Abstract
Background:
Bone metastases represent a common and severe complication in breast cancer, and the involvement of cancer stem cells (CSCs) in the promotion of bone metastasis is currently under discussion. Here, we used a human-in-mice model to study bone metastasis formation due to primary breast CSCs-like colonisation.
Methods:
Primary CD44+CD24− breast CSCs-like were transduced by a luciferase-lentiviral vector and injected through subcutaneous and intracardiac (IC) routes in non-obese/severe-combined immunodeficient (NOD/SCID) mice carrying subcutaneous human bone implants. The CSCs-like localisation was monitored by in vivo luciferase imaging. Bone metastatic CSCs-like were analysed through immunohistochemistry and flow cytometry, and gene expression analyses were performed by microarray techniques.
Results:
Breast CSCs-like colonised the human-implanted bone, resulting in bone remodelling. Bone metastatic lesions were histologically apparent by tumour cell expression of epithelial markers and vimentin. The bone-isolated CSCs-like were CD44−CD24+ and showed tumorigenic abilities after injection in secondary mice. CD44−CD24+ CSCs-like displayed a distinct bone tropism signature that was enriched in genes that discriminate bone metastases of breast cancer from metastases at other organs.
Conclusion:
Breast CSCs-like promote bone metastasis and display a CSCs-like bone tropism signature. This signature has clinical prognostic relevance, because it efficiently discriminates osteotropic breast cancers from tumour metastases at other sites.
Keywords: bone metastasis, breast cancer, cancer stem cell, human-in-mice model, osteotropism
Breast tumours are characterised by phenotypic heterogeneity. They are composed of different groups of cells that display a remarkable variability in many phenotypic traits, including different metastatic abilities and survival ability after therapy. The cancer stem cell (CSC) hypothesis, which suggests that the variety between functionally important properties of individual cells arises from differences in their differentiation status, may explain the intra-tumour heterogeneity seen in breast tumours (Campbell and Polyak, 2007; Shackleton et al, 2009). Cancer stem cells are defined as a subset of tumour-initiating cells that share surface markers with somatic stem/progenitor cells of their tissue of origin. They are able to self-renew, proliferate and differentiate to give rise to a heterogeneous tumour cell population (Clarke and Fuller, 2006; Rosen and Jordan, 2009). In breast cancer, CSCs are identified as a rare population of CD44+CD24− epithelial-specific antigenlow cells that show in vivo tumorigenicity based on xenotransplantation assays in non-obese/severe-combined immunodeficient (NOD/SCID) mice (Al-Hajj et al, 2003; Shipitsin et al, 2007).
Breast cancer cells are known to have a peculiar bone tropism; indeed, 70% of breast cancer patients develop bone metastases (Mundy, 2002). The metastatic process sometimes occurs very early in the natural history of the disease but can occur at later stages. In either case, once the tumour has spread to the bone, the cancer is no longer curable (Mundy and Sterling, 2008). Recent reports highlighted the molecular mechanisms underlying the dissemination of breast cancer cells in bone and led to the identification of discrete sets of osteotropic genes associated with bone tropism (Kang et al, 2003; Minn et al, 2005; Liu et al, 2007; van den Hoogen et al, 2010). Evidence that CSCs are responsible for bone metastasis is still not conclusive owing to the difficulties in isolating CSCs and the lack of a valid animal model (Liu et al, 2010; van den Hoogen et al, 2010; Buijs et al, 2011).
This study investigates the ability of non-metastatic human breast CSCs to metastasise to bone. A NOD/SCID-human-in-mice model was utilised to inject CD44+CD24− CSCs-like, isolated from a non-bone metastatic primary breast cancer (Kuperwasser et al, 2005; Roato et al, 2010). These mice were implanted subcutaneously (SC) with a fragment of human bone to establish a human microenvironment. Injected breast CD44+CD24− CSCs-like acquired the ability to metastasise the implanted bone and switched to a CD44−CD24+ phenotype while maintaining tumorigenic abilities in vivo. The CD44−CD24+ CSCs isolated from bone expressed a signature of significant enrichment of sets of genes biologically relevant in the process of cancer dissemination that persisted at subsequent passages in the absence of surrounding bone tissue (Subramanian et al, 2005). This signature derived from CSCs is relevant for clinical prognosis because it efficiently discriminates osteotropic breast cancers from tumours metastasising to other sites.
Materials and methods
Breast CSCs
Breast CSCs were previously isolated and characterised for the expression of stemness markers, showing a CD44+CD24− phenotype, and for tumorigenic potential (Bussolati et al, 2009). Tumour specimens were obtained from a consenting patient, according to the ethics committee of the San Giovanni Battista Hospital of Torino, Italy. The histological analysis showed a HER2-negative lobular-infiltrating carcinoma of the pleomorphic type, expressing the oestrogen receptor in ∼60% of cells. As previously described (Bussolati et al, 2009), the cell suspension obtained from the tumour tissue was processed, and mammospheres were derived. These mammospheres were studied for their ability to metastasise to bone as patients with breast cancer frequently develop bone metastases (Smid et al, 2008). CD44+CD24− CSCs-like were transduced with a luciferase-lentiviral expression vector, originating Luc-CSCs-like. Both breast CSCs-like and Luc-CSCs-like were injected in mice to rule out possible modifications of the tumorigenic phenotype due to lentiviral transduction.
Human-in-mice model of bone metastasis induced by breast CSCs
Experimental animals were treated in compliance with the actual national and international guidelines (the Italian legislative decree 116/92 and the European Community Directive 86/609 CEE) and in accordance with the authorisation provided by the Italian Ministry of Health (as of DM 44/1994-A and subsequent integrations). All of the orthotopic xenograft models were established in NOD/SCID mice as previously described (Roato et al, 2010). A fresh fragment of human bone obtained from the discarded femoral head of an adult patient submitted to total joint replacement (after the patient's informed consent) was transplanted SC in the left flank of 15 NOD/SCID 5-week-old female mice (Charles River Laboratories Italia, Calco, Italy). In six 5-week-old mice, breast CSCs-like (three mice) and Luc-CSCs-like (three mice) were injected SC close to the bone implant. In another six mice, CSCs-like (three mice) and Luc-CSCs-like (three mice) were injected by an IC route to demonstrate the actual capability of CSCs to metastasise to bone. For SC injections, 1.5 × 105 breast CSCs were resuspended in PBS and Matrigel 1 : 3 (BD Biosciences, Bedford, MA, USA), and injected in a volume of 40 μl using a 25-gauge needle. For the IC route, 1 × 103 CSCs-like were injected in the left ventricle. The development of tumour masses and metastases to bone and other organs was monitored by in vivo imaging system (IVIS) for 45 days after the injection. In order to evaluate secondary tumour generation, mammospheres obtained from the metastatic lesion in human-implanted bone were cultured for 1 week. Maintenance of the CSC phenotype was confirmed by flow cytometry for the expression of CD44 and CD24, and then the cells were re-injected into another six mice (three SC and three IC) with the same experimental conditions indicated above. The presence of circulating human IgG in mice sera was demonstrated by human IgG ELISA, purchased by ICL Inc. (Ankara, Turkiye).
Lentivirus production and breast CSCs-like transduction
Vector stocks were produced by transient transfection of the luciferase transfer plasmid, the packaging plasmids pMDLg/pRRE and pRSV.REV, and the vesicular stomatitis virus (VSV) envelope plasmid pMD2.VSV-G (15, 6.5, 2.5 and 3.5 μg, respectively, for 10-cm dishes) in 293T. After 12–14 h, the transfection solution was removed, and the cells were washed twice with 1 × PBS. The media was replaced and collected after 24 h. The viral supernatants were filtered using a 0.45-μm low-protein-binding filter flask (Millipore, Vimodrone, Italy), and viral particles were concentrated by ultracentrifugation as described (Follenzi et al, 2000). Determination of the viral p24 antigen concentration was done by HIV-1 p24 Core profile ELISA (PerkinElmer Life, Waltham, MA, USA).
Breast CSCs were resuspended in Opti-MEM in the presence of 8 μg ml−1 polybrene (Sigma-Aldrich, St. Louis, MO, USA) and plated in 96-well round-bottom plates. Then, high-titer lentiviruses (210 ng of p24 gag equivalent particles) were added to the cells, and the cells were incubated for 4 h.
Breast Luc-CSCs-like were tested for luciferase expression by using the Luciferase Assay System (Promega Corp., Madison, WI, USA), as described in the protocol kit. Briefly, the cells were rinsed with PBS and lysed with RLB lysis buffer by performing a single freeze-thaw cycle. Twenty microlitres of cell lysate was added to a luminometer tube containing the luciferase assay reagent and read by performing a2-s measurement delay followed by a 10-s measurement read for luciferase activity. Importantly, we did not observe significant differences in tumour phenotypes associated with lentiviral transduction, as demonstrated by histological appearance, flow cytometry analysis (see Supplementary Figure S2A and B) and microarray analysis of parental breast CSCs-like and Luc-CSCs-like.
In vivo bioluminescence imaging
For the bioluminescence imaging (BLI), mice were anaesthetised by isoflurane inhalation and were subsequently intraperitoneally injected with 200 μl of 15 mg ml−1 𝒟-luciferin (Caliper Life Science, Hopkinton, MA, USA). The bioluminescence signals were monitored using the IVIS system 2000 series (Xenogen Corp., Alameda, CA, USA) consisting of a highly sensitive cooled CCD camera. Two kinetic bioluminescent acquisitions were collected between 0 and 20 min after 𝒟-luciferin injection to confirm the peak photon emission, which was recorded as maximum photon efflux per second; imaging times ranged from 1 to 60 s, depending on the amount of luciferase activity. Data were analysed using the total photon flux emission (photons s−1) in the regions of interest defined manually. At various time points after tumour implantation (25, 35 and 45 days), the mice were imaged using the IVIS 2000 system.
Isolation of bone metastatic cells
To isolate metastatic cells from the osteolytic lesions, human-implanted bone was retrieved, finely minced and then digested by incubation for 30 min at 37 °C in DMEM containing collagenase I (Sigma-Aldrich). After collagenase neutralisation, the cells were washed by Hank's balanced saline solution (Lonza, Basel, Switzerland), and red blood cells were lysed with Red Blood cell Lysis Solution (Promega Corp.). The cell suspension was forced through a graded series of meshes to separate the cell components from the stroma and aggregates. After filtration, single cells were plated in a serum-free DMEM-F12 selective medium (Gibco, Invitrogen, Monza, Italy) supplemented with 10 ng ml−1 basic fibroblast growth factor, 20 ng ml−1 epidermal growth factor (EGF; PeproTech, London, UK), 5 μg ml−1 insulin and 0.4% bovine serum albumin (Sigma-Aldrich) to avoid the presence of non-neoplastic contaminating cells. After 1 week, the appearance of non-adherent spherical clusters of cells, that is, mammospheres, was observed. Mammospheres were collected on the bottom of a conical tube by spontaneous precipitation to remove nonliving cells. After 5 days, mammospheres were collected by gentle centrifugation and disaggregated with a Non-enzymatic Cell Dissociation Solution (Sigma-Aldrich). In order to analyse the phenotype of these bone-derived mammospheres, the dissociated cells were counted and stained for flow cytometry analysis with anti-human CD44PE and CD24FITC (BD Pharmingen, Bedford, MA, USA). For every antibody, we also used the relative isotype control. Samples were analysed in a FACs Calibur instrument and elaborated by Flowjo (Tree Star, Ashland, OR, USA).
Immunohistochemistry and histological analysis
Immunohistochemistry was performed on tissues fixed in 10% neutral-buffered formalin, and bone tissues were decalcified with EDTA treatment until soft. Tissues were embedded in paraffin, sections were deparaffinised, rehydrated through graded alcohols and subjected to antigen retrieval for immunohistochemistry. Sections were stained for H&E for morphological study. The presence of human vessels was demonstrated by staining for anti-CD34, clone QbndN/10 (NeoMarkers, Freemont, CA, USA), whereas tumour cells were stained by mouse monoclonal antibodies against low molecular weight cytokeratins (CK AE1/AE3, clone AE1/AE3/PCK26), epithelial membrane antigen (EMA, clone E29) and vimentin (clone R9) from DAKO (Glostrup, Denmark), and CD44 (clone C26) and CD24 (clone ML5) from BD Pharmingen. To identify the collagen fibres on the new bone, a trichrome stain was performed by Gomori's trichrome stain kit (DAKO). TRAP staining was performed to identify osteoclasts, according to the manufacturer's instructions (Roche, Basel, Switzerland).
Cancer stem cell RNA extraction and microarray analysis
Total RNA was extracted with the miRNeasy Mini Kit (Qiagen, Milano, Italy). The cells were collected as a pellet, homogenised directly in QIAzol Lysis Reagent (a maximum of 107 cells in 0.7 ml of reagent) and processed according to the manufacturer's protocol. RNA quality control was performed on a Bioanalyzer 2100 (Agilent, Berkshire, UK). Biotinylated antisense RNA (aRNA) was prepared from 500 ng of total RNA using the Illumina RNA Amplification Kit (Ambion, Austin, TX, USA), according to the manufacturer's directions, and quality control was performed on a Bioanalyzer 2100 (Agilent). Labelled aRNA was hybridised on Illumina HumanHT-12 v4 expression BeadChips for 16 h. Hybridised BeadChips were subsequently washed, stained with streptavidin-Cy3 and scanned on a BeadStation 500 (Illumina, San Diego, CA, USA).
Microarray data analysis
Raw microarray data were processed and analysed with the GenomeStudio software (Illumina), according to standard procedures. Briefly, bead-level data were summarised and cubic spline normalised. Samples were subsequently organised into the following groups: (i) breast CSCs-like, CD44+CD24−, (duplicate); (ii) Luc-CSCs-like, CD44+CD24,(singlicate); (iii) bone-isolated metastatic CSCs-like, CD44−CD24+, (duplicate). Subsequently, the samples were (iv) grown in vitro as spheroids (singlicate) or (v) re-grown in subcutaneous implants (duplicate). The ‘Illumina Custom' variant of the t-test implemented in GenomeStudio was employed to select genes with differential expression between breast CSCs-like and bone-isolated metastatic CD44−CD24+ CSCs-like, (P-value<0.001, fold-change>3). This resulted in 860 probes being selected (of which 710 were upregulated and 150 were downregulated in bone metastases). Subsequently, probes were filtered to remove those showing differential expression (fold-change>1.5) between breast CSCs-like and Luc-CSCs-like. This filter reduced upregulated probes to 480 and downregulated probes to 100. Finally, the up- and downregulated probes were tested for their maintenance of change (more than two-fold) compared with breast CSCs-like and derivatives of bone-isolated metastatic CD44−CD24+ CSCs-like, that is, spheroids and subcutaneous implants. Ninety-six upregulated and 23 downregulated probes corresponding to 88 and 22 genes, respectively, passed this final filter and collectively defined the breast CSCs-like bone tropism signature (see Supplementary Table S3). Hierarchical clustering was performed using the GEDAS software (Fu and Medico, 2007). The breast cell line microarray data set was generated by Kang et al (2003) and obtained as Supplementary Material on the publisher's website. The breast cancer metastasis data set generated by Zhang et al (2009) was retrieved from the Gene Expression Omnibus database (GSE14020). GSE14020 is a composite of two sub-data sets obtained with two different Affymetrix array types (Santa Clara, CA, USA). To homogenise them, the data were cross-mapped based on probe-sets and scaled on median expression. Gene set enrichment analysis (GSEA; Mootha et al, 2003; Subramanian et al, 2005) on our expression data filtered for detection was performed online using the Broad Institute application (http://www.broadinstitute.org/gsea/msigdb/downloads.jsp). Genes represented by multiple probes were collapsed to the probe with the maximum value. From the Molecular Signatures Database of GSEA, we selected the collections C2 (curated gene sets; file: c2.v3.symbols.gmt) and C5 (Gene Ontology; file: c5.v3.symbols.gmt) for analysis. Enrichment statistics were performed with default settings and the Signal_to_Noise metric with 1000 permutations. Gene set enrichment analysis considering the CSCs-like bone tropism signature as a gene set was run on published data sets (Kang et al, 2003; Zhang et al, 2009) to test for enrichment in genes with differential expression between bone metastatic vs non-metastatic cells or tissues. The gene set enrichment score was calculated from the ‘Ratio_of_classes' score for the genes in the set, and the FDR was evaluated with 1000 permutations.
Serum detection of IL-6, IL-8 and CCL20
To analyse the expression of human IL-6, IL-8 and CCL20 in mice, we utilised the multi-analyte detection system Milliplex Map, according to the manufacturer's instructions. Molecule detection was performed with the instrument Luminex 200 (Luminex Corporation, Austin, TX, USA), and data were analysed through the MILLIPLEX analyst software.
Results
Breast CSCs-like show tumorigenic and metastatic characteristics in the human-in-mice model
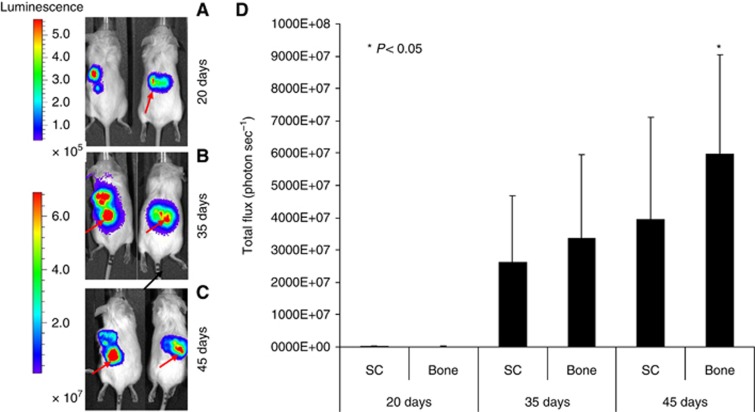
To verify the ability of breast CSCs-like to metastasise bone, we utilised CSCs-like previously isolated from a primary breast tumour. These cells were CD44+CD24−, grew as mammospheres and expressed the stem cells markers Oct-4 and nestin, but not the differentiation markers CK14, CK18 and α-SMA. They grew in SCID mice and were able to generate serial tumours when re-injected into other mice (Bussolati et al, 2009). After isolation from the primary tumour, these breast CSC cells were injected SC or through an IC route in NOD/SCID mice carrying a small piece of human bone previously implanted in a flank. The introduction of human bone in mice allowed us to study in a species-specific manner the interaction between breast CSCs-like and the bone microenvironment. Before the SC and IC injection, we transduced these breast CSCs-like through a luciferase-lentiviral vector (Luc-CSCs-like) in order to monitor their localisation in vivo. The number of mice in the experimental groups, mice survival, percentages of bone engraftment, and bone and lung metastases are indicated in Supplementary Table S1. To analyse the intensity of the luciferase signal released from Luc-CSCs-like, increasing amounts of Luc-CSCs were plated in a multiwell plate and subjected to IVIS imaging. We determined a luciferase signal starting from 5 × 102 cells, and its intensity was roughly proportional to the different cell dilutions (Supplementary Figure S1). After the Luc-CSCs-like SC injection, we monitored the tumour growth at different time points for 45 days. Twenty days after Luc-CSCs-like injection, the primary tumours were macroscopically evident and bone localisations were detectable (Figure 1A). The intensity of the signal in the bone increased progressively over the course of the study, eventually becoming stronger in the bone than in the tumour mass (Figure 1B and C). This result was confirmed by the quantification of the mean luciferase intensity in the bone and in the SC tumour mass, which showed a higher value in the bone than in SC masses, particularly at day 45, P<0.05 (Figure 1D). These data confirm our hypothesis that breast CSCs-like show osteotropism and indicate that the human bone microenvironment is a particularly favourable soil because their number significantly increased in bone. In 5 out of 12 mice, we detected lung metastases at day 20, at which point the animals were killed before the occurrence of bone lesion formation (see also Supplementary Figure S2 and Supplementary Table S1). We also tested the bone metastatic ability of the subpopulation of CD44−/CD24+ cells and found that they did not metastasise to bone (data not shown).
Figure 1.
Bioluminescence imaging (BLI). (A) Tumour growth was examined by BLI. Luc-CSCs-like were injected SC into flank of the SCID mice. Representative images of two animals after different time points (20, 35, and 45 days after SC injection) are shown. (B) At day 20, the SC tumour mass is clearly evident, whereas the bone localisation is detectable only in one mice (arrow). (C) At day 35, an increased tumour volume and invasion to human bone becomes particularly evident (arrow). (D) At 45 days, the signal derived from bone localisation of the CSCs-like is stronger in bone than that in SC tumour mass.
Breast CSCs-like metastasise human bone
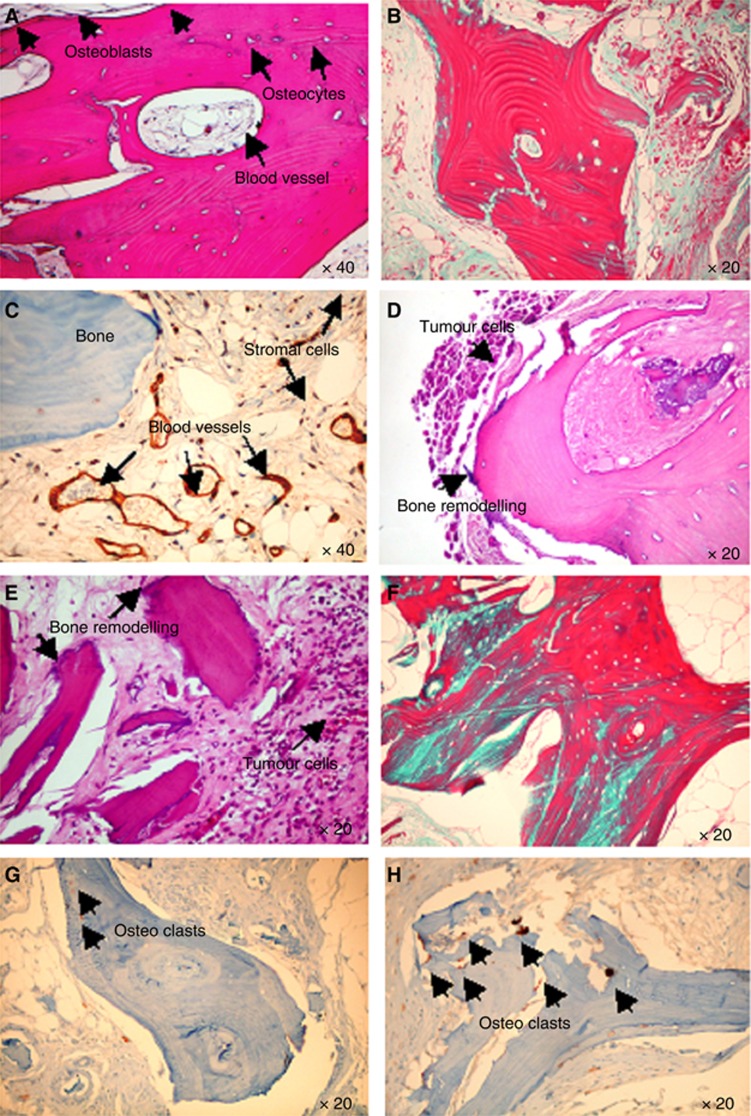
To rule out metastasis of our breast CSCs-like to mice bone, we performed X-rays on the mice, and we did not detect lesions in mice bone (Supplementary Figure S3). To verify the validity of our human-in-mice model, we needed to analyse the viability of the implanted bone in mice. The bone implants were viable because little-to-no necrosis was evident in the grafts and bone marrow cells, mineralised areas and stromal cells, and neo-vascularisation was present (Figure 2A). The engraftment of the human bone was also demonstrated by the newly synthesised bone present in the control (Figure 2B). Neo-vascularisation was present, and vessels expressed human CD34, ruling out the presence of murine vessels in the bone implants (Figure 2C). Breast CSCs-like colonised the human-implanted bone; a large area of invading tumour cells was evident and live bone was remodelled after both IC and SC injection (Figure 2D and E, respectively). The marked increase in bone remodelling due to bone metastases was evaluated through trichrome staining, which showed a large area of new bone apposition (Figure 2F). In detail, the area of neo-apposition was lower (blue stain) in the controls (Figure 2B) than that in the bone invaded by breast CSCs-like (Figure 2F). Moreover, we detected a lower number of TRAP-positive osteoclasts in the controls (Figure 2G) than that in bone with metastatic lesions (Figure 2H). These results demonstrate an increase in bone remodelling due to metastasis of the CSCs-like. To demonstrate the human bone marrow activity in the implanted bone, we measured human IgG in the serum of mice and detected a medium level of 80 μg ml−1 (data not shown).
Figure 2.
Histological analysis of the implanted bone. (A) H&E-stained section in control mice shows the presence of human live bone, blood vessels, newly synthesised bone with osteoblasts lining cells, as indicated by the arrows (magnification × 40). (B) Trichrome staining shows in blue the new collagen fibres in the bone: osteoid originated after the bone implanted in SC of the control mice. (C) Human endothelial cells in the blood vessels are stained for anti-human CD34, as indicated by the arrows. (D and E, respectively) Breast Luc-CSCs-like metastasise the human-implanted bone after IC and SC injection, in both cases area of pathological bone resorption are evident, as indicated by the arrows. (F) A marked neo-bone apposition is evident (osteoid is indicated by the blue staining) in bone invaded by breast CSCs-like. Osteoclasts stained for TRAP in bone of control mice (G) and mice injected with breast CSCs-like (H).
The bone-isolated metastatic cells show a phenotypic switch
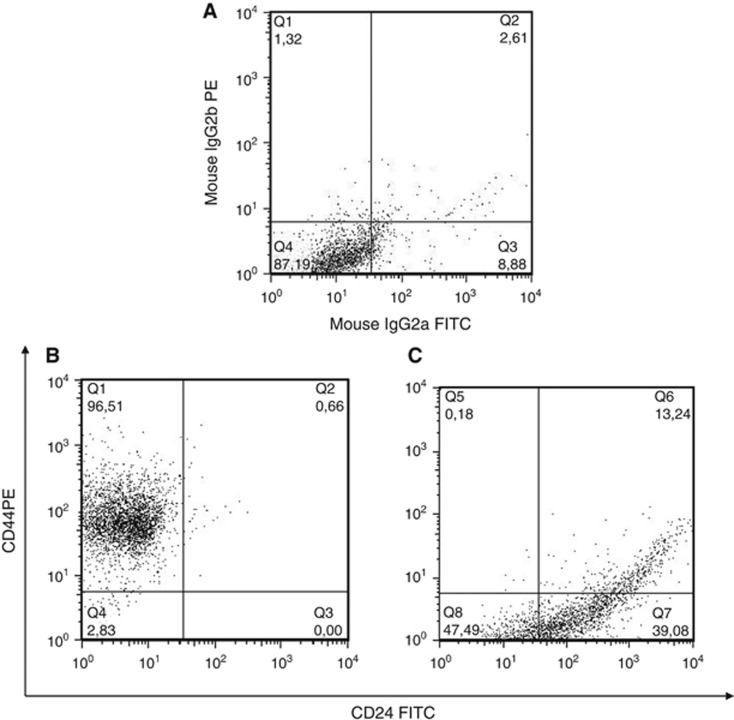
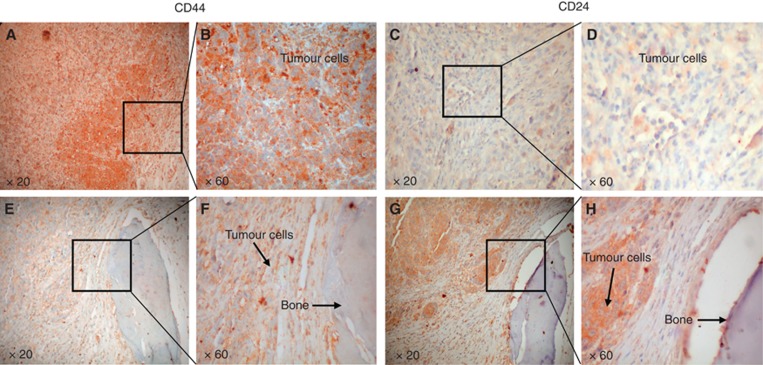
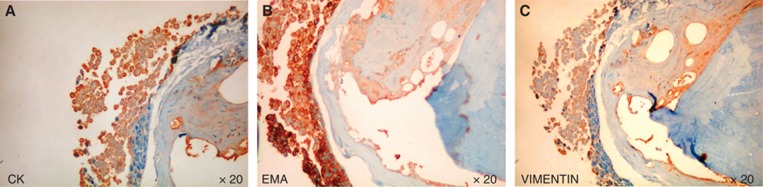
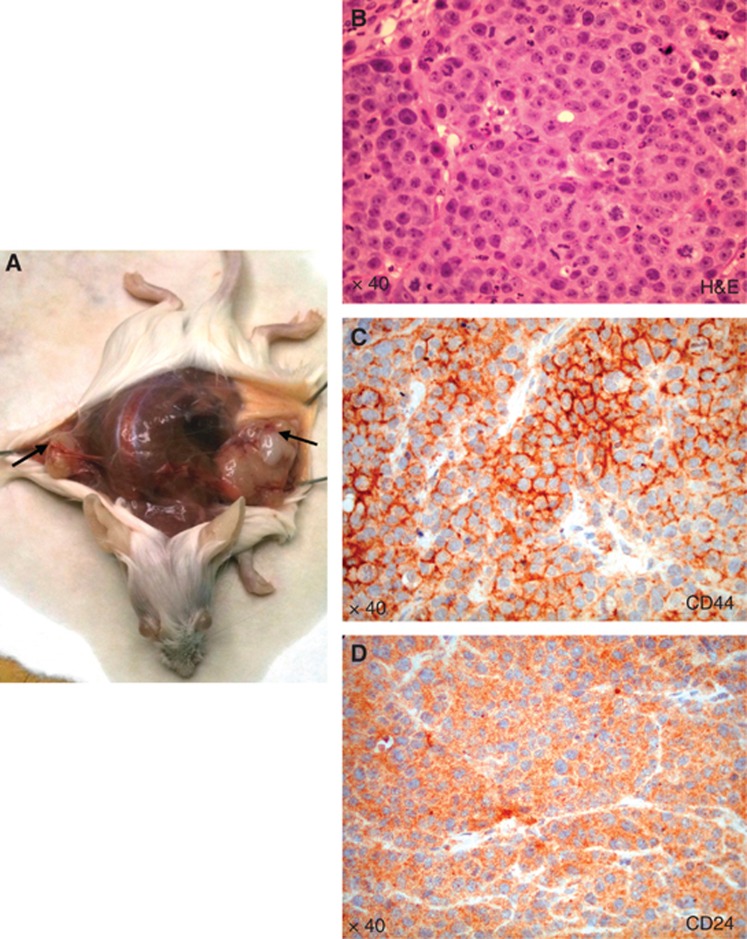
To analyse the phenotype of breast CSCs-like injected in mice and retrieved from tumour masses and bone lesions, we utilised both FACS analysis and histological investigation for the expression of CD44 and CD24. The injected breast CSCs-like were CD44+CD24− (96%±3.2%) (Figure 3B, see also Supplementary Figure S4), but after their bone retrieval they mainly showed a CD44−CD24+ phenotype (40%±4.1% Figure 3C). The analysis of primary tumour masses demonstrated that they were mainly constituted of CD44+CD24− cells (Figure 4A–D), whereas the bone lesions were mainly constituted of CD44−CD24+ cells (Figure 4E–H). These data confirm the FACS results and suggest a phenotypic switch of the injected CD44+CD24− cells, likely induced by the bone microenvironment. The CD44−CD24+ cells present in the bone lesions also expressed low molecular weight CKs, EMA and vimentin (Figure 5A–C). This expression of both epithelial and mesenchymal markers suggests that CD44−CD24+ cells have the capability to perform the epithelial-to-mesenchymal (EMT) and mesenchymal-to-epithelial transition (MET). To verify the tumorigenicity of the CD44−CD24+ bone-isolated metastatic cells, we re-injected them in NOD/SCID mice. They showed a more aggressive growth when injected SC than the parental CD44+CD24− cells, because each mouse developed two or three tumour masses (Figure 6A and B), which were constituted by cellular heterogeneity similar to the original tumour. Cells in the secondary tumour mass expressed both CD44 and CD24 (Figure 6C and D), suggesting the ability of CD44−CD24+ cells to switch their phenotype, likely through an EMT as they reacquired CD44 expression.
Figure 3.
Phenotype of parental breast CSCs-like and bone-isolated CSCs-like. Representative dot plots show the isotipic control (A), the CD44+CD24− breast CSCs-like injected in mice (B) and the bone-isolated CSCs-like, which express a CD44−CD24+ phenotype (C).
Figure 4.
CD44 and CD24 expression in tumour mass and human-implanted bone. IHC staining of tumour masses shows tumour cells with a CD44+CD24− phenotype (A–D). The bone-isolated CSCs-like are mainly CD44−CD24+ (E–H; magnification × 20 and × 60).
Figure 5.
IHC analysis of CD44−CD24+ bone-isolated CSCs-like. Cancer stem cells-like isolated from bone express CKs (A), EMA (B) and vimentin (C). (Magnification × 20).
Figure 6.
Analysis of secondary tumour induced by CD44−CD24+ bone-isolated CSCs-like. Subcutaneous injection of CD44−CD24+ cells results in tumour mass formation, as indicated by the arrows in A. H&E staining of tumour mass (B), CD44 (C) and CD24 (D).
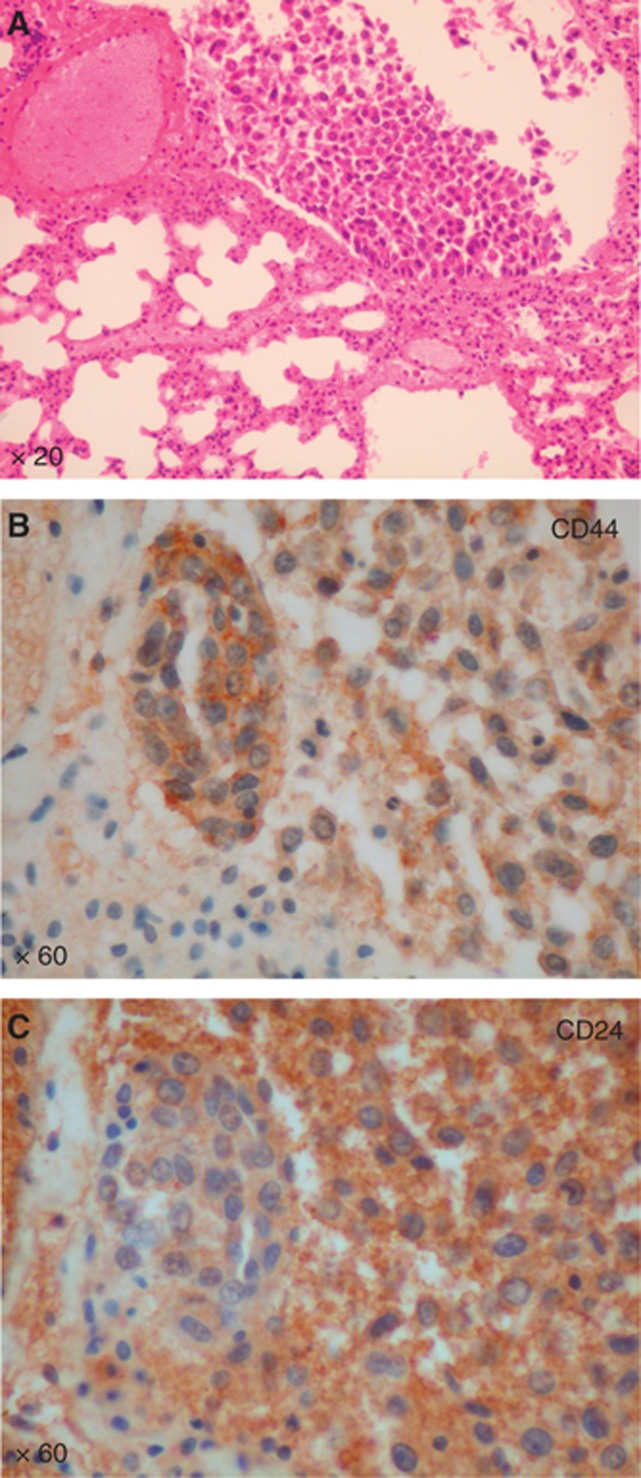
In order to rule out the effect of the metastatic state of these cells on this phenotypic change, we also examined CD44 and CD24 expression in the lung metastases. Tumour cells in the lung lesions expressed both CD44 and CD24 (Figure 7), which indicates a different phenotype compared with the bone metastatic cells. Thus, the phenotypic switch observed in the CD44+CD24− breast CSCs-like seems to depend on the contact with the human-implanted bone, underlying the importance of the bone microenvironment in the metastatic process.
Figure 7.
Analysis of CSCs-like lung metastases. (A) H&E-stained sections of a mouse lung show the presence of a metastatic lesion, (Magnification X 20). (B, C) IHC staining for CD44 and CD24 shows both the markers expressed by the tumor cells. (Magnification X 60).
Identification of a transcriptional signature for CD44−CD24+ breast CSCs-like
Global gene expression profiling was carried out to compare parental breast CSCs-like and Luc-CSCs-like with bone-isolated CSCs-like. To determine whether transcriptional changes associated with the passage in bone were stable, we also profiled the purified CSCs-like bone-isolated metastatic cells that were subsequently grown in vitro as mammospheres or in vivo as subcutaneous secondary implants (see Materials and Methods for details on the analysis). Indeed, a robust 110-gene transcriptional signature (breast CSCs-like bone tropism signature) was found to distinguish bone-isolated CD44−CD24+ CSCs-like from parental CD44+CD24− CSCs-like. This signature persisted after in vitro and in vivo expansion in the absence of surrounding bone tissue. This CSC bone tropism signature is composed of 88 upregulated and 22 downregulated genes (96 and 23 probes, respectively, see Supplementary Table S2), whose expression changed more than two-fold compared with parental CSCs-like in both mammosphere and subcutaneous derivatives of bone-isolated metastatic cells. Seventeen of the genes in the signature were overexpressed >10-fold, including carbonic anhydrase IX, which is known to promote EMT and to be associated with high incidences of metastasis (Shin et al, 2011); two chemokines, CCL20 and IL-8, involved in the tumour metastatic process (Li et al, 2003; Beider et al, 2009); insulin-like growth factor-binding protein 1, which is linked to breast cancer outcome (Goodwin et al, 2002); and transcription factor early growth response-1, which is involved in prostate carcinoma progression and enhances IL-8 transcription and secretion (Ma et al, 2009). We detected CCL20 at higher levels in sera of mice with bone metastases than in that of mice without metastases and controls (Supplementary Figure S5). IL-8 and IL-6 were detectable only in sera of mice with bone metastases (371±65.2 and 826±597.1 pg ml−1, respectively). These chemokines and cytokines might be responsible for the attraction of CSCs-like to bone and might support the phenotypic switch.
Analysis of functional modules in the breast CSCs-like bone tropism signature
To identify differentially expressed functional gene modules (‘gene sets'), we carried out GSEA (Subramanian et al, 2005) utilising CD44+CD24− parental cells (breast CSCs-like, Luc-CSCs-like), and all CD44−CD24+ cell populations derived after bone colonisation, that is, CSCs-like directly isolated from bone lesions (two samples), subsequently grown as mammospheres (one sample) or further implanted SC (two samples). Thus, GSEA allowed us to test for permanent transcriptional features acquired during bone colonisation. In the ‘curated gene set' collection (C2), a total of 93 gene sets were significantly enriched in upregulated genes (FDR<0.25 and P-value<0.01, see also Supplementary Table S3A). In the Gene Ontology collection (C5), we also identified 12 upregulated gene sets (FDR<0.25 and P<0.01). The most relevant upregulated gene sets were condensed into a specific spectrum of functional modules, summarised in Table 1 (see also Supplementary Table S3A and B). These modules include hypoxia response, metabolic stress, growth factors, cytokine stimulation, chemotaxis and acid-base balance, which are all known to be relevant to the biology of the bone metastatic process.
Table 1. Relevant gene sets grouped in functional modules.
| Functional module | Gene set name | Collection | Size | NES | NOM P-value | FDR q-value |
|---|---|---|---|---|---|---|
| MDM4 p53 target |
MARTORIATI_MDM4_TARGETS_FETAL_LIVER_UP |
C2 |
83 |
1.952 |
0.000 |
0.042 |
| |
MARTORIATI_MDM4_TARGETS_NEUROEPITHELIUM_UP |
C2 |
79 |
1.676 |
0.000 |
0.131 |
| Response to growth factor |
AMIT_EGF_RESPONSE_40_HELA |
C2 |
40 |
1.879 |
0.000 |
0.064 |
| |
AMIT_EGF_RESPONSE_480_MCF10A |
C2 |
35 |
1.781 |
0.000 |
0.126 |
| |
ST_ERK1_ERK2_MAPK_PATHWAY |
C2 |
26 |
1.775 |
0.000 |
0.121 |
| |
AMIT_SERUM_RESPONSE_40_MCF10A |
C2 |
27 |
1.749 |
0.000 |
0.116 |
| |
AMIT_EGF_RESPONSE_60_MCF10A |
C2 |
34 |
1.740 |
0.000 |
0.122 |
| |
REACTOME_MAPK_TARGETS_NUCLEAR_EVENTS_MEDIATED_BY_MAP_KINASES |
C2 |
24 |
1.722 |
0.000 |
0.110 |
| Cytokine stimulation |
CYTOKINE_METABOLIC_PROCESS |
C5 |
17 |
1.693 |
0.000 |
0.121 |
| |
CYTOKINE_BIOSYNTHETIC_PROCESS |
C5 |
16 |
1.686 |
0.000 |
0.112 |
| |
CYTOKINE_ACTIVITY |
C5 |
31 |
1.749 |
0.000 |
0.097 |
| Angiogenesis |
VASCULATURE_DEVELOPMENT |
C5 |
31 |
1.813 |
0.000 |
0.079 |
| |
ANGIOGENESIS |
C5 |
26 |
1.666 |
0.000 |
0.132 |
| |
REGULATION_OF_ANGIOGENESIS |
C5 |
15 |
1.748 |
0.000 |
0.083 |
| Hypoxia |
WINTER_HYPOXIA_UP |
C2 |
80 |
1.813 |
0.000 |
0.117 |
| |
MANALO_HYPOXIA_UP |
C2 |
142 |
1.751 |
0.000 |
0.124 |
| |
JIANG_HYPOXIA_NORMAL |
C2 |
184 |
1.734 |
0.000 |
0.107 |
| |
HARRIS_HYPOXIA |
C2 |
61 |
1.731 |
0.000 |
0.106 |
| |
WINTER_HYPOXIA_METAGENE |
C2 |
172 |
1.718 |
0.000 |
0.112 |
| ATM target |
RASHI_RESPONSE_TO_IONIZING_RADIATION_1 |
C2 |
31 |
1.897 |
0.000 |
0.074 |
| Histone |
REACTOME_TELOMERE_MAINTENANCE |
C2 |
55 |
1.710 |
0.000 |
0.115 |
| Chemotaxis |
LOCOMOTORY_BEHAVIOR |
C5 |
31 |
1.595 |
0.000 |
0.176 |
| |
BEHAVIOR |
C5 |
45 |
1.599 |
0.000 |
0.178 |
| |
THEILGAARD_NEUTROPHIL_AT_SKIN_WOUND_UP |
C2 |
59 |
1.735 |
0.000 |
0.119 |
| Stat3 |
DAUER_STAT3_TARGETS_UP |
C2 |
31 |
1.714 |
0.000 |
0.114 |
| Acid-base balance |
CARBON_OXYGEN_LYASE_ACTIVITY |
C5 |
22 |
1.647 |
0.000 |
0.147 |
| HYDRO_LYASE_ACTIVITY | C5 | 18 | 1.637 | 0.000 | 0.136 |
Abbreviations: C2=curated gene collection; C5=gene ontology collection; FDR=false discovery rate; NES=normalised enrichment score.
The table reports the most relevant upregulated gene sets (P-value<0.01 and FDR<0.25) connected with bone metastatic process.
Among the downregulated gene sets, 67 and 19 (for the C2 and C5 collections, respectively) had a P-value<0.01, but none reached an FDR<0.25. However, the C2 gene set with the highest downregulation enrichment was composed of genes upregulated in brain-metastatic breast cancer (SMID_BREAST_CANCER_RELAPSE_IN_BRAIN_UP). The fact that such genes are preferentially downregulated in the CSCs-like bone tropism signature supports organ specificity of the signature rather than a generic metastatic propensity. The C5 gene set with the highest downregulation enrichment was ORGANELLE_ORGANIZATION and BIOGENESIS, with a core constituted by proteins modulating the cytoskeleton.
The breast CSCs-like bone tropism signature discriminates osteotropic breast cancers from tumours metastasising other sites
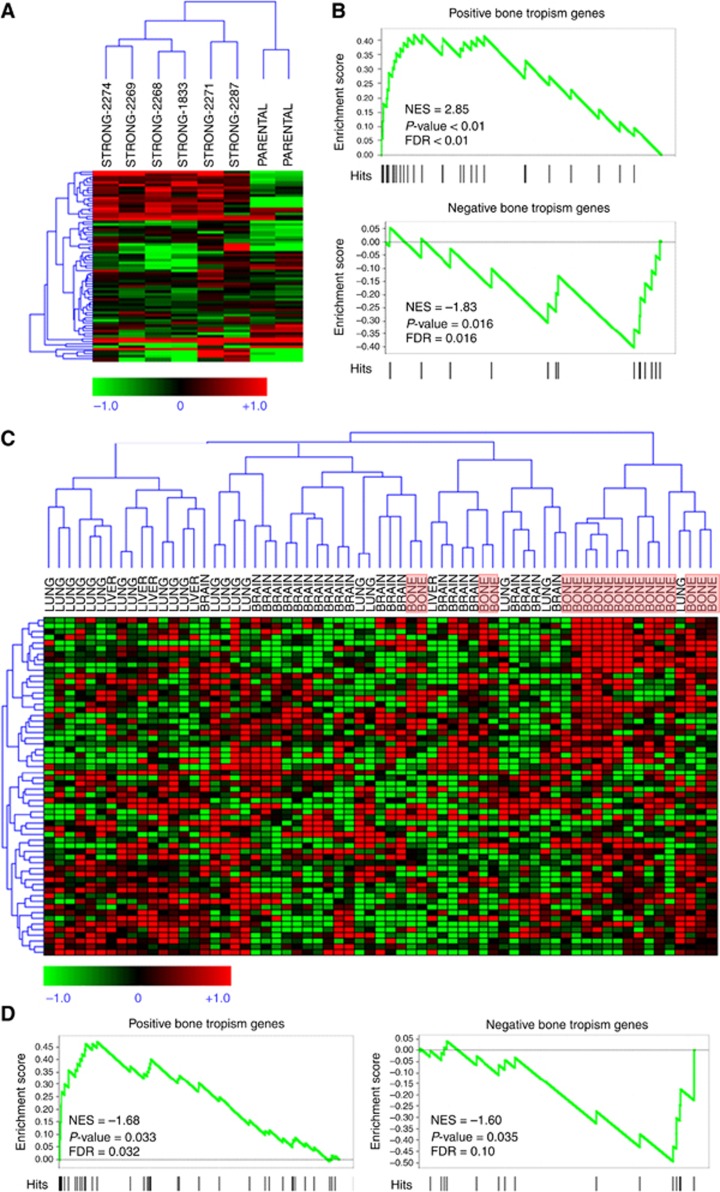
The ability to distinguish cell lines with different bone tropism was tested by hierarchical clustering of a data set generated by Kang et al (2003), in which human breast cancer cells with high bone tropism were obtained by in vivo selection from a starting cell line with lower bone metastatic propensity (Kang et al, 2003). Expression data for 75 probes of this data set mapping to 51 of the 110 bone tropism genes were converted into log(2) ratio against mean expression in all samples. Our signature was able to segregate the strong bone metastatic cell lines from the two parental lines from which they were derived (Figure 8A). The GSEA analysis confirmed that the CSC bone tropism signature is significantly enriched in genes either up- or downregulated in bone metastatic cells vs their parental counterparts (Figure 8B, see also Supplementary Table S2).
Figure 8.
Microarray analysis of breast CSCs-like bone tropism signature. (A) Hierarchical clustering of human breast cancer cell lines with strong bone metastatic potential derived by parental lines in Kang et al (2003) data set, using our CSCs-like bone tropism signature composed of 110 genes (119 probes). (B) Gene set enrichment analysis samples of Kang et al (2003) for enrichment of the CSCs-like bone tropism signature in genes that discriminate bone metastatics from parental lines. Upper panel: positive signature genes are significantly high in cells with strong metastatic potential. Lower panel: negative signature genes are significantly low in strong metastatic potential. (C) Hierarchical clustering of 65 breast cancer samples from metastatic lesions at different sites in a published data set, using our CSCs-like bone tropism signature. For convenience, bone metastasis samples are highlighted by light red boxes. A subset of signature genes with significantly high expression in bone metastases is evident in the top right corner. (D) The GSEA analysis on the metastasis samples for enrichment of the breast CSCs-like bone tropism signature in genes that discriminate bone metastases from lesions at other sites. Left panel: positive signature genes are significantly high in bone metastases. Right panel: negative signature genes are significantly low in bone metastases. FDR, false discovery rate; NES, normalised enrichment score.
To verify whether the CSCs-like bone tropism signature actually discriminates bone metastatic breast cancers from cancers with different metastatic locations, we mapped the signature on a published data set that includes expression profiles of 65 breast cancer samples from metastatic lesions in the bone (n=18), brain (n=22), liver (n=5) and lung (n=20; Zhang et al, 2009). Expression data for 128 probes of this data set mapping to 74 of the 110 bone tropism genes were converted into log2 ratio against mean expression in all samples and hierarchically clustered (Figure 8C). Interestingly, the CSCs-like bone tropism signature was able to aggregate 13 of 18 bone metastases within a cluster where a subset of signature genes was highly expressed (top right part of the heatmap). Metastases at other sites, such as liver, brain and lung, were clustered in other groups. The GSEA analysis on this data set showed significant enrichment of the CSCs-like bone tropism signature in genes discriminating bone metastases from metastases at other organs (Figure 8D, see also Supplementary Table S2).
Discussion
This work analysed the ability of isolated CD44+CD24− breast CSCs-like to metastasise to human bone implanted in immunocompromised mice. This model was chosen because it provides a human microenvironment, which has a key role in the growth of metastatic cells and in their interaction with the specialised niche in the bone marrow (Tzukerman et al, 2003; Guise, 2010).
The results of this study indicate that breast CSCs-like have a mesenchymal and migratory CD44+CD24− phenotype and that they are endowed with a metastatic potential to human-implanted bone. The finding that breast CSCs-like grow in bone suggests that breast CSCs-like encountered a particularly favourable soil in the human-implanted bone. In the tumour masses grown SC and in the bone lesions, the CSCs-like expressed both epithelial and mesenchymal markers. These observations support the existence of a migrating CSC that possesses features of both stemness and mobility that may allow it to undergo an epithelial-to-mesenchymal transition (EMT) and migrate to bone. CSCs-like isolated from the bone lesion were CD44−CD24+, indicating that they likely performed a MET after reaching the bone, with a consequent phenotypical switch from CD44+CD24− to CD44−CD24+, driven by the interaction with the bone microenvironment. Moreover, after re-injection in mice, the CD44−CD24+ bone-isolated CSCs-like formed tumour masses constituted mainly by CD44+CD24− cells, indicating another phenotypic transition. In different carcinomas, the importance of EMT for the induction of stem characteristics has been demonstrated (Wellner et al, 2009; Kong et al, 2010). The transitions between different states observed in our CSCs-like could also be explained by a quantitative Markov model of cell-state interconversion. According to this model, breast cancers include discrete populations that randomly perform transitions between states without increasing their proliferation rate to reach a progressive equilibrium proportion (Harris et al, 2008; Gupta et al, 2011). Thus, the capability of the CD44−CD24+ bone-isolated CSCs-like to generate subcutaneous and heterogeneous tumour masses with a high percentage of CD44+CD24− cells could be consistent with the attempt of these CSCs-like to recapitulate the differentiation state heterogeneity present in the parental tumour from which they were derived.
The phenotypic switch of breast CSCs-like could also be due to their interaction with the haematopoietic stem cell niche. Supporting this view is the recent report that prostate cancer cells are able to occupy the haematopoietic stem cell niche in the bone marrow, which is important for tumour dissemination and has a central role in bone metastases (Shiozawa et al, 2011).
In this study, we identified for the first time a CSCs-like bone tropism signature that can distinguish CD44−CD24+ bone-isolated CSCs-like from parental breast CD44+CD24− CSCs-like. This signature represents a characterisation of a genetic/transcriptional programme by which CSCs-like isolated from a primary breast tumour promote bone metastasis in a human-implanted bone. Moreover, the signature can be a proof of concept of the phenotypic switch observed in CSCs-like after the passage in bone. The CSCs-like bone tropism signature is characterised by genes that are strongly upregulated, such as CCL20, IL-8 and IL-6. These human markers were also detected in the sera of mice with bone metastases, but not in mice without bone colonisation by CSCs-like or control mice. It is reasonable to assume that these chemokines and cytokines have a role in attracting CSCs-like to the bone microenvironment and then in inducing the phenotypic switch of the CSCs-like. Indeed, they are upregulated during the bone invasion and retained after in vitro and in vivo passages. The microarray analysis does highlight a strong cytokine/growth factor transcriptional module in the CSCs-like bone tropism signature. The upregulation of a number of functional gene modules involved in various aspects of the bone metastatic process was observed. A central role seems to be performed by hypoxia, which promotes angiogenesis, bone metastasis (Dunn et al, 2009; Lu et al, 2010), metabolic stress adaptation (Winter et al, 2007), motility and invasiveness (Pennacchietti et al, 2003). Another set of genes of potential interest are those involved in telomere maintenance, which are reported to promote cancer stemness through the expression of EGFR (Pantel and Brakenhoff, 2004; Beck et al, 2011). In fact, the results showed upregulation of multiple target gene sets and the Ras/ERK pathway downstream from EGFR, which also contributes to the maintenance and acquisition of stem-like hallmarks (Tabu et al, 2010). The significant upregulation of ERK/MAP kinase targets suggests an activation of proliferation, and it is likely flanked by concomitant upregulation of genes of the MDM4 pathway, reflecting a possible functional block of p53 (Abdel-Fatah et al, 2010). The upregulation of STAT3 targets has also been identified, supporting the role of STAT3 signalling in the growth of breast CSCs-like (Marotta et al, 2011). Overall, these results suggest concomitant acquisition of proliferation and stemness potential by bone-isolated CSCs-like.
In conclusion, we demonstrated the ability of primary breast CSCs-like to metastasise to bone by showing their direct involvement in the pathogenesis of breast cancer bone metastasis. Anticipating a risk of bone metastasis is mandatory because bone metastatic recurrence often follows a period of tumour dormancy. Identification of this CSCs-like bone tropism signature in breast cancer patients may be a relevant prognostic factor for clinical outcomes and may allow modification of therapeutic regimens according to the breast cancer characteristics.
Acknowledgments
We thank Dr Flavio Cristofani, Chief of the Stabulation Facility at Molecular Biotechnology Center in Turin, and Roberta Porporato and Tommaso Renzulli for their technical assistance with microarray data generation and analysis. We specially thank Professor Luigi Naldini and Roberta Faccio for the critical discussion of the manuscript. This work was supported by Compagnia di San Paolo and by the Italian Ministry of Health: Ricerca Sanitaria Finalizzata e Giovani Ricercatori 2009 (GR 2009-1584485).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Abdel-Fatah TM, Powe DG, Agboola J, Adamowicz-Brice M, Blamey RW, Lopez-Garcia MA, Green AR, Reis-Filho JS, Ellis IO. The biological, clinical and prognostic implications of p53 transcriptional pathways in breast cancers. J Pathol. 2010;220 (4:419–434. doi: 10.1002/path.2663. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100 (7:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck S, Jin X, Sohn YW, Kim JK, Kim SH, Yin J, Pian X, Kim SC, Nam DH, Choi YJ, Kim H. Telomerase activity-independent function of TERT allows glioma cells to attain cancer stem cell characteristics by inducing EGFR expression. Mol Cells. 2011;31 (1:9–15. doi: 10.1007/s10059-011-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beider K, Abraham M, Begin M, Wald H, Weiss ID, Wald O, Pikarsky E, Abramovitch R, Zeira E, Galun E, Nagler A, Peled A. Interaction between CXCR4 and CCL20 pathways regulates tumor growth. PloS one. 2009;4 (4:e5125. doi: 10.1371/journal.pone.0005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijs JT, van der Horst G, van den Hoogen C, Cheung H, de Rooij B, Kroon J, Petersen M, van Overveld PG, Pelger RC, van der Pluijm G. The BMP2/7 heterodimer inhibits the human breast cancer stem cell subpopulation and bone metastases formation. Oncogene. 2011;31 (17:2164–2174. doi: 10.1038/onc.2011.400. [DOI] [PubMed] [Google Scholar]
- Bussolati B, Grange C, Sapino A, Camussi G. Endothelial cell differentiation of human breast tumour stem/progenitor cells. J Cell Mol Med. 2009;13 (2:309–319. doi: 10.1111/j.1582-4934.2008.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution. Cell Cycle. 2007;6 (19:2332–2338. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124 (6:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Dunn LK, Mohammad KS, Fournier PG, McKenna CR, Davis HW, Niewolna M, Peng XH, Chirgwin JM, Guise TA. Hypoxia and TGF-beta drive breast cancer bone metastases through parallel signaling pathways in tumor cells and the bone microenvironment. PloS one. 2009;4 (9:e6896. doi: 10.1371/journal.pone.0006896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat Genet. 2000;25 (2:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- Fu L, Medico E. FLAME, a novel fuzzy clustering method for the analysis of DNA microarray data. BMC Bioinformatics. 2007;8:3. doi: 10.1186/1471-2105-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin PJ, Ennis M, Pritchard KI, Trudeau ME, Koo J, Hartwick W, Hoffma B, Hood N. Insulin-like growth factor binding proteins 1 and 3 and breast cancer outcomes. Breast Cancer Res Treat. 2002;74 (1:65–76. doi: 10.1023/a:1016075709022. [DOI] [PubMed] [Google Scholar]
- Guise T. Examining the metastatic niche: targeting the microenvironment. Semin Oncol. 2010;37 (Suppl 2:S2–14. doi: 10.1053/j.seminoncol.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146 (4:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Harris MA, Yang H, Low BE, Mukherjee J, Guha A, Bronson RT, Shultz LD, Israel MA, Yun K. Cancer stem cells are enriched in the side population cells in a mouse model of glioma. Cancer Res. 2008;68 (24:10051–10059. doi: 10.1158/0008-5472.CAN-08-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3 (6:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Kong D, Banerjee S, Ahmad A, Li Y, Wang Z, Sethi S, Sarkar FH. Epithelial to mesenchymal transition is mechanistically linked with stem cell signatures in prostate cancer cells. PLoS One. 2010;5 (8:e12445. doi: 10.1371/journal.pone.0012445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuperwasser C, Dessain S, Bierbaum BE, Garnet D, Sperandio K, Gauvin GP, Naber SP, Weinberg RA, Rosenblatt M. A mouse model of human breast cancer metastasis to human bone. Cancer Res. 2005;65 (14:6130–6138. doi: 10.1158/0008-5472.CAN-04-1408. [DOI] [PubMed] [Google Scholar]
- Li A, Dubey S, Varney ML, Dave BJ, Singh RK. IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J Immunol. 2003;170 (6:3369–3376. doi: 10.4049/jimmunol.170.6.3369. [DOI] [PubMed] [Google Scholar]
- Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, Wen S, Chang YF, Bachmann MH, Shimono Y, Dalerba P, Adorno M, Lobo N, Bueno J, Dirbas FM, Goswami S, Somlo G, Condeelis J, Contag CH, Gambhir SS, Clarke MF. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci USA. 2010;107 (42:18115–18120. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356 (3:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- Lu X, Yan CH, Yuan M, Wei Y, Hu G, Kang Y. In vivo dynamics and distinct functions of hypoxia in primary tumor growth and organotropic metastasis of breast cancer. Cancer Res. 2010;70 (10:3905–3914. doi: 10.1158/0008-5472.CAN-09-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Ren Z, Ma Y, Xu L, Zhao Y, Zheng C, Fang Y, Xue T, Sun B, Xiao W. Targeted knockdown of EGR-1 inhibits IL-8 production and IL-8-mediated invasion of prostate cancer cells through suppressing EGR-1/NF-kappaB synergy. J Biol Chem. 2009;284 (50:34600–34606. doi: 10.1074/jbc.M109.016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ, Choudhury SA, Maruyama R, Wu Z, Gonen M, Mulvey LA, Bessarabova MO, Huh SJ, Silver SJ, Kim SY, Park SY, Lee HE, Anderson KS, Richardson AL, Nikolskaya T, Nikolsky Y, Liu XS, Root DE, Hahn WC, Frank DA, Polyak K. The JAK2/STAT3 signaling pathway is required for growth of CD44CD24 stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121 (7:2723–2735. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Kang Y, Serganova I, Gupta GP, Giri DD, Doubrovin M, Ponomarev V, Gerald WL, Blasberg R, Massague J. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest. 2005;115 (1:44–55. doi: 10.1172/JCI22320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34 (3:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Mundy GR. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer. 2002;2 (8:584–593. doi: 10.1038/nrc867. [DOI] [PubMed] [Google Scholar]
- Mundy GR, Sterling J.2008Metastatic solid tumors to bone Primer on the metabolic bone diseases and disorders of mineral metabolism7th ednVol. 79ASBMR: Washington [Google Scholar]
- Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer. 2004;4 (6:448–456. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- Pennacchietti S, Michieli P, Galluzzo M, Mazzone M, Giordano S, Comoglio PM. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer cell. 2003;3 (4:347–361. doi: 10.1016/s1535-6108(03)00085-0. [DOI] [PubMed] [Google Scholar]
- Roato I, Caldo D, Godio L, D'Amico L, Giannoni P, Morello E, Quarto R, Molfetta L, Buracco P, Mussa A, Ferracini R. Bone invading NSCLC cells produce IL-7: mice model and human histologic data. BMC Cancer. 2010;10:12. doi: 10.1186/1471-2407-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324 (5935:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138 (5:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Shin HJ, Rho SB, Jung DC, Han IO, Oh ES, Kim JY. Carbonic anhydrase IX (CA9) modulates tumor-associated cell migration and invasion. J Cell Sci. 2011;124 (Pt 7:1077–1087. doi: 10.1242/jcs.072207. [DOI] [PubMed] [Google Scholar]
- Shiozawa Y, Pedersen EA, Havens AM, Jung Y, Mishra A, Joseph J, Kim JK, Patel LR, Ying C, Ziegler AM, Pienta MJ, Song J, Wang J, Loberg RD, Krebsbach PH, Pienta KJ, Taichman RS. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J Clin Invest. 2011;121 (4:1298–1312. doi: 10.1172/JCI43414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, Halushka MK, Sukumar S, Parker LM, Anderson KS, Harris LN, Garber JE, Richardson AL, Schnitt SJ, Nikolsky Y, Gelman RS, Polyak K. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11 (3:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, Foekens JA, Martens JW. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68 (9:3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102 (43:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabu K, Kimura T, Sasai K, Wang L, Bizen N, Nishihara H, Taga T, Tanaka S. Analysis of an alternative human CD133 promoter reveals the implication of Ras/ERK pathway in tumor stem-like hallmarks. Mol Cancer. 2010;9:39. doi: 10.1186/1476-4598-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzukerman M, Rosenberg T, Ravel Y, Reiter I, Coleman R, Skorecki K. An experimental platform for studying growth and invasiveness of tumor cells within teratomas derived from human embryonic stem cells. Proc Natl Acad Sci USA. 2003;100 (23:13507–13512. doi: 10.1073/pnas.2235551100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Lippitt JM, Guzman-Ramirez N, Hamdy FC, Eaton CL, Thalmann GN, Cecchini MG, Pelger RC, van der Pluijm G. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70 (12:5163–5173. doi: 10.1158/0008-5472.CAN-09-3806. [DOI] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11 (12:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- Winter SC, Buffa FM, Silva P, Miller C, Valentine HR, Turley H, Shah KA, Cox GJ, Corbridge RJ, Homer JJ, Musgrove B, Slevin N, Sloan P, Price P, West CM, Harris AL. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007;67 (7:3441–3449. doi: 10.1158/0008-5472.CAN-06-3322. [DOI] [PubMed] [Google Scholar]
- Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, Foekens JA, Massague J. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16 (1:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.