Abstract
Lotus root is a popular wetland vegetable which produces edible rhizome. At the molecular level, the regulation of rhizome formation is very complex, which has not been sufficiently addressed in research. In this study, to identify differentially expressed genes (DEGs) in lotus root, four libraries (L1 library: stolon stage, L2 library: initial swelling stage, L3 library: middle swelling stage, L4: later swelling stage) were constructed from the rhizome development stages. High-throughput tag-sequencing technique was used which is based on Solexa Genome Analyzer Platform. Approximately 5.0 million tags were sequenced, and 4542104, 4474755, 4777919, and 4750348 clean tags including 151282, 137476, 215872, and 166005 distinct tags were obtained after removal of low quality tags from each library respectively. More than 43% distinct tags were unambiguous tags mapping to the reference genes, and 40% were unambiguous tag-mapped genes. From L1, L2, L3, and L4, total 20471, 18785, 23448, and 21778 genes were annotated, after mapping their functions in existing databases. Profiling of gene expression in L1/L2, L2/L3, and L3/L4 libraries were different among most of the selected 20 DEGs. Most of the DEGs in L1/L2 libraries were relevant to fiber development and stress response, while in L2/L3 and L3/L4 libraries, major of the DEGs were involved in metabolism of energy and storage. All up-regulated transcriptional factors in four libraries and 14 important rhizome formation-related genes in four libraries were also identified. In addition, the expression of 9 genes from identified DEGs was performed by qRT-PCR method. In a summary, this study provides a comprehensive understanding of gene expression during the rhizome formation in lotus root.
Introduction
Lotus root (Nelumbo nucifera Gaertn), which originated from India and China, is an aquatic herb vegetable and a member of the family Nelumbonaceae [1]. It is one of the oldest dicot plants in the world with many features of monocot plants, and has been widely cultivated in China, Japan, and other Southeast Asian counties for multiple purposes [2]. The products of lotus root such as fresh, salted and boiled rhizomes, lotus root starch, drinks, teas, and lotus seeds are very popular in the daily diet because of its richness in nutrients including starch, proteins, vitamins, and mineral substances [3], [4]. China is already exporting the processed products of lotus root to Japan, Korea, Europe, and the United States as a kind of off-season vegetable. In addition, Nodus nelumbinis rhizomatis, germ, stamens, and lotus root stems are also used as important ingredients in the traditional medicine [5], [6], [7].
With the unique characteristics, the rhizome of lotus root is formed underground, and it grows in size after sprouting in the shallow water, such as the pools, water gardens, tanks or tubs of the greenhouse, which indicates that the plant has developed mechanisms of surviving in the submerged environment. Then some floating leaves emerge from the nodes of enlarged and elongated rhizomes. Lotus root produces several rhizomes in a single growing season with average length of 10–20 cm each. Actually, similar with other wetland vegetable (corm, tuber, and bulbs), rhizome is a kind of underground stems, and work as storage organs. These are storage units for food that provide the plants with the energy for growth, blooming, and completing their lifecycle.
Development of storage organ (rhizome) can be classified into four stages: induction (stolon stage), initial swelling, middle swelling, and later swelling stage [8]. Stolon tips grow radically in the induction stage. In the second stages, longitudinal growth of stolon stops and its tip swells [9], [10]. At the middle swelling and later swelling stages, some important carbohydrates are synthesized in the storage organ. For example, accumulation of starch greatly increases in these stages. Environmental factors affect above four stages through triggering signal molecules or gene regulation.
Development of storage organs have been extensively studied, especially in tuber and corm, and great changes have been found in genetic and morphometric processes [12], [13]. Short days (SDP) condition promotes the formation of storage organ, while, long days (LDP) prolongs this process. For example, Masuda et al. (2007) have found that rhizome enlargement was brought about under 8,10 and 12 h photoperiods [14]. The leaf receives photoperiodic signal, and then transports it to the underground stolon tips via the phloem, which promotes the transition of storage organ. CONSTANS, SFT family protein, GIGANTEA, and cycling dof factor are believed to participate in the signal transduction of photoperiodic control, and expression of these genes affects the formation of storage organ [15], [16], [17], [18]. In addition, under SDP condition, StBEL5 represses the gibberellin StGA20 ox1 biosynthesis, which promotes formation of the storage organ [19]. At the same time, the expression of StBEL5 is enhanced by miR172, suggesting that long distance transport of RNA signal also participates in the formation of underground storage organ [20]. In addition, phytochrome B is involved in the response of plants to photoperiodic control, and the formation of storage organ is affected by PHYB in SD. Decreasing the levels of PHYB in transgenic plants promotes the formation of storage organ both in SD and LD, while non-transgenic plants form storage organ only in SD [21]. These results suggest that plants lose the inhibitory effect on formation of storage organ caused by LD when expression of PHYB is down-regulated [15]. It is believed that high content of sucrose is required as a necessary condition during the formation of storage organs [15], and sucrose transporters can trigger the formation of storage organ at the early stages [22]. Therefore, sucrose plays an important role at the initial swelling stages of the formation of storage organ [23].
Hormones including: cytokinin, jasmonic acid (JA), gibberellic acid (GA), abscisic acid (ABA), and ethylene are also involved in the initiation and regulation of growth in these storage organs [24], [25], [26]. It has been reported that exogenous application of GA can act as an inhibitor from stolon to induction stage. Transgenic potato plants with GA oxidase gene postpone the storage organ formation. Whereas, decreased expression of this gene results in an earlier formation than non-transgenic plants [27]. Cytokinin and jasmonic acid promote the induction and elongation of storage organ [28]. Bhat et al 29. found that exogenous cytokinin is necessary to induce formation of storage organ in ginger due to improvement in photosynthesis. ABA shows high correlation to tuber formation because ABA-deficient potato plants show retarded tuberization [30]. Exogenous application of auxin on the decapitated peas and potatoes inhibits the formation of axillary buds [31]. Ethylene, produced by almost all plants mediates a variety of developmental processes in plants, such as seed germination, lateral bud stimulation, adventitious rooting, overcoming dormancy, and organ senescence and abscission [26], [32]. Exogenous ethylene is believed as an inducer for the storage organ and root bulking in carrots [33].
Dynamics of rhizome enlargement is relatively poorly known in the lotus root, because detailed investigation of underground rhizome growth is time and labour consuming. Therefore, lotus can be suitable as a model plant to study rhizome growth [14]. Just like the other storage organs, rhizome is also an important edible product, and the developmental processes of this kind of storage organ are regulated by genes [34]. Expressions of these genes affect rhizome formation. Although much work in other species has partially described the above processes, and many storage organ-related genes have been documented [35], but the expression of genes which affect rhizome formation in lotus root have not been studied in detail. Tag sequencing technique has been established as an efficient approach to study gene expression in different environmental conditions [36], [37]. In some vegetable plants, a lot of important genes involved in plant critical metabolisms have been successfully identified by this technique [38], [39], [40], [41]. In this study, DEGs from four developmental stages of lotus root rhizome were sequenced and analyzed with aim to comprehensively understand the processes of rhizome formation and development at molecular level.
Materials and Methods
Plant Materials
‘MeiRen Hong’, a wildly cultivated species in China, was planted in the field at the water depth of 20–25 cm in spring with average temperature 30°C/day and 20°C/night during the whole growth season. Three or more stolons were developed and elongated in proper order in each plant. When plants grew up to 4–5 leaves stage (about 90–100 days after plantation), formation of rhizome started at stolon tips. For the analysis of tag-sequencing and gene expression, rhizomes of four developmental stages (stolon, initial swelling, middle swelling and later swelling stage) (Fig. S1) from the plants (three tips from different plants were combined for each stage) were used. To get the materials of different developmental stages, lotus was (rhizome lotus, flower lotus and seed lotus) were cultivated in a field (non-private), located in the South-Eastern China. The permission for sample collection was taken from the Department of Horticulture of YangZhou University, China. No specific permissions were required for the location and the field studies, because the experiments did not involve any endangered or protected species.
Screening DEGs
Rhizome transcriptome from the above four development stages was analyzed. Stolon tips, rhizomes in the initial swelling, middle swelling and later swelling stages were collected and ground, and the RNA was isolated from the ground samples using RNA extraction mini kit (QIAGEN, Germany). DNaseI was added to eliminate DNA contamination. Sequencing of transcripts in the form of special constructs was completed by Beijing Institute of Genomics (BIG).
To screen the DEGs, transcriptome from these four stages was analyzed with the aspirations to track the major changes in metabolism. RNA was isolated from the materials of these four stages. The DEG libraries of four samples were determined in parallel using Illumina gene expression sample preparation kits. Briefly, the total RNA from four stages was used for mRNA capture with magnetic oligo (dT) beads. The first and second strand cDNA were synthesized, and bead-bound cDNA was subsequently digested with NlaIII.
The 3′-cDNA fragments attached to the oligo (dT) beads were ligated to the Illumina GEX NlaIII adapter 1, which contained a recognition site for the endonuclease MmeI for cutting 17 bp downstream of the recognition site (CATG) to produce tags with adapter 1. After removing 3′ fragment via magnetic beads precipitation, an Illumina GEX adapter 2 was introduced at the site of MmeI cleavage. The resulting adapter-ligated cDNA tags were amplified using PCR-primers that were annealed to the adaptor ends for 15 cycles.
The 85 base fragments were purified and recovered by 6% polyacrylamide Trisborate-EDTA gel. The final quality of the tagged sequences was checked by an Agilent 2100 Bioanalyzer. The four tag libraries constructed underwent Illumina proprietary sequencing chip for cluster generation through in situ amplification and were deep-sequenced using Illumina Genome Analyzer. For the raw data, we filtered adaptor sequences, low quality tags (tags with unknown nucleotides N), empty reads and tags that were too short or too long, and tags with only one copy to get clean tags. The types of clean tags were represented as the distinct clean tags. Subsequently, we classified the clean tags and distinct clean tags according to their copy number in the library, and showed their percentage in the total clean and distinct tags, and analyzed saturation of the four libraries.
For annotation, all tags were mapped to the reference sequence of NCBI database (http://www.ncbi.nlm.nih.gov/), and no more than 1-bp nucleotide mismatch was allowed. The alignment procedures were conducted essentially by following the protocols described in the online documentation (http://maq. sourceforge.net) and adopting the default parameter values. To monitor mapping events on both strands, both sense and complementary antisense sequences were included in the mapping process. The tags mapped to reference sequences from multiple genes were filtered [38].
Identification of DEGs
The transcriptome of the lotus root from the above four stages was used as reference for the screening and analysis of the DEGs due to unavailability of the existing data. All expressed genes were monitored, and the gene functions were explored by using database annotations like nr, Swiss-Prot, KEGG, and COG with following criteria: for the gene annotations, blastx alignment (evalue <0.00001) between unigenes and protein databases, such as Swiss-Prot, KEGG, and COG were performed. Best aligning results were used to decide sequence direction and functions of the unigenes.
In case of any conflict in the results from different databases, a priority order of nr, Swiss-Prot, KEGG, and COG was followed when deciding sequence direction of unigenes. When a unigene happened to be unaligned with none of the above databases, ESTscan was used to predict its coding regions as well as to decide its sequence direction. All of the expressed unigenes were classified according to their functions in metabolism processes. For screening the differentially expressed genes, “FDR≤0.001” and the absolute value of “log2 Ratio ≥1” were used as a threshold to judge the significance of difference in expression of unigenes. Important genes related to rhizome formation were reposited in NCBI database (TSA: BioProject ID is PRJNA196449; Accession number is GAHV01000000; BioSample is SAMN02028153; Sequence read archive is SRR82669).
Gene Expression Analysis by qRT-PCR
To carry out the study of gene expression in different species, the cultivation conditions of lotus (rhizome lotus, flower lotus and seed lotus) were kept same as described above. Quantitative RT-PCR analysis was performed to quantify the transcriptional level of nine novel genes with rhizome lotus at stolon stage, initial stage, middle swelling stage and later swelling stage to evaluate the results of tag-sequencing. In addition, expression of 18 important genes at stolon stage, initial stage and middle swelling stage (no obvious later swelling stage in flower and seed lotus) relevant to rhizome formation were also studied in rhizome lotus, flower lotus and seed lotus. Total RNA was extracted from stolon tips, rhizomes of initial swelling, middle swelling and later swelling stage respectively, using RNA extraction mini kit (QIAGEN, Germany). DNaseI was used to digest DNA during the RNA extraction process to eliminate DNA contamination. A total of 1–2 µg of RNA was used in cDNA synthesis according to the manufacturer’s instructions (Promega, USA). The quantitative RT-PCR reaction was performed with the Mx 3000P machine (STRATAGENE, http://www.stratagene.com). The SYBR Green Master Mix was used to identify mRNA level according to the manufacturer’s instructions (Tiangen, China). According to the sequencing results, the primers were designed for the genes that enhanced transcriptional level during rhizome formation which were listed in Table 1. β-Actin was used as internal standard and amplified with the primers, forward: 5′-AACCTCCTCCTCATCGTACT-3′, and reverse: 5′-GACAGCATCAG CCATGTTCA-3′. Amplification was performed in a 20 µl reaction mixture, containing 0.16 mM dNTPs, 0.1 µM forward and reversed primers respectively, 1 mM MgCl2, 0.4 U Taq polymerase (Tiangen, China), and 1 µl cDNA. The PCR program consisted of 30 cycles: 94°C for 10 min; 94°C for 1 s; 56–60°C for 30 s; 72°C for 60 s, and the final extension at 72°C for 10 min. Triplicate samples were used for quantitative RT-PCR.
Table 1. Detailed information about primers used for qRT-PCR variation.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Tm (°C) | Product (bp) |
| CALM | GAATTGAAGGAAGCGTTTAA | TGGCCAGCATCATTCTCACA | 58 | 200 |
| SUS | GCTGATGCTGAATGATCGGA | CATCAGGTATTAAACGGGTC | 56 | 357 |
| GA20ox | TGTCTCCTAATGGATTCAAC | CTCGTCACCGTTGAGGAATC | 52 | 225 |
| ERF | GGAATGGAAGGGTCCGATGT | CATGAGATACACTTTCTAACT | 52 | 354 |
| bZIP | CGCCACCTTGAGTCAGCTGT | CAGTGTCTGGGAGTCGGTCA | 55 | 276 |
| WRKY | CTCTTTATCCAGCATCTTTGA | GGTTCTCTCTGCCGTTTCTG | 54 | 348 |
| SAUR | ATGGCTCTCAGGAAATCAAA | ATAGTGAGACCCATGTCGTG | 53 | 266 |
| CBF | AGAGGAGTCAGGGAGAGGA | GCAACCTTCTCTTCGCTCTC | 57 | 329 |
| GBSS | TGGGGAACACTTCAATATCT | CTCCCGAAGAGGAAGACCA | 52 | 307 |
| CONSTANS | GGCATTCCTTGCGATTTCTG | TCGGAGCTGCAATTGTCG | 55 | 182 |
| GIGANTEA | AGAACTCAAGCCACCTACTA | GGTAGCAGTTTCATAGCGAG | 53 | 153 |
| MADS | CTGAGGTCGCTGTGATCGT | GCCTTAAGTTTGGTGTGTTC | 52 | 178 |
| Dof | CAATCTTTCCCAACCTCG | ACGAAGACAAGGACGATGA | 53 | 168 |
| HD | CGCCGTCTGCTCCAACAA | CGATGAAAGTTCCTGTGGCA | 60 | 200 |
| Lipoxygenase | CCCTTTGCCTCAAGTTATCC | GCAGCCCATTCAGGTTATTC | 55 | 206 |
| GP | AGGTTTGGGCACTACTATGG | TTAGGGTATCATCATGCGTG | 54 | 173 |
| PHYB | GTGGAATGCAGCAATGGA | ATCCACATATTTTCCTTTCC | 51 | 211 |
| APETALA | GTCAGTCGGAACCTCTTGC | CCGCCCAAATAAACCTGC | 54 | 175 |
All primers used for qPCR were designed according to CDS obtained in this study in lotus root.
Results
2.1 Transcriptome of Rhizome
Four stages of rhizome development: stolon stage, initial swelling, middle swelling and later swelling stages were selected to construct libraries using the Illumina sequencing platform to investigate the genes involved in rhizome formation. A total of 49053 genes which included 39497 (accounted for 80.52%) CATG site genes used as reference genes were obtained after the transcriptome of above four stages were preprocessed.
About 5 million raw tags in each library were obtained with 327019, 312178, 363183, and 385558 raw distinct tags respectively. To get clean tags, all the raw tags were filtered with reference sequences, and 4542104, 4474755, 4777919, and 4750348 clean tags including 151282, 137476, 215872, and 166005 distinct tags in L1, L2, L3, and L4 libraries were obtained respectively. All data of tags for each library is given in Table 2. To testify whether or not the sequenced tags were sufficient to cover the whole transcriptome, the analysis of sequencing saturation was also performed in the four libraries. The number of detected genes increased until the sequencing tags reached 3 million or more, which indicated that the identified expressed tags were enough to reflect the whole transcriptional information in genome of lotus root (Fig. S2).
Table 2. Categorization and abundance of tags.
| L1 | L2 | L3 | L4 | ||
| Raw Tag | Total number | 4722270 | 4654357 | 4931007 | 4975127 |
| Distinct Tag | 327019 | 312178 | 363183 | 385558 | |
| Clean Tag | Total number | 4542104 | 4474755 | 4777919 | 4750348 |
| Distinct Tag number | 151282 | 137476 | 215872 | 166005 | |
| All Tag Mappingto Gene | Total number | 3593944 | 3550079 | 3833530 | 4043129 |
| Total percentage of clean tag | 79.13% | 79.34% | 80.23% | 85.11% | |
| Distinct Tag number | 91450 | 81334 | 115876 | 108718 | |
| Distinct Tag percentage of clean tag | 60.45% | 59.16% | 53.68% | 65.49% | |
| Unambiguous TagMapping to Gene | Total number | 2918451 | 2919531 | 3315975 | 3300501 |
| Total percentage of clean tag | 64.25% | 65.24% | 69.40% | 69.48% | |
| Distinct Tag number | 73493 | 64898 | 93692 | 87374 | |
| Distinct Tag percentage of clean tag | 48.58% | 47.21% | 43.40% | 52.63% | |
| All Tag-mappedGenes | Total number | 26261 | 24551 | 29314 | 27666 |
| Percentage of reference genes | 53.54% | 50.05% | 59.76% | 56.40% | |
| Unambiguous Tag-mapped Genes | Total number | 20471 | 18785 | 23448 | 21778 |
| Percentage of reference genes | 41.73% | 38.30% | 47.80% | 44.40% | |
| Unknown Tag | Total number | 948160 | 924676 | 944389 | 707219 |
| Total percentage of clean tag | 20.87% | 20.66% | 19.77% | 14.89% | |
| Distinct Tag number | 59832 | 56142 | 99996 | 57287 | |
| Distinct Tag percentage of clean tag | 39.55% | 40.84% | 46.32% | 34.51% |
About 5.0 million tags were sequenced, and clean tags and distinct tags were obtained after removal of low quality tags from each library. Some distinct tags were unambiguous tags after mapping to the reference genes, and some were unambiguous tag-mapped genes.
Functions of gene were annotated by comparison against existing NCBI database.
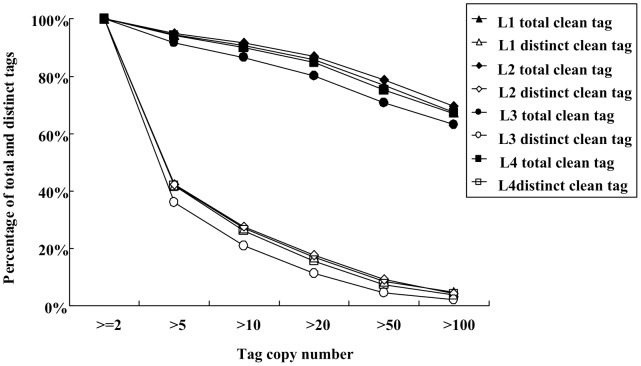
Among which, more than 41% of all distinct sequences in four libraries showed an above cut-off BLAST result, and about 59% could not match with the known genes. These identified genes were classified into 26 catalogues according to their functions. One catalogue containing the largest number of genes predicted functions, and the smallest one was relevant to nuclear structure (Fig.S3). The copy distribution of total and distinct clean tags in four libraries showed the same tendency with about 4% of distinct clean tags higher than 100 copies and more than 11% tags being in 20–50 counts. The number of distinct clean tags between 2–5 copies (approximately 40%) was high as compared with that of others (Fig.1).
Figure 1. Distribution of total clean tags and distinct clean tags count from the four libraries.
2.2 DEGs in Four Libraries
Gene expression profiles during rhizome formation
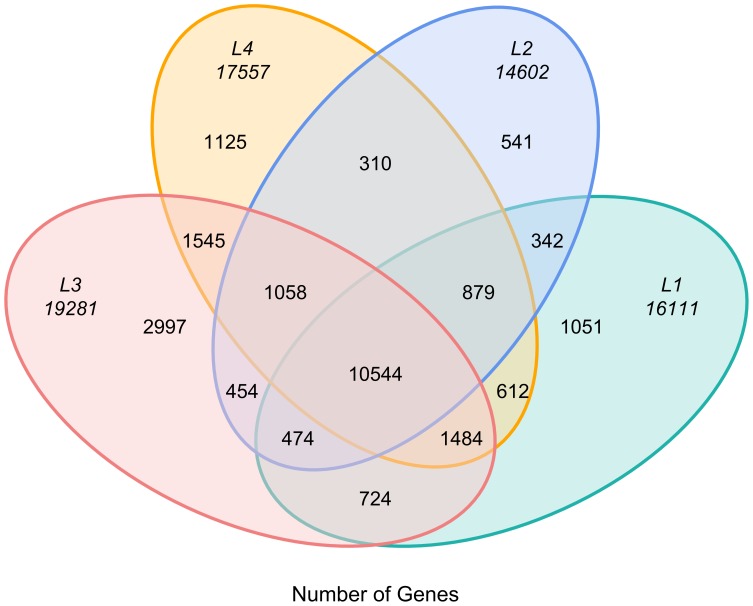
Gene expression profiles during rhizome development in four libraries were examined. Total 16111, 14602, 19281, and 17557 transcripts were identified from L1, L2, L3, and L4 libraries respectively. We found that 10544 genes were expressed in all four libraries, and 1051, 541, 2997, and 1125 were especially expressed in L1, L2, L3, and L4 libraries respectively (Fig. 2).
Figure 2. Analysis of tag mapped genes among four libraries.
Differentially expressed genes were identified from these four different developmental stages during rhizome formation to uncover the changes in metabolism at molecular level. The abundance of transcripts in four libraries was counted by the number of genes per million clean tags (TPM). FDR (false discovery rates) <0.001 and the absolute value of |log2 Ratio|≥1 were used as a threshold to judge whether gene expression was significant. A total of 1714 genes were found to change their transcriptional level in L1/L2 libraries. From which, 759 were up-regulated (Table S1) and 955 were down-regulated. In L2/L3 libraries, 4013 genes, including 2157 up-regulated genes and 1856 down-regulated genes were found. In L3/L4 libraries, 2344 genes were observed to change their transcription including 1429 up-regulated genes and 915 down-regulated genes (Table 3). According to the gene expression profile in L1/L2, L2/L3, and L3/L4 libraries, a large number of genes showed altered mRNA transcriptional levels in L2/L3 libraries. However, compared with other libraries, number of transcripts in L1/L2 libraries showed a little change.
Table 3. DEGs across all libraries.
| L1:L2 | L1:L3 | L2:L3 | L3:L4 | L1:L4 | L2:L4 | |
| Total | 1714 | 4339 | 4013 | 2344 | 2115 | 1667 |
| Up-regulated | 759 | 2052 | 2157 | 1429 | 1069 | 1059 |
| Down-regulated | 955 | 2287 | 1856 | 915 | 1046 | 608 |
All the genes mapped to the reference sequence and genome sequences were examined for their expression differences across different libraries. Numbers of differentially expressed genes represent transcripts, using threshold values FDR≤0.001 and |log2 Ratio|≥1 for controlling false discovery rates. L1, L2, L3, and L4 represent the samples which were collected at stolon stage, initial swelling stage, middle swelling stage, and later swelling stage of rhizome, respectively.
Most DEGs in each library
The variation in expression was observed in 1714, 4013, and 2344 genes in L1/L2, L2/L3, and L3/L4 libraries, respectively. Therefore, 20 DEGs with higher levels of changes were selected respectively, to monitor changes in metabolism during rhizome formation. We found that the most DEGs in L1/L2 libraries were Pectinesterase, a ubiquitous cell-wall-associated enzyme that facilitates plant cell wall modification and subsequent breakdown. DEGs in L1/L2 libraries could be mainly classified into 3 catalogues which belonged to hormone response (SAUR family protein and Ethylene responsive transcription factor), fiber synthesis (Glycerophosphoryl diester phosphodiesterase, Receptor protein kinase-like protein, G-type lectin S-receptor-like serine, Fiber expressed protein, Calcium-dependent protein kinase, Sucrose synthase) and stress response proteins (Ethylene responsive transcription factor, Calcium-dependent protein kinase, Peroxidase, SAUR family protein). In addition, an extensin and vacuolar-processing enzyme was also found to be involved in the process from stolon stage to initial swelling stage (Table 4). DEGs in L2/L3 libraries could be mainly classified into four catalogues. One catalogue was associated with energy metabolism (NADH dehydrogenase, Malate dehydrogenase [NADP], ATP synthase subunit beta). The second was relevant to synthesis and transport of matter (Starch-branching enzyme I, Sugar transporter, 3-ketoacyl-CoA synthase 4, 2-methyl-6-phytylbenzoquinone methyltranferase, Vacuolar H+-pyrophosphatase),DEGs in third catalogue were involved in hormone metabolism and response (Auxin response factor 2, Ethylene-responsive transcription factor ERF114, ACC oxidase, Xanthine dehydrogenase). The fourth catalogue of DEGs covered was stress response genes (Transcription factor bHLH80, Vacuolar H+-pyrophosphatase). In L3/L4 libraries, DEGs were classified into three major catalogues, such as synthesis of matter (Basic 7S globulin, UDP-glycosyltransferase 89B2, Sucrose synthase, UDP-glycosyltransferase, Glutathione S-transferase, Fiber expressed protein, Patatin group), hormone synthesis and response (Gibberellin-regulated protein 6, Gibberellin 20 oxidase, Ethylene-responsive transcription factor ERF025, ERF3 transcription factor) and swelling related proteins (Alpha-expansin 3, Extensin). In addition, we also found ATPase subunit enhanced transcriptional level in later swelling stage. A lot of genes were down regulated in L1/L2, L2/L3, and L3/L4 libraries, most of which in L1/L2 libraries were involved into synthesis of matter, and others in L2/L3 and L3/L4 libraries were relevant to cell growth and differentiation, translation, RNA process, and modification.
Table 4. 20 most differentially expressed annotated genes in L1/L2, L2/L3, and L3/L4 libraries based on expressed tag frequency.
| Gene ID | Relative abundant (TMP ratio) | Function annotation | Fragment (bp) |
| L1/L2 libraries | |||
| L21966 | 15.747 | Pectinesterase-2 precursor | 1068 |
| L6372 | 12.111 | Non-specific lipid-transfer protein | 558 |
| L4842 | 11.182 | Primary amine oxidase-like | 355 |
| L19738 | 10.569 | Inositol oxygenase | 924 |
| L5740 | 9.611 | MLP-like protein | 28 456 |
| L5559 | 9.525 | Glycerophosphoryl diester phosphodiesterase | 489 |
| L9661 | 9.26 | Extensin (class I) | 198 |
| L4093 | 9.23 | Formamidase | 291 |
| L11656 | 9.12 | Vacuolar-processing enzymeprecursor | 1464 |
| L14179 | 8.94 | Receptor protein kinase-like protein | 402 |
| L24462 | 8.93 | Chaperone protein dnaJ 8 | 234 |
| L19479 | 8.81 | CytochromeP45082C4 | 1479 |
| L22249 | 8.8 | Peroxidase 64-like isoform 2 | 933 |
| L9369 | 8.73 | Rop guanine nucleotide exchange factor 2 | 219 |
| L13340 | 8.65 | G-type lectinS-receptor-likeserine | 1484 |
| L15631 | 8.41 | Fiber expressed protein | 345 |
| L16154 | 8.33 | Ethylene responsive transcription factor | 927 |
| L7272 | 8.29 | SAUR family protein | 234 |
| L3989 | 8.28 | Calcium-dependent protein kinase | 783 |
| L2349 | 8.20 | Sucrose synthase | 519 |
| L2/L3 libraries | |||
| L9592 | 13.40 | Cytochrome P450 86A1 | 1530 |
| L3363 | 12.66 | Phytylbenzoquinone methyltranferase | 810 |
| L19686 | 11.81 | 3-ketoacyl-CoA synthase4 | 1499 |
| L24558 | 10.83 | CASP-like protein RCOM_1174750-like | 480 |
| L14401 | 10.58 | GDSL esterase/lipase At2g23540-like | 743 |
| L13523 | 9.98 | Transcription factor bHLH80 | 450 |
| L9063 | 9.97 | Auxin response factor 2 | 711 |
| L11556 | 9.67 | E3 ubiquitin-protein ligase UPL4-like | 699 |
| L3219 | 9.43 | Starch-branching enzymeI | 2205 |
| L6212 | 9.19 | NADH dehydrogenase | 291 |
| L4985 | 9.14 | Sugar transporter | 1470 |
| L2123 | 8.98 | Calcineurin subunit B isoform 1 | 231 |
| L2369 | 8.97 | Vacuolar H+-pyrophosphatase | 300 |
| L21145 | 8.96 | ABC transporter family protein | 420 |
| L4307 | 8.95 | Ethylene-responsive transcription factor ERF114; | 804 |
| L18002 | 8.90 | Xanthine dehydrogenase | 486 |
| L26121 | 8.67 | Malate dehydrogenase [NADP] | 254 |
| L6411 | 8.44 | ATP synthase subunit beta | 1461 |
| L7701 | 8.40 | Calcium-dependent protein kinase 30 | 1005 |
| L378 | 8.30 | 1-aminocyclopropane-1-carboxylate oxidase | 822 |
| L3/L4 libraries | |||
| L22026 | 13.72 | Basic 7S globulin | 962 |
| L18835 | 12.626 | Omega-hydroxypalmitate O-feruloyl Transferase | 1361 |
| L134 | 11.97 | Alpha-expansin 3 | 750 |
| L4954 | 11.91 | Extensin | 513 |
| L6449 | 11.01 | Gibberellin-regulated protein 6 | 315 |
| L3691 | 10.98 | Gibberellin 20 oxidase | 289 |
| L3566 | 10.56 | Ethylene-responsive transcription factor ERF025 | 393 |
| L19429 | 10.48 | ATPase subunit | 285 |
| L24010 | 10.41 | F-box family protein | 1266 |
| L4407 | 10.40 | Heat shock protein70 | 1188 |
| L15631 | 10.39 | Fiber expressed protein | 345 |
| L7453 | 10.12 | Glutathione S-transferase | 411 |
| L8862 | 10.09 | UDP-glycosyltransferase89B2 | 1107 |
| L2616 | 9.62 | Sucrose synthase | 1131 |
| L3393 | 9.56 | Cell wall-associated hydrolase | 495 |
| L309 | 9.39 | ERF3 transcription factor | 498 |
| L15819 | 9.19 | UDP-glycosyltransferase 88A | 1407 |
| L23091 | 9.15 | EF-hand calcium-binding protein | 516 |
| L8032 | 8.97 | WD repeat-containing protein 18 | 627 |
| L10302 | 8.75 | Patatin group A-3 | 1014 |
| L1/L2 libraries | |||
| L16154 | 8.29 | Ethylene-responsive transcription factorERF017 | 348 |
| L1375 | 8.18 | CBF/DREB3 transcription factor | 609 |
| L17409 | 8.04 | Transcription factor bHLH35 | 726 |
| L22245 | 7.81 | AP2 domain class transcription factor | 923 |
| L5201 | 7.80 | WRKY transcription factor 6-like | 666 |
| L161 | 7.31 | KNAT3-like transcription factor | 747 |
| L7327 | 6.81 | bZIP transcription factor 60 | 495 |
| L3810 | 6.47 | GRAS family transcription factor | 308 |
| L959 | 6.46 | Transcription factor hy5 | 435 |
| L24021 | 6.06 | MYB transcription factor | 594 |
| L5723 | 6.05 | MADS-box transcription factor | 744 |
| L16316 | 5.52 | Transcription factor TCP9 | 1215 |
| L4285 | 5.49 | Heat stress transcription factor A-4b | 960 |
| L10423 | 5.48 | NAC domain-containing protein | 471 |
| L2/L3 libraries | |||
| L13523 | 9.98 | Transcription factor bHLH80 | 450 |
| L4307 | 8.95 | Ethylene-responsive transcription factor ERF114 | 804 |
| L7947 | 8.19 | Homeobox protein BEL1 homolog | 336 |
| L3540 | 8.100 | Trihelix transcription factor GT-1 | 972 |
| L9587 | 7.97 | NAC domain protein NAC1 | 710 |
| L1325 | 7.71 | Transcription factor ICE1 | 477 |
| L7126 | 6.70 | Transcription factor MYB59-like | 345 |
| L3348 | 6.98 | Transcription factor SPATULA | 801 |
| L21444 | 5.55 | NF-X1-type zinc finger protein NFXL1-like | 2277 |
| L6302 | 5.54 | KANADI like transcription factor | 834 |
| L545 | 5.52 | WRKY transcription factor 44 | 1125 |
| L4641 | 5.2 | Scarecrow-like transcription factor PAT1-like | 1644 |
| L20454 | 4.71 | BZIP domain class transcription factor 60 | 495 |
| L1140 | 4.70 | Calmodulin-binding transcription factor SR3L | 345 |
| L25749 | 3.92 | GATA domain class transcription factor | 279 |
| L12403 | 3.39 | HSF transcription factor | 384 |
| L6635 | 3.36 | HD domain class transcription factor | 258 |
| L5314 | 2.35 | Transcription factor KAN2 | 447 |
| L25842 | 1.34 | Zinc finger protein CONSTANS-LIKE | 894 |
| L3/L4 libraries | |||
| L3566 | 10.56 | Ethylene-responsive transcription factor ERF025 | 393 |
| L5181 | 10.40 | HSP transcription factor | 384 |
| L309 | 9.39 | ERF3 transcription factor | 498 |
| L18531 | 8.55 | Transcription factor HEC1 | 699 |
| L6220 | 7.19 | WRKY transcription factor 70 | 951 |
| L5632 | 6.98 | Transcription factor AS1 | 1071 |
In this study, some important transcription factors involved in rhizome formation were also identified. A total of 14, 19, and 20 transcription factors were found in L1/L2, L2/L3, and L3/L4 libraries, respectively (Table 5). In L1/L2 libraries, some of transcription factors were ethylene-responsive which include: ERF017, CBF/DREB3, bHLH35, TCP9, and WRKY transcription factors, and AP2 domain class transcription factor, KNAT3-like transcription factor, bZIP transcription factor 60, GRAS family transcription factor, transcription factor hy5, MYB transcription factor, MADS-box transcription factor, heat stress transcription factor A-4b, and NAC domain-containing protein. In L2/L3 libraries, expression levels of many important regulators (e.g. transcription factor bHLH80, homeobox protein BEL1 homolog, ethylene-responsive transcription factor ERF114, trihelix transcription factor GT-1, NAC domain protein, transcription factor ICE1, transcription factor MYB59-like, transcription factor SPATULA, NF-X1-type zinc finger protein NFXL1-like, KANADI like transcription factor, WRKY transcription factor 44, scarecrow-like transcription factor PAT1-like, BZIP domain class transcription factor, calmodulin-binding transcription factor SR3L, GATA domain class transcription factor, HSF domain class transcription factor, HD domain class transcription factor, transcription factor KAN2, and zinc finger protein CONSTANS-LIKE) were enhanced. In L3/L4, most of the genes were found to be involved in matter metabolism and stress response.
Table 5. Expression abundance of up-regulated transcriptional factors during rhizome formation.
| L3982 | 6.97 | GATA transcription factor | 621 |
| L18475 | 6.71 | Dof zinc finger protein | 630 |
| L14316 | 6.70 | NF-X1-type zinc finger protein NFXL1 | 2277 |
| L1375 | 6.38 | CRT/DRE binding factor 1 | 681 |
| L2135 | 5.98 | NAC transcription factor | 471 |
| L19946 | 5.96 | BZIP transcription factor 107 | 81 |
| L21009 | 5.39 | ZF-HD homeobox protein | 705 |
| L5026 | 5.38 | Transcription factor PIF3 | 2184 |
| L5723 | 5.37 | MADS-box transcription factor | 744 |
| L12725 | 4.32 | NAC domain protein | 471 |
| L6472 | 3.11 | MYB transcription factor MYB2 | 411 |
| L22833 | 2.51 | BEL1-like home domain protein 1 | 1244 |
| L2868 | 2.44 | Scarecrow-like protein 4 | 1818 |
| L13679 | 1.37 | Transcription initiation factor TFIID | 587 |
Genes related to rhizome formation
To test whether the transcription patterns in this study had coverage of the well-defined genes, the data sets of this experiment were compared to previous reports. We found 14 identified genes relevant to rhizome formation. The expression of these genes and their biological functions are listed in Table 6. Among which, 8 genes including zinc finger CONSTANS-like protein, GIGANTEA (clock-regulated protein), BEL1-like HD transcription factor, sucrose synthase, ca2+/Calmodulin-like protein, lipoxygenase, glucose pyrophosphorylase, granule-bound starch synthase, phytochrome B enhanced their transcriptional levels, and 3 genes (SFT family protein, cycling dof Factor) did not show any significant change in its expression in all the four libraries. Three genes [MADS-box transcription factor, GA 20-oxidase (aside from L3/L4 stages), APETALA1)] directly decreased their expression during the rhizome development.
Table 6. Expressed abundance of some rhizome formation-related genes identified previously.
| Gene ID | TPM-L1 | TPM-L2 | TPM-L3 | TPM-L4 | Function annotation | References | ||
| L20523 | 5.28 | 7.82 | 20.93 | 10.95 | Zinc finger CONSTANS-like protein | [17] | ||
| L21782 | 0.88 | 2.01 | 4.6 | 1.89 | GIGANTEA (clock-regulated protein) | [20] | ||
| L6549 | 27.08 | 21.05 | 18.42 | 20.0 | MADS-box transcription factor | [84] | ||
| L9854 | 1.1 | 1.45 | 2.26 | 1.26 | SFT family protein | [124] | ||
| L22473 | 7.93 | 6.7 | 6.28 | 6.74 | Cycling Dof Factor | [125] | ||
| L29236 | 1.32 | 2.46 | 7.53 | 3.37 | BEL1-like HD transcription factor | [126] | ||
| L2349 | 0.01 | 0.45 | 3.35 | 0.01 | Sucrose synthase | [90] | ||
| L16799 | 0.01 | 1.79 | 1.26 | 1.26 | Calmodulin-like protein | [93] | ||
| L18271 | 0.44 | 2.01 | 9 | 7.37 | Lipoxygenase | [127] | ||
| L5098 | 76.18 | 73.63 | 135.62 | 154.09 | Glucose pyrophosphorylase | [128] | ||
| L19795 | 0.01 | 0.45 | 11.93 | 1.68 | Granule-Bound Starch Synthase | [129] | ||
| L16933 | 0.01 | 0.89 | 3.98 | 2.74 | Phytochrome B | [130] | ||
| L18298 | 0.44 | 0.22 | 0.12 | 1.45 | GA 20-oxidase | [27] | ||
| L4398 | 0.44 | 0.03 | 0.09 | 0.04 | APETALA | [131] | ||
TMP, transcripts per million clean tags.
2.3 Expression Analysis of Nine Genes through qRT-PCR
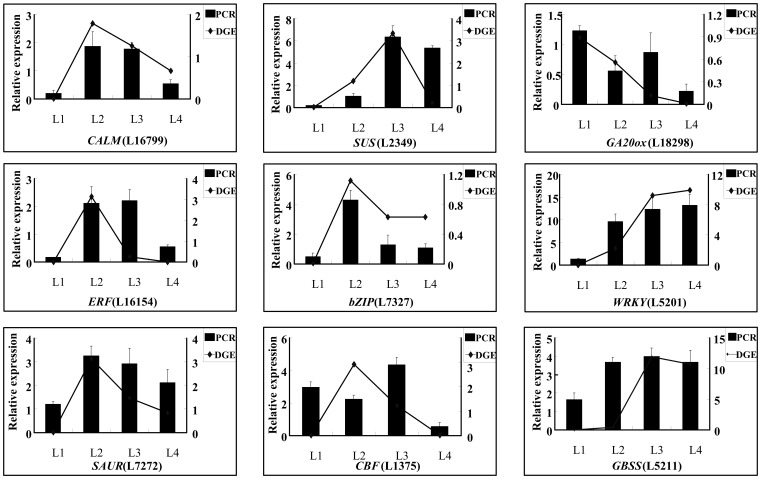
Nine genes including CALM, SUS, GA20ox, ERF, bZIP, WRKY, SAUR, CBF, and GBSS involved in rhizome formation were studied by quantitative RT-PCR method. From the data of results, expression profiling of seven genes in four developmental stages i.e. stolon, initial swelling, middle swelling, and later swelling stage by qRT-PCR showed similar tendency as found in Tag-sequencing, which indicates a correspondence of the results from qRT-PCR analysis with the Tag sequencing analysis. Only two genes (CBF and GBSS) were observed with some difference in L2/L3 libraries in transcriptional level between Tag-sequencing analysis and qRT-PCR data (Fig. 3). In addition, detailed study for the expression of 18 genes in rhizome lotus, flower lotus and seed lotus was also carried out with qRT-PCR method. The results showed that many genes including bZIPs, WRKY, GBSS, CONSTANS, GIGANTEA, Dof, HD, PHYB, Lipoxygenase and GP showed different expression profiles in three phenotypes of lotus. Among which, expression of CONSTANS, GIGANTEA, Dof, HD, PHYB, which involved in photoperiodic signals was higher in flower lotus and seed lotus than that of rhizome. Whereas, GBSS and Lipoxygenase bZIPs, WRKY and GP showed higher transcriptional level in rhizome as compared with that of flower lotus and seed lotus (Fig. S4).
Figure 3. Validation of tag-mapped genes from three stages of lotus root with quantitative RT–PCR.
Discussion
Tags Identified in Four Stages of Lotus Rhizome
Recently, a lot of regulatory mechanisms have been identified by high throughout tag-sequencing technique [38], [39], [41], [42]. Although a large number of genes have not been annotated, tag-mapped genes have been believed to fully cover the whole plant genome [39]. It is an evidence that enlargement of lotus rhizome (Nelumbo nucifera) is strictly regulated by environmental factors, especially photoperiodic response can affect rhizome morphogenesis [14]. Therefore, in this experiment, lotus root was cultivated in an open field with normal environmental factors. In addition, rhizomes at four obvious differential developmental stages including stolon stage, initial stage, middle swelling stage, and later swelling stage were used due to few reports on the rhizome transition. Genes with differential expression during rhizome formation were identified using this technique. Approximately 5.0 million tags were identified per library, 4542104, 4474755, 4777919, and 4750348 clean tags were obtained. Only 20471, 18785, 23448, and 21778 genes from L1, L2, L3, and L4 libraries respectively, could be annotated, due to unavailability of complete genome of lotus root (Table 2). We found that many genes significantly changed their expression during rhizome development. The higher number of DEGs in L2/L3 libraries (Table 3) suggests that regulation from the initial swelling stage to middle swelling stage is more complex as compared with that of other developmental stages.
Identification of DEGs in Four Libraries
DEGs in four libraries were involved in the transport signal transduction, small molecular biosynthesis, transcription, cell cycle, response to the stimuli, organelle organization, anatomical structure development, cell differentiation, translation, organ development, cellular macromolecular metabolic processes, energy, and cellular component organization (Fig. S3). In L1/L2 libraries, 20 most DEGs are involved in hormone response, fiber synthesis, and response to stresses. In L2/L3 libraries, however, the expression profiles of major DEGs were relevant to the energy metabolism, synthesis and transport of storage, hormone metabolism and response, stress response and energy metabolism. In L3/L4 libraries, most DEGs were involved in storage synthesis, hormone synthesis and response. All genes which showed enhanced expression in these four libraries were listed in Table S1 (Table S1).
Adaptation to Submergence Stress
Adaptation to submergence is very important for the survival of lotus root. In this study, we found that several genes which included (ADH (L15773), MYB (L24021), NADH dehydrogenase (L6212), SOD (L3610), Ethylene-responsive transcription factor (L16154), Calcium-dependent protein kinase (L3989), ATPase subunit (L6411), heat shock protein (L4407) related to submergence enhanced their expression during rhizome formation (Table 1 and Table S1). Some species such as soybean, barley, tomato, and corn are also known to be sensitive to flooding [43], [44], [45], [46], and their productivity is seriously affected due to exposure to anaerobic stress. However, most aquatic plants (including lotus root) exposed to submergence show high resistance [45], [47]. According to the results of present study, the regulation of gene expression to submergence adaptation is essential for formation of storage organ, such as ERF, ADH, MYB, SNF like protein gene, SOS2 like protein kinase gene, SNF-related kinase gene, superoxide dismutase gene, G-box binding protein gene, and heat shock protein gene [45]. ADH (alcohol dehydrogenase) plays an important role in low-oxygen tolerance. Expression and activity of ADH are thought to be an indicator of oxygen limitation [48], [49], [50], [51], [52], [53]. Therefore, ADH enhanced their expression during rhizome development which is essential for lotus root to adapt to submergence condition.
Ethylene not only mediates a range of different biotic and abiotic stress responses 54. but is also involved in storage organ development and submergence adaptation [55]. It is considered to be a critical hormone in ethylene-mediated growth promotion which also helps to confront anoxia [56]. The signal transduction to oxygen shortage during stem development is dependent on ethylene, and elongation of stem in response to ethylene is a widely spread adaptation to anaerobic stress [56]. Ethylene responsive element binding factor (ERF) belongs to AP2 which regulate plant metabolism process. Many ERF genes were found to be responsive to abiotic stresses such as cold, wounding or drought stress, and also induced by some kinds of hormones such as ethylene, JA, and SA [57]. In four developmental stages of lotus root, we also observed enhanced mRNA level of several ERFs during development of storage organ, which indicates that regulations of ethylene also affects the growth and development of lotus root. In Arabidopsis, ERFs have been studied and their functions were also identified in recent years [58]. Three ethylene response-factor (ERF) genes were identified to be located on an sus1 locus, which was considered as a submergence tolerance QTL in rice [59], [60], [61]. In this study, we found increase in the transcriptional level of many members of ERF family during growth of lotus root, suggesting that ERFs also aid to lotus root survival in anaerobic stress.
It was reported that energy regulation was a measure to alleviate the hurt acquired because of submergence, and at the same time, many critical processes for storage organ formation also need energy consumption [62], [63], [64], [65]. ATP is normally generated from glycolysis in plant cells to sustain plant growth. For generation of ATP, ATP/ADP translocator (L23655) is thought to be a gated pore through which ADP and ATP are exchanged between the mitochondrial matrix and the cytoplasm. ATP/ADP translocator is an inner membrane protein which is observed to enhance the number and morphology of storage organ in transgenic plants. For lotus root, an ATP/ADP translocator was observed to improve the transcriptional level during rhizome development, suggesting that enhanced ATP level is helpful for survival and plant growth [66]. Evidence shows that delivery of NADPH/NADH reduced ferredoxin, and ATP in the specific cellular compartments occurs for energy-consuming reactions under stress condition. Generally, NADPH/NADH, and reduced ferredoxin are not directly transported across bio-membranes, therefore, indirect transport of reducing equivalents is achieved by malate/oxaloacetate shuttles, involving malate dehydrogenase (MDH) and NADP-MDH (Isoenzyme of MDH) for the interconversion of oxaloacetic acid and malate [67], [68]. MDH is one of the more active enzymes in peroxisomes, mitochondria, chloroplast, glyoxysomes, and cytoplasm. The activities of the enzymes of malate and NADP-MDH valves are changed when plants are subjected to conditions such as high light, high CO2, nutrition or stress. A lot of evidences show that NADP-MDH is involved in response to environmental factors. When plants are subjected to stress conditions, which require changed activities of the enzymes of malate valves, changed expression levels of MDH isoforms can be observed [69], [70], [71], [72] . The expression of ATP/ADP translocator (L23655), MDH (L8940) and NADP-MDH (L26121) was enhanced in L1/L2, L2/L3, and L3/L4 libraries. Therefore, these genes might be an adaptive response for lotus root in submergence conditions during storage development.
Carbohydrate/storage Metabolism
Starch is considered as an important component in the storage organ (rhizome, corm, tuber, and bulb). Simultaneous swelling of storage organ and accumulation of starch have already been testified [73], suggesting that synthesis of starch shows high coordination with formation of storage organ [74]. In our study, a gene encoding granule-bound starch synthase was found to enhance mRNA level during the rhizome development (Table 4). Granule-bound starch synthase is believed to have a higher amounts of starch synthesized in plant storage organs [75]. It is similar to the report of Hanashiro et al. (2008) [76]. in this study, GBSS enhanced the transcriptional level in L1/L2 and L2/L3 libraries, which indicates that the expression profile of GBSS has high correlation with the development of rhizome.
Patatin, which is identical to glycoprotein, is usually believed as a major storage protein in rhizome, tuber, corm, and bulb. We found that expression of patatin (L10302) decreased in L1/L2 libraries and L2/L3 libraries, but enhanced in L3/L4 libraries, suggesting that patatin accumulation increases in the later swelling stage. Compared with other storage proteins, patatin is more stable because no degradation products are detected during tuber development. Evidence shows that patatin might be involved in metabolism because acyl hydrolase has been found to be encoded by this gene [77], [78]. Another study shows that patatin is involved in pollen development [79], [80], which is further testified by Vancanneyt et al. 81. Further study shows that patatin is associated with early events of formation of underground storage organ according to its expression profile [82], [83]. In stolon tips of non-induced plants, only a small amount of patatin is found, which rapidly accumulates during storage organ formation [84]. Accumulation of patatin has high correlation with the swelling of storage organ because it is observed to be synthesized only in stolon and storage organ and induced by sucrose [85], [86], [87]. For lotus root, patatin enhances the expression only in L3/L4 libraries, which obviously showed that patatin was accumulated in later swelling stages of rhizome. High expression pattern might have some relationship with rhizome development, although the data of present result did not show any correlation between patatin and formation of storage organ.
Sugar can provide enough materials and energy for plant to complete some important activities during its life cycle [88]. The processes of sugar synthesis, transport, consumption, and storage have been widely studied in the past decades [89]. Evidence shows that soluble carbohydrates, most notably sucrose are considered to be the strong inducers of underground storage organ formation, because increasing concentration of sucrose in medium during cultivation leads to higher numbers of tubers [90]. Sucrose plays its role as an inducing signal molecule, and enhancing the level of sucrose in stolons results in an increased number of initiated storage organs [91]. A gene (L2349) encoding sucrose improved its transcriptional level in L1/L2 libraries (Table 4), suggesting that accumulation of sucrose is helpful for rhizome formation of lotus root. Further evidence also shows that SNF1 kinase (the sucrose non-fermenting-1) is involved in sugar-signaling pathways to regulate metabolism of carbohydrate or other storage proteins [92]. mRNA level of SNF1of lotus root was enhanced in L1/L2 (log2 ratio (L2/L1: 1.2) and L2/L3 (log2 ratio (L2/L1: 6.39) and L3/L4 (log2 Ratio (L2/L1: 2.08) libraries. From the characteristics of gene expression, SNF1 (L10886) undoubtedly promoted the formation and development of rhizome (Table S1).
Up-regulation of Transcription Factors during Rhizome Formation
Transcription factors regulate gene expression during cell metabolism processes. Transcriptional levels of 14, 20, and 20 regulators were enhanced in L1/L2, L2/L3, and L3/C4 libraries, respectively (Table 5). For these transcription factors, we found that CaM-binding protein (L1140) and AP2 domain class transcription factors (L1140) have been identified to perform critical roles in the formation of underground storage organ. Ca2+ has been testified to perceive endogenous and exogenous signals as a second messenger before system responses [93]. Calmodulin is a sensor of Ca2+, and many cellular processes are modulated after Ca2+ binds to CaM [94]. Ca2+/CaM have already been identified to be involved in the formation of storage organs [95]. Further study shows that transgenic potato plants with constitutive expression of a CaM gene (PCM1) produces more elongated tuber [96]. CaM-binding proteins have been found in many plants and their functions have also been considered for the development of storage organs [93], [97].
A potato CaM-binding protein (PCBP) is found to play an important role in signal transduction during tuber formation according to characteristics of its expression [98]. We observed that calmodulin-like protein (L16799) and calmodulin binding protein (L22245) have similar expression profiles in L1/L2, L2/L3, and L3/L4 libraries. From stolon to initial swelling stage of rhizome, expression levels of these two genes improved, and decreased from initial swelling stage to middle swelling stage and middle swelling stage to swelling stage, indicating that Ca2+/CaM might be involved in processes from stolon stage to initial swelling stage of rhizome.
In this study, the expression level of ethylene responsive factor was found to be enhanced (Table 5). From the characteristics of expression, we could conclude that this gene play an important role in rhizome formation. Ethylene is not only involved in a range of biotic and abiotic stress responses, especially to help wetland plants to adapt to anaerobic condition, but also mediates swelling of underground storage organs [99]. At the same time, AP2 domain class transcription factor, which is unique in plant kingdom also shows multi-functions in metabolism from response to stresses to regulation of plant development [100]. Overexpression of an AP2 domain class transcription factor, confers potato plant more resistance to abiotic stress, whereas down-regulation leads to a series of other effects such as decreased cell size, plant height, hypocotyl elongation and fertility [101]. Several AP2 domain class transcription factors are found to control flowering time [102]. It is reported that a putative AP2 domain class transcription factor (WRI1), regulates the storage metabolism in Arabidopsis seed. Constitutive expression of this gene leads to improvement of the seed oil content and triacylglycerols [103]. Transgenic rice plant with an AP2 domain class transcription factor increases the expression of waxy gene, which directly results in a change of storage content [104]. In addition, AP2 domain class transcription factors have been found to be induced by abscisic acid (ABA) and sugar, suggesting that this gene might be involved in ABA and sugar signal transduction pathway [105]. Therefore, future studies about role of AP2 domain class transcription factor in rhizome formation should be carried out with molecular engineering techniques.
Hormonal Regulation
Swelling of underground storage organ is affected by various environmental and endogenous factors. Short photoperiods, low temperatures, low nitrogen, and hormone favor the formation of storage organ [90]. In literature, many reports describe the importance of gibberellic acid (GA), JA, ethylene for the formation of storage organ. GA content is enhanced in transgenic plants by overexpression of GA oxidase gene, and elevation of GA content leads to transgenic potato plants that require a longer duration of short-day photoperiods to form storage organ. Whereas, inhibition of this enzyme activity results in earlier formation of these organs as compared with that of non transgenic plants [27]. Decrease in GA content in a dwarf mutant of S. tuberosum ssp. Andigena showed lead to formation of storage organ both in LD and SD condition. However, the storage organ is also inhibited in SD when GA biosynthesis is inhibited [9]. These results suggest that GA is probably involved in the photoperiodic induction to regulate the formation of storage organ. In addition, Xu et al. 106. observed that high content of GA promotes stolon elongation and inhibit storage organ formation. In this study, the expression of GA 20-oxidase gene (L18298) involved in GA biosynthesis pathway is high in L1 libraries (stolon stage), whereas it decreased in L2 and L3, but increased in L4 stages (data not shown). Enhancing GA level in stolon probably benefited the elongation of stolon and decreased GA level in L2 and L3 might have benefited the rhizome development. As we all known, in later development stages of lotus root, few new rhizomes will be formed again, and this is the reason why we observed increase in the expression of gibberellin 20 oxidase in the later swelling stage.
JA is widely distributed in plants and exerts a critical function in plant development and growth [107]. Furthermore, JA also plays a significant role in stress response [108]. Several studies have been performed to study the effects of JA on the swell of underground storage organ. Endogenous JA is believed to be a strong inducer of storage organ in dicot potato plants and monocot yam plants [109]. Ravnikar et al (1993) reports that exogenous JA plays an important role in the formation of garlic bulbs [110]. Zeal 111. found that application of exogenous JA not only induces the formation of storage organ, but simultaneously enhances the storage organ number. In addition, tuber formation of Pterostylis sanguinea is also promoted by exogenous JA [112]. In the four stages of rhizome development, we found that many JA-induced genes (MYB, transcription factor bHLH, bZIP, and AP2/ERF domain-containing transcription factor, lipoxygenase) enhanced transcriptional level during rhizome development [113], [114], [115], [116], [117]. These JA-induced genes, which promote storage organ formation, have already been testified in the past decades. Therefore, JA and JA responsive genes are helpful for rhizome formation. In addition, some storage organ-related genes were also documented in this study (Table 6), and the relationship between gene expression and storage organ formation has been testified in literature [118], [119], [120], [121], [122], [123]. Overall, expression profiles of these genes show that formation of rhizome is very complex and regulated by multiple genes.
Conclusions
Using high-throughput tag-sequencing (based on Solexa Genome Analyzer Platform), four expression libraries were constructed and analyzed from different developmental stages for rhizome formation. Based on the results of tag-sequencing, after comparison with the existing databases, overall 20471, 18785, 23448, and 21778 tags were annotated from four developmental stages (stolon stage, initial swelling stage, middle swelling stage, and swelling stage) respectively. In addition, several DEGs relevant to rhizome formation were also identified from these four libraries. Expression of nine genes was studied by qRT-PCR to verify the results of tag-sequencing. The qRT-PCR results revealed that gene expression showed high correlation with the tag sequencing analysis.
Supporting Information
Developmental stages of lotus rhizome including stolon stage, initial stage, middle swelling stage and later swelling stage.
(TIF)
Sequencing saturation analysis of three libraries. L1: tag-sequencing for stolon stage; L2: tag-sequencing for initial swelling stage; L3: tag-sequencing for middle swelling stage; L4: tag-sequencing for later swelling stage.
(TIF)
GO analysis of genes expressed during the rhizome formation. All the genes identified in L1/L2, L2/L3, and L3/L4 libraries were classified into 26 classifications according to gene functions.
(TIF)
Expression profiles of 18 genes at stolon stage, initial swelling stage and middle swelling stage in rhizome lotus, flower lotus, and seed lotus.
(TIF)
Genes were found to enhance their expression in L1/L2, L2/L3, and L3/L4 libraries. These genes were listed in descending order according to alteration of expression during rhizome formation.
(XLS)
Acknowledgments
Authors thank Javeed Hussain and Liben Xiong for his language check. We also extend our thanks to some members of BIG for their cooperation in obtaining high throughout sequence assembly of the lotus root.
Funding Statement
This work was supported by the Special Fund for Agro-scientific Research in the Public Interest (200903017-02), China Postdoctoral Science Foundation (2012M511805) and Jiangsu Postdoctoral Science Foundation (1102144C). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Xue JH, Dong WP, Chen T, Zhou SL (2012) Nelumbonaceae: Systematic position and species diversification revealed by the complete chloroplast genome. of Systematics and Evolution50: 477–487. [Google Scholar]
- 2.Sakamoto Y (1977) Lotus. Tokyo: Hosei University Press [in Japanese]. [Google Scholar]
- 3. Liu J, Zhang M, Wang S (2010) Processing characteristics and flavour of full lotus root powder beverage. J Sci Food Agric 90: 2482–2489. [DOI] [PubMed] [Google Scholar]
- 4.Slocum PD, Robinson P (1996). Water gardening, water lilies and lotuses. Portland, OR: Timber Press. [Google Scholar]
- 5. Borgi W, Ghedira K, Chouchane N (2007) Antiinflammatory and analgesic activities of zizyphus lotus root barks. Fitoterapia. 78: 16–19. [DOI] [PubMed] [Google Scholar]
- 6. Renato BRAZ, Hechenleitner AAW, Cavalcanti OA (2007) Extraction, structural modification and characterization of lotus roots polysaccharides (Nelumbo nucifera Gaertn). Excipient with potential application in modified drug delivery systems. Lat. Am. J. Pharm 26: 706–710. [Google Scholar]
- 7.Terashima M, Awano K, Honda Y, Yoshino N, Mori T, et al.. (2011) “Arteries within the artery” after kawasaki diease-A lotus root appearance by intravascular ultrasound. Circulation DOI: 10.1161/01.CIR.0000030708.86783.92 [DOI] [PubMed]
- 8. Vartapetian BB, Jackson M (1997) Plant adaptations to anaerobic stress. Ann of Bot 79: 3–20. [Google Scholar]
- 9. Vreugdenhil D, Struik PC (1989) An integrated view of the hormonal regulation of tuber formation in potato (Solanum tuberosum). Physiol Plant 75: 525–531. [Google Scholar]
- 10. Hannapel DJ (1991) Characterization of the early events of potato tuber development. Physiol. Plant 83: 568–573. [Google Scholar]
- 11. MacLeod RAF, Dirks WG, Matsuo Y, Kaufmann M, Milch H, et al. (1999) Widespread intraspecies cross-contamination of human tumor cell lines arising at source. Int J Cancer. 83: 555–563. [DOI] [PubMed] [Google Scholar]
- 12. Paiva E, Lister RM, Park WD (1983) Induction and accumulation of major tuber proteins of potato in stems and petioles. Plant Physiol 71: 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park WM (1983) Tuber proteins of potato- a new and surprising molecular system. Plant Mol Biol Rep 1: 61–66. [Google Scholar]
- 14. Masuda JI, Ozaki YK, ·Okubo HS (2007) Rhizome transition to storage organ is under phytochrome control in lotus (Nelumbo nucifera). . Planta 226: 909–915. [DOI] [PubMed] [Google Scholar]
- 15. Jackson SD (1999) Multiple signaling pathways control tuber induction in potato. Plant Physiol 119: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Inui H, Ogura Y, Kiyosue T (2010) Overexpression of Arabidopsis thaliana LOV KELCH REPEAT PROTEIN 2 promotes tuberization in potato (Solanum tuberosum cv. May Queen). FEBS Lett. 584: 2393–2396. [DOI] [PubMed] [Google Scholar]
- 17. Martinez-Garcia JF, Virgos-Soler A, Prat S (2002) Control of photoperiod-regulated tuberization in potato by the Arabidopsis flowering-time gene constans . PNAS 99: 15211–15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, et al. (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nat 449: 356–360. [DOI] [PubMed] [Google Scholar]
- 19. Chen H, Banerjee AK, Hannnapel DJ (2004) The tandem complex of BEL and Knox partners is required for transcriptional repression of ga20ox1. Plant J. 38: 276–284. [DOI] [PubMed] [Google Scholar]
- 20. Abelenda JA, Navarro C, Prat S (2011) From the model to the crop: genes controlling tuber formation in potato. Cur Opin in Biotech 22: 287–292. [DOI] [PubMed] [Google Scholar]
- 21. Jackson S, Heyer A, Dietze J, Prat S (1996) Phytochrome B mediates the photoperiodic control of tuber formation in potato. Plant J 9: 159–166. [Google Scholar]
- 22. Alisdair RF, Willmitzer L (2001) Molecular and biochemical triggers of potato. Plant Physiol 127: 1459–1465. [PMC free article] [PubMed] [Google Scholar]
- 23. Appeldoorn NJG, de Bruijn SM, Koot-Gronsveld EAM, Visser RGF, Vreugdenhil D, et al. (1997) Developmental changes of enzymes involved in conversion of sucrose to hexose-phosphate during early tuberization of potato. Planta 202: 220–226. [Google Scholar]
- 24. Jasik J, de Klerk GJ (2006) Effect of methyl jasmonate on morphology and dormancy development in lily bulblets regenerated in vitro . J Plant Growth Regu 25: 45–51. [Google Scholar]
- 25. Kim KJ, Kim KS (2005) Changes of endogenous growth substances during bulb maturation after flowering in Lilium oriental hybrid ‘Casa Blanca’ Acta Hort. 570: 661–667. [Google Scholar]
- 26. Usha PR, Rajasekaran RL, Claude DC, Samuel KA, Kevin JS (2011) Role of ethylene and jasmonic acid on rhizome induction and growth in rhubarb (Rheum rhabarbarum L.). Plant Cell Tiss Organ Cult 105: 253–263. [Google Scholar]
- 27. Carrera E, Bou J, Garcia-Martinez JL, Prat S (2000) Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant J 22: 247–256. [DOI] [PubMed] [Google Scholar]
- 28. Galis I, Macas J, Vlasak J, Ondrej M, van Onckelen HA (1995) The effect of an elevated cytokinin level using the ipt gene and N-6 benzyladenine on a single node and intact potato plant tuberization in vitro . J Plant Growth Regul 14: 143–150. [Google Scholar]
- 29. Bhat SR, Chandel KPS, Kacker A (1994) In vitro induction of rhizomes in zinger (Zingiber officinale Rosc.). Indian J Expt Biol 32: 340–344. [Google Scholar]
- 30. Vreugdenhil D, Bindels P, Reinhoud P, Klocek J, Hendriks T (1994) Use of the growth retardant tetcyclacis for potato tuber formation in vitro. . J Plant Growth Regul 14: 257–265. [Google Scholar]
- 31. himann KV, Skoog F (1934) On the inhibition of bud development and other functions of growth substance in Vicia faba. Proc. R. Soc. Lond. B Biol. Sci 114: 317–339. [Google Scholar]
- 32.Abeles FB, Morgan PW, Saltveit Jr ME (1992) Ethylene in Plant Biology. San Diego: Academic Press. [Google Scholar]
- 33. Neuteboom CE, Lada RL, Caldwell CD, Eaton L, Havard P (2002) Ethephon and spermidine enhance bulking in dicer carrots (Daucus carota var. sativus). PGRSAQ 30: 77. [Google Scholar]
- 34. Hu FY, Wang D, Zhao XQ, Zhang T, Sun HX, et al. (2011) Identification of rhizome-specific genes by genome-wide differential expression Analysis in Oryza longistaminata. BMC Plant Biology 11: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mingo-Castel AM, Smith OE, Kumamoto J (1976) Studies on the carbon dioxide promotion and ethylene inhibition of tuberizaton in potato explants cultured in vitro . Plant Physiol 57: 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bentley DR (2006) Whole-genome re-sequencing. Curr Opin Genet Dev 16: 545–552. [DOI] [PubMed] [Google Scholar]
- 37. Ansorge WJ (2009) Next-generation DNA sequencing techniques. New Biotechnol 25: 195–203. [DOI] [PubMed] [Google Scholar]
- 38.Qi XH, Xu XW, Lin XJ, Zhang WJ, Chen XH (2011) Identification of differentially expressed genes in cucumber (Cucumis sativus L.) root under waterlogging stress by digital gene expression profile. Genomics. doi:10.1016/j.ygeno 2011.12.008 [DOI] [PubMed]
- 39. Wang QQ, Liu F, Chen XS, Ma XJ, Zeng HQ, et al. (2010) Transcriptome profiling of early developing cotton fiber by deep-sequencing reveals significantly differential expression of genes in a fuzzless/lintless mutant. Genomics 96: 369–376. [DOI] [PubMed] [Google Scholar]
- 40.Mach J (2011) Unpureeing the tomato layers of information revealed by microdissection and high-throughput transcriptome sequencing. Plant Cell. doi: 10. 1105/tpc 111.231111 [DOI] [PMC free article] [PubMed]
- 41.Wang FD, Li LB, Li HY, Liu LF, Zhang YH, et al.. (2012) Transcriptome analysis of rosette and folding leaves in Chinese cabbage using high-throughput RNA sequencing. Genomics. doi: 10.1016/j.ygeno.02.005 [DOI] [PubMed]
- 42. Wu T, Qin ZW, Zhou XY, Feng Z, Du YL (2010) Transcriptome profile analysis of floral sex determination in cucumber. J Plant Physiol 167: 905–913. [DOI] [PubMed] [Google Scholar]
- 43. Perata P, Alpi A (1993) Plant responses to anaerobiosis. Plant Sci. 93: 1–7. [Google Scholar]
- 44. Subbaiah CC, Zhang J, Sachs MM (1994) Involvement of intracellular calcium in anaerobic gene expression and survival of maize seeding. Plant Physiolo 105: 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vartapetian BB, Jackson MB (1997) Plant adaptations to anaerobic stress. Ann. Bot 79: 3–30. [Google Scholar]
- 46. Ishizawa A, Murakami S, Kawakami Y, Kuramochi H (1999) Growth and energy status of arrowhead tubers, pondweed turions and rice seeding under anoxic conditions. Plant Cell Environ. 22: 505–514. [Google Scholar]
- 47. Crawford RMM, Braendle R (1996) Oxygen deprivation stress in a changing environment. J Exp Bot 47: 145–159. [Google Scholar]
- 48. Jung SH, Lee JY, Lee DH (2003) Use of SAGE technology to reveal changes in gene expression in Arabidopsis leaves undergoing cold stress. Plant Mol Biol 52: 553–567. [DOI] [PubMed] [Google Scholar]
- 49. Shiao TL, Ellis MH, Dolferus R, Dennis ES, Doran PM (2002) Overexpression of alcohol dehydrogenase or pyruvate decarboxylase improves growth of hairy roots at reduced oxygen concentrations. Biotechnol Bioeng 77: 455–461. [DOI] [PubMed] [Google Scholar]
- 50. Gibbs J, Greenway H (2003) Mechanisms of anoxia tolerance in plants. I. Growth, survival and anaerobic catabolism. Funct. Plant Biol 30: 1–47. [DOI] [PubMed] [Google Scholar]
- 51. Baxter-Burrell A, Chang R, Springer P, Bailey-Serres J (2003) Gene and enhancer traptransposable elements reveal oxygen deprivation-regulated genes and their complex patterns of expression in Arabidopsis. Ann. Bot 91: 129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xie ZD, Li DM, Wang LJ, Sack FD, Grotewold E (2010) Role of the stomatal development regulators FLP/MYB88 in abiotic stress responses. Plant J 64: 731–739. [DOI] [PubMed] [Google Scholar]
- 53. Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES (1998) Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 149: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grichko VP, Glick BR (2001) Ethylene and flooding stress in plants. Plant Physiol. Biochem 39: 1–9. [Google Scholar]
- 55. Suge H, Kusanagi T (1975) Ethylene and carbon dioxide: regulation of growth in two perennial aquatic plants, arrowhead and pondweed. Plant and Cell Physiol 16: 65–72. [Google Scholar]
- 56. Jackson MB (2008) Ethylene-promoted Elongation: an Adaptation to Submergence Stress. Ann. Bot 101: 229–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Singh KB, Foley RC, Sánchez LO (2002) Transcription factors in plant defense and stress responses. Cur Opin in Plant Biol 5: 430–436. [DOI] [PubMed] [Google Scholar]
- 58. Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vij S, Tyagi AK (2007) Emerging trends in the functional genomics of the abiotic stress response in crop plants. Plant Biotech J (5) 361–380. [DOI] [PubMed] [Google Scholar]
- 60. Fukao T, Xu K, Ronald PC, Bailey-Serres J (2006) A Variable Cluster of Ethylene Response Factor–Like Genes Regulates Metabolic and Developmental Acclimation Responses to Submergence in Rice. 18: 2021–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhou SF, lü SH 2, Fu FL, Lan H, Zhang ZM, et al. (2011) Over-expression of Sub1A, a submergence tolerance gene from rice, confers enhanced hypoxic stress tolerance in transgenic tobacco plants African J Biotech. 10: 17934–17939. [Google Scholar]
- 62. Bailey-Serres J, Voesenek L ACJ (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339. [DOI] [PubMed] [Google Scholar]
- 63. Pederson O, Rich SM, Colmer TD (2009) Surviving floods: leaf gas films improve O2 and CO2 exchange, root aeration, and growth of completely submerged rice. Plant J 58: 147–156. [DOI] [PubMed] [Google Scholar]
- 64. Rocha M, Licausi F, Araujo WL, Nunes-Nesi A, Sodek L, et al. (2010) Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of lotus japonicus. Plant Physiol 152: 1501–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Geigenberger P, Stitt M, Fernie AR (2004) Metabolic control analysis and regulation of the conversion of sucrose to starch in growing potato tubers. Plant Cell Environ 27: 655–673. [Google Scholar]
- 66. Tjaden J, Mohlmann T, Kampfenkel K, Henrichs G, Neuhaus HE (1998) Altered plastidic ATP/ADP-transporter activity influences potato tuber morphology, yield and composition of starch. Plant J 16: 531–540. [Google Scholar]
- 67. Gietl C (1992) Malate dehydrogenase isoenzymes: cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochem Biophys Acta 1100: 217–234. [DOI] [PubMed] [Google Scholar]
- 68. Scheibe R (2004) Malate valves to balance cellular energy supply. Physiol Plant 120: 21–26. [DOI] [PubMed] [Google Scholar]
- 69. Hameister S, Becker B, Holtgreffe S, Strodtkotter I, Linke V, et al. (2007) Transcriptional regulation of the NADP dependent malate dehydrogenase: comparative genetics and identification of DNA-binding proteins. J Mol Evol 65: 437–455. [DOI] [PubMed] [Google Scholar]
- 70. Becker B, Holtgrefe S, Jung S, Wunrau C, Kandlbinder A, et al. (2006) Influence of the photoperiod on redox regulation and stress responses in Arabidopsis thaliana L. (Heynh.) plants under long- and short-day conditions. Planta 224: 380–393. [DOI] [PubMed] [Google Scholar]
- 71. Cushman JC (1993) Molecular cloning and expression of chloroplast NADP-malate dehydrogenase during Crassulacean acid metabolism induction by salt stress. Photosynth Res 35: 15–27. [DOI] [PubMed] [Google Scholar]
- 72. Sahi C, Singh A, Kumar K, Blumwald E, Grover A (2006) Salt stress response in rice: genetics, molecular bilogy, and comparative genomics. Func Inter Genomics 6: 263–284. [DOI] [PubMed] [Google Scholar]
- 73. Burton WG, Van Es A, Hartmans KJ (1992) The physics and physiology of storage. The Potato Crop 2: 608–727. [Google Scholar]
- 74. Geigenberger P, Merlo L, Reimholz R, Stitt M (1994) When growing potato tubers are detached from their mother plant there is a rapid inhibition of starch synthesis, involving inhibition of ADP-glucose pyrophosphorylase. Planta 193: 486–493. [Google Scholar]
- 75. Nakamura T, Vrinten P, Hayakawa K, Ikeda J (1998) Characterization of a Granule-Bound Starch Synthase Isoform Found in the Pericarp of Wheat. Plant Physiol. 118: 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hanashiro I, Itoh K, Kuratomi Y, Yamazaki M, Igarashi T, et al. (2008) Granule-Bound Starch Synthase I is Responsible for Biosynthesis of Extra-Long Unit Chains of Amylopectin in Rice. Plant Cell Physiol 49: 925–933. [DOI] [PubMed] [Google Scholar]
- 77. Racusen D (1985) Patatin and esterase in desire potato tuber. Food Ind 9: 61–367. [Google Scholar]
- 78. Andrews DL. Beames B, Summers MD, Park WD (1988) Characterisation of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochem J 252: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sawhney VK, Nave EB (1986) Enzymatic changes in post-meiotic anther development in Petunia hybrida. 11. Histochemical localisation of esterase, peroxidase, malate- and alcohol dehydrogenase. J Plant Physiol 125: 467–473. [Google Scholar]
- 80. Delvallee I, Dumas C (1988) Anther development in Zea mays : Changes in protein, peroxidase, and esterase patterns. J Plant Physiol 132: 210–217. [Google Scholar]
- 81.Vancanneyt G, Sonnewald U, Hofgen R, Willmitzer (1989) Expression of a patatin-like protein in the anthers of potato and sweet pepper flowers. Plant Cell 533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Perl A, Aviv D, Willmitzer L, Galun E (1991) In vitro tuberization in transgenic potatoes harboring 13-glucuronidase linked to a patatin promoter: effects of sucrose levels and photoperiods. Plant Sci 73: 87–95. [Google Scholar]
- 83. Bamfalvi Z, Kostyal Z, Barta E (1994) Solanum brevidens possesses a non-sucrose-inducible patatin gene. Mol Gen Genet 245: 517–522. [DOI] [PubMed] [Google Scholar]
- 84. Hannapel DJ, Miller J C, Park WD (1985) Regulation of Potato Tuber Protein Accumulation by Gibberellic Acid. Plant Physiol 78: 700–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Stupar RM, Beaubien KA, Jin WW, Song JQ, Lee MK, et al. (2006) Structural Diversity and Differential Transcription of the Patatin Multicopy Gene Family During Potato Tuber Development. Genetics 172: 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Rocha-Sosa M, Sonnewald U, Frommer W, Stratmann M, Schell J (1989) Both developmental and metabolic signals activate the promoter of a class I patatin gene. EMBO J 8: 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Grierson C, Du JS, de Torres ZM, Beggs K, Smith C (1994) Separate cis sequences and trans factors direct metabolic and developmental regulation of a potato tuber storage protein gene. Plant J 5: 815–826. [DOI] [PubMed] [Google Scholar]
- 88. Gibson SI (2005) Control of plant development and gene expression by sugar signaling. Cur Opin in Plant Biol 8: 93–102. [DOI] [PubMed] [Google Scholar]
- 89. Sheen J, Zhou L, Jang JC (1999) Sugars as signaling molecules. Cur Opin in Plant Biol 2: 410–418. [DOI] [PubMed] [Google Scholar]
- 90. Fernie AR, Willmitzer L (2001) Molecular and biochemical triggers of potato tuber development. Plant Physiol 127: 1459–1465. [PMC free article] [PubMed] [Google Scholar]
- 91. Dijkwel PP, Huijser C, Weisbeek PJ, Chua NH, Smeekens SCM (1997) Sucrose control of phytochrome A signaling in Arabidopsis . Plant Cell 9: 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tiessen A, Prescha K, Branscheid A, Palacios N, Mckibbin R, et al. (2003) Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signaling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J 35: 490–500. [DOI] [PubMed] [Google Scholar]
- 93. Kim MC, Chung WS, Yun DJ, Cho M J (2009) Calcium and calmodulin-mediated regulation of gene expression in plants. Mol Plant 2: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hoeflich KP, Ikura M (2002) Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 108: 739–742. [DOI] [PubMed] [Google Scholar]
- 95. Balamani V, Veluthambi K, Poovaiah BW (1986) Effect of calcium on tuberization in potato (Solanum tuberosum L.). Plant Physiol 80: 856–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Poovaiah BW, Takezawa D, An G, Han TJ (1996) Regulated expression of a calmodulin isoform alters growth and development in potato. J. Plant Physiol 149: 553–558. [DOI] [PubMed] [Google Scholar]
- 97. Bouché N, Scharlat A, Snedden W, Bouchez D, Fromm H (2002) A novel family of calmodulin-binding transcription activators in multicellular organisms. J Biol Chem 277: 21851–21861. [DOI] [PubMed] [Google Scholar]
- 98. Reddy ASN, Safadi F, Narasimhulu SB, Golovkin M, Hu X (1996) Isolation of a novel calmodulin-binding protein from Arabidopsis tha/iana. . J Biol Chem 271: 7052–7060. [DOI] [PubMed] [Google Scholar]
- 99.Suge H, Kusanagi T (1975) Ethylene and carbon dioxide: regulation of growth in two perennial aquatic plants, arrowhead and pondweed. Plant Cell Physiol 16, 65–72. [Google Scholar]
- 100. Krizek BA (2009) Aintegumenta and aintegumentalke6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol 150: 1916–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Dietz KJ, Vogel MO, Viehhauser A (2010) AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signaling. Protoplasma 245: 3–14. [DOI] [PubMed] [Google Scholar]
- 102. Feng JX, Liu D, Pan Y, Gong W, Ma LG, et al. (2005) An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol Biol 59: 853–868. [DOI] [PubMed] [Google Scholar]
- 103. Cernac A, Benning C (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585. [DOI] [PubMed] [Google Scholar]
- 104. Zhu Y, Cai XL, Wang ZY, Hong MM (2003) An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wax gene. J Biol Chem 278: 47803–47811. [DOI] [PubMed] [Google Scholar]
- 105. Rook F, Corke F, Card R, Munz G, Smith C, et al. (2001) Impaired Suc-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J 26: 421–433. [DOI] [PubMed] [Google Scholar]
- 106. Xu X, van Lammeren AAM, Vermeer E, Vreugdenhil D (1998) The role of gibberellin, abscisic acid, and sucrose in the regulation of potato tuber formation in vitro. Plant Physiol 117: 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Sembdner G, Parthier B (1993) The biochemistry and the physiological and molecular actions of jasmonates. Annu. Rev. Plant Physiol 44: 569–589. [Google Scholar]
- 108. Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. 48: 355–381. [DOI] [PubMed] [Google Scholar]
- 109. Koda Y (1992) The role ofjasmonic acid and related compounds in the regulation of plant development. Int Rev Cytol 135: 155–199. [DOI] [PubMed] [Google Scholar]
- 110. Ravnikar M, Zel J, Plaper I, Spacapan A (1993) Jasmonic Acid Stimulates Shoot and Bulb Formation of Garlic In Vitro. J Plant Growth Regul 12: 73–77. [Google Scholar]
- 111. Zel J, Debeljak N, Ucman R, Ravnikar M (1997) The effect of jasmonic acid, sucrose and darkness on garlic (Allium sativum L.cv. ptujski jesenski) bulb formation in vitro. In Vitro Cell Dev. Biol – Plant 33: 231–235. [Google Scholar]
- 112. Debeljak N, Regvar M, Dixon KW, Sivasithamparam K (2002) Induction of tuberisation in vitro with jasmonic acid and sucrose in an Australian terrestrial orchid, Pterostylis sanguinea . Plant Growth Regul 36: 253–260. [Google Scholar]
- 113. Lee MW, Qi M, Yang Y (2001) A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death. Mol Plant Microbe Interact 14: 527–35. [DOI] [PubMed] [Google Scholar]
- 114. Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 3: 701–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Meng XB; Zhao WS, Lin RM, Wang M, Peng YL (2005) Identification of a novel rice bZIP-type transcription factor gene, OsbZIP1, involved in response to infection of Magnaporthe grisea . Plant Mol Biol Rep 23: 301–302. [Google Scholar]
- 116. Zhang HW, Lu XY, Huang DF, Huang RF (2004) The ethylene-, jasmonate-, abscisic acid- and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box-containing genes and salt tolerance in tobacco. Planta 220: 262–270. [DOI] [PubMed] [Google Scholar]
- 117. Mueller MJ (1997) Enzymes involved in jasmonic acid biosynthesis. Physiol Plant 100: 653–663. [Google Scholar]
- 118. Müller-Röber B, Sonnewald U, Willmitzer L (1992) Inhibition of the ADP-glucose pyrophosphorylase in transgenic potatoes leads to sugar-storing tubers and influences tuber formation and expression of tuber storage protein genes. EMBO 11: 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kuipers AGJ, Jacobsen E, Visser RGF (1994) Formation and Deposition of Amylose in the Potato Tuber Starch Granule Are Affected by the Reduction of Granule-Bound Starch Synthase Gene Expression. Plant Cell 6: 1 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Jackson SD, Heyer A, Dietze J, Prat S (1996) Phytochrome B mediates the photoperiodic control of tuber formation in potato. Plant J 9: 159–166. [Google Scholar]
- 121. Carrera E, Bou J, García-Martínez JL, Prat S (2000) Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants.Plant J. 22: 247–256. [DOI] [PubMed] [Google Scholar]
- 122. Zeevaart JAD (2008) Leaf-produced floral signals. Cur Opin Plant Biol 11: 541–547. [DOI] [PubMed] [Google Scholar]
- 123. Baudry A, Ito S, Song YH, Strait AA (2010) Takatoshi Kiba,Sheen Lu, Rossana Henriques, José L. Pruneda-Paz, Nam-Hai Chua, Elaine M. Tobin, F-Box Proteins FKF1 and LKP2 Act in Concert with ZEITLUPE to Control Arabidopsis Clock Progression. Plant Cell 22: 606–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Krieger U, Lippman ZB, Zamir D (2010) The flowering gene single flower truss drives heterosis for yield in tomato. Nat Genet 42: 459–463. [DOI] [PubMed] [Google Scholar]
- 125. Imaizumi T, Kay SA (2006) Photoperiodic control of flowering: not only by coincidence. Trands in Plant Sci 11: 550–558. [DOI] [PubMed] [Google Scholar]
- 126. Banerjee AK, Chatterjee M, Yu Y, Suh SG, Miller WA, Hannapek DJ (2006) Dynamics of a mobile a RNA of potato involved in a long distance signaling pathway. Plant Cell 18: 3443–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kolomiets1 MV, Hannapel DJ, Chen H, Tymeson M, Gladon RJ (2001) Lipoxygenase is involved in the control of potato tuber development. Plant Cell 13: 613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Müller-Röber B, Sonnewald U, Willmitzer L (1992) Inhibition of the ADP-glucose pyrophosphorylase in transgenic potatoes leads to sugar-storing tubers andinfluences tuber formation and expression of tuber storage protein genes. EMBO 11: 1229–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kuipers AGJ, Jacobsen E, Visser RGF (1994) Formation and Deposition of Amylose in the Potato Tuber Starch Granule Are Affected by the Reduction of Granule-Bound Starch Synthase Gene Expression. Plant Cell 6: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Jackson SD (1996) Phytochrome B mediates the photoperiodic control of tuber formation in potato. Plant J 2: 159–166. [Google Scholar]
- 131. Zeevaart JA (2008) Leaf-produced floral signals. Curr. Opin. Plant Biol. 11: 541–547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Developmental stages of lotus rhizome including stolon stage, initial stage, middle swelling stage and later swelling stage.
(TIF)
Sequencing saturation analysis of three libraries. L1: tag-sequencing for stolon stage; L2: tag-sequencing for initial swelling stage; L3: tag-sequencing for middle swelling stage; L4: tag-sequencing for later swelling stage.
(TIF)
GO analysis of genes expressed during the rhizome formation. All the genes identified in L1/L2, L2/L3, and L3/L4 libraries were classified into 26 classifications according to gene functions.
(TIF)
Expression profiles of 18 genes at stolon stage, initial swelling stage and middle swelling stage in rhizome lotus, flower lotus, and seed lotus.
(TIF)
Genes were found to enhance their expression in L1/L2, L2/L3, and L3/L4 libraries. These genes were listed in descending order according to alteration of expression during rhizome formation.
(XLS)