Abstract
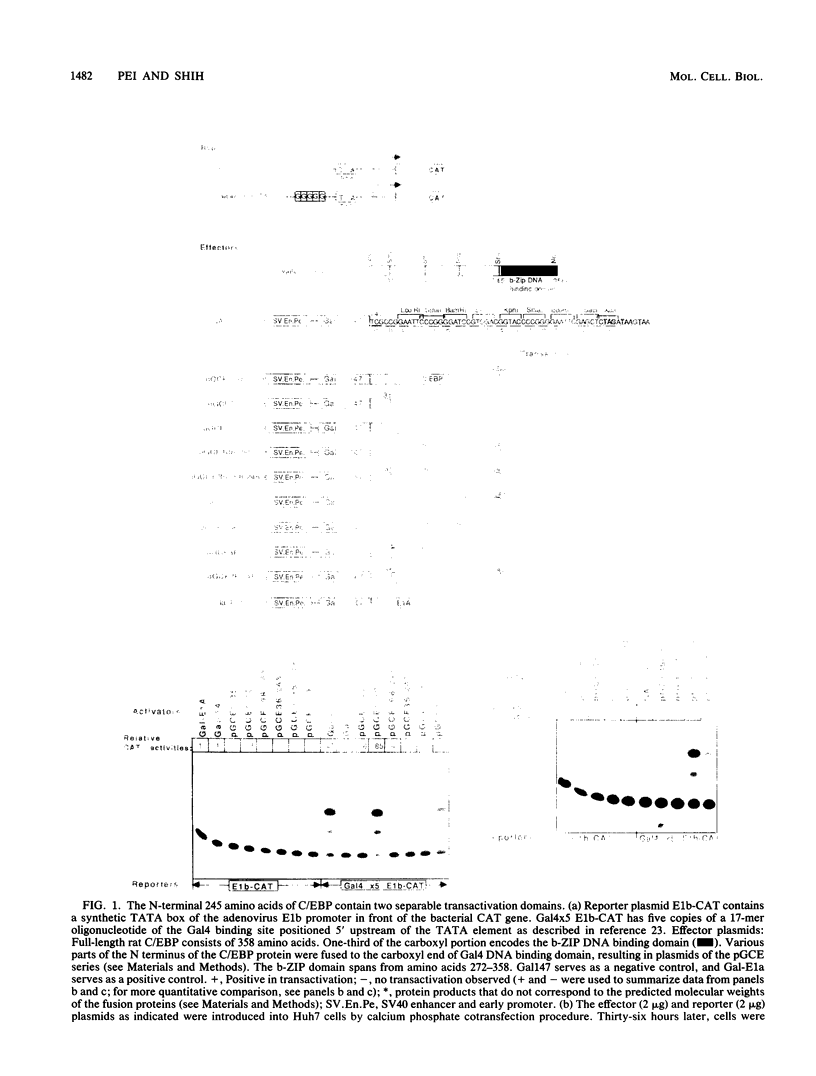
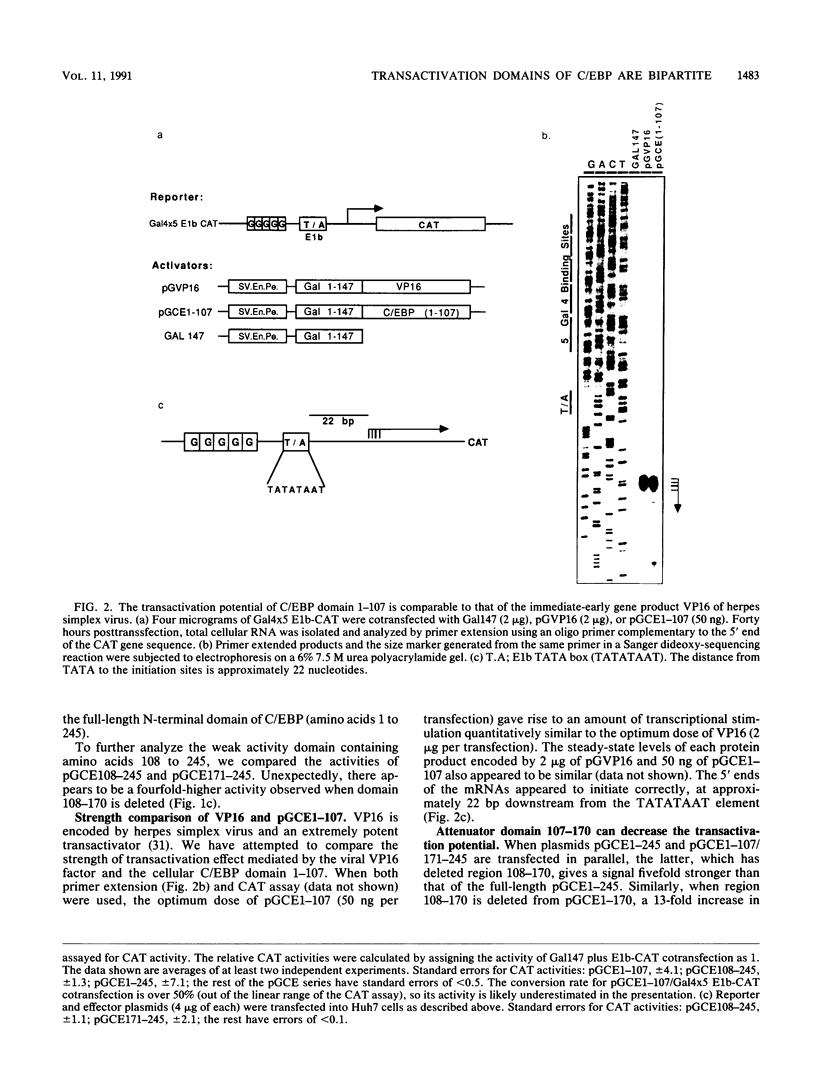
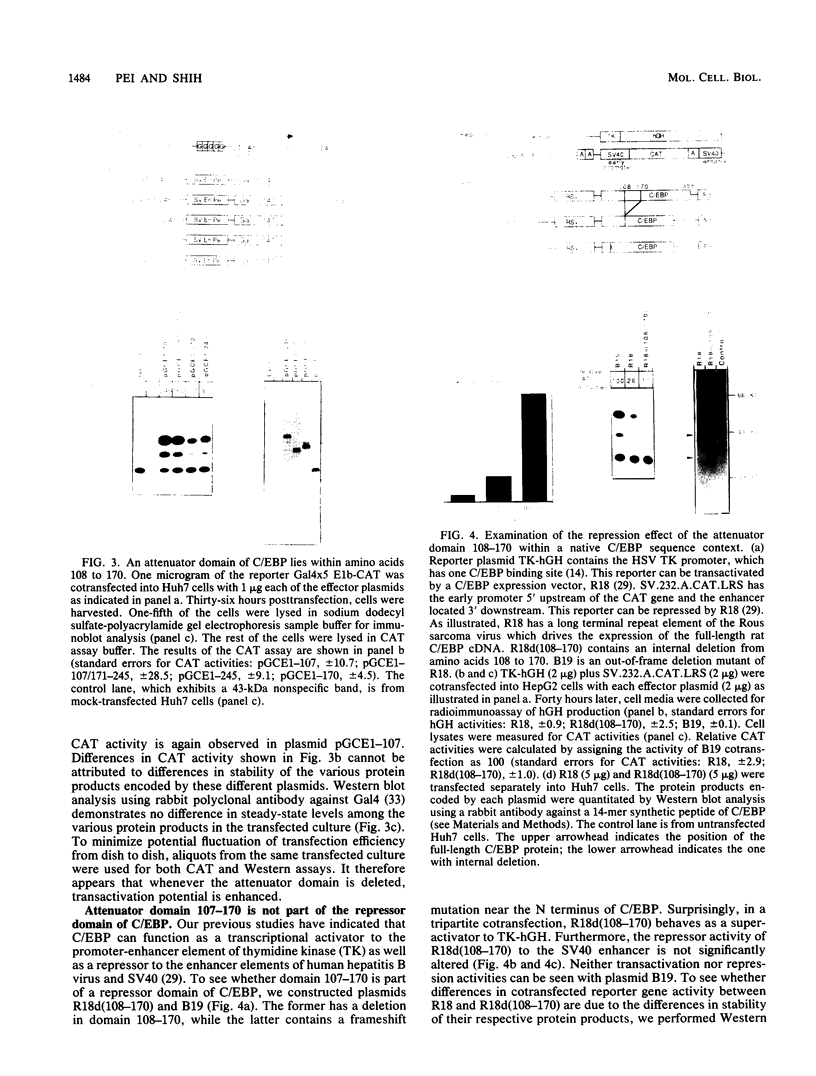
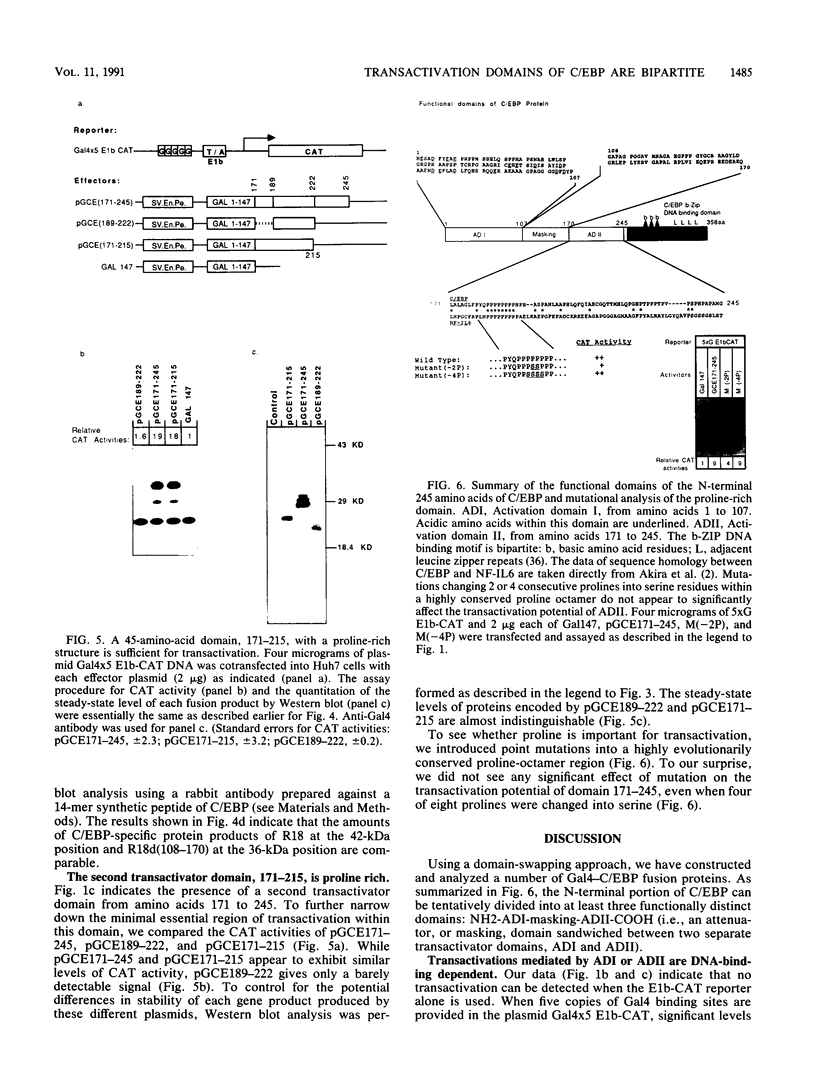
C/EBP is a rat liver DNA-binding protein which can act as a transcription factor. Its N-terminal portion contains three distinct domains. The first domain (amino acids 1 to 107) appears to be a highly potent transactivator. The second domain (amino acids 107 to 170) does not appear to exhibit either activation or repression activity. This domain is defined as an "attenuator domain" because its presence under four different sequence contexts reproducibly decreases the effect of transactivation of C/EBP. The third domain (amino acids 171 to 245) is a relatively weaker transactivator with a striking proline-rich motif. Deletional analysis of this third domain has shown that a 45-amino-acid region is sufficient for transactivation. This region (amino acids 171 to 215) contains 12 proline, 6 histidine, and mainly hydrophobic or noncharged amino acids. Further mutational analysis of a highly conserved proline-octamer region within this domain indicates that a specific proline content is not crucial for transactivation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aden D. P., Fogel A., Plotkin S., Damjanov I., Knowles B. B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979 Dec 6;282(5739):615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., Nakajima T., Hirano T., Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990 Jun;9(6):1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann D., Tjian R. Biochemical analysis of transcriptional activation by Jun: differential activity of c- and v-Jun. Cell. 1989 Nov 17;59(4):709–717. doi: 10.1016/0092-8674(89)90017-2. [DOI] [PubMed] [Google Scholar]
- Bruzik J. P., Van Doren K., Hirsh D., Steitz J. A. Trans splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature. 1988 Oct 6;335(6190):559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Christy R. J., Yang V. W., Ntambi J. M., Geiman D. E., Landschulz W. H., Friedman A. D., Nakabeppu Y., Kelly T. J., Lane M. D. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev. 1989 Sep;3(9):1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- Costa R. H., Grayson D. R., Xanthopoulos K. G., Darnell J. E., Jr A liver-specific DNA-binding protein recognizes multiple nucleotide sites in regulatory regions of transthyretin, alpha 1-antitrypsin, albumin, and simian virus 40 genes. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3840–3844. doi: 10.1073/pnas.85.11.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Crossley M., Brownlee G. G. Disruption of a C/EBP binding site in the factor IX promoter is associated with haemophilia B. Nature. 1990 May 31;345(6274):444–446. doi: 10.1038/345444a0. [DOI] [PubMed] [Google Scholar]
- Friedman A. D., Landschulz W. H., McKnight S. L. CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells. Genes Dev. 1989 Sep;3(9):1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- Friedman A. D., McKnight S. L. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990 Aug;4(8):1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski K., Carneiro M., Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986 Dec 5;47(5):767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- Graves B. J., Johnson P. F., McKnight S. L. Homologous recognition of a promoter domain common to the MSV LTR and the HSV tk gene. Cell. 1986 Feb 28;44(4):565–576. doi: 10.1016/0092-8674(86)90266-7. [DOI] [PubMed] [Google Scholar]
- Heard J. M., Herbomel P., Ott M. O., Mottura-Rollier A., Weiss M., Yaniv M. Determinants of rat albumin promoter tissue specificity analyzed by an improved transient expression system. Mol Cell Biol. 1987 Jul;7(7):2425–2434. doi: 10.1128/mcb.7.7.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., Landschulz W. H., Graves B. J., McKnight S. L. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987 Apr;1(2):133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., Adashi E. Y., Graves B. J., McKnight S. L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988 Jul;2(7):786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Green M. R. Transcription activation by the adenovirus E1a protein. Nature. 1989 Mar 2;338(6210):39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- López-Cabrera M., Letovsky J., Hu K. Q., Siddiqui A. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci U S A. 1990 Jul;87(13):5069–5073. doi: 10.1073/pnas.87.13.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. A new class of yeast transcriptional activators. Cell. 1987 Oct 9;51(1):113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Nakabayashi H., Taketa K., Miyano K., Yamane T., Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982 Sep;42(9):3858–3863. [PubMed] [Google Scholar]
- Nieto-Sotelo J., Wiederrecht G., Okuda A., Parker C. S. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell. 1990 Aug 24;62(4):807–817. doi: 10.1016/0092-8674(90)90124-w. [DOI] [PubMed] [Google Scholar]
- Pei D. Q., Shih C. H. Transcriptional activation and repression by cellular DNA-binding protein C/EBP. J Virol. 1990 Apr;64(4):1517–1522. doi: 10.1128/jvi.64.4.1517-1522.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Roychoudhury S., Shih C. cis rescue of a mutated reverse transcriptase gene of human hepatitis B virus by creation of an internal ATG. J Virol. 1990 Mar;64(3):1063–1069. doi: 10.1128/jvi.64.3.1063-1069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell. 1990 Aug 24;62(4):793–805. doi: 10.1016/0092-8674(90)90123-v. [DOI] [PubMed] [Google Scholar]
- Triezenberg S. J., Kingsbury R. C., McKnight S. L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988 Jun;2(6):718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]