Abstract
With the exception of viral proteins E1 and E2, papillomaviruses depend heavily on host replication machinery for replication of their viral genome. E1 and E2 are known to recruit many of the necessary cellular replication factors to the viral origin of replication. Previously, we reported a physical interaction between E1 and the major human single-stranded DNA (ssDNA)-binding protein, replication protein A (RPA). E1 was determined to bind to the 70-kDa subunit of RPA, RPA70. In this study, using E1-affinity coprecipitation and enzyme-linked immunosorbent assay-based interaction assays, we show that E1 interacts with the major ssDNA-binding domain of RPA. Consistent with our previous report, no measurable interaction between E1 and the two smaller subunits of RPA was detected. The interaction of E1 with RPA was substantially inhibited by ssDNA. The extent of this inhibition was dependent on the length of the DNA. A 31-nucleotide (nt) oligonucleotide strongly inhibited the E1-RPA interaction, while a 16-nt oligonucleotide showed an intermediate level of inhibition. In contrast, a 10-nt oligonucleotide showed no observable effect on the E1-RPA interaction. This inhibition was not dependent on the sequence of the DNA. Furthermore, ssDNA also inhibited the interaction of RPA with papillomavirus E2, simian virus 40 T antigen, human polymerase alpha-primase, and p53. Taken together, our results suggest a potential role for ssDNA in modulating RPA-protein interactions, in particular, the RPA-E1 interactions during papillomavirus DNA replication. A model for recruitment of RPA by E1 during papillomavirus DNA replication is proposed.
The Papillomavirinae are small, nonenveloped viruses with double-stranded, circular DNA genomes. With the exception of two virally encoded proteins, E1 and E2, papillomavirus (PV) DNA replication is carried out entirely by the host cellular replication machinery (reviewed in references 14, 42, and 68). A partially reconstituted cell-free system of replication has allowed the identification of many of these cellular factors (34, 49, 55, 57). Among them are cellular factors that were previously found to be necessary and sufficient for simian virus 40 (SV40) DNA replication, including DNA polymerase alpha-primase (pol-prim) and replication protein A (RPA) (reviewed in references 13, 51, and 67). However, unlike SV40, additional cellular factors, some of which have yet to be identified, are required for efficient PV DNA replication (44, 46, 49). Nevertheless, many parallels have been drawn between the two viral systems, particularly regarding the initiation and elongation of DNA replication.
The PV E1 protein is an ATP-dependent viral DNA helicase (77); E2 is a multifunctional regulator of viral transcription and replication (18, 26, 35, 48). PV DNA replication is initiated by the assembly of E1 as a double hexamer at the origin of replication, resulting in a localized melting of the DNA template (21, 63). The sequence-specific binding of E1 to the origin is directed by its interactions with E2 (45, 54, 61). Subsequent formation of E1 double hexamers is promoted by the protein-folding activities of cellular chaperone proteins (44), which also displace E2 from the E1-origin complex (39). After its release from E2, E1 further catalyzes an extensive unwinding of the double-stranded viral DNA template bidirectionally from the origin (29, 63, 64, 79).
The activities of cellular RPA and pol-prim are required to synthesize the RNA-DNA primers necessary to initiate DNA replication (reviewed in reference 73). Both these cellular proteins are essential for PV DNA replication and are presumably recruited to the origin through direct interactions with viral replication proteins E1 and E2. RPA and pol-prim are similarly required for SV40 DNA replication and are recruited to the origin through direct interactions with large T antigen (Tag), the functional equivalent of PV E1 with regards to DNA replication (67). The interactions among Tag, RPA, and pol-prim are highly specific (15, 50) and essential for primer synthesis during SV40 DNA replication (75, 76). Similarly to Tag, E1 has been reported to interact with the p180 and p68 subunits of pol-prim (9, 16, 47, 57). Like Tag, E1 also binds RPA (27).
Human RPA is a single-stranded DNA (ssDNA)-binding protein with complex and essential functions in many aspects of DNA metabolism, including DNA replication, repair, and recombination (reviewed in references 30, 52, and 78). It is a highly structured, heterotrimeric complex consisting of three subunits: a 70-kDa subunit (RPA70), a 32-kDa subunit (RPA32), and a 14-kDa subunit (RPA14). Homologs of all three RPA subunits have been identified in Saccharomyces cerevisiae and are known to be essential genes (11). As a complex, RPA demonstrates a strong affinity for ssDNA, but binding is not strictly sequence specific. The two strongest ssDNA-binding motifs of RPA, DBD-A and DBD-B, are located within the central region of RPA70 (4, 58, 59). Together, DBD-A and DBD-B form the major ssDNA-binding domain of RPA (Fig. 1) and exhibit ssDNA-binding affinity similar to that of the heterotrimer (23, 24). The occluded binding site for this domain is approximately 8 nucleotides (nt) on ssDNA (4). A third ssDNA-binding motif, DBD-C, is located within the carboxyl terminus of RPA70, along with a zinc finger structure that has been found to be important for structural stability and the ssDNA-binding activity of RPA (7, 10, 20, 74). RPA32 also binds ssDNA via a fourth ssDNA-binding motif, DBD-D (3, 6, 59). When all four DNA-binding domains are bound to ssDNA, the occluded binding site for RPA is approximately 30 nt (32, 65). In addition to interactions with viral replication initiation proteins E1 and Tag, RPA also associates with a number of proteins important for DNA replication, repair, and/or recombination, including p53, pol-prim, Rad52, XPA, VP16, and uracil-DNA glycosylase (reviewed in references 30 and 78).
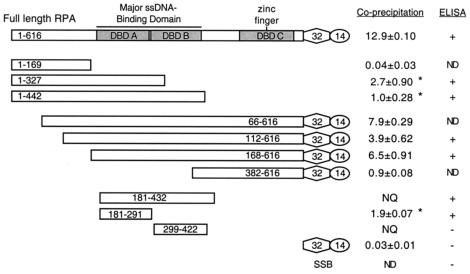
FIG. 1.
Summary of the RPA-E1 interaction mapping studies. Full-length RPA and RPA70 truncation mutants were tested for their ability to interact with BPV1 E1 in GST affinity coprecipitation and ELISA-based protein interaction assays. In this figure, RPA70 is shown as long rectangles, RPA32 is shown as hexagons, and RPA14 is shown as circles. The ssDNA-binding domains within RPA70 are shaded and indicated as DBD-A, DBD-B, and DBD-C. Together, DBD-A and DBD-B form the primary ssDNA-binding region within RPA. A fourth ssDNA-binding domain, DBD-D, is located within RPA32. The zinc finger domain, which is embedded in DBD-C, is also indicated. Since the carboxyl terminus of RPA70 is essential for heterotrimer assembly, mutants containing the carboxyl terminus were expressed in a complex with RPA32 and RPA14. The results for the coprecipitation assays were taken from at least three separate experiments. The numbers indicate the percentages of 35S-RPA precipitated with GST-E1 beads (± standard deviation), after the subtraction of background (binding of that protein to GST Sepharose beads). Binding to E1 that could not be assessed due to excessive proteolysis of the 35S-labeled RPA protein is labeled as NQ (not quantifiable). (*, two N-terminal RPA domain constructs, RPA70(1-327) and RPA70(1-442), clearly showed much higher levels of coprecipitation with GST-E1 than did the RPA70(1-169) construct, but the levels of coprecipitation were lower than those for the C-terminal constructs. This was attributable to a relatively higher degree of proteolysis for the N-terminal domain constructs than for the C-terminal domain constructs.) Results from the ELISA are summarized as binding comparable to that of full-length RPA (+) and binding comparable to that of the negative control, E coli SSB (−). Mutant proteins that were not tested in a particular assay are noted as ND (not done).
Recently, Tag was reported to bind to RPA through the major ssDNA-binding domain in RPA70 and the carboxyl terminus of RPA32 (30, 37). In studying the recruitment of RPA by the PV E1 protein, we demonstrated that E1 interacts with the 70-kDa subunit of RPA (27). In this study, using E1-affinity coprecipitation and enzyme-linked immunosorbent assay (ELISA)-based protein interaction assays, we show that E1 interacts with the major ssDNA-binding domain of RPA. Consistent with our previous report, we did not detect a stable interaction between E1 and the two smaller subunits of RPA. The interaction between E1 and RPA is substantially inhibited by ssDNA. This inhibition is dependent on the length of the DNA but does not appear to be dependent on the sequence of the DNA. In addition, ssDNA binding by RPA also inhibits the interactions between RPA and PV E2, SV40 Tag, human pol-prim, and p53 proteins. These results indicate that ssDNA may play a general role in modulating RPA-protein interactions, including that between RPA and E1 during PV DNA replication.
MATERIALS AND METHODS
Materials, plasmids, and proteins.
The ssDNA oligonucleotides used in this study were synthesized by either Integrated DNA Technologies, Inc., or Gibco Invitrogen BRL. These consisted of three oligonucleotides consisting of alternating thymine and guanine residues (31, 16, and 10 nt), a 31-nt oligonucleotide consisting entirely of thymine, and a 31-nt nonspecific oligonucleotide consisting of the sequence 5′-TTCAGTGTCAGGATTTGACCTCATACAAGGC-3′. Where indicated, the oligonucleotides were end labeled with 32P by using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (Amersham Pharmacia). The RPA32-specific monoclonal antibody was described previously (19), while the glutathione S-transferase (GST)-specific polyclonal antibody was obtained from Amersham Pharmacia. The horseradish peroxidase-conjugated anti-goat and anti-mouse antibodies were both obtained from Amersham Pharmacia, and 3,3′-5,5′-tetramethylbenzidine (TMB) was from Sigma.
The expression vectors for full-length RPA and the majority of the RPA truncation mutants were kindly provided by Marc S. Wold. The proteins were expressed in Escherichia coli with the T7 expression system and purified to near homogeneity as described previously (12, 23, 24, 28). The expression vectors and purified proteins for the major ssDNA-binding domain of RPA and the two subdomains, DBD-A and DBD-B, were kindly provided by Alexey Bochkarev (4). Purified full-length RPA was shown to bind to ssDNA and to support SV40 DNA replication in vitro (data not shown). In contrast, the purified RPA truncation proteins were reported to have varied affinities for ssDNA (4, 23, 24). Among the RPA truncations used in this study, only those with N-terminal deletions in RPA70 up to or less than 168 amino acids were capable of supporting SV40 DNA replication in vitro (23, 24). Where indicated, these full-length and truncated RPA proteins were also expressed as 35S-labeled proteins by using the STP3 in vitro transcription-translation kit (Novagen) and [35S]methionine (New England Nuclear), in accordance with the manufacturer's instructions.
The GST-tagged bovine PV type 1 (BPV1) E1 fusion protein, untagged E1, and BPV1 E2 proteins were expressed in E coli and purified as described previously (49, 62). Jen-Sing Liu and Shu-Ru Kuo kindly provided the expression vectors for both the E1 and E2 fusion proteins of human PV type 11 (HPV11). HPV11 E1 was expressed as a fusion protein tagged with a glutamate-rich epitope (EE) at the amino terminus and purified from recombinant baculovirus-infected Sf9 cells (34, 43, 44). The E1 proteins were purified to apparent homogeneity as analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. These proteins were shown to bind to both single-stranded and double-stranded DNA and to support DNA replication in cell-free PV DNA replication assays (data not shown). The HPV11 E2 cDNA was excised from pET-11E2 with NdeI and HindIII and ligated into the XhoI and HindIII sites of pET-BB, creating the plasmid pET-BB-11E2, which expresses the HPV11 E2 fusion protein with a BB epitope (RKTPRVTG) on the amino terminus. The plasmid pET-BB-11E2 was transformed into BL21(DE3), and the bacteria were grown to an optical density of 0.6 at 600 nm before being induced with IPTG (isopropyl-β-d-thiogalactopyranoside) at 0.1 mM overnight at 18°C. The bacteria were pelleted and resuspended in 1× phosphate-buffered saline (1 liter of 1× phosphate-buffered saline: 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, 0.24 g of KH2PO4; pH adjusted to 7.4 with HCl) with 5 μg of lysozyme/ml. Following incubation on ice for 30 min, the bacteria were again pelleted and resuspended in buffer A (20 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride). The bacteria were lysed by sonication (setting 6, three 30-s intervals on ice), and the resultant lysates were clarified by centrifugation at 12,500 × g for 15 min at 4°C. The supernatant was applied to a Q-Sepharose column, and the unbound fraction was applied directly to a HiTrap SP-Sepharose column (Amersham Pharmacia). BB-E2 was eluted from the SP column with buffer A with the use of a 100 to 500 mM NaCl gradient. The E2 proteins were purified to a single polypeptide when subjected to SDS-PAGE and Coomassie blue staining, according to published protocols shown to produce E2 capable of binding to its cognate DNA-binding site (49, 62). The purified E2 was shown to be functional in E2-stimulated PV DNA replication in vitro (49).
SV40 Tag was expressed in Sf9 cells infected with a recombinant baculovirus expression vector and purified from lysates by immunoaffinity chromatography (36, 56, 66, 69). Tag was purified to apparent homogeneity as analyzed by SDS-PAGE and Coomassie blue staining. The purified Tag was able to support SV40 DNA replication in vitro (data not shown). Human pol-prim was purified from human 293 S100 extracts as described previously (31, 71, 72). Only the four constituent polypeptides of pol-prim were detected when the purified protein was subjected to SDS-PAGE and Coomassie blue staining. The purified protein was able to synthesize DNA with a variety of primed and unprimed ssDNA templates (data not shown). Purified p53 protein was kindly provided by Maria L. Avantagiatti. The p53 protein provided is a point mutant (amino acid 245) that shows wild-type levels of binding to RPA. Purified E coli SSB was purchased from U.S. Biochemical Corp.
Coprecipitation of 35S-labeled RPA proteins.
GST and GST-E1 were bacterially expressed and bound to glutathione Sepharose as described previously (27, 76). The beads were washed and resuspended as a 50% (vol/vol) suspension in buffer S (20 mM Tris-HCl [pH 7.4], 100 mM NaCl, 10% glycerol, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride) and stored at 4°C for up to a week. For each experiment, the levels of GST and GST-E1 bound to the beads were evaluated to ensure that the control beads (GST) had levels of bound protein equal to or greater than those of the GST-E1 beads. For every experiment, at least twice as much GST as GST-E1 was bound to the matrix.
The coprecipitation assays were performed in microcentrifuge tubes that had been blocked overnight with 0.2% (wt/vol) bovine serum albumin (BSA) in phosphate-buffered saline. Following the removal of BSA, approximately 0.8 μCi of the 35S-labeled RPA protein was added to 25 μl of the Sepharose bead suspension. The final volume was adjusted to 0.5 ml with buffer SN (buffer S with an additional 0.05% [vol/vol] NP-40) supplemented with 1 mM CaCl2, 1 mM MgCl2, and 20 U of micrococcal nuclease/ml. The tubes were rocked at 4°C for 3 h, and the beads were precipitated by centrifugation. The precipitates were washed in buffer SN and boiled in 20 μl of 2× SDS-PAGE sample buffer (100 mM Tris-HCl [pH 7.5], 3 M β-mercaptoethanol, 40 mg of SDS/ml, 2 mg of bromophenol blue/ml, 20% [vol/vol] glycerol), and the proteins were subjected to gel electrophoresis. The proteins on the gel were detected by Coomassie blue staining, followed by autoradiography. The radioactive products were analyzed with a Bio-Rad FX Molecular Imager system, with Quantity One software.
For coprecipitation assays performed in the presence of oligonucleotides, the procedures described above were modified to omit micrococcal nuclease from buffer SN during the incubation. The oligonucleotides were diluted in TE (10 mM Tris-HCl [pH 7.6], 1 mM EDTA [pH 8.0]) to the necessary concentration, and a standard volume of 5 μl of the oligonucleotide-TE solution was added to the precipitation reaction mixtures.
ELISA-based protein interaction assays.
ELISAs were carried out in 96-well vinyl plates at room temperature as described previously (27). Briefly, wells were coated for 1 h with purified proteins (as indicated in the figure legends) in 50 μl of TBS (25 mM Tris-HCl [pH 7.4], 150 mM NaCl). The wells were then washed with TBST (TBS with 0.1% [vol/vol] NP-40 or Triton X-100) and blocked with 190 μl of 5% (wt/vol) dry milk and 2% (vol/vol) calf serum in TBST for an hour. After being washed with TBST, various amounts of the secondary proteins (as indicated in the figure legends) were added to the wells in 50 μl of TBST supplemented with 1 mM MgCl2, 1 mM CaCl2, and 20 U of micrococcal nuclease/ml and incubated for 1 h. The plates were washed in TBST and incubated for 1 h with the appropriate primary antibody specific for the challenging protein (as indicated in the figure legends), diluted in 50 μl of TBST with 0.5% (wt/vol) dry milk and 1% (vol/vol) calf serum. The plates were then washed in TBST and incubated for 1 h with the appropriate horseradish peroxidase-conjugated secondary antibody diluted in 50 μl of TBST with 0.5% (wt/vol) dry milk and 1% (vol/vol) calf serum. After being washed five times with TBST and four times with TBS, the wells were incubated with 50 μl of visualization buffer (110 mM sodium acetate [pH 5.5] containing 0.02 mg of TMB/ml and 0.0075% [vol/vol] hydrogen peroxide). After 10 min, the reactions were stopped by the addition of an equal volume of 2 M sulfuric acid. The assays were then quantified spectrophotometrically by absorbance at 450 nm.
For ELISAs performed in the presence of oligonucleotides, the procedures described above were modified to include an additional step. The secondary proteins were preincubated at 25°C for 15 min with various amounts of oligonucleotide (as indicated in the figures) before being added to the blocked wells. For oligonucleotide-containing assay mixtures, MgCl2, CaCl2, and micrococcal nuclease were omitted from the buffers.
Electrophoretic mobility shift assays (EMSAs).
Oligonucleotides of various lengths were labeled with [γ-32P]ATP by using T4 polynucleotide kinase (New England Biolabs) in accordance with the manufacturer's instructions. For each reaction, 2 fmol of the radiolabeled oligonucleotide was incubated with various amounts of protein(s) (as indicated in the figure legends) in 25 mM Tris (pH 7.5)-10% (vol/vol) glycerol-5 mg of acetylated BSA/ml-100 mM NaCl-1 mM dithiothreitol for 15 min at room temperature. Glutaraldehyde was then added to a final concentration of 0.2% (vol/vol), and the reaction mixture was incubated for an additional 15 min. The final products were resolved by electrophoresis on a discontinuous (5 and 15% [wt/vol] 16:1 acrylamide/bis-acrylamide) native polyacrylamide gel in 0.5× Tris-borate-EDTA electrophoresis buffer (TBE) at 4°C. The resultant gels were dried, and the radioactive products were analyzed with a Bio-Rad FX Molecular Imager system, with Quantity One software. Each assay was repeated at least six times. The figures depict data from representative experiments.
RESULTS
E1 interacts with the major ssDNA-binding domain of RPA.
We have previously reported that, like the SV40 large Tag, the papillomavirus E1 protein also interacts with RPA. Of the three subunits of RPA, we detected an interaction only between E1 and the largest subunit of RPA, RPA70 (27). To better understand the role of this interaction in viral DNA replication, we have further mapped the region within RPA70 that is required for the interaction with E1. Full-length and truncated RPA proteins were purified and tested for their ability to interact with E1 in coprecipitation and ELISA-based protein interaction assays. Since the carboxyl terminus of RPA70 is required for the assembly of the RPA heterotrimer, mutants with truncations in the carboxyl-terminal domain of RPA70 were expressed in the absence of RPA32 and RPA14 (23, 24). Previous studies have also shown that the expression of the carboxyl terminus of RPA70 in the absence of either RPA32 or RPA14 renders the protein insoluble (24). Mutants that retained the carboxyl-terminal domain of RPA70 were therefore coexpressed with RPA32 and RPA14.
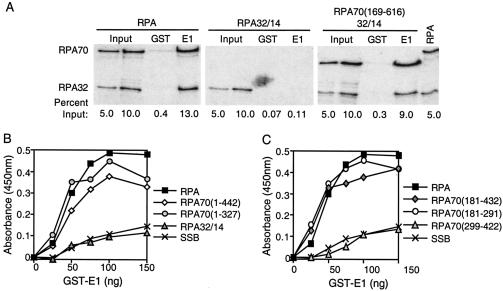
For the coprecipitation assays, bacterially expressed GST-E1 or GST proteins were purified on glutathione Sepharose beads. These beads were then incubated with various 35S-labeled RPA proteins at 4°C in the presence of micrococcal nuclease to ensure that the interactions were not mediated by DNA. The precipitates from these incubations were washed, separated by gel electrophoresis, and analyzed by phosphorimager analysis. Results from the coprecipitation assays are summarized in Fig. 1. Using GST-E1 Sepharose beads, we were able to coprecipitate comparable amounts of full-length RPA and RPA mutants with truncations within the first 168 amino acids of RPA70 (Fig. 2A). In contrast, we were unable to coprecipitate significant amounts of any of the RPA proteins with GST Sepharose beads. Neither the GST nor the GST-E1 Sepharose beads were capable of precipitating RPA32/14, a mutant that lacks RPA70. RPA mutants with truncations of the carboxyl terminus of RPA70 (and without the RPA32 and RPA14 subunits) were especially sensitive to degradation during the coprecipitation process (data not shown). Hence, the efficiencies with which these proteins were coprecipitated by GST-E1 were difficult to quantitate. Nonetheless binding to GST-E1 by proteins expressed from any construct encoding amino acid residues 181 to 291 was substantially higher than binding of the negative controls (Fig. 1).
FIG. 2.
E1 interacts with DBD-A of RPA's major ssDNA-binding domain. (A) Bacterially expressed GST or GST-E1 proteins were purified on glutathione Sepharose beads. These beads were incubated with 0.8 to 1.0 μCi of radiolabeled full-length or truncated RPA proteins in the presence of micrococcal nuclease for 3 h at 4°C. The beads were precipitated by centrifugation, washed, and subjected to SDS-PAGE. RPA retained by the beads was visualized by phosphorimager analysis, and the intensity of the bands was quantitated using Quantity One software. Each assay was repeated at least three times. The results depicted were taken from a representative experiment. The lanes marked “Input” consist of 5 or 10% of radiolabeled RPA used in each precipitation that was directly subjected to SDS-PAGE without precipitation. The lanes marked “GST” contain proteins precipitated using GST-bound glutathione Sepharose beads. The lanes marked “E1” contain proteins precipitated using GST-E1-coated glutathione Sepharose beads. The lane marked “RPA” contains the radiolabeled full-length RPA, which was directly subjected to SDS-PAGE. The 70- and 32-kDa subunits of full-length RPA are labeled RPA70 and RPA32, respectively. Due to its small size and low level of labeling, RPA14 is not shown. The percentages on the bottom of each figure are the percent radioactive RPA that was loaded or precipitated in that lane compared to the total RPA added to each reaction mixture. (B) Full-length RPA, RPA mutants with truncations of the RPA70 carboxyl terminus, RPA32/14, and SSB were tested for their ability to bind to BPV1 E1 in ELISA-based protein interaction assays. The carboxyl terminus of RPA70 was truncated up to amino acid residues 442 and 327 in the RPA mutants RPA70(1-442) and RPA70(1-327), respectively. These mutants were expressed without RPA32 or RPA14. The proteins were immobilized in ELISA wells in equimolar amounts (4 pmol). The wells were blocked and then challenged with increasing concentrations of GST-E1. The wells were washed, and the retained GST-E1 was measured as described in Materials and Methods. (C) Full-length RPA; RPA mutants consisting of the major ssDNA-binding domain of RPA [RPA70(181-432)], DBD-A [RPA70(181-291)], or DBD-B [RPA70(299-422)]; and SSB were also tested for their ability to bind to BPV1 E1 in ELISA-based protein interaction assays. Four picomoles of each protein was immobilized in ELISA plate wells. The wells were blocked and then challenged with increasing concentrations of GST-E1. The wells were washed, and the retained GST-E1 protein was detected as described above. Each of these assays was performed at least three times. Panels B and C each depict data from a representative experiment.
A number of the RPA truncation mutants were also tested for their ability to interact with E1 in ELISA-based protein interaction assays. Results from these assays are also summarized in Fig. 1. Equimolar amounts of purified full-length or truncated RPA proteins were immobilized in the wells of ELISA plates, which were then blocked. A GST-tagged BPV1 E1 fusion protein was then titrated into the wells in the presence of micrococcal nuclease to ensure the removal of trace DNA that might mediate protein interactions. Unbound protein was removed by washing, and the GST-E1 that remained bound to RPA was detected using a GST-specific polyclonal antibody. Binding of GST-E1 to the negative control, E coli ssDNA-binding protein (SSB), remained near background levels (Fig. 2B). In contrast, the amount of GST-E1 retained by full-length RPA increased as a function of the GST-E1 concentration. The levels of interaction between GST-E1 and RPA mutants with truncations of either the amino or carboxyl terminus of RPA70 were similar to that observed between GST-E1 and full-length RPA (Fig. 2B; summarized in Fig. 1). In contrast, GST-E1 showed a level of binding to RPA32/14 that was equivalent to that observed with the negative control, E coli SSB. Purified GST was also tested for its ability to bind to full-length RPA, RPA truncation mutants, and SSB but showed no appreciable binding to any of these proteins. E1 protein affinity chromatography was also used to evaluate which RPA truncations were capable of binding to E1. Results of the affinity chromatography assay were consistent with those shown for the ELISA and coprecipitation studies (data not shown).
The results from both the coprecipitation assay and the ELISA confirm our previous finding that E1 interacts with RPA through the 70-kDa subunit of RPA. They also suggest that neither the amino nor the carboxyl terminus of RPA70 is required for the interaction with E1. Upon closer inspection, the major ssDNA-binding domain of RPA spans almost the entire length of the region involved in the interaction with E1 (Fig. 1). This domain is composed of two ssDNA-binding motifs, DBD-A and DBD-B (4, 23, 58, 59). To determine if the entire domain or if either of the two motifs is sufficient for the interaction with E1, three RPA70 truncation mutants were tested for their ability to interact with GST-E1 in ELISAs. The amount of GST-E1 retained by RPA70(181-432) increased as a function of GST-E1 concentration (Fig. 2C). The level of interaction was similar to that observed between GST-E1 and full-length RPA. GST-E1 also showed binding to RPA70(181-291). In contrast, GST-E1 showed a level of binding to RPA70(299-422) that was equivalent only to that observed with the negative control, E coli SSB. Together, our data indicate that E1 interacts with RPA through the major ssDNA-binding domain of RPA and also suggest that DBD-A alone is sufficient for binding to E1.
Tag, the functional equivalent of E1 in SV40 DNA replication, has been similarly reported to interact with the major ssDNA-binding domain of RPA (27, 30). However, it has also been reported that interactions exist between Tag and the 32-kDa subunit of RPA (30, 37). In our experiments, no measurable interaction between E1 and the two smaller subunits of RPA was ever detected (Fig. 1 and 2) (27). It is possible that such an interaction might exist but is beyond the limits of detection with the methods used in this study.
RPA binding to PV E1 is inhibited by ssDNA.
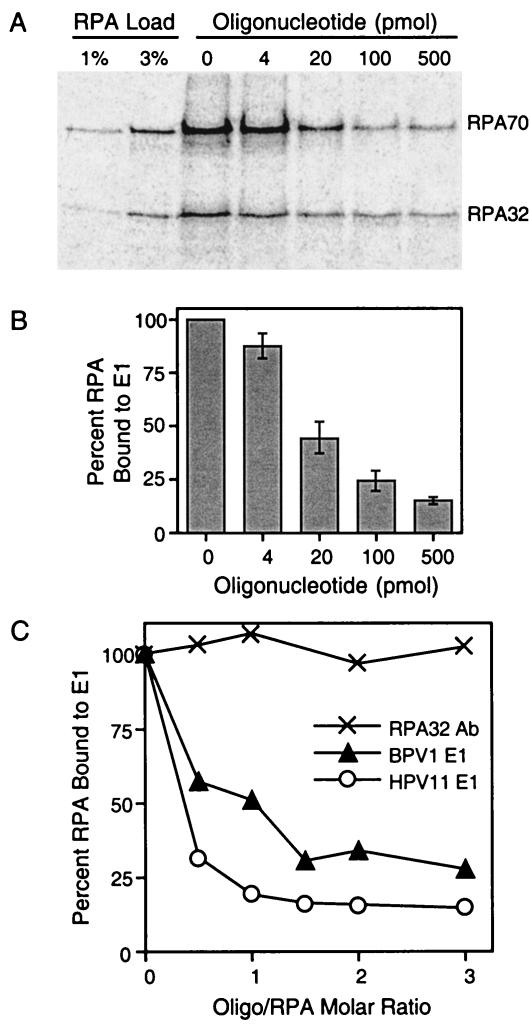
Since BPV1 E1 interacts with the major ssDNA-binding domain of RPA, we addressed whether RPA could bind to ssDNA and E1 simultaneously. Interactions among E1, RPA, and ssDNA were examined using a modified coprecipitation assay. 35S-labeled full-length RPA was incubated with GST-E1 Sepharose beads in the presence of increasing concentrations of a 31-nt oligonucleotide of arbitrary sequence. The beads were washed and subjected to SDS-PAGE. The RPA retained by the beads was then detected using phosphorimager analysis. The amount of 35S-RPA that was precipitated by GST-E1 Sepharose beads decreased as a function of the oligonucleotide concentration (Fig. 3A). Quantitation (as described in Materials and Methods) showed a more than 50% decrease in the amount of 35S-RPA precipitated by GST-E1 when the reactions were carried out in the presence of 40 nM (20 pmol) oligonucleotide (Fig. 3B).
FIG. 3.
RPA binding to ssDNA interferes with the E1-RPA interaction. (A) An 0.8-μCi amount of 35S-labeled RPA was incubated with GST-E1 Sepharose beads and various concentrations of a 31-nt oligonucleotide for 3 h at 4°C. The beads were washed and subjected to SDS-PAGE. The RPA retained by the beads was detected using phosphorimager analysis. Only the RPA70 and RPA32 subunits are shown. For lanes 1 and 2 (RPA input), approximately 1 and 3% of the total RPA used in each precipitation reaction were subjected to SDS-PAGE directly, omitting the E1 precipitation step. (B) The results from panel A were subjected to phosphorimager analysis and quantified with the Quantity One software. The radioactivity of both RPA32 and RPA70 from each lane was quantified and compared to the radioactivity of the RPA that was precipitated by GST-E1 in the absence of oligonucleotide (panel A, lane 3), which was assigned a value of 100%. The error bars depict the standard deviation for at least three separate experiments. (C) Purified BPV1 E1 or HPV11 E1 (150 ng) was immobilized in ELISA wells. The wells were blocked and then challenged with 4 pmol of RPA that had been preincubated with increasing concentrations of a 31-nt ssDNA consisting of an arbitrary sequence. The wells were washed, and the retained RPA was detected with a monoclonal antibody to RPA32. As a control, RPA that had been preincubated with the 31-nt oligonucleotide was also immobilized in the ELISA wells and detected with the antibody to RPA32. The assay was repeated three times with similar results. Results of a representative experiment are shown.
Similar results were obtained using a modified ELISA-based interaction assay. Purified BPV1 E1 was immobilized in the wells of ELISA plates, which were then blocked. These wells were then incubated with purified full-length RPA that had been preincubated with various concentrations of a 31-nt oligonucleotide of arbitrary sequence. Unbound RPA was removed by extensive washing. RPA that was retained by E1 was detected with an RPA32-specific monoclonal antibody. The amount of RPA retained by BPV1 E1 decreased as a function of oligonucleotide concentration (Fig. 3C). To determine if this oligonucleotide-dependent inhibition of RPA binding is limited to the E1 protein from BPV1, we also evaluated E1 from HPV11 for its ability to bind oligonucleotide-bound RPA. The amount of RPA retained by immobilized HPV11 E1 decreased with oligonucleotide concentration in a similar fashion (Fig. 3C). ELISAs were also performed where either the oligonucleotide-RPA preincubation step was omitted or the oligonucleotide was introduced only after the E1-coated wells had been incubated with RPA. In the former case, the RPA retained by E1 still decreased as a function of the oligonucleotide concentration, although to a slightly lesser degree. In the latter case, addition of oligonucleotide had a negligible effect on RPA that was already bound to E1 (data not shown). These results indicate that oligonucleotide binding by RPA interferes with the E1-RPA interaction. A series of important control experiments were also performed. First, RPA was preincubated with various amounts of 32P-labeled oligonucleotide and directly immobilized in ELISA plate wells. The wells were then washed and individually subjected to liquid scintillation counting. The level of 32P detected in these wells increased with increasing oligonucleotide concentration (data not shown). In contrast, little or no 32P was detected in wells that had been incubated with the 32P-labeled oligonucleotide in the absence of RPA. This indicates that the oligonucleotide is still retained by RPA after immobilization but does not bind to the wells of ELISA plates alone, as expected. Next, RPA was preincubated with the unlabeled oligonucleotide and then directly immobilized in ELISA plate wells. These wells were then blocked, and the immobilized RPA was detected with the RPA32-specific monoclonal antibody. The level of RPA detected remained constant despite preincubation with all levels of oligonucleotide used in these studies (Fig. 3C), indicating that this oligonucleotide-dependent effect was not caused by decreased recognition of the oligonucleotide-bound RPA by the antibody used.
To verify that these oligonucleotide-dependent effects were not caused by double-stranded DNA or secondary structures within the oligonucleotide, the ELISAs were also performed with a 31-nt oligonucleotide consisting entirely of alternating thymines and guanines (TG). Preincubation of RPA with this oligonucleotide also efficiently inhibited the E1-RPA interaction (Fig. 4).
FIG. 4.
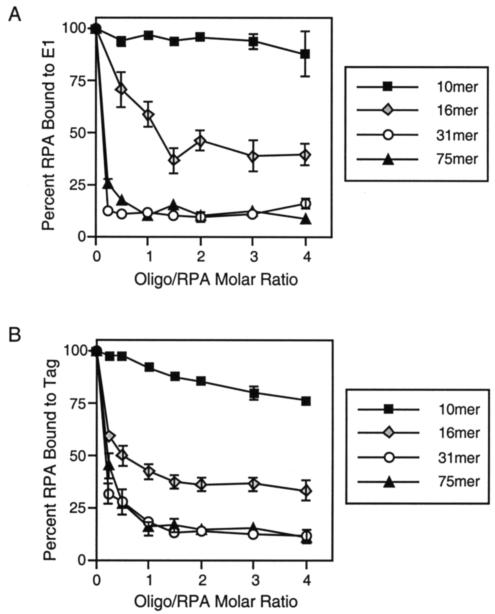
The ssDNA-dependent inhibition of the E1-RPA interaction varies with oligonucleotide length. Either purified BPV1 E1 (A) or SV40 Tag (B) was immobilized in ELISA wells. The wells were blocked and incubated with RPA that had been preincubated with increasing concentrations of a 10-, 16-, 31-, or 75-nt oligonucleotide consisting of alternating thymines and guanines. The wells were washed, and the retained RPA was detected with a monoclonal antibody to RPA32. The bars depict the standard deviation calculated from at least four separate experiments (symbols without bars indicate that standard deviations were less than the size of the symbol).
Mounting evidence suggests that RPA can adopt different conformations when bound to ssDNA, resulting in each RPA occupying either 8 or 30 nt on ssDNA (reviewed in references 30, 52, and 78). We therefore questioned whether the E1-RPA interaction is inhibited to different extents by ssDNA of different lengths. When RPA was preincubated with increasing concentrations of a 10-nt alternating TG oligonucleotide, RPA binding to E1 appeared to be only mildly inhibited by the ssDNA (Fig. 4A). In contrast, use of a 16-nt alternating TG oligonucleotide resulted in an intermediate level of inhibition, while a 75-nt alternating TG oligonucleotide had an inhibitory effect similar to that observed with the 31-nt oligonucleotide. Together, our results suggest that the oligonucleotide-dependent inhibition of the E1-RPA interaction varies with the length of the oligonucleotide but appears not to be affected by the sequence of the ssDNA. The interaction between SV40 Tag and RPA was also inhibited by ssDNA in a length-dependent manner similar to that observed with E1 (Fig. 4B).
RPA competes with E1 for binding to a 31-nt ssDNA oligonucleotide.
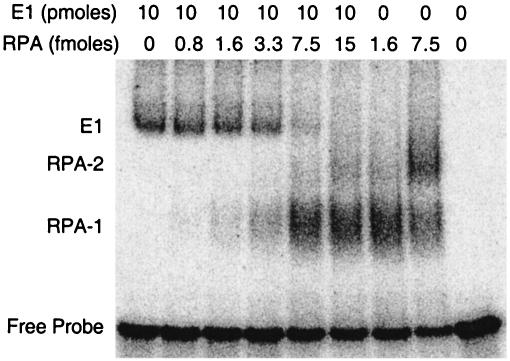
Having demonstrated that ssDNA inhibits the interaction between E1 and RPA, we wanted to verify whether RPA binding of E1 and that of ssDNA are mutually exclusive. We further questioned whether RPA could compete with E1 for the binding of ssDNA. To address this, the complex interactions among RPA, E1, and ssDNA were further explored using ssDNA EMSAs. An oligonucleotide 31 nt in length was radiolabeled with 32P and incubated with purified E1 and/or RPA. The proteins and the oligonucleotide probe were allowed to associate briefly before being incubated with glutaraldehyde at a final concentration of 0.2% for 10 min at room temperature. The products were resolved on native polyacrylamide gels, and the radioactive complexes were analyzed by autoradiography. Glutaraldehyde is an amine-amine cross-linker that was added to the reaction mixtures to capture transient complexes. The binding of BPV1 E1 to the radiolabeled probe resulted in a radioactive complex of lower mobility (Fig. 5, E1). The addition of increasing concentrations of RPA to this complex resulted in the gradual disappearance of the E1-probe complex, with the concomitant appearance of a faster-migrating RPA-probe complex (RPA-1). The addition of higher concentrations of RPA resulted in the appearance of a second RPA-probe complex (RPA-2) and the complete disappearance of the E1-probe complex. At even higher concentrations of RPA, the RPA-1 complex gradually decreased, while the RPA-2 complex increased in intensity. The components of each of the complexes were confirmed with the addition of antibodies. The addition of the polyclonal antibody to GST further retarded the mobility of the E1-probe complex but did not have any observable effect on the mobility of the RPA-probe complexes. Conversely, the addition of the monoclonal antibody to RPA32 further retarded the mobility of both the RPA-1 and RPA-2 complexes but did not have any observable effect on the mobility of the E1-probe complex (data not shown).
FIG. 5.
RPA outcompetes E1 for the binding of ssDNA. A radiolabeled 31-nt oligonucleotide (2 fmol) was incubated at 25°C for 20 min with purified BPV1 E1 (10 pmol), RPA (ranging from 0 to 15 fmol), or both proteins, as indicated. The reaction mixtures were cross-linked with glutaraldehyde and subjected to nondenaturing PAGE and phosphorimager analysis. E1 (on the left) indicates the migration of the probe with E1 alone. RPA-1 indicates the migration of the probe with RPA alone, while RPA-2 indicates a second shift of the probe with RPA when higher concentrations of RPA were used. The migration of the probe alone is also indicated (Free Probe).
These results indicate that RPA does indeed compete with E1 for binding to the 31-nt probe. Note that far less RPA (femtomole levels) was required to compete with E1 (picomole levels) for binding to the probe. This is presumably due to RPA's high affinity for ssDNA and/or the possibility that E1 might have to multimerize to bind ssDNA. Despite the addition of glutaraldehyde to the reaction mixtures, we were unable to detect the presence of a three-component RPA-E1-probe complex. These data show that E1 and RPA cannot simultaneously occupy a 31-nt stretch of ssDNA. The lack of a three-component RPA-E1-probe complex also suggests that E1 and RPA cannot bind to each other while one is bound to ssDNA. This is consistent with the results from Fig. 3 and 4, showing that RPA binding to ssDNA prevents the E1-RPA interaction. However, we cannot completely rule out the possibility that a transient RPA-E1-ssDNA complex exists but is beyond the limits of detection with EMSA, even with the addition of glutaraldehyde. Curiously, RPA consistently showed preferential binding to E1-bound probe, since the E1-probe complex was competed by RPA before the free probe was fully depleted. This suggests that E1 may play a role in loading RPA onto ssDNA. Similar results were seen with both an oligonucleotide with an arbitrary sequence and an oligonucleotide consisting of alternating thymines and guanines (data not shown).
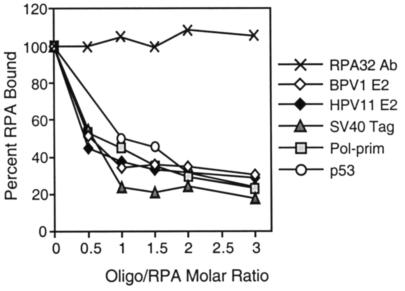
RPA binding to PV E2 proteins, SV40 Tag, human pol-prim, and p53 is also inhibited by ssDNA.
Many proteins that interact with RPA were shown to bind to RPA outside the major ssDNA-binding domain. Hence, RPA was evaluated for its ability to bind to a number of cellular and viral proteins in the presence of the 31-nt oligonucleotide by the modified interaction ELISA. Purified E2 from BPV1 and HPV11, Tag from SV40, human pol-prim, and p53 were each immobilized in ELISA wells. These wells were blocked and challenged with RPA that had been preincubated with increasing concentrations of oligonucleotide. Bound RPA was again detected using the monoclonal antibody to RPA32. RPA that was retained by immobilized p53 decreased as a function of oligonucleotide concentration (Fig. 6), consistent with results previously published by Miller et al. (53). Although the effects varied slightly in degree, the amount of RPA that remained bound to the other immobilized proteins also decreased as a function of oligonucleotide concentration. Similar results were observed when the assay was performed with an oligonucleotide of arbitrary sequence or an oligonucleotide consisting of alternating thymines and guanines (data not shown). This suggests that, like the interaction of RPA with E1 or p53, the binding of ssDNA also interferes with RPA's ability to interact with PV E2, SV40 Tag, and human pol-prim. As shown previously, the binding of RPA by the RPA32 monoclonal antibody was not inhibited by the presence of the oligonucleotide.
FIG. 6.
ssDNA binding prevents RPA interactions with multiple proteins. Purified BPV1 E2, HPV11 E2, SV40 Tag, human pol-prim, and p53 (150 ng) were immobilized in ELISA wells. The wells were blocked and challenged with RPA that had been preincubated with increasing concentrations of a 31-nt oligonucleotide. The wells were washed, and the bound RPA was detected with a monoclonal antibody to RPA32. As a control, RPA that had been preincubated with ssDNA was also immobilized in the ELISA wells and detected with the antibody to RPA32. Detection of control RPA remained constant despite increasing amounts of ssDNA bound. This assay was performed at least three times. This figure depicts data from a representative experiment.
DISCUSSION
In this study, we demonstrated that E1 binds to the major ssDNA-binding domain of RPA, which is located within the central region of RPA70 (23, 24, 33, 40). Using modified coprecipitation assays and ELISAs, we showed that the binding of E1 by RPA is inhibited by ssDNA. It is unclear at this time how the interaction between E1 and RPA might be disrupted by ssDNA. Two likely possibilities are that ssDNA and E1 both compete for a similar region within RPA and, as such, ssDNA binding may physically obstruct E1 from binding to RPA and that ssDNA binding may alter RPA's conformation in such a way as to render it less efficient in binding to E1.
The idea that ssDNA binding might physically block E1 from binding to RPA is consistent with results from the EMSAs. Despite the use of glutaraldehyde, no E1-RPA-oligonucleotide complexes were ever observed. This suggests that RPA cannot bind to E1 and oligonucleotide simultaneously. However, from our experiments, we also know that the ssDNA-induced inhibition of the E1-RPA interaction is dependent on oligonucleotide length. Shorter stretches of oligonucleotides were less efficient in disrupting the interaction than were longer stretches of oligonucleotides. Since E1 binds to the major ssDNA-binding domain of RPA, and since this domain of RPA is known to occupy approximately 8 nt of ssDNA when bound, 10-, 16-, 31-, and 75-nt oligonucleotides should be equally effective at obstructing E1's access to its binding site on RPA. As this is not the case, we believe that the inhibition of the E1-RPA interaction is likely not caused by the mere obstruction of the E1-binding site by ssDNA. However, as the difference in inhibition by the four oligonucleotides could be explained by RPA's stronger affinity for longer oligonucleotides, we cannot completely rule out the possibility that ssDNA binding by RPA obstructs the E1-binding site on RPA.
We believe the second explanation to be more likely, that ssDNA binding by RPA alters RPA's conformation such that it no longer binds E1 efficiently. Consistent with this hypothesis, RPA that is free of ssDNA exhibits a proteolysis profile that differs significantly from that of RPA that is bound to ssDNA, suggesting that RPA is capable of adopting different conformations when it is associated with ssDNA (2, 8, 22). Computer modeling of ssDNA-bound RPA based on its crystal structures also predicts conformational changes in RPA upon ssDNA binding (5, 8). In addition, RPA was shown to bind to ssDNA in two distinct modes in EMSAs: one involving a minimum binding site on ssDNA of approximately 8 nt and a second mode involving a minimum binding site on ssDNA of 30 nt (1, 2). Together, these studies suggest that ssDNA binding by RPA can indeed alter RPA's conformation and that these alterations are dependent on the length of the ssDNA bound by RPA.
From our experiments, we know that ssDNA-bound RPA is less efficient in binding E1 than is RPA not bound to ssDNA. The efficiency with which the interaction is inhibited depends on the length of the ssDNA. RPA's interaction with Tag, which likewise binds to the major DNA-binding domain of RPA, is also inhibited by ssDNA. RPA was also evaluated for its ability to interact with pol-prim, p53, and PV E2 in the presence of ssDNA. Although an interaction between E2 and RPA has been demonstrated (38), the E2-binding site within RPA has yet to be mapped. In contrast, pol-prim and p53 interact with different regions of RPA that do not all coincide with ssDNA-binding domains of RPA (9, 16, 40, 57). But like E1 and Tag, they all show decreased binding to RPA in the presence of ssDNA. It is unlikely that ssDNA can physically obstruct all these sites on RPA. Therefore, it seems likely that the reduced ability of RPA to interact with E1 and these other proteins is due to conformational changes induced by ssDNA binding.
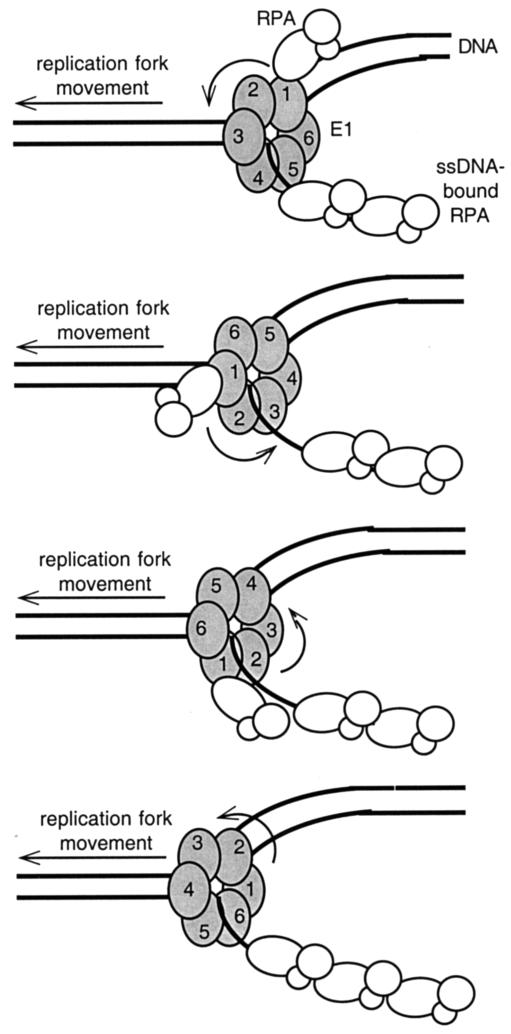
Based on our results, we propose the following model for RPA recruitment by the PV E1 protein (Fig. 7). We have shown that RPA that is not bound to ssDNA can be bound by E1 (Fig. 3). During viral DNA replication, E1 may bind to this free RPA, thereby helping target RPA to either a viral origin of replication or an active replication fork. It has been proposed that hexameric DNA helicases rotate as they unwind duplex DNA (60). Rotation of the E1 hexamer would as a natural consequence bring any E1-bound RPA into greater proximity with the exposed ssDNA generated behind the E1 helicase. As RPA binds to ssDNA, its conformation becomes altered, and we have shown that RPA bound to ssDNA does not bind well to E1 (Fig. 3). This could result in RPA's release from the moving E1. Additional free RPA may be simultaneously bound and recruited by the other subunits of E1, allowing for the continuous but specific placement of RPA by the E1 helicase on the ssDNA generated at active replication forks. E1 would thus play an active role in regulating RPA's involvement and activities in PV DNA replication. Consistent with this model, RPA appears to bind preferentially to E1-bound radiolabeled ssDNA probes in EMSAs despite the ready availability of unbound probe (Fig. 5). In further investigation of this model, our laboratory is currently evaluating whether E1 has to assemble into a hexamer before interacting with RPA. Since the interaction between RPA and Tag, the functional equivalent of E1 in SV40 DNA replication, is similarly disrupted by ssDNA, we propose that Tag may also function to recruit RPA in much the same way as suggested in our model for E1.
FIG. 7.
Proposed model of RPA recruitment by E1 or Tag during viral DNA replication. During viral DNA replication, free RPA is in a conformation that favors interaction with the viral helicase. It associates with the viral helicase and is brought into closer contact with ssDNA (lagging strand) as the helicase rotates during the unwinding of duplex DNA. As RPA binds to the exposed ssDNA generated by the unwinding of the duplex DNA, its conformation is altered, which results in its release from the moving helicase. As the first RPA is placed on the ssDNA at the replication fork, additional free RPA can be simultaneously bound and recruited by the other subunits of the helicase, allowing for continuous recruitment and placement of RPA at the replication fork by the viral DNA helicase complex.
Our results also suggest that changes in RPA's conformations induced by ssDNA binding can modulate RPA's ability to interact with other proteins, including E1 and Tag during PV and SV40 DNA replication. However, it appears that this inhibition of RPA's interactions is not universal, since the binding of ssDNA by RPA has also been shown to stimulate RPA's association with and phosphorylation by the human DNA-dependent protein kinase (2) and by the related checkpoint kinase ATR/ATRIP (17, 81). Indeed, there is increasing evidence suggesting that many ssDNA-binding proteins may have evolved to respond to ssDNA binding in much the same way. For instance, during the SOS response in E coli, stalled replication forks induced by DNA damage promote ssDNA binding by the RecA protein, resulting in its activation (41). Active RecA is then available to bind to inactive LexA, thereby promoting the self-cleavage and activation of LexA. Activated LexA can then relieve the transcriptional repression normally imposed on SOS genes, allowing for the increased production of DNA repair factors. In the same system, DinI, a RecA regulator, has been shown to interact with activated RecA but not free RecA (80). In bacteriophage RB69, it is suggested that ssDNA binding by phage SSB induces a conformational change in SSB that allows it to associate specifically with adjacent SSB molecules and with its DNA polymerase, gp43 (70). These specific interactions are thought to be important for bacteriophage DNA replication. Similarly to the PV DNA replication model that we propose here, the ssDNA-binding protein of herpes simplex virus type 1, ICP-8, is released from the origin-binding protein and helicase, OBP, only when it associates with ssDNA (25). Together, these findings suggest that ssDNA binding may be a common mechanism for regulating the activities and protein-protein interactions of ssDNA-binding proteins in many different systems.
Acknowledgments
This work was supported by National Institutes of Health grant GM56406 (to T.M.) and a Mark Diamond Student Research Grant (to Y.-M.L.). T.M. was also supported by an NIH career development award, AI01686.
We thank A. Bochkarev and M. Wold for the RPA expression vectors and proteins, J.-S. Liu and S.-R. Kuo for the HPV11 E1 and E2 expression vectors, and M. Avantagiatti for the p53 protein. We thank J.-S. Liu and W. Zhang for technical assistance in purifying some of the proteins used in this work. We thank M. Sutton for critical reading of the manuscript. We also thank J.-S. Liu, S.-R. Kuo, E. Fanning, L. A. Henricksen, J. C. Fisk, J. Huberman, G. Brown, and members of the Witebsky Center for Microbial Pathogenesis and Immunology for helpful scientific discussions.
REFERENCES
- 1.Blackwell, L. J., and J. A. Borowiec. 1994. Human replication protein A binds single-stranded DNA in two distinct complexes. Mol. Cell. Biol. 14:3993-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackwell, L. J., J. A. Borowiec, and I. A. Mastrangelo. 1996. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol. Cell. Biol. 16:4798-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bochkarev, A., E. Bochkareva, L. Frappier, and A. M. Edwards. 1999. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 18:4498-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochkarev, A., R. A. Pfuetzner, A. M. Edwards, and L. Frappier. 1997. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 385:176-181. [DOI] [PubMed] [Google Scholar]
- 5.Bochkareva, E., V. Belegu, S. Korolev, and A. Bochkarev. 2001. Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J. 20:612-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bochkareva, E., L. Frappier, A. M. Edwards, and A. Bochkarev. 1998. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J. Biol. Chem. 273:3932-3936. [DOI] [PubMed] [Google Scholar]
- 7.Bochkareva, E., S. Korolev, and A. Bochkarev. 2000. The role for zinc in replication protein A. J. Biol. Chem. 275:27332-27338. [DOI] [PubMed] [Google Scholar]
- 8.Bochkareva, E., S. Korolev, S. P. Lees-Miller, and A. Bochkarev. 2002. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 21:1855-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonne-Andrea, C., S. Santucci, P. Clertant, and F. Tillier. 1995. Bovine papillomavirus E1 protein binds specifically DNA polymerase alpha but not replication protein A. J. Virol. 69:2341-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brill, S. J., and S. Bastin-Shanower. 1998. Identification and characterization of the fourth single-stranded-DNA binding domain of replication protein A. Mol. Cell. Biol. 18:7225-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brill, S. J., and B. Stillman. 1991. Replication factor A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 5:1589-1600. [DOI] [PubMed] [Google Scholar]
- 12.Brill, S. J., and B. Stillman. 1989. Yeast replication factor A functions in the unwinding of the SV40 origin of DNA replication. Nature 342:92-95. [DOI] [PubMed] [Google Scholar]
- 13.Bullock, P. A. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32:503-568. [DOI] [PubMed] [Google Scholar]
- 14.Chow, L. T., and T. R. Broker. 1994. Papillomavirus DNA replication. Intervirology 37:150-158. [DOI] [PubMed] [Google Scholar]
- 15.Collins, K. L., and T. J. Kelly. 1991. Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase alpha-primase. Mol. Cell. Biol. 11:2108-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conger, K. L., J. S. Liu, S. R. Kuo, L. T. Chow, and T. S. F. Wang. 1999. Human papillomavirus DNA replication—interactions between the viral E1 protein and two subunits of human DNA polymerase alpha/primase. J. Biol. Chem. 274:2696-2705. [DOI] [PubMed] [Google Scholar]
- 17.Costanzo, V., D. Shechter, P. J. Lupardus, K. A. Cimprich, M. Gottesman, and J. Gautier. 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell 11:203-213. [DOI] [PubMed] [Google Scholar]
- 18.del Mar Pena, L. M., and L. A. Laimins. 2002. Regulation of human papillomavirus gene expression in the vegetative life cycle, p. 31-52. In D. J. McCance (ed.), Human papilloma viruses, 1st ed., vol. 8. Elsevier Science B.V., Amsterdam, The Netherlands.
- 19.Din, S., S. J. Brill, M. P. Fairman, and B. Stillman. 1990. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev. 4:968-977. [DOI] [PubMed] [Google Scholar]
- 20.Dong, J., J. S. Park, and S. H. Lee. 1999. In vitro analysis of the zinc-finger motif in human replication protein A. Biochem. J. 337:311-317. [PMC free article] [PubMed] [Google Scholar]
- 21.Fouts, E. T., X. Yu, E. H. Egelman, and M. R. Botchan. 1999. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274:4447-4458. [DOI] [PubMed] [Google Scholar]
- 22.Gomes, X. V., L. A. Henricksen, and M. S. Wold. 1996. Proteolytic mapping of human replication protein A: evidence for multiple structural domains and a conformational change upon interaction with single-stranded DNA. Biochemistry 35:5586-5595. [DOI] [PubMed] [Google Scholar]
- 23.Gomes, X. V., and M. S. Wold. 1996. Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry 35:10558-10568. [DOI] [PubMed] [Google Scholar]
- 24.Gomes, X. V., and M. S. Wold. 1995. Structural analysis of human replication protein A. Mapping functional domains of the 70-kDa subunit. J. Biol. Chem. 270:4534-4543. [DOI] [PubMed] [Google Scholar]
- 25.Gustafsson, C. M., M. Falkenberg, S. Simonsson, H. Valadi, and P. Elias. 1995. The DNA ligands influence the interactions between the herpes simplex virus 1 origin binding protein and the single strand DNA-binding protein, ICP-8. J. Biol. Chem. 270:19028-19034. [DOI] [PubMed] [Google Scholar]
- 26.Ham, J., N. Dostatni, J. M. Gauthier, and M. Yaniv. 1991. The papillomavirus E2 protein: a factor with many talents. Trends Biochem. Sci. 16:440-444. [DOI] [PubMed] [Google Scholar]
- 27.Han, Y., Y. M. Loo, K. T. Militello, and T. Melendy. 1999. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol. 73:4899-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henricksen, L. A., C. B. Umbricht, and M. S. Wold. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269:11121-11132. (Erratum, 269:16519.) [PubMed] [Google Scholar]
- 29.Hughes, F. J., and M. A. Romanos. 1993. E1 protein of human papillomavirus is a DNA helicase/ATPase. Nucleic Acids Res. 21:5817-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iftode, C., Y. Daniely, and J. A. Borowiec. 1999. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34:141-180. [DOI] [PubMed] [Google Scholar]
- 31.Kenny, M. K., S. H. Lee, and J. Hurwitz. 1989. Multiple functions of human single-stranded-DNA binding protein in simian virus 40 DNA replication: single-strand stabilization and stimulation of DNA polymerases alpha and delta. Proc. Natl. Acad. Sci. USA 86:9757-9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, C., R. O. Snyder, and M. S. Wold. 1992. Binding properties of replication protein A from human and yeast cells. Mol. Cell. Biol. 12:3050-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, D. K., E. Stigger, and S. H. Lee. 1996. Role of the 70-kDa subunit of human replication protein A (I). Single-stranded DNA binding activity, but not polymerase stimulatory activity, is required for DNA replication. J. Biol. Chem. 271:15124-15129. [DOI] [PubMed] [Google Scholar]
- 34.Kuo, S. R., J. S. Liu, T. R. Broker, and L. T. Chow. 1994. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J. Biol. Chem. 269:24058-24065. [PubMed] [Google Scholar]
- 35.Laimins, L. A. 1998. Regulation of transcription and replication by human papillomaviruses, p. 201-223. In D. J. McCance (ed.), Human tumor viruses. ASM Press, Washington, D.C.
- 36.Lanford, R. E. 1988. Expression of simian virus 40 T antigen in insect cells using a baculovirus expression vector. Virology 167:72-81. [DOI] [PubMed] [Google Scholar]
- 37.Lee, S. H., and D. K. Kim. 1995. The role of the 34-kDa subunit of human replication protein A in simian virus 40 DNA replication in vitro. J. Biol. Chem. 270:12801-12807. [DOI] [PubMed] [Google Scholar]
- 38.Li, R., and M. Botchan. 1993. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell 73:1207-1221. [DOI] [PubMed] [Google Scholar]
- 39.Lin, B. Y., A. M. Makhov, J. D. Griffith, T. R. Broker, and L. T. Chow. 2002. Chaperone proteins abrogate inhibition of the human papillomavirus (HPV) E1 replicative helicase by the HPV E2 protein. Mol. Cell. Biol. 22:6592-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, Y. L., C. Chen, K. F. Keshav, E. Winchester, and A. Dutta. 1996. Dissection of functional domains of the human DNA replication protein complex replication protein A. J. Biol. Chem. 271:17190-17198. [DOI] [PubMed] [Google Scholar]
- 41.Little, J. W. 1984. Autodigestion of lexA and phage λ repressors. Proc. Natl. Acad. Sci. USA 81:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, J.-S., and T. Melendy. 2002. Human papillomavirus DNA replication, p. 53-70. In D. J. McCance (ed.), Human papilloma viruses, 1st ed., vol. 8. Elsevier Science B.V., Amsterdam, The Netherlands.
- 43.Liu, J. S., S. R. Kuo, T. R. Broker, and L. T. Chow. 1995. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell-free DNA replication. J. Biol. Chem. 270:27283-27291. [DOI] [PubMed] [Google Scholar]
- 44.Liu, J. S., S. R. Kuo, A. M. Makhov, D. M. Cyr, J. D. Griffith, T. R. Broker, and L. T. Chow. 1998. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J. Biol. Chem. 273:30704-30712. [DOI] [PubMed] [Google Scholar]
- 45.Lusky, M., J. Hurwitz, and Y. S. Seo. 1993. Cooperative assembly of the bovine papilloma virus E1 and E2 proteins on the replication origin requires an intact E2 binding site. J. Biol. Chem. 268:15795-15803. [PubMed] [Google Scholar]
- 46.Ma, T., N. Zou, B. Y. Lin, L. T. Chow, and J. W. Harper. 1999. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc. Natl. Acad. Sci. USA 96:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masterson, P. J., M. A. Stanley, A. P. Lewis, and M. A. Romanos. 1998. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit. J. Virol. 72:7407-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McBride, A. A., H. Romanczuk, and P. M. Howley. 1991. The papillomavirus E2 regulatory proteins. J. Biol. Chem. 266:18411-18414. [PubMed] [Google Scholar]
- 49.Melendy, T., J. Sedman, and A. Stenlund. 1995. Cellular factors required for papillomavirus DNA replication. J. Virol. 69:7857-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Melendy, T., and B. Stillman. 1993. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J. Biol. Chem. 268:3389-3395. [PubMed] [Google Scholar]
- 51.Melendy, T., and B. Stillman. 1992. SV40 DNA replication, p. 129-158. In F. Eckstein and D. M. J. Lilly (ed.), Nucleic acids and molecular biology, vol. 6. Springer Verlag, Berlin, Germany.
- 52.Mer, G., A. Bochkarev, W. J. Chazin, and A. M. Edwards. 2000. Three-dimensional structure and function of replication protein A. Cold Spring Harbor Symp. Quant. Biol. LXV:193-200. [DOI] [PubMed] [Google Scholar]
- 53.Miller, S. D., K. Moses, L. Jayaraman, and C. Prives. 1997. Complex formation between p53 and replication protein A inhibits the sequence-specific DNA binding of p53 and is regulated by single-stranded DNA. Mol. Cell. Biol. 17:2194-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mohr, I. J., R. Clark, S. Sun, E. J. Androphy, P. MacPherson, and M. R. Botchan. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699. [DOI] [PubMed] [Google Scholar]
- 55.Muller, F., Y. S. Seo, and J. Hurwitz. 1994. Replication of bovine papillomavirus type 1 origin-containing DNA in crude extracts and with purified proteins. J. Biol. Chem. 269:17086-17094. [PubMed] [Google Scholar]
- 56.Murphy, C. I., B. Weiner, I. Bikel, H. Piwnica-Worms, M. K. Bradley, and D. M. Livingston. 1988. Purification and functional properties of simian virus 40 large and small T antigens overproduced in insect cells. J. Virol. 62:2951-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park, P., W. Copeland, L. Yang, T. Wang, M. R. Botchan, and I. J. Mohr. 1994. The cellular DNA polymerase alpha-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc. Natl. Acad. Sci. USA 91:8700-8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfuetzner, R. A., A. Bochkarev, L. Frappier, and A. M. Edwards. 1997. Replication protein A. Characterization and crystallization of the DNA binding domain. J. Biol. Chem. 272:430-434. [DOI] [PubMed] [Google Scholar]
- 59.Philipova, D., J. R. Mullen, H. S. Maniar, J. Lu, C. Gu, and S. J. Brill. 1996. A hierarchy of SSB protomers in replication protein A. Genes Dev. 10:2222-2233. [DOI] [PubMed] [Google Scholar]
- 60.Putnam, C. D., S. B. Clancy, H. Tsuruta, S. Gonzalez, J. G. Wetmur, and J. A. Tainer. 2001. Structure and mechanism of the RuvB Holliday junction branch migration motor. J. Mol. Biol. 311:297-310. [DOI] [PubMed] [Google Scholar]
- 61.Sanders, C. M., and A. Stenlund. 1998. Recruitment and loading of the E1 initiator protein: an ATP-dependent process catalysed by a transcription factor. EMBO J. 17:7044-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sedman, J., and A. Stenlund. 1995. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 14:6218-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sedman, J., and A. Stenlund. 1998. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 72:6893-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seo, Y. S., F. Muller, M. Lusky, and J. Hurwitz. 1993. Bovine papilloma virus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc. Natl. Acad. Sci. USA 90:702-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seroussi, E., and S. Lavi. 1993. Replication protein A is the major single-stranded DNA binding protein detected in mammalian cell extracts by gel retardation assays and UV cross-linking of long and short single-stranded DNA molecules. J. Biol. Chem. 268:7147-7154. [PubMed] [Google Scholar]
- 66.Simanis, V., and D. P. Lane. 1985. An immunoaffinity purification procedure for SV40 large T antigen. Virology 144:80-100. [DOI] [PubMed] [Google Scholar]
- 67.Simmons, D. T. 2000. SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res. 55:75-134. [DOI] [PubMed] [Google Scholar]
- 68.Stenlund, A. 1996. Papillomavirus DNA replication, p. 679-697. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 69.Stillman, B., R. D. Gerard, R. A. Guggenheimer, and Y. Gluzman. 1985. T antigen and template requirements for SV40 DNA replication in vitro. EMBO J. 4:2933-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun, S., and Y. Shamoo. 2003. Biochemical characterization of interactions between DNA polymerase and single-stranded DNA-binding protein in bacteriophage RB69. J. Biol. Chem. 278:3876-3881. [DOI] [PubMed] [Google Scholar]
- 71.Tsurimoto, T., T. Melendy, and B. Stillman. 1990. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature 346:534-539. [DOI] [PubMed] [Google Scholar]
- 72.Tsurimoto, T., and B. Stillman. 1989. Multiple replication factors augment DNA synthesis by the two eukaryotic DNA polymerases, alpha and delta. EMBO J. 8:3883-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 74.Walther, A. P., X. V. Gomes, Y. Lao, C. G. Lee, and M. S. Wold. 1999. Replication protein A interactions with DNA. 1. Functions of the DNA-binding and zinc-finger domains of the 70-kDa subunit. Biochemistry 38:3963-3973. [DOI] [PubMed] [Google Scholar]
- 75.Weisshart, K., H. Forster, E. Kremmer, B. Schlott, F. Grosse, and H. P. Nasheuer. 2000. Protein-protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system. J. Biol. Chem. 275:17328-17337. [DOI] [PubMed] [Google Scholar]
- 76.Weisshart, K., P. Taneja, and E. Fanning. 1998. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J. Virol. 72:9771-9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilson, V. G., M. West, K. Woytek, and D. Rangasamy. 2002. Papillomavirus E1 proteins: form, function, and features. Virus Genes 24:275-290. [DOI] [PubMed] [Google Scholar]
- 78.Wold, M. S. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61-92. [DOI] [PubMed] [Google Scholar]
- 79.Yang, L., I. Mohr, E. Fouts, D. A. Lim, M. Nohaile, and M. Botchan. 1993. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 90:5086-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yasuda, T., K. Morimatsu, R. Kato, J. Usukura, M. Takahashi, and H. Ohmori. 2001. Physical interactions between DinI and RecA nucleoprotein filament for the regulation of SOS mutagenesis. EMBO J. 20:1192-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]