Abstract
The Vif protein of human immunodeficiency virus type 1 (HIV-1) is essential for viral evasion of the host antiviral protein APOBEC3G, also known as CEM15. Vif mutant but not wild-type HIV-1 viruses produced in the presence of APOBEC3G have been shown to undergo hypermutations in newly synthesized viral DNA upon infection of target cells, presumably resulting from C-to-U modification during minus-strand viral DNA synthesis. We now report that HIV-1 Vif could induce rapid degradation of human APOBEC3G that was blocked by the proteasome inhibitor MG132. The efficiency of Vif-induced downregulation of APOBEC3G expression depended on the level of Vif expression. A single amino acid substitution in the conserved SLQXLA motif reduced Vif function. Vif proteins from distantly related primate lentiviruses such as SIVagm were unable to suppress the antiviral activity of human APOBEC3G or the packaging of APOBEC3G into HIV-1 Vif mutant virions, due to a lack of interaction with human APOBEC3G. In the presence of the proteasome inhibitor MG132, virion-associated Vif increased dramatically. However, increased virion packaging of Vif did not prevent virion packaging of APOBEC3G when proteasome function was impaired, and the infectivity of these virions was significantly reduced. These results suggest that Vif function is required during virus assembly to remove APOBEC3G from packaging into released virions. Once packaged, virion-associated Vif could not efficiently block the antiviral activity of APOBEC3G.
The Vif protein, which modulates viral infectivity (8, 11, 13, 15, 19, 27, 32, 40, 45, 53, 56, 57, 60-62) and pathogenicity (7, 19, 20, 24, 25, 35), is present in nearly all lentiviruses, including human immunodeficiency virus type 1 (HIV-1). It is believed to act during the late stages of virus assembly by enabling the establishment of integrated provirus in new target cells. Since Vif mutant virions show severely impaired infectivity, Vif must regulate one or more of the molecules found in virions. Immunofluorescence analysis of infected cells has demonstrated that Vif and the major structural protein, Gag, colocalize in the cytoplasm (51). Furthermore, Vif cosediments with some of the intracellular preassembly complexes of Gag, but not with the more mature forms of these assembly complexes (50). These data suggest that Vif and Gag may be transiently found together in a complex during virus assembly. However, the failure of Gag and Vif to coimmunoprecipitate tends to argue against a direct interaction of Vif with assembling Gag particles (50), although the involvement of additional bridging molecules has not been ruled out. For example, HIV-1 Vif has been shown to interact with viral genomic RNA (8, 30, 65), and viral RNA could serve as a linker between Gag and Vif.
Analysis of Vif mutant virions during entry into target cells has suggested that either the stabilization of the viral nucleoprotein complex is compromised (51) or the processivity of reverse transcription is impaired (41, 57, 62). Studies examining biochemical differences between wild-type and Vif mutant virions have provided little compelling evidence for Vif-mediated modification or altered incorporation of the virion-associated proteins Gag, Pol, and Env (1, 12, 43, 62) or RNA components (8, 14). However, some studies have found that detergent-treated Vif mutant virions are defective in de novo reverse transcription (8, 10, 17, 41, 44), and treatment of Vif mutant virions with high concentration of deoxynucleoside triphosphates partially restores virus infectivity (10).
The requirement for Vif in viral replication exhibits a striking cell type dependence. For example, Jurkat, CEM-SS, and SupT1 cells do not require Vif for HIV-1 replication (permissive cells); for H9 cells, CEM cells, and primary blood-derived monocytes, however, Vif is essential (nonpermissive cells). In the case of nonhuman lentiviruses, primary blood-derived monocytes derived from the appropriate animals fail to support the replication of Vif mutant viruses (10, 19, 45, 61). Cell fusion experiments with permissive and nonpermissive cells have indicated that the nonpermissive phenotype is dominant (36, 52), leading to the concept that there exist specific cellular factors that act as inhibitors of lentiviral replication and which Vif must overcome (36, 52).
Recently, CEM15 (also called APOBEC3G, and hereafter referred to by this name), which is present only in nonpermissive cells, has been identified as a mediator of anti-HIV-1 activity, and its activity has been shown to be suppressed by Vif (48). APOBEC3G belongs to a family of proteins that have cytidine deaminase activity (26, 48, 63), but its cellular function is still unknown (26, 48, 63). When expressed in Escherichia coli, proteins from this family have been shown to induce DNA mutations (22) and may show a preference for single-stranded DNA (46). Vif mutant but not wild-type HIV-1 viruses produced in the presence of APOBEC3G undergo hypermutations in newly synthesized viral DNA (21, 31, 37, 38, 66), presumably due to C-to-U modification during minus-strand viral DNA synthesis (21, 31, 37, 38, 66). Although it was shown initially that APOBEC3G was incorporated efficiently into wild-type and Vif mutant virions (48), recent studies have shown that HIV-1 Vif could efficiently prevent virion incorporation of APOBEC3G (29, 38, 39, 49, 59, 64).
In this study, we report that unlike HIV-1 Vif, Vif proteins from distantly related primate lentiviruses such as SIVagm are unable to interact with and to suppress the antiviral activity of human APOBEC3G and cannot prevent APOBEC3G packaging into HIV-1 virions. In the case of HIV-1, a single amino acid substitution in the highly conserved lentiviral Vif motif (SLQXLA) reduced the ability of HIV-1 Vif to exclude virion packaging of APOBEC3G and to suppress the antiviral activity of APOBEC3G. Experiments in the presence of proteasome inhibitors indicated that the majority of the Vif molecules were removed from released virions through proteasome activity. When proteasome activity was compromised, more molecules of Vif than of APOBEC3G were packaged in the released virions. However, the infectivity of these virions was significantly impaired, suggesting that the mere presence of Vif in virions was not sufficient to overcome the APOBEC3G activity.
MATERIALS AND METHODS
Plasmid construction.
The parental wild-type HIV-1 (HXB2) and Vif mutant (HXB2ΔVif) have been described previously (8). HXB2VifΔSLQ was constructed by changing amino acids 144 to 146 from SLQ to AAA. HXB2VifΗΑ (64) and HXB2Hygro (33) have been described previously. The VR1012 vector (23) was generously provided by Vical Inc. (San Diego, Calif.). The expression vector for HIV-1 Vif (pHIV-1 Vif) was constructed by PCR amplification of HXB2 Vif with forward primer 5′-GTCGACAACATGGAAAACAGATGGCAGG-3′ and reverse primer 5′-GCGGCCGCCTAGTGTCCATTCATTGTGTG-3′, which contains SalI and NotI sites, on the end, respectively. The PCR products were cloned into VR1012 to generate pHIV-1Vif. pHIV-1VifΔS was generated from pHIV-1Vif by changing S144 to A in Vif.
The expression vector for SIVagmTan Vif was constructed by PCR amplification of SIVagmTan Vif with pSIVagmTan-1 (AIDS Research and Reference Reagents Program, Division of AIDS, NIAID, National Institutes of Health) (55) as the template and the following forward primer 5′-GTCGACACCATGGAGAGAGAAAAACTGTG and reverse primer 5′-GCGGCCGCCTAAAGATCTTCTTCTGATATGAGTTTTTGTTCTAAGCCCTTTCTATC-3′,containing SalI and NotI sites, respectively. The PCR products were cloned into VR1012 to generate pTanVif.
Human APOBEC3G-HA was amplified by reverse transcription-PCR with mRNA from H9 cells with the forward primer 5′-CTCGAGACCATGAAGCCTCACTT-3′ and reverse primer 5′-GAATTCTCACGCGTAATCTGGGACGTCGTAAGGGTAGTTTTCCTGATTCTGGAG-3′, containing XhoI and EcoRI sites, respectively. The PCR product was cloned into pcDNA3.1(−) to generate pAPOBEC3G-HA. Phoenix retrovirus vector expressing APOBEC3G-HA (pBMN-I-APOBEC3G) was constructed by inserting the APOBEC3G-HA gene into the pBMN-Z-I-Neo vector (from Garry Nolan, Stanford University) at the EcoRI site.
Antibodies and cells.
The following antibodies were used for this study: polyclonal rabbit anti-Vif antibody (AIDS Research and Reference Reagents Program, Division of AIDS, NIAID, National Institutes of Health. catalog number 2221) (16), anti-p24 monoclonal antibody (AIDS Research and Reference Reagents Program, Division of AIDS, NIAID, National Institutes of Health, catalog number 1513) (5), anti-p17MA monoclonal antibody (AIDS Research and Reference Reagents Program, Division of AIDS, NIAID, National Institutes of Health, catalog number286) (6), mouse antihemagglutinin (anti-HA) monoclonal antibody (Covance, catalog numberMMS-101R-10000), and anti-human ribosomal P antigen (Immunovision, catalog number HP0-0100) antibodies. 293T, COS-7, and MAGI-CCR5 cells (AIDS Research and Reference Reagents Program, Division of AIDS, NIAID, National Institutes of Health. catalog number 3522) (3) were maintained in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) with 10% fetal bovine serum and penicillin-streptomycin (D-10 medium) and passaged upon confluence. Jurkat and SupT1 cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum and penicillin/streptomycin (R-10 medium).
Transfection, infection, and virus purification.
DNA transfection was carried out with Lipofectamine 2000 (Invitrogen) as described by the manufacturer. To obtain 293T/APOBEC3G cells, the pABOBEC3G-HA plasmid was transfected into 293T cells and selected with 1 mg of G418 (Invitrogen) per ml for 2 weeks. Expression of APOBEC3G was detected by immunoblotting with the anti-HA monoclonal antibody. A Moloney murine leukemia virus-derived retroviral expression vector, pBMN-Z-I-Neo, which expresses both lacZ and neo under the control of the retroviral long terminal repeat, was used to construct a retroviral expression vector for APOBEC3G by replacing lacZ. The APOBEC3G expression vector, pBMN-APO-3G, was transfected into Phoenix helper cells (from Garry Nolan) to generate infectious retroviruses. At 48 h after transfection, retroviral supernatant from the transfected helper cell culture was used to transduce Jurkat or SupT1 cells. Cells were selected in the presence of neomycin for 2 weeks to generate SupT1/APOBEC3G and Jurkat/APOBEC3G cells.
COS-7 cells were transfected with HXB2 or HXB2ΔVif for 48 h. Supernatants of viruses (normalized by p24 level) were used to infect SupT1/APOBEC3G or Jurkat/APOBEC3G cells. Input viruses were removed by washing cells twice with Hanks' balanced salt solution (Invitrogen), and infected cells were maintained in fresh R-10 medium. HIV-1 replication was determined by measuring the amount of p24 in the culture supernatant with an HIV-1 p24 enzyme-linked immunosorbent assay kit (PerkinElmer Life Sciences, Inc.). Virion-associated viral proteins were prepared from cell culture supernatants and separated from cellular debris by centrifugation at 3,000 rpm for 30 min in a Sorvall RT 6000B centrifuge and filtration through a 0.2-μm-pore-size membrane. Virus particles were concentrated by centrifugation through a 30% sucrose cushion at 100,000 × g for 2 h in a Sorvall Ultra80 ultracentrifuge. For virus purification in the presence of proteasome inhibitor, culture media were replaced with fresh D-10 medium containing 2.5 μM MG132. Media from the control cell cultures were replaced with D-10 medium containing dimethyl sulfoxide. Cells were treated for 16 h, and virions were purified as described above.
MAGI assay.
Viral infection was determined by MAGI assay (3) as follows. MAGI-CCR-5 cells were prepared in six-well plates in D-10 medium 1 day before infection, and the cells were at 30 to 40% confluence on the day of infection. Cells were infected by removing medium from each well and adding dilutions of virus in a total volume of 500 μl of complete DMEM with 20 μg of DEAE-dextran per ml. After a 2-h incubation at 37°C in a 5% CO2 incubator, 2 ml of complete DMEM was added to each well. The cells were incubated for 48 h under the same conditions. Supernatants were removed, and 800 μl of fixing solution (1% formaldehyde, 0.2% glutaraldehyde in phosphate-buffered saline [PBS]) was added. After a 5-min incubation, cells were washed twice with PBS. The staining solution (20 μl of 0.2 M potassium ferrocyanide, 20 μl of 0.2 M potassium ferricyanide, 2 μl of 1 M MgCl2, and 10 μl of 40-mg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactosidaseactopyranoside [X-Gal]) was added. Cells were incubated for 2 h at 37°C in a non-CO2 incubator. Staining was stopped by removing the staining solution, and the cells were thoroughly washed twice with PBS. Positive blue dots were counted, and viral infectivity was determined after normalizing the amount of input virus in terms of p24 antigen content.
Immunoblot analysis.
Cells were collected 48 h after transfection. Cell and viral lysates were prepared as previously described (9). Cells (105) were lysed in 1x loading buffer (0.08 M Tris, pH 6.8, with 2.0% sodium dodecyl sulfate, 10% glycerol, 0.1 M dithiothreitol, and 0.2% bromophenol blue). The samples were boiled for 10 min, and proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
For virion lysates, cell culture supernatants were collected 72 h after transfection by removal of cellular debris through centrifugation at 3,000 rpm for 10 min in a Sorvall RT 6000B and filtration through a 0.2-μm-pore-size membrane. Virus particles were concentrated by centrifugation through a 30% sucrose cushion at 100,000 × g for 2 h in a Sorvall Ultra80 ultracentrifuge. Proteins were transferred onto two separate nitrocellulose membranes by passive diffusion for 16 h, producing identical mirror-image blots. Membranes were probed with various primary antibodies against proteins of interest. Secondary antibodies were alkaline phosphatase-conjugated anti-human, anti-rabbit, anti-mouse, or anti-goat IgG (Jackson Immunoresearch, Inc), and staining was carried out with 5-bromo-4-chloro-3-indolylphosphate (BCIP) and nitroblue tetrazolium (NBT) solutions prepared from chemicals obtained from Sigma. The staining of HA-tagged APOBEC3G in virion samples was performed with the ECL plus kit (Amersham, RPN2132) with mouse anti-HA monoclonal antibody as the primary antibody and horseradish peroxidase-conjugated anti-mouse IgG as the secondary antibody.
Immunoprecipitation.
For APOBEC3G-HA immunoprecipitation, transfected 293T cells were harvested and washed twice with cold PBS and lysed with PBS containing 0.5% Triton X-100 and protease inhibitor cocktail (Roche, Basel, Switzerland) at 4°C for 1 h. Cell lysates were clarified by centrifugation at 10,000 × g for 30 min at 4°C. Anti-HA agarose (Roche) was mixed with the precleared cell lysates and incubated at 4°C for 3 h. The reaction mixture was then washed three times with cold PBS and eluted with 0.1 M glycine-HCl buffer, pH 2.0. The eluted materials were subsequently analyzed by immunoblotting.
Pulse-chase experiments.
293T cells were transfected with VR1012Vif-Myc or empty vector VR1012 and human pAPOBEC3G-HA at a 1:1 ratio. Two days posttransfection, the cells were washed twice with PBS and then starved for 30 min in methionine- and cysteine-free medium supplemented with 2% fetal bovine serum, after which the cells were suspended and labeled for 15 min in medium containing 200 μCi of [35S]methionine and 200 μCi of [35S]cysteine (PerkinElmer Life Sciences). The pulse was ended by adding warm DMEM supplemented with 10% fetal bovine serum and 0.5 mM cycloheximide (Sigma), together with 5 mM unlabeled methionine and cysteine. After various chase periods, the cells were transferred to ice and washed twice with ice-cold PBS. For pulse-chase experiments in the presence of proteasome inhibitor, labeling and chase media were supplemented with 10 μM MG132. For immunoprecipitation, the cells were lysed, and APOBEC3G-HA was immunoprecipitated with anti-HA monoclonal antibody (Roche). The proteins were separated by SDS-PAGE and quantified on a phosphorimager.
RESULTS
HIV-1 infection reduced intracellular levels of APOBEC3G.
To study the function of APOBEC3G in HIV-1 replication, we transduced permissive Jurkat and SupT1 cells with retroviral vectors expressing human APOBEC3G. Expression of transduced APOBEC3G converted these cells from the permissive to the nonpermissive phenotype for HIV-1 Vif mutant viruses. While wild-type HXB2 replicated efficiently in these transduced cells, a Vif deletion mutant virus, HXB2ΔVif (8), did not generate productive infection (Fig. 1A). These results are consistent with the recent observation that APOBEC3G can convert permissive CEM-SS cells to the nonpermissive phenotype (48), suggesting that APOBEC3G is a major antiviral factor that determines the permissive versus nonpermissive phenotype for HIV-1 Vif mutant viruses.
FIG. 1.
Influence of HIV-1 replication on APOBEC3G expression. (A) Wild-type HXB2 and Vif mutant HXB2ΔVif replication in Jurkat/APOBEC3G and SupT1/APOBEC3G cells. Jurkat cells and SupT1 cells were transduced with a Moloney murine leukemia virus-derived retroviral expression vector expressing human APOBEC3G. HIV-1 propagation was monitored in the transduced cells by measuring p24 in the supernatant. (B) Immunoblot of cell lysates from HXB2- and HXB2ΔVif-infected cells. Cell lysates (26 days after infection) were prepared and separated by SDS-PAGE, transferred to nitrocellulose membranes, and reacted with HIV-1-positive human sera (top panel), a rabbit anti-HIV-1 Vif antibody, a monoclonal antibody to the HA tag for the detection of APOBEC3G-HA, and an antibody to ribosomal P antigens.
Immunoblotting of cell lysates from infected cells (at days 26 after infection) indicated that HIV-1-specific proteins were detected only in wild-type HXB2-infected Jurkat/Apo-3G and SupT1/Apo-3G cells (Fig. 1B, lanes 2 and 4), and not in HXB2ΔVif-infected Jurkat/Apo-3G (Fig. 1B, lane 3) or SupT1/Apo-3G cells (Fig. 1B, lane 5). Interestingly, the levels of APOBEC3G in HXB2-infected Jurkat/Apo-3G (Fig. 1B, lane 2) and SupT1/Apo-3G cells (Fig. 1B, lane 4) were significantly lower than those in the mock-infected (Fig. 1B, lanes 1 and 6) and Vif mutant-infected cells (Fig. 1B, lanes 3 and 5). These results suggest that one of the viral components was responsible for the reduced intracellular level of APOBEC3G. Since the Vif mutant virus could not replicate in these cells, it was difficult to determine whether a viral component(s) other than Vif was responsible for the reduced intracellular level of APOBEC3G.
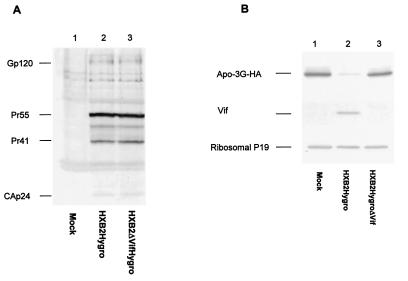
To address this question, we established 293T cells that expressed APOBEC3G together with wild-type HIV-1 or the Vif mutant virus. Stable expression of APOBEC3G was achieved by selection with G418. Stable expression of wild-type HIV-1 or the Vif mutant virus in 293T/APOBEC3G cells was achieved with HXB2Hygro or HXB2ΔVifHygro by selection with hygromycin. HIV-1 viral protein expression was comparable in wild-type- (Fig. 2A, lane 2) and Vif mutant-transfected cells (Fig. 2A, lane 3). As expected, Vif was detected in the cells expressing wild-type HIV-1 (Fig. 2B, lane 2) but not the Vif deletion mutant (Fig. 2B, lane 3). The level of APOBEC3G in wild-type HIV-1-transfected cells (Fig. 2B, lane 2) was significantly lower than that in the untransfected cells (Fig. 2B, lane 1) or in cells transfected with the Vif mutant HIV-1 (Fig. 2B, lane 3). The expression of cellular protein ribosomal P19 antigen was not altered by HIV-1 Vif (Fig. 2B). These results indicated that the expression level of APOBEC3G could be altered by Vif expression but not other viral components.
FIG. 2.
Detection of HIV-1 protein and APOBEC3G expression in transfected 293T/APOBEC3G cells. 293T cells stably expressing APOBEC3G and HIV-1 proteins were obtained by selection with G418 and hygromycin as described in Materials and Methods. (A) Expression of HIV-1 proteins in 293T/APOBEC3G cells, as demonstrated by immunoblotting with HIV-1-positive human sera. (B) Expression of APOBEC3G detected by immunoblotting. Cell lysates from transfected 293T/APOBEC3G were prepared and separated by SDS-PAGE, transferred to nitrocellulose membranes, and reacted with a rabbit anti-HIV-1 Vif antibody, a monoclonal antibody to the HA tag for the detection of APOBEC3G-HA, and an antibody to ribosomal P antigens.
Expression of HIV-1 Vif alone reduced intracellular levels of APOBEC3G.
To address the question of whether HIV-1 Vif alone can reduce the expression of APOBEC3G, we constructed a Vif expression vector with VR1012. Cotransfection of this Vif expression vector with the HIV-1 Vif-deletion mutant construct was sufficient to rescue the Vif defect in the presence of APOBEC3G (Fig. 3A). Virus infectivity was evaluated with viruses from transfected 293T cells and tested with the MAGI-CCR5 cell assay (3). MAGI-CCR5 cells contain one integrated copy of the HIV-1 long terminal repeat linked to the β-galactosidase gene, and productive HIV-1 infection will induce the expression of the β-galactosidase gene product. Therefore, HIV-1 infectivity can be measured by counting the number of cells that are positive for β-galactosidase expression.
FIG. 3.
Influence of HIV-1 Vif on APOBEC3G expression. (A) Expression of HIV-1 Vif in trans rescued the Vif mutant defect. Wild-type HIV-1 (HXB2) and Vif mutant (HXB2ΔVif) viruses were produced from 293T/APOBEC3G cells, and their infectivity was examined with MAGI-CCR5 cells. Virus input was normalized by the level of p24. The infectivity of HXB2 produced from 293T/APOBEC3G cells was set at 100%. Cotransfection of the Vif expression vector with HXB2ΔVif restored virus infectivity to the wild-type HXB2 levels. (B) Influence of HIV-1 Vif on APOBEC3G expression. APOBEC3G expression vector was cotransfected with a control vector (lane 4) or the Vif expression vector (lanes 1 and 3) into 293T cells. The DNA concentrations of APOBEC3G expression vector and the Vif expression vectors used for these experiments are indicated as micrograms per transfection at the top. The total DNA concentration per transfection was 6 μg. Cell lysates from transfected cells were prepared and separated by SDS-PAGE, transferred to nitrocellulose membranes, and reacted with an monoclonal antibody to the HA tag for the detection APOBEC3G-HA and with antibody to ribosomal P antigens.
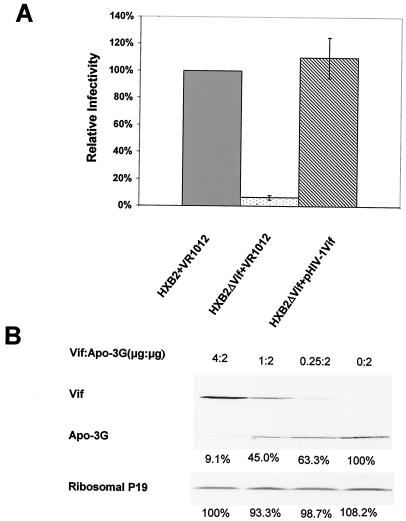
For these assays, the amount of input virus was normalized according to the p24 antigen concentration, and the infectivity of the wild-type HIV-1 produced from 293T/APOBEC3G cells was established as 100%. When viruses were produced in the presence of APOBEC3G (from 293T/APOBEC3G cells), virion infectivity of the Vif mutant virus was reduced by more than 90% (Fig. 3A, lane 2) compared to that of the wild-type virions (Fig. 3A, lane 1). Cotransfection of the Vif expression vector with the HIV-1 Vif mutant construct restored virus infectivity (Fig. 3A, lane 3) to near that of the wild-type HXB2 virus (Fig. 3A, lane 1).
We next examined the effect of Vif expression on the level of APOBEC3G. 293T cells were cotransfected with the APOBEC3G expression vector and either the Vif expression vector or the control VR1012 vector. In this transient transfection system, HIV-1 Vif alone significantly reduced the expression of APOBEC3G in a dose-dependent manner (Fig. 3B). Efficient reduction of APOBEC3G was observed at 2:4 (Fig. 3B, lane 3), 2:1 (Fig. 3B, lane 2), and 2:0.25 ratios of APOBEC3G to Vif DNA concentration. The level of cellular protein ribosomal P19 antigen was not affected by Vif expression (Fig. 3).
HIV-1 Vif but not Vif mutants blocked virion packaging of APOBEC3G.
It has been reported that APOBEC3G can be efficiently incorporated into wild-type HIV-1 virions (48). However, we observed that APOBEC3G was detected in the released HIV-1 Vif deletion mutant (HXB2ΔVif) virions (Fig. 4A, lane 3), but little APOBEC3G was detected in the released wild-type HIV-1 virions (Fig. 4A, lane 2). This observation is more consistent with recent reports that HIV-1 Vif efficiently excludes APOBEC3G from released HIV-1 virions (29, 38, 39, 49, 59, 64). Increased packaging of APOBEC3G in released Vif mutant virions correlated well with reduced virus infectivity, as measured by the MAGI-CCR5 assay (Fig. 4B). We also observed that point mutations (SLQ to AAA, HXB2VifΔSLQ) in the most highly conserved protein motif (SLQ) in the Vif proteins of all lentiviruses also resulted in increased packaging of APOBEC3G into released mutant virions (Fig. 4A, lane 4) compared to the wild-type virus (Fig. 4A, lane 2). Virion incorporation of APOBEC3G into HXB2VifΔSLQ virions was slightly lower than that into the Vif deletion mutant HXB2ΔVif, suggesting that the SLQ mutant Vif may still have some residual activity. However, released HXB2VifΔSLQ mutant viruses had a significantly reduced infectivity that was similar to that of HXB2ΔVif (Fig. 4B). Like HXB2ΔVif, HXB2VifΔSLQ could not replicate in nonpermissive H9 cells (Fig. 4C). As expected, all Vif mutants maintained virus infectivity when produced from 293T cells in the absence of APOBEC3G (Fig. 4B).
FIG. 4.
HIV-1 Vif but not mutant Vif blocked virion incorporation of APOBEC3G. (A) APOBEC3G incorporated into HIV-1 Vif mutant virions. 293T/APOBEC3G cells were transfected with HXB2, HXB2ΔVif, or HXB2VifΔSLQ. Comparable amounts of virions were used, as indicated by immunoblotting with the anti-p24 monoclonal antibody (lanes 2 to 4). The amount of APOBEC3G detected in HIV-1 virions was analyzed by immunoblotting with the anti-HA monoclonal antibody. Lane 1, mock; lane 2, HXB2 virions; lane 3, HXB2ΔVif virions; lane 4, HXB2VifΔSLQ virions. (B) Incorporation of APOBEC3G into HIV-1 Vif mutant virions reduced virus infectivity. HXB2, HXB2ΔVif, and HXB2VifΔSLQ viruses recovered from transfected 293T or 293T/APOBEC3G cells were tested with MAGI-CCR5 cells. Virus input was normalized by the level of p24. The infectivity of HXB2 produced from 293T cells was set at 100%. (C) Replication of HXB2, HXB2ΔVif, and HXB2VifΔSLQ in H9 cells. (D) APOBEC3G incorporated into HIV-1 Vif mutant virions. 293T/APOBEC3G cells were transfected with HXB2ΔVif plus control vector (lane 2), wild-type Vif expression vector (lane 3), or S144/A mutant Vif expression vector (lane 4). Comparable amounts of virions were used, as indicated by immunoblotting with the anti-p24 monoclonal antibody (lanes 2 to 4). The amount of APOBEC3G detected in HIV-1 virions was analyzed by immunoblotting with the anti-HA monoclonal antibody. (E) Single S144 mutation in Vif reduced virus infectivity. Viruses recovered from transfected 293T/APOBEC3G cells as described for panel D were tested in MAGI-CCR5 cells. Virus input was normalized by the level of p24. The infectivity of HXB2ΔVif plus wild-type Vif expression vector was set at 100%.
Although the SLQ motif is highly conserved among the Vif proteins of various HIV-1 subtypes, variants of HIV-1 with changes of S to other amino acids in Vif were observed (18). We next addressed whether the S144 amino acid is important for Vif function in HXB2. 293T/APOBEC3G cells were cotransfected with HXB2ΔVif plus wild-type Vif (pHIV-1Vif) or mutant Vif(pHIV-1VifΔS). Expression of mutant Vif(pHIV-1VifΔS) resulted in more APOBEC3G packaging (Fig. 4D, lane 4) than did the wild-type Vif (Fig. 4D, lane 2). However, the APOBEC3G level in HXB2ΔVif plus pHIV-1 VifΔS virions was lower than that in the Vif deletion mutant HXB2ΔVif alone (Fig. 4D, lane 3), suggesting that the S144 to A mutant Vif maintained some functional activity. The infectivity of HXB2ΔVif plus pHIV-1VifΔS virions was significantly lower than that of HXB2ΔVif plus wild-type Vif, pHIV-1Vif (Fig. 4D), suggesting that at least in the context of HXB2 Vif, amino acid S144 is required for full Vif function.
Vif from distantly related primate lentiviruses could not block APOBEC3G packaging into HIV-1 Vif mutant virions.
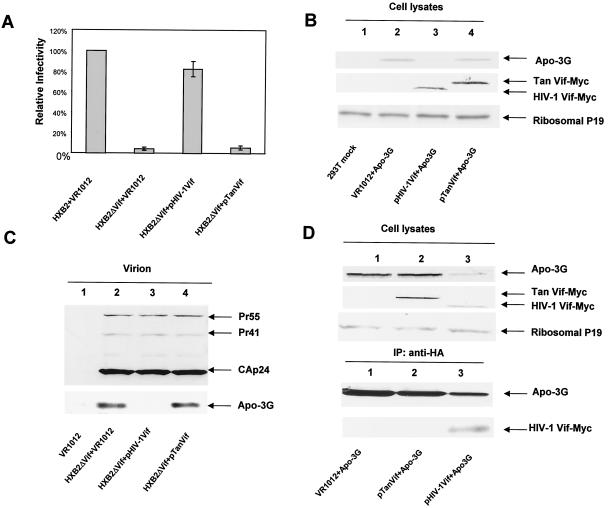
Previous studies have shown that HIV-1 Vif defect can be complemented in trans by HIV-1 or HIV-2/SIVmac Vif but not by SIVagm or nonprimate lentiviral Vif in human cells (54). Using the MAGI-CCR5 cell assay system, we observed that unlike HIV-1 Vif, Vif from SIVagmTan could not suppress the antiviral activity of human APOBEC3G in 293T cells (Fig. 5A). Unlike HIV-1 Vif, SIVagmTan Vif could not reduce the intracellular expression of human APOBEC3G (Fig. 5B), despite efficient intracellular expression (Fig. 5B).
FIG. 5.
SIVagmTan Vif was functionally inactive against human APOBEC3G. (A) SIVagmTan Vif could not suppress the antiviral activity of human APOBEC3G, as indicated by the MAGI-CCR5 assay. (B) SIVagmTan Vif did not induce intracellular degradation of human APOBEC3G. Both HIV-1 Vif and SIVagmTan Vif were tagged with the Myc epitope and detected with an anti-Myc antibody. APOBEC3G-HA was detected by an anti-HA antibody. (C) SIVagmTan Vif did not prevent virion packaging of human APOBEC3G. 293T/APOBEC3G cells were transfected with HXB2ΔVif plus a control vector VR1012, HXB2ΔVif plus pHIV-1Vif, or HXB2ΔVif plus pTanVif. Comparable amounts of virions were used, as indicated by immunoblotting with the anti-p24 antibody (lanes 2 to 4). The amount of APOBEC3G detected in HIV-1 virions was analyzed by immunoblotting with the anti-HA antibody. (D) SIVagmTan Vif could not interact with human APOBEC3G, as indicated by coimmunoprecipitation analysis. 293T cells were transfected with the APOBEC3G-HA and pHIV-1Vif-Myc or pTanVif-Myc expression vectors as indicated. Cell lysates were analyzed by immunoblotting with an anti-HA antibody for the detection of APOBEC3G-HA, an anti-Myc antibody for the detection of HIV-1Vif-Myc or SIVagmTanVif-Myc, or with an antibody to ribosomal P antigens as a control. For immunoprecipitation analysis, equal amounts of cell extract were immunoprecipitated with the anti-HA antibody and analyzed by immunoblotting with antibodies against HA (for detection of APOBEC3G-HA, upper panel) or Myc (for detection of HIV-1Vif-Myc or SIVagmTanVif-Myc, lower panel).
To determine whether SIVagm Vif could alter virion incorporation of APOBEC3G, 293T/APOBEC3G cells were cotransfected with HXB2ΔVif plus a control vector, pHIV-1Vif or pTanVif. Comparable levels of virus were detected in the supernatants of transfected cells (Fig. 5C). When virion proteins were examined, it was clear that HIV-1 Vif blocked the packaging of APOBEC3G into released virions (Fig. 5, lane 3), but SIVagm Vif did not (Fig. 5, lane 4). Again, APOBEC3G virion incorporation correlated well with virus infectivity (Fig. 5A). A possible reason for the lack of degradation and virion exclusion of human APOBEC3G by SIVagmTan Vif is the lack of interaction between APOBEC3G and SIVagmTan Vif, as indicated by our coimmunoprecipitation experiments (Fig. 5D). Interaction between APOBEC3G and HIV-1 Vif was readily detected under the same conditions (Fig. 5D).
Influence of proteasome inhibitor on virion packaging of APOBEC3G and Vif.
The reduced intracellular expression and virion packaging of APOBEC3G in the presence of Vif could be a consequence of protein degradation mediated by proteasome. This question was addressed by pulse-chase experiments. In the absence of Vif, APOBEC3G was relatively stable during the chase period (Fig. 6A). In the presence of Vif, APOBEC3G rapidly disappeared (Fig. 6A). However, when cells were treated with the proteasome inhibitor MG132, APOBEC3G became stable even in the presence of Vif (Fig. 6A). The effect of MG132 on intracellular expression of Vif was also evaluated. 293T/APOBEC3G cells were transfected with HXB2 or HXB2ΔVif and treated with dimethyl sulfoxide or MG132. The intracellular expression of APOBEC3G was reduced by Vif expressed from HXB2 (Fig. 6B, lane 2) compared to that in the mock- (lane 1) or HXB2ΔVif-transfected cells (lane 3). In the presence of MG132, the Vif-induced reduction of APOBEC3G was blocked (Fig. 6B, lane 5). Interestingly, in the presence of MG132, the intracellular level of Vif also increased significantly (Fig. 6B, lane 5) compared to the dimethyl sulfoxide-treated control (Fig. 6B, lane 2). Since both APOBEC3G and Vif were tagged with the HA epitope, the relative ratio of Vif and APOBEC3G expression in the cell and virion packaging could be compared with the anti-HA antibody.
FIG. 6.
Influence of proteasome inhibitor on degradation of APOBEC3G and virion packaging of Vif and APOBEC3G. (A) Effects of Vif and proteasome inhibitor MG132 on degradation of APOBEC3G with pulse-chase analysis. Cells were metabolically labeled for 15 min and chased for the indicated times. APOBEC3G-HA was immunoprecipitated, separated by SDS-PAGE, and analyzed by fluorography. The amount of APOBEC3G-HA at time zero was normalized to 100%. (B) Immunoblotting of intracellular APOBEC3G and Vif from transfected 293T/APOBEC3G cells. 293T/APOBEC3G cells were transfected with HXB2VifHA or HXB2ΔVif. At 24 h after transfection, cells were treated with dimethyl sulfoxide or 2.5 μM MG132 for 16 h, and cell lysates were prepared and analyzed by immunoblotting with an anti-HA antibody. Both Vif and APOBEC3G were tagged with the HA epitope. (C) Increased virion packaging of APOBEC3G and Vif in the presence of MG132. 293T/APOBEC3G cells were transfected with HXB2VifHA or HXB2ΔVif. At 24 h after transfection, cells were treated with dimethyl sulfoxide or 2.5 μM MG132 for 16 h, and viruses were purified and analyzed by immunoblotting. Viral Gag proteins were detected with an anti-p24 antibody. APOBEC3G-HA and Vif-HA were detected with the anti-HA antibody. Viral infectivity was assayed in MAGI-CCR5 cells, and the infectivity of HXB2VifHA treated with dimethyl sulfoxide was set at 100%.
Next, we evaluated the effect of the proteasome inhibitor MG132 on virion packaging of APOBEC3G and Vif. It has previously been reported that a high concentration of proteasome inhibitors (10 μM MG132) reduced HIV-1 release by threefold and resulted in aberrant Gag processing (47). We therefore treated cells with a lower concentration of MG132 (2.5 μM). Under these conditions, intracellular Gag processing was not significantly affected by MG132 (data not shown), and virus release was minimally affected (Fig. 6C). Virions were collected from transfected cells, and concentrations were normalized according to p24 content. Examination of released virions revealed that MG132 had a significant effect on APOBEC3G packaging. In the absence of MG132, APOBEC3G was detected in the released HIV-1 Vif deletion mutant (HXB2ΔVif) virions (Fig. 6C, lane 3). APOBEC3G was significantly reduced in the wild-type HXB2 virions (Fig. 6C, lane 2). The level of APOBEC3G was not significantly affected in HXB2ΔVif virions by the presence of MG132 (Fig. 6C, lane 6). However, in the presence of MG132, APOBEC3G was substantially increased in the released wild-type HXB2 virions (Fig. 6C, lane 5). Surprisingly, treatment with MG132 also drastically increased the virion packaging of Vif in released HXB2 virions (Fig. 6, lane 5) compared to the dimethyl sulfoxide-treated HXB2 virions (lane 2).
Since Vif and APOBEC3G were both tagged with the HA epitope and detected with the anti-HA epitope antibody, it seemed that there were more Vif molecules than APOBEC3G in the released HXB2 virions in the presence of MG132 (Fig. 6C, lane 5). However, the net effect of increased packaging of Vif and APOBEC3G in the presence of MG132 resulted in reduced virus infectivity (Fig. 6C, compares lane 5 and lane 2). We have previously observed that treatment with MG132 in the absence of APOBEC3G under the same conditions does not significantly affect HXB2 virus infectivity (64). Therefore, a low concentration of MG132 selectively blocked virus infectivity in the presence of APOBEC3G.
DISCUSSION
Accumulating evidence indicates that APOBEC3G is a major cellular antiviral factor that needs to be neutralized by HIV-1 Vif if successful virus production is to occur (21, 29, 31, 37-39, 48, 49, 59, 64, 66). Virion-packaged APOBEC3G may negatively influence the reverse transcription complex through its RNA components and/or protein components. It may also induce instability in the newly synthesized viral DNA by causing a C-to-U modification in the minus strand viral DNA (21, 31, 37, 38, 66). Finally, base substitutions in the viral DNA (21, 31, 37, 38, 66) may also alter translation (e.g., through premature stop codons) and the functions of translated proteins (e.g., through amino acid substitutions). Therefore, prevention of APOBEC3G virion packaging by HIV-1 Vif is critical for the maintenance of viral infectivity (29, 38, 39, 49, 59, 64).
Recent observations showed that HIV-1 Vif recruits E3 ubiquitin ligase, consisting of Cullin5, Elongin B, Elongin C, and Rbx1 (64), to induce polyubiquitination of APOBEC3G (39, 49, 64). The ubiquitinated APOBEC3G is subsequently degraded in a proteasome-dependent fashion (39, 49, 59, 64), resulting in the removal of APOBEC3G from released virions (29, 38, 39, 49, 59, 64). Interference with the function of Cul5-containing E3 ubiquitin ligase resulted in the inhibition of Vif-induced APOBEC3G polyubiquitination and degradation, efficient incorporation of APOBEC3G in the presence of Vif, and compromised viral infectivity (64).
An HIV-1 Vif mutant containing amino acid substitutions only in the SLQ motif failed to prevent APOBEC3G incorporation into released virions; this SLQ motif is the most conserved region in the Vif proteins of all lentiviruses (42). The SLQ mutant Vif remains competent for interaction with APOBEC3G (39, 64) but has a reduced ability to interact with Cul5, Elongin B, and Elongin C (64). Therefore, recognition of APOBEC3G and efficient interaction with Cul5-containing E3 ubiquitin ligase are both required for Vif function. Although the SLQ motif is highly conserved among various M group HIV-1 isolates, HIV-1 variants such as HIV-1mn, which contains an S to P substitution in Vif, were observed (18). We have shown that mutation of S144 in HXB2 Vif significantly reduces Vif's ability to overcome the antiviral activity of APOBEC3G (Fig. 4). This mutant Vif could not exclude APOBEC3G as efficiently as wild-type HIV-1 Vif (Fig. 4). These results suggest that at least in the context of HXB2 Vif, amino acid S144 is required for complete Vif function. Further study will be required to elucidate the function of HIV-1mn Vif and identify the reason for the substitution of the conserved S144 in HIV-1mn Vif.
Vif from distantly related primate lentiviruses such as SIVagm and nonprimate lentiviruses is unable to complement the HIV-1 Vif defect in human cells (54). In the present study, we found that SIVagmTan Vif did not induce human APOBEC3G degradation or virion exclusion of APOBEC3G (Fig. 5). Consequently, Vif from SIVagmTan could not suppress the antiviral activity of human APOBEC3G against HIV-1 Vif deletion mutant viruses (Fig. 5). Although interaction between HIV-1 Vif and human APOBEC3G could be detected (Fig. 5), interaction between SIVagmTan Vif and human APOBEC3G was not detected (Fig. 5). This finding could be the reason for the lack of degradation and virion exclusion of human APOBEC3G by SIVagmTan Vif. We also observed that Vif from SIVagmSab did not induce human APOBEC3G degradation or virion exclusion of APOBEC3G (data not shown). The lack of SIVagm Vif function in human cells may be partially responsible for the lack of SIVagm transmission to humans.
The activity of both APOBEC3G and Vif is required in virus-producing cells but not in target cells, suggesting that interaction between Vif and APOBEC3G, either directly or indirectly, is likely to occur during virus assembly. HIV-1 Vif has been shown to interact with viral genomic RNA in virus-infected cells (8, 30, 65). APOBEC3G is potentially an RNA-binding protein, or, as is the case for APOBEC1, it may associate with other RNA-binding proteins (26, 63). Affinity for RNA molecules is consistent with the observations that APOBEC3G is incorporated into unrelated retroviruses (21, 37, 38). Since Vif and APOBEC3G are likely to be present at the site of virus assembly, removal of APOBEC3G by Vif-induced polyubiquitination and subsequent proteasome-mediated degradation will occur more efficiently during virus assembly.
It is still controversial whether Vif is efficiently incorporated into released virions (2, 4, 9, 12, 28, 30, 34, 58). The proteasome inhibitor experiments presented here indicated that the majority of HIV-1 Vif molecules which were competent for packaging were removed during virus assembly by the proteasome pathway (Fig. 6C). In the presence of MG132, both Vif and APOBEC3G levels were increased in the released virions (Fig. 6C). Actually, more Vif molecules than APOBEC3G molecules were detected in these released virions (Fig. 6C). However, the net effect of increased incorporation of APOBEC3G and Vif in the presence of MG132 was compromised virus infectivity (Fig. 6C). Therefore, a major functional activity of Vif is required during virus assembly and not in the released virions.
Acknowledgments
We thank R. Garten, T. Sarkis, R. Markham, and Markus Dettenhofer for useful discussions and Vical, Inc., for the VR1012 plasmid.
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: antiserum to HIV-1 Vif (Dana Gabuzda), monoclonal antibody to HIV-1 p24 (Bruce Chesebro and Hardy Chen), pSIVagmTan-1 (Marcelo Soares and Beatrice Hahn), and MAGI-CCR-5 (Michael Emerman).
REFERENCES
- 1.Bouyac, M., M. Courcoul, G. Bertoia, Y. Baudat, D. Gabuzda, D. Blanc, N. Chazal, P. Boulanger, J. Sire, R. Vigne, and B. Spire. 1997. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J. Virol. 71:9358-9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camaur, D., and D. Trono. 1996. Characterization of human immunodeficiency virus type 1 Vif particle incorporation. J. Virol. 70:6106-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chackerian, B., E. M. Long, P. A. Luciw, and J. Overbaugh. 1997. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J. Virol. 71:3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterji, U., C. K. Grant, and J. H. Elder. 2000. Feline immunodeficiency virus Vif localizes to the nucleus. J. Virol. 74:2533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66:6547-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deminie, C. A., and M. Emerman. 1994. Functional exchange of an oncoretrovirus and a lentivirus matrix protein. J. Virol. 68:4442-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desrosiers, R. C., J. D. Lifson, J. S. Gibbs, S. C. Czajak, A. Y. Howe, L. O. Arthur, and R. P. Johnson. 1998. Identification of highly attenuated mutants of simian immunodeficiency virus. J. Virol. 72:1431-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dettenhofer, M., S. Cen, B. A. Carlson, L. Kleiman, and X. F. Yu. 2000. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J. Virol. 74:8938-8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dettenhofer, M., and X. F. Yu. 1999. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 73:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dornadula, G., S. Yang, R. J. Pomerantz, and H. Zhang. 2000. Partial rescue of the Vif-negative phenotype of mutant human immunodeficiency virus type 1 strains from nonpermissive cells by intravirion reverse transcription. J. Virol. 74:2594-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, A. G., B. Ensoli, L. Ivanoff, M. Chamberlain, S. Petteway, L. Ratner, R. C. Gallo, and F. Wong-Staal. 1987. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science 237:888-893. [DOI] [PubMed] [Google Scholar]
- 12.Fouchier, R. A., J. H. Simon, A. B. Jaffe, and M. H. Malim. 1996. Human immunodeficiency virus type 1 Vif does not influence expression or virion incorporation of Gag-, Pol-, and Env-encoded proteins. J. Virol. 70:8263-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabuzda, D. H., K. Lawrence, E. Langhoff, E. Terwilliger, T. Dorfman, W. A. Haseltine, and J. Sodroski. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66:6489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaddis, N. C., E. Chertova, A. M. Sheehy, L. E. Henderson, and M. H. Malim. 2003. Comprehensive investigation of the molecular defect in vif-deficient human immunodeficiency virus type 1 virions. J. Virol. 77:5810-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of SIVmac mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retrovir. 10:607-616. [DOI] [PubMed] [Google Scholar]
- 16.Goncalves, J., P. Jallepalli, and D. H. Gabuzda. 1994. Subcellular localization of the Vif protein of human immunodeficiency virus type 1. J. Virol. 68:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goncalves, J., Y. Korin, J. Zack, and D. Gabuzda. 1996. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J. Virol. 70:8701-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurgo, C., H. G. Guo, G. Franchini, A. Aldovini, E. Collalti, K. Farrell, F. Wong-Staal, R. C. Gallo, and M. S. Reitz, Jr. 1988. Envelope sequences of two new United States HIV-1 isolates. Virology 164:531-536. [DOI] [PubMed] [Google Scholar]
- 19.Harmache, A., P. Russo, F. Guiguen, C. Vitu, M. Vignoni, M. Bouyac, C. Hieblot, M. Pepin, R. Vigne, and M. Suzan. 1996. Requirement of caprine arthritis encephalitis virus vif gene for in vivo replication. Virology 224:246-255. [DOI] [PubMed] [Google Scholar]
- 20.Harmache, A., P. Russo, C. Vitu, F. Guiguen, J. F. Mornex, M. Pepin, R. Vigne, and M. Suzan. 1996. Replication in goats in vivo of caprine arthritis-encephalitis virus deleted in vif or tat genes: possible use of these deletion mutants as live vaccines. AIDS Res. Hum. Retrovir. 12:409-411. [DOI] [PubMed] [Google Scholar]
- 21.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 22.Harris, R. S., S. K. Petersen-Mahrt, and M. S. Neuberger. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10:1247-1253. [DOI] [PubMed] [Google Scholar]
- 23.Hartikka, J., M. Sawdey, F. Cornefert-Jensen, M. Margalith, K. Barnhart, M. Nolasco, H. L. Vahlsing, J. Meek, M. Marquet, P. Hobart, J. Norman, and M. Manthorpe. 1996. An improved plasmid DNA expression vector for direct injection into skeletal muscle. Hum. Gene Ther. 7:1205-1217. [DOI] [PubMed] [Google Scholar]
- 24.Inoshima, Y., M. Kohmoto, Y. Ikeda, H. Yamada, Y. Kawaguchi, K. Tomonaga, T. Miyazawa, C. Kai, T. Umemura, and T. Mikami. 1996. Roles of the auxiliary genes and AP-1 binding site in the long terminal repeat of feline immunodeficiency virus in the early stage of infection in cats. J. Virol. 70:8518-8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoshima, Y., T. Miyazawa, and T. Mikami. 1998. The roles of vif and ORF-A genes and AP-1 binding site in in vivo replication of feline immunodeficiency virus. Arch. Virol. 143:789-795. [DOI] [PubMed] [Google Scholar]
- 26.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 27.Kan, N. C., G. Franchini, F. Wong-Staal, G. C. DuBois, W. G. Robey, J. A. Lautenberger, and T. S. Papas. 1986. Identification of HTLV-III/LAV sor gene product and detection of antibodies in human sera. Science 231:1553-1555. [DOI] [PubMed] [Google Scholar]
- 28.Kao, S., H. Akari, M. A. Khan, M. Dettenhofer, X. F. Yu, and K. Strebel. 2003. Human immunodeficiency virus type 1 Vif is efficiently packaged into virions during productive but not chronic infection. J. Virol. 77:1131-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan, M. A., C. Aberham, S. Kao, H. Akari, R. Gorelick, S. Bour, and K. Strebel. 2001. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 75:7252-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 32.Lee, T. H., J. E. Coligan, J. S. Allan, M. F. McLane, J. E. Groopman, and M. Essex. 1986. A new HTLV-III/LAV protein encoded by a gene found in cytopathic retroviruses. Science 231:1546-1549. [DOI] [PubMed] [Google Scholar]
- 33.Lee, Y. M., and X. F. Yu. 1998. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology 243:78-93. [DOI] [PubMed] [Google Scholar]
- 34.Liu, H., X. Wu, M. Newman, G. M. Shaw, B. H. Hahn, and J. C. Kappes. 1995. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J. Virol. 69:7630-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lockridge, K. M., M. Chien, G. A. Dean, K. Stefano Cole, R. C. Montelaro, P. A. Luciw, and E. E. Sparger. 2000. Protective immunity against feline immunodeficiency virus induced by inoculation with vif-deleted proviral DNA. Virology 273:67-79. [DOI] [PubMed] [Google Scholar]
- 36.Madani, N., and D. Kabat. 1998. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 72:10251-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broda antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed]
- 38.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 39.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed]
- 40.Michaels, F. H., N. Hattori, R. C. Gallo, and G. Franchini. 1993. The human immunodeficiency virus type 1 (HIV-1) vif protein is located in the cytoplasm of infected cells and its effect on viral replication is equivalent in HIV-2. AIDS Res. Hum. Retrovir. 9:1025-1030. [DOI] [PubMed] [Google Scholar]
- 41.Nascimbeni, M., M. Bouyac, F. Rey, B. Spire, and F. Clavel. 1998. The replicative impairment of Vif mutants of human immunodeficiency virus type 1 correlates with an overall defect in viral DNA synthesis. J. Gen. Virol. 79:1945-1950. [DOI] [PubMed] [Google Scholar]
- 42.Oberste, M. S., and M. A. Gonda. 1992. Conservation of amino-acid sequence motifs in lentivirus Vif proteins. Virus Genes 6:95-102. [DOI] [PubMed] [Google Scholar]
- 43.Ochsenbauer, C., T. Wilk, and V. Bosch. 1997. Analysis of vif-defective human immunodeficiency virus type 1 (HIV-1) virions synthesized in ‘non-permissive’ T lymphoid cells stably infected with selectable HIV-1. J. Gen. Virol. 78:627-635. [DOI] [PubMed] [Google Scholar]
- 44.Ohagen, A., and D. Gabuzda. 2000. Role of Vif in stability of the human immunodeficiency virus type 1 core. J. Virol. 74:11055-11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, I. W., K. Myrick, and J. Sodroski. 1994. Effects of vif mutations on cell-free infectivity and replication of simian immunodeficiency virus. J. Acquir. Immune Defic. Syndr. 7:1228-1236. [PubMed] [Google Scholar]
- 46.Petersen-Mahrt, S. K., and M. S. Neuberger. 2003. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1). J. Biol. Chem. 278:19583-19586. [DOI] [PubMed] [Google Scholar]
- 47.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 49.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 11:1404-1407. [DOI] [PubMed]
- 50.Simon, J. H., E. A. Carpenter, R. A. Fouchier, and M. H. Malim. 1999. Vif and the p55Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J. Virol. 73:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon, J. H., R. A. Fouchier, T. E. Southerling, C. B. Guerra, C. K. Grant, and M. H. Malim. 1997. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 71:5259-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simon, J. H., N. C. Gaddis, R. A. Fouchier, and M. H. Malim. 1998. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4:1397-1400. [DOI] [PubMed] [Google Scholar]
- 53.Simon, J. H., and M. H. Malim. 1996. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 70:5297-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simon, J. H., D. L. Miller, R. A. Fouchier, M. A. Soares, K. W. Peden, and M. H. Malim. 1998. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 17:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soares, M. A., D. L. Robertson, H. Hui, J. S. Allan, G. M. Shaw, and B. H. Hahn. 1997. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology 228:394-399. [DOI] [PubMed] [Google Scholar]
- 56.Sodroski, J., W. C. Goh, C. Rosen, A. Tartar, D. Portetelle, A. Burny, and W. Haseltine. 1986. Replicative and cytopathic potential of HTLV-III/LAV with sor gene deletions. Science 231:1549-1553. [DOI] [PubMed] [Google Scholar]
- 57.Sova, P., and D. J. Volsky. 1993. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J. Virol. 67:6322-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sova, P., D. J. Volsky, L. Wang, and W. Chao. 2001. Vif is largely absent from human immunodeficiency virus type 1 mature virions and associates mainly with viral particles containing unprocessed Gag. J. Virol. 75:5504-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 60.Strebel, K., D. Daugherty, K. Clouse, D. Cohen, T. Folks, and M. A. Martin. 1987. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature 328:728-730. [DOI] [PubMed] [Google Scholar]
- 61.Tomonaga, K., J. Norimine, Y. S. Shin, M. Fukasawa, T. Miyazawa, A. Adachi, T. Toyosaki, Y. Kawaguchi, C. Kai, and T. Mikami. 1992. Identification of a feline immunodeficiency virus gene which is essential for cell-free virus infectivity. J. Virol. 66:6181-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wedekind, J. E., G. S. Dance, M. P. Sowden, and H. C. Smith. 2003. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19:207-216. [DOI] [PubMed] [Google Scholar]
- 64.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed]
- 65.Zhang, H., R. J. Pomerantz, G. Dornadula, and Y. Sun. 2000. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J. Virol. 74:8252-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]