Summary
Intellectual disability is a prevalent disorder that remains incurable. Mutations of the X-linked protein PHF6 cause the intellectual disability disorder Börjeson–Forssman–Lehmann syndrome (BFLS). However, the biological role of PHF6 relevant to BFLS pathogenesis has remained unknown. We report that knockdown of PHF6 profoundly impairs neuronal migration in the mouse cerebral cortex in vivo, leading to the formation of white matter heterotopias displaying neuronal hyperexcitability. We find that PHF6 physically associates with the PAF1 transcription elongation complex, and inhibition of PAF1 phenocopies the PHF6 knockdown-induced migration phenotype in vivo. We also identify Neuroglycan C/Chondroitin sulfate proteoglycan 5 (NGC/CSPG5), a potential schizophrenia susceptibility gene, as a critical downstream target of PHF6 in the control of neuronal migration. These findings define PHF6, PAF1, and NGC/CSPG5 as components of a novel cell-intrinsic transcriptional pathway that orchestrates neuronal migration in the brain, with important implications for the pathogenesis of developmental disorders of cognition.
Introduction
Intellectual disability is a common developmental disorder affecting 1 to 3 percent of the general population (Bhasin et al., 2006). The economic costs of intellectual disability are enormous. No effective treatments are available for intellectual disability, and thus there is an urgent need for improved understanding of these disorders. In recent years, mutations in many genes have been identified that cause intellectual disability, but how these mutations trigger intellectual disability remains largely to be elucidated.
Mutations of the X-linked gene encoding the protein PHF6 cause the Börjeson–Forssman–Lehmann syndrome (BFLS), characterized by moderate to severe intellectual disability associated variably with seizures (Lower et al., 2002). However, the function of PHF6 relevant to BFLS pathogenesis has remained unknown. Cognitive dysfunction is evident from a very early age, suggesting that abnormal brain development contributes to intellectual disability in BFLS patients (Turner et al., 2004). Therefore, understanding PHF6’s role during brain development should provide important insights into the pathogenesis of BFLS.
In this study, we have discovered a novel function for the X-linked intellectual disability protein PHF6 in the development of the cerebral cortex in vivo. Loss of PHF6 profoundly impairs neuronal migration, and importantly, this impairment persists beyond the developmental stage of corticogenesis, leading to white matter heterotopias with aberrant neuronal activity. We have also identified an intimate link between PHF6 and the PAF1 transcription elongation complex that plays an essential role in neuronal migration in the cerebral cortex. Finally, we have identified Neuroglycan C/Chondroitin sulfate proteoglycan 5 (NGC/CSPG5) as a downstream target of PHF6 and the PAF1 complex in the control of neuronal migration. Our findings define a novel pathophysiologically relevant cell-intrinsic transcriptional pathway that orchestrates neuronal migration in the cerebral cortex.
Results
PHF6 plays an essential role in radial neuronal migration in the cerebral cortex in vivo
To interrogate PHF6 function in the mammalian brain, we first characterized the expression of PHF6 in the developing cerebral cortex. We found that PHF6 was highly expressed during early phases of development in primary cortical neurons and in the developing mouse brain (Figures S1A and S1B). PHF6 was broadly expressed in the mouse cerebral cortex at embryonic day 17 (E17) (Figure S1C). The temporal profile of PHF6 expression raised the possibility that PHF6 might play a role in cortical development.
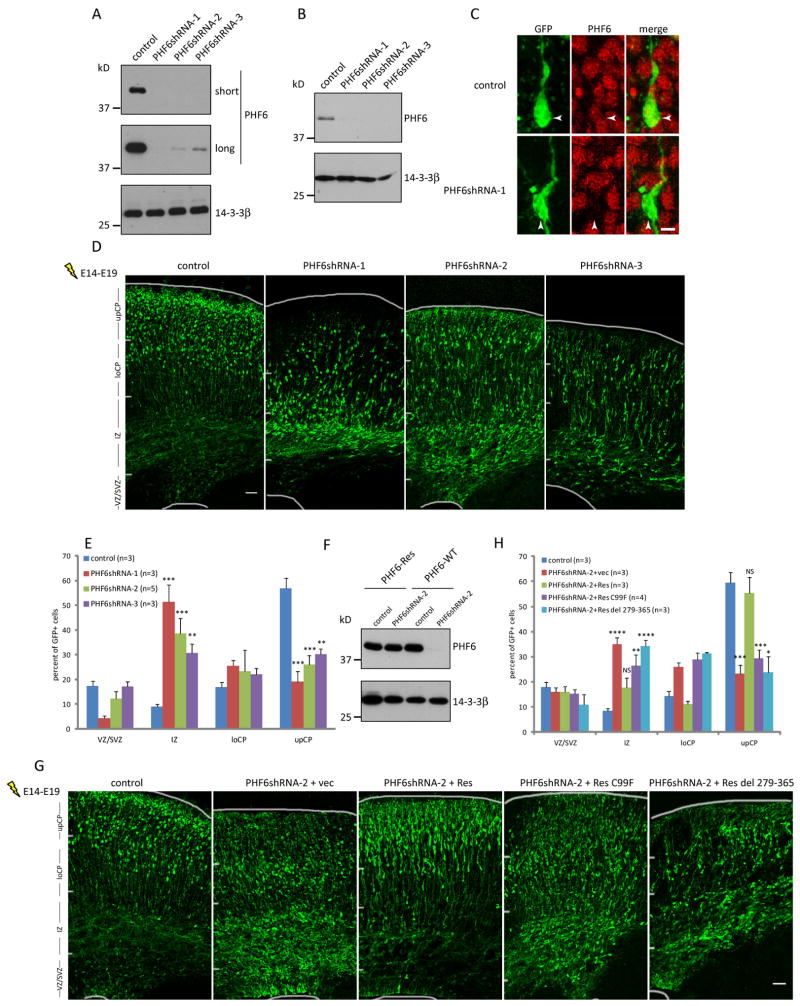
To determine PHF6 function in cortical development, we used a plasmid-based method of RNA interference (RNAi) to acutely knockdown PHF6 in the developing cerebral cortex (Gaudilliere et al., 2002). Expression of three short hairpin RNAs (shRNAs) targeting distinct regions of PHF6 mRNA induced knockdown of exogenous PHF6 protein in 293T cells and endogenous PHF6 in primary mouse cortical neurons (Figures 1A, 1B, S1D, and S1E).
Fig. 1. PHF6 plays an essential role in radial neuronal migration in the cerebral cortex in vivo.
(A) Lysates of 293T cells transfected with an expression plasmid encoding FLAG-PHF6 together with an RNAi plasmid encoding PHF6 shRNAs or control scrambled shRNAs were immunoblotted with the FLAG and 14-3-3β antibodies. Two exposures (short and long) are shown for PHF6. (B) Lysates of primary mouse cortical neurons infected with lentivirus expressing PHF6 shRNAs or control shRNAs were immunoblotted with the PHF6 and 14-3-3β antibodies. (C) E14 mouse embryos electroporated with an RNAi plasmid encoding PHF6 shRNAs or control shRNAs together with a GFP expression plasmid were allowed to develop until E19. The cerebral cortex from the E19 embryos was subjected to immunohistochemical analyses with the PHF6 and GFP antibodies. PHF6 RNAi induced robust downregulation of PHF6 in cortical neurons in vivo (arrow heads). (D) E14 mouse embryos were subjected to in utero electroporation as in (C) and the cerebral cortex of E19 embryos was analyzed by immunohistochemistry using the GFP antibody. The upper cortical plate (upCP), lower cortical plate (loCP), intermediate zone (IZ), ventricular zone/sub-ventricular zone (VZ/SVZ) are indicated. (E) Quantification of the percentage of GFP positive neurons in distinct regions of the cerebral cortex in E19 mouse embryos treated as in (D) is presented as mean + SEM. (F) Lysates of 293T cells transfected with an expression plasmid encoding FLAG-rat PHF6 (PHF6-Res) or FLAG-mouse PHF6 (PHF6-WT) together with the PHF6 RNAi or control RNAi plasmid were immunoblotted with the FLAG and 14-3-3β antibodies. (G) E14 embryos were electroporated and analyzed as in (D) with the PHF6 RNAi or control RNAi plasmid together with an expression plasmid encoding wild type or patient mutant (C99F or deletion 279-365) PHF6-Res protein or the corresponding control vector. vec, control vector; Res, PHF6-Res: del, deletion. (H) Quantification of the percentage of GFP positive neurons in distinct regions of the cerebral cortex in E19 mouse embryos treated as in (G) is presented as mean + SEM. Expression of PHF6-Res significantly reduced the percentage of neurons stalled in the intermediate zone and significantly increased the percentage of neurons that reached the upper cortical plate in the background of PHF6 RNAi in the cerebral cortex in vivo (ANOVA, p<0.05). The C99F mutation or deletion of amino acids 279-365 blocked the ability of PHF6-Res to drive neuronal migration. See also Figure S1. Scale bars: 3μm in (C); 50μm in (D) and (G).
We next employed an in utero electroporation method to induce knockdown of PHF6 in the developing mouse cerebral cortex in vivo. The PHF6 RNAi plasmids were electroporated together with a plasmid encoding GFP in the developing cortex in mice at E14, when superficial layer neurons are generated. Embryos were allowed to develop in utero until E19, and brains were harvested and subjected to immunohistochemical analyses. We first confirmed that PHF6 RNAi triggered the downregulation of endogenous PHF6 in the cerebral cortex in vivo (Figures 1C, S1F). Upon characterizing the consequences of PHF6 knockdown on cortical development, we found a striking migration phenotype. Neurons in control animals differentiated and migrated properly to the superficial layers of the cortical plate. By contrast, cortical neurons in PHF6 knockdown animals failed to migrate to the proper location in the upper cortical plate (Figures 1D and 1E). PHF6 RNAi reduced the percentage of neurons reaching the upper cortical plate by 2- to 3-fold, and increased the percentage of neurons in the intermediate zone by 3- to 5-fold. The extent of the migration defect correlated with the degree of PHF6 knockdown (Figure 1A). In control analyses, PHF6 knockdown had little or no effect on neuronal differentiation (Figure S1H, and data not shown), progenitor cell proliferation (Figure S1I), general cell health (Figures S1J and S1K), or morphology of radial glial cells (data not shown). Together, these results suggest that PHF6 knockdown specifically impairs the radial migration of neurons in the developing cerebral cortex.
To determine whether the PHF6 RNAi-induced migration phenotype is due to specific knockdown of PHF6, we performed a rescue experiment. Expression of rat PHF6 (PHF6-Res), which contains two mismatches with shRNA-2 targeting mouse PHF6, was refractory to PHF6 RNAi (Figure 1F). Importantly, expression of PHF6-Res in PHF6 knockdown animals largely restored the normal migration pattern of cortical neurons (Figures 1G and 1H). These data indicate that the PHF6 RNAi-induced migration phenotype is the result of specific knockdown of PHF6.
We next asked whether the PHF6 knockdown-induced migration phenotype is relevant to intellectual disability. The patient mutation C99F targets a conserved cysteine residue in PHF6’s first PHD domain (Lower et al., 2002), and impaired the ability of PHF6-Res to drive migration in the cerebral cortex in vivo in the background of PHF6 RNAi (Figures 1G and 1H). Likewise, deletion of the C-terminal 86 amino acids, which also causes BFLS (Berland et al., 2011), impaired PHF6-dependent neuronal migration (Figures 1G and 1H). These data suggest that impaired neuronal migration might underlie the pathogenesis of BFLS.
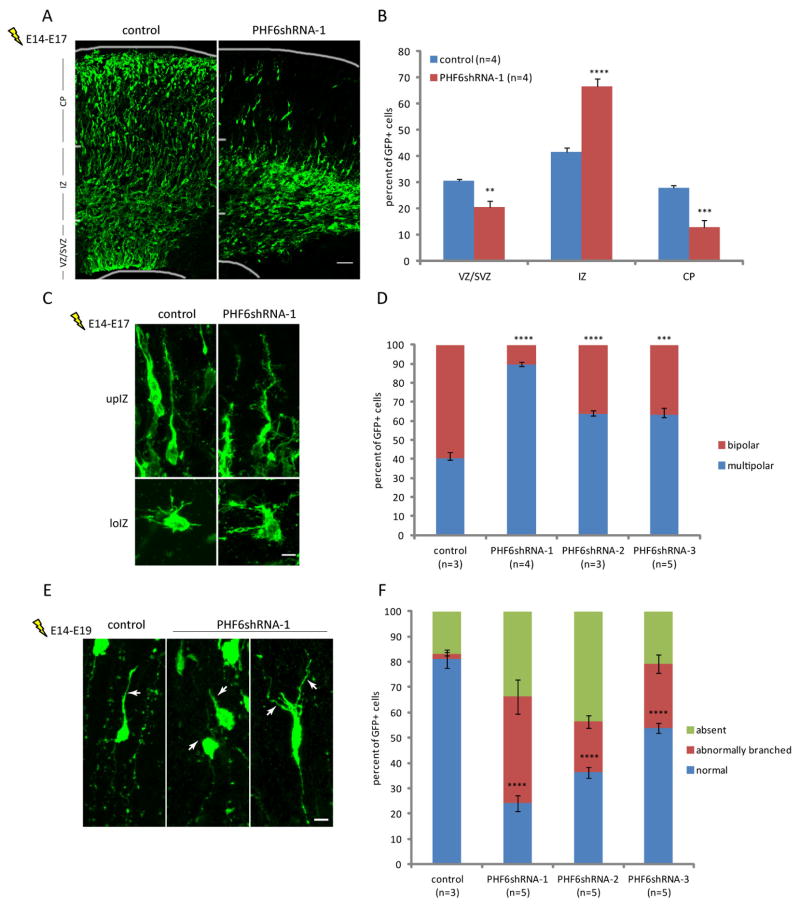
Having established a critical role of PHF6 in neuronal migration, we next asked how PHF6 functions in neurons at the cellular level. Radial neuronal migration in the cerebral cortex results from major cell morphological rearrangements, including the transition from multipolar to bipolar neuronal morphology in the intermediate zone and extension of the leading process toward the pia (Ayala et al., 2007; Kriegstein and Noctor, 2004; Nadarajah et al., 2001). Knockdown of PHF6 substantially increased the number of multipolar neurons and concomitantly reduced the number of bipolar neurons in the intermediate zone in the cerebral cortex in E17 embryos (Figures 2A, 2B and 2D). The remaining bipolar neurons in the PHF6 knockdown embryos harbored a thick leading process with numerous filopodia-like protrusions (Figure 2C). In other analyses, PHF6 knockdown dramatically increased the percentage of migrating neurons that lacked a leading process or that had a short, poorly developed, or aberrantly-branched leading process (Figures 2E and 2F). These data suggest that PHF6 plays a critical role in the multipolar-to-bipolar transition and the morphogenesis of the leading process in migrating neurons.
Fig. 2. PHF6 is critical for morphological changes during neuronal migration in the cerebral cortex in vivo.
(A) E14 embryos were electroporated as in Figure 1D and allowed to develop until E17. The cerebral cortex was then subjected to immunohistochemistry with the GFP antibody. CP, cortical plate. (B) Quantification of the percentage of GFP positive neurons in distinct regions of the cerebral cortex in E17 mouse embryos treated as in (A) is presented as mean + SEM. (C) E14 embryos were electroporated and analyzed as in (A). Representative images for neurons in the lower or upper intermediate zone are shown. loIZ, lower intermediate zone; upIZ, upper intermediate zone. (D) Quantification of the percentage of GFP positive neurons in the intermediate zone exhibiting bipolar or multipolar morphology in E17 mouse embryos treated as in (C) is presented as mean ± SEM. (E) E14 embryos were electroporated as in (A) and allowed to develop until E19. The cerebral cortex was then subjected to immunohistochemistry with the GFP antibody. Representative images of migrating neurons in the lower cortical plate are shown. The leading process is indicated by arrows. (F) Quantification of the percentage of GFP positive cells exhibiting different leading process morphologies in E19 embryos treated as in (E) is presented as mean ± SEM. Scale bar: 50μm in (A), 5μm in (C), 10μm in (E).
The PAF1 complex interacts with PHF6 and plays a critical role in radial migration of neurons in the cerebral cortex in vivo
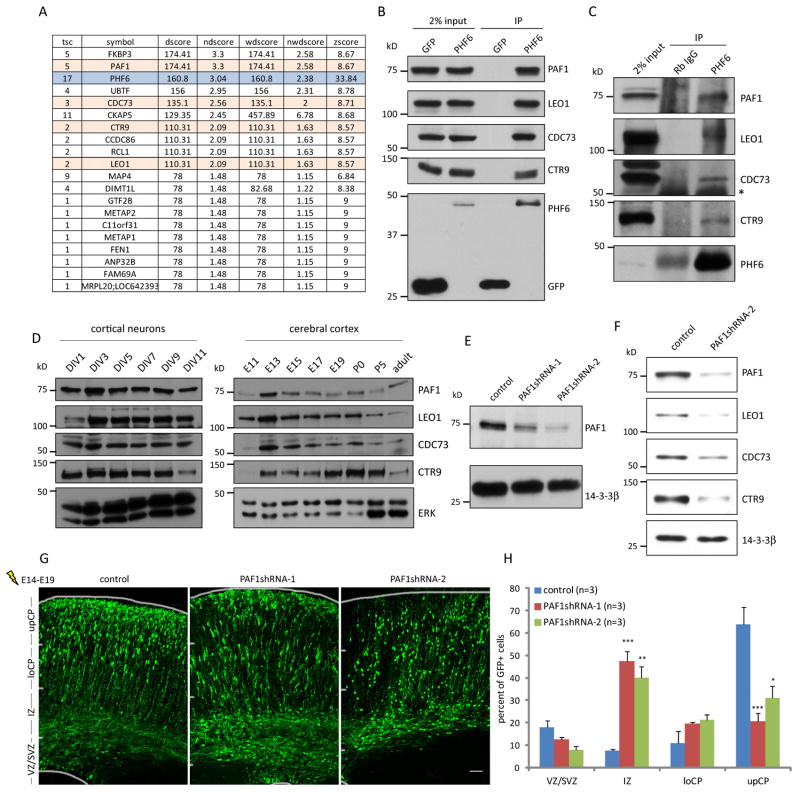
We next determined the mechanism by which PHF6 orchestrates neuronal migration. Immunocytochemical analyses suggested that PHF6 protein localized to the nucleoplasm in primary cortical neurons, consistent with the possibility that PHF6 might regulate transcription (Figure S1G) (Lower et al., 2002). Since PHF6 is not known to harbor enzymatic activity, we reasoned that PHF6 might associate with other factors that in turn regulate transcription. To identify proteins that physically associate with PHF6, we used an approach of immunoprecipitation followed by mass spectrometry (IP/MS). We used a rigorous proteomics method that compares a specific IP/MS dataset against a large set of unrelated parallel IP/MS datasets, thus distinguishing high-confidence candidate interacting proteins (HCIPs) from background proteins (Behrends et al., 2010; Litterman et al., 2011; Sowa et al., 2009). Remarkably, all four core components of the PAF1 transcription elongation complex, PAF1, LEO1, CDC73 and CTR9, were found as robust HCIPs of PHF6 (Figure 3A).
Fig. 3. The PAF1 complex interacts with PHF6 and plays a critical role in radial migration of neurons in the cerebral cortex in vivo.
(A) Lysates of 293T cells stably expressing N-terminally tagged HA-PHF6 were immunoprecipitated with the HA beads and subjected to proteomic analyses using LC-MS/MS. Peptide spectra were analyzed and scored using the CompPASS software program. High-confidence candidate interacting proteins (HCIPs) of PHF6 are shown. Rows in pink denote components of the PAF1 complex. (B) Lysates of 293T cells expressing HA-PHF6 or HA-GFP were subjected to immunoprecipitation with the HA beads followed by immunoblotting with the HA, PAF1, LEO1, CDC73 and CTR9 antibodies. Input was also immunoblotted with the antibodies. (C) Lysates of E17 mouse cerebral cortex were immunoprecipitated with the PHF6 antibody or rabbit IgG followed by immunoblotting with the PAF1, LEO1, CDC73, CTR9 and PHF6 antibodies. Asterisk denotes a non-specific band. Rabbit IgG signal from the PHF6 immunoblot is non-specific immunoreactivity from the rabbit heavy chain. (D) Lysates of primary rat cortical neurons or mouse cerebral cortex were immunoblotted with the PAF1, LEO1, CDC73, CTR9 and ERK antibodies. (E) Lysates of mouse cortical neurons infected with lentivirus expressing PAF1 shRNAs or control shRNAs were immunoblotted with the PAF1 and 14-3-3β antibodies. (F) Lysates of Neuro2A cells infected with lentivirus expressing PAF1 shRNAs or control shRNAs were immunoblotted with the PAF1, LEO1, CDC73, CTR9 and 14-3-3β antibodies. (G) E14 embryos were electroporated with an RNAi plasmid encoding PAF1 shRNAs or control shRNAs, and analyzed as in Figure 1D. (H) Quantification of the percentage of GFP positive neurons in distinct regions of the cerebral cortex in E19 mouse embryos treated as in (G) is presented as mean + SEM. Scale bar: 50μm in (G).
We validated the interaction of HA-PHF6 and the endogenous PAF1 complex in co-immunoprecipitation analyses in cells (Figure 3B). Importantly, we also found that endogenous PHF6 associated with all four components of the endogenous PAF1 complex in mouse cerebral cortex at E17, coinciding temporally with migration of neurons to the superficial layers (Figure 3C). These data suggest that the PAF1 complex might represent a physiological interacting partner of PHF6.
The PAF1 transcription elongation complex was first identified in yeast as an RNA polymerase II-associated complex, and plays a critical role in efficient transcriptional elongation along chromatin (Kim et al., 2010; Rondon et al., 2004; Shi et al., 1996). All four components of the complex were highly expressed during early development in primary cortical neurons and the cerebral cortex in vivo (Figure 3D). Intriguingly, the role of the PAF1 complex in the brain has remained unexplored.
We asked whether the PHF6-PAF1 interaction is functionally relevant in neuronal migration. We reasoned that if PHF6 acts via the PAF1 complex to regulate neuronal migration, loss of PAF1 would be predicted to disrupt neuronal migration. Consistent with this prediction, PAF1 knockdown by two distinct shRNAs substantially impaired neuronal migration in the cerebral cortex in vivo, phenocopying the effect of PHF6 knockdown (Figures 3E, 3F, 3G and 3H). Notably, knockdown of PAF1 led to downregulation of the other components of the PAF1 complex (Figure 3F) (Chen et al., 2009), suggesting that the intact PAF1 complex is required for proper neuronal migration. Collectively, these data suggest that PHF6 physically associates with the PAF1 complex and thereby drives neuronal migration.
The Neuroglycan C/Chondroitin sulfate proteoglycan 5 (NGC/CSPG5) gene is a key downstream target of PHF6 and PAF1 in the control of neuronal migration in vivo
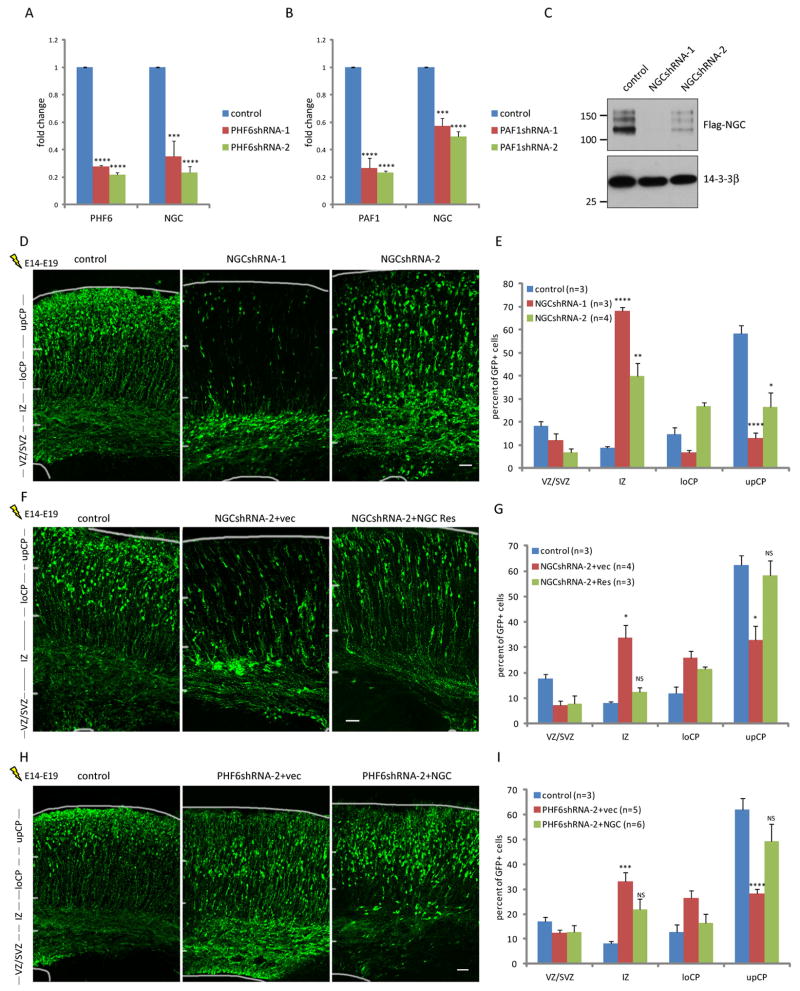
The finding that PHF6 interacts with the PAF1 transcription elongation complex and thereby promotes cortical neuronal migration led us to determine whether PHF6 exerts its function via regulation of gene expression. Because the PAF1 complex promotes transcription, we reasoned that PHF6 might stimulate the expression of genes that trigger neuronal migration. We performed microarray analyses of control and PHF6 knockdown primary cortical neurons. A large number of genes were downregulated in cortical neurons upon PHF6 knockdown (Table S1). Among downregulated genes that were validated by RT-PCR analyses, NGC/CSPG5 was the most consistently and robustly downregulated gene in PHF6 knockdown neurons (Figures 4A and S2A). NGC/CSPG5 was also robustly downregulated by PAF1 knockdown in primary cortical neurons, suggesting that NGC/CSPG5 is coordinately regulated by PHF6 and the PAF1 transcription elongation complex (Figures 4B and S2B).
Fig. 4. The Neuroglycan C/Chondroitin sulfate proteoglycan 5 (NGC/CSPG5) gene is a key downstream target of PHF6 and PAF1 in the control of neuronal migration in vivo.
(A) Quantitative RT-PCR analyses of primary rat cortical neurons infected with lentivirus expressing PHF6 shRNAs or control shRNAs. Levels of PHF6 and NGC/CSPG5 were normalized to GAPDH levels (n=3). (B) Quantitative RT-PCR analyses of primary rat cortical neurons infected with lentivirus expressing PAF1 shRNAs or control shRNAs. Levels of PAF1 and NGC/CSPG5 were normalized to GAPDH levels (n=3). (C) Lysates of 293T cells transfected with an expression vector encoding FLAG-NGC/CSPG5 together with an RNAi plasmid encoding NGC/CSPG5 shRNAs or control shRNAs were immunoblotted with the FLAG and 14-3-3β antibodies. (D) E14 mouse embryos were electroporated with an RNAi plasmid encoding NGC/CSPG5 shRNAs or control shRNAs and analyzed as in Figure 1D. (E) Quantification of the percentage of GFP positive neurons in distinct regions of the cerebral cortex in E19 mouse embryos treated as in (D) is presented as mean + SEM. Knockdown of NGC/CSPG5 also inhibited proliferation of cortical precursor cells (see Figure S2F). (F) E14 mouse embryos were electroporated with the NGC/CSPG5 RNAi or control RNAi plasmid together with an expression plasmid encoding NGC-Res or its corresponding control vector, and analyzed as in (D). (G) Quantification of the percentage of GFP positive neurons in distinct regions of the cerebral cortex in E19 mouse embryos treated as in (F) is presented as mean + SEM. Expression of NGC-Res significantly increased the percentage of neurons reaching the upper cortical plate in the background of NGC/CSPG5 RNAi in the cerebral cortex in vivo (ANOVA, p<0.05). (H) E14 mouse embryos were electroporated with the PHF6 RNAi or control RNAi plasmid together with an expression plasmid encoding NGC/CSPG5 or its corresponding control vector, and analyzed as in (D). (G) Quantification of the percentage of GFP positive neurons in distinct regions of the cerebral cortex in E19 mouse embryos treated as in (H) is presented as mean + SEM. Expression of NGC/CSPG5 significantly increased the percentage of neurons reaching the upper cortical plate in the background of PHF6 RNAi in the cerebral cortex in vivo (ANOVA, p<0.01). See also Figure S2 and Table S1. Scale bars: 50μm in (D), (F) and (G).
The NGC/CSPG5 gene is expressed in the brain (Figure S2C), and encodes a transmembrane chondroitin sulfate glycoprotein that is a member of the neuregulin family of proteins, which is implicated in neuronal migration (Kinugasa et al., 2004; Rio et al., 1997). Interestingly, the NGC/CSPG5 gene is a potential susceptibility locus in schizophrenia, in which impaired neuronal migration is thought to play a role (Impagnatiello et al., 1998; So et al., 2010). These observations raised the possibility that NGC/CSPG5 might represent a physiologically relevant downstream target of the PHF6-PAF1 pathway in the control of neuronal migration.
Knockdown of NGC/CSPG5 in mouse embryos using two distinct shRNAs impaired neuronal migration in the cerebral cortex in vivo (Figures 4C, 4D, 4E and S2F), phenocopying the PHF6 knockdown phenotype. The extent of the migration defect correlated with the efficiency of NGC/CSPG5 knockdown. Importantly, expression of an RNAi-resistant rescue form of NGC/CSPG5 suppressed the NGC/CSPG5 RNAi-induced phenotype, suggesting that the RNAi-induced migration defect is the result of specific knockdown of NGC/CSPG5 (Figures 4F, 4G and S2D). Remarkably, in epistasis analyses, expression of exogenous NGC/CSPG5 in PHF6 knockdown animals largely restored the normal migration pattern in the cerebral cortex in vivo (Figures 4H, 4I, and S2E). Together, our data suggest that NGC/CSPG5 represents a key target of PHF6 in the control of cortical neuronal migration in vivo.
PHF6 knockdown triggers the formation of white matter heterotopias with aberrant neuronal activity
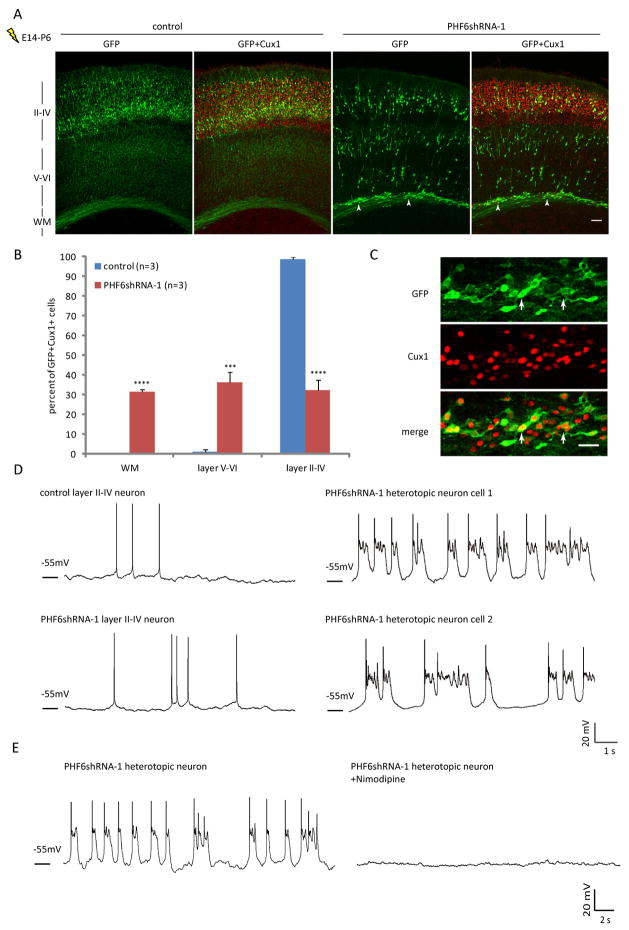
Having elucidated a mechanism by which PHF6 orchestrates neuronal migration in the developing cerebral cortex in vivo, we next addressed the question of how loss of PHF6 might contribute to the pathogenesis of BFLS. We asked whether consequences of impaired migration upon PHF6 knockdown persist beyond the formation of the cerebral cortex. We electroporated E14 mouse embryos, and examined animals at postnatal day 6 (P6). In these analyses, almost all transfected neurons in control animals resided in layers II–IV and expressed Cux1, a marker of superficial layer neurons (Nieto et al., 2004). Strikingly, neurons in PHF6 knockdown animals at P6 formed heterotopias in the white matter and were also found ectopically in layers V–VI (Figure 5A). Quantification revealed that 98% of Cux1 positive, transfected cortical neurons reached layers II–IV in control animals, whereas only 32% of Cux1 positive, transfected neurons reached the superficial layers in PHF6 knockdown animals (Figure 5B). The failure of PHF6 knockdown neurons to reach layers II–IV in P6 animals was not due to impaired differentiation, as heterotopic neurons expressed Cux1, but not the layer V marker Ctip2 (Figures 5C, S3A and S3B). These data suggest that loss of PHF6 triggers the formation of heterotopia in the cerebral cortex in vivo.
Fig. 5. PHF6 knockdown triggers the formation of white matter heterotopias with aberrant neuronal activity.
(A) E14 mouse embryos were electroporated with the RNAi plasmid encoding PHF6 shRNAs or control shRNAs together with the GFP expression plasmid and allowed to develop until P6. The cerebral cortex from the P6 mouse pups was subjected to immunohistochemical analyses with the GFP and Cux1 antibodies. Arrowheads denote heterotopic neurons in PHF6 knockdown animals. WM, white matter. (B) Quantification of the percentage of GFP positive, Cux1 positive neurons in distinct regions of the cerebral cortex in P6 mouse pups treated as in (A) is presented as mean + SEM. (C) Representative image of heterotopic neurons in the white matter in PHF6 knockdown animals. GFP positive neurons are positive for Cux1 (arrows). (D) E14 mouse embryos were electroporated as in (A) and allowed to develop until P10. Representative images for whole-cell recordings in current clamp of GFP positive neuron in cortical slices from P10 PHF6 knockdown or control mouse pups are shown. (E) Whole-cell recordings were performed as in (D). Abnormal firing patterns of heterotopic neurons are blocked by bath application of 10μM nimodipine. Left and right panels are recordings from the same neuron before and after adding the inhibitor. See also Figure S3. Scale bars: 100μm in (A); 30μm in (C).
We next examined the electrophysiological properties of transfected neurons in acute cortical slices prepared from P10 control or PHF6 knockdown animals. Under current clamp configuration, we observed an aberrant pattern of activity in 70% of heterotopic neurons, but not in neurons that reached layers II–IV, in PHF6 knockdown animals (Figure 5D). The membrane potential of heterotopic neurons oscillated, leading to frequent action potentials. Spontaneous excitatory postsynaptic currents (sEPSCs) were observed in layer II–IV neurons in control or PHF6 knockdown animals but markedly reduced in heterotopic neurons in PHF6 knockdown animals, suggesting that the membrane potential of heterotopic neurons may oscillate in the absence of synaptic inputs (Figures S3C, S3D and S3E). The membrane potential oscillation in heterotopic neurons was blocked by nimodipine, suggesting that calcium currents might underlie the spontaneous depolarization (Figure 5E). In other experiments, knockdown of NGC/CSPG5 in E14 mouse embryos led to the formation of white matter heterotopias in P10 mouse pups, which harbored a similar pattern of neuronal activity as heterotopias in PHF6 knockdown animals (Figure S3F). Together, these data suggest that inhibition of the PHF6 pathway triggers the formation of heterotopias in which neurons are hyperexcitable.
Collectively, we have identified a transcriptional pathway whereby the X-linked intellectual disability protein PHF6 forms a complex with the PAF1 transcription elongation complex and thereby induces the expression of NGC/CSPG5, leading to the migration of cortical neurons in the cerebral cortex. Deregulation of this pathway may play a critical role in the pathogenesis of intellectual disability and epilepsy in BFLS.
Discussion
In this study, we have discovered an essential function for the intellectual disability protein PHF6 in the development of the cerebral cortex. Loss of PHF6 impairs neuronal migration and leads to formation of heterotopia, accompanied by aberrant neuronal activity patterns. We have also uncovered the mechanism by which PHF6 orchestrates neuronal migration in the cerebral cortex. PHF6 physically associates with the PAF1 transcription elongation complex, and the PAF1 complex is required for neuronal migration. We have also identified NGC/CSPG5, a potential susceptibility gene for schizophrenia, as a critical downstream target of PHF6 and the PAF1 complex that mediates PHF6-dependent neuronal migration. Together, our data define PHF6, the PAF1 complex, and NGC/CSPG5 as components of a novel cell-intrinsic transcriptional pathway that promotes neuronal migration in the cerebral cortex with pathophysiological relevance to intellectual disability and epilepsy.
The identification of PHF6 function in neuronal migration in the cerebral cortex advances our understanding of the pathogenesis of intellectual disability in the Börjeson–Forssman–Lehmann syndrome (BFLS). We have found that the consequences of impaired migration upon PHF6 inhibition persist beyond corticogenesis. Consistent with our findings, rare neuropathological studies of BFLS patients have revealed cortical dysplasia, absent lamination, and white matter heterotopia (Brun et al., 1974). Therefore, impaired neuronal migration and associated heterotopias may play a key role in the pathogenesis of intellectual disability in BFLS. In view of our findings, it will be important to perform detailed imaging to characterize potential heterotopias in BFLS patients. Heterotopias are associated with epilepsy (Ackman et al., 2009). Therefore, the hyperexcitability of heterotopic PHF6 knockdown neurons suggests the possibility that heterotopias may also contribute to epilepsy in BFLS.
The finding that the PAF1 complex interacts with PHF6 and promotes neuronal migration in the cerebral cortex illuminates a novel biological role for the PAF1 complex. Regulation of transcriptional elongation is a fundamental aspect of gene expression control (Levine, 2011; Muse et al., 2007). Our findings suggest that the control of transcriptional elongation may represent a critical point of regulation in neuronal migration, with relevance to intellectual disability and epilepsy. Both PHF6 and the PAF1 complex have been implicated in the pathogenesis of leukemia (Muntean et al., 2010; Van Vlierberghe et al., 2010). Thus, our findings linking PHF6 and the PAF1 complex may also have ramifications in cancer biology.
The identification of NGC/CSPG5 as a key target gene of PHF6 and PAF1 has implications for the biology of both NGC/CSPG5 and PHF6. Since NGC/CSPG5 might represent a potential locus for schizophrenia (So et al., 2010), our findings raise the possibility that mutations of PHF6 may contribute to the pathogenesis of neuropsychiatric disorders. Conversely, it will be interesting to determine whether deregulation of NGC/CSPG5 might play a role in intellectual disability and epilepsy. Intriguingly, NGC/CSPG5 may have an additional function in neural progenitor cell proliferation (Figure S2F), though whether this potential function is regulated independently of PHF6 or relevant to brain disorders remains to be determined. Because NGC/CSPG5 is a transmembrane protein and a member of the neuregulin family that can directly bind ErbB3 and transactivate ErbB2 (Kinugasa et al., 2004), NGC/CSPG5 signaling might represent an attractive drug target in the treatment of intellectual disability.
Experimental Procedures
Animals
Timed pregnant CD-1 mice were purchased from Charles River Laboratories. All animal experiments were conducted under the institutional guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC).
Plasmids
The PHF6 and NGC/CSPG5 expression plasmids were generated by PCR using mouse or rat cortical neuron cDNA. The shRNA plasmid targeting sequences are described in the supplementary information.
In utero electroporation
In utero electroporations were performed as described (Ge et al., 2010). Briefly, venus plasmid (final concentration 0.5ug/ul) was co-injected with the RNAi plasmid (final concentration 2ug/ul) into the lateral ventricle of E14 mouse embryos within the uterine sac, and electroporation was performed (35V for 50ms, with 950ms intervals, 6 pulses). The uterine sac was then returned to the abdominal cavity and the abdominal wall was sutured. E19 embryos were harvested and fixed in 4% paraformaldehyde for 1hr. P6 brains were fixed by intracardiac perfusion using 4% paraformaldehyde followed by post-fixation in 4% paraformaldehyde for 2hr. After fixation, brains were subjected to cryoprotection in 30% sucrose in PBS overnight. Afterwards, brains were embedded in OCT and flash-frozen, and 20μm coronal sections were prepared. At least 3 animals were analyzed for each condition. Immunohistochemical analyses were then performed at indicated time points as described (Ge et al., 2010).
Immunoprecipitation/mass spectrometry
To identify specific PHF6 interactors, 293T cells stably expressing N-terminally HA-tagged PHF6 were generated using lentivirus followed by antibiotic selection. Four confluent 15cm dishes of stable PHF6-expressing cells were lysed in 0.5% Nonidet P40 and subjected to immunoprecipitation using anti-HA resin (clone HA-7, Sigma). Immunoprecipitated PHF6 was eluted using HA peptide (Anaspec), and the eluted proteins were precipitated with trichloroacetic acid and digested with trypsin. The resulting tryptic peptides were desalted over C18 resin, and then loaded onto an LTQ linear ion trap mass spectrometer (Thermo Finnigan) for LC-MS/MS analyses. MS/MS spectra were searched using SEQUEST against a target-decoy database of tryptic peptides, and protein assignments were analyzed using the CompPASS software platform (Behrends et al., 2010; Litterman et al., 2011; Sowa et al., 2009). Using CompPASS, the uniqueness, abundance, and reproducibility of each protein assignment was compared across parallel sets of MS data generated from multiple unrelated immunoprecipitations to distinguish high-confidence protein interactors (HCIPs) from non-specific interacting proteins. HCIPs have WDN-scores >1.0.
Immunoblotting
Immunoblotting was performed as described (Lehtinen et al., 2006).
RT-PCR
Primers used for RT-PCR are described in the supplementary information.
Lentivirus production
shRNA sequences were cloned into pLentiLox 3.7 vector and lentivirus production was performed as described (Rubinson et al., 2003).
Microarray
Microarray analyses were performed using Affymetrix Rat Gene 1.1 ST Array Strip and Affymetrix GeneAtlas microarray system. All procedures were performed according to manufacturers’ manuals. Analysis was performed as described (Wakamatsu et al., 2013). GEO accession number GSE45953.
Electrophysiology
Acute cortical slices (350μm) were prepared from the cerebral cortex of P10-P12 mice. Acute slice preparation and whole-cell patch clamp recording were preformed as previously described (Debanne et al., 2008).
Statistics
Statistical analyses were performed using the Excel software program. Pairwise comparisons within two groups only were done using the t-test. Pairwise comparisons within multiple groups were done by analysis of variance (ANOVA) followed by the Fisher’s PLSD post hoc test. Data are presented as the mean + SEM. ****: p<0.001; ***: p<0.005; **: p<0.01; *: p<0.05; NS, not significant. Asterisks in the figures denote t-test comparisons between experimental group and control in each experiment.
Supplementary Material
Highlights.
The intellectual disability protein PHF6 drives neuronal migration in the brain.
PHF6 forms a complex with the PAF1 transcription elongation complex.
NGC/CSPG5, a potential schizophrenia gene, mediates PHF6-dependent neuron migration.
Inhibition of PHF6 causes white matter heterotopias, which harbor ectopic activity.
Acknowledgments
We thank the members of the Bonni laboratory for helpful discussions and critical reading of the manuscript. This work was supported by NIH grants NS041021 (A.B.), GM054137 (J.W.H) and AG011085 (J.W.H).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackman JB, Aniksztejn L, Crepel V, Becq H, Pellegrino C, Cardoso C, Ben-Ari Y, Represa A. Abnormal network activity in a targeted genetic model of human double cortex. J Neurosci. 2009;29:313–327. doi: 10.1523/JNEUROSCI.4093-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berland S, Alme K, Brendehaug A, Houge G, Hovland R. PHF6 deletions may cause Borjeson-Forssman-Lehmann syndrome in females. Mol syndromol. 2011;1:294–300. doi: 10.1159/000330111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin TK, Brocksen S, Avchen RN, Van Naarden Braun K. Prevalence of four developmental disabilities among children aged 8 years--Metropolitan Atlanta Developmental Disabilities Surveillance Program, 1996 and 2000. MMWR Surveill Summ. 2006;55:1–9. [PubMed] [Google Scholar]
- Brun A, Borjeson M, Forssman H. An inherited syndrome with mental deficiency and endocrine disorder. A patho-anatomical study. J Ment Defic Res. 1974;18:317–325. doi: 10.1111/j.1365-2788.1974.tb01243.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009;23:2765–2777. doi: 10.1101/gad.1834709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debanne D, Boudkkazi S, Campanac E, Cudmore RH, Giraud P, Fronzaroli-Molinieres L, Carlier E, Caillard O. Paired-recordings from synaptically coupled cortical and hippocampal neurons in acute and cultured brain slices. Nat Protoc. 2008;3:1559–1568. doi: 10.1038/nprot.2008.147. [DOI] [PubMed] [Google Scholar]
- Gaudilliere B, Shi Y, Bonni A. RNA interference reveals a requirement for myocyte enhancer factor 2A in activity-dependent neuronal survival. J Biol Chem. 2002;277:46442–46446. doi: 10.1074/jbc.M206653200. [DOI] [PubMed] [Google Scholar]
- Ge X, Frank CL, Calderon de Anda F, Tsai LH. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron. 2010;65:191–203. doi: 10.1016/j.neuron.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impagnatiello F, Guidotti AR, Pesold C, Dwivedi Y, Caruncho H, Pisu MG, Uzunov DP, Smalheiser NR, Davis JM, Pandey GN, et al. A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc Natl Acad Sci U S A. 1998;95:15718–15723. doi: 10.1073/pnas.95.26.15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinugasa Y, Ishiguro H, Tokita Y, Oohira A, Ohmoto H, Higashiyama S. Neuroglycan C, a novel member of the neuregulin family. Biochem Biophys Res Commun. 2004;321:1045–1049. doi: 10.1016/j.bbrc.2004.07.066. [DOI] [PubMed] [Google Scholar]
- Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–399. doi: 10.1016/j.tins.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litterman N, Ikeuchi Y, Gallardo G, O’Connell BC, Sowa ME, Gygi SP, Harper JW, Bonni A. An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning. PLoS Biol. 2011;9:e1001060. doi: 10.1371/journal.pbio.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower KM, Turner G, Kerr BA, Mathews KD, Shaw MA, Gedeon AK, Schelley S, Hoyme HE, White SM, Delatycki MB, et al. Mutations in PHF6 are associated with Borjeson-Forssman-Lehmann syndrome. Nat Genet. 2002;32:661–665. doi: 10.1038/ng1040. [DOI] [PubMed] [Google Scholar]
- Muntean AG, Tan J, Sitwala K, Huang Y, Bronstein J, Connelly JA, Basrur V, Elenitoba-Johnson KS, Hess JL. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17:609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah B, Brunstrom JE, Grutzendler J, Wong RO, Pearlman AL. Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci. 2001;4:143–150. doi: 10.1038/83967. [DOI] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II–IV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- Rio C, Rieff HI, Qi P, Khurana TS, Corfas G. Neuregulin and erbB receptors play a critical role in neuronal migration. Neuron. 1997;19:39–50. doi: 10.1016/s0896-6273(00)80346-3. [DOI] [PubMed] [Google Scholar]
- Rondon AG, Gallardo M, Garcia-Rubio M, Aguilera A. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 2004;5:47–53. doi: 10.1038/sj.embor.7400045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol Cell Biol. 1996;16:669–676. doi: 10.1128/mcb.16.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So HC, Fong PY, Chen RY, Hui TC, Ng MY, Cherny SS, Mak WW, Cheung EF, Chan RC, Chen EY, et al. Identification of neuroglycan C and interacting partners as potential susceptibility genes for schizophrenia in a Southern Chinese population. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:103–113. doi: 10.1002/ajmg.b.30961. [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G, Lower KM, White SM, Delatycki M, Lampe AK, Wright M, Smith JC, Kerr B, Schelley S, Hoyme HE, et al. The clinical picture of the Borjeson-Forssman-Lehmann syndrome in males and heterozygous females with PHF6 mutations. Clin Genet. 2004;65:226–232. doi: 10.1111/j.0009-9163.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N, De Moerloose B, Philippe J, Gonzalez-Garcia S, Toribio ML, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42:338–342. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu E, Mathis D, Benoist C. Convergent and divergent effects of costimulatory molecules in conventional and regulatory CD4+ T cells. Proc Natl Acad Sci U S A. 2013;110:1023–1028. doi: 10.1073/pnas.1220688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.