Abstract
Pigmentation, defined as the placement of pigment in skin, hair, and eyes for coloration, is distinctive because the location, amount, and type of pigmentation provides a visual manifestation of genetic heterogeneity in pathways regulating the pigment-producing cells, melanocytes. The scope of this genetic heterogeneity in humans ranges from normal to pathological pigmentation phenotypes. Clinically normal human pigmentation encompasses a variety of skin and hair color as well as with punctate pigmentation such as melanocytic nevi (moles) or ephelides (freckles), while clinically abnormal human pigmentation exhibits markedly reduced or increased pigment levels, known as hypopigmentation and hyperpigmentation, respectively. Elucidation of the molecular genetics underlying pigmentation has revealed genes important for melanocyte development and function. Furthermore, many pigmentation disorders show additional defects in cells other than melanocytes, and identification of the genetic insults in these disorders has revealed pleiotropic genes, where a single gene is required for various functions, often in different cell types. Thus unravelling the genetics of easily visualized pigmentation disorders has identified molecular similarities between melanocytes and less visible cell types/tissues, revealing a common cellular origin and/or common genetic regulatory pathways. Herein we discuss notable human pigmentation disorders and their associated genetic alterations, focusing on the fact that the developmental genetics of pigmentation abnormalities is instructive for understanding normal pathways governing development and function of melanocytes.
Introduction
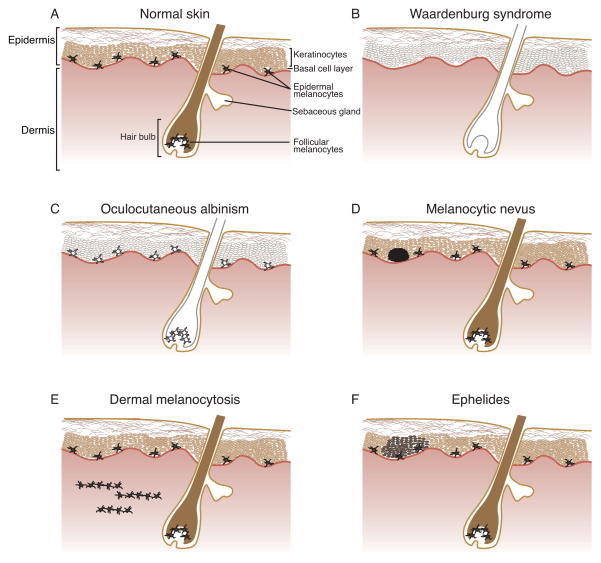
Pigmentation has long fascinated the human race, with developmental melanocyte abnormalities reported by ancient Greeks, Romans, and Egyptians.1 The melanins responsible for mammalian pigmentation include reddish-hued pheomelanin and brown/black-colored eumelanin, and these melanins are produced by melanocytes through a process known as melanogenesis. Melanin provides coloration of hair and skin as well as photoprotection from ultraviolet radiation (UVR), and concordant with these functions, most human melanocytes are located in the epidermis or hair follicles of the skin. The epidermal and follicular melanocyte populations have distinctive anatomical distribution and proliferation. Epidermal melanocytes reside within the basal layer of the epidermis at the junction of the epidermis and dermis; these melanocytes extend dendritic processes that interact with surrounding keratinocytes and function in pigment deposition and cell signaling (Fig. 1A). The proliferation/regeneration of epidermal melanocytes has not yet been fully characterized. Follicular melanocytes reside within the hair follicle, and here their proliferation from stem cells and transfer of melanin into hair are synchronized with the hair cycle. Additionally, melanocytes are located within the uvea of the eye, cochlea and vestibular region of the ear, leptomeninges of the brain, and ventricular septum and valves of the heart.
Figure 1.
Diagrammatic representation of congenital human cutaneous pigmentation abnormalities. A) Normal skin shows individual, dendritic melanocytes located at the basal cell layer of the epidermis and in the bulb of the hair follicle. Epidermal melanocytes provide pigment to surrounding interfollicular keratinocytes, and follicular melanocytes provide pigment to hair shaft keratinocytes. B) In Waardenburg syndrome, developmental anomalies in genes crucial for pigment cell development cause regionalized absence of melanocytes, resulting in areas of hypopigmented skin and hair. C) In oculocutaneous albinism, melanocytes are retained, but they contain a genetic defect that prevents them from producing normal levels of melanin. Illustrated is OCA1A, where complete absence of pigment results from loss of function of the melanogenic enzyme TYR. Other forms of oculocutaneous albinism typically show some residual pigmentation. D) Melanocytic nevi show benign overgrowth of epidermal melanocytes that have lost their dendritic form and are organized in defined nests. E) In dermal melanocytosis, melanocytes retaining dendritic morphology are found within the dermis, in a scattered array often parallel to the skin’s surface. Their deeper, dermal location gives them a characteristic blue tint, with a superficial dermal location typical for nevus of Ito or nevus of Ota, and a deeper dermal location for Mongolian spots. F) In ephelides, normal numbers of melanocytes produce localized patches of increased melanin in surrounding keratinocytes. Formation of ephelides is directly correlated with UV irradiation levels.
Melanocytes develop from the neural crest (NC), a multipotent vertebrate cell population that emerges along the dorsal surface of the neural folds, undergoes an epithelial to mesenchymal transition, and becomes highly migratory, differentiating into a wide variety of cell types throughout the body.2 In addition to melanocytes, NC cells give rise to neurons and glia of the peripheral nervous system (PNS), endocrine cells, and cranio-facial structures. NC cells of the trunk migrate along two primary routes: ventromedial, between the neural tube and somites, and dorsolateral, between the epidermis and somites. Embryonic melanocyte precursors, herein termed melanoblasts, are the sole progeny of dorsolaterally migrating NC cells, while ventromedially migrating NC cells give rise to melanoblasts as well as Schwann cells, adrenomedullary cells, and sensory and sympathetic neurons.3 During later stages of embryonic development, melanoblasts migrate through the dermis into the overlying epidermis, enter into developing hair follicles, undergo extensive proliferation and initiate melanin production.4,5
The wide variation in basal human skin color and tanning response results from differences in amount and/or cellular distribution of melanin rather than differing melanocyte numbers.6 Interestingly, many of the genes implicated in human hypo- and hyperpigmentation disorders also have allelic variants associated with normal phenotypic variation of basal skin color, including tyrosinase (TYR), tyrosinase-related protein 1 (TYRP1), solute carrier family 45, member 2 (SLC45A2), solute carrier family 24, member 5 (SLC24A5), solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 (SLC24A4), oculocutaneous albinism II (OCA2), agouti signaling protein (ASIP), KIT ligand (KITLG), interferon regulatory factor 4 (IRF4), two pore segment channel 2 (TPCN2), and melanocortin 1 receptor (MC1R).6 MC1R is the best characterized of these genes, and it encodes a cell surface receptor that regulates eumelanin/pheomelanin production by melanocytes. The extensive allelic variance of MC1R directly correlates with the presence of various red hair and fair skin phenotypes, and also includes a MC1R null allele, demonstrating that absence of function for this important signaling receptor still falls within the normal clinical spectrum.7,8
The inherent visibility of pigmentation anomalies has allowed extensive characterization of the causative gene mutations as well as the normal functions of the encoded proteins (Table 1). For example, proteins that catalyze/regulate melanin production are fundamental to all pigment cells, and thus mutations in genes encoding these proteins (TYR, TYRP1, OCA2, SLC45A2) globally affect skin, hair, and eyes. In another example, mutation of genes required for melanocyte development—including paired box 3 (PAX3), microphthalmia-associated transcription factor (MITF), SRY (sex determining region Y)-box 10 (SOX10), endothelin receptor type B (EDNRB), and endothelin 3 (EDN3)—result in patchy hypopigmentation along with deafness, caused by absence of both cutaneous and otic melanocytes necessary for proper ear development and function. Of note, some of these mutations also affect the retinal pigmented epithelium of the eye, a structure composed of melanin-producing cells that are of neuroectodermal rather than NC developmental origin yet exhibit overlapping gene expression with cutaneous melanocytes.9 In a third example, mutations in genes that regulate development of multiple NC cell types (SOX10, EDNRB/EDN3) cause coincident developmental defects in melanocytes and neurons/glia of the enteric nervous system. In a fourth example, mutation of genes associated with Hermansky-Pudlak syndrome or Griscelli syndrome (see Table 1) alter subcellular trafficking of melanosomes (melanin-containing organelles) within melanocytes, as well as storage granules within hematopoietic cells, showing that these two cell types with different developmental origins share genetic pathways that regulate similar subcellular processes.
Table 1.
Congenital pigmentation disorders with cloned and characterized genetic components.1
| Disease | Gene | Encoded Protein Function |
|---|---|---|
| Piebaldism | KIT | Receptor tyrosine kinase; ligand is KITLG |
| SNAI2 | Transcription factor | |
| Waardenburg syndrome 1 | PAX3 | Transcription factor |
| Waardenburg syndrome 2 | MITF | Transcription factor |
| SOX10 | Transcription factor | |
| SNAI2 | Transcription factor | |
| Waardenburg syndrome 3 | PAX3 | Transcription factor |
| Waardenburg syndrome 4 | SOX10 | Transcription factor |
| EDNRB | Transmembrane receptor; ligand is EDN3 | |
| EDN3 | Secreted growth factor | |
| Peripheral demyelinating neuropathy, central dysmyelinating leukodystrophy, Waardenburg syndrome, and hirschsprung disease | SOX10 | Transcription factor |
| Tietz syndrome | MITF | Transcription factor |
| Yemenite deaf-blind hypopigmentation syndrome | SOX10 | Transcription factor |
| Albinism, black lock, cell migration disorder of the neurocytes of the gut, and deafness syndrome | EDNRB | Transmembrane receptor; ligand is EDN3 |
| Oculocutaneous albinism 1 | TYR | Melanogenic, rate-limiting enzyme |
| Oculocutaneous albinism 2 | OCA2 | Melanosome membrane protein |
| Oculocutaneous albinism 3 | TYRP1 | Melanogenic enzyme |
| Oculocutaneous albinism 4 | SLC45A2 | Solute transporter |
| Hermansky-Pudlak syndrome | HPS1 | Member of BLOC-3 complex; regulates lysosome-related organelle movement |
| AP3B1 | Member of AP3 complex; regulates protein sorting | |
| HPS3 | Member of BLOC-2 complex; regulates lysosome-related organelle movement | |
| HPS4 | Member of BLOC-3 complex; regulates lysosome-related organelle movement | |
| HPS5 | Member of BLOC-2 complex; regulates lysosome-related organelle movement | |
| HPS6 | Member of BLOC-2 complex; regulates lysosome-related organelle movement | |
| DTNBP1 | Member of BLOC-1 complex; regulates lysosome-related organelle movement | |
| BLOC1S3 | Member of BLOC-1 complex; regulates lysosome-related organelle movement | |
| PLDN | Member of BLOC-1 complex; regulates lysosome-related organelle movement | |
| Chediak-Higashi syndrome | LYST | Function unknown; may regulate lysosome-related organelle size and trafficking |
| Griscelli syndrome | MYO5A | Myosin motor regulating melanosome transport |
| RAB27A | RAS-associated protein that interacts with MLPH to regulate melanosome transport | |
| MLPH | Interacts with MYO5A to regulate melanosome transport | |
| LEOPARD syndrome | PTPN11 | MAPK signal transduction pathway |
| RAF1 | MAPK signal transduction pathway | |
| BRAF | MAPK signal transduction pathway | |
| Noonan syndrome | PTPN11 | Cytoplasmic protein tyrosine phosphatase; modulates RAS activity |
| RAF1 | Serine threonine kinase; downstream effector of RAS | |
| BRAF | Serine threonine kinase; downstream effector of RAS | |
| SHOC2 | Leucine-rich protein; modulates RAS activity | |
| KRAS | Monomeric GTPase; modulates RAS activity | |
| SOS1 | Guanine nucleotide exchange factor; modulates RAS activity | |
| NRAS | Monomeric GTPase; modulates RAS activity | |
| Cardio-facio-cutaneous syndrome | KRAS | Monomeric GTPase; modulates RAS activity |
| BRAF | Serine threonine kinase; downstream effector of RAS | |
| MAP2K1 | Dual specificity kinase; acts downstream of RAS as RAF effector | |
| MAP2K2 | Dual specificity kinase; acts downstream of RAS as RAF effector | |
| CBL-mutation associated syndrome | CBL | E3 ubiquitin ligase; acts downstream of receptor tyrosine kinases |
| Legius syndrome | SPRED1 | Sprouty domain protein; tyrosine kinase substrate; modulates RAS activity |
| Dyschromatosis symmetrica hereditaria | ADAR | RNA specific adenosine deaminase; regulates RNA editing |
| GIST with dysplastic nevi and lentigines | KIT | Receptor tyrosine kinase; ligand is KITLG |
| Peutz-Jeghers syndrome | STK11 | Serine threonine kinase |
| Carney complex | PRKAR1A | Protein kinase regulatory subunit |
| Neurofibromatosis | NF1 | RAS GTPase, modulates RAS activity |
| Neurofibromatosis-Noonan syndrome | NF1 | RAS GTPase, modulates RAS activity |
| Familial progressive hyperpigmentation 2 | KITLG | Growth factor; ligand for KIT |
| Familial progressive hyper- and hypopigmentation | KITLG | Growth factor; ligand for KIT |
| Fanconi anemia | Heterogeneous see OMIM #227650 | Associated with mutation of at least 15 loci, which encode proteins that regulate various DNA repair processes |
This table is not an exhaustive list of all known congenital pigmentation disorders. For extensive descriptions of published human pigmentation disorders, see Further Reading/Resources.
Human pigmentation disorders can be classified by phenotype, including hypo- or hyperpigmentation, congenital or acquired presentation, effects on melanocyte number (hypo- or hyperplasia), or effects on melanocyte function. However, defining the underlying genetic anomalies explains the etiology of human pigmentation disorders, thus allowing correlation of developmental genetics with clinical phenotypes. The combined description of pigmentation disorders by both phenotype and genetic alteration sheds new light on the molecular pathways regulating human pigmentation. In this review, the current knowledge of human pigmentation abnormalities and their associated genetic defects are described.
CONGENITAL PIGMENTATION ABNORMALITIES: HYPOPIGMENTATION FROM MELANOCYTE HYPOPLASIA
When a genetic insult causes the loss of melanocytes during embryonic/fetal development, congenital disorders of hypopigmentation result. These rare disorders display regions of hypopigmentation caused by a localized reduction in melanocytes (Fig. 1B), often accompanied by deafness caused by loss of cochlear melanocytes. In humans, most of these hypopigmentation disorders display dominant inheritance.
One of the best-characterized congenital hypopigmentation syndromes is Piebaldism, associated with loss of function/deletion mutations in the tyrosine kinase receptor KIT or in the transcription factor SNAI2. In Piebaldism, sharply demarcated regions of hypopigmented skin and hair occur, most frequently on the head, ventral trunk, and extremities, with a white forelock of hair being a notable phenotype. Often, small macules of hyperpigmentation occur within or at the borders of the hypopigmented regions.
The disorders collectively described as Waardenburg Syndrome (WS) display cutaneous hypopigmentation and ocular pigmentation anomalies paired with congenital hearing loss, as well as additional phenotypes in various cell types. These coincident phenotypes reflect a common NC origin for the affected cell types, along with pleiotropy of the mutated genes. Mutation of PAX3 in WS1 and WS3 reveals the pleiotropic effects of PAX3 protein on melanocyte and craniofacial/limb development, as these WS forms display the additional phenotypes of dystopia canthorum and upper limb abnormalities. Similarly, SOX10, EDNRB, and EDN3 regulate both melanocyte and enteric ganglia development, and mutations of the genes encoding each of these proteins can cause WS4, which displays the additional phenotype of abnormal enteric ganglia formation known as Hirschsprung disease.
Identification of the heterogeneous genetic defects causing WS has resulted in many examples where distinctive clinical phenotypes of congenital hypopigmentation can be related to each other on a molecular level. For example, Tietz syndrome is associated with mutation of MITF, and is thus allelic with WS2 that is caused by mutation of MITF, even though Tietz syndrome displays the distinctive phenotypes of extensive hypopigmentation and gradual accrual of pigmentation later in life. A mild form of Yemenite deaf-blind hypopigmentation syndrome (YDBS), characterized by deafness with regional hypo- and hyperpigmentation, is associated with a missense mutation in SOX10,10,11 thus demonstrating that mild YDBS and SOX10-associated WS2 are allelic disorders. The syndrome Peripheral demyelinating neuropathy, Central dysmyelinating leukodystrophy, Waardenburg syndrome, and Hirschsprung disease (PCWH), which shows neurological phenotypes combined with WS4 phenotypes, also results from deletion/mutation of SOX10. Similarly, the descriptively named Albinism, Black lock, Cell migration disorder of the neurocytes of the gut, and Deafness syndrome (ABCDS) is caused by homozygosity for an EDNRB mutation, thus showing allelism between ABCDS and EDNRB-associated WS4.
These variable congenital hypopigmentation phenotypes arising from perturbation of the same gene may reflect mutation of different functional domains in the encoded protein. For example, mutations that alter the nuclear localization signal of MITF (missense or in-frame deletion) are associated with the more severe Tietz syndrome, and are suggested to confer a dominant negative phenotype whereby mutant MITF sequesters normal protein in the cytoplasm; conversely, MITF null alleles are proposed to lead to haploinsufficiency, thus resulting in the milder phenotypes of WS2.12,13 Similarly, mutations affecting SOX10 C-terminal domains have been proposed to allow escape from nonsense-mediated decay, creating mutant proteins with dominant negative effects that cause the severe PCWH phenotypes, while N-terminal domain mutations are proposed to result in null alleles associated with the milder WS2 and WS4 phenotypes.14
However, functional domains defined by protein secondary structure do not fully correlate with the phenotypic variability associated with mutation of a single pigmentation gene. Exceptions exist to the correlations of SOX10 mutation location with PCWH or WS phenotypes, and recent work on SOX10 missense mutations suggests SOX10 subcellular localization may be relevant to mutation effects.15,16. Also, WS-associated PAX3 mutations can be categorized into two groups based upon their effect on nuclear localization and mobility, yet these categories show no clear correlation with protein functional domains or DNA binding activity.17
CONGENITAL PIGMENTATION ABNORMALITIES: HYPOPIGMENTATION FROM ALTERED MELANOCYTE FUNCTION
Mutation in a gene that regulates melanin production or distribution does not change melanocyte number, but instead causes reduction and/or altered distribution of melanin pigment (Fig. 1C). These mutations lead to rare, recessively inherited hypopigmentation disorders where normal numbers of melanocytes are retained, but show severe defects in the location or amount of melanin. Some of these disorders also show defects attributable to altered distribution/trafficking of cytoplasmic organelles other than melanosomes, thus revealing pleiotropy of the mutated genes.
The best known of these types of hypopigmentation disorders are oculocutaneous albinisms (OCAs), in which defective melanin production causes severe reduction to complete absence of all skin, hair, and eye pigmentation. Additional ocular phenotypes may include nystagmus, reduced visual acuity, and misrouting of optic nerves. The various OCA forms are classified based upon their associated genetic mutations. OCA1A and OCA1B are caused by mutation of the melanogenic enzyme TYR, with the A form displaying a complete lack of TYR protein function, and the B form showing reduced function. The essential role of TYR in melanin synthesis explains why OCA1 displays the most severe phenotypes of all OCAs. OCA2 is caused by mutation/deletion of the OCA2 gene, encoding a membrane protein proposed to transport TYR, with patients retaining some brown pigment. OCA3 is caused by mutation of the gene encoding the melanogenic enzyme TYRP1, with patients showing reddish hair and freckling. OCA4 is caused by mutation in the gene encoding the transporter protein SLC45A2, with patients retaining modest amounts of brown pigment, similar to OCA2.
Hermansky-Pudlak syndrome (HPS) is a genetically heterogeneous disorder (Table 1), in which cutaneous hypopigmentation results from abnormal melanosome formation, movement, or transfer to keratinocytes. Additionally, HPS patients exhibit phenotypes caused by defects in cytoplasmic organelles in other cell types, including platelet-dense granules, leading to bleeding defects, and lysosomes, leading to phenotypes of pulmonary fibrosis or colitis.18,19 HPS-associated loci encode components of the BLOC-1, -2, or -3 protein complexes, which regulate organelle movement, or components of the AP1 protein complex, which regulates protein sorting.
A disorder with phenotypic similarity to HPS is Chediak-Higashi syndrome (CHS), where patients exhibit hypopigmentation of hair and eyes, bleeding disorders, susceptibility to infection and, in later disease stages, distinctive lymphoma and neurological defects. CHS is caused by mutation of lysosomal trafficking regulator (LYST), and shows subcellular phenotypes of enlarged organelles, including melanosomes (which show clumping in hairs), and giant granules in eosinophils, basophils, and monocytes. A decrease in platelet dense bodies is also present. While the cellular function of LYST protein is unclear, these mutant phenotypes suggest LYST regulates organelle size and/or organelle trafficking.18
Another disorder involving hypopigmentation as a result of abnormal melanosome trafficking is Griscelli syndrome (GS). Patients with GS manifest cutaneous hypopigmentation, neurological defects, immunological defects, and overactive histiocytes and lymphocytes. GS has been associated with mutation of 3 genes, myosin VA (MYO5A), RAB27A, member RAS oncogene family (RAB27A), and melanophilin (MLPH), whose protein products direct transport of melanosomes from microtubules to actin at the cell periphery in melanocytes, and also regulate organelle trafficking in immune cells.
CONGENITAL HYPERPIGMENTATION ABNORMALITIES: MELANOCYTIC HYPERPIGMENTATION
Normal human epidermis exhibits an orderly 3-dimensional cellular arrangement (Fig. 1A), in which individual melanocytes are widely spaced apart from one another along the epidermal basal layer. In this arrangement, melanocytes are able to communicate with nearby keratinocytes and likely other cell types as well, including Langerhans cells, fibroblasts, vascular cells, and nerve endings.20,21 This structural and functional grouping of a single melanocyte with a large number of surrounding keratinocytes has long been appreciated, and is called the epidermal melanin unit (EMU), with the average EMU melanocyte to nucleated keratinocyte ratio maintained at 1:36, regardless of regional variations in melanocyte density.22 Alteration of the signaling that directs the precise architecture of the EMU can cause regional, benign overgrowth of melanocytes (known as melanocytic hyperpigmentation), resulting in sharply demarcated spots of pigment. The presence of moderate numbers of these hyperpigmented spots at birth or in early childhood is very common, thus obscuring somewhat the boundary between normal and pathological human hyperpigmentation. Nevertheless, abnormal congenital hyperpigmentation is typically defined as presentation of significantly greater amounts of hyperpigmentation than that of the general population at or soon after birth.23,24 Below, we first introduce various forms of common melanocytic lesions, and then describe notable examples of congenital hyperpigmentation that illustrate phenotype-genotype correlations.
Common forms of melanocytic hyperpigmentation
The most common melanocytic hyperpigmented lesions are: lentigines, dark brown to black, flat hyperpigmented regions 2–20mm in diameter, caused by expansion of the epidermis into elongated rete ridges along with a corresponding increase in melanocytes along the dermal-epidermal border; nevocellular or melanocytic nevi, pigmented spots consisting of benign overgrowth of melanocytes that have lost their dendritic form and are organized in defined nests (Fig. 1D); and dysplastic (atypical, Clark) nevi, pigmented spots distinct from melanocytic nevi by their larger size, irregular border, and variable pigmentation.25 Clinical distinction of these lesions from other melanocytic proliferations, including melanoma, the cancerous growth of melanocytes, is difficult and often requires application of specialized technology or histological analysis.24,26,27 The molecular genetics regulating these various benign melanocyte overgrowths are not well understood, however genetic factors clearly influence their morphology, frequency, and location.28–32 Furthermore, numerous melanocytic nevi correlates with increased melanoma risk,33,34 suggesting the genetic alterations that lead to benign melanocytic hyperplasia relate to melanomagenesis.
Congenital forms of melanocytic hyperpigmentation
Congenital melanocytic nevi (CMN) are distinguished from melanocytic nevi by their presence at or soon after birth and their deep histological location, including involvement of vascular and other neighboring areas.35 CMN are categorized by size, ranging from small (<1cM) to extremely large (>20cM), and their size appears to directly correlate with melanoma risk.36 Genetic factors that influence CMN are unknown, although heritability has been suggested for large CMN, also known as giant pigmented hairy nevus (GPHN).
Lentigo simplex (LS) is characterized by extensive lentigines in skin, nails, or mucous membranes in newborns or early childhood. The genetics of LS are not well understood, however it appears genetically distinct from melanocytic nevi or solar lentigo (see below).37 Causative gene mutations for LS have been identified in phenotypically complex disorders, where LS occurs in combination with other abnormalities of NC-derived tissues, such as LEOPARD syndrome (multiple lentigines, electrocardiographic conduction abnormalities, ocular hypertelorism, pulmonic stenosis, abnormal genitalia, retardation of growth, and sensorineural deafness). Mutations in the RAS/MAPK signal transduction pathway genes protein tyrosine phosphatase, non-receptor type 11 (PTPN11), v-raf-1 murine leukemia viral oncogene homolog 1 (RAF1), and v-raf murine sarcoma viral oncogene homolog B1 (BRAF) are associated with LEOPARD syndrome; however, mutations in these three genes are also associated with Noonan syndrome, a phenotypically variable disorder characterized primarily by craniofacial and cardiac anomalies with infrequent hyperpigmentation. 38–41 In Leopard syndrome, LS is often preceded by café-au-lait spots, light brown, ovoid, hyperpigmented spots <1cm–30 cm in diameter that typically retain normal melanocyte number rather than being melanocytic; in Noonan syndrome, café-au-lait spots are the most common form of hyperpigmentation. The molecular genetics underlying these phenotypic differences in melanocytes (and other tissues) remains unclear, however recent biochemical studies have begun to associate mutations in distinct protein domains of PTPN11 with various phenotypes.42–44 In another disorder, dyschromatosis symmetrica hereditaria, patchy hyper- and hypopigmentation (including lentigines) on dorsal extremities is caused by mutation of the adenosine deaminase, RNA specific (ADAR) gene; how mutation of this ubiquitous protein, which contributes to RNA editing, leads to coincident yet opposite pigmentation anomalies remains to be determined.
Melanocytic hyperpigmentation that occurs in the dermis rather than the epidermis is known as dermal melanocytosis (Fig. 1E). These lesions are distinctive from other melanocytic growths because their melanocytes typically retain their dendritic morphology, and their deeper, dermal location causes differential light scattering and absorption by melanin pigments, generating a characteristic blue-gray tint. 45 Dermal melanocytoses that can be considered congenital include nevus of Ito (localized on the neck/shoulders), nevus of Ota (localized on the face), and Mongolian spots. Mongolian spots are present at birth on the dorsal surface, most commonly in sacral/gluteal areas,46 then regress in childhood, suggesting they may arise from a transient overgrowth and/or retention of melanocytes in the dermis during development. Although little is known about the molecular genetics of these congenital dermal melanocytoses, potential genetic influences on Mongolian spots are suggested by their familial inheritance and their prevalence in populations of Asian or African descent. Additionally, the cranial nerve locations of nevus of Ota and nevus of Ito suggest they could originate from nerve-associated, medioventral pathway melanoblasts.3
Melanocytic hyperpigmentation is reminiscent of tumorigenic cell growth, and several disorders exist that display melanocyte overgrowth along with tumors of other cell types. These autosomal dominant disorders begin to show abnormal phenotypes during juvenile development, and are all caused by mutation of signaling pathway genes. They include: gastrointestinal stromal tumors paired with dysplastic nevi and lentigines (GIST), associated with activating mutations in the gene encoding the receptor tyrosine kinase KIT;47,48 Peutz-Jeghers syndrome, where hyperpigmented spots on lips, oral mucosa, and fingers occur along with gastrointestinal polyps and additional tumor susceptibility, associated with mutation of the gene encoding the serine/threonine kinase STK11; Carney Complex, exhibiting mucosal membrane lentigines (and additional pigmentatary abnormalities) along with cardiac, endocrine, cutaneous, and neural myxomatous tumors, caused by mutation in the protein kinase A regulatory subunit-1-alpha gene (PRKAR1A);49 and neurofibromatosis (NF), where benign and malignant tumor growth arises in conjunction with axial freckling and melanocytic café-au-lait spots, most commonly associated with mutation of the gene neurofibromin (NF1), whose protein product regulates RAS signaling. NF1 mutations are also associated with neurofibromatosis-Noonan syndrome, a syndrome exhibiting a unique combination of NF phenotypes along with defects of other NC-derived tissues (craniofacial and cardiac), short stature and motor delay.
CONGENITAL HYPERPIGMENTATION ABNORMALITIES: MELANOTIC HYPERPIGMENTATION
Congenital hyperpigmentation abnormalities that increase melanin levels without increasing melanocyte number (known as melanotic hyperpigmentation) reflect an alteration in the signalling cascades that regulate melanogenesis. These can present with pigmentation phenotypes alone, without affecting any other organ system. Of note, common café-au-lait spots (occurring in moderate numbers in approximately 2% of Caucasians) are lesions of melanotic hyperpigmentation that are not considered pathological25 and whose histology differs from that of melanocytic café-au-lait spots of NF.50,51 One rare disorder of melanotic hyperpigmentation is familial progressive hyperpigmentation (FPH2), characterized by general hyperpigmentation and associated with gain-of-function mutations in KITLG, whose protein product functions as a signaling molecule that regulates melanin production through the receptor KIT. The closely related disorder familial progressive hyper- and hypopigmentation is also associated with mutation of KITLG.52 Another example is Legius syndrome, which shows phenotypes of café-au-lait molecules, occasional freckling, and craniofacial and cognitive defects. Legius syndrome is associated with mutation of sprouty-related, EVH1 domain containing 1 (SPRED1), which encodes a regulator of RAS signaling. Therefore, the RAS-MAPK pathway links the phenotypically distinct hyperpigmentary defects of Legius syndrome, NF1, LEOPARD syndrome, and Noonan syndrome. These four syndromes are included in the RASopathies, a varied group of human disorders that all arise from dysregulation of RAS-MAPK signaling. Additional RASopathies with occasional pigmentary anomalies include CBL-mutation associated syndrome and cardio-facio-cutaneous syndrome, caused by mutation of KRAS, BRAF, MAP2K1 or MAP2K2.53–55
Many other congenital disorders exhibit either localized or general melanotic hyperpigmentation that arises in a reticulated pattern. These are typically seen along with other systemic disorders, and are caused by defects in ubiquitous proteins and/or basal cellular processes.56 The presence of hyperpigmentation resulting from altered basal cell processes suggests melanocytes are a cell type with great sensitivity to such perturbations. A notable example is Fanconi Anemia, a genetically heterogeneous disorder affecting DNA repair, characterized by varied phenotypes affecting all organ systems.
ACQUIRED PIGMENTATION DISORDERS
Acquired disorders are clinically defined as those occurring after fetal stages of development. Because this review is focused on developmental genetics, examples of acquired hypo- and hyperpigmentation are briefly summarized, and emphasis is placed on heritable genetic influences on these disorders.
Acquired hypopigmentation
The hypopigmentation disorder vitiligo shows regional, progressive epidermal melanocyte loss that results in severe skin depigmentation. Vitiligo is relatively common, with an estimated frequency of 1–2%, and is categorized into two clinically distinct forms: the more common symmetrical, general form (GV) and the rare, asymmetrical, segmental form. The molecular genetics of vitiligo involves multiple loci and pathways, with recent genome-wide association studies confirming the long-held hypothesis that autoimmune responses directed towards melanocytes are involved in GV progression.57 Other suggested mechanisms contributing to vitiligo include mosaicism, sensory neuron-directed abnormalities, or region-specific, microvascular homing of cytotoxic T-cells to the skin.58–60
Acquired Benign Hyperpigmentation
Exposure to UVR has a strong influence on the appearance of acquired forms of benign hyperpigmentation, and is associated with formation of the common hyperpigmented lesions ephelides, melanocytic nevi, and solar lentigines (SLs). Ephelides, which are localized patches of melanotic hyperpigmentation (Fig. 1F), are also genetically influenced, as their frequency is correlated with allelic variants of MC1R.61 Melanocytic nevi also appear to be influenced both by genetics (described in Melanocytic Hyperpigmentation section) and childhood exposure to UVR.62–65 In contrast, SLs are defined by their appearance on sun-exposed skin, and thus clearly affected by UVR, but genetic influences on their formation are not yet known. Candidate pathways in which allelic variance could regulate SL susceptibility are: the KITL pathway, as SL formation is associated with upregulated KITL, and the fibroblast growth factor receptor 3 (FGFR3) and phosphoinositide-3-kinase, catalytic, alpha polypeptide (PIK3CA) pathways, as SLs can harbor FGFR3 or PIK3CA mutations.37,66 In addition, rarer types of acquired, benign hyperpigmentation include: dysplastic nevus syndrome, characterized by extensive numbers of dysplastic nevi; Spitz nevi, small, symmetric, juvenile nevi that arise singly, often on the face or lower extremities, which clinically resemble melanoma, but are nonmalignant and have a distinctive mutation profile; and acquired blue nevi, a heterogenous group of blue-black dermal lesions,45 which display gene mutation profiles distinct from those of cutaneous melanocytic nevi and Spitz nevi, including the association of some forms with mutations in guanine nucleotide binding protein (G protein), q polypeptide (GNAQ) and guanine nucleotide binding protein (G protein), alpha 11 (GNA11).67,68
Hormonal and inflammatory influences
Acquired hypo- or hyperpigmentation can result from a wide variety of systemic alterations. Changes in hormones can be quite influential on pigmentation, including pregnancy-related estrogen increases (causing melasma and linea nigra), hyperthyroidism, overproduction of adrenocorticotropic hormone (seen in Cushing’s disease), and adrenal gland insufficiency (seen in Addison disease).69 Other systemic alterations that can trigger hypo- or hyperpigmentation are post-inflammatory responses to various external insults, such as infection, drug response, or contact dermatitis.70 A genetic predisposition to development of these anomalies is likely to be complex and/or multifactorial.
Animal models of human cutaneous melanocyte development and function.
Mouse, chick, frog, and zebrafish animal models have been valuable tools for identifying candidate genes regulating human melanocyte development and function, because many important genes that regulate pigmentation are highly conserved among these divergent species. Striking phenotypic parallels occur when orthologous genes are mutated, and the ease of experimental manipulation afforded by animal models has often directed discovery of pathological human gene mutations as well as revealed details of gene function. Each animal has inherent advantages and disadvantages for modeling human melanocytes. For example, zebrafish reproduce rapidly and allow precise pigment cell lineage tracing, but have pigment cell types that are divergent from those of mammals. Chicken and frog models facilitate studies of melanoblasts newly emerging from the NC, but are not amenable to genomic manipulation. Mouse models are readily altered genetically, but display significantly different epidermal architecture: while melanocytes occur in human epidermis, melanocytes are absent from mouse epidermis (excepting tail and ear regions), residing solely in hair follicles, which in mouse are present at much greater density. Of note, this difference has been addressed by creation of transgenic mice in which altered growth factor expression causes retention of melanocytes in the epidermis or dermis.71,72 While newly emerging technologies such as 3-dimensional skin models73 or large-scale genome sequencing will facilitate direct analysis of human melanocyte function and gene expression, animal models will remain essential tools to verify functional data gathered using these new technologies, thus keeping animal models at the forefront of pigmentation research.
Conclusion
Much remains to be discovered regarding the developmental genetic pathways that govern both normal and abnormal human pigmentation. Many human pigmentation disorders have a genetic locus associated with a subset of patients, while additional, phenotypically similar patients harbor no mutations at these identified genes and in some cases map the disease locus to a novel genomic region. These genetically heterogeneous disorders that await further characterization include Piebaldism,74 WS2,75 YDBS,11 Carney complex, and FPH1. For other disorders, no underlying genetic mutations are known, including: Albinism deafness syndrome, exhibiting a piebald-like pigmentation phenotype, congenital deafness and heterochromia irides,76 dyschromatosis universalis hereditaria (DUH1, DUH2), exhibiting extensive hypo- and hyperpigmented patches, and congenital diffuse melanosis, displaying widespread dermal/epidermal hyperpigmentation and neurological abnormalities.77,78
Model organisms (see sidebar) will remain essential tools to aid in discovery and understanding of genes associated with human pigmentation anomalies, their allelic variance, and additional multifactorial effects. Additionally, melanocytes in model organisms and humans may harbor genetic differences associated with their anatomical location or developmental origin, and characterization of these intrinsic features may explain phenotypes of various pigmentation anomalies.32 For example, human skin has been proposed to contain regional mosaicism that is developmentally determined and potentially associated with neuronal architecture. 22,79 Various patterns of hyperpigmented lesions have been proposed to correlate with these defined cutaneous subregions.80 The molecular details of regional mosaicism, and how these regional differences contribute to the pathology of melanocytic hyperpigmentation remains an open field of study, which may yield many discoveries via whole genome sequence analysis or expression profiling. Furthermore, detailed sequence analysis of various melanocytic lesions will help determine the contributions of somatic and/or germline mutations to their proliferation. Additional research also remains in the field of melanocyte stem cell research, as dermal stem cells competent to differentiate into human epidermal melanocytes were recently discovered.81 The pathways governing proliferation of these stem cells need to be discerned, as well as the possibility that aberrations in these stem cells may play an integral role in human pigmentation anomalies.82 Future genetic analyses will allow greater integration of both phenotypes and genotypes of melanocyte abnormalities, thus furthering understanding of melanocyte function and potentiating novel treatment of pigmentation disorders.
Acknowledgments
We thank Darryl Leja and Julia Fekecs for graphics assistance, and members of the Pavan lab for helpful discussions and careful reading of this manuscript. This research was supported by the Intramural Research Program of the NIH, NHGRI.
Footnotes
The authors declare no conflicts of interest
Contributor Information
Laura L. Baxter, Mouse Embryology Section, Genetic Disease Research Branch, National Human Genome, Research Institute, National Institutes of Health, Bethesda, MD, USA
William J. Pavan, Mouse Embryology Section, Genetic Disease Research Branch, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA.
References
- 1.Thomas I, Kihiczak GG, Fox MD, Janniger CK, Schwartz RA. Piebaldism: an update. Int J Dermatol. 2004;43:716–719. doi: 10.1111/j.1365-4632.2004.02114.x. [DOI] [PubMed] [Google Scholar]
- 2.Thomas AJ, Erickson CA. The making of a melanocyte: the specification of melanoblasts from the neural crest. Pigment cell & melanoma research. 2008;21:598–610. doi: 10.1111/j.1755-148X.2008.00506.x. [DOI] [PubMed] [Google Scholar]
- 3.Adameyko I, Lallemend F, Aquino JB, Pereira JA, Topilko P, Müller T, Fritz N, Beljajeva A, Mochii M, Liste I, Usoskin D, Suter U, Birchmeier C, Ernfors P. Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell. 2009;139:366–379. doi: 10.1016/j.cell.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 4.Kelsh RN, Harris ML, Colanesi S, Erickson CA. Stripes and belly-spots -- a review of pigment cell morphogenesis in vertebrates. Semin Cell Dev Biol. 2009;20:90–104. doi: 10.1016/j.semcdb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luciani F, Champeval D, Herbette A, Denat L, Aylaj B, Martinozzi S, Ballotti R, Kemler R, Goding CR, De Vuyst F, Larue L, Delmas V. Biological and mathematical modeling of melanocyte development. Development. 2011;138:3943–3954. doi: 10.1242/dev.067447. [DOI] [PubMed] [Google Scholar]
- 6.Sturm RA. Molecular genetics of human pigmentation diversity. Hum Mol Genet. 2009;18:R9–17. doi: 10.1093/hmg/ddp003. [DOI] [PubMed] [Google Scholar]
- 7.Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet. 1995;11:328–330. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- 8.Beaumont KA, Shekar SN, Cook AL, Duffy DL, Sturm RA. Red hair is the null phenotype of MC1R. Hum Mutat. 2008;29:E88–94. doi: 10.1002/humu.20788. [DOI] [PubMed] [Google Scholar]
- 9.Bharti K, Nguyen M-TT, Skuntz S, Bertuzzi S, Arnheiter H. The other pigment cell: specification and development of the pigmented epithelium of the vertebrate eye. Pigment Cell Res. 2006;19:380–394. doi: 10.1111/j.1600-0749.2006.00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hennekam RC, Gorlin RJ. Confirmation of the Yemenite (Warburg) deaf-blind hypopigmentation syndrome. Am J Med Genet. 1996;65:146–148. doi: 10.1002/(SICI)1096-8628(19961016)65:2<146::AID-AJMG13>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 11.Bondurand N, Kuhlbrodt K, Pingault V, Enderich J, Sajus M, Tommerup N, Warburg M, Hennekam RC, Read AP, Wegner M, Goossens M. A molecular analysis of the yemenite deaf-blind hypopigmentation syndrome: SOX10 dysfunction causes different neurocristopathies. Hum Mol Genet. 1999;8:1785–1789. doi: 10.1093/hmg/8.9.1785. [DOI] [PubMed] [Google Scholar]
- 12.Izumi K, Kohta T, Kimura Y, Ishida S, Takahashi T, Ishiko A, Kosaki K. Tietz syndrome: unique phenotype specific to mutations of MITF nuclear localization signal. Clin Genet. 2008;74:93–95. doi: 10.1111/j.1399-0004.2008.01010.x. [DOI] [PubMed] [Google Scholar]
- 13.Pingault V, Ente D, Dastot-Le Moal F, Goossens M, Marlin S, Bondurand N. Review and update of mutations causing Waardenburg syndrome. Hum Mutat. 2010;31:391–406. doi: 10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- 14.Inoue K, Khajavi M, Ohyama T, Hirabayashi S-I, Wilson J, Reggin JD, Mancias P, Butler IJ, Wilkinson MF, Wegner M, Lupski JR. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet. 2004;36:361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- 15.Bondurand N, Dastot-Le Moal F, Stanchina L, Collot N, Baral V, Marlin S, Attie-Bitach T, Giurgea I, Skopinski L, Reardon W, Toutain A, Sarda P, Echaieb A, Lackmy-Port-Lis M, Touraine R, Amiel J, Goossens M, Pingault V. Deletions at the SOX10 gene locus cause Waardenburg syndrome types 2 and 4. Am J Hum Genet. 2007;81:1169–1185. doi: 10.1086/522090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaoui A, Watanabe Y, Touraine R, Baral V, Goossens M, Pingault V, Bondurand N. Identification and functional analysis of SOX10 missense mutations in different subtypes of waardenburg syndrome. Hum Mutat. 2011;32:1436–49. doi: 10.1002/humu.21583. [DOI] [PubMed] [Google Scholar]
- 17.Corry GN, Hendzel J, Underhill DA. Subnuclear localization and mobility are key indicators of PAX3 dysfunction in Waardenburg syndrome. Hum Mol Genet. 2008;17:1825–37. doi: 10.1093/hmg/ddn076. [DOI] [PubMed] [Google Scholar]
- 18.Huizing M, Helip-Wooley A, Westbroek W, Gunay-Aygun M, Gahl WA. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu Rev Genomics Hum Genet. 2008;9:359–386. doi: 10.1146/annurev.genom.9.081307.164303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cullinane AR, Curry JA, Carmona-Rivera C, Summers CG, Ciccone C, Cardillo ND, Dorward H, Hess RA, White JG, Adams D, Huizing M, Gahl WA. A BLOC-1 mutation screen reveals that PLDN is mutated in Hermansky-Pudlak Syndrome type 9. Am J Hum Genet. 2011;88:778–787. doi: 10.1016/j.ajhg.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Plonka PM, Passeron T, Brenner M, Tobin DJ, Shibahara S, Thomas A, Slominski A, Kadekaro AL, Hershkovitz D, Peters E, Nordlund JJ, Abdel-Malek Z, Takeda K, Paus R, Ortonne JP, Hearing VJ, Schallreuter KU. What are melanocytes really doing all day long...? Exp Dermatol. 2009;18:799–819. doi: 10.1111/j.1600-0625.2009.00912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nordlund JJ. The melanocyte and the epidermal melanin unit: an expanded concept. Dermatologic Clinics. 2007;25:271–81. vii. doi: 10.1016/j.det.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Quevedo WC. Epidermal Melanin Units: Melanocyte-Keratinocyte Interactions. Am Zoologist. 1972;12:35–41. [Google Scholar]
- 23.Taieb A, Boralevi F. Hypermelanoses of the newborn and of the infant. Dermatologic Clinics. 2007;25:327–36. viii. doi: 10.1016/j.det.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Schaffer JV. Pigmented lesions in children: when to worry. Curr Opin Pediatr. 2007;19:430–440. doi: 10.1097/MOP.0b013e32825b0788. [DOI] [PubMed] [Google Scholar]
- 25.Fistarol SK, Itin PH. Disorders of pigmentation. Journal of the German Society of Dermatology. 2010;8:187–201. doi: 10.1111/j.1610-0387.2009.07137.x. [DOI] [PubMed] [Google Scholar]
- 26.Rajpara SM, Botello AP, Townend J, Ormerod AD. Systematic review of dermoscopy and digital dermoscopy/artificial intelligence for the diagnosis of melanoma. Br J Dermatol. 2009;161:591–604. doi: 10.1111/j.1365-2133.2009.09093.x. [DOI] [PubMed] [Google Scholar]
- 27.Elder DE. Precursors to melanoma and their mimics: nevi of special sites. Mod Pathol. 2006;19 (Suppl 2):S4–20. doi: 10.1038/modpathol.3800515. [DOI] [PubMed] [Google Scholar]
- 28.McGregor B, Pfitzner J, Zhu G, Grace M, Eldridge A, Pearson J, Mayne C, Aitken JF, Green AC, Martin NG. Genetic and environmental contributions to size, color, shape, and other characteristics of melanocytic naevi in a sample of adolescent twins. Genet Epidemiol. 1999;16:40–53. doi: 10.1002/(SICI)1098-2272(1999)16:1<40::AID-GEPI4>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 29.Zhu G, Duffy DL, Eldridge A, Grace M, Mayne C, O’Gorman L, Aitken JF, Neale MC, Hayward NK, Green AC, Martin NG. A major quantitative-trait locus for mole density is linked to the familial melanoma gene CDKN2A: a maximum-likelihood combined linkage and association analysis in twins and their sibs. Am J Hum Genet. 1999;65:483–492. doi: 10.1086/302494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wachsmuth RC, Gaut RM, Barrett JH, Saunders CL, Randerson-Moor JA, Eldridge A, Martin NG, Bishop TD, Newton Bishop JA. Heritability and gene-environment interactions for melanocytic nevus density examined in a U.K. adolescent twin study. J Invest Dermatol. 2001;117:348–352. doi: 10.1046/j.0022-202x.2001.01415.x. [DOI] [PubMed] [Google Scholar]
- 31.Falchi M, Bataille V, Hayward NK, Duffy DL, Bishop JAN, Pastinen T, Cervino A, Zhao ZZ, Deloukas P, Soranzo N, Elder DE, Barrett JH, Martin NG, Bishop DT, Montgomery GW, Spector TD. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat Genet. 2009;41:915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whiteman DC, Pavan WJ, Bastian BC. The melanomas: a synthesis of epidemiological, clinical, histopathological, genetic, and biological aspects, supporting distinct subtypes, causal pathways, and cells of origin. Pigment cell & melanoma research. 2011;24:879–97. doi: 10.1111/j.1755-148X.2011.00880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bauer J, Garbe C. Acquired melanocytic nevi as risk factor for melanoma development. A comprehensive review of epidemiological data. Pigment Cell Res. 2003;16:297–306. doi: 10.1034/j.1600-0749.2003.00047.x. [DOI] [PubMed] [Google Scholar]
- 34.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Abeni D, Boyle P, Melchi CF. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur J Cancer. 2005;41:28–44. doi: 10.1016/j.ejca.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 35.Zayour M, Lazova R. Congenital melanocytic nevi. Clin Lab Med. 2011;31:267–280. doi: 10.1016/j.cll.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Hale EK, Stein J, Ben-Porat L, Panageas KS, Eichenbaum MS, Marghoob AA, Osman I, Kopf AW, Polsky D. Association of melanoma and neurocutaneous melanocytosis with large congenital melanocytic naevi--results from the NYU-LCMN registry. Br J Dermatol. 2005;152:512–517. doi: 10.1111/j.1365-2133.2005.06316.x. [DOI] [PubMed] [Google Scholar]
- 37.Hafner C, Stoehr R, van Oers JMM, Zwarthoff EC, Hofstaedter F, Landthaler M, Hartmann A, Vogt T. FGFR3 and PIK3CA mutations are involved in the molecular pathogenesis of solar lentigo. Br J Dermatol. 2009;160:546–551. doi: 10.1111/j.1365-2133.2008.08963.x. [DOI] [PubMed] [Google Scholar]
- 38.Nordlund JJ, Lerner AB, Braverman IM, McGuire JS. The multiple lentigines syndrome. Arch Dermatol. 1973;107:259–261. [PubMed] [Google Scholar]
- 39.Digilio MC, Conti E, Sarkozy A, Mingarelli R, Dottorini T, Marino B, Pizzuti A, Dallapiccola B. Grouping of multiple-lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am J Hum Genet. 2002;71:389–394. doi: 10.1086/341528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, Pogna EA, Schackwitz W, Ustaszewska A, Landstrom A, Bos JM, Ommen SR, Esposito G, Lepri F, Faul C, Mundel P, López Siguero JP, Tenconi R, Selicorni A, Rossi C, Mazzanti L, Torrente I, Marino B, Digilio MC, Zampino G, Ackerman MJ, Dallapiccola B, Tartaglia M, Gelb BD. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 41.Sarkozy A, Carta C, Moretti S, Zampino G, Digilio MC, Pantaleoni F, Scioletti AP, Esposito G, Cordeddu V, Lepri F, Petrangeli V, Dentici ML, Mancini GMS, Selicorni A, Rossi C, Mazzanti L, Marino B, Ferrero GB, Silengo MC, Memo L, Stanzial F, Faravelli F, Stuppia L, Puxeddu E, Gelb BD, Dallapiccola B, Tartaglia M. Germline BRAF mutations in Noonan, LEOPARD, and cardiofaciocutaneous syndromes: molecular diversity and associated phenotypic spectrum. Hum Mutat. 2009;30:695–702. doi: 10.1002/humu.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 43.Stewart RA, Sanda T, Widlund HR, Zhu S, Swanson KD, Hurley AD, Bentires-Alj M, Fisher DE, Kontaridis MI, Look AT, Neel BG. Phosphatase-dependent and -independent functions of Shp2 in neural crest cells underlie LEOPARD syndrome pathogenesis. Dev Cell. 2010;18:750–762. doi: 10.1016/j.devcel.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tartaglia M, Gelb BD, Zenker M. Noonan syndrome and clinically related disorders. Best Pract Res Clin Endocrinol Metab. 2011;25:161–179. doi: 10.1016/j.beem.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zembowicz A, Mihm MC. Dermal dendritic melanocytic proliferations: an update. Histopathology. 2004;45:433–451. doi: 10.1111/j.1365-2559.2004.01975.x. [DOI] [PubMed] [Google Scholar]
- 46.Reza AM, Farahnaz GZ, Hamideh S, Alinaghi SAS, Saeed Z, Mostafa H. Incidence of Mongolian spots and its common sites at two university hospitals in Tehran, Iran. Pediatr Dermatol. 2010;27:397–398. doi: 10.1111/j.1525-1470.2010.01168.x. [DOI] [PubMed] [Google Scholar]
- 47.Robson ME, Glogowski E, Sommer G, Antonescu CR, Nafa K, Maki RG, Ellis N, Besmer P, Brennan M, Offit K. Pleomorphic characteristics of a germ-line KIT mutation in a large kindred with gastrointestinal stromal tumors, hyperpigmentation, and dysphagia. Clin Cancer Res. 2004;10:1250–1254. doi: 10.1158/1078-0432.ccr-03-0110. [DOI] [PubMed] [Google Scholar]
- 48.Campbell T, Felsten L, Moore J. Disappearance of lentigines in a patient receiving imatinib treatment for familial gastrointestinal stromal tumor syndrome. Arch Dermatol. 2009;145:1313–1316. doi: 10.1001/archdermatol.2009.263. [DOI] [PubMed] [Google Scholar]
- 49.Rothenbuhler A, Stratakis CA. Clinical and molecular genetics of Carney complex. Best Pract Res Clin Endocrinol Metab. 2010;24:389–399. doi: 10.1016/j.beem.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Okazaki M, Yoshimura K, Suzuki Y, Uchida G, Kitano Y, Harii K, Imokawa G. The mechanism of epidermal hyperpigmentation in café-au-lait macules of neurofibromatosis type 1 (von Recklinghausen’s disease) may be associated with dermal fibroblast-derived stem cell factor and hepatocyte growth factor. Br J Dermatol. 2003;148:689–697. doi: 10.1046/j.1365-2133.2003.05283.x. [DOI] [PubMed] [Google Scholar]
- 51.De Schepper S, Boucneau J, Vander Haeghen Y, Messiaen L, Naeyaert J-M, Lambert J. Café-au-lait spots in neurofibromatosis type 1 and in healthy control individuals: hyperpigmentation of a different kind? Arch Dermatol Res. 2006;297:439–449. doi: 10.1007/s00403-006-0644-6. [DOI] [PubMed] [Google Scholar]
- 52.Amyere M, Vogt T, Hoo J, Brandrup F, Bygum A, Boon L, Vikkula M. KITLG mutations cause familial progressive hyper- and hypopigmentation. J Invest Dermatol. 2011;131:1234–1239. doi: 10.1038/jid.2011.29. [DOI] [PubMed] [Google Scholar]
- 53.Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19:230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niemeyer CM, Kang MW, Shin DH, Furlan I, Erlacher M, Bunin NJ, Bunda S, Finklestein JZ, Sakamoto KM, Gorr TA, Mehta P, Schmid I, Kropshofer G, Corbacioglu S, Lang PJ, Klein C, Schlegel P-G, Heinzmann A, Schneider M, Starý J, van den Heuvel-Eibrink MM, Hasle H, Locatelli F, Sakai D, Archambeault S, Chen L, Russell RC, Sybingco SS, Ohh M, Braun BS, Flotho C, Loh ML. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet. 2010;42:794–800. doi: 10.1038/ng.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pérez B, Mechinaud F, Galambrun C, Ben Romdhane N, Isidor B, Philip N, Derain-Court J, Cassinat B, Lachenaud J, Kaltenbach S, Salmon A, Désirée C, Pereira S, Menot ML, Royer N, Fenneteau O, Baruchel A, Chomienne C, Verloes A, Cavé H. Germline mutations of the CBL gene define a new genetic syndrome with predisposition to juvenile myelomonocytic leukaemia. J Med Genet. 2010;47:686–691. doi: 10.1136/jmg.2010.076836. [DOI] [PubMed] [Google Scholar]
- 56.Vachiramon V. Approach to reticulate hyperpigmentation. Clin Exp Dermatol. 2011;36:459–466. doi: 10.1111/j.1365-2230.2011.04100.x. [DOI] [PubMed] [Google Scholar]
- 57.Spritz RA. Recent progress in the genetics of generalized vitiligo. J Genet Genomics. 2011;38:271–278. doi: 10.1016/j.jgg.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koga M, Tango T. Clinical features and course of type A and type B vitiligo. Br J Dermatol. 1988;118:223–228. doi: 10.1111/j.1365-2133.1988.tb01778.x. [DOI] [PubMed] [Google Scholar]
- 59.van den Boorn JG, Konijnenberg D, Dellemijn TAM, van der Veen JPW, Bos JD, Melief CJM, Vyth-Dreese FA, Luiten RM. Autoimmune destruction of skin melanocytes by perilesional T cells from vitiligo patients. J Invest Dermatol. 2009;129:2220–2232. doi: 10.1038/jid.2009.32. [DOI] [PubMed] [Google Scholar]
- 60.Sandoval-Cruz M, García-Carrasco M, Sánchez-Porras R, Mendoza-Pinto C, Jiménez-Hernández M, Munguía-Realpozo P, Ruiz-Argüelles A. Immunopathogenesis of vitiligo. Autoimmun Rev. 2011;10:762–765. doi: 10.1016/j.autrev.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 61.Bastiaens M, Huurne ter J, Gruis N, Bergman W, Westendorp R, Vermeer BJ, Bouwes Bavinck JN. The melanocortin-1-receptor gene is the major freckle gene. Hum Mol Genet. 2001;10:1701–1708. doi: 10.1093/hmg/10.16.1701. [DOI] [PubMed] [Google Scholar]
- 62.English DR, Armstrong BK. Melanocytic nevi in children. I. Anatomic sites and demographic and host factors. Am J Epidemiol. 1994;139:390–401. doi: 10.1093/oxfordjournals.aje.a117011. [DOI] [PubMed] [Google Scholar]
- 63.Fritschi L, McHenry P, Green A, Mackie R, Green L, Siskind V. Naevi in schoolchildren in Scotland and Australia. Br J Dermatol. 1994;130:599–603. doi: 10.1111/j.1365-2133.1994.tb13106.x. [DOI] [PubMed] [Google Scholar]
- 64.Harrison SL, MacKie RM, MacLennan R. Development of melanocytic nevi in the first three years of life. J Natl Cancer Inst. 2000;92:1436–1438. doi: 10.1093/jnci/92.17.1436. [DOI] [PubMed] [Google Scholar]
- 65.Whiteman DC, Brown RM, Purdie DM, Hughes M-C. Melanocytic nevi in very young children: the role of phenotype, sun exposure, and sun protection. J Am Acad Dermatol. 2005;52:40–47. doi: 10.1016/j.jaad.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 66.Hattori H, Kawashima M, Ichikawa Y, Imokawa G. The epidermal stem cell factor is over-expressed in lentigo senilis: implication for the mechanism of hyperpigmentation. J Invest Dermatol. 2004;122:1256–1265. doi: 10.1111/j.0022-202X.2004.22503.x. [DOI] [PubMed] [Google Scholar]
- 67.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, Simpson EM, Barsh GS, Bastian BC. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, Sozen MM, Baimukanova G, Roy R, Heguy A, Dolgalev I, Khanin R, Busam K, Speicher MR, O’Brien J, Bastian BC. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stulberg DL, Clark N, Tovey D. Common hyperpigmentation disorders in adults: Part II. Melanoma, seborrheic keratoses, acanthosis nigricans, melasma, diabetic dermopathy, tinea versicolor, and postinflammatory hyperpigmentation. Am Fam Physician. 2003;68:1963–1968. [PubMed] [Google Scholar]
- 70.Ruiz-Maldonado R, Orozco-Covarrubias ML. Postinflammatory hypopigmentation and hyperpigmentation. Semin Cutan Med Surg. 1997;16:36–43. doi: 10.1016/s1085-5629(97)80034-x. [DOI] [PubMed] [Google Scholar]
- 71.Kunisada T, Yoshida H, Yamazaki H, Miyamoto A, Hemmi H, Nishimura E, Shultz LD, Nishikawa S, Hayashi S. Transgene expression of steel factor in the basal layer of epidermis promotes survival, proliferation, differentiation and migration of melanocyte precursors. Development. 1998;125:2915–2923. doi: 10.1242/dev.125.15.2915. [DOI] [PubMed] [Google Scholar]
- 72.Kunisada T, Yamazaki H, Hirobe T, Kamei S, Omoteno M, Tagaya H, Hemmi H, Koshimizu U, Nakamura T, Hayashi SI. Keratinocyte expression of transgenic hepatocyte growth factor affects melanocyte development, leading to dermal melanocytosis. Mech Dev. 2000;94:67–78. doi: 10.1016/s0925-4773(00)00308-7. [DOI] [PubMed] [Google Scholar]
- 73.Li L, Fukunaga-Kalabis M, Herlyn M. The three-dimensional human skin reconstruct model: a tool to study normal skin and melanoma progression. J Vis Exp. 2011 doi: 10.3791/2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murakami T, Hosomi N, Oiso N, Giovannucci-Uzielli ML, Aquaron R, Mizoguchi M, Kato A, Ishii M, Bitner-Glindzicz M, Barnicoat A, Wilson L, Tsukamoto K, Ueda H, Mancini AJ, Suzuki T, Riley J, Miertus J, Camargo M, Santoro-Zea A, Atkin J, Fukai K. Analysis of KIT, SCF, and initial screening of SLUG in patients with piebaldism. J Invest Dermatol. 2005;124:670–672. doi: 10.1111/j.0022-202X.2005.23637.x. [DOI] [PubMed] [Google Scholar]
- 75.Toriello HV. Pigmentary anomalies and hearing loss. Adv Otorhinolaryngol. 2011;70:50–55. doi: 10.1159/000322471. [DOI] [PubMed] [Google Scholar]
- 76.Zlotogora J. X-linked albinism-deafness syndrome and Waardenburg syndrome type II: a hypothesis. Am J Med Genet. 1995;59:386–387. doi: 10.1002/ajmg.1320590321. [DOI] [PubMed] [Google Scholar]
- 77.Bashiti HM, Blair JD, Triska RA, Keller L. Generalized dermal melanocytosis. Arch Dermatol. 1981;117:791–793. [PubMed] [Google Scholar]
- 78.Schwartz RA, Cohen-Addad N, Lambert MW, Lambert WC. Congenital melanocytosis with myelomeningocele and hydrocephalus. Cutis. 1986;37:37–39. [PubMed] [Google Scholar]
- 79.Happle R. What is a nevus? A proposed definition of a common medical term. Dermatology (Basel) 1995;191:1–5. doi: 10.1159/000246468. [DOI] [PubMed] [Google Scholar]
- 80.Torrelo A, Baselga E, Nagore E, Zambrano A, Happle R. Delineation of the various shapes and patterns of nevi. Eur J Dermatol. 2005;15:439–450. [PubMed] [Google Scholar]
- 81.Li L, Fukunaga-Kalabis M, Yu H, Xu X, Kong J, Lee JT, Herlyn M. Human dermal stem cells differentiate into functional epidermal melanocytes. J Cell Sci. 2010;123:853–860. doi: 10.1242/jcs.061598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cramer SF. Stem cells for epidermal melanocytes--a challenge for students of dermatopathology. Am J Dermatopath. 2009;31:331–341. doi: 10.1097/DAD.0b013e31819cd0cb. [DOI] [PubMed] [Google Scholar]
Further Reading/Resources
- 1.Nordlund JJ, Boissy RE, Hearing VJ, King RA, Oetting WS, Ortonne JP. The Pigmentary System: Physiology and Pathophysiology. Blackwell Publishing; Malden, Massachusetts: 2006. [Google Scholar]
- 2.The Online Metabolic and Molecular Bases of Inherited Disease
- 3.Online Mendelian Inheritance in Man