Abstract
To ensure their stable inheritance by daughter cells during cell division, bacterial low copy-number plasmids make simple DNA segregating machines that use an elongating protein filament between sister plasmids. In the ParMRC system of Escherichia coli R1 plasmid, ParM, an actin-like protein, forms the spindle between ParRC complexes on sister plasmids. Using a combination of structural work and total internal reflection fluorescence microscopy, we show that ParRC bound and could accelerate growth at only one end of polar ParM filaments, mechanistically resembling eukaryotic formins. The architecture of ParM filaments enabled two ParRC-bound filaments to associate in an antiparallel orientation, forming a bipolar spindle. The spindle elongated as a bundle of at least two antiparallel filaments, thereby pushing two plasmid clusters towards the poles.
During bacterial cell division, equal distribution of replicated plasmids to daughter cells ensures their stable inheritance. Low copy-number plasmids encode the simplest known DNA segregation machines to perform this task. They comprise a nucleotide-driven (cytomotive) protein filament and a centromere-like DNA region, linked by an adaptor protein. The ParMRC segregation system of E. coli R1 plasmid consists of ParM, an actin-like cytomotive protein (1) that forms polar, left-handed double-helical filaments (2), ParR, an adaptor protein, and parC, a centromeric region (3, 4). Dynamic instability of ParM filaments enables plasmid segregation by a ‘search and capture’ mechanism (5, 6), with ParRC (7, 8) stabilizing the filaments. It has been reported that ParRC binds to both ends of a single ParM filament (9, 10). This leads to a conundrum - how does ParRC bind to two different ends of a polar ParM filament?
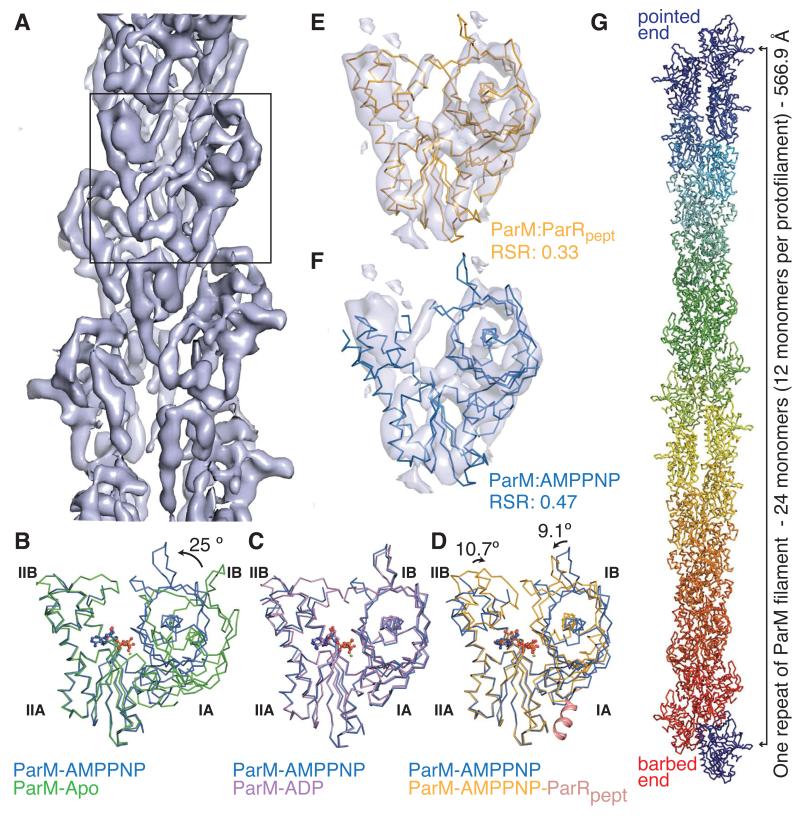
Here, we provide a comprehensive description of ParM in the monomeric and filament states. An electron cryomicroscopy (cryoEM) reconstruction (11) (Fig. 1A) provided a subnanometer-resolution map of the polar filament of ParM (resolution of 8.5 Å at FSC 0.5; Fig. S1A-D), polymerized in the presence of adenylyl-imidodiphosphate (AMPPNP; a non-hydrolyzable ATP analog). We determined the crystal structures of a non-polymerizing mutant, ParM(L163R) bound to AMPPNP, and ParM(L163A) bound to the C-terminal 17-residue-peptide corresponding to the ParM-interacting region of ParR (8, 10) (ParRpept) and AMPPNP (11). Comparison of the crystal structures, including the previously reported apo- and ADP-bound forms (1), revealed no large differences between the ATP and ADP states of ParM monomers (Figs. 1B-C). In contrast, domains IB and IIB showed a rotation of 9.1° and 10.7° towards the nucleotide-binding pocket (Fig. 1D) in the ParM:ParRpept structure, compared to the free monomer. The domain rotations were reminiscent of the transition from G-actin to F-actin (12, 13). Fitting the monomeric structures of ParM into the cryoEM reconstruction showed that the ParM:ParRpept structure fits best (Figs. 1E-F, S1E; Movie S1). Thus, the filament conformation of ParM is very similar to ParM:ParRpept. The best-fit provided a quasi-atomic model of the ParM filament (Fig. 1G), and implies that binding of ParR or ParRC locks ParM monomers in the filament-like conformation.
Fig. 1. Conformational cycle of ParM.
A) CryoEM reconstruction of the ParM filament. Box: a monomeric segment.
B-D) Superposition (using Cα atoms of domain IIA) of the conformations of ParM with the ParM-AMPPNP state (blue; PDB ID: 4A61) B) unliganded ParM (green; (1); PDB ID: 1MWK) C) ParM-ADP state (magenta; (1); PDB ID: 1MWM) D) ParM with ParRpept and AMPPNP (orange; ParRpept in pink cartoon representation; PDB ID: 4A62). Domain rotations are indicated.
E-F) Rigid body fit of ParM monomeric states into the cryoEM reconstruction. Domain I residues were fitted using Chimera (24). Real-space R-factors (RSR) against the map for ParM:AMPPNP (E) and ParM:ParRpept (F) quantifies the best-fit (Movie S1 and Fig. S1E).
G) A repeat unit of the ParM filament (EMDB: EMD-1980, PDB ID: 4A6J).
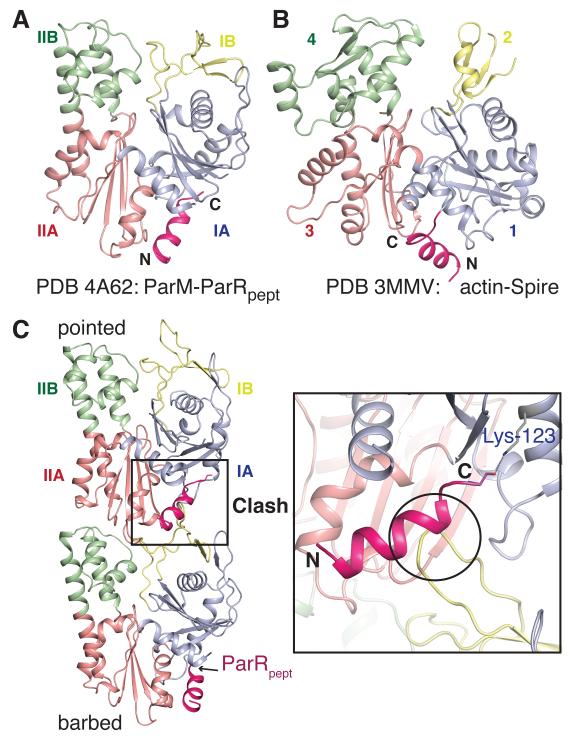
ParRpept was bound in a hydrophobic pocket between domains IA and IIA of ParM (Figs. 2A, S2). The ParM-ParRpept interaction resembled proteins that bind at the barbed-end of actin (Figs. 2A-B). Many actin-binding proteins, including formins (14) and WH2 domain-containing proteins such as Spire (15), insert a helix between the corresponding subdomains 1 and 3 of actin (16). The ParM:ParRpept structure modeled onto the ParM filament highlighted a significant clash between ParRpept and residues 37 to 46 of the next ParM monomer in the protofilament (Fig. 2C). Thus, the ParR-binding site lies within the polymerization interface of ParM, and bound ParR requires to be displaced during elongation. Overlap in the binding site of ParRpept and the polymerization interface occludes all the ParRC-binding sites on the ParM filament except those at the barbed-end, implying that ParRC binds exclusively to this end.
Fig. 2. ParRC binds at the barbed-end of ParM filaments.
A, B) The ParRpept-binding site corresponds to that of Spire on actin. The helices of the interacting proteins are shown in pink, with the rest in grey. A) ParM:ParRpept (PDB ID: 4A62), B) actin:Spire (PDB ID: 3MMV).
C) ParRpept binds at the interprotofilament interface of ParM. Two subunits of the ParM filament with hypothetical ParRpept at the binding sites are shown. In that position, ParRpept clashes with loop 37-46 from domain IB of the adjacent subunit.
Our ParM-ParRpept structure supports a formin-like mechanism for the processive movement of ParRC (Fig. S3). Ten ParR dimers bind parC repeats (7, 8), forming a scaffold that allows a formin-like stair-stepping mechanism (Fig. S3B, C) (17). The C-terminal helices from the twenty ParRs are localized in a confined area. This probably facilitates ParM filament nucleation by ParRC (18). It also ensures ParRC-bound ParM monomer recruitment upon ParR displacement from the filament (Fig. S3D), explaining end-tracking of ParM filaments by ParRC through insertional polymerization (6, 19). The ParRC cap locks the terminal monomers in the filament-conformation, thus protecting the filament from dynamic instability.
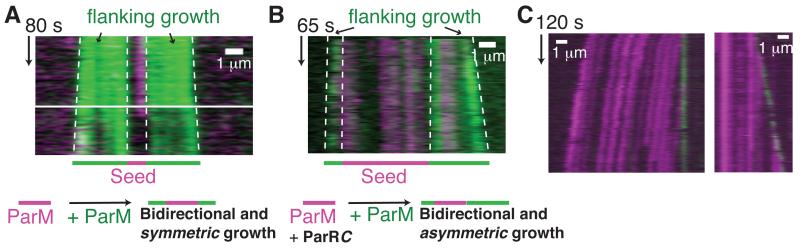
To confirm the proposed single-end binding of ParRC, we examined the effect of ParRC on ParM filament elongation using TIRF microscopy (11). The experiments were performed with non-hydrolyzable AMPPNP to prevent dynamic instability. ParM-AMPPNP elongated symmetrically at both ends from an initial seed of the ParM filament (Fig. 3A; Movie S2) (5). In the presence of unlabeled ParRC, one end of the filaments grew faster, resulting in asymmetric growth (Figs. 3B, S4A; Movie S3). The rates of growth were 9.4 ± 4.1 and 2.5 ± 1.9 monomers/s, for fast and slow growing ends (n = 32; Table S3). Experiments with labeled ParRC showed that ParRC accelerated growth and recruited ParM monomers at the ParRC-bound filament end only (Figs. 3C, S4B; Movie S4), reconfirming insertional polymerization (6). The unidirectional elongation by ParRC leads us to the key question – what enables bipolar plasmid segregation?
Fig. 3. ParRC accelerates growth at one end of ParM filaments.
A-B) TIRF microscopy kymographs of filaments in dual-label experiments (11); A) without ParRC B) with ParRC. Growth of ParM filaments without ParRC is bidirectional and symmetric, with equal slopes on both ends of the kymograph, while addition of ParRC results in asymmetric growth, with unequal slopes. The relevant boundaries are highlighted.
C) Monomers are recruited to the ParRC-bound end of ParM filament. Kymograph from a filament labeled with Alexa-568 (magenta) and YOYO-1 labeled ParRC (green) are shown.
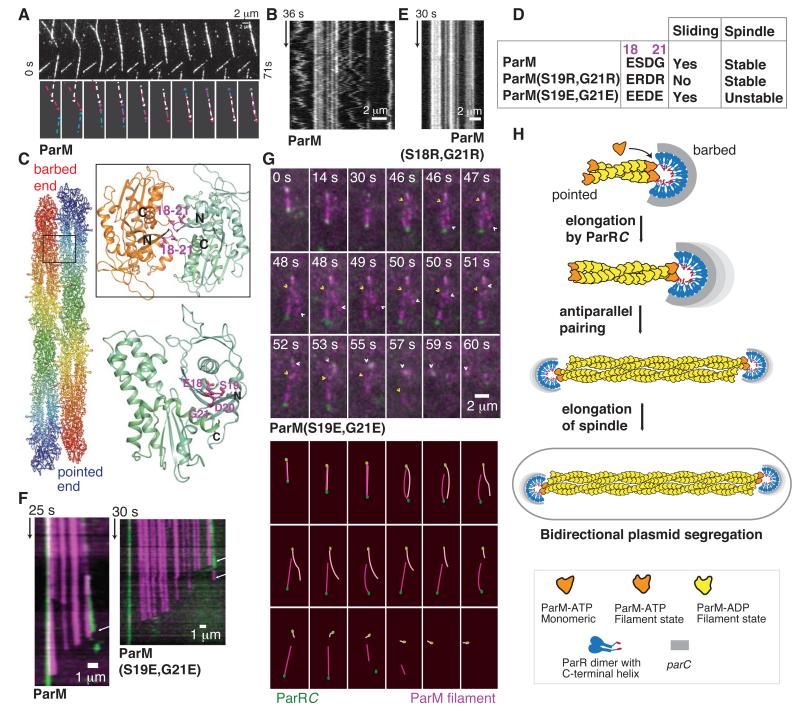
Frequent condensation events were observed between ParM filaments, both with ATP and AMPPNP (Fig. 4A, Movies S5–S8). Furthermore, continuous motion due to interfilament sliding occurred within filament bundles (Figs. 4B, S4C; Movies S9–S11). A molecular model for the filament sliding and condensation was generated based on two monomers in the crystal packing of ParM:ParRpept (Fig. S5A). ParM filament subunits, when superposed sequentially onto the ParM monomers, produced sliding and an antiparallel packing of filaments (Figs. 4C, S5A). Mutation of residues within loop 18-21 (S19R, G21R) at the proposed antiparallel filament interface (Figs. 4C-D, S5B) prevented interfilament sliding (Movie S12; Figs. 4E, S4D), due to stronger interactions between filaments caused by alternating charges.
Fig. 4. A bipolar spindle comprises at least two antiparallel ParM filaments.
A) ParM-AMPPNP filaments condensing into a bundle (Movie S7).
B) Kymograph of a bundle of ParM. Static filaments yield straight lines, while concerted zigzag motion shows sliding filament movement.
C) Atomic model of an antiparallel filament pair. Inset: interface involving loop 18–21 in the antiparallel arrangement.
D) Interface mutations and their effect on sliding and spindle stability.
E) Kymograph of a bundle of ParM(S19R,G21R) filaments that are static due to stronger interfilament interactions.
F) Kymographs of disassembling spindles of ParM and ParM(S19E,G21E). Arrows highlight the slopes of disassembly.
G) Montage of spindle disassembly events in ParM(S19E,G21E) mutant (Movie S17). The spindle splits apart due to repulsive surface charges, demonstrating that the bipolar spindle of ParM (magenta) is formed of more than one filament. The constituent filaments with ParRC (green) bound at one end begin disassembly from the pointed-ends. The pointed-ends are highlighted using arrowheads.
H) Schematic representation of the bipolar spindle model (see also Fig. S6).
The helical geometry of ParM filament, with 12 subunits per turn, is compatible with a hexagonal or square packing of ParM filaments in a bundle (Fig. S5C) (4), in contrast to non-bundling actin filaments with a 13-monomer repeat. Bundles of ParM filaments have been observed in E. coli cells expressing ParM at wild-type levels (20) and during plasmid partitioning in vivo (21). Also, bundles of antiparallel ParM filaments have been described in vitro (22).
Antiparallel pairing explains observations of ParRC at both ends of ParM filaments (9, 10). Of the 826 filaments we counted, 104, 540 and 182 filaments were observed with ParRC at 0, 1 and 2 ends, respectively, consistent with single-end binding and pairing. Bipolar spindles of ParM were observed by TIRF microscopy using ATP. Stable filaments were seen only as elongating spindles with ParRC at both ends, pushing plasmids apart at a rate of 22.6 ± 4.8 monomers/s (n = 40; Movies S13-S14; Table S3). Upon loss of ParRC at limiting concentrations of ParM (350 nM – 500 nM), dynamic instability caused spindle disassembly at a rate of 100.3 ± 18.7 monomers/s (n = 61; Figs. 4F, S4E; Movies S15-S16; Table S3).
To demonstrate that the spindles comprised at least two antiparallel filaments, we introduced negatively charged residues within loop 18-21 (S19E, G21E), (Fig. 4D) to weaken the interfilament interaction through repulsive electrostatic forces. We observed spindles (n = 65) that split into the constituent filaments (Movie S17, Fig. 4G). This confirms that spindles are not formed by a single ParM filament, with ParRC attached at both ends (9, 10) (Fig. 4H). ParM filaments in a spindle were protected from dynamic instability by binding of ParRC at the barbed-end and pairing with another ParRC-bound filament at the pointed-end (Movies S15-S19; Figs. 4F-H, S4E-F). The trigger for disassembly was either the loss of ParRC (Movies S15–S16) or exposure of pointed-ends due to unwinding of the antiparallel bundle (Movies S17–S19).
These observations, and previously published work (3, 4), provide a comprehensive model of ParMRC-mediated plasmid segregation (Fig. S6): a critical concentration of ATP-bound monomers in the cell nucleates ParM filaments (or nucleation is ParRC-mediated) (18). ParRC–binding rescues the dynamic filaments from disassembly at the barbed-end only. ParRC speeds up the growth at the barbed-end by a formin-like mechanism. The free pointed-end remains prone to disassembly unless it pairs up antiparallel with another ParM filament bound to ParRC, probably shortly after plasmid replication. Thus, a bipolar spindle of two antiparallel filaments is the minimum unit in plasmid segregation. R1 plasmid has a copy number of about six, which is approximately the number of filaments within bundles in plasmid-segregating cells (20). ParM bundles are stronger than single filaments, which is advantageous when pushing plasmids through the cytoplasm. A single bundle will also ensure concerted segregation of all sister plasmids, as observed in vivo (21).
The lateral interaction among dynamic filaments, as in ParMRC, may also facilitate contraction in other cytomotive filament systems such as FtsZ in bacterial cell division (23). Our model of antiparallel actin-like ParM filaments, without the necessity of bundling factors or motor proteins provides an attractive conceptual precursor for actin contractile systems, such as muscle.
One Sentence Summary: A bipolar spindle, formed by antiparallel actin-like filaments, pushes sister plasmids apart during bacterial plasmid segregation.
Supplementary Material
Acknowledgments
We acknowledge N. Barry and C. Johnson (MRC-LMB, help with TIRF microscopy and ITC), beamline support at ID29, ID14-2 (ESRF, France) and I02 (Diamond Light Source, UK), L. Amos, S. Bullock, A. Carter and R. Gasper-Schönenbrücher (MRC-LMB, critical reading of the manuscript), K. Gerdes (Newcastle University) and A. Toste Rêgo (MRC-LMB) (provision of reagents). Support by the Medical Research Council (U105184326 to JL), Grants-in-Aid for Scientific Research (21227006 to KN), and FP7-IIF-2008 fellowship (PG) is acknowledged.
Footnotes
Protein Data Bank (PDB) accession codes: ParM-AMPPNP (4A61), ParM-AMPPNP-ParRpept (4A62). The EM reconstruction has been deposited in the EMDB (EMD-1980), fitted coordinates in the PDB (4A6J).
Supplementary Materials: Materials and Methods
Figs. S1 to S6
Tables S1 to S3
Movies S1 to S19
References (24 – 41)
References
- 1.van den Ent F, Møller-Jensen J, Amos LA, Gerdes K, Löwe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Popp D, et al. Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability. EMBO J. 2008;27:570. doi: 10.1038/sj.emboj.7601978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141:927. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 4.Salje J, Gayathri P, Löwe J. The ParMRC system: molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol. 2010;8:683. doi: 10.1038/nrmicro2425. [DOI] [PubMed] [Google Scholar]
- 5.Garner EC, Campbell CS, Mullins RD. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science. 2004;306:1021. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- 6.Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Møller-Jensen J, Ringgaard S, Mercogliano CP, Gerdes K, Löwe J. Structural analysis of the ParR/parC plasmid partition complex. EMBO J. 2007;26:4413. doi: 10.1038/sj.emboj.7601864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumacher MA, et al. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature. 2007;450:1268. doi: 10.1038/nature06392. [DOI] [PubMed] [Google Scholar]
- 9.Choi CL, Claridge SA, Garner EC, Alivisatos AP, Mullins RD. Proteinnanocrystal conjugates support a single filament polymerization model in R1 plasmid segregation. J. Biol. Chem. 2008;283:28081. doi: 10.1074/jbc.M803833200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salje J, Löwe J. Bacterial actin: architecture of the ParMRC plasmid DNA partitioning complex. EMBO J. 2008;27:2230. doi: 10.1038/emboj.2008.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.See Supplementary materials in Science Online.
- 12.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 13.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 14.Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 15.Hertzog M, et al. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117:611. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- 16.Dominguez R. Actin-binding proteins--a unifying hypothesis. Trends Biochem. Sci. 2004;29:572. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 2007;76:593. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 18.Møller-Jensen J, Jensen RB, Löwe J, Gerdes K. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 2002;21:3119. doi: 10.1093/emboj/cdf320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Møller-Jensen J, et al. Bacterial Mitosis:: ParM of Plasmid R1 Moves Plasmid DNA by an Actin-like Insertional Polymerization Mechanism. Mol. Cell. 2003;12:1477. doi: 10.1016/s1097-2765(03)00451-9. [DOI] [PubMed] [Google Scholar]
- 20.Salje J, Zuber B, Löwe J. Electron cryomicroscopy of E. coli reveals filament bundles involved in plasmid DNA segregation. Science. 2009;323:509. doi: 10.1126/science.1164346. [DOI] [PubMed] [Google Scholar]
- 21.Campbell CS, Mullins RD. In vivo visualization of type II plasmid segregation: bacterial actin filaments pushing plasmids. J. Cell Biol. 2007;179:1059. doi: 10.1083/jcb.200708206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popp D, et al. Concerning the dynamic instability of actin homolog ParM. Biochem. Biophys. Res. Commun. 2007;353:109. doi: 10.1016/j.bbrc.2006.11.130. [DOI] [PubMed] [Google Scholar]
- 23.Sun SX, Walcott S, Wolgemuth CW. Cytoskeletal cross-linking and bundling in motor-independent contraction. Curr Biol. 2010;20:R649. doi: 10.1016/j.cub.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 25.Stock D, Perisic O, Löwe J. Robotic nanolitre protein crystallisation at the MRC Laboratory of Molecular Biology. Prog Biophys Mol Biol. 2005;88:311. doi: 10.1016/j.pbiomolbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 26.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62:48. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 27.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 32.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 33.Yonekura K, Braunfeld MB, Maki-Yonekura S, Agard DA. Electron energy filtering significantly improves amplitude contrast of frozen-hydrated protein at 300kV. J Struct Biol. 2006;156:524. doi: 10.1016/j.jsb.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 35.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 36.Yin J, et al. Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15815. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin J, Lin AJ, Golan DE, Walsh CT. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat Protoc. 2006;1:280. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 38.Dam M, Gerdes K. Partitioning of plasmid R1. Ten direct repeats flanking the parA promoter constitute a centromere-like partition site parC, that expresses incompatibility. J. Mol. Biol. 1994;236:1289. doi: 10.1016/0022-2836(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 39.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using microManager. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1420s92. Chapter 14, Unit 14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.