Abstract
The circuitry of the phage λ genetic switch determining the outcome of lytic or lysogenic growth is well-integrated and complex, raising the question as to how it evolved. It is plausible that it arose from a simpler ancestral switch with fewer components that underwent various additions and refinements, as it adapted to vast numbers of different hosts and conditions. We have recently identified a new class of genetic switches found in mycobacteriophages and other prophages, in which immunity is dependent on integration. These switches contain only three genes (integrase, repressor and cro) and represent a major departure from the λ-like circuitry, lacking many features such as xis, cII and cIII. These small self-contained switches represent an unrealized, elegant circuitry for controlling infection outcome. In this addendum, we propose a model of possible events in the evolution of a complex λ-like switch from a simpler integration-dependent switch.
Keywords: genetic switch, genetic circuits, bistable, integration-dependent immunity, lytic and lysogenic growth
Introduction
The genetic circuitry controlling the outcome of infection by phage λ is remarkably complex, but presumably arose from an ancestral system that was simpler and had fewer components.1,2 In addition, the genetic switches of other phages that have been studied in some detail show them to be equally complex with many shared and some additional features.3-7 Such complexity could arise from additions and refinements to an ancestral switch that make it work ‘better’ under particular selective pressures.2,8-10 This idea is supported by experimental data showing the non-essential nature of many features of the λ genetic switch and to some extent those of other phages. These include the differential affinities of CI and Cro for OR operators,11 positive autoregulation of cI,12 cooperativity of CI binding to operator sites,13 negative autoregulation of cI14 and repression of PRM by Cro.15,16 Many of these features contribute to the efficiency of the switch, but not its core function, suggesting that they are refinements to earlier simpler systems (for review see ref. 2).
As the increase in phage genome sequencing continues, it is expected that genetic switches that have undergone dramatically different evolutionary paths of feature addition and refinement in relation to that of phage λ and similar phages—possibly representing a system closer to the ancestral switch—might be discovered. Such switches may contain all or some of the core components of λ-like switches, while other features may be absent, reflecting use of alternative circuitries to achieve the same biological end-points: lytic and lysogenic outcomes from a bacteriophage infection.
In this addendum, we will briefly describe the genetic circuits used in the integration-dependent immunity systems published recently,17 and then speculate on the evolutionary relationships between these relatively simple circuits and the more complex ones found in λ and its relatives.
Integration-Dependent Immunity
We recently described a new class of lytic-lysogenic switches present in several unrelated mycobacteriophages (represented by three types found in mycobacteriophages BPs, Brujita and Charlie) and prophages of various bacteria, in which phage immunity is directly dependent on integration.17 The direct involvement of integration in deciding the outcome of infection is in contrast to its role in other systems (including λ), where it confers prophage stability, but is not directly involved in determining the outcome of infection. Furthermore, it is attractive as an early evolutionary state, because what biological process is better suited to generate two alternative outcomes than site-specific recombination that yields two mutually exclusive DNA states?
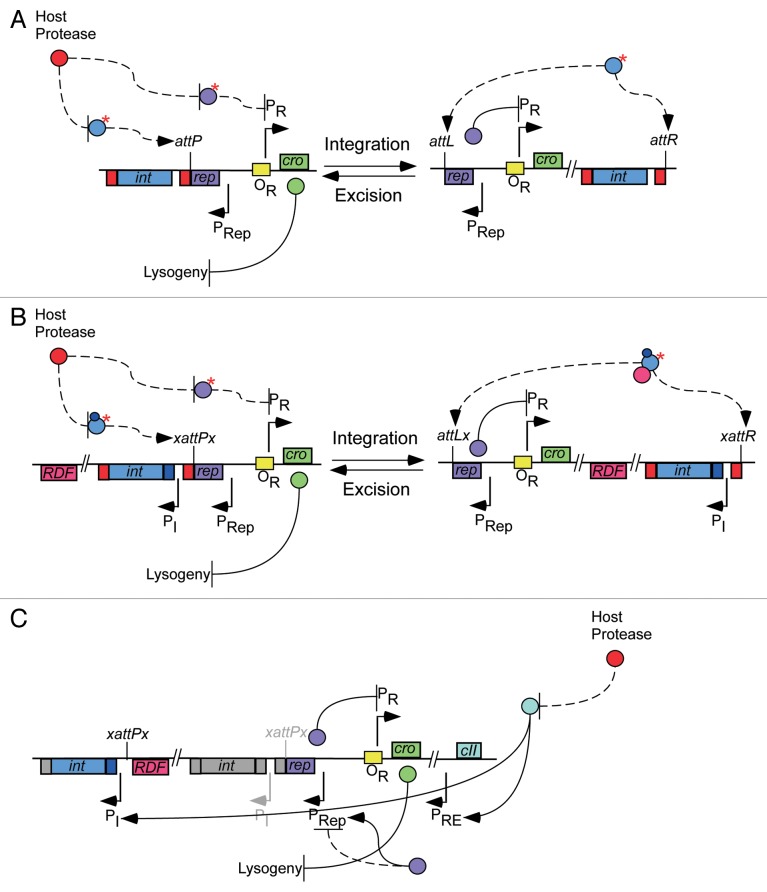
The integration-dependent switches are contained in small ~2 kbp units containing only three genes: integrase (int), repressor (rep) and cro (Fig. 1A); thus, they do not include other components of λ-like switches including xis, cII and cIII. The organization of these genes is such that rep and int are transcribed leftwards, with the putative cro gene transcribed rightwards. Between rep and cro there is a ~200 bp intergenic control region that contains the divergently transcribing promoters PRep (leftwards) and PR (rightwards), and in BPs a 12 bp operator (OR), that overlaps PR, to which repressor binds and downregulates lytic gene expression (Fig. 1A). Surprisingly, the attP site for integration lies within the rep coding sequence such that different forms of Rep are expressed from viral and prophage genomes (Fig. 1A). This is critical to the function of the switch, since Rep is C-terminally tagged for proteolytic degradation, and integration results in removal of the C-terminal tag and expression of a stable and active form of the repressor. Additionally, Int is also C-terminally tagged for proteolytic degradation rendering it responsive to host cell input signals (proteases such as ClpXP). Integration is thus not only required for establishment of lysogeny, but is the sensor for the central decision-making event in infection. These features are well supported experimentally. For example, a phage mutant with a single amino acid substitution that stabilizes the viral form of the repressor has a much higher frequency of lysogeny than wild-type.17 Conversely, a phage mutant with a catalytically defective integrase is completely defective in lysogeny.17
Figure 1. Evolution of a complex genetic switch from a simple integration-dependent genetic switch. (A) Circuitry diagram of an integration-dependent genetic switch composed of three genes: int, rep and cro. Int (blue circles) and Rep (purple circles) are C-terminally tagged (red boxes on 3′ of the genes, red asterisk on proteins) for degradation by host proteases (red circles). The PRep promoter drives leftward transcription of two lysogenic genes rep and int, while PR drives rightward transcription of lytic genes to the right of the switch, the first of which is cro. The attP sequence, used for site-specific homologous recombination into the bacterial chromosome, is located within the open reading frame of rep. Integration stabilizes Rep by removing the C-terminal degradation tag. Rep has a negative effect on PR expression by binding to the operator OR. Cro (green circles) has a negative effect on lysogeny by an unknown mechanism. Int alone is capable of both integrative and excisive recombination and it is the expression of Int that controls directionality of recombination. (B) Addition of a new promoter (PI) between rep and int leads to higher expression of int. This leads to prophage instability. Stability is restored by addition of a recombination directionality factor (RDF) followed by subsequent acquisition of an additional DNA binding domain in Int (dark blue box at the 5′ of the int gene, dark blue circle on Int) and arm-type binding sites in attP (xattPx). Control of directionality of recombination is now dependent on expression of Int and the RDF (pink circle). (C) A duplication event leads to a second copy of the attP-int cassette in the phage genome, leading to further evolution of the newly added cassette and loss of the int and attP functions in the original (grayed). With the addition of an RDF, direction of recombination is less responsive to Int concentrations and the C-terminal degradation tag on Int is lost (grayed box on 3′ of gene). As dependency on integration for immunity is lost, new means to control rep expression arise. Rep gains the ability to bind to RNA Polymerase and activate its own expression at PRep. Following this, the C-terminal degradation tag on Rep is lost (grayed box on 3′ of gene). This may lead to very high levels of Rep, which is downregulated by mutations that make repressor dependent on autoactivation. This leads to low lysogenization frequencies that are restored to normal levels by addition of a repressor establishment promoter (PRE) that is activated by a host transcriptional activator (CII, turquoise circle), which is later acquired by the phage; CII can then acquire the ability to control int expression through PI. Additions of negative autoregulation of Rep and cooperativity of Rep binding further fine-tune the switch. The final product is a genetic switch with circuitry resembling that of λ and no longer involving integration to determine the outcome of infection.
A rather surprising observation is that Int seems to be able to mediate both integrative and excisive recombination without any apparent requirement for a recombination directionality factor (RDF). In a plasmid transformation assay in which selection of antibiotic resistance is dependent on integration, transformation is very inefficient because of targeted degradation of Int. But stabilization of Int—which can be accomplished with a single amino acid substitution at its C-terminus—leads to dramatically higher levels of integration and thus higher transformation efficiency of these plasmids. When the stability of the integrated plasmids is examined, we find that transformants generated with the wild-type unstable Int are very stable, and we see no loss after many generations of unselected growth. In contrast, transformants recovered with the stabilized Int show substantial levels of plasmid loss after unselected growth.17 As the only other phage-encoded component in this assay is repressor, the simplest interpretation is that Int is able to catalyze excisive recombination without need for an RDF. We therefore must assume that prophage stability in a lysogen is largely dependent on low levels of int expression coupled with degradation. This raises the question then as to how excision ever occurs. It is plausible that it is simply a stochastic event and that within a given population of cells, there are some that will accumulate sufficient Int to catalyze excision, leading to reconstruction of the inactive viral form of the repressor, and spontaneous induction of lytic growth. An alternative explanation is that expression is induced from adjacent host genes, although no conditions, including mitomycin C exposure, have been identified that lead to efficient prophage induction.
One unresolved question about these integration-dependent immunity systems is how Cro works. We have not been able to delete the putative cro gene, and we presume that it is required for lytic growth. When expressed from a plasmid, it greatly reduces lysogenization frequencies, but does not seem to downregulate the PRep promoter.17 How Cro works and how repressor synthesis is shut down during lytic growth therefore remain important unanswered questions.
Evolution of Genetic Switch Complexity
The compact, self-contained nature of these integration-dependent switches makes them attractive as ancestral forms of more complex phage genetic switches. Although they perhaps represent different lineages of switch development from those more closely resembling λ, they also help to understand potential origins of switch complexity. There are of course numerous possible individual pathways of evolutionary development from one switch type to another, and we will just focus here on some scenarios that could plausibly contribute to development of complex λ-like switches from the simpler integration-dependent immunity systems.
Integrase expression and addition of a recombination directionality factor
Consider the potential impact of a mutation that enhances the expression of int by forming a new and highly active promoter between int and rep, PI (Fig. 1B). This would increase the efficiency of both integrative and excisive recombination, potentially leading to prophage instability under some conditions. Prophage stability could then be enhanced by a mutation that makes Int dependent on a host protein for excision, perhaps through protein-protein interactions (Fig. 1B). This gene could then be acquired by the phage to act as the recombination directionality factor (RDF), followed by subsequent acquisition of an additional DNA binding domain to Int, and arm-type binding sites to attP such that recombination is now regulated by the DNA binding properties of the RDF (Fig. 1B). A scenario of this type is supported by the observation that serine-integrases use simple sites that lack the equivalent of arm-type binding sites in the lambda system, and the RDF acts through direct subunit-subunit interactions with Int.18,19 Several RDFs of serine-integrases have been described that are unrelated at the sequence level, but these include relatives of the family that includes lambda Xis.20 At least in one example, this RDF is capable of DNA binding even though DNA binding is not required for its action.21 These thus represent plausible examples of intermediate stages in the evolution of tyrosine integrase systems typified by phage λ.
Relocation of int and attP
The events described above—including separation of int from the PRep promoter—could be facilitated by a duplication event that inserts a second copy of the attP-int cassette elsewhere in the phage genome (in λ, int is separated by over 8 kbp from cI) (Fig. 1C). Such a rearrangement could itself be mediated by aberrant Int-mediated recombination, and we note that there is at least one example of an apparent re-location of an integration cassette in mycobacteriophage Giles.22 Such a duplication event is attractive because it would enable the further evolution of the newly added cassette, and development of a repressor system that is independent of recombination as a central control mechanism. Furthermore, with addition of a RDF and mechanistic separation of integration and excision events, these would be less responsive to the integrase concentrations, and reduce dependency on the rate of degradation of Int from its C-terminal tag, leading to its loss (Fig. 1C). However, this would also demand an alternative sensor for determining the outcomes of infection, which would then require an alternative means of regulating repressor expression.
Regulation of repressor and separation of lysogeny establishment and maintenance functions
With loss of dependence on integration for immunity, other processes for control of repressor expression must be acquired, as well as a separation of mechanisms for expressing repressor for lysogenic establishment, as opposed to maintenance. One scenario is to imagine a circumstance in which the ancestral proteolytically tagged repressor regains function by the ability to bind to RNA Polymerase and activate its own expression (Fig. 1C). This is attractive, as we have demonstrated that overexpression of the viral form of the BPs repressor at least partially restores immunity.17 This could then lead to loss of the C-terminal proteolytic tag on repressor, as this would no longer serve a critical role (Fig. 1C). However, loss of the tag generating a stable form of the repressor that activates its own expression could give rise to runaway autoactivation and accumulation of very high levels of repressor in the cell. An important consequence of this is that spontaneous induction of lytic growth would now be reduced to very low levels, and as virtually all temperate phages undergo readily detectable levels of spontaneous induction, we assume that this is a biologically important event.
The selective pressures to downregulate repressor synthesis could involve numerous mechanisms, but an attractive one is the introduction of mutations that now make repressor synthesis dependent on autoactivation (Fig. 1C). This phage would grow fine lytically, but be capable of only very low lysogenization frequencies, although the lysogens formed would be quite stable. Elevation back to normal lysogenization frequencies could be accomplished by mutations that generate a promoter (equivalent to λ PRE) that is activated by a host transcriptional activator followed by acquisition of that activator from the host (equivalent to cII) (Fig. 1C). This activator could later gain control of the PI promoter driving int expression (Fig. 1C). Negative autoregulation and cooperativity of repressor binding could be added to further fine-tune the switch (Fig. 1C).
Putting the pieces together
Combined into one genomic context, the above-mentioned refinements would make up a genetic circuitry resembling the λ switch in many respects (Fig. 1C). Because of the mosaic nature of phage genomes and the prevalence of recombination within and between genomes,23-25 evolutionary events leading to complex genetic circuits are unlikely to have occurred in a single genomic context. It seems more likely that events would take place within a large variety of different phage genomes with different hosts, but in a scenario in which all phage genomes have access to a large common gene pool, albeit at different frequencies.26 There would be plenty of opportunity for these components to find themselves in common genomes, especially given the vast scale and dynamic nature of the phage population.27
Concluding Remarks
Because of the prevalence of the temperate lifestyle among phages and their vast diversity, it is likely that as more phage genomes are sequenced, we will gain a greatly improved understanding of the evolution of genetic switches. At this point, we have very few examples of the variety of genetic switches in existence and the circuitry of most have only been studied to a partial level. Furthermore, we have a limited knowledge of the biology of phages in the context of selective pressures phages encounter outside of a laboratory setting. However, as our knowledge of the ecology of phages and the types of natural genetic switches expands, and as efforts to engineer synthetic genetic switches leads to new insights,28-30 our understanding of how complex genetic circuits evolve and the variety of circuitries possible will be greatly enhanced. This will further allow us to control gene expression to a level of sophistication that will open many possibilities for research, medical and industrial use of finely tuned genetic switches.
Acknowledgments
We thank Valerie Villanueva and Lauren Oldfield for comments on the manuscript. This work was supported by NIH grants GM093901 and AI059114.
Disclosure of Potential Conflicts of Interest
The authors verify that there is no conflict of interest concerning this manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/24186
References
- 1.Court DL, Oppenheim AB, Adhya SL. A new look at bacteriophage lambda genetic networks. J Bacteriol. 2007;189:298–304. doi: 10.1128/JB.01215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Little JW. Evolution of complex gene regulatory circuits by addition of refinements. Curr Biol. 2010;20:R724–34. doi: 10.1016/j.cub.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Campbell A. Comparative molecular biology of lambdoid phages. Annu Rev Microbiol. 1994;48:193–222. doi: 10.1146/annurev.mi.48.100194.001205. [DOI] [PubMed] [Google Scholar]
- 4.Knight KL, Bowie JU, Vershon AK, Kelley RD, Sauer RT. The Arc and Mnt repressors. A new class of sequence-specific DNA-binding protein. J Biol Chem. 1989;264:3639–42. [PubMed] [Google Scholar]
- 5.Nilsson AS, Haggård-Ljungquist E. Evolution of P2-like phages and their impact on bacterial evolution. Res Microbiol. 2007;158:311–7. doi: 10.1016/j.resmic.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Susskind MM, Botstein D. Molecular genetics of bacteriophage P22. Microbiol Rev. 1978;42:385–413. doi: 10.1128/mr.42.2.385-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krause HM, Higgins NP. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986;261:3744–52. [PubMed] [Google Scholar]
- 8.Bhattacharyya RP, Reményi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–80. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 9.Jacob F. Evolution and tinkering. Science. 1977;196:1161–6. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 10.Ptashne M, Gann A. Imposing specificity by localization: mechanism and evolvability. Curr Biol. 1998;8:R897. doi: 10.1016/s0960-9822(07)00557-x. [DOI] [PubMed] [Google Scholar]
- 11.Little JW, Shepley DP, Wert DW. Robustness of a gene regulatory circuit. EMBO J. 1999;18:4299–307. doi: 10.1093/emboj/18.15.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michalowski CB, Little JW. Positive autoregulation of cI is a dispensable feature of the phage lambda gene regulatory circuitry. J Bacteriol. 2005;187:6430–42. doi: 10.1128/JB.187.18.6430-6442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babić AC, Little JW. Cooperative DNA binding by CI repressor is dispensable in a phage lambda variant. Proc Natl Acad Sci U S A. 2007;104:17741–6. doi: 10.1073/pnas.0602223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodd IB, Shearwin KE, Perkins AJ, Burr T, Hochschild A, Egan JB. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev. 2004;18:344–54. doi: 10.1101/gad.1167904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atsumi S, Little JW. Role of the lytic repressor in prophage induction of phage lambda as analyzed by a module-replacement approach. Proc Natl Acad Sci U S A. 2006;103:4558–63. doi: 10.1073/pnas.0511117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schubert RA, Dodd IB, Egan JB, Shearwin KE. Cro’s role in the CI Cro bistable switch is critical for lambda’s transition from lysogeny to lytic development. Genes Dev. 2007;21:2461–72. doi: 10.1101/gad.1584907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broussard GW, Oldfield LM, Villanueva VM, Lunt BL, Shine EE, Hatfull GF. Integration-dependent bacteriophage immunity provides insights into the evolution of genetic switches. Mol Cell. 2013;49:237–48. doi: 10.1016/j.molcel.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh P, Wasil LR, Hatfull GF. Control of phage Bxb1 excision by a novel recombination directionality factor. PLoS Biol. 2006;4:e186. doi: 10.1371/journal.pbio.0040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khaleel T, Younger E, McEwan AR, Varghese AS, Smith MC. A phage protein that binds φC31 integrase to switch its directionality. Mol Microbiol. 2011;80:1450–63. doi: 10.1111/j.1365-2958.2011.07696.x. [DOI] [PubMed] [Google Scholar]
- 20.Bibb LA, Hatfull GF. Integration and excision of the Mycobacterium tuberculosis prophage-like element, phiRv1. Mol Microbiol. 2002;45:1515–26. doi: 10.1046/j.1365-2958.2002.03130.x. [DOI] [PubMed] [Google Scholar]
- 21.Bibb LA, Hancox MI, Hatfull GF. Integration and excision by the large serine recombinase phiRv1 integrase. Mol Microbiol. 2005;55:1896–910. doi: 10.1111/j.1365-2958.2005.04517.x. [DOI] [PubMed] [Google Scholar]
- 22.Morris P, Marinelli LJ, Jacobs-Sera D, Hendrix RW, Hatfull GF. Genomic characterization of mycobacteriophage Giles: evidence for phage acquisition of host DNA by illegitimate recombination. J Bacteriol. 2008;190:2172–82. doi: 10.1128/JB.01657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatfull GF, Cresawn SG, Hendrix RW. Comparative genomics of the mycobacteriophages: insights into bacteriophage evolution. Res Microbiol. 2008;159:332–9. doi: 10.1016/j.resmic.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- 25.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, et al. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–82. doi: 10.1016/S0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 26.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci U S A. 1999;96:2192–7. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatfull GF, Hendrix RW. Bacteriophages and their genomes. Curr Opin Virol. 2011;1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avlund M, Dodd IB, Sneppen K, Krishna S. Minimal gene regulatory circuits that can count like bacteriophage lambda. J Mol Biol. 2009;394:681–93. doi: 10.1016/j.jmb.2009.09.053. [DOI] [PubMed] [Google Scholar]
- 29.Bonnet J, Subsoontorn P, Endy D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc Natl Acad Sci U S A. 2012;109:8884–9. doi: 10.1073/pnas.1202344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420:224–30. doi: 10.1038/nature01257. [DOI] [PubMed] [Google Scholar]