Abstract
Bacteriophage (phage) are among the most diverse and abundant life forms on Earth. Studies have recently used phage diversity to identify novel antimicrobial peptides and proteins. We showed that one such phage protein, Staphylococcus aureus (Sau) phage G1 gp67, inhibits cell growth in Sau by an unusual mechanism. Gp67 binds to the host RNA polymerase (RNAP) through an interaction with the promoter specificity σ subunit, but unlike many other σ-binding phage proteins, gp67 does not disrupt transcription at most promoters. Rather, gp67 prevents binding of another RNAP domain, the α-C-terminal domain, to upstream A/T-rich elements required for robust transcription at rRNA promoters. Here, we discuss additional biochemical insights on gp67, how phage promoters escape the inhibitory function of gp67, and methodological advancements that were foundational to our work.
Keywords: Staphylococcus aureus, RNA polymerase, bacteriophage, gp67, transcription
Introduction
Staphylococcus aureus (Sau) is a gram-positive bacterium of significant clinical importance.1 Differences in transcriptional profiles drive the switch from commensal to pathogenic growth profiles, and these changes have been studied extensively using genetic and high-throughput approaches.1-5 However, relatively few studies have examined transcription in Sau using mechanistic, biochemical and structural tools.6-8
Studies on bacteriophage (phage) have been fundamental to our understanding of molecular biology in prokaryotes. Early studies using phage elucidated many of the mechanisms of transcription and replication, in addition to understanding how phage modulate these critical processes to favor viral production over host cell functions.9-18 Due to the rise of antibiotic resistance in Sau, recent studies have examined the use of Sau specific phage as a platform to design novel therapeutics, or even for direct use as therapeutic agents.19,20 These studies have largely used high-throughput techniques to identify proteins or peptides with antimicrobial effects, but have failed to perform the structural and mechanistic analyses required to evaluate whether the host targets would be accessible by traditional drug design processes.19,20
Our recent work examined the mechanism of one such phage protein, Sau phage G1 gp67.21 Gp67 was identified as a putative RNA polymerase (RNAP) inhibitor and subsequently shown to bind to Sau, but not Eco, RNAP.19 Dehbi et al.19 showed that gp67 interacts with domain 4 of the housekeeping sigma factor (σA4) in Sau. Biochemical analysis suggested that gp67 blocked −35 recognition, a mechanism of RNAP inhibition known to be exploited by other phage proteins.19,22 Dehbi et al.19 used well-characterized Eco proteins and promoters in their biochemical studies.
We sought to understand the mechanism through which gp67 blocked RNAP activity and cell growth by solving its structure in complex with σA4. However, the structure showed that gp67 did not appear to block promoter DNA recognition or the interaction between core RNAP and σA that is required for promoter-specific RNAP activity. Our subsequent biochemical analysis showed that a native Sau transcription system was absolutely required to the inhibitory effect of gp67. We showed, using in vitro biochemical and in vivo approaches, that gp67 does not block −35 recognition but rather modulates the binding of the α subunit C-terminal domain (α-CTD) to upstream A/T rich sequences (UP-elements).23 Gp67 therefore targets only UP-element dependent promoters, including the rRNA promoters. Blocking rRNA transcription prevents logarithmic growth in Eco, and we provided the first evidence that this is the case in Sau as well.21
Our studies showed that gp67 inhibits Sau RNAP and subsequently Sau growth by an unusual mechanism. The structural data, in combination with the development of a native Sau in vitro transcription system that used Sau RNAP and Sau promoters were critical to our ability to examine the mechanism of inhibition by gp67. Additionally, our work on this phage protein allowed us to identify novel promoters in Sau and evaluate rRNA transcription, and its regulation, in this pathogenic organism.21 Further biochemical and structural work in gram positive organisms should bear in mind the importance of using native components in in vitro experiments, despite the relative ease of using more well-developed model organisms such as Eco.
In this article, we will expand on our work on gp67 and provide additional biochemical detail on this protein and its interactions, discuss how phage promoters likely escape gp67 function, and extend our discussion on the methodological advancements required to study gp67.
Structure of apo-gp67
After solving the X-ray crystal structure of the complex between Sau σA4 and gp67, we attempted to solve the structure of gp67 alone. While the protein expresses well and is easily purified to homogeneity, extensive screening for crystallization conditions did not yield any hits.
To determine whether gp67 was well folded, we performed limited proteolysis on gp67 alone and in complex with full-length σA and σA4. While gp67 resisted proteolysis to relatively high protease concentration in the presence of its binding partner (either σA or σA4), gp67 was readily cleaved even at low protease concentration in the absence of σ (Fig. 1). Gp67 alone in solution is likely poorly structured and undergoes a significant conformational change upon binding to RNAP. Along with the extended network of interactions between gp67 and σA4,21 this may explain the tight binding between the proteins and the fact that we found evidence for gp67 bound to Sau RNAP even at promoters that were not directly inhibited.21 Experimental evaluation of binding kinetics could confirm this hypothesis.
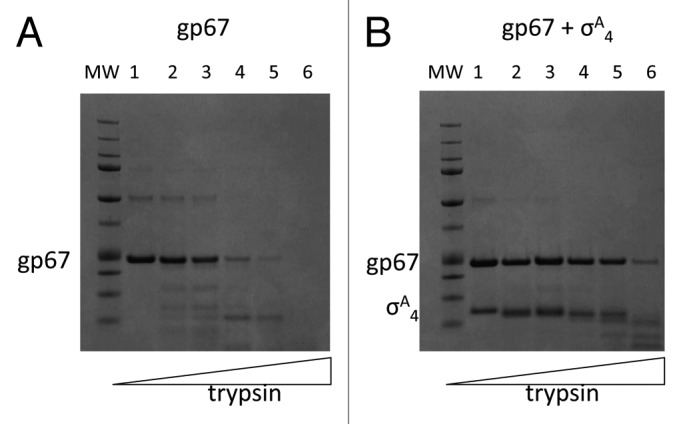
Figure 1. gp67 is conformationally stabilized by its interaction with σ. Limited proteolysis of (A) gp67 alone and (B) gp67 in complex with σA4. The gp67/σA4 complex or gp67 alone, was incubated on ice in 1× proteolysis buffer at 5 μM prior to incubation at 30°C for 20 min with protein:trypsin concentrations of 1:0, 1000:1, 100:1, 50:1, 10:1, 5:1. After the incubation, reactions were stopped by the addition of 1mM PMSF and run on a 4–12% SDS-PAGE gel.
Conservation of gp67 in Phage Genomes
BLAST searches using the sequence of phage G1 gp67 found five homologs with an E-score of < 0.1. All of the putative homologs are found in phage that infect gram-positive organisms, including phage specific to Bacillus, Enterococcus and Listeria. Gp67 also has a homolog in the Sau phage Twort. We showed that expression of the Twort homolog of gp67 in Sau cells also inhibits logarithmic cell growth, arguing for functional conservation between these two proteins.21
Using the program Consurf,24 we used the structure of phage G1 gp67, and an alignment of the gp67 homologs, to map conservation onto the crystal structure. All but one of the universally conserved residues in gp67 are hydrophobic amino acids in the core of the protein, evidence of structural conservation between gp67 homologs. Additional regions of conservation map to the binding surface with the conserved region of σA4, arguing that all gp67 homologs bind similarly to the host RNAP (Fig. 2).
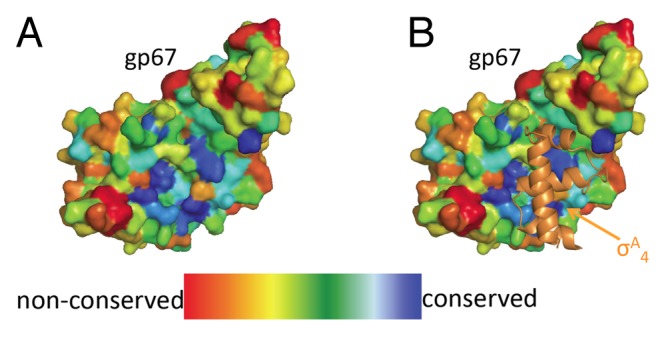
Figure 2. Structural conservation of gp67. (A) gp67 from the 2.0 Å co-crystal structure with σA4 colored by conservation. The structural conservation map was made using ConSurf using five available gp67 sequences. Highly conserved residues are shown in blue and poorly conserved residues in red. (B) Highly conserved gp67 residues map to the σA4 binding site. gp67 from the 2.0 Å co-crystal structure is shown as in (A) and σA4 from the co-crystal structure is shown in orange as a cartoon representation. The most highly conserved surface residues in gp67 map to the interaction between gp67 and σA4.
Our work shows the gp67 alone is sufficient to block normal Sau growth by blocking robust rRNA transcription.21 However, other phage proteins that bind to host RNAP are known to also interact with additional phage proteins to specifically recruit RNAP to phage promoters.25-27 Performing pull-downs with tagged gp67 or RNAP in phage infected Sau cells could easily identify any gp67-binding partners of phage or host origin. We find it likely that gp67 expressing phages, which do not encode their own RNAP, have complex coordination of transcription throughout the phage life cycle. Subsequent studies on G1, phage Twort and other phages may reveal these mechanisms of transcriptional regulation.
Phage Promoters that Control Expression of gp67
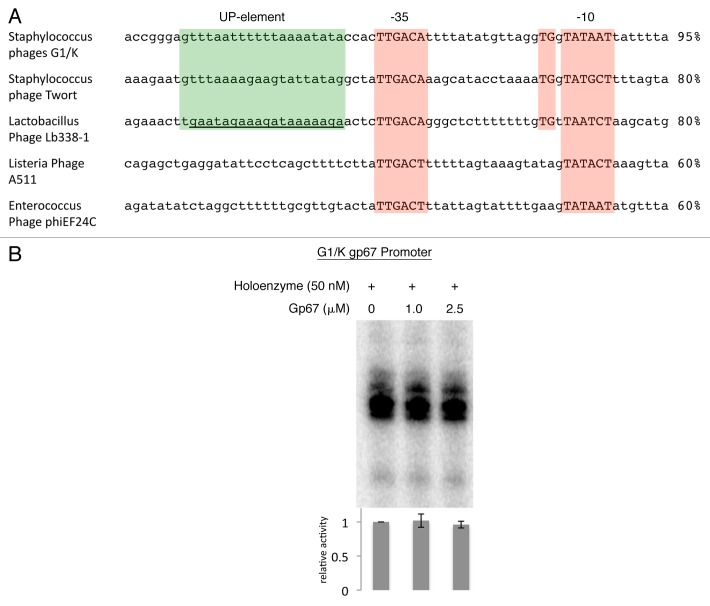
The G1 phage ORF67 (encoding gp67) is located downstream of a perfect consensus −10/−35 promoter and we therefore expect that it is one of the phage genes that is initially expressed upon injection of the dsDNA phage genome into the host cell. Gp67 is then translated and modifies the host RNAP, which affects the transcription of host genes, including the important rrn promoters.21 However, phage promoters that contain UP-elements could also be inhibited by gp67. Furthermore, many G1 promoters contain clear evidence for an UP-element, with A/T sequences (above the already A/T-rich Sau genome) in the region upstream of the −35 element that contains UP-elements (up to 100% A/T; Fig. 3A).
Figure 3. Sequence of promoters that drive gp67 expression in phage genomes. (A) −35 and −10 elements are highlighted in red, as are the extended −10 elements (TGn immediately upstream of the 10 element). The region expected to act as a putative UP-element is highlighted in green and the percent A/T richness of this region is shown to the right of the sequences. (B) gp67 does not inhibit Sau RNAP at the phage G1 gp67 promoter. RNAP holoenzyme (50 nM) was incubated with gp67 at the indicated concentration and promoter DNA (50 nM) and reactions were initiated with 200 nM CTP/GTP/UTP and 50 nM ATP with 0.1 μL α-P32-ATP. After 10 min reactions were stopped with 2× formamide buffer, boiled, electrophoresed on a 12% Urea-PAGE gel and visualized by autoradioagraphy. Results from three independent experiments were quantified, normalized to the signal in the absence of gp67, and averaged, and are expressed as a mean in the graph below each lane (error bars represent one standard deviation above and below the mean).
The Eco phage T4 encodes an anti-σ factor, AsiA, that binds to σ704 and blocks recognition of the −35 element.22 Work on T4 promoters showed that the presence of either an extended −10 element or an UP-element allowed promoters to escape AsiA inhibition, and therefore early phage promoters could still drive the expression of phage proteins in the presence of AsiA.28 All of the phages that contain a gp67 homolog infect firmicutes, also characterized by high genomic A/T content. Of the five phages that contain clear gp67 homologs, three are expressed from promoters that might be targeted by gp67 function (based on high A/T content; Fig. 3A). These promoters, however, likely escape gp67 inhibition through the presence of an extended −10 element. Phage G1 gp67 does not inhibit transcription from its own promoter in vitro (Fig. 3B). Two additional promoters contain no extended −10 elements; however, these promoters do not contain evidence for an upstream A/T rich region indicative of an UP-element (Fig. 3A). Therefore, it appears that phage encoding gp67 have evolved mechanisms through which early phage promoters can escape the effect of gp67, allowing the protein to target Sau growth without modulating the transcription of phage proteins. Further in vitro studies on phage promoters and an in vivo examination of transcription in phage-infected cells could test these hypotheses.
Gp67 Effect on Promoter Stability
rRNA promoters in E. coli form characteristically unstable open promoter complexes (OPCs). Therefore, protein factors that modulate the stability of OPCs can affect the output at rRNA promoters alone, while not modulating the output at other promoters. Gp67 blocks the α-CTD from forming functional interactions with UP-element sequences. Therefore, we would not expect gp67 to modulate the stability of OPCs. We tested the effect of gp67 on promoter stability at Sau (aag, rrnA) promoters and a G1 phage (gp67) promoter. At all promoters we tested, we observed no evidence for a strong effect of gp67 on promoter stability (Fig. 4).
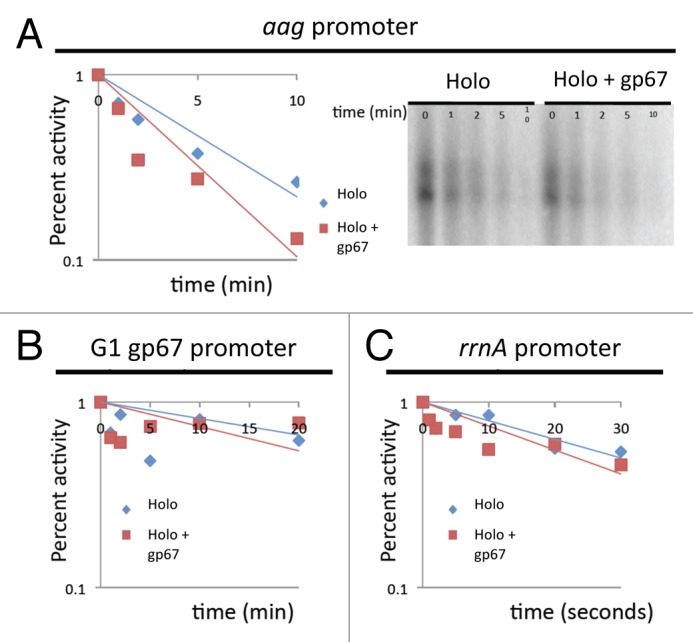
Figure 4. gp67 does not alter the stability of Sau RNAP at Sau or phage promoters. OPCs were formed by incubating Sau RNAP holoeznyme, in the presence or absence of gp67, with linear promoter fragments. After 10 min, complexes were challenged with the addition of 20-fold excess FullCon promoter fragment. At different time points after the challenge, transcription was initiated or complexes pipetted onto filter paper and the percent complexes remaining were quantified by phosphoimagery. (A) Sau aag promoter fragment monitored by transcription output (12% Urea-PAGE gel shown in right panel, quantification shown in left panel). (B) gp67 does not alter the stability of the phage G1 gp67 promoter as monitored by filter binding. (C) gp67 does not alter the stability of the Sau rrnA promoter as monitored by transcription output.
Use of a Native Sau in vitro Transcription System
The initial biochemical work on gp67 suggested that it blocked −35 recognition.19,20 In contrast, our results show that gp67 does not affect −35 binding, as this would lead to inhibition at all promoters that require this interaction.21 The initial studies were performed using Eco RNAP and Eco phage promoters.19 We attempted to reproduce these results but were unable to do so21; additional experiments using Eco RNAP and promoters also produced inconclusive results. Even using Sau RNAP on well-studied Eco RNAP promoters did not produce consistent evidence for inhibition by gp67 (Hochschild A, personal communication). Sau is an A/T-rich gram-positive organism, and the DNA topology at promoters and kinetics of transcription initiation may not be the same as in Eco. It was only when we examined Sau promoters, and in particular the Sau rrn promoters, that we saw clearly reproducible effects of gp67 on RNAP output.21 Using RNA-seq to identify additional gp67-sensitive promoters that could be tested in vitro was also critical to forming our mechanistic hypotheses.21 Differences have also been described in transcription initiation between E. coli and Bacillus subtilis29,30 and recent work has examined the basis for the differences in promoter stability and initiation between E. coli and the thermophilic bacteria.31,32 Based on these results and our work, we suggesting using fully native transcription systems whenever possible, except when direct comparisons have been made between E. coli and the organism of interest, as described.33
The Use of Heparin as a DNA Competitor in in vitro Transcription Assays
To test RNAP activity in a single-round assay, or to isolate kinetic steps in the transcription cycle, competitor must be used to prevent RNAP re-binding to the promoter element. For decades, heparin has been used as a non-specific competitor to block RNAP/DNA interactions.34-38 More recent work has used large excesses of tight-binding dsDNA promoter elements identified by in vitro selection for RNAP binding using σS (FullCon promoter).39 In addition to competing away RNAP that has dissociated from the test promoter element after elongation or due to RNAP disassociation, heparin has been documented to actively destroy RNAP/promoter complexes.35,40
In Sau, we found that using heparin in our in vitro transcription system severely decreased transcriptional output. In fact, at most Sau promoters, the presence of heparin in the reaction led to little or no detectible transcription. However, when we used the FullCon promoter construct as competitor,39 at 20-fold excess, we were able to detect RNA products, measure single round transcription levels, and determine open-promoter complex lifetimes (Supp. Materials and Methods; Fig. 4). In many organisms, the use of heparin in in vitro transcription assays may be ill advised, and the use of more gentle methods to block RNAP rebinding, such as using competing dsDNA promoter fragments, may be preferable.
Concluding Remarks
Gp67 illustrates the diversity of biological functions utilized by phage. This small protein has no sequence or structural homology to any known protein or fold. Our initial hypothesis, based on previously published work,19,20 was that gp67 inhibited RNAP using a mechanism previously ascribed to phage-encoded anti-σ factors. However, our structural and biochemical work quickly challenged these assumptions. In the end, gp67 functions by binding to σ but modulating the binding state of the RNAP α-CTD to upstream promoter elements.21
The phage T4 encodes a protein that ADP-ribsolyates the α-CTD of Eco RNAP at R265, the residue responsible for the interaction with the minor groove of UP-elements (Gourse R, personal communication). This effectively eliminates the ability of the α-CTD to interact productively with UP-elements, blocking robust rRNA transcription. Gp67 acts by a similar mechanism but does not covalently modify RNAP.21 Rather, it forms a stable ternary complex with RNAP through its interaction with σ. Based on our structural modeling, gp67 likely only blocks the proximal α-CTD/UP-element interaction, which appears to be sufficient for RNAP inhibition at rrn promoters.21 The molecular detail of the interaction between σ, the Sau α-CTD and gp67 in the context of promoter DNA is of great interest. Crystallization of ternary complexes containing DNA may reveal the details of these interactions.
Studying phage biology has contributed to our understanding of many central mechanisms of transcription and DNA replication in prokaryotic cells.13,17,41-47 Relatively little work has been done on transcription in Sau using biochemical and structural tools. Our research clearly shows that the use of common in vitro model systems (such as Eco) can lead to spurious results when studying other bacterial species. We also developed an RNA-seq based transcriptome tool to evaluate transcriptional differences in Sau upon the expression of a transcription factor with single-nucleotide resolution (Submitted). We consider it likely that species specific differences in promoter sequences and transcription regulation, which have been described between Bsub and Eco,48 are present in other species as well. The tools developed in our work will be of great use in the continued examination of the basic mechanisms of transcription in Sau, and for further evaluation of the differences in transcription regulation between Eco and other bacteria of clinical significance.49
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/bacteripphage/article/24767
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/24767
References
- 1.Tuchscherr L, Medina E, Hussain M, Völker W, Heitmann V, Niemann S, et al. Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol Med. 2011;3:129–41. doi: 10.1002/emmm.201000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Truong-Bolduc QC, Dunman PM, Eidem T, Hooper DC. Transcriptional profiling analysis of the global regulator NorG, a GntR-like protein of Staphylococcus aureus. J Bacteriol. 2011;193:6207–14. doi: 10.1128/JB.05847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reyes D, Andrey DO, Monod A, Kelley WL, Zhang G, Cheung AL. Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J Bacteriol. 2011;193:6020–31. doi: 10.1128/JB.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu HH, Trawick JD, Haselbeck RJ, Forsyth RA, Yamamoto RT, Archer R, et al. Staphylococcus aureus TargetArray: comprehensive differential essential gene expression as a mechanistic tool to profile antibacterials. Antimicrob Agents Chemother. 2010;54:3659–70. doi: 10.1128/AAC.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossolini GM, Mantengoli E, Montagnani F, Pollini S. Epidemiology and clinical relevance of microbial resistance determinants versus anti-Gram-positive agents. Curr Opin Microbiol. 2010;13:582–8. doi: 10.1016/j.mib.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds J, Wigneshweraraj S. Molecular insights into the control of transcription initiation at the Staphylococcus aureus agr operon. J Mol Biol. 2011;412:862–81. doi: 10.1016/j.jmb.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 7.Deora R, Misra TK. Characterization of the primary sigma factor of Staphylococcus aureus. J Biol Chem. 1996;271:21828–34. doi: 10.1074/jbc.271.36.21828. [DOI] [PubMed] [Google Scholar]
- 8.Rao L, Karls RK, Betley MJ. In vitro transcription of pathogenesis-related genes by purified RNA polymerase from Staphylococcus aureus. J Bacteriol. 1995;177:2609–14. doi: 10.1128/jb.177.10.2609-2614.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siebenlist U, Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980;77:122–6. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siebenlist U. RNA polymerase unwinds an 11-base pair segment of a phage T7 promoter. Nature. 1979;279:651–2. doi: 10.1038/279651a0. [DOI] [PubMed] [Google Scholar]
- 11.Pribnow D. Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc Natl Acad Sci U S A. 1975;72:784–8. doi: 10.1073/pnas.72.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahmsdorf HJ, Pai SH, Ponta H, Herrlich P, Roskoski RJ, Jr., Schweiger M, et al. Protein kinase induction in Escherichia coli by bacteriophage T7. Proc Natl Acad Sci U S A. 1974;71:586–9. doi: 10.1073/pnas.71.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinkle DC, Ring J, Chamberlin MJ. Studies of the binding of Escherichia coli RNA polymerase to DNA. 3. Tight binding of RNA polymerase holoenzyme to single-strand breaks in T7 DNA. J Mol Biol. 1972;70:197–207. doi: 10.1016/0022-2836(72)90533-5. [DOI] [PubMed] [Google Scholar]
- 14.Ouhammouch M, Adelman K, Harvey SR, Orsini G, Brody EN. Bacteriophage T4 MotA and AsiA proteins suffice to direct Escherichia coli RNA polymerase to initiate transcription at T4 middle promoters. Proc Natl Acad Sci U S A. 1995;92:1451–5. doi: 10.1073/pnas.92.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouhammouch M, Orsini G, Brody EN. The asiA gene product of bacteriophage T4 is required for middle mode RNA synthesis. J Bacteriol. 1994;176:3956–65. doi: 10.1128/jb.176.13.3956-3965.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shamoo Y, Friedman AM, Parsons MR, Konigsberg WH, Steitz TA. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376:362–6. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 17.Allison DP, Ganesan AT, Olson AC, Snyder CM, Mitra S. Electron microscopic studies of bacteriophage M13 DNA replication. J Virol. 1977;24:673–84. doi: 10.1128/jvi.24.2.673-684.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Echols H. Developmental pathways for the temperate phage: lysis vs lysogeny. Annu Rev Genet. 1972;6:157–90. doi: 10.1146/annurev.ge.06.120172.001105. [DOI] [PubMed] [Google Scholar]
- 19.Dehbi M, Moeck G, Arhin FF, Bauda P, Bergeron D, Kwan T, et al. Inhibition of transcription in Staphylococcus aureus by a primary sigma factor-binding polypeptide from phage G1. J Bacteriol. 2009;191:3763–71. doi: 10.1128/JB.00241-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, et al. Antimicrobial drug discovery through bacteriophage genomics. Nat Biotechnol. 2004;22:185–91. doi: 10.1038/nbt932. [DOI] [PubMed] [Google Scholar]
- 21.Osmundson J, Montero-Diez C, Westblade LF, Hochschild A, Darst SA. Promoter-specific transcription inhibition in Staphylococcus aureus by a phage protein. Cell. 2012;151:1005–16. doi: 10.1016/j.cell.2012.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert LJ, Wei Y, Schirf V, Demeler B, Werner MH. T4 AsiA blocks DNA recognition by remodeling sigma70 region 4. EMBO J. 2004;23:2952–62. doi: 10.1038/sj.emboj.7600312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estrem ST, Ross W, Gaal T, Chen ZWS, Niu W, Ebright RH, et al. Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase alpha subunit. Genes Dev. 1999;13:2134–47. doi: 10.1101/gad.13.16.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–33. doi: 10.1093/nar/gkq399. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minakhin L, Severinov K. Transcription regulation by bacteriophage T4 AsiA. Protein Expr Purif. 2005;41:1–8. doi: 10.1016/j.pep.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 26.Hinton DM, March-Amegadzie R, Gerber JS, Sharma M. Characterization of pre-transcription complexes made at a bacteriophage T4 middle promoter: involvement of the T4 MotA activator and the T4 AsiA protein, a sigma 70 binding protein, in the formation of the open complex. J Mol Biol. 1996;256:235–48. doi: 10.1006/jmbi.1996.0082. [DOI] [PubMed] [Google Scholar]
- 27.Pande S, Makela A, Dove SL, Nickels BE, Hochschild A, Hinton DM. The bacteriophage T4 transcription activator MotA interacts with the far-C-terminal region of the sigma70 subunit of Escherichia coli RNA polymerase. J Bacteriol. 2002;184:3957–64. doi: 10.1128/JB.184.14.3957-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orsini G, Igonet S, Pène C, Sclavi B, Buckle M, Uzan M, et al. Phage T4 early promoters are resistant to inhibition by the anti-sigma factor AsiA. Mol Microbiol. 2004;52:1013–28. doi: 10.1111/j.1365-2958.2004.04038.x. [DOI] [PubMed] [Google Scholar]
- 29.Artsimovitch I, Svetlov V, Anthony L, Burgess RR, Landick R. RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J Bacteriol. 2000;182:6027–35. doi: 10.1128/JB.182.21.6027-6035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whipple FW, Sonenshein AL. Mechanism of initiation of transcription by Bacillus subtilis RNA polymerase at several promoters. J Mol Biol. 1992;223:399–414. doi: 10.1016/0022-2836(92)90660-C. [DOI] [PubMed] [Google Scholar]
- 31.Mekler V, Minakhin L, Kuznedelov K, Mukhamedyarov D, Severinov K. RNA polymerase-promoter interactions determining different stability of the Escherichia coli and Thermus aquaticus transcription initiation complexes. Nucleic Acids Res. 2012;40:11352–62. doi: 10.1093/nar/gks973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miropolskaya N, Ignatov A, Bass I, Zhilina E, Pupov D, Kulbachinskiy A. Distinct functions of regions 1.1 and 1.2 of RNA polymerase σ subunits from Escherichia coli and Thermus aquaticus in transcription initiation. J Biol Chem. 2012;287:23779–89. doi: 10.1074/jbc.M112.363242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weixlbaumer A, Leon K, Landick R, Darst SA. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–41. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schäfer R, Zillig W, Zechel K. A model for the initiation of transcription by DNA-dependent RNA polymerase from Escherichia coli. Eur J Biochem. 1973;33:207–14. doi: 10.1111/j.1432-1033.1973.tb02671.x. [DOI] [PubMed] [Google Scholar]
- 35.Pfeffer SR, Stahl SJ, Chamberlin MJ. Binding of Escherichia coli RNA polymerase to T7 DNA. Displacement of holoenzyme from promoter complexes by heparin. J Biol Chem. 1977;252:5403–7. [PubMed] [Google Scholar]
- 36.Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287:2497–500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 37.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–90. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 38.Robb NC, Cordes T, Hwang LC, Gryte K, Duchi D, Craggs TD, et al. The transcription bubble of the RNA polymerase-promoter open complex exhibits conformational heterogeneity and millisecond-scale dynamics: implications for transcription start-site selection. J Mol Biol. 2013;425:875–85. doi: 10.1016/j.jmb.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaal T, Ross W, Estrem ST, Nguyen LH, Burgess RR, Gourse RL. Promoter recognition and discrimination by EsigmaS RNA polymerase. Mol Microbiol. 2001;42:939–54. doi: 10.1046/j.1365-2958.2001.02703.x. [DOI] [PubMed] [Google Scholar]
- 40.Dayton CJ, Prosen DE, Parker KL, Cech CL. Kinetic measurements of Escherichia coli RNA polymerase association with bacteriophage T7 early promoters. J Biol Chem. 1984;259:1616–21. [PubMed] [Google Scholar]
- 41.Bronfenbrenner JJ, Korb C. Studies on the Bacteriophage of D'herelle: I. Is the Lytic Principle Volatile? J Exp Med. 1925;41:73–9. doi: 10.1084/jem.41.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chamberlin M, McGrath J, Waskell L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature. 1970;228:227–31. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- 43.Hinkle DC, Chamberlin MJ. Studies of the binding of Escherichia coli RNA polymerase to DNA. II. The kinetics of the binding reaction. J Mol Biol. 1972;70:187–95. doi: 10.1016/0022-2836(72)90532-3. [DOI] [PubMed] [Google Scholar]
- 44.Hinkle DC, Chamberlin MJ. Studies of the binding of Escherichia coli RNA polymerase to DNA. I. The role of sigma subunit in site selection. J Mol Biol. 1972;70:157–85. doi: 10.1016/0022-2836(72)90531-1. [DOI] [PubMed] [Google Scholar]
- 45.Hinkle DC, Mangel WF, Chamberlin MJ. Studies of the binding of Escherichia coli RNA polymerase to DNA. IV. The effect of rifampicin on binding and on RNA chain initiation. J Mol Biol. 1972;70:209–20. doi: 10.1016/0022-2836(72)90534-7. [DOI] [PubMed] [Google Scholar]
- 46.Ray DS, Dueber J, Suggs S. Replication of bacteriophage M13 IX. Requirement of the Escherichia coli dnaG function for M13 duplex DNA replication. J Virol. 1975;16:348–55. doi: 10.1128/jvi.16.2.348-355.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zillig W, Fujiki H, Blum W, Janeković D, Schweiger M, Rahmsdorf H, et al. In vivo and in vitro phosphorylation of DNA-dependent RNA polymerase of Escherichia coli by bacteriophage-T7-induced protein kinase. Proc Natl Acad Sci U S A. 1975;72:2506–10. doi: 10.1073/pnas.72.7.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krásný L, Gourse RL. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004;23:4473–83. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, et al. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–22. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.