The advent of massively parallel sequencing (MPS) has revolutionized genetic testing for deafness by enabling personal genomics in diagnosis (for a comprehensive review, see Shearer et al1). This technology has drastically increased the throughput of genetic testing but concomitantly has exponentially increased the amount of genetic data generated. To address this deluge of data and to streamline analysis, we have developed a custom variant prioritization pipeline incorporating data from a patient’s genome and phenome (the patient’s phenotype). In aggregate, the patient’s phenome is his or her constellation of phenotypic traits, which for hearing loss includes the patien’s audioprofile (pattern of hearing loss on audiogram), temporal bone anatomy (imaging), and ocular pathology (fundoscopy). Here we present 3 cases to illustrate how knowledge of a patient’s phenome can assist variant prioritization by corroborating likely pathogenic variants and excluding variants of unknown significance (VUS).
Methods
Patients and Phenomic Data
All methods were approved by the institutional review board at the University of Iowa. Clinical data were collected from all patients, including audiometric data, family history, and temporal bone imaging. Included patients segregated autosomal dominant hearing loss, and environmental causes were ruled out. Audiometric profiling (using air conduction thresholds) was performed using AudioGene v4.0, freely available at http://audiogene.eng.uiowa.edu, which includes 3246 audiograms from 1445 patients that map to 34 loci.2,3
Genomic Data and Bioinformatics Analysis
DNA was extracted from peripheral blood, and OtoSCOPE testing (http://morl-otoscope.org) was performed as described previously.4 All exons of all genes implicated in nonsyndromic hearing loss (NSHL) and NSHL mimic genes (those causing Usher and Pendred syndromes) were targeted for enrichment and sequenced on the Illumina HiSeq (Illumina, Inc, San Diego, California). Sequencing reads were mapped with BWA and variants were called using the GATK Unified Genotyper. Variants were filtered against (1) a list of all known pathogenic changes to identify any previously implicated deafness-causing mutations (http://deafnessvariationdatabase.org), (2) publicly available population-scale genomic databases (ESP, 1000Genomes), and (3) an internal variant list that includes data from all prior OtoSCOPE runs. On average, <10 VUSs remain per patient. Clinically relevant variants were confirmed with Sanger sequencing.
Results
Patient 1
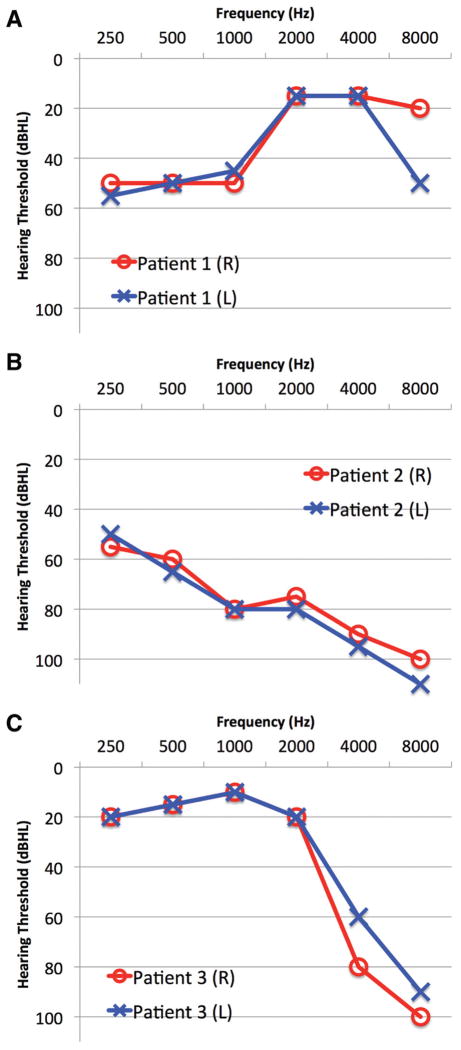
Patient 1 is a 66-year-old man with dominant NSHL and a progressive, “up-sloping” audioprofile (Table 1 and Figure 1A). OtoSCOPE variant analysis identified 1 reported pathogenic variant (WFS1 p.R685P) and 2 high-quality VUSs. AudioGene predicted DFNA6 (WFS1) as the most likely affected loci. Filtering the OtoSCOPE-generated variant list against AudioGene data corroborated this reported variant, WFS1 p.R685P, as causative and allowed the exclusion of the other VUSs.
Table 1.
Patients in This Study: Type of Hearing Loss, Inheritance, Genes Screened, Audiogene Prediction, and Final Result
| Patient | Inheritance | Type of Hearing Loss | OtoSCOPE Result: Genes with VUS | AudioGene Prediction | Final Result |
|---|---|---|---|---|---|
| Patient 1 | Dominant | Up-sloping | DSPP, LRTOMT, WFS1 | DFNA6 (WFS1), DFNA11 (MYO7A), DFNA8/12 (TECTA) | WFS1 p.R685P |
| Patient 2 | Dominant | Down-sloping | CDH23, GPR98, KCNQ4, WFS1 | DFNA2 (KCNQ4), DFNA10 (EYA4), DFNA9 (COCH) | KNCQ4 p.L281S |
| Patient 3 | Dominant | Sharply down-sloping | MYO1A, MYO7A, OTOA, PCDH15, TRIOBP (2) | DFNA2 (KCNQ4), DFNA9 (COCH), DFNA25 (SLC17A8) | MYO7A p.R1743W |
Abbreviation: VUS, variants of unknown significance.
Figure 1.
Audiograms from patients described in this study. (A) Patient 1 presented with a characteristic up-sloping audioprofile associated with a mutation in WFS1 (DFNA6), (B) patient 2 presented with a down-sloping audiogram associated with a mutation in KCNQ4 (DFNA2), and (C) patient 3 presented with a sharply down-sloping audioprofile with a causative mutation in MYO7A (DFNA11).
Patient 2
Patient 2 is a 57-year-old man with dominant NSHL displaying a down-sloping audioprofile (Table 1 and Figure 1B). Genetic testing with OtoSCOPE identified 3 VUSs and a reported pathogenic mutation in KCNQ4 (p.L281S). AudioGene predicted DFNA2 (KCNQ4) as the most likely cause of the patient’s audioprofile. Concurrence between OtoSCOPE and AudioGene data allowed exclusion of VUSs.
Patient 3
Patient 3 is a 55-year-old woman with dominant NSHL and a sharply down-sloping audiogram (Table 1 and Figure 1C). Genetic testing with OtoSCOPE identified 6 VUSs. AudioGene predicted DFNA2 (KCNQ4), DFNA9 (COCH), and DFNA25 (SLC17A8) as likely causes of hearing loss. No coding variants were identified in KCNQ4 or COCH; however, 1 VUS was identified in MYO7A. The MYO7A variant, p.R1743W, is novel and predicted to be pathogenic by several in silico pathogenicity prediction algorithms (BLOSUM62, SIFT, PolyPhen2, and PhyloP). DFNA11 was not predicted by audiogene to be causative. However, this may reflect a limitation to the support vector machine (SVM): it has been trained on a set of audioprofiles and is inherently limited by the number of individual audioprofiles per genotype and phenotype. Upon manual comparison with a previously published DFNA11 mutation, the audioprofiles closely matched.5 As we ascertain more audioprofiles, the SVM will improve in accuracy. Sanger sequencing confirmed the presence of the MYO7A p.R1743W variant in patient 3.
Discussion
The capacity of massively parallel sequencing technology is vast, as a single run on the Illumina HiSeq generates more sequencing data than the entire human genome project did per year using the previous generation of sequencing technology. As such, data interpretation has replaced sequencing as a bottleneck in genomic analysis. OtoSCOPE, for example, targets only 66 genes but yields an average of 252 genomic variants that must be categorized and prioritized. A step toward facilitating this interpretation is integrating the genome with the phenome.
AudioGene and OtoSCOPE provide a focused example of this integration. We have begun expanding our phenotypic categorization of patients with hearing loss and have used AudioGene to identify specific audioprofiles, which aid in genomic variant interpretation and can help achieve a genetic diagnosis. We demonstrate that the AudioGene tool can streamline cumbersome variant analysis by corroborating reported variants and allowing exclusion of others.
As costs decrease, comprehensive screening with multigene panels will replace individual gene testing. Our lab offers both individual gene testing and comprehensive testing with the OtoSCOPE platform, which includes 66 genes. Tiered genetic testing of individuals with apparent recessive deafness by first screening GJB2 prior to completing a multigene screen using a panel such as OtoSCOPE may be more cost-effective currently. However, as costs continue to decrease, comprehensive testing will supplant individual gene testing for deafness.
Conclusion and Future Directions
AudioGene facilitates variant prioritization in the context of deafness. For that reason, we are expanding AudioGene to include recessive forms of deafness and additional phenotypic data, including temporal bone imaging and funduscopy, which will aid in identifying pathogenic variants in Usher and Pendred syndromes.
Acknowledgments
Sponsorships: None.
Funding source: NIH, NIDCD Grant: RO1DC03544.
Footnotes
Reprints and permission: sagepub.com/journalsPermissions.nav
Author Contributions
Robert W. Eppsteiner, designed research, analyzed data, wrote manuscript, performed research; A. Eliot Scherer, designed research, analyzed data, wrote manuscript, performed research; Michael S. Hildebrand, designed research, analyzed data, performed research; Kyle R. Taylor, designed research, analyzed data, performed research; Adam P. DeLuca, designed research, analyzed data, performed research; Steve Scherer, performed research; Patrick Huygen, designed research, analyzed data; Todd E. Scheetz, designed research, analyzed data; Terry A. Braun, designed research, analyzed data; Thomas L. Casavant, designed research, analyzed data; Richard J. H. Smith, designed research, analyzed data, wrote manuscript.
Competing interests: None.
References
- 1.Shearer AE, Hildebrand MS, Sloan CM, Smith RJ. Deafness in the genomics era. Hear Res. 2011;282:1–9. doi: 10.1016/j.heares.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hildebrand MS, Tack D, McMordie SJ, et al. Audioprofile-directed screening identifies novel mutations in KCNQ4 causing hearing loss at the DFNA2 locus. Genet Med. 2008;10:797–804. doi: 10.1097/GIM.0b013e318187e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hildebrand MS, DeLuca AP, Taylor KR, et al. A contemporary review of AudioGene audioprofiling: a machine-based candidate gene prediction tool for autosomal dominant nonsyndromic hearing loss. Laryngoscope. 2009;119:2211–2215. doi: 10.1002/lary.20664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shearer AE, DeLuca AP, Hildebrand MS, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luijendijk MW, Van Wijk E, Bischoff AM, et al. Identification and molecular modelling of a mutation in the motor head domain of myosin VIIA in a family with autosomal dominant hearing impairment (DFNA11) Hum Genet. 2004;115:149–156. doi: 10.1007/s00439-004-1137-3. [DOI] [PubMed] [Google Scholar]