Abstract
Five cases of orf virus infection in Korean black goats were diagnosed in our laboratory between 2010 and 2011. One orf virus (ORF/2011) was isolated from an ovine testis cell line (OA3.Ts) for use as a vaccine candidate. Sequences of the major envelope protein and orf virus interferon resistance genes were determined and compared with published reference sequences. Phylogenetic analyses revealed that orf viruses from Korean black goats were most closely related to an isolate (ORF/09/Korea) from dairy goats in Korea. This result indicates that the orf viruses might have been introduced from dairy goats into the Korean black goat population.
Keywords: goat, isolation, orf virus, phylogenetic analysis
Members of the genus Parapoxvirus from the Poxviridae family are oval, enveloped viruses containing a double strand (ds)DNA genome [13]. This group includes the orf virus (ORFV), pseudocowpox virus, and bovine papular stomatitis virus that infect cattle along with the parapoxvirus of red deer in New Zealand. The major viral envelope protein B2L and virus interferon resistance (VIR) genes have been used to measure genetic variation among parapoxvirus since these genes are a highly conserved [2,4,8,9,10]. Contagious ecthyma (contagious pustular dermatitis) is a highly contagious disease that primarily affects sheep, goats, and wild ruminants, and is characterized by the formation of papules, nodules, or vesicles that develop into thick crusts or heavy scabs on the lips, gingiva, and tongue [4,5,13]. Patients with zoonotic ORFV infection develop nodular and papillomatous lesions mainly on the hands, face, and mouth [5,6,13]. The importance of ORFV has increased recently due to emergence of this virus in new territories as well as the occurrence of interspecies infection and reinfection of animals previously infected [7].
An outbreak of ORFV infection among dairy goats in 2009 was reported in Korea [12]. Infection with this virus in black goats had not been previously observed in Korea prior to the import of dairy goats from abroad. In the present study, five ORFVs were isolated from Korean black goats and characterized to evaluate the genetic relationships between these ORFVs and other strains of this virus. The complete B2L and VIR genes were sequenced, and phylogenetic trees were subsequently constructed.
Five suspected cases of contagious ecthyma in Korean black goats were reported in different geographic areas of Korea between 2010 and 2011 (Table 1). The affected goats were 6~12 months old. Heavy scabs on the lips were observed in all the animals. Dried scabs were collected from the infected goats and stored at -70℃ until analysis.
Table 1.
Details of the orf viruses isolated from Korean black goats
PCR with OVS- and OVA-specific primers was performed as previously described [10]. The OVS and OVA primer sequences were 5'-AGGCGGTGGAATGGA AAGA-3' and 5'-CCAGCAGGTATGCCAGGATG-3', respectively. Briefly, total viral DNA was extracted from scab samples using a QIAamp DNA Mini kit (Qiagen, Germany) according to the manufacturer's instructions. Amplification was performed using a GeneAmp PCR System 2700 (Applied Biosystems, USA) under the following conditions: preliminary heating at 94℃ for 5 min; 10 cycles of denaturation at 94℃ for 30 seconds, annealing at 55℃ for 45 seconds, and elongation at 72℃ for 1 min; 20 cycles of denaturation at 94℃ for 30 seconds, annealing at 57℃ for 45 seconds, and elongation at 72℃ for 1 min; and a final elongation step at 72℃ for 5 min.
Virus was isolated from the clinical scab samples of Korean black goats. The scab samples were homogenized mechanically in PBS in a tube using a pellet pestle device (Kimble Chase, USA). Homogenized samples were centrifuged in 4℃ at 3,000 × g for 5 min. The supernatant was then passed through a 0.45-µM filter (Millipore) and used to inoculate ovine testis cells (OA3.Ts) (ATCC). The cell cultures were incubated in Dulbeccos modified Eagles medium (Invitrogen, Carlsbad, CA) at 37℃ in 5% CO2 and three blind passages were performed until cytopathic effects (CPEs) were observed. The ORFV was isolated in OA3.Ts cells and propagated in Madin-Darby bovine kidney (MDBK) cells (ATCC).
Also, the homogenates were centrifuged in 4℃ at 3,000 × g for 5 min. The supernatant was then collected and negatively stained with 2% phosphotungstic acid for electron microscopy.
PCR for the phylogenetic analysis was performed using previously described primers [4,8]. The amplified products were purified using an Agarose Gel DNA Extraction kit (INtRON, Korea) and cloned into a pGEM-T vector (Promega, USA) according to the manufacturer's instructions. Automated nucleotide sequencing of the VP60 gene insert was performed using an ABI 3130XL Genetic Analyzer (Applied Biosystems, USA) with a BigDye Terminator Cycle Sequencing kit (Applied Biosystems, USA). The sequences were confirmed by three or more independent reactions in both directions. Published parapoxvirus sequences were retrieved from GenBank (National Center for Biotechnology Information, USA) for phylogenetic analyses.
B2L and VIR gene sequences of the Korean ORFV strains were aligned with the corresponding parapoxvirus sequences using Bioedit software (Ibis Biosciences, USA). A phylogenetic analysis was conducted using Bioedit software and Molecular Evolutionary Genetics Analysis (MEGA) 4.0 software with bootstrap values calculated from 1,000 replicates [14]. The neighbor-joining method was used for tree construction.
Cases of ORFV infection at five black goat farms were diagnosed and confirmed by PCR and DNA sequencing. The amplified PCR products were separated by electrophoresis and visualized by ethidium bromide staining. The size of the amplified PCR product was 708 bp. One ORFV strain (ORF/2011) was isolated from clinical samples from the infected black goats. CPEs were first observed 6 days after the cell cultures were passed twice, and typical parapoxvirus CPE patterns, indicated by cell rounding, pyknosis, and cell detachment, were observed.
Negatively stained ORFV particles were examined by electron microscopy. The particles had an oval-shaped morphology characteristic of parapoxviruses. Viral particle size was approximately 150~200 nm (data not shown).
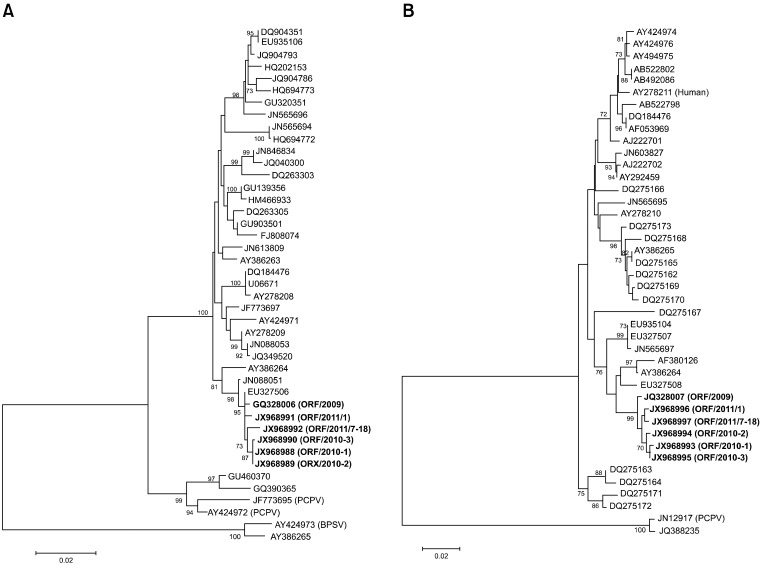
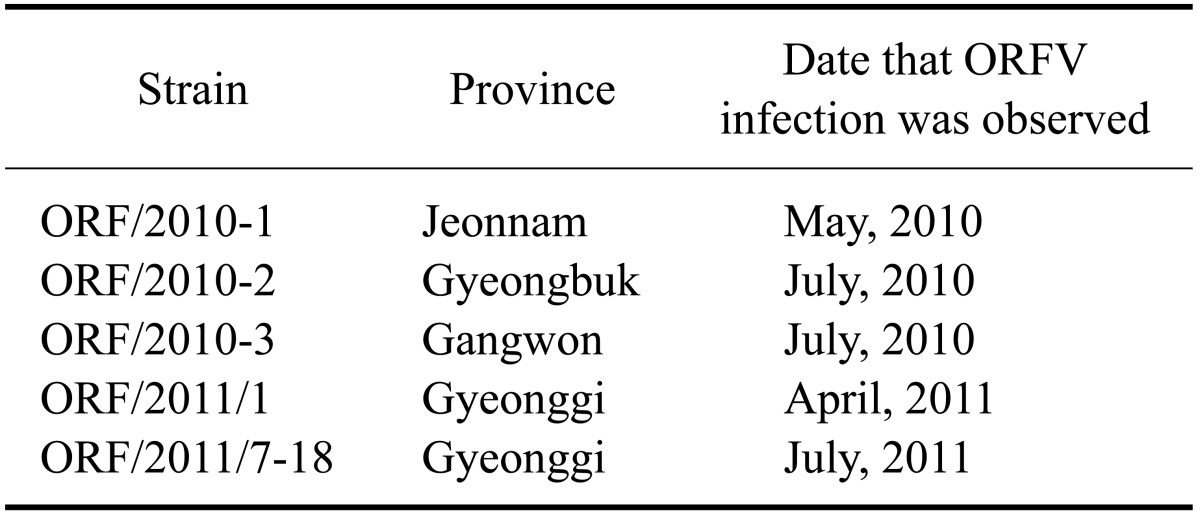
An additional PCR assay was performed to obtain the full-length B2L and partial VIR gene fragments for sequencing. The amplified PCR products were 1206 bp for the full-length B2L gene and 552 bp for the partial VIR gene. The DNA was purified and sequenced for comparative analysis. Sequence comparison of the five Korean ORFV strains (contained one ORFV isolate) identified from Korean black goats demonstrated a high degree of identity between each virus and a previously reported ORF/09/Korea strain [12]. Results of the phylogenetic analysis based on the complete B2L gene (Fig. 1A) demonstrated that the five ORFV strains were most closely related to the ORF/09/Korea strain identified in 2009 and the Taiwan EU327506 strain. The five ORFV strains and ORF/09/Korea strain had a 99.2~99.5% and 98.4~99.2% similarity at the nucleotide and predicted amino acid level, respectively. The phylogenetic analysis based on the partial VIR gene revealed that the five ORFV strains were most closely related to the ORF/09/Korea strain identified in 2009 (Fig. 1B). The five ORFV strains and ORF/09/Korea strain showed a 99.0~99.2% and 98.7% similarity at the nucleotide and amino acid level, respectively.
Fig. 1.
Phylogenetic analysis of different parapoxviruses based on nucleotide sequences of the B2L gene (A) and partial virus interferon resistance gene (B). Nucleotide sequences of different orf viruses were aligned using the Bioedit program and Mega 4 software. One thousand bootstrap replicates were used to determine nucleotide sequence distance. A consensus phylogenetic tree that was created using the neighbor-joining methods is shown.
ORFV is a highly contagious transboundary pathogen. If the virus is introduced into disease-free areas, it infects small ruminants and spreads easily [1]. ORFV infection among the Korean black goat population was not reported until dairy goats were brought into Korea. Dairy goats were initially imported from abroad during the 1990s to produce liquid and powdered milk. The dairy goat population in Korea has increased over the past several years. Several causes for ORFV outbreaks in Korean black goats exist, and a phylogenetic analysis may indicate the hypothetical origin of the viral strains involved.
B2L and VIR gene sequences have been widely used for molecular epidemiological studies of parapoxviruses. These are epidemiologically relevant sequences and a significant quantity of sequence data has been deposited in GenBank [1-4,9,10]. In the present study, the Korean ORFV strains were most closely related to the ORF/09/Korea strain from dairy goats according to the phylogenetic analysis based on the B2L and VIR genes. This result indicates that ORFVs from dairy goats might have been introduced into the Korean black goat population. Based on a molecular analysis alone, it is difficult to determine the precise route by which the ORFV from dairy goats was transmitted to Korean black goats. However, data from the current study suggest that introduction of ORFV into the Korean black goat population might have occurred through dairy goats imported for meat production or improvement of stock genetics.
The vaccine using orf virus isolated from goat may potentially provide better protection for goats than for sheep [11]. We isolated one ORFV as a vaccine candidate from a large-scale outbreak of ORFV infection in Korean goats. A vaccine will be developed in a future study. Additional epidemiological data concerning the distribution of ORFVs among dairy and Korean black goats in Korea will also contribute to the prevention of further ORFV outbreaks.
Acknowledgments
This study was financially supported by the Animal and Plant Quarantine Agency, Ministry of Agriculture, Food and Rural Affairs, Korea.
References
- 1.Bora DP, Barman NN, Das SK, Bhanuprakash V, Yogisharadhya R, Venkatesan G, Kumar A, Rajbongshi G, Khatoon E, Chakraborty A, Bujarbaruah KM. Identification and phylogenetic analysis of orf viruses isolated from outbreaks in goats of Assam, a northeastern state of India. Virus Genes. 2012;45:98–104. doi: 10.1007/s11262-012-0740-y. [DOI] [PubMed] [Google Scholar]
- 2.Chan KW, Lin JW, Lee SH, Liao CJ, Tsai MC, Hsu WL, Wong ML, Shih HC. Identification and phylogenetic analysis of orf virus from goats in Taiwan. Virus Genes. 2007;35:705–712. doi: 10.1007/s11262-007-0144-6. [DOI] [PubMed] [Google Scholar]
- 3.Chan KW, Yang CH, Lin JW, Wang HC, Lin FY, Kuo ST, Wong ML, Hsu WL. Phylogenetic analysis of parapoxviruses and the C-terminal heterogeneity of viral ATPase proteins. Gene. 2009;432:44–53. doi: 10.1016/j.gene.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 4.Guo J, Zhang Z, Edwards JF, Ermel RW, Taylor C, Jr, de la Concha-Bermejillo A. Characterization of a North American orf virus isolated from a goat with persistent, proliferative dermatitis. Virus Res. 2003;93:169–179. doi: 10.1016/s0168-1702(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 5.Haig DM, Mercer AA. Ovine diseases. Orf. Vet Res. 1998;29:311–326. [PubMed] [Google Scholar]
- 6.Haig DM, Thomson J, Mclnnes C, McCaughan C, Imlach W, Mercer A, Fleming S. Orf virus immuno-modulation and the host immune response. Vet Immunol Immunopathol. 2002;87:395–399. doi: 10.1016/s0165-2427(02)00087-9. [DOI] [PubMed] [Google Scholar]
- 7.Hosamani A, Scagliarini A, Bhanuprakash V, McInnes CJ, Singh RK. Orf: an update on current research and future perspectives. Expert Rev Anti infect Ther. 2009;7:879–893. doi: 10.1586/eri.09.64. [DOI] [PubMed] [Google Scholar]
- 8.Hosamani M, Bhanuprakash V, Scagliarini A, Singh RK. Comparative sequence analysis of major envelope protein gene (B2L) of Indian orf viruses isolated from sheep and goats. Vet Microbiol. 2006;116:317–324. doi: 10.1016/j.vetmic.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 9.Kottaridi C, Nomikou K, Teodori L, Savini G, Lelli R, Markoulatos P, Mangana O. Phylogenetic correlation of Greek and Italian orf virus isolates based on VIR gene. Vet Microbiol. 2006;116:310–316. doi: 10.1016/j.vetmic.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Mondal B, Bera AK, Hosamani M, Tembhurne PA, Bandyopadhyay SK. Detection of orf virus from an outbreak in goats and its genetic relation with other parapoxviruses. Vet Res Commun. 2006;30:531–539. doi: 10.1007/s11259-006-3270-z. [DOI] [PubMed] [Google Scholar]
- 11.Musser JMB, Taylor CA, Guo J, Tizard IR, Walker JW. Development of a contagious ecthyma vaccine for goats. Am J Vet Res. 2008;69:1366–1370. doi: 10.2460/ajvr.69.10.1366. [DOI] [PubMed] [Google Scholar]
- 12.Oem JK, Roh IS, Lee KH, Lee KK, Kim HR, Jean YH, Lee OS. Phylogenetic analysis and characterization of Korean orf virus from dairy goats: case report. Virol J. 2009;6:167. doi: 10.1186/1743-422X-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson AJ, Balassu TC. Contagious pustular dermatitis (orf) Vet Bull. 1981;51:771–782. [Google Scholar]
- 14.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]