Abstract
Cleavage of the rotavirus spike protein, VP4, is required for rotavirus-induced membrane permeability and viral entry into cells. The VP5* cleavage product selectively permeabilizes membranes and liposomes and contains an internal hydrophobic domain that is required for membrane permeability. Here we investigate VP5* domains (residues 248 to 474) that direct membrane binding. We determined that expressed VP5 fragments containing residues 248 to 474 or 265 to 474, including the internal hydrophobic domain, bind to cellular membranes but are not present in Triton X-100-resistant membrane rafts. Expressed VP5 partitions into aqueous but not detergent phases of Triton X-114, suggesting that VP5 is not integrally inserted into membranes. Since high-salt or alkaline conditions eluted VP5 from membranes, our findings demonstrate that VP5 is peripherally associated with membranes. Interestingly, mutagenesis of residue 394 (W→R) within the VP5 hydrophobic domain, which abolishes VP5-directed permeability, had no effect on VP5's peripheral membrane association. In contrast, deletion of N-terminal VP5 residues (residues 265 to 279) abolished VP5 binding to membranes. Alanine mutagenesis of two positively charged residues within this domain (residues 274R and 276K) dramatically reduced (>95%) binding of VP5 to membranes and suggested their potential interaction with polar head groups of the lipid bilayer. Mutations in either the VP5 hydrophobic or basic domain blocked VP5-directed permeability of cells. These findings indicate that there are at least two discrete domains within VP5* required for pore formation: an N-terminal basic domain that permits VP5* to peripherally associate with membranes and an internal hydrophobic domain that is essential for altering membrane permeability. These results provide a fundamental understanding of interactions between VP5* and the membrane, which are required for rotavirus entry.
Rotaviruses are the most important cause of severe dehydrating viral gastroenteritis in infants and young children (34). Rotaviruses are members of the Reoviridae, containing 11 double-stranded RNA (dsRNA) gene segments within an icosahedral triple-layer particle (TLP) (21). The outer capsid is composed of two proteins, VP4 and VP7. VP7 is the predominant outer capsid surface glycoprotein which is recognized by neutralizing antibodies and a determinant of rotavirus serotype G (21). The virion contains 780 copies of VP7, which are assembled into calcium-dependent trimers (49). Calcium chelation causes a conformational change in VP7 and converts TLPs into transcriptionally active double-layered particles (DLPs) (8, 14, 16). The TLP-to-DLP transition can be accomplished in vitro and has been reported to occur following exposure of particles to low intracellular calcium ion concentrations (8, 52, 53). DLPs are incapable of crossing the cellular membrane and are noninfectious unless transfected into cells (2). DLPs contain viral dsRNA and the dsRNA-dependent RNA polymerase and are capable of extruding capped mRNAs into the cytoplasm of cells through pores in the DLP (36, 50). The calcium-dependent uncoating step activates rotavirus transcription in the cytoplasm of cells following viral entry.
The other outer capsid surface protein, VP4, forms 60 dimeric spikes on the virion's surface with viral attachment, entry, and neutralization functions (21, 48, 49). VP4 is a determinant of viral virulence, host range, and protective immunity, and its proteolytic cleavage is required for viral infectivity (20, 22, 28, 43, 44). Trypsin cleaves VP4 into two fragments, an N-terminal VP8* polypeptide and a C-terminal VP5* polypeptide (residues 248 to 776), both of which are recognized by neutralizing antibodies (22, 40, 43, 57). VP4 cleavage stabilizes the spike protein, and Fab fragments directed toward both VP8* and VP5* bind to the head of the VP4 spike, indicating that both proteins remain virion associated after cleavage (10, 48, 60). The VP8* protein of many rotaviruses binds sialic acid on the surfaces of the cells and is the viral hemagglutinin (25, 32, 39). However, rotavirus interactions with α2β1 and αvβ3 integrins have been reported, and motifs on VP5* or VP7 have been implicated in rotavirus-integrin interactions (1, 9, 29, 31, 38, 61). The VP5* portion of VP4 has also been shown to permeabilize membranes, providing a means for rotavirus outer capsid proteins to direct the attachment and entry processes (13, 17, 23, 42).
However, the means by which nonenveloped rotaviruses enter cells is still poorly understood. Rotavirus reportedly enter cells with a half-life of <10 min via a mechanism consistent with direct membrane penetration (33, 59). Nontrypsinized particles are not infectious and enter into endosomes where they are degraded (33, 35, 58). Rotaviruses have also been suggested to enter cells via early endocytic vesicles in a process where high extracellular calcium in the vesicles is depleted to permit virion uncoating and membrane disruption (46, 52, 53). However, there is still controversy about this process, in part because it is difficult to distinguish uncoating, membrane disruption, and membrane permeability during viral entry (6, 11, 17, 46). In either case, the role of VP5* in permeabilizing membranes is central to the rotavirus entry process (13, 17).
Trypsin cleavage of VP4 activates membrane-destabilizing properties of the virus and the viral outer capsid proteins (13, 23, 51). Expressed VP5 and N-terminal VP5 fragments have been shown to permeabilize model and cellular membranes, whereas uncleaved VP4 or VP8* have no permeabilizing activities (13, 17). As a result, trypsin cleavage, which is required for viral infectivity, provides a means for exposing membrane-interactive portions of VP5* and activating the virus for cellular entry. Permeability directed by VP5 is blocked by neutralizing monoclonal antibodies, and mutations within the VP5 hydrophobic domain have been shown to abolish liposome or cell membrane permeability (13, 17, 40). Residues 265 to 404 of VP5 are required for permeability and contain the internal hydrophobic domain (residues 385 to 404) necessary for permeabilizing membranes (13, 17). Interestingly, VP5 directs the formation of transient and size selective pores and fails to lyse the model or cellular membranes that it permeabilizes (13, 17). The means by which VP5 interacts with cellular membranes and forms membrane pores remain to be defined.
In this study, we investigated the nature of VP5 interactions with mammalian cell membranes. We demonstrate that permeability-competent VP5 fragments are peripherally and not integrally associated with membranes and that at least two domains are necessary for interactions between VP5 and the membrane. Mutations in the VP5 hydrophobic domain failed to inhibit VP5 membrane association, although these mutations blocked VP5-directed cell permeability. In contrast, deletion of or mutations in an N-terminal basic domain of VP5 inhibited VP5 binding to membranes and also blocked VP5-directed membrane permeability. These findings indicate that VP5 contains two discrete domains required for permeability: one domain that directs VP5's peripheral membrane association and a second hydrophobic domain that is required for pore formation.
MATERIALS AND METHODS
Plasmids, cells, and transfection.
Primers to rotavirus VP5 coding sequences were synthesized with 5′ BamHI and 3′ XhoI sites. PCR products containing VP5N248 (residues 248 to 474), VP5N265 (residues 265 to 474), VP5N280 (residues 280 to 474), and the W394R hydrophobic domain mutant VP5N248(394R) were generated using previously described pET-6His constructs by standard methods (13, 17). The VP5N248 mutant containing R274A and K276A, VP5N248(274A,276A), was generated by oligonucleotide-directed mutagenesis as previously described (13, 17). DNA was cloned into the mammalian expression vector pcDNA4/HisC (Invitrogen) containing a downstream six-His tag by standard methods.
HEK293 cells or COS7 cells were grown at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium supplemented with antibiotics and 10% fetal bovine serum (57). Cells were transfected with 5 μg of plasmid DNA by an enhanced calcium phosphate method as previously described (45).
Membrane flotation analysis of expressed VP5 proteins.
HEK293 or COS7 cells were harvested, homogenized, and subjected to sucrose gradient centrifugation 42 to 46 h posttransfection (62). Briefly, cells were collected by centrifugation (1,000 × g, 2 min) and resuspended in ice-cold homogenization buffer containing 10% (wt/vol) sucrose, 10 mM Tris-HCl (pH 7.4), 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride (2 × 106 cells per 0.2 ml). Cells were sonicated, and nuclei and whole cells were removed by centrifugation (3,000 × g, 10 min, 4°C). Sucrose was added to the supernatants until they were 80% sucrose, and these supernatants were overlaid with equal volumes of 65 and 10% sucrose in ultracentrifuge tubes. Sucrose gradients were centrifuged 50,000 rpm for 2.5 h at 4°C in an SW55 rotor (Beckman).
Visible membrane bands at the 10% sucrose-65% sucrose interface were harvested, diluted threefold in 10 mM Tris-HCl-1 mM EDTA, and pelleted by ultracentrifugation (35,000 rpm [SW55 rotor], 1 h, 4°C). Membranes were treated with 0.4 to 2.0 M NaCl (high-salt treatment) or 0.1 to 1.0 M Na2CO3 (pH 11.5) (high pH treatment) for 45 min at 4°C or not treated and repelleted (12, 26). The resulting membrane pellets were applied to a second sucrose gradient.
Gradient fractions were collected and diluted with 5 ml of guanidine buffer (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole [pH 8.0]). His-tagged proteins were precipitated with Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) as previously described (13). Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotted using anti-HisG primary antibody (Invitrogen). His-tagged proteins were precipitated and detected by Western blotting as described above. Endogenous caveolin present in Triton X-100-resistant fractions from COS7 cells was detected by Western blotting using a primary antibody directed against caveolin 1 (Santa Cruz).
Triton X-114 phase partitioning.
Total cellular membrane pellets were resuspended in a solution consisting of 10 mM Tris-HCl (pH 7.4) and 150 mM NaCl supplemented with 1% Triton X-114 (3, 62). Samples were incubated for 60 min at 4°C to extract detergent-soluble proteins and clarified by centrifugation (35,000 rpm [SW55 rotor], 1 h, 4°C). The supernatant was incubated for 5 min at 30°C for Triton-X114 clouding. Aqueous and detergent phases of each sample were separated by centrifugation (300 × g, 5 min, 22°C) through a 6% (wt/vol) sucrose cushion containing 0.06% Triton X-114, 10 mM Tris-HCl (pH 7.4), and 150 mM NaCl. Aqueous and detergent phases were diluted with guanidine buffer and six-His-tagged proteins were precipitated and identified by Western blotting as described above.
Analysis of plasma membrane permeability to divalent cations.
Free cellular cytosolic [Ca2+] was measured with fluo-3 (Sigma) as previously described (2). To load cells, a modified version of the method of Chen et al. (7) was used. Briefly, 42 to 46 h posttransfection, HEK293 cells were washed with KRH buffer minus divalent cations (125 mM NaCl, 5 mM KCl, 1.2 mM KH2PO4, 6 mM glucose, 25 mM HEPES [pH 7.4]) and incubated in the same buffer at 37°C for 45 min. Cells were detached from culture dishes by pipetting and pelleted by centrifugation (1,000 × g, 3 min, 22°C) (7).
Cells (2 × 106/ml) were loaded with fluo-3 (4 to 6 μM) in KRH buffer containing 1.2 mM MgCl2, 2 mM CaCl2, 0.1% bovine serum albumin, and 250 μM sulfinpyrazone at 22°C for 30 min. Cells were washed (three times) and resuspended in the same buffer to remove unincorporated fluo-3. Aliquots of fluo-3-loaded cells were added to 1.8-ml portions of KRH buffer, and the fluorescence intensity of fluo-3 (F) at 526 nm was measured at 22°C in a Perkin-Elmer LS-5B luminescence spectrometer following excitation with 506-nm-wavelength light. Fluorescence signals were calibrated at saturating Ca2+ concentrations with 0.1% Triton X-100 or 50 mM EDTA to obtain maximum (Fmax) or minimum fluorescence signals (Fmin), respectively. The average intracellular calcium concentration ([Ca2+]i) was calculated from the fluo-3 fluorescence intensity using the following formula: [Ca2+]i = Kd(F − Fmin)/(Fmax − F), where Kd is 400 nM for intracellular fluo-3 dye (56). The plasma membrane permeability of HEK293 cells was evaluated by measuring the change in [Ca2+]i in cells. [Ca2+]i changes were monitored after adding CaCl2 to cell suspensions, transiently increasing extracellular [Ca2+] 5 to 8 mM for 30 to 60 s in KRH buffer. To evaluate Ni2+ entry, 2.5 mM NiCl2 was added to cells and the ability of Ni2+ to quench fluo-3 signals was defined by the fluorescence measurements discussed above (19). The relative fluorescence intensities in the presence of Ni2+ were calculated as (FNi2+/Fo) × 100%, where Fo is fluo-3 fluorescence in the presence of 2 mM Ca2+ and FNi2+ is fluo-3 fluorescence of the sample after 2.5 mM NiCl2 was added (19).
RESULTS
VP5 and VP5 hydrophobic domain mutants are membrane associated.
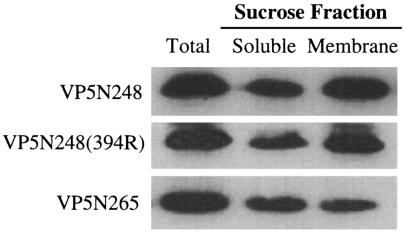
We previously determined that expressed VP5N248 and VP5N265 proteins were capable of permeabilizing model and cellular membranes in the absence of other viral proteins and that mutations in an internal hydrophobic domain of VP5, VP5N248(394R), abolished membrane permeability (13, 17). In order to define fundamental interactions of VP5 domains with membranes, we analyzed membrane binding of wild-type and mutant VP5 proteins. VP5N248, VP5N265, and VP5N248(394R) were expressed from plasmids following transfection of HEK293 or COS7 cells. Expressed VP5 proteins were evaluated for their ability to bind cellular membranes using sucrose gradient flotation. We observed that approximately half of the expressed VP5N248, VP5N248(394R), and VP5N265 proteins were associated with cellular membranes (Fig. 1). Interestingly, inserting a charged residue into the VP5 hydrophobic domain had no effect on VP5 binding to membranes, while this mutation blocks VP5-directed membrane permeability (17). These findings indicate that the internal hydrophobic domain does not determine VP5 membrane association and suggested that a discrete integral or peripheral membrane-interactive domain is present within VP5.
FIG. 1.
Membrane association of the rotavirus VP5 proteins. HEK293 cells were transfected with pcDNA4 constructs expressing VP5N248, VP5N248(394R), or VP5N265. Cellular membranes were fractionated by sucrose gradient centrifugation. His-tagged VP5N248, VP5N248(394R), or VP5N265 proteins were precipitated from total cell lysates (Total), membrane fractions (Membrane), or soluble fractions (Soluble) using Ni-NTA agarose. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotted using an anti-HisG antibody.
VP5 membrane association is independent of membrane rafts.
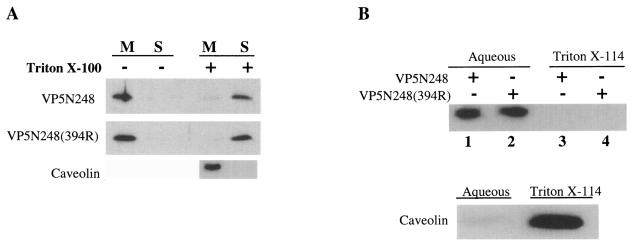
Rafts have been reported to promote the assembly and targeting of rotaviruses during maturation, and as a result, the VP5 portion of VP4 could mediate interactions with rafts (54). Rafts are Triton X-100 resistant at 4°C, and caveolin is a protein known to be present within membrane rafts (4, 24, 30, 37, 55). To determine whether VP5N248 proteins were present in membrane rafts, we treated membrane fractions with 1% Triton X-100 at 4°C and fractionated Triton X-100-resistant and -sensitive components on sucrose gradients. Triton X-100 treatment of membrane fractions containing VP5N248 caused a change in the association of both mutant and wild-type VP5 proteins from membrane to soluble fractions (Fig. 2A). Untreated VP5N248 remained associated with membranes following reflotation. In contrast to VP5 proteins, the raft-associated protein, caveolin, remained associated with Triton X-100-resistant membrane fractions at the top of the gradient (Fig. 2A). Although HEK293 cells do not have endogenous caveolin, both HEK293 cells and COS7 cells form lipid rafts (30, 55). There was no apparent association of VP5N248 in Triton X-100-resistant lipid rafts from either COS7 or HEK293 cells, indicating that the protein was not associated with rafts or caveolin. These findings indicate that VP5N248 is present in Triton X-100-sensitive membrane fractions or that expressed VP5 is eluted from membranes by Triton X-100 treatment.
FIG. 2.
(A) Triton X-100 treatment of VP5-containing membrane fractions. COS7 cells were transfected with plasmids expressing VP5N248 or VP5N248(394R), and membranes containing VP5N248 or VP5N248(394R) proteins were fractionated by sucrose gradient centrifugation and treated with 1% Triton X-100 at 4°C (+) or not treated (−). Samples were subjected to a second sucrose gradient centrifugation, and caveolin was detected by Western blotting using anti-caveolin 1 antibody. VP5N248 and VP5N248(394R) proteins from the membrane (M) or soluble (S) fraction were precipitated and analyzed as described in the legend to Fig. 1. (B) Triton X-114 phase partitioning of VP5. COS7 cells were transfected with plasmid expressing VP5N248 or VP5N248(394R) and fractionated by sucrose gradient centrifugation. Membrane fractions containing VP5N248 (lanes 1 and 3) or VP5N248(394R) (lanes 2 and 4) were subjected to Triton X-114 phase partitioning, and aliquots were analyzed for caveolin by Western blotting. VP5 proteins from aqueous (lanes 1 and 2) or detergent (lanes 3 and 4) phases were analyzed and detected as described in the legend to Fig. 1.
VP5* is peripherally associated with cellular membranes.
The presence of only one potential membrane-spanning domain within VP5 and the fact that inserting a charged residue within this domain did not prevent VP5-membrane interactions suggested that VP5 was not likely to be integrally associated with membranes and that additional domains might mediate membrane interactions (40). In order to distinguish integral and peripheral membrane associations of VP5, we determined the ability of VP5N248 to partition between detergent (integral) and aqueous (peripheral) phases of Triton X-114 (3, 62). Membrane fractions containing VP5N248 or VP5N248(394R) protein resulted in the exclusive partitioning of both proteins into the aqueous phase (Fig. 2B). In contrast, the integral membrane protein caveolin was exclusively present in the detergent phase of Triton X-114 (Fig. 2B).
Since integral membrane proteins could fail to partition into the Triton X-114 detergent phase, we used alternative approaches to determine whether VP5 was integrally or peripherally associated with membranes. High-salt or alkaline conditions disrupt electrostatic interactions which associate peripheral membrane proteins with lipid bilayers (12, 26). Membrane fractions containing VP5N248 or VP5N248(394R) were extracted with high salt or high pH and then subjected to sucrose flotation. As shown in Fig. 3A, approximately 80% of wild-type and mutant proteins were dissociated from membranes after 1 M NaCl treatment. VP5N248 was similarly released from membrane fractions following treatment with either 0.4 or 2 M NaCl (data not shown). In contrast, caveolin was not extracted from membranes by high salt concentrations (Fig. 3A). Treatment of VP5N248 and VP5N248(394R) proteins at pH 11.5 dissociated expressed VP5 proteins from membrane fractions (Fig. 3B). The conversion of closed vesicles to open membrane sheets, reported during alkaline treatment, is consistent with the complete release of VP5N248 from membranes following alkaline, but not NaCl, treatment. Collectively, these findings indicate that both VP5N248 and VP5N248(394R) proteins are peripherally and not integrally associated with cellular membranes.
FIG. 3.
(A) High-salt treatment of membrane fractions containing VP5. COS7 cells were transfected with plasmids expressing VP5N248 or VP5N248(394R). Membrane fractions containing VP5N248 or VP5N248(394R) were extracted with 1 M NaCl for 45 min at 4°C (+) or not treated (−). Following treatment, membranes were refloated on a second sucrose gradient, and caveolin was detected by Western blotting as described in the legend to Fig. 2. VP5N248 or VP5N248(394W→R) proteins from the membrane (M) or soluble (S) fractions were analyzed as described in the legend to Fig. 1. (B) Alkaline treatment of VP5-containing membrane fractions. HEK293 cells were transfected with plasmid expressing VP5N248 or VP5N248(394R). Membrane (M) fractions containing VP5N248 (+) or VP5N248(394R) (+) were treated with 0.1 M Na2CO3 (pH 11.5) for 45 min at 4°C (+) or not treated (−). Treated or untreated membrane fractions were refloated on a second sucrose gradient, and membrane fractions were Western blotted for VP5N248 or VP5N248(394R) protein.
N-terminal residues confer VP5 peripheral membrane association.
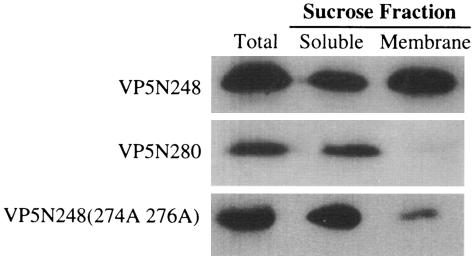
We previously reported that deleting residues 265 to 279 from the VP5 N terminus abolished VP5 membrane permeability similar to mutating or deleting the internal hydrophobic domain (17). Interestingly, this region is highly conserved and contains two basic residues (274R and 276K) that could participate in electrostatic interactions with the lipid bilayer. The VP5N280 truncation was expressed and analyzed by sucrose flotation assay as described above. The VP5N280 truncation was not associated with membrane fractions and instead was present entirely in the soluble fraction (Fig. 4). This finding prompted us to mutate residues R274 and K276 to alanine and evaluate membrane interactions of the VP5 mutant. There was a dramatic change in the association of the VP5N248(274A,276A) mutant with membranes, since >95% of the mutant protein was present in the soluble fraction (Fig. 4). This finding suggests that basic residues within the region of VP5 from residues 265 to 279 direct the peripheral membrane association of the proteins. Although we cannot exclude the possibility that residues 274 and 276 contribute to conformations of VP5N248 (15) which permit peripheral membrane association via discrete VP5N248 domains, our results are consistent with the direct membrane interaction of basic residues at the VP5* N terminus.
FIG. 4.
Influence of N-terminal mutations on membrane association of VP5 proteins. HEK293 cells were transfected with plasmids expressing VP5N248, VP5N280, or VP5N248(274A,276A). Cellular membranes were fractionated by sucrose gradient centrifugation, and six-His-tagged VP5 proteins were precipitated from total cell lysates (Total), membrane fractions (Membrane), or soluble fractions (Soluble) using Ni-NTA agarose. Precipitated proteins were analyzed by Western blotting.
VP5-membrane interactions alter membrane integrity.
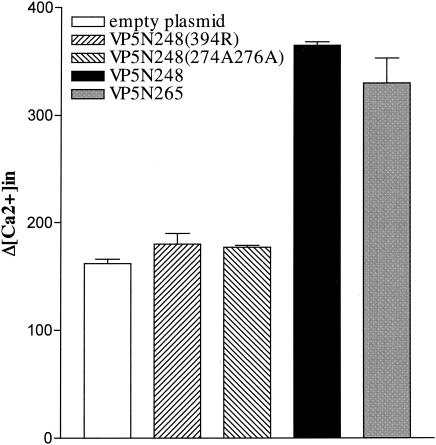
We have previously shown that VP5-membrane interactions increase membrane permeability to small molecules, including divalent cations (13, 17). In order to determine the effects of N-terminal basic residue mutations on VP5N248-directed membrane permeability, we determined the change in intracellular calcium induced by expressing VP5 and mutant VP5 proteins. We expressed VP5N248, VP5N265, VP5N248(394R), and VP5N248(274A,276A) in mammalian cells, loaded cells with the calcium-sensitive fluorophore fluo-3, and monitored fluorescence changes in response to changes in extracellular [Ca2+] as a measure of membrane permeability (7). VP5N248 expression in mammalian cells does not lyse or kill cells, and consistent with this, there was no significant change in [Ca2+]i at normal extracellular calcium concentrations (2 mM). However, when extracellular calcium concentrations were transiently increased (30 to 60 s), [Ca2+]i in VP5N248-expressing cells was significantly higher than that in control cells (Fig. 5). Both VP5N248 and VP5N265 permeabilized the plasma membrane, increasing intracellular calcium levels in response to the increased extracellular calcium gradient. This finding is consistent with the ability of these proteins to permeabilize liposomes and bacteria (13, 17). VP5-induced permeability was not calcium specific, since Ni2+ similarly crossed membranes where it inhibited fluo-3 fluorescence by 43% (19). In contrast, expression of either the hydrophobic domain mutant, VP5N248(394R), the VP5N280 deletion (not shown), or the basic domain mutant, VP5N248(274A,276A), abolished membrane permeability (Fig. 5). Each mutant resulted in intracellular calcium levels identical to untransfected or mock-transfected controls (Fig. 5). These findings demonstrate that the hydrophobic domain and the N-terminal basic domain of VP5 are required for VP5-directed membrane permeability.
FIG. 5.
VP5-directed changes in intracellular Ca2+ concentration. HEK293 cells were transfected with plasmids expressing VP5N248, VP5N265, VP5N248(394R), or VP5N248(274A,276A) or not transfected. Cells were loaded with the calcium-sensitive fluorophore fluo-3, and fluorescence was monitored at 526 nm after extracellular [Ca2+] was increased to 5 to 8 mM for 30 to 60 s. Intracellular (in) [Ca2+] was calculated as described in Materials and Methods. The averages ± standard errors (error bars) of three separate measurements are shown.
DISCUSSION
Early reports demonstrated that rotavirus entry into cells did not occur through a classical endosome acidification route (33, 35, 58, 59). During these studies, it was shown that trypsin-activated rotavirus is inaccessible to neutralizing antibodies 5 to 10 min after the temperature of prebound virus was increased to 37°C (33). These findings demonstrated that entry of infectious, trypsinized rotavirus was rapid and consistent with direct membrane penetration by the virion (33). However, massive conformational changes in the virion also occur as part of the entry process, since the outer capsid is uncoated by calcium chelation and the viral transcriptase is activated following exposure to low intracellular calcium levels (8, 52). Since high extracellular calcium levels surrounding virions cannot be lowered unless viral particles are compartmentalized away from extracellular ions, the presence of rotaviruses in small early endocytic vesicles has been suggested as a requirement of virion uncoating (6, 46). The entry of infectious rotaviruses via early endosomes is consistent with both the rapid inaccessibility of virions to neutralizing antibodies and the requirement for lowering calcium ion concentrations surrounding the virion for uncoating (6, 33, 46, 52).
However, it is still unclear how nonenveloped rotaviruses cross cellular membranes during infection. A tenet of rotavirus entry is that infectious rotaviruses require proteolytic cleavage of VP4 into VP8* and VP5* fragments in order to infect cells (22, 23, 27, 33). It was initially shown that trypsin-cleaved outer capsid proteins were able to permeabilize or fuse membranes in the absence of the viral particle (27). It was further shown that expressed VP5 and VP5 fragments permeabilize but do not lyse model and cellular membranes forming transient size-selective pores that convey small molecules across the membrane (13, 17). As a result, trypsin cleavage appears to trigger the exposure of membrane-interactive domains of VP5* to the lipid bilayer, and membrane-interactive VP5* domains facilitate membrane permeability. Mutagenesis of an internal hydrophobic domain of VP5 abolished membrane permeability and further indicated that the hydrophobic domain directed pore formation and membrane permeability (17, 40).
It has been suggested that VP5*'s role in the entry process is the direct disruption of membranes following virion uncoating, thereby permitting virion access to the cytoplasm (46). However, the lack of VP5*-directed membrane lysis argues against this process (17). Instead, the selective membrane permeability directed by expressed VP5 fosters a role for viral VP5* in lowering early endocytic calcium levels surrounding virions to effect uncoating (17). This suggests that VP5* initiates calcium efflux from the early endosomes through transient size-selective pores. Although it is still unclear how rotaviruses cross the plasma membrane, the importance of VP5* in virion membrane interactions is clear and the interactions that direct VP5* binding to membranes need to be defined.
In order to evaluate the physical interactions of VP5 with membranes, we initiated studies of fundamental VP5 membrane contacts. However, our initial findings suggested that the hydrophobic domain was not solely responsible for VP5-membrane interactions, even though it was required for pore formation. Our findings demonstrate that inserting a charged residue into the VP5* hydrophobic domain had no effect on the association of VP5 with membrane fractions, although it disrupted membrane permeability directed by VP5. Since this is the only hydrophobic, potentially membrane-spanning domain within VP5, our results suggest that the protein is not integrally inserted into membranes and that additional domains are likely to be involved in the association of the protein with membranes. Our findings demonstrate that VP5 is peripherally, and not integrally, associated with mammalian cell membranes and the peripheral membrane association of VP5 is not altered by the addition of a charged residue to the hydrophobic domain.
We previously demonstrated that deletion of residues 265 to 279 of VP5 also blocked VP5-directed membrane permeability (17). In addition, the VP5 domain from residues 265 to 279 is highly conserved, and we hypothesized that basic residues within this domain could direct VP5 interactions with negatively charged polar head groups of the lipid bilayer. Our results indicate that deleting residues 265 to 279 completely abolished VP5's association with membranes and that alanine mutagenesis of the two basic residues within this domain reduced VP5's peripheral membrane association by >95% (Fig. 4). Both deletion and mutation of this region also abolished VP5-directed membrane permeability. These findings suggest that basic residues (residues 274 and 276) within theVP5 N-terminal domain from residues 265 to 279 direct VP5 membrane binding. Although we cannot exclude the possibility that these residues cause structural changes in VP5 conformation (15) required for membrane binding, our results are consistent with the direct interaction of these residues with negatively charged polar head groups of the lipid bilayer. These findings suggest a primary role for the N-terminal basic domain of VP5 in membrane interactions that precede and direct interactions of the hydrophobic domain and pore formation.
Our findings also suggest that VP5-induced permeability is fundamentally different from that provided by ion-specific channels which are integrally inserted into membranes. The ability of peripherally associated membrane proteins, like VP5, to permeabilize membranes has been demonstrated (5, 18, 47). Peripherally associated polypeptides have been shown to bind bilayers via amphiphilic sheets or helices and cause membrane permeabilization without significant penetration of the lipid bilayer (18, 47). Alternatively, peripherally associated polypeptides may adopt additional secondary and tertiary structures when the polypeptides are associated with membranes that expose previously hidden hydrophobic regions to the lipophilic membrane interior. These “lipidic” pores are localized, semistable structural defects in the lipid bilayer that flicker open and closed, termed fusion pore flickering, and the initial sizes of these pores are similar to the size of ion channels (5, 41).
VP5-induced permeability and its peripheral membrane association are consistent with the formation of lipid pores. VP5 is likely to cause pore flickering, since VP5-directed permeability is transient, VP5 is peripherally associated with membranes, and VP5 does not lyse cellular or model membranes (13, 17). The N-terminal basic domain of VP5 appears to direct its peripheral membrane association and may effect protein conformational changes that permit the VP5 hydrophobic domain to impact the bilayer. Our results demonstrate that both domains are required for membrane permeability.
The interaction of VP4 with membrane rafts has recently been presented and hypothesized to mediate the assembly and targeting of rotaviruses within cells (54). In contrast to caveolin controls, our findings indicate that expressed VP5 (residues 248 to 474 of VP4) does not associate with Triton X-100-resistant lipid rafts and further demonstrate the peripheral rather than integral association of VP5 with membranes. Reported VP4-raft interactions were speculated to occur through the hydrophobic domain or a putative caveolin binding sequence at residues 287 to 296 of VP4 (54). The VP5N248 fragment we expressed contains both these regions, but we did not detect any interaction of VP5N248 with rafts and demonstrated that caveolin-containing rafts were present in cells. As a result, neither the hydrophobic domain nor the suggested caveolin binding domain appears to mediate the interactions of VP5 with rafts or caveolin. This confirms our previous report demonstrating that VP5 permeabilization of model membranes did not require cholesterol and suggests that if VP4 is raft associated, domains outside the region from residues 248 to 474 may mediate these interactions (13). This does not preclude the possibility that other rotavirus strains may have altered mechanisms for binding membranes. The suggested integrin binding domain is also present in the expressed VP5N248 protein (residues 302 to 315) and although it is not required for VP5 interactions with membranes, VP5-integrin interactions could facilitate the binding of adjacent N-terminal basic domain residues to the bilayer and enhance pore formation (1, 9).
Our findings demonstrate that at least two domains within VP5 direct membrane interactions. These data suggest that VP5 contains an N-terminal basic domain that mediates its peripheral membrane association and an internal hydrophobic domain that is required for membrane permeability but does not appear to direct peripheral membrane interactions. It is likely that trypsin cleavage of VP4 (following residues 231, 241, and 247) permits the association of adjacent N-terminal basic residues (residues 274 and 276) of VP5 with early endosomal membranes and that this association permits permeabilizing interactions of the hydrophobic domain. These findings suggest a functional role for two discrete VP5 domains in binding to and permeabilizing early endosomes in order to facilitate virion uncoating and viral entry.
Acknowledgments
We thank Yu Son Chong and Karen Endriss for technical assistance and Tracy Raymond and Adrish Sen for critical reading of the manuscript.
This work was supported in part by a Merit Award from the Veterans Administration and by NIH grants AI047873 and AI044917.
REFERENCES
- 1.Arias, C. F., P. Isa, C. A. Guerrero, E. Mendez, S. Zarate, T. Lopez, R. Espinosa, P. Romero, and S. Lopez. 2002. Molecular biology of rotavirus cell entry. Arch. Med. Res. 33:356-361. [DOI] [PubMed] [Google Scholar]
- 2.Bass, D. M., M. R. Baylor, C. Chen, E. R. Mackow, M. Bremont, and H. B. Greenberg. 1992. Liposome-mediated transfection of intact viral particles reveals that plasma membrane penetration determines permissivity of tissue culture cells to rotavirus. J. Clin. Investig. 90:2313-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 4.Brown, D. A., and E. London. 2000. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275:17221-17224. [DOI] [PubMed] [Google Scholar]
- 5.Chanturiya, A., L. V. Chernomordik, and J. Zimmerberg. 1997. Flickering fusion pores comparable with initial exocytotic pores occur in protein-free phospholipid bilayers. Proc. Natl. Acad. Sci. USA 94:14423-14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chemello, M. E., O. C. Aristimuno, F. Michelangeli, and M. C. Ruiz. 2002. Requirement for vacuolar H+-ATPase activity and Ca2+ gradient during entry of rotavirus into MA104 cells. J. Virol. 76:13083-13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S. R., X. Li, K. Ebisawa, and L. Zhang. 1997. Functional characterization of the recombinant type 3 Ca2+ release channel (ryanodine receptor) expressed in HEK293 cells. J. Biol. Chem. 272:24234-24246. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, J., J. Laporte, A. Charpilienne, and R. Scherrer. 1979. Activation of rotavirus RNA polymerase by calcium chelation. Arch. Virol. 60:177-186. [DOI] [PubMed] [Google Scholar]
- 9.Coulson, B. S., S. L. Londrigan, and D. J. Lee. 1997. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc. Natl. Acad. Sci. USA 94:5389-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford, S. E., S. K. Mukherjee, M. K. Estes, J. A. Lawton, A. L. Shaw, R. F. Ramig, and B. V. Prasad. 2001. Trypsin cleavage stabilizes the rotavirus VP4 spike. J. Virol. 75:6052-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuadras, M. A., C. F. Arias, and S. Lopez. 1997. Rotaviruses induce an early membrane permeabilization of MA104 cells and do not require a low intracellular Ca2+ concentration to initiate their replication cycle. J. Virol. 71:9065-9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.den Boon, J. A., J. Chen, and P. Ahlquist. 2001. Identification of sequences in Brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denisova, E., W. Dowling, R. LaMonica, R. Shaw, S. Scarlata, F. Ruggeri, and E. R. Mackow. 1999. Rotavirus capsid protein VP5* permeabilizes membranes. J. Virol. 73:3147-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dormitzer, P. R., and H. B. Greenberg. 1992. Calcium chelation induces a conformational change in recombinant herpes simplex virus-1-expressed rotavirus VP7. Virology 189:828-832. [DOI] [PubMed] [Google Scholar]
- 15.Dormitzer, P. R., H. B. Greenberg, and S. C. Harrison. 2001. Proteolysis of monomeric recombinant rotavirus VP4 yields an oligomeric VP5* core. J. Virol. 75:7339-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dormitzer, P. R., H. B. Greenberg, and S. C. Harrison. 2000. Purified recombinant rotavirus VP7 forms soluble, calcium-dependent trimers. Virology 277:420-428. [DOI] [PubMed] [Google Scholar]
- 17.Dowling, W., E. Denisova, R. LaMonica, and E. R. Mackow. 2000. Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J. Virol. 74:6368-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durell, S. R., I. Martin, J. M. Ruysschaert, Y. Shai, and R. Blumenthal. 1997. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol. Membr. Biol. 14:97-112. [DOI] [PubMed] [Google Scholar]
- 19.Egger, M., A. Ruknudin, P. Lipp, P. Kofuji, W. J. Lederer, D. H. Schulze, and E. Niggli. 1999. Functional expression of the human cardiac Na+/Ca2+ exchanger in Sf9 cells: rapid and specific Ni2+ transport. Cell Calcium 25:9-17. [DOI] [PubMed] [Google Scholar]
- 20.Espejo, R. T., S. Lopez, and C. Arias. 1981. Structural polypeptides of simian rotavirus SA11 and the effect of trypsin. J. Virol. 37:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Estes, M. K. 1996. Rotaviruses and their replication, p. 1625-1655. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Press, Philadelphia, Pa.
- 22.Estes, M. K., D. Y. Graham, and B. B. Mason. 1981. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 39:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falconer, M. M., J. M. Gilbert, A. M. Roper, H. B. Greenberg, and J. S. Gavora. 1995. Rotavirus-induced fusion from without in tissue culture cells. J. Virol. 69:5582-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Field, F. J., E. Born, S. Murthy, and S. N. Mathur. 1998. Caveolin is present in intestinal cells: role in cholesterol trafficking? J. Lipid Res. 39:1938-1950. [PubMed] [Google Scholar]
- 25.Fiore, L., H. B. Greenberg, and E. R. Mackow. 1991. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology 181:553-563. [DOI] [PubMed] [Google Scholar]
- 26.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert, J., and H. Greenberg. 1998. Cleavage of rhesus rotavirus VP4 after arginine 247 is essential for rotavirus-like particle-induced fusion from without. J. Virol. 72:5323-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenberg, H. B., J. Flores, A. R. Kalica, R. G. Wyatt, and R. Jones. 1983. Gene coding assignments for growth restriction, neutralization and subgroup specificities of the Wa and DS-1 strains of human rotavirus. J. Gen. Virol. 64:313-320. [DOI] [PubMed] [Google Scholar]
- 29.Guerrero, C. A., E. Mendez, S. Zarate, P. Isa, S. Lopez, and C. F. Arias. 2000. Integrin αvβ3 mediates rotavirus cell entry. Proc. Natl. Acad. Sci. USA 97:14644-14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guzzi, F., E. Celano, G. Levi, and M. Parenti. 2000. Interaction between HIV-1 NEF and Go proteins in transfected COS-7 cells. Biochem. Biophys. Res. Commun. 270:570-575. [DOI] [PubMed] [Google Scholar]
- 31.Hewish, M. J., Y. Takada, and B. S. Coulson. 2000. Integrins α2β1 and α4β1 can mediate SA11 rotavirus attachment and entry into cells. J. Virol. 74:228-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalica, A. R., J. Flores, and H. B. Greenberg. 1983. Identification of the rotaviral gene that codes for hemagglutination and protease-enhanced plaque formation. Virology 125:194-205. [DOI] [PubMed] [Google Scholar]
- 33.Kaljot, K. T., R. D. Shaw, D. H. Rubin, and H. B. Greenberg. 1988. Infectious rotavirus enters cells by direct cell membrane penetration, not by endocytosis. J. Virol. 62:1136-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapikian, A. Z., and R. M. Chanock. 1996. Rotaviruses, p. 1657-1708. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa.
- 35.Keljo, D. J., M. Kuhn, and A. Smith. 1988. Acidification of endosomes is not important for the entry of rotavirus into the cell. J. Pediatr. Gastroenterol. Nutr. 7:257-263. [DOI] [PubMed] [Google Scholar]
- 36.Lawton, J. A., M. K. Estes, and B. V. Prasad. 1997. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat. Struct. Biol. 4:118-121. [DOI] [PubMed] [Google Scholar]
- 37.London, E., and D. A. Brown. 2000. Insolubility of lipids in Triton X-100: physical origin and relationship to sphingolipid/cholesterol membrane domains (rafts). Biochim. Biophys. Acta 1508:182-195. [DOI] [PubMed] [Google Scholar]
- 38.Londrigan, S. L., M. J. Hewish, M. J. Thomson, G. M. Sanders, H. Mustafa, and B. S. Coulson. 2000. Growth of rotaviruses in continuous human and monkey cell lines that vary in their expression of integrins. J. Gen. Virol. 81:2203-2213. [DOI] [PubMed] [Google Scholar]
- 39.Mackow, E. R., J. W. Barnett, H. Chan, and H. B. Greenberg. 1989. The rhesus rotavirus outer capsid protein VP4 functions as a hemagglutinin and is antigenically conserved when expressed by a baculovirus recombinant. J. Virol. 63:1661-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackow, E. R., R. D. Shaw, S. M. Matsui, P. T. Vo, M. N. Dang, and H. B. Greenberg. 1988. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc. Natl. Acad. Sci. USA 85:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melikov, K. C., V. A. Frolov, A. Shcherbakov, A. V. Samsonov, Y. A. Chizmadzhev, and L. V. Chernomordik. 2001. Voltage-induced nonconductive pre-pores and metastable single pores in unmodified planar lipid bilayer. Biophys. J. 80:1829-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nandi, P., A. Charpilienne, and J. Cohen. 1992. Interaction of rotavirus particles with liposomes. J. Virol. 66:3363-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Offit, P. A., and G. Blavat. 1986. Identification of the two rotavirus genes determining neutralization specificities. J. Virol. 57:376-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Offit, P. A., H. F. Clark, G. Blavat, and H. B. Greenberg. 1986. Reassortant rotaviruses containing structural proteins vp3 and vp7 from different parents induce antibodies protective against each parental serotype. J. Virol. 60:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Mahoney, J. V., and T. E. Adams. 1994. Optimization of experimental variables influencing reporter gene expression in hepatoma cells following calcium phosphate transfection. DNA Cell Biol. 13:1227-1232. [DOI] [PubMed] [Google Scholar]
- 46.Perez, J. F., M. C. Ruiz, M. E. Chemello, and F. Michelangeli. 1999. Characterization of a membrane calcium pathway induced by rotavirus infection in cultured cells. J. Virol. 73:2481-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polozov, I. V., G. M. Anantharamaiah, J. P. Segrest, and R. M. Epand. 2001. Osmotically induced membrane tension modulates membrane permeabilization by class L amphipathic helical peptides: nucleation model of defect formation. Biophys. J. 81:949-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prasad, B. V., J. W. Burns, E. Marietta, M. K. Estes, and W. Chiu. 1990. Localization of VP4 neutralization sites in rotavirus by three-dimensional cryo-electron microscopy. Nature 343:476-479. [DOI] [PubMed] [Google Scholar]
- 49.Prasad, B. V., and W. Chiu. 1994. Structure of rotavirus. Curr. Top. Microbiol. Immunol. 185:9-29. [DOI] [PubMed] [Google Scholar]
- 50.Prasad, B. V., R. Rothnagel, C. Q. Zeng, J. Jakana, J. A. Lawton, W. Chiu, and M. K. Estes. 1996. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature 382:471-473. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz, M. C., S. R. Alonso-Torre, A. Charpilienne, M. Vasseur, F. Michelangeli, J. Cohen, and F. Alvarado. 1994. Rotavirus interaction with isolated membrane vesicles. J. Virol. 68:4009-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruiz, M. C., A. Charpilienne, F. Liprandi, R. Gajardo, F. Michelangeli, and J. Cohen. 1996. The concentration of Ca2+ that solubilizes outer capsid proteins from rotavirus particles is dependent on the strain. J. Virol. 70:4877-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruiz, M. C., J. Cohen, and F. Michelangeli. 2000. Role of Ca2+ in the replication and pathogenesis of rotavirus and other viral infections. Cell Calcium 28:137-149. [DOI] [PubMed] [Google Scholar]
- 54.Sapin, C., O. Colard, O. Delmas, C. Tessier, M. Breton, V. Enouf, S. Chwetzoff, J. Ouanich, J. Cohen, C. Wolf, and G. Trugnan. 2002. Rafts promote assembly and atypical targeting of a nonenveloped virus, rotavirus, in Caco-2 cells. J. Virol. 76:4591-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scherer, P. E., R. Y. Lewis, D. Volonte, J. A. Engelman, F. Galbiati, J. Couet, D. S. Kohtz, E. van Donselaar, P. Peters, and M. P. Lisanti. 1997. Cell-type and tissue-specific expression of caveolin-2. Caveolins 1 and 2 co-localize and form a stable hetero-oligomeric complex in vivo. J. Biol. Chem. 272:29337-29346. [DOI] [PubMed] [Google Scholar]
- 56.Sei, Y., K. L. Gallagher, and A. S. Basile. 1999. Skeletal muscle type ryanodine receptor is involved in calcium signaling in human B lymphocytes. J. Biol. Chem. 274:5995-6002. [DOI] [PubMed] [Google Scholar]
- 57.Shaw, R. D., P. T. Vo, P. A. Offit, B. S. Coulson, and H. B. Greenberg. 1986. Antigenic mapping of the surface proteins of rhesus rotavirus. Virology 155:434-451. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki, H., S. Kitaoka, T. Konno, T. Sato, and N. Ishida. 1985. Two modes of human rotavirus entry into MA 104 cells. Arch. Virol. 85:25-34. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki, H., S. Kitaoka, T. Sato, T. Konno, Y. Iwasaki, Y. Numazaki, and N. Ishida. 1986. Further investigation on the mode of entry of human rotavirus into cells. Arch. Virol. 91:135-144. [DOI] [PubMed] [Google Scholar]
- 60.Tihova, M., K. A. Dryden, A. R. Bellamy, H. B. Greenberg, and M. Yeager. 2001. Localization of membrane permeabilization and receptor binding sites on the VP4 hemagglutinin of rotavirus: implications for cell entry. J. Mol. Biol. 314:985-992. [DOI] [PubMed] [Google Scholar]
- 61.Zarate, S., R. Espinosa, P. Romero, C. A. Guerrero, C. F. Arias, and S. Lopez. 2000. Integrin α2β1 mediates the cell attachment of the rotavirus neuraminidase-resistant variant nar3. Virology 278:50-54. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, J., and R. A. Lamb. 1996. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology 225:255-266. [DOI] [PubMed] [Google Scholar]