Abstract
RNA viruses are remarkably adaptable to changing environments. This is medically important because it enables pathogenic viruses to escape the immune response and chemotherapy and is of considerable theoretical interest since it allows the investigation of evolutionary processes within convenient time scales. A number of earlier studies have addressed the dynamics of adapting RNA virus populations. However, it has been difficult to monitor the trajectory of molecular changes in RNA genomes in response to selective pressures. To address the problem, we developed a novel in vitro evolution system based on a recombinant double-stranded RNA bacteriophage, φ6, containing a β-lactamase (bla) gene marker. Carrier-state bacterial cells are resistant to ampicillin, and after several passages, they become resistant to high concentrations of another β-lactam antibiotic, cefotaxime, due to mutations in the virus-borne bla gene. We monitored the changes in bla cDNAs induced by cefotaxime selection and observed an initial explosion in sequence variants with multiple mutations throughout the gene. After four passages, a stable, homogeneous population of bla sequences containing three specific nonsynonymous mutations was established. Of these, two mutations (E104K and G238S) have been previously reported for β-lactamases from cefotaxime-resistant bacterial isolates. These results extend our understanding of the molecular mechanisms of viral adaptation and also demonstrate the possibility of using an RNA virus as a vehicle for directed evolution of heterologous proteins.
RNA virus replication is catalyzed by RNA-dependent polymerases that apparently lack a proofreading function (9). This makes RNA copying an intrinsically erroneous process. Viruses with RNA genomes are often envisioned as living on the edge of error catastrophe, with an inevitable fitness loss in small populations (11, 28). Despite these challenges, RNA viruses cause over 75% of all viral diseases and include major human pathogens (9). In fact, the success of RNA viruses is most probably a result of their high mutation rates.
The quasispecies model describes populations of RNA replicons as clouds of distinct but closely related genotypes (10, 14). Such an organization allows RNA viruses to rapidly adapt to new environments, as a number of potentially advantageous mutations are already present in the population at the onset of selective pressure. Indeed, many RNA viruses, including human immunodeficiency virus and hepatitis C virus, are known to efficiently escape host immune responses and medical treatment by promptly accumulating resistant mutants (12, 15, 16). Understanding the principles of viral population dynamics is thus of paramount importance for designing new antiviral drugs and efficient treatment regimens.
The remarkable plasticity of RNA viruses has also drawn the attention of evolutionary biologists. A number of studies have been carried out in which RNA viruses were used as convenient experimental models to address fundamental questions of population genetics and evolutionary theory (6, 26, 36). In a currently preferred experimental format, a virus population is propagated under various conditions, and its fitness (the ability to produce viable progeny) is compared with that of a reference virus to derive trajectories of population changes as a function of time (9, 27). This approach, despite its well-documented utility, is limited to assessing the overall fitness phenotype, which is usually determined by numerous (and often undefined) genetic components. In principle, it is possible to determine the complete sequence of a representative number of viral genomes in order to follow the genotype dynamics of the evolving population. However, given the complexity of viral populations and the substantial lengths of their genomes, this task is too laborious and resource intensive for practical use.
The above methodology has revealed that several discrete steps are normally required for viral populations to regain fitness that was initially reduced by bottlenecking (9). Indeed, RNA viruses adapted to novel environments may contain a number of changes scattered throughout their genomes, implying that several rounds of genetic change and fixation had occurred. The adaptation process can therefore be imagined as a walk through the sequence space from an unfit genotype to one of maximum fitness. Although sequence information is often available for the endpoints, the trajectories of adaptive walks have not been studied at the genotype level.
Toward this end, we developed a novel evolution system in which a compact heterologous marker introduced into an RNA virus genome is employed as a selection target. Bacteriophage φ6 is a well-characterized double-stranded RNA (dsRNA) virus that infects the bacterium Pseudomonas syringae and contains three genomic segments, namely L, M, and S (20, 21). We constructed a recombinant φ6 containing a TEM β-lactamase (bla) gene integrated into the M segment. P. syringae cells carrying this construct were resistant to ampicillin (AMP). Selection pressure was applied to the carrier-state bacteria by substituting AMP with the related antibiotic cefotaxime (CTX). CTX is not cleaved by wild-type (wt) β-lactamase, but several specific amino acid changes that enable CTX hydrolysis have been described (5). Indeed, carrier-state cells that are resistant to high CTX concentrations appeared within several days of growth on the selective medium and were accompanied by mutations in the virus-encoded bla gene. Because bla is <0.9 kb long, it was feasible to sequence a number of bla cDNA clones that were isolated at different time points. We observed an initial increase in sequence variance at the onset of CTX selection. Genetic homogeneity of the population was regained within several passages after the acquisition of relevant mutations in the bla gene. These results expand our understanding of virus population biology and offer a new tool for directed evolution of heterologous sequences.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Plasmid pEM35 was produced by inserting the neomycin phosphotransferase (npt) cassette from pUC4K (Pharmacia) into the PstI site of pLM656 in the sense orientation (30). To construct pEM37, the TfiI-XbaI fragment of pLM656, containing the φ6 M segment, was inserted into the pSU18 vector (with a chloramphenicol resistance marker [4]) at the HindIII and XbaI sites, the TfiI- and HindIII-generated ends having been blunted using the Klenow fragment of DNA polymerase I. To produce pEM38, the β-lactamase (bla) gene was amplified from pUC18 by using the primers 5′-TTCACTGCAGATGCATAAGGAAGCATATGAGTATTCAACATTTCCGT-3′ and 5′-CAAACTGCAGAAGCTTACCAATGCTTAATCAGTGAGGCA-3′ (restriction sites are shown in italics) and Pfu DNA polymerase (Stratagene). The resulting PCR fragment was inserted at the PstI site of pEM37 in the sense orientation.
Construction of φ6 npt carrier-state cells.
φ6 npt was constructed as previously described for the φ6 K1 derivative (31). Briefly, purified recombinant φ6 procapsids (PCs) were packaged in vitro with recombinant m+ (a single-stranded sense copy of the φ6 M segment) containing the npt gene (a T7 transcript from pEM35 treated with XbaI and mung bean nuclease) and with wt l+ and s+ (single-stranded sense copies of L and S). The packaged single-stranded RNAs were replicated into dsRNAs within the PCs, and the particles were coated with φ6 P8 protein to obtain infectious nucleocapsids (3). These were used to produce recombinant virus plaques on a P. syringae HB10Y lawn. Material from one of the plaques was streaked onto Luria-Bertani (LB) agar plates containing 30 μg of kanamycin (KAN)/ml to select carrier-state bacteria HB10Y(φ6-npt) bearing the recombinant virus. These could be stably propagated on KAN-containing LB agar or in LB medium without losing the npt gene, as judged by agarose gel electrophoresis of viral dsRNA and by reverse transcriptase (RT)-PCR with npt-specific primers 5′-CAAGGAATTCCATGGGCCATATTCAACGGGAAA-3′ and 5′-CCAGGATCCTTTAAAAAAACTCATCGAGCATCAAATGAAACT-3′.
Construction of φ6 bla carrier-state cells.
Electrocompetent HB10Y(φ6-npt) cells were prepared as described previously (19). These (40 μl) were electroporated with 0.1 mg of pEM38 plasmid (with the bla gene within the m+ segment and no Pseudomonas-specific origin of replication) per ml. The cell suspension was diluted with 1 ml of LB broth containing 1 mM MgSO4, incubated at 28°C for 2 h, and plated onto LB agar containing 150 μg of AMP/ml. AMP-resistant colonies of P. syringae containing φ6 bla virus HB10Y(φ6-bla) formed after 48 to 72 h of incubation at 28°C. The φ6 bla carrier-state cells could be stably propagated in the presence of AMP.
Preparation of total RNA from carrier-state bacteria.
Bacterial cells that were pooled from 20 to 40 carrier-state colonies or pelleted from 1.5-ml liquid cultures were resuspended in 300 μl of a mixture containing 50 mM Tris-HCl (pH 8.0), 100 mM EDTA, and 8% (wt/vol) sucrose. Lysozyme was added to a concentration of 1 mg/ml, and the mixture was incubated for 5 min at room temperature. Cells were lysed with 1% sodium dodecyl sulfate (SDS) for 3 to 5 min. SDS and most of the chromosomal DNA were precipitated with 1.5 M potassium acetate, pH 7.5, on ice. RNA was precipitated from the supernatant fraction by the addition of 0.7 volume of isopropanol. The RNA pellet was dissolved in 400 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA), extracted successively with equal volumes of phenol-chloroform and chloroform, and reprecipitated with ethanol. The pellet was washed with 70% ethanol and dissolved in 100 μl of sterile water.
RT-PCR and cloning of the bla gene.
To obtain cDNA copies of the virus-encoded bla gene, we mixed total RNAs (1 to 5 μg) from carrier-state bacteria with 10 pmol of a reverse transcription primer (5′-CTATCGAGCACAGCGCCAACT-3′), denatured them by boiling for 1 min, and chilled them on ice. Reverse transcription was performed by using avian myeloblastosis virus RT (Sigma) at 45°C for 1 h as recommended. The bla cDNA was PCR amplified by using a mixture of Pfu and Taq DNA polymerases and the primers 5′-CCGAATTCATAAGGAAGCATATGAGTATTCA-3′ and 5′-CAACTTTTACGCTGGTGCTATACAACGACT-3′. HindIII-EcoRI-cut PCR products were ligated with a similarly treated pSU18 vector and transformed into Escherichia coli DH5α. Cloned bla sequences were determined by using a commercial automated sequencing facility (MWG-Biotech). Throughout the paper, amino acid numbering is given according to the accepted TEM β-lactamase nomenclature (1), which exceeds the physical number by 2.
RESULTS
Generating carrier-state P. syringae cells.
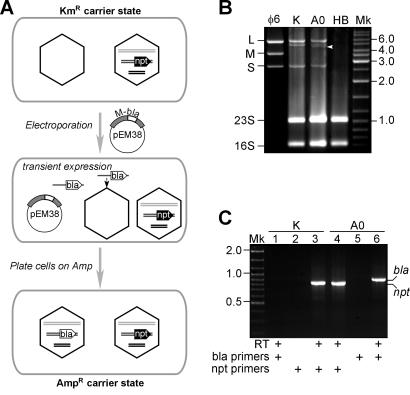
The infection of P. syringae HB10Y with wt φ6 culminates in cell lysis and the release of viral progeny (20). However, when the KAN resistance marker npt is inserted into the φ6 M segment, it is possible to select carrier-state bacteria on KAN-containing medium (31). We repeated this experiment to obtain a KAN-resistant strain, HB10Y(φ6-npt). As expected, the dsRNA segment M of the φ6 npt virus (M-npt) was longer than wt M, whereas the φ6 npt L and S segments had regular lengths (Fig. 1B, lanes φ6 and K).
FIG. 1.
Generation of carrier-state P. syringae cells containing φ6 bla. (A) Schematic diagram of the technology used (see text for details). (B) Agarose gel electrophoresis of total RNAs from the following strains: K, KAN-resistant HB10Y(φ6-npt); A0, AMP-resistant HB10Y(φ6-bla); HB, noninfected HB10Y. Lane φ6, dsRNA segments L, M, and S extracted from the wt φ6 (positions are indicated on the left, along with the positions of P. syringae 23S and 16S rRNAs); lane Mk, dsDNA markers. Marker lengths in kilobase pairs are shown on the right. The arrowhead shows the new segment, M-bla, which appears in AMP-resistant cells. (C) RT-PCR analysis with npt- and bla-specific primers was performed with RNAs from HB10Y(φ6-npt) (K) and HB10Y(φ6-bla) (A0). The reverse transcription step was omitted in reactions 2 and 5. Different PCR primers were used as specified under the panel. The positions of the npt- and bla-specific PCR fragments are marked on the right. dsDNA marker (Mk) lengths are shown on the left.
The construction of φ6 npt involved manipulations with purified RNAs and viral PCs in vitro, followed by spheroplast infection (3). To avoid these technical difficulties when preparing the φ6 bla virus, we used a plasmid-based strategy (Fig. 1A) first suggested by Mindich (22). HB10Y(φ6-npt) cells were transformed with plasmid pEM38, which encodes the φ6 M segment containing the AMP resistance marker bla. pEM38 cannot replicate in P. syringae, but it can direct transient expression of the recombinant M segment, as was previously shown for other E. coli plasmids (22). Some of the RNA transcripts can be packaged by PCs present in the HB10Y(φ6-npt) cytoplasm, giving rise to the φ6 bla virus. Indeed, AMP-resistant colonies (101 to 102 μg−1 of DNA) appeared on pEM38- but not mock-transformed plates. One of the AMP-resistant clones (no. 7) was used for subsequent experiments. Electrophoretic analysis of the φ6 bla dsRNA genomic segments revealed the presence of two M species, M-npt and a new segment, M-bla, migrating between M-npt and wt M (Fig. 1B, lane A0).
Carrier-state bacteria contain RNA-encoded antibiotic resistance genes.
We carried out RT-PCR analysis to ensure that the bla gene was indeed encoded by φ6 bla rather than by host DNA. The bla PCR product was readily detectable when the nucleic acid extracted from HB10Y(φ6-bla) was reverse transcribed and amplified with bla-specific primers (Fig. 1C, lane 6). However, no product appeared for the control reaction, in which the reverse transcription step was performed without RT (lane 5). Notably, no PCR product was detected without the reverse transcription step when the assay was repeated with the samples containing readily detectable amounts of chromosomal DNA (not shown). This strongly suggests the RNA nature of the bla gene. Using npt-specific primers, we also found that HB10Y(φ6-bla) bacteria retain detectable amounts of the npt gene (lane 4), consistent with the electrophoretic analysis of HB10Y(φ6-bla) RNA. As expected, HB10Y(φ6-npt) contained only an RNA-encoded npt gene (lanes 1 to 3).
P. syringae carrying φ6- but not DNA-encoded bla quickly adapts to CTX.
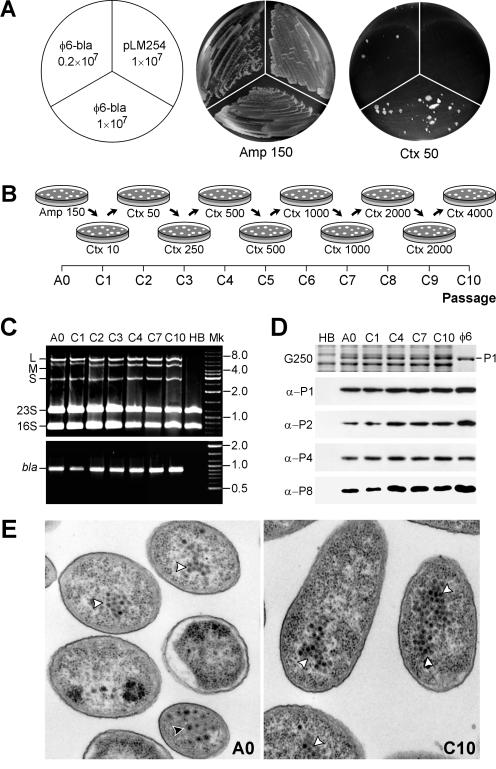
The wt TEM β-lactamase encoded by φ6 bla hydrolyzes penicillin β-lactam antibiotics (e.g., AMP) but cannot efficiently cleave some cefalosporins such as CTX. Since several CTX-resistant β-lactamase variants have been reported (5, 32), we checked whether these could be selected in the carrier-state bacteria. HB10Y(φ6-bla) cells were plated onto LB agar containing either 150 μg of AMP/ml or 50 μg of CTX/ml and were incubated at 28°C. As a control, we used HB10Y cells transformed with a broad-host-range plasmid, pLM254, whose bla gene is identical to that inserted into φ6 bla (23). Both HB10Y(φ6-bla) and HB10Y(pLM254) grew equally well on AMP medium (Fig. 2A). On CTX medium, HB10Y(φ6-bla) formed slowly growing colonies of various sizes, with an average frequency of ∼4 CFU per 106 CFU on AMP medium. No colonies were detected in the case of HB10Y(pLM254) after 96 h of incubation (Fig. 2A). Because the abundance of pLM254 within the cells was comparable to that of M-bla (not shown), we concluded that CTX-resistant mutants appear considerably more often when bla is encoded by φ6 RNA rather than plasmid DNA.
FIG. 2.
φ6 bla carrier cells rapidly adapt to CTX. (A) HB10Y(φ6-bla) carrier-state cells (0.2 × 107 to 1 × 107) were plated onto LB agar containing either 150-μg/ml AMP (Amp150) or 50-μg/ml CTX (Ctx50). CTX-resistant colonies appeared after 3 days of incubation at 28°C. No colonies were detected at this time on the sector inoculated with 107 HB10Y(pLM254) cells, which contain a plasmid encoding the bla gene. (B) Schematic diagram of the CTX adaptation experiment. Cells were cultivated on LB agar containing increasing CTX concentrations (in micrograms per milliliter), as shown below the diagrams. Twenty to 40 of the largest colonies were pooled after each passage and were used for subsequent rounds of selection. (C) Agarose gel analysis of RNAs extracted from carrier-state cells at passages A0, C1, C2, C3, C4, C7, and C10 (top panel). HB, RNA from uninfected HB10Y cells. Also shown are RT-PCR products generated with bla-specific primers (bottom panel). Other designations are defined as for Fig. 1. (D) SDS-polyacrylamide gel electrophoresis analysis (29) of carrier-state cells from different passages (A0, C1, C4, C7, and C10) or of purified φ6 virus (φ6). HB, uninfected HB10Y cells. Panels: G250, a Coomassie G250-stained gel fragment showing the band of protein P1; α-P1, α-P2, α-P4, and α-P8, immunoblots produced with antibodies specific to corresponding φ6 nucleocapsid proteins and visualized by ECL detection (Pierce Biotechnology). (E) Transmission electron micrograph of osmium tetroxide- and uranyl acetate-stained thin cell sections from A0 and C10 passages produced as described previously (2). Black arrowhead, enveloped virions; white arrowheads, nucleocapsids or PC particles.
HB10Y(φ6-bla) can gradually adapt to high CTX concentrations.
When the above experiment was repeated with ≥100 μg of CTX/ml, no growth was detected even on the plates with HB10Y(φ6-bla). We therefore tested the possibility that increased CTX resistance can be developed by gradually increasing the concentration of CTX and selecting the best growers. HB10Y(φ6-bla) cells were passaged 10 times, with the CTX concentration increased from 10 to 4,000 μg/ml, as shown in Fig. 2B. The initial HB10Y(φ6-bla) stock was referred to as A0, and the cells obtained from different CTX passages were called C1, C2,…, C10. On average, 107 to 108 CFU from AMP medium were plated onto several petri dishes, and the 20 to 40 largest colonies were picked and pooled after 48 h of incubation. After a brief propagation (8 to 12 h at 28°C) in LB medium containing CTX at 1/4 the plate concentration, the cells were subjected to the next round of selection. By repeating this procedure several times, it was possible to obtain P. syringae colonies that were resistant to 4,000 μg of CTX/ml.
Several analyses were used to verify the presence of φ6 bla throughout the adaptation process. First, cellular RNA was studied by agarose gel electrophoresis and RT-PCR using bla-specific primers (Fig. 2C). M segments of increased mobility were clearly present in all samples, from C1 to C10, which correlated with the presence of the bla PCR fragment. M-bla was relatively sparse in C1 cells, as judged by the reproducibly weak RT-PCR signal and the dominance of M-npt over M-bla on the RNA gel (lane C1). However, the amount of M-bla in C2 to C10 cells was notably higher than that in A0 cells. The M-npt band disappeared from the RNA pattern for C2 cells.
In the second analysis, cellular proteins were separated by SDS-polyacrylamide gel electrophoresis and subjected to immunoblotting with polyclonal antisera against φ6 proteins P1, P2, P4, and P8, which are components of φ6 nucleocapsids (Fig. 2D). Corresponding protein bands were detected for A0 and C1 to C10 cells. The major φ6 capsid protein, P1, was also visible on Coomassie-stained gels.
Finally, when carrier-state bacteria were examined by electron microscopy, φ6 subviral particles and enveloped virions were observed in the cytoplasm of A0, C1, C4, C7, and C10 cells, but not in the HB10Y control cells (Fig. 2E; also data not shown).
bla from CTX-adapted carrier state P. syringae cells confers CTX resistance in E. coli.
For characterization of the possible effect of CTX selection on the β-lactamase gene, bla cDNAs from the A0, C1 to C4, C7, and C10 passages were cloned into pSU18 (E. coli plasmid containing a chloramphenicol [CHL] resistance marker) under the control of the lac promoter. E. coli DH5α was transformed with the resulting plasmid libraries and was plated onto CHL medium. Because existing CTX-specific β-lactamases are also resistant to AMP (5), we used plates with a low AMP concentration (50 μg/ml) to screen the libraries for clones containing the bla insert. A sufficient amount of β-lactamase was produced from the lac promoter without induction. Plasmids from the AMP-resistant clones (isolated from the master CHL plates) always contained the bla inserts. Conversely, several randomly selected clones that were resistant to CHL but not to AMP were the same size as the pSU18 vector.
We next examined whether E. coli containing pSU18 with bla inserts is also resistant to CTX. For this purpose, ∼106 cells were transferred from colonies grown on CHL to plates containing 5 or 10 μg of CTX/ml. Of the 50 to 100 colonies analyzed for each library, 22% of the C1-derived bla clones were indeed resistant to 5-μg/ml CTX. In the cases of the C2-, C3-, C4-, C7-, and C10-derived libraries, the fractions of CTX-resistant bla clones were 72, 81, 93, 100, and 100%, respectively, with most of the clones growing in the presence of 5- and 10-μg/ml CTX. No CTX-resistant colonies were detected in the A0-derived library.
Changes in bla sequence during adaptation to CTX.
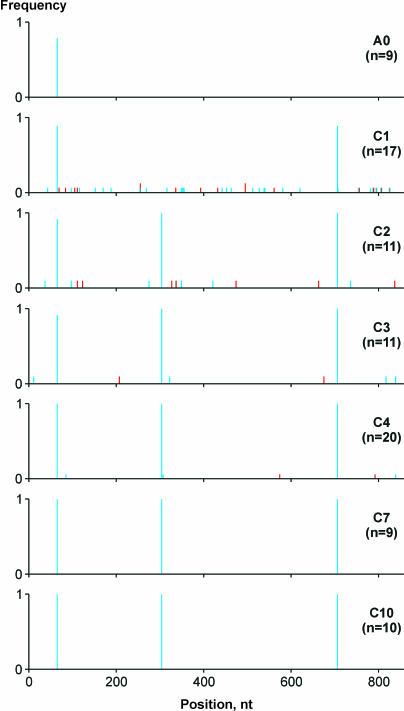
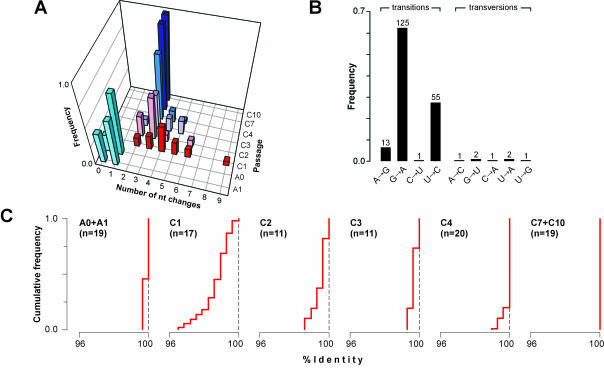
Complete bla sequences from several CTX-resistant clones were determined for each library (Fig. 3). Two bla alleles were found in the A0 library. One of these was the wt allele, occurring at an apparent frequency of 0.22, while the other contained a single U→C mutation that changed F24 to S and occurred at an apparent frequency of 0.78. Surprisingly, multiple mutations were found in the bla sequences from the initial CTX passages, with one segment often containing several substitutions (up to 9 in C1 cells) (Fig. 4A). Most of the changes were transitions, with a dramatic prevalence of G→A, U→C, and A→G (Fig. 4B).
FIG. 3.
Sequence changes in bla in response to CTX selection. The graphs show normalized point mutation frequencies at the indicated nucleotide (nt) positions summed for n bla sequences from each passage. Blue bars, nonsynonymous substitutions; red bars, synonymous substitutions.
FIG. 4.
Population dynamics of bla sequences during adaptation. (A) Normalized frequencies of bla alleles containing a given number of mutations as a function of passage number. Cyan, passages A0 and A1; red, C1; pink, C2; purple, C3; blue, C4; dark blue, C7 and C10. (B) Distribution of different mutation types in bla sequences from passages C1, C2, and C3. (C) Percent identity plots showing genetic variance in bla populations from different passages. Plots (red lines) are cumulative distribution functions of identities between every pair within n sequences, for which the vertical axis represents the fraction of data points with a value as low as or lower than a given identity value. More heterogeneous sequence populations give plots that deviate more from the 100% identity asymptote (dashed line). Data for related passages A0 and A1 and also for passaged C7 and C10 were combined to improve the statistics. Plots were created with GeneDoc (http://www.psc.edu/biomed/genedoc/).
In addition to clone-specific mutations, two point mutations, F24S and a G→A substitution causing the G238S mutation, were detected in most bla sequences from C1 and subsequent passages. Beginning at the C2 passage, all sequences contained yet another common substitution, G→A, that changed E104 to K (compare C1 and C2 in Fig. 3). Interestingly, most clones from the C4 passage and all clones from the C7 and C10 passages contained only the F24S, E104K, and G238S mutations, with no other mutations being detected (Fig. 3).
To ensure that the accumulation of bla mutants after the antibiotic change was a specific effect of CTX, we carried out a mock selection experiment. A0 cells were plated onto dishes containing 150 mg of AMP/ml and were incubated for 48 h at 28°C (passage A1). dsRNA purified from 40 pooled colonies was used to construct an RT-PCR library in E. coli, as described above. No CTX-resistant clones were found, and no other alleles were detected besides the wt and F24S alleles (with frequencies of 0.4 and 0.6, respectively).
Since 78% of the AMP-resistant clones from the C1 library failed to grow in the presence of CTX, we determined the bla sequences from seven CTX-sensitive clones. All of these sequences contained one or several mutations compared to the wt and F24S alleles, with the overall picture being similar to that for CTX-resistant clones (not shown). The only difference was that none of the CTX-sensitive clones contained the G238S mutation. We conclude that the E104K and G238S mutations are critical for enabling CTX hydrolysis. Indeed, both mutations map to the enzyme active site and are often observed in CTX-resistant bacteria (5, 32). When the CTX evolution experiment was repeated, four randomly selected C4 clones contained F24S, E104K, and G238S, thus confirming the adaptive nature of these changes.
The overall dynamics of the bla population adapting to CTX are apparent from percent identity plots (Fig. 4C). A relatively homogenous population in the A0 (and A1) passage was diversified dramatically in the C1 and C2 passages. After the appropriate mutations appeared, the population regained homogeneity in the C4 to C7 passages. Further passages did not change the genetic structure of the population. Importantly, the genetic heterogeneity levels in the C2 and C3 passages were clearly higher than that in the A0 passage, and the M-bla segment was more abundant in the C2 and C3 passages than in the A0 passage (Fig. 2C). Therefore, possible effects of RT-PCR-derived mutations can be ruled out.
DISCUSSION
An understanding of the mechanisms of adaptation is among the most important problems of modern biology. RNA viruses have been convenient models for studying evolutionary processes due to their potentials for rapid genetic change (27). Here we introduced a β-lactamase (bla) gene marker into the genome of the dsRNA bacteriophage φ6, propagated this recombinant virus in carrier-state bacterial cells in the presence of β-lactam antibiotics, and monitored changes in the bla sequences as the carrier-state population adapted from one type of antibiotic resistance to another.
The use of a heterologous marker to study viral adaptation on the molecular level has several advantages as follows. (i) Selection is focused on the target gene and is easy to modulate. In contrast, when using virus-specific genes, it is often difficult to limit the selection pressure to a single specific gene. (ii) It is unlikely that the use of a heterologous gene for selection affects the virus mutation rate. Conversely, many antiviral drugs that target the virus replication machinery do alter the RNA polymerization fidelity (8). (iii) A short heterologous marker is highly desirable to facilitate the sequencing of sufficient numbers of cDNA clones.
Our data suggest that, under conditions of gradual increases in selective pressure, a viral population may respond to environmental challenges by a transiently increasing genetic variance (Fig. 3 and 4). This process can be imagined as exploring multiple adaptation routes to find a fitness optimum. However, it remains to be tested whether separate bla lineages converge to the common fitness optimum or if only one lineage reaches the optimum while others become extinct.
According to the quasispecies theory, genetic diversification upon CTX challenge would result from selection for a number of advantageous peripheral members (and against the centrally located wt and F24S alleles) of the initial AMP-adapted population rather than from the generation of mutations de novo. The decreased abundance of M-bla in the C1 passage supports this conclusion (Fig. 2C).
The role of the F24S mutation, first appearing in the A0 passage, is currently unknown. Being adjacent to the β-lactamase leader peptide, it may have some effect on protein secretion into the P. syringae periplasm. The key beneficial mutation featured in the C1 passage is G238S. Indeed, this mutation is known to improve eightfold the CTX hydrolysis by the wt β-lactamase (35). The second advantageous change, E104K, was first seen in the C2 passage and acts synergistically with G238S, with double mutants displaying a 267-fold increase in the MIC versus the wt enzyme (35).
Because viral populations obey the principle of competitive exclusion (10), it is not surprising that only one genetic variant (F24S, E104K, and G238S) reached dominance after several passages at increasing CTX concentrations. What is surprising is that only three nonsynonymous mutations were fixed, despite the overflow of changes in the C1, C2, and C3 passages. Perhaps this can be explained by the strong selective pressure that disfavors even slightly deleterious mutations, such as those affecting codon usage, RNA secondary structure, or protein stability (37). Notably, RNA recombination has been described for φ6 (24, 33). This might have operated to deter the M-bla sequence from less favorable changes.
A β-lactamase containing only the E104K and G238S mutations is not the most optimal catalyst for CTX hydrolysis. Additional changes, such as M182T, can further improve the efficiency of this process (32, 34). However, the adapting population did not display these mutations even when challenged with very high CTX concentrations. The relatively low numbers of carrier-state P. syringae cells (107 to 108) used for each round of CTX selection and the apparent preference for G→A mutations (Fig. 4B) are among possible explanations.
It is unlikely that the observed prevalence of the G→A substitutions is due to the CTX selection, as many of these changes are silent (Fig. 3 and data not shown). G→A changes have been reported for some other RNA viruses (for examples, see reference 17); however, one cannot formally exclude that the G→A changes in the φ6 plus-strand reflect biased C→U mutations during minus-strand synthesis. In any case, mutation bias might be a reason for the difference in published estimates of per-nucleotide mutation rates of φ6, ranging from ∼1 × 10−5 to 2.7 × 10−6 depending on the method used (7, 13).
This study reports, for the first time, that an RNA virus can be used for directed evolution of a heterologous protein. Just two passages, of 48 h each, were sufficient for recombinant φ6 to discover a successful combination of three mutations that changed the β-lactamase substrate specificity. Previous bla evolution experiments using E. coli mutator strains lasted several weeks (18). Therefore, one may envision the future development of φ6-based replicons into a system for rapidly changing proteins with selectable properties. Recombinant φ6 can be easily constructed within several days by using the plasmid-based strategy depicted in Fig. 1A. An obvious advantage of φ6 over eukaryotic RNA viruses is the low cost and relative ease of propagation. Furthermore, a number of φ6-related bacteriophages (Cystoviridae) are known, some of which have been shown to infect E. coli in addition to Pseudomonas hosts (25).
Clearly, many questions will need to be addressed, including the following. What is the maximal tolerated size of heterologous inserts? Is it possible to reduce the G→A bias? Will the use of larger numbers of carrier-state cells enable a search for remote fitness peaks rather than for local optima? Further experiments will reveal to what extent one can harness the remarkable evolutionary potential of RNA viruses.
Acknowledgments
We thank Riitta Tarkiainen for technical assistance.
This work was supported by the Academy of Finland (“Finnish Centre of Excellence Program 2000-2005” grants 164298 and 172621).
REFERENCES
- 1.Ambler, R. P., A. F. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bamford, D. H., and L. Mindich. 1980. Electron microscopy of cells infected with nonsense mutants of bacteriophage φ6. Virology 107:222-228. [DOI] [PubMed] [Google Scholar]
- 3.Bamford, D. H., P. M. Ojala, M. Frilander, L. Walin, and J. K. H. Bamford. 1995. Isolation, purification, and function of assembly intermediates and subviral particles of bacteriophages PRD1 and φ6, p. 455-474. In K. W. Adolph (ed.), Methods in molecular genetics, vol. 6. Academic Press, San Diego, Calif.
- 4.Bartolome, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, P. A. 2001. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao, L. 1990. Fitness of RNA virus decreased by Muller's ratchet. Nature 348:454-455. [DOI] [PubMed] [Google Scholar]
- 7.Chao, L., C. U. Rang, and L. E. Wong. 2002. Distribution of spontaneous mutants and inferences about the replication mode of the RNA bacteriophage φ6. J. Virol. 76:3276-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domingo, E., C. K. Biebricher, M. Eigen, and J. J. Holland. 2001. Quasispecies and RNA virus evolution: principles and consequences. Landes Bioscience, Georgetown, Tex.
- 10.Domingo, E., C. Escarmis, N. Sevilla, A. Moya, S. F. Elena, J. Quer, I. S. Novella, and J. J. Holland. 1996. Basic concepts in RNA virus evolution. FASEB J. 10:859-864. [DOI] [PubMed] [Google Scholar]
- 11.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 12.Domingo, E., L. Menendez-Arias, M. E. Quinones-Mateu, A. Holguin, M. Gutierrez-Rivas, M. A. Martinez, J. Quer, I. S. Novella, and J. J. Holland. 1997. Viral quasispecies and the problem of vaccine-escape and drug-resistant mutants. Prog. Drug Res. 48:99-128. [DOI] [PubMed] [Google Scholar]
- 13.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eigen, M. 1996. On the nature of virus quasispecies. Trends Microbiol. 4:216-218. [DOI] [PubMed] [Google Scholar]
- 15.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 16.Harrigan, P. R., and C. S. Alexander. 1999. Selection of drug-resistant HIV. Trends Microbiol. 7:120-123. [DOI] [PubMed] [Google Scholar]
- 17.Kurath, G., M. E. Rey, and J. A. Dodds. 1992. Analysis of genetic heterogeneity within the type strain of satellite tobacco mosaic virus reveals variants and a strong bias for G to A substitution mutations. Virology 189:233-244. [DOI] [PubMed] [Google Scholar]
- 18.Long-McGie, J., A. D. Liu, and V. Schellenberger. 2000. Rapid in vivo evolution of a beta-lactamase using phagemids. Biotechnol. Bioeng. 68:121-125. [DOI] [PubMed] [Google Scholar]
- 19.Lyra, C., H. Savilahti, and D. H. Bamford. 1991. High-frequency transfer of linear DNA containing 5′-covalently linked terminal proteins: electroporation of bacteriophage PRD1 genome into Escherichia coli. Mol. Gen. Genet. 228:65-69. [DOI] [PubMed] [Google Scholar]
- 20.Mindich, L. 1988. Bacteriophage φ6: a unique virus having a lipid-containing membrane and a genome composed of three dsRNA segments. Adv. Virus Res. 35:137-176. [DOI] [PubMed] [Google Scholar]
- 21.Mindich, L. 1999. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage φ6. Microbiol. Mol. Biol. Rev. 63:149-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mindich, L. 1999. Reverse genetics of dsRNA bacteriophage φ6. Adv. Virus Res. 53:341-353. [DOI] [PubMed] [Google Scholar]
- 23.Mindich, L., G. MacKenzie, J. Strassman, T. McGraw, S. Metzger, M. Romantschuk, and D. Bamford. 1985. cDNA cloning of portions of the bacteriophage φ6 genome. J. Bacteriol. 162:992-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mindich, L., X. Qiao, S. Onodera, P. Gottlieb, and J. Strassman. 1992. Heterologous recombination in the double-stranded RNA bacteriophage φ6. J. Virol. 66:2605-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mindich, L., X. Qiao, J. Qiao, S. Onodera, M. Romantschuk, and D. Hoogstraten. 1999. Isolation of additional bacteriophages with genomes of segmented double-stranded RNA. J. Bacteriol. 181:4505-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miralles, R., P. J. Gerrish, A. Moya, and S. F. Elena. 1999. Clonal interference and the evolution of RNA viruses. Science 285:1745-1747. [DOI] [PubMed] [Google Scholar]
- 27.Moya, A., S. F. Elena, A. Bracho, R. Miralles, and E. Barrio. 2000. The evolution of RNA viruses: a population genetics view. Proc. Natl. Acad. Sci. USA 97:6967-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichol, S. 1996. RNA viruses. Life on the edge of catastrophe. Nature 384:218-219. [DOI] [PubMed] [Google Scholar]
- 29.Olkkonen, V. M., and D. H. Bamford. 1989. Quantitation of the adsorption and penetration stages of bacteriophage φ6 infection. Virology 171:229-238. [DOI] [PubMed] [Google Scholar]
- 30.Olkkonen, V. M., P. Gottlieb, J. Strassman, X. Y. Qiao, D. H. Bamford, and L. Mindich. 1990. In vitro assembly of infectious nucleocapsids of bacteriophage φ6: formation of a recombinant double-stranded RNA virus. Proc. Natl. Acad. Sci. USA 87:9173-9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onodera, S., V. M. Olkkonen, P. Gottlieb, J. Strassman, X. Y. Qiao, D. H. Bamford, and L. Mindich. 1992. Construction of a transducing virus from double-stranded RNA bacteriophage φ6: establishment of carrier states in host cells. J. Virol. 66:190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orencia, M. C., J. S. Yoon, J. E. Ness, W. P. Stemmer, and R. C. Stevens. 2001. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat. Struct. Biol. 8:238-242. [DOI] [PubMed] [Google Scholar]
- 33.Qiao, X., J. Qiao, and L. Mindich. 1997. An in vitro system for the investigation of heterologous RNA recombination. Virology 227:103-110. [DOI] [PubMed] [Google Scholar]
- 34.Sideraki, V., W. Huang, T. Palzkill, and H. F. Gilbert. 2001. A secondary drug resistance mutation of TEM-1 beta-lactamase that suppresses misfolding and aggregation. Proc. Natl. Acad. Sci. USA 98:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stemmer, W. P. 1994. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370:389-391. [DOI] [PubMed] [Google Scholar]
- 36.Turner, P. E., and L. Chao. 1999. Prisoner's dilemma in an RNA virus. Nature 398:441-443. [DOI] [PubMed] [Google Scholar]
- 37.Wang, X., G. Minasov, and B. K. Shoichet. 2002. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol. 320:85-95. [DOI] [PubMed] [Google Scholar]