Abstract
Objective
To investigate the GCM2 gene in three siblings with congenital hypoparathyroidism and perform functional analysis.
Materials and methods
We sequenced the GCM2 gene by PCR and analyzed the functional consequence of the mutation by transient transfection studies. Haplotype analysis was performed.
Results
We identified a nucleotide change, c.408C>A, in exon 3 that is predicted to truncate the Gcm2 protein (p.Tyr136Ter). All three affected siblings were homozygous and both parents were heterozygous for the mutation. Transfection studies revealed the mutant mRNA but not expression of the Gcm2 protein. Haplotype analysis revealed that the two mutant GCM2 alleles shared genotypes on chromosome 6p24.2.
Conclusions
We describe the first GCM2 mutation in exon 3 in patients with severe congenital hypoparathyroidism. Informative genetic markers could not exclude identity by descent for the mutant alleles. Gcm2 protein was not detected after transfection, suggesting that complete lack of Gcm2 action accounts for severe hypoparathyroidism.
Keywords: GCMB, GCM2, isolated hypoparathyroidism, mutation
Introduction
Early neonatal hypocalcemia is often mild, transient and resolves with minimal medical intervention. Severe neonatal hypocalcemia due to permanent hypoparathyroidism is rare and can cause permanent mental and physical deterioration when accompanied by seizures, laryngospasm and delayed treatment (1). Permanent hypoparathyroidism can be due to several genetic causes: mutation or hemizygosity of TBX1 as part of the 22q11deletion in DiGeorge sequence (2); loss of function mutations in TCBE in Kenny-Caffey syndrome (3); loss of function mutation of GATA3 in hypoparathyroidism, deafness, and renal dysplasia (HDR) syndrome (4); activating mutations of CASR in autosomal dominant hypercalciuric, hypocalcemia (5); and loss of function mutations of parathyroid hormone (PTH) in isolated hypoparathyroidism (6).
Recently, our group and others have demonstrated that either recessive amorphic mutations (7–11) or dominant inhibitor mutations (12, 13), in the Glial Cell Missing 2 (GCM2 , formerly GCMB) gene located on chromosome 6p24.2 are the most common cause of isolated hypoparathyroidism (Figure 1). GCM2 encodes a 506-amino acid transcription factor that is expressed principally in the developing and mature parathyroid glands. Genetic ablation of Gcm2 in mice results in hypoparathyroidism because of aplasia of the parathyroid glands, which establishes Gcm2 as critically important for parathyroid gland homeostasis. The human GCM2 gene contains five coding exons, and all previous small mutations have been identified in exons 2 and 5 (8–13). These mutations interfere with DNA binding and nuclear localization or transactivation, respectively. We report the first mutation in exon 3 in a nonconsanguineous family with autosomal recessive isolated hypoparathyroidism (Table 1). Functional analysis showed that HEK293 cells transfected with the mutant cDNA did not contain Gcm2 protein, suggesting that a complete lack of Gcm2 led to severe hypoparathyroidism and absence of circulating PTH in the affected subjects.
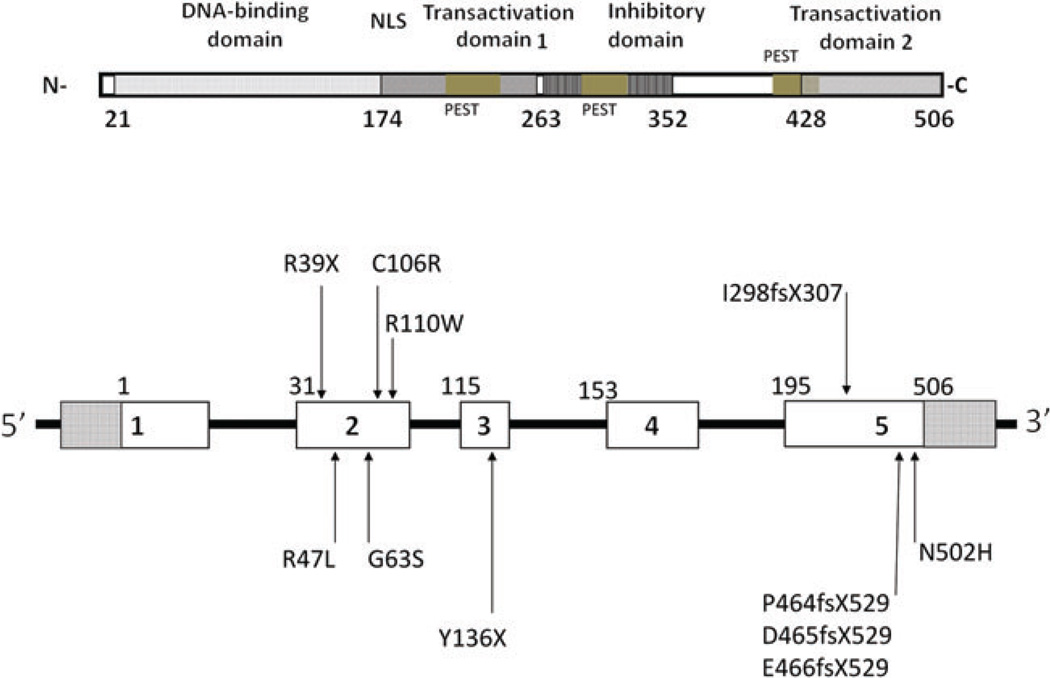
Figure 1.
Schematic illustration of the genomic structure of the GCM2 gene. The upper portion illustrates the location of the various domains of the gene and the approximate number of amino acids encoded in each domain, as well as the cumulative number of 506 amino acids encoded by the GCM2 gene. The protein contains a GCM DNA-binding domain, two transactivation domains, an inhibitory domain and a predicted nuclear localization signal (NLS) at residues 176–191. The lower portion illustrates the known mutations for isolated hypoparathyroidism and the location of each with respect to the five codons in the gene. Mutations in exon 2 cause recessive hypoparathyroidism while many of the mutations in exon 5 are associated with dominant hypoparathyroidism. Putative mutational hotspots would include the R47L missense and the I298FS307 frameshift mutation; both of which arose non-consanguineously [after Ref. (11)].
Table 1.
Currently recognized GCMB mutations that result in reduced expression or function of protein.
| Mutation | Inheritance | Location | Study |
|---|---|---|---|
| R39X | AR | Exon 2 | Bowl et al. (11) |
| R47L | AR | Exon 2 | Baumber et al. (9) |
| G63S | AR | Exon 2 | Thomee et al. (10) |
| C106R | AD | Exon 2 | Yi et al. (14) |
| R110W | AR | Exon 2 | Bowl et al. (11) |
| Y136X | AR | Exon 3 | Doyle et al. (current report) |
| 1298fsX307 | AR | Exon 5 | Bowl et al. (11) |
| P464fsX529 | AD | Exon 5 | Mannstadt et al. (13) |
| D465fsX529 | AD | Exon 5 | Mannstadt et al. (13) |
| E466fsX529 | AD | Exon 5 | Canaff et al. (12) |
| N502H | AD | Exon 5 | Mirczuk et al. (15) |
AR, autosomal recessive; AD, autosomal dominant.
Patients and methods
The Institutional Review Board of the Alfred I. duPont Children’s Hospital for Children approved the study protocol, and written informed consent was obtained from both parents on behalf of their younger children and assent obtained from the older brother (age 9 years).
The oldest affected sister was the product of an uncomplicated full-term pregnancy and spontaneous vaginal delivery to a 28-year-old mother who had recently immigrated to the United States from Guatemala. She was discharged home on a standard infant formula and presented at 7 days of age with a generalized tonic-clonic seizure and difficulty breathing. Her initial serum calcium was 5.6 mg/dL with a serum phosphorus of 10.3 mg/dL; serum intact PTH (iPTH) was undetectable. Fluorescence in situ hybridization (FISH) analysis for DiGeorge sequence showed two copies of the 22q11 locus. She was hospitalized and received intravenous (IV) calcium gluconate and was started on calcitriol by mouth twice daily. On the second day of hospitalization, she was transitioned to oral calcium supplements and was switched to a low-phosphorus infant formula (PM60:40, Mead Johnson). She maintained normal serum concentrations of calcium and phosphorus throughout childhood while treated with oral calcium and calcitriol, and developmental and cognitive milestones were all normal. Subsequent testing for a mutation in the CaSR gene was negative.
The second sister had her serum calcium evaluated at birth, and it was normal at 8.8 mg/dL. Subsequent testing showed asymptomatic hypocalcemia (6.3 mg/dL) on day-of-life 3 with undetectable iPTH. Molecular analysis of the 22q11 locus and CaSR gene was normal. A third sister also had asymptomatic hypocalcemia (7.3 mg/dL) when tested on day-of-life 2 with undetectable iPTH. Both sisters were treated with oral calcium and calcitriol and have done well.
The mother, father and an older brother of the affected children had normal serum levels of calcium and iPTH (Table 2). The parents had both immigrated to the United States from Guatemala and reported that while they were living in Guatemala, they had a 7-monthold son who had died of intractable seizures. They did not know if the deceased child had ever had serum calcium measured. The parents denied consanguinity.
Table 2.
Laboratory values for the three affected siblings at the time of diagnosis as neonates and of their parents.
| Family member |
iPTH, pg/mL |
Ca, mg/dL |
Phos., mg/dL |
AlkP | 250HD, ng/mL |
1.250HD, pg/mL |
|---|---|---|---|---|---|---|
| Mother | 36 | 9.8 | – | – | 29 | 61 |
| Father | 40.7 | 9.7 | – | – | – | – |
| Sibling 1 | <3 | 5.1 | 10.4 | 270 | 23 | 78 |
| Sibling 2 | <3 | 7.2 | 9.8 | 151 | 24 | 80 |
| Sibling 3 | <3 | 7.0 | 10.9 | 303 | 41 | 104 |
–, not done. HD, hydroxyl; iPTH, intact parathyroid hormone.
DNA analysis
DNA was extracted from peripheral blood leukocyctes using standard protocols (ArchivePure DNA Isolation Kit; 5Prime, Gaithersburg, MD, USA). Oligonucleotide primers were designed to amplify the five exons and adjacent intronic regions from the GCM2 (OMIM 603716) gene. The polymerase chain reaction (PCR) was performed with five primer pairs (PCR conditions and primer sequences are available upon request), and amplicons were sequenced bidirectionally using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) on an ABI 3130x1 Genetic Analyzer. Nucleotide sequences were compared with the control reference sequence (NG_008970) by MacVector and Assembler Software (Cary, NC, USA).
Biochemical studies
Site-directed mutagenesis was performed to introduce the single nucleotide replacement into the wild type Gcm2 cDNA as previously described (7), and verified by sequence analysis. To analyze the mutant Gcm2 protein, we performed transient transfection of HEK293T cells with mutant and wild type Gcm2 cDNAs that were native or contained FLAG epitope tags. Briefly, HEK293T cells were seeded at 60%–80% confluence in six-well dishes, and 24 h later they were transfected with 1.0 µg of cDNA per well using Lipofectamine Plus. Cells were cultured in DMEM (Life Technologies, Rockville, MD, USA), supplemented with 10% fetal bovine serum, penicillin (100 units/mL) and streptomycin (100 µg/mL) in a humidified (95%) atmosphere of 5% carbon dioxide at 37°C. Approximately 48 h after transfection, the culture medium was removed and RNA or total protein was extracted as previously described (7). Cell extracts were subjected to SDS-PAGE on 8% gels and immunoblot analysis was performed using an affinity-purified rabbit anti-human GCM2 polyclonal antiserum (1:3000) as previously described (7) or M2 anti-FLAG monoclonal antibody (Sigma-Aldrich, St. Louis, MO, USA). Antibody binding was detected by use of peroxidase-labeled secondary antibody and was visualized using enhanced chemiluminescence. Reverse transcriptase-PCR for Gcm2 was performed as previously described (7).
Haplotype analysis
Primers were designed to amplify 11 loci on Chromosome 6 within and flanking the GCM2 gene, including eight short tandem repeat (STR) loci (DS1640, D6S309, D6S470, rs6149425, D6S1721, D6S429, D6S1653, D6S260) and three intragenic single nucleotide polymorphisms (SNPs) (rs9461258, rs16870746, and rs2275387). The STR targets were amplified with unlabeled primers, run on an agarose gel to confirm their size, purified, and sequenced to confirm locus specificity. Genotyping of the family members was performed using 6-FAM labeled primers and an Applied Biosystems 3130xl Genetic Analyzer.
Results
Sequence analysis of the GCM2 gene revealed a novel nucleotide change, c.408C>A, in exon 3 (Figure 2). The proband and her affected sisters were all homozygous for this mutation, while her parents were heterozygous carriers. The proband’s older brother was homozygous for wild type GCM2 alleles. The single base transversion replaces tyrosine with a premature stop at codon 136 (p.Tyr136Ter). We assessed the functional consequence(s) of this nonsense mutation by transient transfection of HEK293T cells. Cells that had been transfected with wild type Gcm2 cDNA expressed a recombinant Gcm2 protein of expected 55-kDa molecular weight. By contrast, no Gcm2 protein was detected using either the FLAG antibody or anti-Gcm2 antibody in cells that had been transfected with the Gcm2 c.408C>A cDNA. Finally, cells that had been transfected with equal amounts of mutant Gcm2 cDNA plus wild type Gcm2 containing a FLAG epitope tag expressed only the wild type Gcm2 protein and in expected quantities. Reverse transcription (RT)-PCR demonstrated equivalent expression of transcripts corresponding to wild type and c.408C>A forms of Gcm2 in transfected cells.
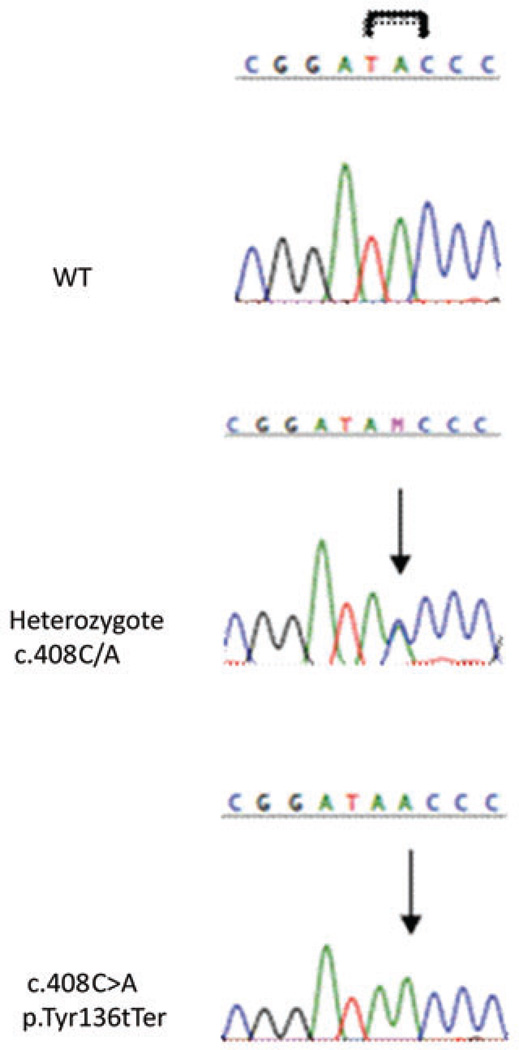
Figure 2.
Sequence chromatograms of exon 3 region showing the GCM2 Y136X mutations. Affected individuals show the presence of a C>A transition leading to a stop codon within exon 3. Each unaffected heterozygote parent carries the change within one copy of their GCM2 gene.
Haplotype analysis is summarized on the pedigree shown in Figure 3. Seven markers were informative, and spanned a 9.7 Mb region on 6p24.2 that included the GCM2 locus. All three affected siblings shared the same genotype. The informative intragenic SNP markers found in both mutated parental chromosomes that were passed on to the affected siblings were identical, and thus we cannot exclude that the two parents shared a common ancestor and that these two mutant alleles were identical by descent.
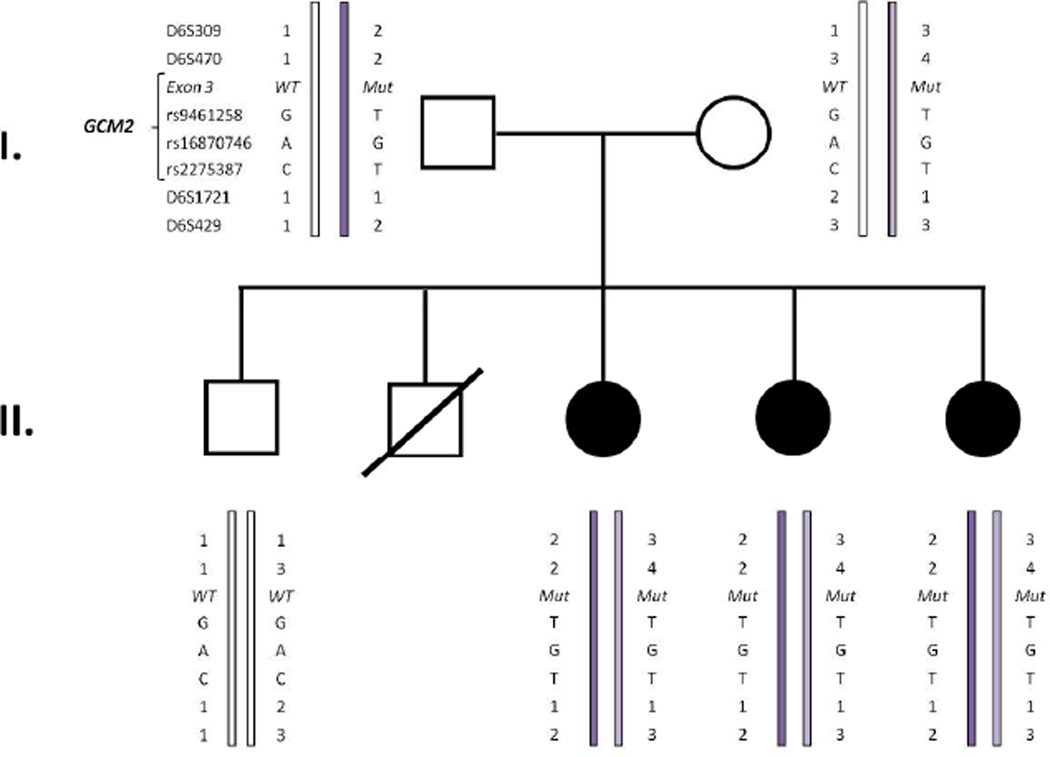
Figure 3.
Haplotype analysis using chromosome 6p23-24 loci and the GCM2 mutation in the reported AR-HPT family. The genotypes at the chromosome 6p23-24 microsatellite polymorphic loci and SNPs were ascertained. The paternal haplotypes are on the left and the maternal haplotypes are on the right. The two parental haplotypes share genotypes at 5 loci, thereby indicating that the c.408C>Amutation may have arisen in a shared ancestor of the father and mother.
Discussion
Gcm2 is a transcription factor that is crucial for parathyroid gland homeostasis (16). In transgenic mice, loss of Gcm2 results in absence of the parathyroids, and in humans spontaneous mutations of GCM2 are associated with congenital hypoparathyroidism (17). We report a novel point mutation in exon 3 of the GCM2 gene that is associated with a null-protein defect and severe early onset hypoparathyroidism. The proband presented with tetany and a hypocalcemic seizure at 7 days of age. She has, thus far, had normal developmental and cognitive milestones throughout early childhood on calcium and calcitriol supplementation. Her two affected siblings were also ascertained within the first few days of life, and have similarly done well with calcitriol therapy. The three affected subjects had undetectable levels of serum intact PTH, which differs from reports of low serum levels of PTH in previously reported patients who carried small mutations that lead to Gcm2 variants with reduced function or that mislocalize within the cell (8–13). Thus, it is conceivable that residual Gcm2 action allows development of small amounts of parathyroid tissue that can secrete detectable but physiologically insignificant amounts of PTH into the circulation.
The c.408C>A null-protein mutation follows a predictable autosomal recessive mode of inheritance with three siblings who each have a homozygous GCM2 mutation and two heterozygous, non-consanguineous parents. The parents are both of Guatemalan ethnicity, which is similar to the first reported human cases of isolated hypoparathyroidism caused by a different, large intragenic deletion of GCM2 (18). In both the previous family and the family reported here, molecular analysis demonstrated that the mutant alleles shared similar haplotypes and thus likely arose within common founders. By contrast, two other GCM2 mutations – the R47L missense (9, 11, 12) and the I298fsX307 frameshift (11) – have each been identified in three unrelated kindreds, and thus may represent potential mutational ‘hotspots’.
Mutational hotspots are often associated with repetitive sequences, such as homonucleotide runs, direct and inverted repeats and microsatellite repeats (18). For these mutation hotspots, the exact DNA sequence is not critical but only the fact that a sequence motif is repeated. Alternatively, mutation hotspots can depend on nucleotide sequence context (mutable motifs, subsequences) nucleotide substitutions or deletions within homonucleotide repeats, or during the repair of mismatches caused by heteroduplex formation between imperfect direct repeats (18). Although our haplotype analysis suggests identity by descent for the c.408C>A mutation that we have identified, this substitution occurs within a four cytosine homonucleotide repeat, and it is conceivable that the C to A transversion could have arisen independently.
We expressed the c.408C>A Gcm2 cDNA in mammalian cells to assess the biochemical consequences of the mutation. The mutation introduces a premature stop codon (p.Try136Ter) within the DNA-binding domain or GCM domain that is proximal to the nuclear localization signal and transactivation domain 1 (Figure 1) (9).
GCM2 is a member of a small family of GCM transcription factors that are present in vertebrates and some invertebrates (11, 19), and which share a conserved zinc-coordinating DNA-binding domain (20) that binds to the consensus motif 5-ATGCGGGT-3 (21, 22). Mutations in the GCM domain are predicted to cause structurally significant effects that have been shown to interfere with DNA binding or transactivational activity (10, 11).
To date, previously described small mutations in GCM2 have been described in exons 2 or 5 and have disrupted or eliminated sequences required for DNA binding, nuclear localization or transactivation. Accordingly, mutations in exon 2 have been associated with hypomorphic alleles and recessive hypoparathyroidism, while mutations in exon 5 have been associated with inhibitory function and dominant hypoparathyroidism (Figure 1).
Theoretically, the p.Tyr136Ter mutation could lead to nonsense-mediated decay of transcripts, a truncated protein that lacks the nuclear localization signal as well as the transcriptional activation domains, or an unstable protein that is rapidly degraded. Biochemical studies performed in our laboratory showed that Gcm2 transcripts but not protein were expressed in HEK293T cells that had been transfected with the Gcm2 p.Try136Ter cDNA.
Hence, we suggest that severe hypoparathyroidism occurred in the three affected subjects we studied because the premature termination reduced stability of the Gcm2 protein, further enhancing the rapid rate of degradation (23) and leading to a null-protein phenotype.
In conclusion, this is the first report of a mutation in exon 3 of the GCM2 gene and a mutation that leads to a null-protein phenotype. All three affected children presented with profound hypocalcemia and hyperphosphatemia during the neonatal period, and intact PTH was undetectable in the circulation. These results increase our understanding of the spectrum of GCM2 mutations that cause autosomal recessive isolated hypoparathyroidism and suggest that minimal GCM2 activity may allow development of sufficient parathyroid tissue to account for small amounts of circulating PTH.
Acknowledgements
We are grateful to the subjects and parents for participating in this study. We thank the Center for Applied Clinical Genomics for continued support and the Nemours Bimolecular Core Laboratory for technical support. This work was supported in part by grant #2P20RR020173-06A1 under the COBRE Program of the National Center for Research Resources, a component of the National Institutes of Health to Dr. Sol-Church, and NIH R01 DK079970 to Dr. Levine.
Footnotes
Conflict of interest statement
The authors have nothing to disclose.
Contributor Information
Daniel Doyle, Division of Endocrinology, Nemours/Alfred I. duPont Hospital for Children, P.O. Box 269, Wilmington, DE 19803, USA.
Susan M. Kirwin, Nemours/Alfred I. duPont Hospital for Children, Wilmington, DE, USA
Katia Sol-Church, Nemours/Alfred I. duPont Hospital for Children, Wilmington, DE, USA.
Michael A. Levine, The Children’s Hospital of Philadelphia and Department of Pediatrics, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA
References
- 1.Doyle DA. Hypoparathyroidism. In: Kleigman RM, Stanton BF, Schor NF, St Geme JW, Behrman RE, editors. Nelson Textbook of Pediatrics. 19th ed. Philadelphia, PA: Elsevier Saunders; 2011. pp. 1916–1919. [Google Scholar]
- 2.Greig F, Paul E, DiMartino-Nardi J, Saenger P. Transient congenital hypoparathyroidism: resolution and recurrence in chromosome 22q11 deletion. J Pediatr. 1996;128:563–567. doi: 10.1016/s0022-3476(96)70372-4. [DOI] [PubMed] [Google Scholar]
- 3.Parvari R, Hershkovitz E, Grossman N, Gorodischer R, Loeys B, et al. HRD/Autosomal Recessive Kenny-Caffey Syndrome Consortium. Mutation of TBCE causes hypoparathyroidism-retardation-dysmorphism and autosomal recessive Kenney-Caffey syndrome. Nat Genet. 2002;32:448–452. doi: 10.1038/ng1012. [DOI] [PubMed] [Google Scholar]
- 4.Nesbit MA, Bowl MR, Harding B, Ali A, Ayala A, et al. Characterization of GATA3 mutations in the hypoparathyroidism, deafness and renal dysplasia (HDR) syndrome. J Biol Chem. 2004;279:22624–22634. doi: 10.1074/jbc.M401797200. [DOI] [PubMed] [Google Scholar]
- 5.Pearce SH, Williamson C, Kifor O, Bai M, Coulthard MG, et al. A familial syndrome of hypocalcemia with hypercalciuria due to mutations in the calcium-sensing receptor. N Engl J Med. 1996;335:1115–1122. doi: 10.1056/NEJM199610103351505. [DOI] [PubMed] [Google Scholar]
- 6.Arnold A, Horst SA, Gardella TJ, Baba H, Levine MA, et al. Mutation of the signal peptide-encoding region of the preproparathyroid hormone gene in familial isolated hypoparathyroidism. J Clin Invest. 1990;86:1084–1087. doi: 10.1172/JCI114811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maret A, Ding C, Kornfield SL, Levine MA. Analysis of the GCM2 gene in isolated hypoparathyroidism: a molecular and biochemical study. J Clin Endocrinol Metab. 2008;93:1426–1432. doi: 10.1210/jc.2007-1783. [DOI] [PubMed] [Google Scholar]
- 8.Sticht H, Hashemolhosseini S. A common structural mechanism underlying GCMB mutations that cause hypoparathyroidism. Med Hypotheses. 2006;67:482–487. doi: 10.1016/j.mehy.2006.01.062. [DOI] [PubMed] [Google Scholar]
- 9.Baumber L, Tufarelli C, Patel S, King P, Johnson CA, et al. Identification of a novel mutation disrupting the DNA binding activity of GCM2 in autosomal recessive familial isolated hypoparathyroidism. J Med Genet. 2005;42:443–448. doi: 10.1136/jmg.2004.026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomee C, Schubert SW, Parma J, Le PQ, Hashemolhosseini S, et al. GCMB mutation in familial isolated hypoparathyroidism with residual secretion of parathyroid hormone. J Clin Endocrinol Metab. 2005;90:2487–2492. doi: 10.1210/jc.2004-2450. [DOI] [PubMed] [Google Scholar]
- 11.Bowl MR, Mirczuk SM, Grigorieva IV, Piret SE, Cranston T, et al. Identification and characterization of novel parathyroid-specific transcription factor glial cells missing homolog B (GCMB) mutations in eight families with autosomal recessive hypoparathyroidism. Hum Mol Genet. 2010;19:2028–2038. doi: 10.1093/hmg/ddq084. [DOI] [PubMed] [Google Scholar]
- 12.Canaff L, Zhou X, Mosesova I, Cole DE, Hendy GN. Glial cells missing-2 (GCM2) transactivates the calcium-sensing receptor gene: effect of a dominant-negative GCM2 mutant associated with autosomal dominant hypoparathyroidism. Hum Mutat. 2009;30:85–92. doi: 10.1002/humu.20827. [DOI] [PubMed] [Google Scholar]
- 13.Mannstadt M, Bertrand G, Muresan M, Weryha G, Leheup B, et al. Dominant-negative GCMB mutations cause an autosomal dominant form of hypoparathyroidism. J Clin Endocrinol Metab. 2008;93:3568–3576. doi: 10.1210/jc.2007-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi HS, Eom YS, Park leB, Lee S, Hong S, et al. Identification and characterization of C106R, a novel mutation in the DNA-binding domain of GCMB, in a family with autosomal-dominant hypoparathyroidism. Clin Encodrinol. 2012;76:625–633. doi: 10.1111/j.1365-2265.2011.04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mirczuk SM, Bowl MR, Nesbit MA, Cranston T, Fratter C, et al. A missence glial cells missing homolog B (GCMB) mutation, Asn502His, causes autosomal dominant hypoparathyroidism. J Clin Endocrinol. 2010;95:3512–3516. doi: 10.1210/jc.2009-2532. [DOI] [PubMed] [Google Scholar]
- 16.Ding C, Buckingham B, Levine MA. Familial isolated hypoparathyroidism caused by a mutation in the gene for the transcription factor GCMB. J Clin Invest. 2001;108:1215–1220. doi: 10.1172/JCI13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones BW, Fetter RD, Tear G, Goodman CS. Glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell. 1995;82:1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- 18.Rogozin IB, Pavlov YI. Theoretical analysis of mutation hotspots and their DNA sequence context specificity. Mutat Res. 2003;544:65–85. doi: 10.1016/s1383-5742(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 19.Akiyama Y, Hosoya T, Poole AM, Hotta Y. The gcm-motif: a novel DNA-binding motif conserved in Drosophila and mammals. Proc Natl Acad Sci USA. 1996;93:14912–14916. doi: 10.1073/pnas.93.25.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen SX, Moulin M, Schilling O, Meyer-Klaucke W, Schreiber J, et al. The GCM domain is a Zn-coordinating DNA-binding domain. FEBS Lett. 2002;528:95–100. doi: 10.1016/s0014-5793(02)03257-x. [DOI] [PubMed] [Google Scholar]
- 21.Cohen SX, Moulin M, Hashemolhosseini S, Kilian K, Wegner M, et al. Structure of the GCM domain-DNA complex: a DNA-binding domain with a novel fold and mode of target site recognition. EMBO J. 2003;22:1835–1845. doi: 10.1093/emboj/cdg182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuerk EE, Schreiber J, Wegner M. Protein stability and domain topology determine the transcriptional activity of the mammalian glial cells missing homolog, GCMB. J Biol Chem. 2000;275:4774–4782. doi: 10.1074/jbc.275.7.4774. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, Jones BW, Zock C, Chen Z, Wang H, et al. Isolation and characterization of mammalian homologs of the Drosophilia gene glial cells missing. Proc Natl Acad Sci USA. 1998;95:12364–12369. doi: 10.1073/pnas.95.21.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]