Abstract
During advanced atherosclerosis, chronic activation of the endoplasmic reticulum (ER) stress pathway, otherwise known as the unfolded protein response (UPR), is strongly associated with atherosclerotic plaque destabilization, the precursor to acute myocardial infarction and sudden death. Destabilized or vulnerable plaques are characterized by features that include thinning of a protective collagenous cap at the interface between the plaque and the vascular lumen and expansion of the necrotic core, a lipid-rich graveyard of dead macrophages. The cell biology of advanced plaque progression is complex and includes multiple cellular stressors that combine to promote chronic inflammation and progressive plaque deterioration. Several of these stressors converge at the ER, leading to activation of the UPR in multiple cell types. In macrophages, prolonged UPR activation triggers apoptosis, which when coupled to defective phagocytic clearance of these dying cells, leads to secondary necrosis and expansion of the plaque necrotic core. Though much insight has been gained recently on the role of the UPR in atherosclerosis, future studies are warranted to determine the cell-type specific contributions of ER stress to athero-progression and the therapeutic potential of UPR modulation. For such objectives to be met, reliable and standardized methodology must be utilized and developed. This chapter summarizes our current understanding of ER stress-induced macrophage apoptosis in atheromata and outlines both in vitro and in vivo methodologies to quantify the UPR in the context of experimental murine-advanced atherosclerosis.
1. Introduction to ER Stress in Atherosclerosis
The adaptive cellular response known as the unfolded protein response (UPR) occurs under conditions that perturb endoplasmic reticulum (ER) homeostasis and induce ER stress (Ron and Walter, 2007). Three ER resident proteins sense and differentially regulate the UPR. These include inositol-requiring enzyme-1 (IRE-1; Cox et al., 1993), activating transcription factor-6 (ATF6; Yoshida et al., 1998), and PKR-like eukaryotic initiation factor 2 kinase (PERK; Harding et al., 1999). The function of the UPR or integrated stress response (ISR) is to protect the ER from normal and pathophysiological perturbations that include increased protein synthesis, disruption of ER calcium homeostasis, changes in redox potential, and alterations in the physical properties of the ER membrane bilayer (Hossain et al., 2003; Ma and Hendershot, 2001). In response to interference with ER homeostasis, the UPR initiates transcription of ER chaperones, oxidoreductases, and ER-associated degradation (ERAD) proteins (Travers et al., 2000). The UPR also exerts translational control by phosphorylating eif2α, which is activated by PERK (Harding et al., 2000). This leads to generalized inhibition of protein translation, except in the case of ATF4, which is selectively upregulated. ATF4 induces the proapoptotic transcription factor CHOP (GADD153; Zinszner et al., 1998). In the case of ATF6, its activation by intramembrane proteolysis leads to the production of protein-folding chaperones. This is partially accomplished through ATF6-mediated transcription of XBP-1 (Yoshida et al., 2001). XBP-1 is also activated by IRE-1. IRE-1, through its endoribonuclease domain, promotes XBP-1 mRNA splicing. Spliced XBP-1 targets genes that enhance ER protein folding capacity (Lee, 2003). IRE-1 is also implicated in nonadaptive proapoptotic signaling that includes ribonuclease-mediated mRNA decay and proapoptotic c-Jun kinase (JNK) signaling (Urano, 2000). If the UPR is successful in alleviating ER stress, negative feedback signaling leads to UPR suppression (Merksamer et al., 2008). If on the other hand, these adaptive responses are not sufficient to restore ER equilibrium, the UPR triggers programmed cell death (Lin et al., 2007). Over the last 10 years, ER-stress signaling has emerged to be a key factor in events that promote atherosclerosis. Recent evidence in experimental mice and humans support a scenario whereby chronic ER stress in the vascular wall promotes atherosclerotic progression and events that directly contribute to plaque vulnerability, the precursor to acute myocardial infarction and sudden death.
During atherosclerosis, ER stress is activated in multiple cell types of the vascular wall. Though little is known about the role of ATF6 signaling during atherosclerotic development, the other arms of the UPR, that is, IRE-1 and PERK, have been shown to play a significant role in endothelial cells and macrophages of the vascular wall. In endothelial cells, the potentially pro-atherogenic molecule homocysteine causes activation of ER stress-induced growth arrest (Outinen et al., 1999). Also in endothelial cells, IRE-1-mediated XBP-1 splicing leads to endothelial apoptosis and atherosclerosis development in response to disturbed arterial flow (Zeng et al., 2009). The PERK pathway, through its induction of ATF4 and CHOP, appears to be especially important during atherosclerotic progression. Several studies have documented increased mRNA and protein of CHOP in both experimental animals and in human coronary artery plaques. Myoishi et al. reported a strong correlation between expression of CHOP and plaque necrosis and rupture in human coronary lesions (Myoishi et al., 2007). Furthermore, deficiency of CHOP in both the Ldlr–/– and Apoe–/– models of atherosclerosis reduces advanced lesional macrophages apoptosis and plaque necrosis (Thorp et al., 2009). Thus, the activation of multiple arms of ER stress in distinct cell types combines to affect disparate events that advance atherosclerotic progression. The aforementioned reports shed light on the physiological relevance of ER stress during atherosclerosis; however, these few studies have only scratched the surface. Future studies are needed to test the significance of UPR modulation in atherosclerosis and its amenability to therapeutic intervention. This chapter discusses experimental methods for the interrogation and characterization of one of the best characterized UPR pathways in atheromata, that which occurs in the macrophage during ER stress-mediated apoptosis.
2. Significance of Macrophage ER Stress and Apoptosis in Advanced Atherosclerosis
There is sufficient evidence that the UPR is chronically activated in cells of the atherosclerotic vascular wall, including macrophages (Tabas, 2009). Macrophages are a central protagonist of both early and advanced atherosclerotic development. Early atherogenesis is characterized by an inflammatory response to retained apolipoproteins in the subendothelium (Tabas et al., 2007; Williams and Tabas, 1995). This involves infiltration of monocytes that later differentiate into macrophages and ingest the retained lipids. During atherosclerotic progression, lesions exhibit evidence of significant accelerated macrophage death (Ball et al., 1995). Increased macrophage apoptosis, in tandem with defective phagocytic clearance of apoptotic macrophages, contributes to expansion of the necrotic core (Tabas, 2005). This destabilizes plaques and in turn promotes plaque disruption and acute thrombotic events, including myocardial infarction and stroke (Kolodgie et al., 2003; Tabas, 2005; Virmani et al., 2006). Work by Feng et al. and later Austin and colleagues showed that ER stress was significantly elevated in lesional macrophages (Feng et al., 2003a; Zhou et al., 2005). These studies also provided evidence for increased expression of the UPR effector CHOP in advanced lesions. These data in experimental mice were later confirmed in studies of human carotid endarterectomy specimens. In these studies, robust CHOP expression and lesional apoptosis was only identified in advanced vulnerable plaques (Myoishi et al., 2007). Together, the experimental evidence supports a working model whereby ER stress-induced macrophage apoptosis is a key event in plaque destabilizing necrotic core generation.
The mechanisms of ER-stressed macrophage apoptosis have been explored in detail using cultured macrophage models. These studies indicate that robust apoptotic induction requires the CHOP pathway and that this is upstream of canonical Fas and mitochondrial death pathways (Timmins et al., 2009). CHOP promotes apoptosis by activating the transcription of ER oxidase, which in turn promotes ER calcium release through the IP3 receptor (Harding et al., 2003; Li et al., 2009). Increased cytosolic calcium activates calcium/calmodulin-dependent protein kinase II, which then triggers proapoptotic pathways that include Fas, mitochondrial release of apoptogenic factors, signal transducer and activator of transcription-1 (STAT1) and NADPH oxidase-mediated reactive oxygen species (ROS; Lim et al., 2008). In vitro, 70% of macrophages from Chop–/– mice are protected from ER stress-induced apoptosis (Feng et al., 2003a). This is consistent with the report described above in which Chop–/– mice are protected from macrophage apoptosis and necrosis in plaque. Thus, the combination of mechanistic data from primary cultured cells with in vivo genetic tests of causality have uncovered key roles for the UPR in advanced lesional macrophage death and plaque necrosis (Feng et al., 2003a,b). Below we describe (1) in vitro methods to measure ER stress-induced apoptosis in macrophages under conditions that model the in vivo milieu and later (2) in vivo methodologies for monitoring ER stress and macrophage apoptosis in atherosclerotic lesions.
3. Modeling Atherosclerosis-Relevant ER Stress-Induced Apoptosis
Mechanistic insight from in vitro studies has proven essential to explain the biology of atherosclerotic progression in vivo. Such studies have implicated multiple ER-stress pathways that may contribute to macrophage apoptosis during atherosclerosis. Athero-relevant inducers are identified from molecules and processes within athermoata that can lead to UPR activation. These include intracellular unesterified cholesterol, oxysterols, oxidant stress, hypoxia, and peroxynitrite (Seimon and Tabas, 2009). For example, in early lesions, aggregated lipoprotein-derived cholesterol is trafficked to the macrophage ER after phagocytic internalization, where acyl-coenzyme A-cholesterol acyltransferase (ACAT) esterifies cholesterol to its cholesteryl acyl ester (Brown et al., 1980). However, in advanced atherosclerosis, macrophages accumulate large amounts of unesterified “free” cholesterol (FC; Katz et al., 1976). In vitro, FC accumulation in macrophages activates the UPR and promotes apoptosis (Feng et al., 2003a). Robust ER stress can also be achieved with 7-ketocholesterol (7-KC). 7-KC is an ER stressor that is the most abundant oxysterol in human atherosclerotic lesions (Myoishi et al., 2007). Another scenario of athero-relevant ER stress-induced apoptosis is triggered by the combination of a low-dose ER stressor and a “second hit,” each of which are unable to induce apoptosis by themselves (Seimon et al., 2006). For the second hit, activation of combinatorial pattern recognition receptors (PRR) triggers apoptosis through NADPH oxidase and an ROS pathway in combination with suppression of cell survival pathways. Below, we outline the methodology for measuring athero-relevent ER stress-induced apoptosis in primary macrophages, specifically FC-induced macrophage apoptosis.
3.1. Method: Athero-relevant ER stress-induced apoptosis in primary macrophages
3.1.1. Materials
3.1.1.1. Reagents for eliciting peritoneal macrophages by methyl-BSA or concanavalin A
Concanavalin A can be obtained from Sigma (L-7647). Make a 2-mg/ml stock in sterile PBS and store aliquots at –70 °C. If eliciting by mBSA injection: Stock methyl-BSA solution: Dissolve 20-mg methyl-BSA (Sigma A1009) in 10-ml sterile 0.9% NaCl. Methyl-BSA in Complete Freund's adjuvant (CFA DIFCO 231131): Place 2 ml of CFA that has been shaken well before use in a syringe. Fill 2 ml of methyl-BSA solution in another syringe. Connect the 2 syringes to a double-hub microemulsifying needle (20 × 2–7/8 from Popper & Sons, Inc. #7972) and push back and forth ~30 times. The mixture becomes very viscous. Methyl-BSA in Incomplete Freund's adjuvant (IFA: DIFCO 263910). Prepare methyl-BSA-IFA solution same as CFA solution above. Methyl-BSA in PBS: mix 1-ml methyl-BSA solution above with a 9-ml sterile PBS.
3.1.1.2. Media for primary macrophage culture
Tissue culture media, cell culture reagents, and heat-inactivated fetal bovine serum/FBS (GIBCO BRL) can be purchased from invitrogen. Macrophage medium is Dulbecco's modified eagle's medium (DMEM) supplemented with 10% FBS and 20% L-cell conditioned medium. L-cells (L929) are from ATCC. L-cell conditioned medium is collected 24 h after adding fresh DMEM 10% FBS to L-cells at 80% confluency.
3.1.1.3. Preparation of LDL by density ultracentrifugation
Isolate low-density lipoprotein/LDL (d, 1.020–1.063 g/ml) from freshly pooled human plasma (never frozen) by ultracentrifugation. The density of the plasma is assumed to be 1.006; however, the actual density can be measured by weighing 1 ml of solution on a precision scale. Adjust the density to 1.019 with NaBr. The amount to be added is calculated by the following equation: Grams of NaBr is equal to Vi(Df – Di)/1 – (V′Df) where Vi = initial volume in ml, Df = final density, and Di = initial density, and V′ = 0.24 when Df = 1.019, V′ = 0.244 when Df = 1.063, and V′ = 0.246 when Df = 1.09. Spin 45 k in 50.2 Beckman centrifuge rotor or equivalent for ~24 h at 10 °C. The VLDL is now in the top fraction and can be discarded. Harvest the orange layer of each tube and then pool together. Adjust the density to 1.063 with NaBr. To the remaining plasma, adjust the density to 1.063. Spin 45 k in a 50.2 Beckman centrifuge rotor for ~24 h at 10°C. After spin, harvest the top orange LDL layer of each tube and pool together. Adjust the density to 1.09 with NaBr. Spin 35 k in a 50.3 Beckman centrifuge rotor for ~16 h. Harvest the top layers and dialyze against LP buffer (150-mM NaCl, 1-mM EDTA pH 7.4). Sterilize with a 0.45 millipore syringe filter and store under argon gas.
3.1.1.4. Acetylation of LDL
Acetyl-LDL is prepared from a reaction with acetic anhydride and used at a concentration of 50–100 μg/ml in all experiments. In a clean 3-ml stirring vial (Vial—Wheaton, Cat. no. 986257; stirring bar, VWR, Cat. no. 58949-272), place 20–22 mg of LDL. Add an equal volume of saturated sodium acetate. Add LP buffer (formulation below) to a final concentration of 18 mg/ml LDL. Stir briefly at room temperature to mix and take to a 4 °C cold room. Determine the amount of acetic anhydride (Sigma A6404) to add. Volume of acetic anhydride required = 2 × amount in mg of LDL (e.g., 20-mg LDL required 40 μl of acetic anhydride). Aliquot more than the desired total amount of acetic anhydride and keep on ice. Every 10 min add 1/6 of the acetic anhydride in a dropwise manner to the diluted LDL (rinse the pipette tip in the protein solution). After the second or third addition, the solution should become cloudy and a precipitate will form. If the solution becomes too viscous, then add more LP buffer. Stir for an additional hour in the cold room. Place in a dialysis sac (12,000–14,000 MW cutoff) and dialyze against approximately 2 l of LP buffer. Change three times. Collect acetylated LDL and spin in a 1.5-ml microfuge tube for 3 min. Filter the supernatant through a 0.45-μm filter. Determine the protein concentration by a Lowry assay. LP buffer: dissolve 175 g of NaCl (Fisher S271-3) and 2.5 g of EDTA (BP121–500) in 20-l DI water. Adjust pH to 7.4 and chill to 4 °C before use. It is important to extensively dialyze after acetylation using multiple bath changes. Keep stored under argon gas and at 4°C. We typically use AcLDL that is less than 0.01 units of endotoxin as measured by Lonza LAL endotoxin assay.
3.1.1.5. Additional Reagents
The ACAT inhibitor is 58035 (3-[decyldi-methylsilyl]-N-[2-(4-methylphenyl)-1-phenylethyl] propanamide (Ross et al., 1984). Prepare a 10-mg/ml stock in DMSO and use at a concentration of 10 μg/ml in all experiments. CHOP antibody: GADD153 (B-3), Mouse monoclonal IgG1, 200 μl/mg from Santa Cruz Biotechnology, Cat. no. sc-7351 (we have been using Lot # B2603). The Vybrant Annexin V/Propidium Iodide Apoptosis Assay kit can be purchased from molecular probes.
3.1.2. Method
3.1.2.1. Elicitation of primary macrophages
Macrophages are obtained from 8 to 10-week-old female C57BL/6J mice. Harvest macrophages either 3 days after intraperitoneal (i.p.) injection of concanavalin A or 4 days after immunization with methyl-BSA (Cook et al., 2003). For the methyl-BSA elicitation, 2 μg/ml methyl-BSA in 0.9% saline is emulsified in an equal volume of CFA (DIFCO). Mice are immunized intradermally with 100 μl of the emulsion. After 14 days, the immunization protocol is repeated, except use IFA instead of CFA. Seven days later, the mice are injected i.p. with 0.5-ml PBS containing 100-μg methyl-BSA and then the macrophages are harvested 4 days after that by peritoneal lavage. For conA: Add 20 μl of stock (40 μg) to 480-μl sterile PBS and inject i.p.
3.1.2.2. Harvesting and culturing peritoneal macrophages
Sacrifice mice by CO2 asphyxiation and pin mice down, abdomen side-up. Do not stretch mice too much when pinning as tightness may result in peritoneal leakiness during harvest. Sterilize abdomen by spraying with 70% EtOH. Lift skin with forceps and with scissors expose peritoneum without puncture. Inject peritoneum with 5 ml of ice-cold PBS and agitate stomach to mix. Hold skin up with forceps and push the pasteur pipette into the abdomen to aspirate and harvest macrophages. Rinse peritoneum to remove residual cells and filter cells with a 40-μm nylon cell strainer. Spin down cells and plate in medium containing 10% FBS and 20% L-cell conditioned medium. Discard any harvests that have contamination from the intestinal lumen or the blood. Wash away nonadherent cells (nonmacrophages) after 60 min of culture. On the day of plating, the wells should be 70–80% confluent with adherent cells and the cells can be used the next day. If the cells are more sparse, they can be grown in a medium for extra days and then used when the monolayers are more confluent. If the cells are plated too sparsely (<50% confluency), they will not grow well and may die. The medium is replaced every 24 h until the cells reached 80–90% confluency. If the cells are >100% confluent (i.e., overcrowded), biological artifacts can occur, but the experiment can be salvaged by washing the cells soon after harvest to dislodge the overcrowded cells.
3.1.2.3. Inducing ER stress by FC- loading for analysis of CHOP expression and apoptosis
When cells are 80–90% confluent, rinse cells with warm PBS and incubate in a macrophage medium containing 50–100 μg/ml acetyl-LDL plus 10 μg/ml compound 58-035. As a control, incubate cells only with acetyl-LDL to generate foam cells which will be readily visible under the microscope. At desired timepoints (see Fig. 16.1), aspirate media and rinse cells in ice-cold PBS, and again aspirate and add sample lysis buffer (Laemmli buffer) to each well. Scrape cells (the lysate will become viscous), collect the lysate in the Eppendorf tube, and boil samples for 5 min. Freeze at –20 °C or load on a 4–20% gel for immunoblotting of CHOP. For apoptosis, macrophages are assayed for early to mid-stage apoptosis by staining with Alexa 488-conjugated Annexin V. Late-stage apoptosis is determined by costaining with propidium iodide. View cells immediately at room temperature on an inverted fluorescent microscope with appropriate imaging software. Enumerate representative fields (five fields containing ~1000 cells) per condition. Count the number of Annexin V- and PI-positive cells and express as a percent of the total number of cells in at least four separate fields from duplicate wells. An example of FC-induced CHOP expression and annexin positivity is shown in Fig. 16.1.
Figure 16.1.
CHOP expression and annexin staining of FC-loaded primary macrophages. Peritoneal macrophages were elicited and incubated in macrophage medium. At 90% confluence, cells were either loaded with AcLDL or alternatively FC-loaded (AcLDL + 58-035) and (A) immunoblotted for CHOP at indicated times. Blots were stripped and reprobed for actin as a protein loading control. (B) Monolayers were in parallel stained with annexin V (green) and propidium iodide (red) at 16 h to detect mid-stage and late-stage apoptosis. The graph is a quantitation of annexin V-positive, propidium iodide-negative (apoptotic) macrophages in AcLDL cholesteryl ester (CE)-loaded, or FC-loaded macrophages. ★p < 0.05.
3.1.3. Notes/troubleshooting
Because apoptosis is a rapid process, perform analysis immediately after staining. If fixation is required, incubate cells with annexin V prior to fixation and then add 2% paraformaldehyde in PBS. Elevated Annexin V and/or PI staining may reflect that apoptosis continues even after removing the plates from temperature-controlled incubators. Therefore, cells stained with Annexin V should not be kept to linger for prolonged times before measurement. Cells that maintain membrane integrity for longer incubation times may become positive for PI, as this dye can slowly permeate and enter intact cells. Thus, add PI solution just before the analysis. Different batches of AcLDL will vary with respect to their potency of UPR induction and apoptosis. This may be due to variability in the donor that may include a drug or serum component that promotes cell survival.
4. The UPR and Macrophage Death at the Murine Aortic Root
Advanced atherosclerotic lesions consist of a heterogeneous population of cell types, each of which is differentially susceptible to ER stress. Precise analysis of UPR gene expression in plaque therefore requires methodology to selectively capture cell-type specific mRNA. Laser-capture microdissection (LCM) is a method to harvest subpopulations of tissue cells by direct microscopic and immunohistochemical identification (Espina et al., 2006). IHC-navigated cell-type identification is complicated by the fact that immunoidentification requires prolonged antibody incubations that expose tissue sections to the activity of exogenous and endogenous RNases. Fisher and colleagues have cleverly utilized rapid immunostaining protocols with high-affinity antibodies. This approach permits detection and capture from the same section with limited loss in RNA integrity and yield (Trogan and Fisher, 2005). However, in some cases, immunodetection of antigen may not be amenable to rapid staining. This may be due to low antibody affinity or low expression levels of antigen. Herein, we describe an alternative approach that couples immunodetection of macrophage markers with laser-capture of UPR mRNA on separate sections. This method allows users to incorporate prestandardized immunohistochemical staining protocols with LCM. IHC is performed in tandem with LCM on separate but closely cut (6 μm) adjacent sections. As UPR stress signaling is closely linked to cellular and macrophage apoptosis, we subsequently outline a protocol for monitoring macrophage apoptosis in ER-stressed lesions by double immunofluorescence staining.
4.1. Method: Laser-capture microdissection of the UPR in atherosclerosis
4.1.1. Materials and Equipment
4.1.1.1. Materials required for harvesting and preparation of lesional sections from the aortic root
Isoflurane and mouse container for anesthesia. Dissecting platform and dissecting microscope for aortic root harvest. Small animal surgical tools, including fine scissors and forceps. A 1-ml syringe for drawing blood from the heart (prefill with 15 μl 0.5 M EDTA pH 8.0). PBS in a 10-mL syringe with a 24-gauge needle for perfusion of organs. Optimal cutting temperature (OCT) embedding medium. Cryostat with disposable microtome blades (we recommend Leica). Fisher Plus coated microscope slides (Fisher, Cat. no. 12-550-15). All experiments should be performed in compliance with NIH guidelines for the care and use of laboratory animals.
4.1.1.2. Reagents for staining and immunohistochemistry of frozen sections
For rapid staining of slides prior to LCM: The AM1935 LCM Staining Kit from Ambion includes reagents for processing 80 slides. This kit avoids exposure of tissue sections to pure water and potential reactivation of endogenous nucleases. The active ingredient during this staining protocol is Cresyl Violet, which permits nuclear staining and visualization of cell morphology from dehydrated tissue. For IHC: Normal goat serum can be used for blocking and can be purchased from Vector Laboratories, Cat. no. S-1000. For detection of F4/80+ phagocytes: Anti-F480 can be purchased from Serotec (biotin-conjugated). Biotin-Alexa 488 is sold by the Molecular Probes division of Invitrogen or alternatively, biotin-FITC may be used.
4.1.1.3. Equipment and reagents during laser-capture microdissection
LCM work station
This protocol was optimized using the Zeiss PALM (Positioning and Ablation with Laser Microbeams) laser microdissection system equipped with fluorescence imaging. Bring gloves and change frequently. Bring RNA lysis buffer, pipette and tips, and dry ice to store the harvested sample. The LCM work station should be situated in a temperature-controlled and low-humidity work space.
4.1.1.4. Reagents for RNA purification and quantitative RT-PCR
From Ambion, AM1931 RNAqueous-Micro Kit for RNA purification downstream of LCM. RNA integrity can be monitored using the RNA 6000 Nano Lab Chip kit from Agilent per manufacturer's instructions. For reverse transcription, you will need standard oligo-dT (from Invitrogen Cat. no. 18418-012) and dNTP mix (Invitrogen, 10 mM dNTP MIX-PCR grade, Cat. no. 18427-013) and lamda superscript II (RNaseH reverse transcriptase # 18064-014). For qPCR: The forward primer for CHOP is CCA CCA CAC CTG AAA GCA GAA. The reverse primer is AGG TGA AAG GCA GGG ACT CA. If using probe: CTG GTC CAC GTG CAG TCA TGG. For control CypA, the forward primer is GGC CGA TGA CGA GCC C and reverse primer: TGT CTT TGG AAC TTT GTC TGC AA. The probe for CypA is TGT CTT TGG AAC TTT GTC TGC AA. The PCR conditions for Chop cDNA detection are 95 °C for 1 min, 58 °C for 30 s, and 72 °C for 30 s, for 40 cycles.
4.1.2. Method
4.1.2.1. Aortic root processing
Feed B6 Ldlr–/– mice (Jackson, stock # 002207) high-fat, high-cholesterol diet from Harlan Teklad for 12 weeks (diet #88137) starting at 8–10 weeks of age. Anesthetize mice under isoflurane and open thorax. It is important to be relatively expedient during the remaining steps to limit RNA degradation. Remove the blood from the left ventricle and perfuse at physiological pressure with PBS. Remove heart and aorta and cut heart in half, parallel to the aortic valve. Place the upper half on the heart in an embedding mold filled with OCT cryoembedding medium. Position the heart with the cut surface on the bottom of the embedding mold. Fill the embedding mold with OCT and freeze on dry ice. Store specimens at –80 °C. Set cryostat temperature at –25 and –30 °C for knife. Clean the sectioning platform with 70% EtOH. Adjust the tissue block surface parallel to the plane that the blade cuts. Cut the thick sections until you see the valves. Section tissue blocks at 6 μm thickness and mount on positively charged slides. We generally cut 50 sections up the aortic root, of which 6 are used for H&E staining and morphometric and area analysis. The remaining 38 sections are used for LCM (half for IHC staining and the other half for capture).
4.1.2.2. Immunodetection of F4/80+ phagocytes
Allow frozen sections to air-dry for 2 min and then fix in cold acetone for 5 min. Block in 3% goat blocking serum (made in PBS) and then incubate slides with antimouse F4/80 IgG (1:100 dilution) in 3% goat serum. Rinse slides in PBS 3 times for 5 min each. Incubate slides with streptavidin FITC antibody in blocking serum for 1.5 h and subsequently rinse slides in PBS 3 times for 5 min each. Image under a fluorescence microscope to identify the phagocyte regions of interest. Capture images from immunostained sections and identify immunopositive regions of interest for later alignment with the immediately adjacent 6 μm serial section. In parallel analyses, we have confirmed that the majority of directly adjacent sections exhibit equivalent immunostaining for F4/80+ regions of interest when cut at 6 μm. A similar approach has been applied for LCM combined with IHC in brain (Kase et al., 2007). For parallel LCM slides, perform rapid staining as described by Ambion (AM1935 LCM Staining Kit) to provide contrast and for alignment with the identified regions of interest. After staining as per the manufacturer's instructions, the slides are dehydrated in increasing concentrations of EtOH and finally, xylene. We typically process 4 slides at a time by incubating 1 min each in 70% EtOH, followed by 95% EtOH, followed by 100% EtOH, and then two 1-min incubations in fresh xylene. Slides are then air-dried for 10 min and immediately taken to the LCM work station. Slides are stored in a slide box with a desiccant.
4.1.2.3. LCM on PALM
To isolate RNA from frozen sections by LCM, it is important to minimize RNA degradation during sample preparation. Capture is performed on a PALM series LCM instrument with fluorescence imaging capability. After marking the region of interest by immunostaining as described above, quickly identify the parallel ROI in the rapid-stained and dehydrated immediate serial section for capture. For laser-capture, this is best done at 40× magnification. Catapult the specimen into a 500-μl tube that is mounted above the slide. The tube should be filled with a RNA catapult buffer (RNA lysis buffer) and later stored on dry ice prior to buffer crystallization. Do not let the tube sit at room temperature longer than 30 min. Alternatively, one can use the PALM zeiss membrane cap and solubilize RNA at a later time. Select the region of interest by demarcating the area as described in the PALM software program. Settings on the PALM must be optimized. We have used the following settings: UV energy -63, Focus, -35. Perform catapult of tissue into the tube and store the specimen in dry ice. After 12 weeks of western diet and capture of intimal F4/80+ regions from the ~20 rapid-stained sections described above, more than enough RNA should be obtained for an analysis of UPR gene expression. For a thorough analysis and gene expression profiling, we recommend that at least 1 × 106 square microns of surface area be collected. RNA yield and integrity will vary as described below.
4.1.2.4. RNA processing
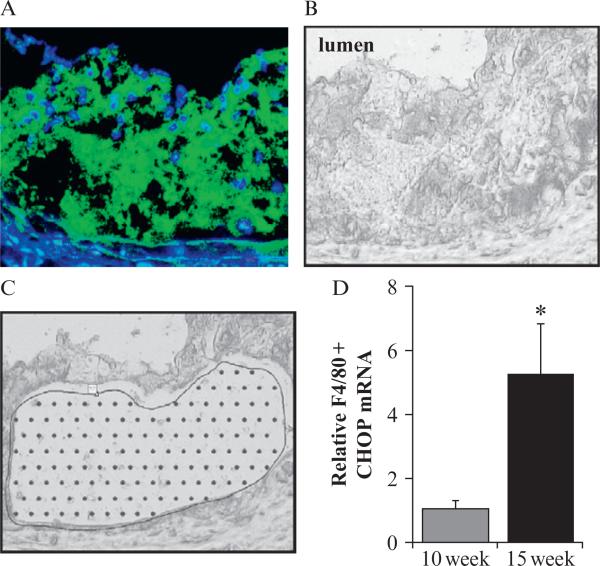
RNA integrity can be examined on a Nano Lab Chip kit from Agilent. Total RNA is isolated using the RNAqueous-Micro Kit from Ambion and reverse-transcribed into cDNA using standard procedures and materials (Invitrogen). Perform quantitative PCR for Chop and control mRNA cyclophilin A (CypA). In some cases, two rounds of linear amplification may be needed prior to qPCR. When designing future primers for PCR of LCM-captured material, you want to limit the size of the amplicon to less than 300 base pairs if possible due to increased RNA degradation. An example of qPCR of CHOP expression from LCM of atherosclerotic lesions, normalized to CypA, is shown in Fig. 16.2.
Figure 16.2.
Laser-capture microdissection of CHOP in F4/80+ phagocytes of atherosclerotic plaque. (A) Aortic root lesions from LDLR–/– mice were stained with F4/80 and (B) a parallel section captured by LCM. Sections were cut at 6-μm thickness. (C) An image of the lesion after capture is shown. (D) CHOP qRT-PCR of targeted F4/80+ areas reveals increased CHOP expression after increased diet duration. ★p < 0.05.
4.1.3. Notes/troubleshooting
Perform the procedure under RNase-free conditions and with RNase decontamination solutions. Change disposable gloves frequently. Note that high humidity or moisture promotes activation of endogenous RNases. Therefore, minimize slide exposure to high room temperature. Keep cryosections at –80 °C to prevent RNA degradation and microdissect cells immediately after staining. Also, change cryostat blade between processing of separate murine samples to avoid sample contamination. Also add RNase inhibitors to staining reagents and minimize the staining time. Expected RNA yield can vary depending on sample integrity. Decreased RNA yield may occur by degradation of RNA before processing of sample for LCM. Therefore, minimize the time required for tissue harvesting and for microdissection at PALM station. A single section should not be exposed to room temperature for longer than 20 min. To test the quality of the RNA before LCM, simply scrape the entire section into a RNA extraction buffer and analyze for 18S/28S RNA. LCM can be performed on paraffin-embedded PFA-fixed sections; however, you will need special extraction techniques for the captured material. This will lower RNA yield and integrity and likely entail a protease treatment to break cross-links formed during fixation.
4.2. Method: Detection of apoptotic macrophages in ER-stressed plaque
4.2.1. Materials
4.2.1.1. Reagents for TUNEL staining
Bovine serum albumin, double-distilled water, DNase solution (~300 U/ml in 50-mM Tris–HCl, pH 7.4, 1 mg/ml BSA) ethanol, GVA (glycerol-vinyl-alcohol) mounting solution, Histopen, Hoechst 33342 (2 μg/ml in PBS), phosphate-buffered saline, protease K (PCR-grade, nuclease-free at 20 μg/ml in 10 mM (Tris–HCl pH 7.4), 0.1% TX-100, Tunel Assay Kit from Roche 12-156-792-910, and xylene.
4.2.1.2. Reagents for macrophage immunohistochemistry after TUNEL staining
Mac-3 is from BD Biosciences, San Diego, California, USA. Rabbit pan antimacrophage antibody (AIA31240) is from Accurate Chemical and Scientific Corporation.
4.2.2. Method
4.2.2.1. Section preparation
This protocol is for paraformaldehyde-fixed, paraffin-embedded lesions, which in our hands have yielded the best results in terms of lesional morphology and preservation of apoptotic DNA. If frozen sections are utilized from slides prepared for LCM above, then we advise first fixing sections with PFA, followed by TUNEL staining below. For paraffin-embedded and PFA-fixed sections: Under chemical hood, dewax samples in xylene 3 times, for 10 min each (alternatively perform 12 sequential dips). Rehydrate samples by dipping in 100% EtOH 2×, followed by 2 dips in 95% EtOH, and finally, 10 dips in clean H2O. Hold the samples in clean H2O. Circle the samples on slides with histopen for downstream TUNEL and IHC.
4.2.2.2. TUNEL staining of atherosclerotic lesions
Add 100 μl of protease K solution to each slide and incubate for 30 min at 37 °C. Then, rinse 2× in PBS and hold in PBS solution. First process positive control: Add a 50-μL DNase solution and leave for 10 min at room temperature. Wash 2× in PBS. Next, dry the area around all samples, including positive and negative control. Prepare Roche TUNEL reaction mix as per the manufacturer's instructions. This is done by mixing 50 μL of vial 1 (TDT enzyme solution) + 450 μL vial 2 (TMR-red dUTP label solution). This is for nine samples and a positive control. TUNEL staining is performed by adding 50 μl of enzyme mixture to each sample and to positive control. Do not add TUNEL reaction mixture to negative control. Incubate all samples plus controls 60 min at 37 °C in a dark, humidified incubator. Rinse the slides in PBS 3× (or 12 dips). Add Hoechst solution to each slide for 5 min. Wash in PBS two times and coverslip using GVA mounting solution. Check for apoptosis per microscopy (excitation 520–560, detection 570–620; may store slides at 4 °C in dark prior).
4.2.2.3. Stain with macrophage-specific antibody
After performing TUNEL labeling, sections are blocked in 1% BSA and 1% normal goat serum and stained with rat antimouse Mac-3 mAb at a dilution of 1:50 dilution overnight at 4 °C h, followed by an incubation of a secondary goat antirat IgG-FITC. Alternatively, macrophages are detected using a rabbit pan antimacrophage antibody (AIA31240). To determine antibody specificity, a preimmune or isotype control should be included in parallel sections.
4.2.2.4. Quantification of TUNEL-positive phagocytes
TUNEL-positive lesions are enumerated as TUNEL-positive nuclei. Apoptotic images are merged using Photoshop CS analysis software (version 8.0; Adobe), and the number of TUNEL- and HOECST-positive cells is counted from each lesion. Apoptotic phagocytes are analyzed by fluorescence microscopy by identification and colocalization of fluorescein and TUNEL-red staining after image overlay in Adobe Photoshop. The percentage of TUNEL-positive phagocytes is determined by counting the number of double-stained phagocytes and total phagocytes per aortic root cross-section. An example of TUNEL-positive macrophage enumeration is shown in Fig. 16.3.
Figure 16.3.
Staining TUNEL-positive macrophages from PFA-fixed paraffin-embedded sections. (A) Representative micrographs show TUNEL-positive signal (red) in nuclei (blue) of aortic root lesions from lesions. Blue is Hoechst nuclear dye. Green is F4/80. (B) Enumeration of TUNEL-positive F4/80+ macrophages from early (4 week Western Diet fed) and advanced (12-week Western+Diet fed) lesions. ★p < 0.05.
4.2.3. Notes/troubleshooting
It is important to avoid nonspecific TUNEL reactivity by optimization of the enzyme concentration or duration of enzyme incubation. Under optimal conditions, TUNEL-positive nuclei in atherosclerotic plaque are typically less than 2% (Kockx and Knaapen, 2000). Optimization of protease K treatment is not only important for accessibility of the TDT enzyme but also for preservation of macrophage epitopes, which can be destroyed with high levels of protease. Finally, apoptosis is a function of lesion maturity. Increased levels of TUNEL and caspase positive cells are found in advanced lesions. Given the heterogeneity of plaque size on the vascular wall, it is important that analyses between two experimental groups are done in lesions at similar stages of development (e.g., equal lesion size). This is critical to avoid measuring differences that manifest simply due to changes in lesion maturation stage.
5. Conclusion and Future Methodological Advances
Future methods will exploit endogenous in vivo ER stress reporters that eliminate processing artifacts that are often the result of sample degradation or nonspecific epitope recognition. For example, a transgenic fluorescent reporter that is induced during ER stress in vivo would eliminate extra secondary detection steps that are required for tissue analysis. In this context, Iwawaki, Miura, and colleagues have created a mouse that is transgenic for a UPR reporter. This consists of the UPR X-box binding protein 1 (XBP-1) fused to venus fluorescent protein and driven by the universal chicken β-actin promoter (Iwawaki et al., 2004). Murine cells that are transgenic for this marker specifically fluoresce after activation of the UPR and therefore can be used to monitor physiological and pathological ER stress in vivo. Thus, information about the level of ER stress during atherosclerosis could be obtained by crossing the ERAI-transgenic mouse with hyperlipidemic mouse models. The combination of in vitro mechanistic studies of ER stress with in vivo models of physiological significance are together key to understanding how ER stress modulates atherosclerotic progression.
ACKNOWLEDGMENTS
These methods were developed in the laboratory of Dr Ira Tabas at Columbia University Medical Center. I thank and credit this manuscript to my training with Dr Tabas. Thanks also to Dr Alain Borczuk for LCM consultation and Manikandan Subramanian for critical reading of the manuscript. Grant Support from NIH NHLBI 1K99HL097021-01.
REFERENCES
- Ball RY, Stowers EC, Burton JH, Cary NR, Skepper JN, Mitchinson MJ. Evidence that the death of macrophage foam cells contributes to the lipid core of atheroma. Atherosclerosis. 1995;114:45–54. doi: 10.1016/0021-9150(94)05463-s. [DOI] [PubMed] [Google Scholar]
- Brown MS, Ho YK, Goldstein JL. The cholesteryl ester cycle in macrophage foam cells. Continual hydrolysis and re-esterification of cytoplasmic cholesteryl esters. J. Biol. Chem. 1980;255:9344–9352. [PubMed] [Google Scholar]
- Cook AD, Braine EL, Hamilton JA. The phenotype of inflammatory macrophages is stimulus dependent: Implications for the nature of the inflammatory response. J. Immunol. 2003;171:4816–4823. doi: 10.4049/jimmunol.171.9.4816. [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Espina V, Wulfkuhle JD, Calvert VS, VanMeter A, Zhou W, Coukos G, Geho DH, Petricoin EF, III, Liotta LA. Laser-capture microdissection. Nat. Protoc. 2006;1:586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- Feng B, Yao PM, Li Y, Devlin CM, Zhang D, Harding HP, Sweeney M, Rong JX, Kuriakose G, Fisher EA, Marks AR, Ron D, et al. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat. Cell Biol. 2003a;5:781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- Feng B, Zhang D, Kuriakose G, Devlin CM, Kockx M, Tabas I. Niemann-Pick C heterozygosity confers resistance to lesional necrosis and macrophage apoptosis in murine atherosclerosis. Proc. Natl. Acad. Sci. USA. 2003b;100:10423–10428. doi: 10.1073/pnas.1732494100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Hossain GS, van Thienen JV, Werstuck GH, Zhou J, Sood SK, Dickhout JG, de Koning AB, Tang D, Wu D, Falk E, Poddar R, Jacobsen DW, et al. TDAG51 is induced by homocysteine, promotes detachment-mediated programmed cell death, and contributes to the development of atherosclerosis in hyperhomocysteinemia. J. Biol. Chem. 2003;278:30317–30327. doi: 10.1074/jbc.M212897200. [DOI] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat. Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- Kase M, Houtani T, Sakuma S, Tsutsumi T, Sugimoto T. Laser microdis-section combined with immunohistochemistry on serial thin tissue sections: A method allowing efficient mRNA analysis. Histochem. Cell Biol. 2007;127:215–219. doi: 10.1007/s00418-006-0241-y. [DOI] [PubMed] [Google Scholar]
- Katz SS, Shipley GG, Small DM. Physical chemistry of the lipids of human atherosclerotic lesions. Demonstration of a lesion intermediate between fatty streaks and advanced plaques. J. Clin. Invest. 1976;58:200–211. doi: 10.1172/JCI108450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kockx MM, Knaapen MW. The role of apoptosis in vascular disease. J. Pathol. 2000;190:267–280. doi: 10.1002/(SICI)1096-9896(200002)190:3<267::AID-PATH523>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Kolodgie FD, Gold HK, Burke AP, Fowler DR, Kruth HS, Weber DK, Farb A, Guerrero LJ, Hayase M, Kutys R, Narula J, Finn AV, et al. Intraplaque hemorrhage and progression of coronary atheroma. N Engl. J. Med. 2003;349:2316–2325. doi: 10.1056/NEJMoa035655. [DOI] [PubMed] [Google Scholar]
- Li G, Mongillo M, Chin KT, Harding H, Ron D, Marks AR, Tabas I. Role of ERO1-alpha-mediated stimulation of inositol 1, 4, 5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J. Cell Biol. 2009;186:783–792. doi: 10.1083/jcb.200904060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WS, Timmins JM, Seimon TA, Sadler A, Kolodgie FD, Virmani R, Tabas I. Signal transducer and activator of transcription-1 is critical for apoptosis in macrophages subjected to endoplasmic reticulum stress in vitro and in advanced atherosclerotic lesions in vivo. Circulation. 2008;117:940–951. doi: 10.1161/CIRCULATIONAHA.107.711275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. The unfolding tale of the unfolded protein response. Cell. 2001;107:827–830. doi: 10.1016/s0092-8674(01)00623-7. [DOI] [PubMed] [Google Scholar]
- Merksamer PI, Trusina A, Papa FR. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell. 2008;135:933–947. doi: 10.1016/j.cell.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myoishi M, Hao H, Minamino T, Watanabe K, Nishihira K, Hatakeyama K, Asada Y, Okada K, Ishibashi-Ueda H, Gabbiani G, Bochaton-Piallat ML, Mochizuki N, et al. Increased endoplasmic reticulum stress in atherosclerotic plaques associated with acute coronary syndrome. Circulation. 2007;116:1226–1233. doi: 10.1161/CIRCULATIONAHA.106.682054. [DOI] [PubMed] [Google Scholar]
- Outinen PA, Sood SK, Pfeifer SI, Pamidi S, Podor TJ, Li J, Weitz JI, Austin RC. Homocysteine-induced endoplasmic reticulum stress and growth arrest leads to specific changes in gene expression in human vascular endothelial cells. Blood. 1999;94:959–967. [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Ross AC, Go KJ, Heider JG, Rothblat GH. Selective inhibition of acyl coenzyme A:cholesterol acyltransferase by compound 58-035. J. Biol. Chem. 1984;259:815–819. [PubMed] [Google Scholar]
- Seimon T, Tabas I. Mechanisms and consequences of macrophage apoptosis in atherosclerosis. J. Lipid Res. 2009;50(Suppl.):S382–S387. doi: 10.1194/jlr.R800032-JLR200. Epub: 2008 Oct 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimon TA, Obstfeld A, Moore KJ, Golenbock DT, Tabas I. Combinatorial pattern recognition receptor signaling alters the balance of life and death in macrophages. Proc. Natl. Acad. Sci. USA. 2006;103:19794–19799. doi: 10.1073/pnas.0609671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I. Consequences and therapeutic implications of macrophage apoptosis in atherosclerosis: The importance of lesion stage and phagocytic efficiency. Arterioscler. Thromb. Vasc. Biol. 2005;25:2255–2264. doi: 10.1161/01.ATV.0000184783.04864.9f. [DOI] [PubMed] [Google Scholar]
- Tabas I. Macrophage apoptosis in atherosclerosis: Consequences on plaque progression and the role of endoplasmic reticulum stress. Antioxid. Redox Signal. 2009;11:2333–2339. doi: 10.1089/ars.2009.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, Williams KJ, Boren J. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: Update and therapeutic implications. Circulation. 2007;116:1832–1844. doi: 10.1161/CIRCULATIONAHA.106.676890. [DOI] [PubMed] [Google Scholar]
- Thorp E, Li G, Seimon TA, Kuriakose G, Ron D, Tabas I. Reduced apoptosis and plaque necrosis in advanced atherosclerotic lesions of Apoe–/– and Ldlr–/– mice lacking CHOP. Cell Metab. 2009;9:474–481. doi: 10.1016/j.cmet.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins JM, Ozcan L, Seimon TA, Li G, Malagelada C, Backs J, Backs T, Bassel-Duby R, Olson EN, Anderson ME, Tabas I. Calcium/calmodulin-dependent protein kinase II links ER stress with Fas and mitochondrial apoptosis pathways. J. Clin. Invest. 2009;119:2925–2941. doi: 10.1172/JCI38857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Trogan E, Fisher EA. Laser capture microdissection for analysis of macrophage gene expression from atherosclerotic lesions. Methods Mol. Biol. 2005;293(221–31):221–231. doi: 10.1385/1-59259-853-6:221. [DOI] [PubMed] [Google Scholar]
- Virmani R, Burke AP, Farb A, Kolodgie FD. Pathology of the vulnerable plaque. J. Am. Coll. Cardiol. 2006;47:C13–C18. doi: 10.1016/j.jacc.2005.10.065. [DOI] [PubMed] [Google Scholar]
- Williams KJ, Tabas I. The response-to-retention hypothesis of early atherogenesis. Arterioscler. Thromb. Vasc. Biol. 1995;15:551–561. doi: 10.1161/01.atv.15.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Yvan-Charvet L, Pagler TA, Seimon TA, Thorp E, Welch CL, Witztum JL, Tabas I, Tall AR. ABCA1 and ABCG1 protect against oxidative stress-induced macrophage apoptosis during efferocytosis. Circ. Res. 2010;106:1861–1869. doi: 10.1161/CIRCRESAHA.110.217281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zampetaki A, Margariti A, Pepe AE, Alam S, Martin D, Xiao Q, Wang W, Jin ZG, Cockerill G, Mori K, Li YS, et al. Sustained activation of XBP1 splicing leads to endothelial apoptosis and atherosclerosis development in response to disturbed flow. Proc. Natl. Acad. Sci. USA. 2009;106:8326–8331. doi: 10.1073/pnas.0903197106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lhotak S, Hilditch BA, Austin RC. Activation of the unfolded protein response occurs at all stages of atherosclerotic lesion development in apolipoprotein E-deficient mice. Circulation. 2005;111:1814–1821. doi: 10.1161/01.CIR.0000160864.31351.C1. [DOI] [PubMed] [Google Scholar]
- Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]