Abstract
Purpose of review
To provide an update on recently discovered human deafness genes and to describe advances in comprehensive genetic testing platforms for deafness, both of which have been enabled by new massively parallel sequencing technologies.
Recent findings
Over the review period, three syndromic and six nonsyndromic deafness genes have been discovered, bringing the total number of nonsyndromic deafness genes to 64. Four studies have shown the utility of massively parallel sequencing for comprehensive genetic testing for deafness. Three of these platforms have been released on a clinical or commercial basis.
Summary
Deafness is the most common sensory deficit in humans. Genetic diagnosis has traditionally been difficult due to extreme genetic heterogeneity and a lack of phenotypic variability. For these reasons, comprehensive genetic screening platforms have been developed with the use of massively parallel sequencing. These technologies are also accelerating the pace of gene discovery for deafness. Because genetic diagnosis is the basis for molecular therapies, these advances lay the foundation for the clinical care of deaf and hard-of-hearing persons in the future.
Keywords: deafness, genetics, genomics, hearing loss, massively parallel sequencing
INTRODUCTION
Deafness is the most common sensory deficit in humans, affecting one in every 500 newborns [1] and 278 million individuals worldwide (http://www.who.int). The majority (about 70%) of congenital hereditary deafness is nonsyndromic (non-syndromic hearing loss, NSHL) with the remaining 30% accounted for by a large number of different syndromes (syndromic hearing loss, SHL). Of NSHL, in developed countries, nearly 80% is due to a genetic cause.
Traditionally, hereditary deafness has been distinguished from nongenetic causes of deafness by medical and family history, physical examination and audiologic testing. In addition, in order to distinguish NSHL from syndromic forms of deafness, other ancillary tests such as temporal bone imaging, urinalysis, thyroid function studies and ECG were often obtained. However, even with this testing workup, a clear distinction between heritable and environmental causes of deafness and NSHL or syndromic deafness could be difficult, often making the diagnostic basis for hereditary deafness exclusionary. Comprehensive genetic testing for NSHL has changed the evaluation paradigm and today, after a history, physical examination and an audiogram, genetic testing should be the next test that is ordered.
Comprehensive genetic diagnosis for deafness has only recently been possible, an advance in clinical care linked to the development of novel genetic testing platforms for deafness based on massively parallel sequencing (MPS; reviewed in [2▪]). Prior to the advent of MPS, standard DNA sequencing techniques such as Sanger sequencing were used. These techniques made it difficult if not impossible to offer clinically useful screening for each of the 64 genes currently implicated in NSHL (http://www.hereditaryhearingloss.org).
MPS has provided clinicians and scientists with the ability to sequence millions or billions of DNA base-pairs simultaneously. Genetic diagnosis for NSHL is uniquely suited to take advantage of these technologies due to its extreme genetic heterogeneity. For example, more than 1200 causative allele variants have been reported in the 64 NSHL genes (http://www.deafnessvariationdatabase.org). As will be reviewed here, MPS permits comprehensive genetic testing for deafness by allowing sequencing of all known deafness genes simultaneously, while also facilitating the discovery of novel genetic causes of Mendelian syndromic and nonsyndromic deafness. As these new genes are added to comprehensive genetic testing platforms, the diagnostic ‘hit’ rate is improved.
Molecular genetic testing unequivocally diagnoses hereditary hearing loss and helps to dictate further patient management. It is the first test a clinician should order after history, physical examination and audiometry. In the next section, we will outline recently identified deafness genes, both syndromic and nonsyndromic, and provide an overview of genomic technologies used for genetic testing for deafness.
NEW DEAFNESS GENES IDENTIFIED
Mutations that cause deafness have been identified that affect almost every part of the organ of Corti: the cellular cytoskeleton – including actins (ACTG1) as well as actin-associated genes (TRIOBP and RDX); myosins (MYO7A, MYO15A, MYO6, MYO1A, MYH9, MYH14); cell–cell junctions (OTOA, CLDN14); cell–cell attachments (CDH23, PCDH15); gap-junctions (GJB2, GJB6); transporters (SLC26A4); and ion channels (KCNQ4). However, of the 102 reported NSHL loci, the causative gene has been identified for only 67, implying that at a minimum 34% of deafness genes have yet to be discovered (http://hereditaryhearingloss.org).
Gene discovery for all Mendelian diseases, including deafness, has accelerated since the advent of MPS technology (Table 1) [3▪–7▪,8▪▪,9▪,10▪,11▪▪]. Traditional methods for disease gene identification required large pedigrees with enough individuals to achieve significance in whole-genome linkage analysis studies. Genes within linked intervals were then sequenced one-by-one in the search for pathogenic variations. Although this method has been immensely successful, it is low-throughput.
Table 1.
Newly discovered syndromic and nonsyndromic deafness genes
| Gene symbol | Gene name | Inheritance | Locus | Phenotype | Methods | Protein function | Mutations /families | Reference |
|---|---|---|---|---|---|---|---|---|
| Syndromic deafness genes | ||||||||
| CACNA1D | Cav1.3 voltage-gated L-type calcium channel | Autosomal recessive | – | Sinoatrial node dysfunction and deafness (SANDD) | Genome-wide linkage, candidate gene screening | L-type calcium channel controls glutamate release in inner ear and SA node calcium release | 1 mutation/ 2 families | [3▪] |
| DNMT1 | DNA methyltransferase 1 | Autosomal recessive | – | Hereditary sensory and autonomic neuropathy type 1 (HSAN 1) | Genome-wide linkage, exome sequencing | Component of DNA replication machinery during S phase of cell cycle | 2 mutations/ 4 families | [4▪] |
| HARS2 | Mitochondrial histidyl tRNA synthetase | Autosomal recessive | – | Ovarian dysgenesis and deafness (Perrault syndrome) | Genome-wide linkage, candidate gene screening, TGE and MPS | Mitochondrial enzyme that catalyzes amino acid to cognate tRNA for translation | 2 mutations/ 1 family | [5▪] |
| Nonsyndromic deafness genes | ||||||||
| CEACAM16 | Carcinoembryonic antigen- related cell-adhesion molecule 16 | Autosomal dominant | DFNA4 | Postlingual, moderate SNHL, flat/downsloping audio profile | Genome-wide linkage, candidate gene screening, exome sequencing | Maintains integrity of tectorial membrane | 1 mutation/ 1 family | [6▪] |
| DIABLO | Direct inhibitor of apoptosis protein binding protein with low pI | Autosomal dominant | DFNA64 | Progressive to severe, onset age 22, flat audiogram | Genome-wide linkage, candidate gene screening | Functions in mitochondrial stress resistance | 1 mutation/ 1 family | [7▪] |
| GIPC3 | GAIP interacting protein, C terminus | Autosomal recessive | DFNB15/95 | Congenital, progressing to profound SNHL | Murine model and congenic gene mapping, genome- wide linkage | Functions in maturation of hair bundle and long-term survival of spiral ganglion | 2 mutations/ 2 families | [8▪▪] |
| ILDR1 | Immunoglobulin-like domain containing receptor 1 | Autosomal recessive | DFNB42 | Prelingual, moderate–profound SNHL | Homozygosity mapping, candidate gene screening | Transmembrane receptor of unknown function | 9 mutations/ 10 families | [9▪] |
| MSRB3 | Methionine sulfoxide reductase B3 | Autosomal recessive | DFNB74 | Prelingual, profound SNHL | Genome-wide linkage, candidate gene screening | Reduces oxidized methionine residues to repair oxidatively damaged proteins | 2 mutations/ 8 families | [10▪] |
| SMPX | Small muscle protein, X-linked | X-linked | DFNX4 | Slowly progressive, onset age 3 years in males, 28 in females, downsloping audiogram | X chromosome linkage, candidate gene screening, TGE and MPS | Cytoskeletal associated, responds to force | 2 mutations/ 2 families | [11▪▪] |
MPS, massively parallel sequencing; SNHL, sensorineural hearing loss; TGE, targeted genomic enrichment.
The development of MPS and the accompanying ability to sequence millions of base-pairs simultaneously has made whole-genome sequencing and whole-exome sequencing (WES, sequencing every exon of every gene in the genome) routine, changing the strategy for disease-gene identification. Whole-genome linkage analysis to identify a candidate genomic region can now be followed by simultaneous sequencing of every gene in the linked interval using targeted genomic enrichment (TGE) followed by MPS. To avoid generating a custom-targeted capture for each newly identified genomic region, WES can be used. This approach has been validated by numerous groups that have used traditional linkage analysis to identify specific genomic regions and then performed WES on one or more affected individuals in the pedigree, restricting the analysis of the WES data to the region of interest. Yet another method in families too small for linkage analysis is to perform WES on multiple affected individuals in a pedigree, filtering data to identify a single shared pathogenic variant (Table 1).
SYNDROMIC HEARING LOSS GENES
A new syndrome, sinoatrial node (SAN) dysfunction and deafness (SANDD) has been described and the causative gene identified as CACNA1D [3▪]. Previously, only Jervell and Lange-Nielsen syndrome had been associated with deafness and impaired cardiac conduction. However, in this study the authors identified two consanguineous families with similar phenotypes: congenital severe hearing loss and SANDD leading to pronounced bradycardia. After genome-wide linkage analysis, candidate gene screening led to the identification of a mutation in the ear-specific and heart-specific isoform of CACNA1D. CACNA1D encodes for the Cav1.3 voltage-gated L-type calcium channel, which tightly controls glutamate release at the inner hair cell ribbon synapse and calcium release in the SAN, explaining the ear and heart phenotypes of SANDD. In-vitro studies have shown that the mutation affects the Ca2+ ion pore, significantly impairing Ca2+ conduction. Without a thorough workup or genetic diagnosis, individuals with SANDD will appear as NSHL, as the occult SAN dysfunction may not be apparent until it manifests as syncope.
Perrault syndrome is a rare autosomal recessive disorder that is genetically and clinically heterogeneous. It is characterized by ovarian dysgenesis in females and progressive sensorineural hearing loss that is variable in onset and severity in affected males and females. Of 11 families identified with Perrault syndrome, mutations in HSD17B4 were found to be the genetic cause in one family [12]; a recent study uncovered mutations in HARS2 as the genetic cause in another family [5▪]. The authors performed whole-genome linkage analysis on an outbred family and identified two variants in HARS2. In addition, the authors used TGE to sequence the linked genomic region to exclude other variants. HARS2 encodes the highly conserved mitochondrial histidyl tRNA synthetase, an enzyme that catalyzes the linkage of amino acids to their cognate tRNAs. Disruption of mitochondrial translation has been shown to cause progressive deafness, as exemplified by mutations in mitochondrial rRNA and tRNA genes associated with syndromic and nonsyndromic deafness. The authors point out that, in the case of Perrault syndrome, patients may be diagnosed with NSHL if only males and prepubertal females are evaluated. This study underscores the importance of genetic diagnosis in evaluation of NSHL, as genetic counseling for Perrault syndrome would be vastly different from NSHL.
Hereditary sensory and autonomic neuropathy type I (HSAN1) is a genetically heterogeneous rare autosomal-dominant neurodegenerative disorder with central and peripheral involvement. A recent study identified pathogenic mutations in DNMT1 in four kindreds with a clinically distinct form of HSAN1 characterized by early onset dementia, sensorineural hearing loss (SNHL) and sensory neuropathy by 20–35 years of age [4▪]. The authors performed whole-genome linkage analysis and identified a significant region of chromosome 19. After WES, the authors focused their analysis on this region and identified a mutation in the gene DNMT1. The same mutation was found in two other kindreds and a different mutation affecting the same residue was found in a fourth kindred. DNMT1 encodes DNA methyltransferase 1, a crucial component of DNA replication during the S phase of the cell cycle. The authors show that mutations affect the proper folding of DNMT1, which in turns decreases overall enzymatic activity with global effects of hypomethylation with localized regions of hypermethylation.
NONSYNDROMIC HEARING LOSS GENES
Two recent studies have used genome-wide linkage to identify novel autosomal dominant NSHL genes (Table 1). In the first study, a mutation in CEA-CAM16 was implicated as the cause of autosomal-dominant DFNA4 deafness [6▪]. The authors of this study used WES to exclude other variants in the genomic region. DFNA4 is characterized by postlingual, moderate SNHL with a flat or slightly down-sloping audio profile. In the second study, a mutation in DIABLO was shown to cause dominant deafness at the DFNA64 locus [7▪]. Affected persons noted progressive hearing loss at an average age of 22 years, with progression to severe hearing loss by middle age. DIABLO was shown to be involved in mitochondrial stress resistance.
The recently discovered genes underlying autosomal-recessive DFNB42 and DFNB72 have been identified as ILDR1 and MSRB3, respectively [9▪,10▪]. ILDR1 was identified as a cause of prelingual moderate-to-profound SNHL by homozygosity mapping in a consanguineous family, a finding verified by identification of causative mutations in ILDR1 in nine other families with similar hearing loss [9▪]. MSRB3, which encodes a protein involved in repairing oxidatively damaged proteins, was identified through genome-wide linkage analysis and candidate gene screening. Mutations in MSRB3 were then shown to cause prelingual profound SNHL in seven additional families [10▪].
To uncover the cause of DFNB15/95 deafness, Charizopoulou et al. [8▪▪] first identified the cause of age-related hearing loss in BSLW mouse strain using congenic gene mapping. The causative gene, GIPC3, was then sequenced in families with significant linkage to that genomic region. In the two identified families, the hearing loss is congenital and rapidly progresses from mild to severe in degree. GIPC3 is hypothesized to function in the maturation of the hair bundle and the survival of the spiral ganglion.
And, finally, SMPX has been identified as the third gene causing X-linked NSHL [11▪▪]. The authors used X chromosome linkage analysis followed by unsuccessful candidate gene screening, and then adopted a targeted sequence capture strategy to analyze all X chromosome genes. A pathogenic mutation was identified in SMPX, which encodes for a protein associated with the cytoskeleton and hypothesized to respond to force. A second family has also been identified with mutations in SMPX. The hearing loss is slowly progressive with onset on average at 3 years of age for males and 28 years of age for females.
The discovery of these genes will aid in diagnosis of deafness, as these genes will be added to comprehensive testing platforms described below and improve the overall diagnostic rate for NSHL.
ADVANCES IN GENETIC TESTING FOR DEAFNESS
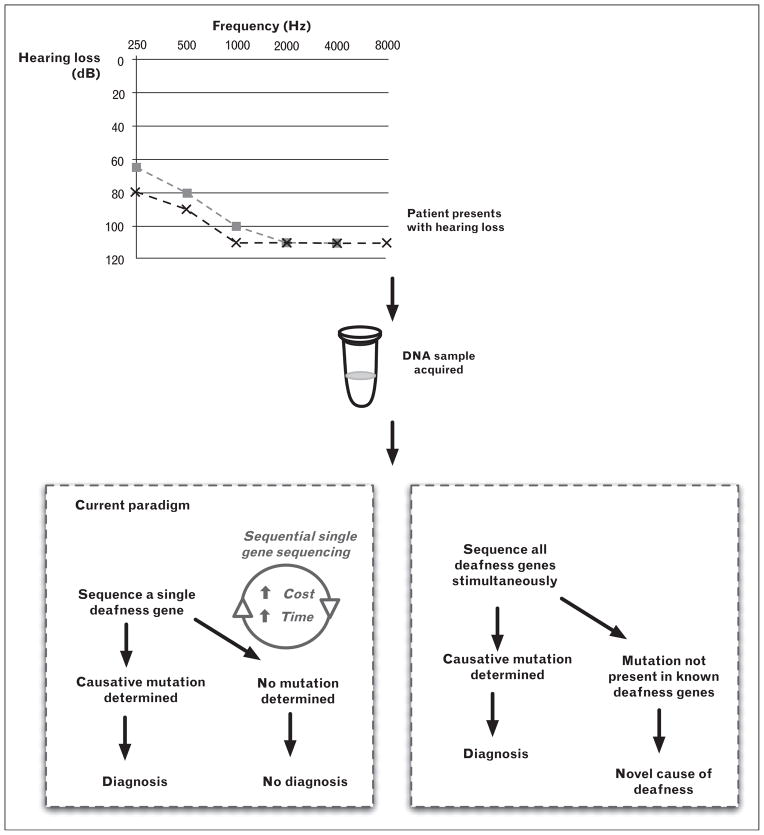
Genetic testing for deafness has been available since the late 1990s; however, it has only been in the past 2 years that it has moved to the forefront in importance in the clinical evaluation of the deaf and hard-of-hearing patient. When first available, the value of genetic testing for deafness was marginal primarily because of limited throughput due to DNA sequencing technologies and high genetic heterogeneity (Fig. 1). Sequential gene screening was neither time nor cost-effective. Attention was focused primarily on GJB2, the most common cause of autosomal recessive NSHL in many world populations; however after GJB2 had been screened, subsequent gene prioritization was difficult [13]. To be more all-inclusive, several groups used nonsequencing-based screening technology such as resequencing arrays and single-primer extension microarrays [14,15]; however, these methods do not provide an unbiased analytical approach, as they focus on reported mutations and/or do not provide comprehensive screening of all deafness genes. The long-term goal, to develop a method to screen all genes causing NSHL simultaneously (Fig. 1), has been realized with the advent of MPS (Table 2) [16,17▪–19▪].
FIGURE 1.
Paradigms for genetic testing for deafness. The current paradigm involves sequential gene screening, whereas new comprehensive genetic screening platforms screen all deafness genes simultaneously.
Table 2.
Massively parallel sequencing genetic testing platforms for deafness
| Reference | Genes screened | Methods: enrichment/ sequencing | Total DNA samples used (controls) | Results | Availability |
|---|---|---|---|---|---|
| [16] | 54 NSHL genes | cRNA solution-phase TGE/Illumina GAIIx | 10 (4) | Identified causative mutation in positive controls and in five of six unknowns | University of Iowa – OtoSCOPE (http://morlotoscope.org) |
| [17▪] | 246 genes causing human and murine deafness | cRNA solution-phase TGE/Illumina HiSeq | 11 (0) | Identified causative mutation in six of 11 unknowns | N/A |
| [18▪] | 5 genes | cDNA array-based TGE/Illumina HiSeq | 12 (12) | Identified causative mutation in all positive controls | OtoGenetics – Oto-DA1 (http://otogenetics.com) |
| [19▪] | 15 most commonly mutated ARNSHL genes | Semi-automated PCR/Roche 454 GS FLX | 5 (1)a | Identified causative mutation in positive control and three of four unknowns | NXTGNT (http://www.nxtgnt.com) |
N/A, not available; TGE, targeted genomic enrichment.
Three of the four unknowns had previous linkage shown to a genomic region containing one of the 15 genes sequenced.
The first study using MPS for diagnosis of NSHL was published in 2010 [16]. This study used TGE to isolate all exons of all NSHL genes, followed by MPS. This method was tested on 10 individuals, including three positive controls and a single negative control. Of the six individuals with idiopathic causes of NSHL, a genetic diagnosis was identified in five. The authors tested the sensitivity and specificity of this method against the current gold standard for genetic testing, Sanger sequencing, and found a sensitivity of 99.72% and specificity of more than 99% for 605 single nucleotide polymorphisms; the platform identified large and small deletions as well. This study showed that these methods hold great promise for providing comprehensive genetic testing for deafness. This platform has since been updated to include 66 genes and is now offered on a clinical basis as OtoSCOPE (http://morl-otoscope.org).
In the second study using TGE and MPS for genetic diagnosis of hearing loss, Brownstein et al. [17▪] sequenced 246 genes simultaneously in 11 probands. The authors chose 82 human SHL and NSHL genes as well as 162 genes known to cause murine deafness, reasoning that the inclusion of mouse deafness genes would speed the discovery of human deafness genes. The 11 probands were negative for mutations in GJB2 and common genetic causes of deafness in either the Palestinian Arab or Jewish Israeli ethnicities. Causative mutations were identified in six probands, all of which were in reported NSHL genes. The authors then screened other deaf persons of the same ethnicity for these mutations and were able to determine the genetic cause of deafness in a further 20 families. This study highlights the potential of TGE and MPS for comprehensive diagnosis and candidate gene screening, while at the same time underscoring the importance of ethnicity-based testing in deafness.
As opposed to using solution-phase custom complementary RNA (cRNA) oligonucleotides for TGE as in the previous two studies, Tang et al. [18▪] describe preliminary data for a commercialized approach to targeted capture for diagnostic screening of known deafness genes using complementary DNA oligonucleotides. The authors developed a method for cost-effective manufacture of custom DNA oligonucleotides from cDNAs of genes to be targeted for sequencing. The TGE baits are created by performing an amplification from cDNAs and these baits are then baked on to an array prior to use for solid-phase TGE. This method precludes the requirement for expensive infrastructure to design and manufacture cRNA TGE bait sets. The authors demonstrate data using this method for only five genes, however, and only used positive controls [18▪]. They were able to detect single nucleotide variants as well as indels. The authors commercialized this platform as a method for screening of all deafness genes, although these data were not shown. This study presents a unique method for cost-effective screening for genetic deafness that is highly customizable and easily adaptable if new genes are discovered; however, questions remain with regard to performance with a large number of samples, detection of copy number variants and scaling to include a greater number of genes. The authors use the same method for their commercial screening platform offered through OtoGenetics (http://www.otogenetics.com).
Taking a different approach to more comprehensive genetic screening for deafness, a recent study used a semi-automated PCR amplification method paired with MPS to simultaneously sequence 15 genes associated with autosomal recessive NSHL [19▪]. In total, amplification required 646 primers pairs and the use of a liquid-handling robot. The authors tested the method on five patients, including a single positive control and three individuals with genetic linkage to one of the 15 genes. The authors identified the causative mutation in the positive control as well as all three individuals with linkage to a sequenced gene, but did not identify the causative mutation in the fifth individual. This method clearly overcomes some of the limitations of TGE, including the high initial cost to design complementary baits and the capture protocol, which involves at least a 24-h hybridization, but it does not sequence all known deafness genes and, therefore, the diagnostic rate is lower. Three of the four methods for more comprehensive genetic testing for deafness described here are available today for clinical and research use (Table 2). These methods will continue to improve and vastly change the clinical evaluation of deaf and hard-of-hearing persons.
CONCLUSION
In the evaluation of NSHL, genetic testing has become the first test that should be ordered after history, physical examination and audiometry. Establishing a genetic diagnosis is valuable because it can preclude further (invasive) diagnostic testing; identify possible medical comorbidities; and provide families with genetic counseling and valuable prognostic information. As we move toward personalized genomic medicine, genetic testing will be the basis for targeted molecular therapies.
KEY POINTS.
Massively parallel sequencing technologies have accelerated the pace of gene discovery for deafness and have made comprehensive genetic testing available for deafness for the first time.
Establishing a genetic diagnosis for deafness is valuable because it can, first, preclude further (invasive) diagnostic testing; second, identify possible medical comorbidities; and, third, provide families with genetic counseling and valuable prognostic information.
Four studies have shown the utility of massively parallel sequencing for comprehensive genetic testing for deafness, and three of these platforms have been released on a clinical or commercial basis.
As we move toward personalized genomic medicine, comprehensive genetic testing for deafness should be the first test ordered after history, physical examination and audiometry and will be the basis for targeted molecular therapies in the future.
Acknowledgments
This work was supported by NIDCD RO1s DC003544, DC002842 and DC012049 to R.J.H.S., and NIDCD 1F30DC011674 to A.E.S.
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest Additional references related to this topic can also be found in the Current World Literature section in this issue (pp. 770–771).
- 1.Morton CC, Nance WE. Newborn hearing screening: a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2▪.Shearer AE, Hildebrand MS, Sloan CM, et al. Deafness in the genomics era. Hear Res. 2011;282:1–9. doi: 10.1016/j.heares.2011.10.001. This review provides an overview of MPS technologies in the context of deafness research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3▪.Baig SM, Koschak A, Lieb A, et al. Loss of Cav1.3 (CACNA1D) function in a human channelopathy with bradycardia and congenital deafness. Nat Neurosci. 2011;14:77–84. doi: 10.1038/nn.2694. This study describes a new syndrome characterized by deafness and profound bradycardia. It is the second syndrome involving deafness and impaired cardiac conduction and underscores the need for a thorough workup and genetic testing for cases of apparent NSHL. [DOI] [PubMed] [Google Scholar]
- 4▪.Klein CJ, Botuyan M, Wu Y, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600. doi: 10.1038/ng.830. This study implicates mutations in the gene DNMT1 as the cause of HSAN1, a rare neurologic syndrome characterized by peripheral and central neuropathy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5▪.Pierce SB, Chisholm KM, Lynch ED, et al. Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc Natl Acad Sci U S A. 2011;108:6543–6548. doi: 10.1073/pnas.1103471108. In this study, the authors identified the second genetic cause of Perrault syndrome, a rare recessive disorder that is characterized by ovarian dysgenesis and hearing loss. If only prepubertal females and males are examined, then cases of Perrault syndrome may be mischaracterized as NSHL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6▪.Zheng J, Miller KK, Yang T, et al. Carcinoembryonic antigen-related cell adhesion molecule 16 interacts with alpha-tectorin and is mutated in autosomal dominant hearing loss (DFNA4) Proc Natl Acad Sci U S A. 2011;108:4218–4223. doi: 10.1073/pnas.1005842108. In this study, the authors identify CEACAM16 as the second autosomal-dominant NSHL gene at the DFNA4 locus. Exome sequencing was used to rule out other variants within the linked region. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7▪.Cheng J, Zhu Y, He S, et al. Functional mutation of SMAC/DIABLO, encoding a mitochondrial proapoptotic protein, causes human progressive hearing loss DFNA64. Am J Hum Genet. 2011;89:56–66. doi: 10.1016/j.ajhg.2011.05.027. In this study, the authors uncovered mutations in DIABLO as the cause of DFNA64 deafness. DIABLO was shown to encode for a protein involved in mitochondrial stress resistance, thereby explaining the progressive nature of this type of deafness. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8▪▪.Charizopoulou N, Lelli A, Schraders M, et al. Gipc3 mutations associated with audiogenic seizures and sensorineural hearing loss in mouse and human. Nat Commun. 2011;2:201. doi: 10.1038/ncomms1200. The authors first identified the cause of age-related hearing loss in mice using congenic mapping and then sequenced the orthologous gene in affected humans to identify a progressive form of autosomal recessive deafness. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9▪.Borck G, Ur Rehman A, Lee K, et al. Loss-of-function mutations of ILDR1 cause autosomal-recessive hearing impairment DFNB42. Am J Hum Genet. 2011;88:127–137. doi: 10.1016/j.ajhg.2010.12.011. The classic methods for disease gene identification, homozygosity mapping and candidate gene screening, were used in this study to identify the ILDR1 as the cause of DFNB42 deafness. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10▪.Ahmed ZM, Yousaf R, Lee BC, et al. Functional null mutations of MSRB3 encoding methionine sulfoxide reductase are associated with human deafness DFNB74. Am J Hum Genet. 2011;88:19–29. doi: 10.1016/j.ajhg.2010.11.010. In this study, the authors used genome-wide linkage analysis and candidate gene screening to implicate MSRB3 as the cause of DFNB74 deafness. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11▪▪.Schraders M, Haas SA, Weegerink NJD, et al. Next-generation sequencing identifies mutations of SMPX, which encodes the small muscle protein, X-linked, as a cause of progressive hearing impairment. Am J Hum Genet. 2011;88:628–634. doi: 10.1016/j.ajhg.2011.04.012. This study describes the discovery of SMPX as the third gene causing X-linked NSHL. The authors turned to TGE and MPS when traditional candidate gene screening failed to identify a causative mutation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce SB, Walsh T, Chisholm KM, et al. Mutations in the DBP-deficiency protein HSD17B4 cause ovarian dysgenesis, hearing loss, and ataxia of Perrault syndrome. Am J Hum Genet. 2010;87:282–288. doi: 10.1016/j.ajhg.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilgert N, Smith RJH, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681:189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kothiyal P, Cox S, Ebert J, et al. High-throughput detection of mutations responsible for childhood hearing loss using resequencing microarrays. BMC Biotechnol. 2010;10:10. doi: 10.1186/1472-6750-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Paris J, Pique L, Colen T, et al. Genotyping with a 198 mutation arrayed primer extension array for hereditary hearing loss: assessment of its diagnostic value for medical practice. PLoS One. 2010;5:e11804. doi: 10.1371/journal.pone.0011804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shearer AE, DeLuca AP, Hildebrand MS, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17▪.Brownstein Z, Friedman LM, Shahin H, et al. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol. 2011;12:R89. doi: 10.1186/gb-2011-12-9-r89. The authors used TGE MPS to sequence 246 genes including human syndromic and nonsyndromic genes as well as genes causing deafness in mouse. This unique approach could speed the discovery of human deafness genes as candidate genes are included on the platform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18▪.Tang W, Qian D, Ahmad S, et al. A low-cost exon capture method suitable for large-scale screening of genetic deafness by the massively-parallel sequencing approach. Genet Test Mol Biomark. 2012;16:536–542. doi: 10.1089/gtmb.2011.0187. The authors of this study developed a unique method for TGE that is flexible and may be more cost-effective, as the targeting baits are manufactured in-house. However, they show data for only five genes and tested only positive controls. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19▪.De Keulenaer S, Hellemans J, Lefever S, et al. Molecular diagnostics for congenital hearing loss including 15 deafness genes using a next generation sequencing platform. BMC Med Genomics. 2012;5:17. doi: 10.1186/1755-8794-5-17. In this study, the authors used large-scale PCR instead of TGE and sequenced on the 454 instead of the more widely used Illumina sequencer. The technique described may be more flexible and cost-effective, but only 15 genes are sequenced simultaneously. [DOI] [PMC free article] [PubMed] [Google Scholar]