D, Rovira II, Schimel D, Kuo CJ, Gutkind JS, Hwang M, Finkel T. Augmented Wnt signaling in a mammalian model of accelerated aging. Science. 2007;317:803–806. doi: 10.1126/science.1143578. [DOI] [PubMed] [Google Scholar]
- Mise-Omata S, Kuroda E, Niikura J, Yamashita U, Obata Y, Doi TS. A proximal kappaB site in the IL-23 p19 promoter is responsible for RelA- and c-Rel-dependent transcription. J Immunol. 2007;179:6596–6603. doi: 10.4049/jimmunol.179.10.6596. [DOI] [PubMed] [Google Scholar]
- Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, Kastelein RA, Sedgwick JD, Cua DJ. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med. 2003;198:1951–1957. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Pendergrass KD, Averill DB, Ferrario CM, Diz DI, Chappell MC. Differential expression of nuclear AT1 receptors and angiotensin II within the kidney of the male congenic mRen2. Lewis rat. Am J Physiol Renal Physiol. 2006;290:F1497–F1506. doi: 10.1152/ajprenal.00317.2005. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, III, Grant PA. Chd1 chromodomain links histone H3 methylation with. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Rao S, Gerondakis S, Woltring D, Shannon MF. c-Rel is required for chromatin remodeling across the IL-2 gene promoter. J Immunol. 2003;170:3724–3731. doi: 10.4049/jimmunol.170.7.3724. [DOI] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]
- Sanjabi S, Hoffmann A, Liou HC, Baltimore D, Smale ST. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc Natl Acad Sci USA. 2000;97:12705–12710. doi: 10.1073/pnas.230436397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Sedivy JM, Banumathy G, Adams PD. Aging by epigenetics–a consequence of chromatin damage? Exp Cell Res. 2008;314:1909–1917. doi: 10.1016/j.yexcr.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard KA, Rose DW, Haque ZK, Kurokawa R, McInerney E, Westin S, Thanos D, Rosenfeld MG, Glass CK, Collins T. Transcriptional activation by NF-kappaB requires multiple coactivators. Mol Cell Biol. 1999;19:6367–6378. doi: 10.1128/mcb.19.9.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci USA. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Lu J, Wei L, Wang X, Xu X, Dong M, Huang B. Histone acetyltransferase activity of p300 enhances the activation of IL-12 p40 promoter. Mol Immunol. 2004;41:1241–1246. doi: 10.1016/j.molimm.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Swift ME, Burns AL, Gray KL, DiPietro LA. Age-related alterations in the inflammatory response to dermal injury. J Invest Dermatol. 2001;117:1027–1035. doi: 10.1046/j.0022-202x.2001.01539.x. [DOI] [PubMed] [Google Scholar]
- Trebilcock GU, Ponnappan U. Evidence for lowered induction of nuclear factor kappa B in activated human T lymphocytes during aging. Gerontology. 1996;42:137–146. doi: 10.1159/000213785. [DOI] [PubMed] [Google Scholar]
- Utsugi M, Dobashi K, Ishizuka T, Kawata T, Hisada T, Shimizu Y, Ono A, Mori M. Rac1 negatively regulates lipopolysaccharide-induced IL-23 p19 expression in human macrophages and dendritic cells and NF-kappaB p65 trans activation plays a novel role. J Immunol. 2006;177:4550–4557. doi: 10.4049/jimmunol.177.7.4550. [DOI] [PubMed] [Google Scholar]
- Van DD, Shaw AC. Toll-like receptors in older adults. J Am Geriatr Soc. 2007;55:1438–1444. doi: 10.1111/j.1532-5415.2007.01300.x. [DOI] [PubMed] [Google Scholar]
- Verma IM, Stevenson JK, Schwarz EM, Van AD, Miyamoto S. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Plevy SE, Smale ST. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity. 1999;11:665–675. doi: 10.1016/s1074-7613(00)80141-7. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Mitchell DM, Sanjabi S, Bradley MN, Hoffmann A, Liou HC, Smale ST. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event Nat. Immunology. 2001;2:51–57. doi: 10.1038/83168. [DOI] [PubMed] [Google Scholar]
- Wen H, Dou Y, Hogaboam CM, Kunkel SL. Epigenetic regulation of dendritic cell-derived interleukin-12 facilitates immunosuppression after a severe innate immune response. Blood. 2008;111:1797–1804. doi: 10.1182/blood-2007-08-106443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikby A, Ferguson F, Forsey R, Thompson J, Strindhall J, Lofgren S, Nilsson BO, Ernerudh J, Pawelec G, Johansson B. An immune risk phenotype, cognitive impairment, and survival in very late life: impact of allostatic load in Swedish octogenarian and nonagenarian humans. J Gerontol A Biol Sci Med Sci. 2005;60:556–565. doi: 10.1093/gerona/60.5.556. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Yung RL, Julius A. Epigenetics, aging, and autoimmunity. Autoimmunity. 2008;41:329–335. doi: 10.1080/08916930802024889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.