Abstract
Cancer is a disease of aberrant gene expression. While the genetic causes of cancer have been intensively studied, it is becoming evident that a large proportion of cancer susceptibility cannot be attributed to variation in protein-coding sequences. This is highlighted by genome-wide association studies in cancer that reveal that more than 80% of cancer-associated SNPs occur in noncoding regions of the genome. In this review, we posit that a significant fraction of the genetic aetiology of cancer is exacted by noncoding regulatory sequences, particularly by long noncoding RNAs (lncRNAs). Recent studies indicate that several cancer risk loci are transcribed into lncRNAs and these transcripts play key roles in tumorigenesis. We discuss the epigenetic and other mechanisms through which lncRNAs function and how they contribute to each stage of cancer progression, understanding of which will be crucial for realising new opportunities in cancer diagnosis and treatment. Long noncoding RNAs play important roles in almost every aspect of cell biology from nuclear organisation and epigenetic regulation to post-transcriptional regulation and splicing, and we link these processes to the hallmarks and genetics of cancer. Finally, we highlight recent progress and future potential in the application of lncRNAs as therapeutic targets and diagnostic markers.
Keywords: GWAS, lncRNA, cancer heritability, regulatory RNA
Cancer is a genetic disease – the result of dysregulation of the gene networks that maintain normal cellular identity, growth and differentiation. Only a small proportion of cancers are attributed to inheritable single-gene disorders, usually involving non-synonymous mutations in the coding sequence of protein-coding genes, such as BRCA1 in familial breast cancer (Miki et al, 1994) and Rb1, which causes familial retinoblastoma (Du and Pogoriler, 2006). The majority of cancers arise via somatic mutations. The susceptibility to develop sporadic cancer is complex, comprising various genetic and environmental factors. The heritable component of cancer susceptibility is dependent on the cancer type and is in many cases considerable. Despite extensive study, the majority of the genetic component of cancer susceptibility has not yet been linked to individual genes, highlighting significant deficiencies in our understanding of the molecular basis of cancer development. A key development in unravelling the complex genetics of cancer may be the shift in focus from looking exclusively at the protein-coding components of the genome to consideration of the role of variation in regulatory elements.
The combination of various genome-wide approaches, best typified by the ENCODE project (Dunham et al, 2012), has stimulated a dramatic reassessment of the information content of the human genome. Rather than islands of protein-coding genes in a sea of junk DNA, it is increasingly apparent that much of the genome, far more than expected, encodes regulatory information. Indeed the ENCODE project recently concluded that although only ∼1.2% of the genome is protein-coding, at least 20% shows biological function and over 80% exhibits biochemical indices of function. The non-protein-coding regions of the genome serve not only as a substrate for DNA-binding proteins that in turn govern both the expression and 3D architecture of the genome, but also as a template for the transcription of vast numbers of noncoding RNAs (Carninci et al, 2005), which exhibit exquisite cell-specific and developmental dynamic expression patterns (Dinger et al, 2008; Mercer et al, 2008, 2010; Pang et al, 2009; Cabili et al, 2011; Khaitan et al, 2011), capable of transacting a wide repertoire of regulatory functions (Amaral et al, 2008; Mercer et al, 2009).
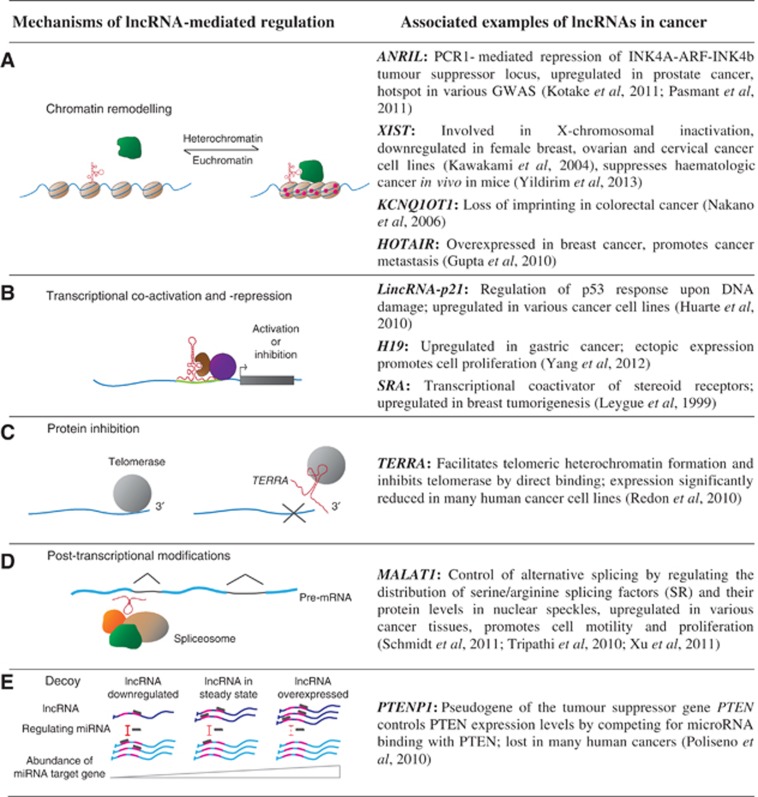
Based on transcript size, these noncoding RNAs can be grouped into two major classes: small noncoding RNAs (<200 bp) and long noncoding RNAs (lncRNAs; >200 bp, up to ∼100 kb). Long noncoding RNAs share many features of mRNAs; they are frequently transcribed by RNA polymerase II, polyadenylated and can show complex splicing patterns. Long noncoding RNAs are found in sense or antisense orientation to protein-coding genes, within introns of protein-coding genes or in intergenic regions of the genome. While many lncRNAs may function in cis (through the act of their transcription), a significant proportion of lncRNAs have intrinsic RNA-mediated functions in trans (Guttman et al, 2011). Although the functions of only a minority have been described, their dynamic and regulated expression suggests that many more may be functional. Long noncoding RNAs can operate through a variety of mechanisms (reviewed by Rinn and Chang, 2012) and their importance in many aspects of cell differentiation and homeostasis is well established. In Figure 1, we have summarised a number of these mechanisms and provide examples for lncRNAs involved in different steps of cancer progression.
Figure 1.
Generalised mechanisms and associated examples of lncRNAs involved in cancer progression. Long noncoding RNAs act through a variety of mechanisms such as remodelling of chromatin (A), transcriptional co-activation or -repression (B), protein inhibition (C), as post-transcriptional modifiers (D) or decoy elements (E). Consequently, mis-expression of lncRNAs can lead to changed expression profiles of various target genes involved in different aspects of cell homeostasis.
Regulatory roles of lncRNAs in cancer
The mechanisms through which lncRNAs contribute to the regulatory networks that underpin cancer development are diverse. Accumulating evidence suggests that a major role of lncRNAs is to guide the site specificity of chromatin-modifying complexes to effect epigenetic changes (Mattick and Gagen, 2001; Mattick et al, 2009). At least 38% of lncRNAs present in several human tissues bind to the polycomb repressive complex 2 or the chromatin-modifying proteins CoREST and SMCX (Khalil et al, 2009). Others bind to trithorax chromatin-activating complexes and/or activated chromatin (Dinger et al, 2008). The well-characterised lncRNAs ANRIL, XIST, HOTAIR and KCNQ1OT1 are able to recruit epigenetic modifiers to specific loci to reprogram the chromatin state. Recent studies have linked their mis-expression to diverse cancers (ANRIL: prostate cancer, XIST: female cancers, HOTAIR: breast cancer, KCNQ1OT1: colorectal cancer) (Figure 1A; Gutschner and Diederichs, 2012).
Other lncRNAs have been found to be key regulators of the protein signalling pathways underlying carcinogenesis. The lncRNA lincRNA-p21 contains binding sites for the tumour suppressor p53 in its promoter and is directly activated by p53 in response to DNA damage. LincRNA-p21 is associated with heterogenous nuclear ribonucleoprotein K and localises this protein to promoters of genes, downregulated in the canonical p53 pathway and p53-mediated apoptosis, to maintain gene repression (Figure 1B; Huarte et al, 2010). Thus, similar to its activator p53, lincRNA-p21 may play an important role in tumour suppression by operating as a transcriptional repressor.
To achieve replicative immortality, cancerous cells need to override the cellular mechanisms inhibiting proliferation. Telomeres are the protective ends of chromosomes, composed of several kilobases of short repeats. These ends are progressively shortened during cell division until they reach a critical length triggering cell death or senescence. However, the majority of cancer cells circumvent this loss by expressing telomerase, an enzyme that adds telomeric repeats to the 3′ end of chromosomes. Recent studies have demonstrated that telomeric ends are transcribed into a lncRNA named TERRA, which binds telomerase, inhibiting its activity in vitro (Redon et al, 2010). In many cancer cells TERRA is downregulated, providing a possible link to the longevity of cancer cells by telomerase-mediated lengthening of chromosomal ends (Ng et al, 2009; Figure 1C).
Some lncRNAs are constituents of macromolecular complexes with roles in RNA processing. The lncRNA MALAT1 is thought to act at a post-transcriptional level (Figure 1D) by controlling alternative splicing of pre-mRNAs. It modulates the levels of active (phosphorylated) serine/arginine (SR) splicing factors (Tripathi et al, 2010). MALAT1 is upregulated in several cancer types and its overexpression has been linked to an increase in cell proliferation and migration in lung and colorectal cancer cells (Schmidt et al, 2011; Xu et al, 2011). A recent study indicated that MALAT1 may also have a role in the regulation of gene expression, but not alternative splicing, in lung metastasis, highlighting the controversial nature of MALAT1's molecular mechanism (Gutschner et al, 2013).
Other lncRNAs can also act as decoys, sequestering biomolecules and preventing them from fulfilling their cellular functions. An example of this mechanism is represented by the tumour suppressor gene PTEN and its pseudogene PTENP1. While PTENP1 has a disrupted open reading frame precluding its ability to encode a functional protein, its 3′UTR region is well-conserved. By ectopic expression, Poliseno et al showed that the PTENP1 3′UTR can increase PTEN expression by binding to microRNAs that downregulate PTEN expression. Thus, PTENP1 plays an important role in cancer biology, restricting cell proliferation by acting as a microRNA decoy for the tumour suppressor PTEN (Poliseno et al, 2010; Figure 1E).
In summary, lncRNAs can act through a number of mechanisms to control cancer state. Despite growing knowledge about the molecular mechanisms of lncRNA functions in cancer, the modes of action of most lncRNAs remain unclear. A broader understanding of the mechanisms of action of lncRNAs, and the regulatory pathways, hierarchies and networks in which they operate, will greatly increase our understanding of their functions in cancer and open new therapeutic avenues to modulate their function.
Genetic association of lncRNAs and cancer
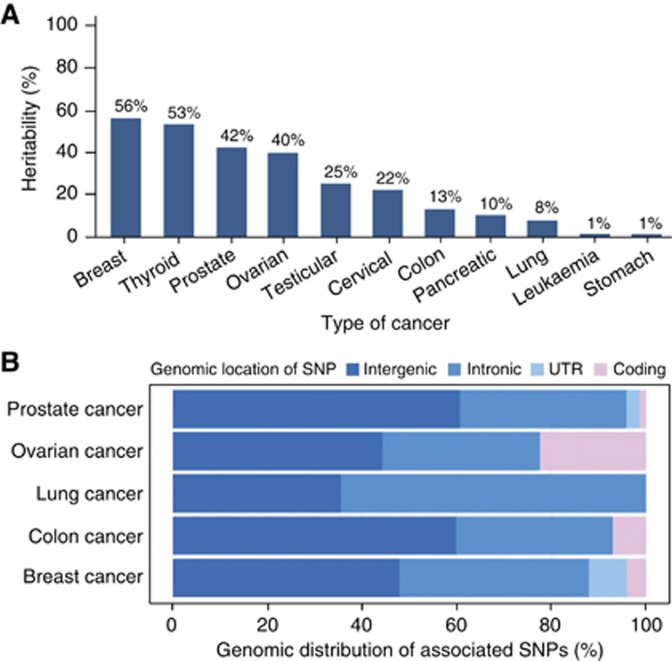
As mentioned above, cancer susceptibility has a considerable heritable component that varies with cancer type (Figure 2A). Surprisingly, the large majority of genome-wide association studies (GWAS) identify cancer risk loci outside of protein-coding regions. We utilised a comprehensive GWAS catalogue (http://www.genome.gov/gwastudies/) to assess the genomic context of SNPs, filtered for association with cancer-related conditions. Of 301 SNPs currently linked to cancer (Supplementary Table 1), only 12 (3.3%) change the protein amino-acid sequence. Most are located in the introns of protein-coding genes (40%) or intergenic regions (44%) (Figure 2B), raising the question of the function of these noncoding loci and their role in cancer development. While some may contribute to cancer risk through cis-regulatory interactions, many of these loci may be transcribed into noncoding RNAs.
Figure 2.
Heritability and genomic distribution of SNPs in cancer. (A) The heritable component of risk for common cancers. Despite many cancers having sizeable genetic components, the identity of most of these heritable risk factors is currently unknown. Table adapted from SNPedia (Cariaso and Lennon, 2012). (B) Genomic distribution (%) of SNPs in selected cancer types. The majority of cancer-related SNPs are located in noncoding regions of the genome (intergenic or intronic) and only a small number are found in coding regions.
Recently several examples of functional lncRNAs transcribed from cancer risk loci have been reported. For example, SNP rs944289 in the 14q13.3 region, which is strongly associated with papillary thyroid carcinoma (PTC), affects the function of a tumour suppressor lncRNA (Jendrzejewski et al, 2012). A thyroid-specific lncRNA, termed PTC susceptibility candidate 3 (PTCSC3), that was strongly downregulated in PTC was identified in this region and it was found that the repression was caused by the associated SNP. The risk allele alters the binding of the C/EBP proteins to the PTCSC3 promoter, reducing gene expression. PTCSC3 exhibits tumour suppressor activity, controlling the expression of genes involved in DNA replication and repair, tumour morphology, cell movement and cell death. This study is to our knowledge the first to link a cancer-associated SNP to a mechanism of action by altering the expression of a tumour suppressor lncRNA.
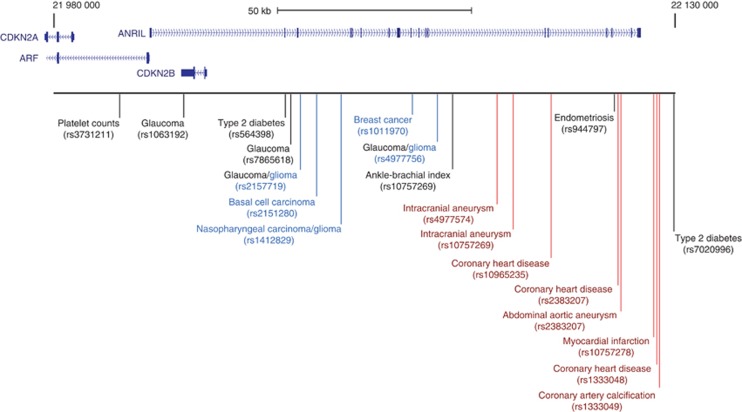
ANRIL, a large lncRNA gene spanning 126 kb adjacent to p14/ARF, is located in a GWAS ‘hot spot' linked to many complex diseases, including type-2 diabetes, coronary artery disease and, recently, cancer (Pasmant et al, 2011). ANRIL interacts with polycomb group proteins and may add repressive histone marks to the p15/CDKN2B-p16/CDKN2A-p14/ARF locus, suppressing cell proliferation. Intriguingly, disease-associated SNPs in ANRIL cluster by disorder; vascular conditions are associated with the 3′ end of the transcript, while cancer susceptibility SNPs map to the 5′ region (Figure 3). This may reflect the mode of action of ANRIL. Polymorphisms in different regions of the ANRIL RNA may affect the RNA–DNA or RNA–protein interactions necessary for ANRIL-induced gene silencing. The region mutated may cause changes in site selectivity of epigenetic programming, with some interactions required for vascular function while other interactions are required for cell cycle maintenance. Further analysis of the structure–function effects of ANRIL polymorphisms may unravel the complexity of this GWAS ‘hot spot'.
Figure 3.
The GWAS ‘hot spot' at the ANRIL locus. Multiple disease-associated SNPs map to the ANRIL locus, including various cancers (blue) and vascular-related conditions (red). These individual polymorphisms may affect ANRIL function differently, resulting in diverse diseases.
It is likely that many more lncRNAs are transcribed from cancer loci, as these loci have typically not been examined in a targeted manner and low-abundance RNAs originating from them may not have been detected or characterised. Even apparent gene desert regions including the extensively studied prostate cancer 8q24 locus produce a lncRNA that may be involved in prostate carcinogenesis (Chung et al, 2011). Recently we described a technology termed RNA Capture-Seq, which is capable of detecting lowly expressed and highly tissue-specific transcripts (Mercer et al, 2012). When applied to gene deserts, whole forests of previously unannotated transcripts were identified. This technique will allow the focused re-examination of cancer-associated regions for novel and rare noncoding transcripts that may be important regulators of carcinogenesis. Combined, these studies suggest that many noncoding cancer risk loci identified by GWAS are transcribed into lncRNAs with important regulatory functions in cancer biology.
Diagnostics and therapeutic potential of lncRNAs in cancer
The discovery that lncRNAs are key regulators in cancer transformation and progression leads to intriguing possibilities of application for diagnostics and therapeutics. Many lncRNAs are expressed in a tissue- and cancer-type restricted manner and have already shown to be useful as prognostic markers. HOTAIR expression, for example, is strongly increased in primary tumours and metastases of breast cancer patients, and expression levels correlate positively with a poor outcome (Gupta et al, 2010). In a recent study, Yang et al proposed that high expression levels of HOTAIR could serve as a biomarker to predict tumour recurrence in patients with hepatocellular carcinomas (Yang et al, 2011).
The use of noncoding RNAs in diagnostics has intrinsic advantages over protein-coding RNAs. Although lncRNAs may require post-transcriptional modifications or protein interactions to function, because the mature product is the functional end-product, measurement of its expression directly represents the levels of the active molecule. In contrast, mRNA levels are only indirectly indicative of the levels of the functional product of coding genes (the encoded protein). Long noncoding RNA levels may have a higher correlation with particular cancer states and thus be more useful diagnostic tools. Noncoding RNAs are often stable in human serum and thus measuring either individual marker RNAs (e.g., by qPCR) or the entire transcriptome (e.g., RNA-seq) may allow the non-invasive generation of reliable and actionable clinical indicators (Tong and Lo, 2006). For example, the lncRNA prostate cancer gene 3 (PCA3) is highly associated with prostate cancer and is routinely used to indicate prostate cancer risk (Progensa PCA3 urine test) in urine samples, thereby avoiding unnecessary prostate biopsies (de la Taille, 2007).
Clinical transcriptomics will greatly impact on the medical treatment of cancer. It is foreseeable that clinicians may use analysis of tumour transcriptomes on initial diagnosis, enabling a personalised treatment regime rather than a generic alternative. This unbiased approach avoids preconceptions about which molecular pathways (coding or noncoding) may underlie the disease. Additionally, this information may allow more accurate prognostic predictions, for example, by determining the expression of molecular markers of prognosis and metastasis (e.g., HOTAIR). Subsequently, progression of the individual cancer may be monitored by transcriptomics to detect progression, recurrence and metastasis. Long noncoding RNAs are typically more cell-type specific than protein-coding genes (Cabili et al, 2011) and may allow estimation of the cellular composition of a tumour by marking a specific cell population (e.g., cancer stem cells) (Chan et al, 2013).
Finally, the cell-type specificity of cancer-associated lncRNAs and their regulatory networks can aid the development of targeted therapies. H19, a lncRNA with oncogenic properties, is upregulated in a wide range of tumours. To treat H19-driven cancer types, a plasmid (BC-819) carrying diphtheria toxin under the control of the H19 regulatory sequence has been developed to target cells overexpressing H19. Intratumoral injection of BC-819 was successfully applied in patients with bladder, ovarian and pancreatic cancer to reduce tumour size (Smaldone and Davies, 2010). Several studies also indicate that the reduction of MALAT1 expression levels by siRNAs can influence the migratory and proliferative potential of lung adenocarcinoma and cervical cancer cells in culture (Guo et al, 2010; Tano et al, 2010). Similarly to H19/BC-819, expression of these lncRNA-specific siRNAs in a tumour-restricted manner will allow precise targeting of further tumour types without excessive harm to healthy tissue.
Conclusion
In summary, lncRNAs play integral roles in the control of cellular growth, division and differentiation. The perturbation of lncRNA expression can contribute to the development and progression of cancer. While a large proportion of cancer susceptibility is heritable, the underlying genetic components are largely unknown. Here we posit that a large proportion of the cancer risk may be explained by lncRNAs transcribed from cancer-associated loci. These RNAs exact functions through a diverse range of mechanisms. Characterisation of these lncRNA genes and their modes of action will allow their use for improved cancer diagnosis, monitoring of progression and targeted therapies.
Acknowledgments
MED and JSM are supported by a Career Development Award and an Australia Fellowship, respectively, from the National Health and Medical Research Council of Australia.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariaso M, Lennon G. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Res. 2012;40:D1308–D1312. doi: 10.1093/nar/gkr798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest ARR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SPT, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schönbach C, Sekiguchi K, CAM Semple, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y, FANTOM Consortium, RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Chan KC, Jiang P, Zheng YW, Liao GJ, Sun H, Wong J, Siu SS, Chan WC, Chan SL, Chan AT, Lai PB, Chiu RW, Lo YM. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- Chung S, Nakagawa H, Uemura M, Piao L, Ashikawa K, Hosono N, Takata R, Akamatsu S, Kawaguchi T, Morizono T, Tsunoda T, Daigo Y, Matsuda K, Kamatani N, Nakamura Y, Kubo M. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011;102:245–252. doi: 10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- de la Taille A. Progensa PCA3 test for prostate cancer detection. Expert Rev Mol Diagn. 2007;7:491–497. doi: 10.1586/14737159.7.5.491. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Pogoriler J. Retinoblastoma family genes. Oncogene. 2006;25:5190–5200. doi: 10.1038/sj.onc.1209651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Dunham I, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kelllis M, Khatun J, Kheradpour P, Kundaje A, Lassman T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SC, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, Ward LD, Whitfield TW, Wilder SP, Wu W, Xi HS, Yip KY, Zhuang J, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, Pazin MJ, Lowdon RF, Dillon LA, Adams LB, Kelly CJ, Zhang J, Wexler JR, Green ED, Good PJ, Feingold EA, Bernstein BE, Birney E, Crawford GE, Dekker J, Elinitski L, Farnham PJ, Gerstein M, Giddings MC, Gingeras TR, Green ED, Guigó R, Hardison RC, Hubbard TJ, Kellis M, Kent WJ, Lieb JD, Margulies EH, Myers RM, Snyder M, Starnatoyannopoulos JA, Tennebaum SA, Weng Z, White KP, Wold B, Khatun J, Yu Y, Wrobel J, Risk BA, Gunawardena HP, Kuiper HC, Maier CW, Xie L, Chen X, Giddings MC, Bernstein BE, Epstein CB, Shoresh N, Ernst J, Kheradpour P, Mikkelsen TS, Gillespie S, Goren A, Ram O, Zhang X, Wang L, Issner R, Coyne MJ, Durham T, Ku M, Truong T, Ward LD, Altshuler RC, Eaton ML, Kellis M, Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Batut P, Bell I, Bell K, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena HP, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Li G, Luo OJ, Park E, Preall JB, Presaud K, Ribeca P, Risk BA, Robyr D, Ruan X, Sammeth M, Sandu KS, Schaeffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Hayashizaki Y, Harrow J, Gerstein M, Hubbard TJ, Reymond A, Antonarakis SE, Hannon GJ, Giddings MC, Ruan Y, Wold B, Carninci P, Guigó R, Gingeras TR, Rosenbloom KR, Sloan CA, Learned K, Malladi VS, Wong MC, Barber GP, Cline MS, Dreszer TR, Heitner SG, Karolchik D, Kent WJ, Kirkup VM, Meyer LR, Long JC, Maddren M, Raney BJ, Furey TS, Song L, Grasfeder LL, Giresi PG, Lee BK, Battenhouse A, Sheffield NC, Simon JM, Showers KA, Safi A, London D, Bhinge AA, Shestak C, Schaner MR, Kim SK, Zhang ZZ, Mieczkowski PA, Mieczkowska JO, Liu Z, McDaniell RM, Ni Y, Rashid NU, Kim MJ, Adar S, Zhang Z, Wang T, Winter D, Keefe D, Birney E, Iyer VR, Lieb JD, Crawford GE, Li G, Sandhu KS, Zheng M, Wang P, Luo OJ, Shahab A, Fullwood MJ, Ruan X, Ruan Y, Myers RM, Pauli F, Williams BA, Gertz J, Marinov GK, Reddy TE, Vielmetter J, Partridge EC, Trout D, Varley KE, Gasper C, Bansal A, Pepke S, Jain P, Amrhein H, Bowling KM, Anaya M, Cross MK, King B, Muratet MA, Antoshechkin I, Newberry KM, McCue K, Nesmith AS, Fisher-Aylor KI, Pusey B, DeSalvo G, Parker SL, Balasubramanian S, Davis NS, Meadows SK, Eggleston T, Gunter C, Newberry JS, Levy SE, Absher DM, Mortazavi A, Wong WH, Wold B, Blow MJ, Visel A, Pennachio LA, Elnitski L, Margulies EH, Parker SC, Petrykowska HM, Abyzov A, Aken B, Barrell D, Barson G, Berry A, Bignell A, Boychenko V, Bussotti G, Chrast J, Davidson C, Derrien T, Despacio-Reyes G, Diekhans M, Ezkurdia I, Frankish A, Gilbert J, Gonzalez JM, Griffiths E, Harte R, Hendrix DA, Howald C, Hunt T, Jungreis I, Kay M, Khurana E, Kokocinski F, Leng J, Lin MF, Loveland J, Lu Z, Manthravadi D, Mariotti M, Mudge J, Mukherjee G, Notredame C, Pei B, Rodriguez JM, Saunders G, Sboner A, Searle S, Sisu C, Snow C, Steward C, Tanzer A, Tapanari E, Tress ML, van Baren MJ, Walters N, Washieti S, Wilming L, Zadissa A, Zhengdong Z, Brent M, Haussler D, Kellis M, Valencia A, Gerstein M, Raymond A, Guigó R, Harrow J, Hubbard TJ, Landt SG, Frietze S, Abyzov A, Addleman N, Alexander RP, Auerbach RK, Balasubramanian S, Bettinger K, Bhardwaj N, Boyle AP, Cao AR, Cayting P, Charos A, Cheng Y, Cheng C, Eastman C, Euskirchen G, Fleming JD, Grubert F, Habegger L, Hariharan M, Harmanci A, Iyenger S, Jin VX, Karczewski KJ, Kasowski M, Lacroute P, Lam H, Larnarre-Vincent N, Leng J, Lian J, Lindahl-Allen M, Min R, Miotto B, Monahan H, Moqtaderi Z, Mu XJ, O'Geen H, Ouyang Z, Patacsil D, Pei B, Raha D, Ramirez L, Reed B, Rozowsky J, Sboner A, Shi M, Sisu C, Slifer T, Witt H, Wu L, Xu X, Yan KK, Yang X, Yip KY, Zhang Z, Struhl K, Weissman SM, Gerstein M, Farnham PJ, Snyder M, Tenebaum SA, Penalva LO, Doyle F, Karmakar S, Landt SG, Bhanvadia RR, Choudhury A, Domanus M, Ma L, Moran J, Patacsil D, Slifer T, Victorsen A, Yang X, Snyder M, White KP, Auer T, Centarin L, Eichenlaub M, Gruhl F, Heerman S, Hoeckendorf B, Inoue D, Kellner T, Kirchmaier S, Mueller C, Reinhardt R, Schertel L, Schneider S, Sinn R, Wittbrodt B, Wittbrodt J, Weng Z, Whitfield TW, Wang J, Collins PJ, Aldred SF, Trinklein ND, Partridge EC, Myers RM, Dekker J, Jain G, Lajoie BR, Sanyal A, Balasundaram G, Bates DL, Byron R, Canfield TK, Diegel MJ, Dunn D, Ebersol AK, Ebersol AK, Frum T, Garg K, Gist E, Hansen RS, Boatman L, Haugen E, Humbert R, Jain G, Johnson AK, Johnson EM, Kutyavin TM, Lajoie BR, Lee K, Lotakis D, Maurano MT, Neph SJ, Neri FV, Nguyen ED, Qu H, Reynolds AP, Roach V, Rynes E, Sabo P, Sanchez ME, Sandstrom RS, Sanyal A, Shafer AO, Stergachis AB, Thomas S, Thurman RE, Vernot B, Vierstra J, Vong S, Wang H, Weaver MA, Yan Y, Zhang M, Akey JA, Bender M, Dorschner MO, Groudine M, MacCoss MJ, Navas P, Stamatoyannopoulos G, Kaul R, Dekker J, Stamatoyannopoulos JA, Dunham I, Beal K, Brazma A, Flicek P, Herrero J, Johnson N, Keefe D, Lukk M, Luscombe NM, Sobral D, Vaquerizas JM, Wilder SP, Batzoglou S, Sidow A, Hussami N, Kyriazopoulou-Panagiotopoulou S, Libbrecht MW, Schaub MA, Kundaje A, Hardison RC, Miller W, Giardine B, Harris RS, Wu W, Bickel PJ, Banfai B, Boley NP, Brown JB, Huang H, Li Q, Li JJ, Noble WS, Bilmes JA, Buske OJ, Hoffman MM, Sahu AO, Kharchenko PV, Park PJ, Baker D, Taylor J, Weng Z, Iyer S, Dong X, Greven M, Lin X, Wang J, Xi HS, Zhuang J, Gerstein M, Alexander RP, Balasubramanian S, Cheng C, Harmanci A, Lochovsky L, Min R, Mu XJ, Rozowsky J, Yan KK, Yip KY, Birney E, ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Li Y, Liu Y, Wang J, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2010;42:224–229. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Hämmerle M, Eißmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Groß M, Zörnig M, MacLeod AR, Spector DL, Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrzejewski J, He H, Radomska HS, Li W, Tomsic J, Liyanarachchi S, Davuluri RV, Nagy R, de la Chapelle A. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci USA. 2012;109:8646–8651. doi: 10.1073/pnas.1205654109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Zhang C, Taniguchi T, Kim CJ, Okada Y, Sugihara H, Hattori T, Reeve AE, Ogawa O, Okamoto K. Characterization of loss-of-inactive X in Klinefelter syndrome and female-derived cancer cells. Oncogene. 2004;23:6163–6169. doi: 10.1038/sj.onc.1207808. [DOI] [PubMed] [Google Scholar]
- Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leygue E, Dotzlaw H, Watson PH, Murphy LC. Expression of the steroid receptor RNA activator in human breast tumors. Cancer Res. 1999;59:4190–4193. [PubMed] [Google Scholar]
- Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol. 2001;18:1611–1630. doi: 10.1093/oxfordjournals.molbev.a003951. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long noncoding RNAs: insights into function. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Nakano S, Murakami K, Meguro M, Soejima H, Higashimoto K, Urano T, Kugoh H, Mukai T, Ikeguchi M, Oshimura M. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer Sci. 2006;97:1147–1154. doi: 10.1111/j.1349-7006.2006.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LJ, Cropley JE, Pickett HA, Reddel RR, Suter CM. Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic Acids Res. 2009;37:1152–1159. doi: 10.1093/nar/gkn1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, Mattick JS. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol. 2009;182:7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LH, Spieker T, Koschmieder S, Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, Wiewrodt R, Muller-Tidow C. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther. 2010;12:607–616. [PubMed] [Google Scholar]
- Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Tong YK, Lo YM. Diagnostic developments involving cell-free (circulating) nucleic acids. Clin Chim Acta. 2006;363:187–196. doi: 10.1016/j.cccn.2005.05.048. [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39:169–175. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.