Abstract
Investigators have made key advances in rheumatoid arthritis (RA) genetics in the past 10 years. Although genetic studies have had limited influence on clinical practice and drug discovery, they are currently generating testable hypotheses to explain disease pathogenesis. Firstly, we review here the major advances in identifying RA genetic susceptibility markers both within and outside of the MHC. Understanding how genetic variants translate into pathogenic mechanisms and ultimately into phenotypes remains a mystery for most of the polymorphisms that confer susceptibility to RA, but functional data are emerging. Interplay between environmental and genetic factors is poorly understood and in need of further investigation. Secondly, we review current knowledge of the role of epigenetics in RA susceptibility. Differences in the epigenome could represent one of the ways in which environmental exposures translate into phenotypic outcomes. The best understood epigenetic phenomena include post-translational histone modifications and DNA methylation events, both of which have critical roles in gene regulation. Epigenetic studies in RA represent a new area of research with the potential to answer unsolved questions.
Introduction
Genetic and epigenetic underpinnings of RA
That genetic and environmental factors participate in mechanisms of rheumatoid arthritis (RA) pathogenesis1 is well established. The overall contribution of genetic factors to RA development has historically been investigated through analysis of family pedigrees. For example, familial clustering—greater disease occurrence in relatives of probands than of healthy controls—has been a consistent observation in RA;2 the relative risk of RA development in first degree relatives of affected individuals is estimated at ~2 or greater.3–5 In addition, disease discordance in monozygotic compared with dizygotic twins suggests that the genetic contribution to RA, or disease heritability, approaches 65%;6 these estimates are, however, based on a relatively small number of twins (23 monozygotic and 10 dizygotic disease-concordant twin pairs). Heritability is the proportion of phenotypic variance that can be attributed to genetic, rather than environmental, causes. Thus, although RA clearly has a considerable genetic component, and few known environmental triggers (such as cigarette smoke),7 many environmental factors remain largely unknown and their contribution to RA aetiology is likely substantial.
Mechanisms that underlie the observed sex-bias (3:1 female to male ratio) in the incidence of RA are also unknown. Investigators have suggested a range of hypotheses including potential roles for sex hormones.8,9 The sex chromosomes have been underinvestigated in genetic studies in RA. An Immunochip study published in 2012,10 described in the ‘Immunochip’ subsection of this manuscript, shows for the first time an association with an X chromosome locus in RA, in this case IRAK1 (encoding interleukin-1 receptor-associated kinase 1). This locus has been shown to escape X-inactivation in humans,11 pointing to a possible epigenetic mechanism underlying the sex bias in RA.
Implicating the MHC
In 1969, researchers noticed that peripheral blood lymphocytes from patients with RA were non-reactive in so-called mixed lymphocyte cultures to cells of the same type from other patients with RA.12 Investigators demonstrated in 1976 that patients with RA tend to share the same HLA genes,13 thus explaining the lack of reactivity in mixed cultures. Serotyping experiments subsequently identified an increased proportion of patients with RA who were positive for the HLA allele HLA-DRw4, in comparison with healthy controls,14 establishing the HLA region as a genetic contributor to RA susceptibility. A decade later, further characterization of the HLA locus identified multiple RA risk alleles within HLA-DRB1, and showed that the molecules they encoded shared a conserved amino acid sequence; this finding led to the ‘shared epitope’ hypothesis.15 HLA molecules that contain this 5-amino-acid sequence, which is encoded by shared epitope alleles and is arranged around the antigen-binding groove, are associated with the development of anti-citrullinated protein antibodies (ACPA), and —mostly—with ACPA-positive RA. Although this feature is thought to influence the affinity of binding to citrullinated peptides, and to modulate T-cell responses, the precise biological implications of the shared epitope are not yet clear;1 as we discuss in this manuscript, new associations with ACPA-negative RA complicate the shared epitope theory.
Non-MHC associations with RA
Outside of the MHC region, candidate gene studies performed prior to 2007 had identified only a handful of RA susceptibility loci, including PTPN22 (encoding tyrosine-protein phosphatase non-receptor type 22),16 protein-arginine deiminase type 4 (PADI4)17 and cytotoxic T-lymphocyte protein 4 (CTLA-4)18 By 2007, genome-wide association studies (GWAS) had become possible due to several major preceding advances, including the completion of the Human Genome Project in 200119 and the initial release of the International HapMap project data in 2003.20 These initiatives enabled the design of single-nucleotide polymorphism (SNP) chips with good coverage of variations that occur across the entire genome. In the past 5 years, these technologies have escalated the rate of discovery of disease-associated variants, and around 60 risk loci for RA are now known in European and Asian populations.10,21,22 This advance has been aided by the attainment of large, well-characterized, and homogeneous (that is, ACPA-positive) collections of samples from patients.
GWAS power and undiscovered associations
By design, GWAS are powered to detect associations with variants that are common in the population (minor allele frequency >5%). Most of the variants identified to date in GWAS in RA, and in other complex diseases, have modest effect sizes, with odds ratios of 1.5 or less.22,23 These associations are potentially caused by causal variants that are in tight linkage disequilibrium with the observed variants. Given that their effect sizes are so modest, each of these alleles individually explains a small fraction of the genetic contribution to RA susceptibility. Currently, all RA genetic risk factors taken together only explain ~16% of the total susceptibility (heritable and environmental).22,24,25 Hundreds of common risk alleles are likely to exist but remain undiscovered to date owing to the limited power of current GWAS. A recent analysis by our group suggests that hundreds of uncharacterized SNP associations throughout the genome, taken together with known risk alleles, in aggregate explain ~36% of RA disease risk.25 SNP associations and known risk alleles therefore account for only about half of the estimated 65% of RA risk that is thought to be heritable. Sequencing experiments in the coming years have the potential to identify causal variants across the entire allele frequency range, including low frequency variants.
Better genotyping—accurate phenotyping
Of prime importance for future genetic studies is stratification of samples by distinct phenotypic subgroups of RA. Although the clinical presentation at disease initiation is very similar between patients with ACPA-positive and ACPA-negative RA, disease course, possibly disease pathogenesis,26 and genetic susceptibility27,28 are different. The association of RA with the shared epitope is, as we have mentioned, different between the two serotype subsets.27 Now, it further seems that non-HLA SNPs associated with RA susceptibility are only partially shared between ACPA-positive and ACPA-negative patients with RA,28 confirming the hypothesis that ACPA-positive and ACPA-negative RA are two genetically different diseases.29 Although patients with ACPA-negative RA included in genetic studies satisfy the 1987 American College of Rheumatology classification criteria for RA,30 concerns remain about misclassification in this subgroup of patients.26,29 Nevertheless, even if ACPA-negative RA represents a heterogeneous disease group, the overall contribution of genetic factors to disease susceptibility in ACPA-negative RA seems to be as high as for ACPA-positive RA.31 Interestingly, we could show that the pattern of association of ACPA-positive susceptibility SNPs with ACPA-negative RA (in terms of effect size or presence or absence of an association) cannot be explained solely by contamination with erroneously characterized ACPA-positive samples, because the ratios of the effect sizes between ACPA-positive and ACPA-negative RA vary widely for different genetic markers.28 Nevertheless, ACPA-negative RA is likely to be subclassified in the future on the basis of further types of autoantibody. In 2011, antibodies against carbamylated proteins (anti-CarP antibodies) were shown to be present in around 20% of patients with ACPA-negative RA.32 Furthermore, anti-CarP antibodies were associated with more severe joint damage in this group.32 Twin studies have also established important differences between ACPA-positive and ACPA-negative disease.31 Although heritability estimates remain similar in both serological strata, the contribution of the HLA-DRB1 shared epitope alleles differs markedly, explaining 18% and 2.4% of RA heritability in ACPA-positive and ACPA-negative patients, respectively.31
Functional implications of risk alleles
One of the biggest challenges for the future will be to elucidate the biological mechanisms in which risk alleles operate. We are already beginning to understand which cells are central to RA pathogenesis. For example, CD4+ effector memory T cells specifically express many of the genes located within RA-associated loci.33 Also, certain pathways seem to be critical for disease pathogenesis; for example, multiple genes within RA loci are involved in signalling downstream of the CD40 molecule (also known as TNF receptor superfamily member 5).34
Although discoveries from GWAS in RA have not yet lead to the direct identification of therapeutic targets, some existing therapies target genes and/or pathways that have been highlighted by such studies. For example, abatacept is a fusion protein made up of cytotoxic T-lymphocyte protein 4 (CTLA-4) and immunoglobulin. CTLA-4, together with other transmembrane receptors expressed on T cells (CD28 and inducible T-cell costimulator [ICOS]), has a crucial role in T-cell co-stimulation, and CTLA-436 (in addition to CD28)37 polymorphisms are associated with RA risk. Nevertheless, the translation of genetic findings into clinical applications is often more challenging than originally postulated.38,39 As we discuss in detail below, an association study published in 201235 examining the entire MHC region could not confirm the frequently-reported association of TNF polymorphisms with disease susceptibility, although anti-TNF treatment has substantially improved quality of life for patients with RA.
MHC genes and risk of RA
The early days and unresolved hypotheses
The discovery of a strong association between HLA-DRB1 and RA was initially made using antibodies to specific MHC class II proteins and thus serotyping individuals according to the surface expression of antigenic molecules on their circulating B cells. Stastny13,14 found that substantially more patients with RA were positive for the B-cell alloantigen DRw4 (later renamed DR4) than were healthy individuals. Subsequently, investigators used cloning and sequencing experiments to characterize different alleles at that gene locus (now called HLA-DRB1). According to the current nomenclature,39 HLA-DRB1*04 denominates the allele group corresponding roughly to the archaic serotypic classification DRw4, while the next appendant digit set defines a specific allele; for example, HLA-DRB1*0401. A decade after Stastny’s discoveries, Gregersen et al.15 showed that molecules encoded by RA-associated HLA-DRB1 alleles share a common amino acid sequence in the third hyper-variable region of the DRβ1 chain—the shared epitope. A T-cell epitope is, by definition, a three-dimensional structure recognized by a paratope (the T-cell receptor [TCR]) and constituted in part by the MHC molecule and in part by the antigenic peptide bound in the groove. The term ‘shared epitope’, therefore, suggests the existence of an autoantigenic peptide that has not unequivocally been identified after two decades of research. As a result, the ‘arthritogenic peptide hypothesis’15,40,41 remains controversial42 and, although shared epitope alleles are established genetic risk factors in RA, the immunological implications of their expression remain uncertain.
Genome-wide linkage scans and the MHC
Between 1998 and 2003, five genome-wide linkage scans in family-based cohorts of people with or without RA demonstrated strong and significant linkage of the disease with the MHC region, but not consistently with any other region in the genome.43–47 As we have mentioned, linkage with the MHC applied only to ACPA-positive, not ACPA-negative, RA.27 Initially, MHC associations with ACPA-positive RA were attributed to HLA genes; however, shared epitope alleles at the HLA-DRB1 locus do not fully explain the association of the MHC region with RA—several studies in which the HLA-DRB1 effect was controlled for have suggested additional independent associations within the MHC.48–50
Until 2012, MHC alleles were thought to be exclusively associated with ACPA-positive RA. Now, several reports from well-powered studies have identified and confirmed the association of the shared epitope with ACPA-negative RA.10,28,51 The role of this association and its possible restriction to specific serotypes or subtypes of ACPA-negative RA remain to be determined.
Revisiting the shared epitope hypothesis
Despite advances in high-throughput SNP genotyping technologies, the application of probe-based genotyping to query HLA genes within the MHC has been limited, owing to the highly polymorphic nature of these genes. Historically, investigation of HLA genes required direct PCR-based genotyping. Twenty-first century advances in statistical genetics have now facilitated imputation of HLA alleles based on SNP data.52,53 Imputation employs a large reference data set from individuals genotyped for classical HLA alleles and HLA SNPs to determine the most likely HLA alleles in individuals for whom SNP data over the HLA region, but not direct HLA genotyping, are available.
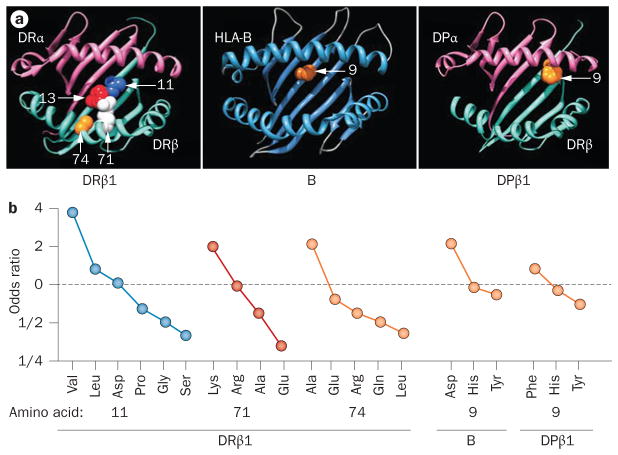
In 2012, we applied this imputation approach to SNP data from the 2010 GWAS meta-analysis by Stahl et al.22 (Figure 1), and demonstrated that the risk of RA associated with the HLA-DRB1 gene correlates most strongly with the amino acid residue in position 11, located at the bottom of the DRβ1 antigen-binding groove.38 Amino acids 71 and 74, whose sidechains constitute the surface of the antigen-binding groove, also correlated independently with susceptibility to RA. In addition, we found independent RA risk alleles in HLA-B and HLA-DPB1; in both cases, signals from these regions were best explained by a variation in a single amino acid site at the bottom of their respective antigen-binding grooves. No further signal of an association with RA was found within the MHC when we controlled for the independent effects mentioned here. That is, these genetic variants in HLA-B, HLA-DRB1 and HLA-DPB1—affecting a total of 5 amino acid positions—almost completely explained the variance in RA risk caused by the MHC region. Although other SNP associations are indeed possible within the MHC and other HLA genes, such variants are likely to have comparatively weak effects in conferring susceptibility to RA. Importantly, no association signal with RA was identified within the TNF region, indicating that frequently reported associations between TNF promoter polymorphisms and RA susceptibility were probably confounded by nearby HLA-B, HLA-DRB1 and HLA-DPB1 gene variants.
Figure 1.
Antigen-binding groove HLA amino acid substitutions and influence on susceptibility to RA. a | Three-dimensional ribbon models for the MHC class I molecule HLA-B and for the MHC class II molecules HLA-DRβ1 and HLA-DPβ1. Direct views of the peptide-binding groove are presented, showing key amino acid positions identified in an association analysis by Raychaudhuri, S. et al. Nat. Genet. 44, 291–296 (2012).38 © NPG b | The odds ratio for association with RA depends on which amino acid is substituted at positions 11, 71 and 74 of HLA-DRβ1, at position 9 of HLA-B or at position 9 of HLA-DPβ1. Abbreviation: RA, rheumatoid arthritis.
Interestingly, when haplotypes of alleles for the 3 RA-associated amino acid positions within the locus were studied and coded using the classical 4-digit HLA-DRB1 allele nomenclature, the hierarchy of HLA alleles associated with risk of, and protection from, RA was consistent with previous classification systems or studies:51,54–60 for example, shared epitope alleles were associated with greatest susceptibility to RA, whereas HLA-DRB1*130154 was part of the most protective haplotype (Table 1).38 A large European meta-analysis in 2010 confirmed HLA-DRB1*1301 as a protective allele for RA.54
Table 1.
Effect-size estimates for HLA-DRB1 haplotypes on rheumatoid arthritis risk
| HLA-DRB1 amino acid at position | Odds ratio‡ | Classical HLA-DRB1 alleles§ | |||
|---|---|---|---|---|---|
| 11 | 13 | 71 | 74 | ||
| Val | His | Lys | Ala | 4.44 | *0401 (Shared epitope) |
| Val | His or Phe | Arg | Ala | 4.22 | *0408, *0405, *0404, *1001 (Shared epitope) |
| Leu | Phe | Arg | Ala | 2.17 | *0102, *0101 (Shared epitope) |
| Pro | Arg | Arg | Ala | 2.04 | *1601 |
| Val | His | Arg | Glu | 1.65 | *0403, *0407 |
| Asp | Phe | Arg | Glu | 1.65 | *0901 |
| Val | His | Glu | Ala | 1.43 | *0402 |
| Ser | Ser | Lys | Ala | 1.04 | *1303 |
| Pro | Arg | Ala | Ala | 1 | *1501, *1502 (Reference haplotype) |
| Gly | Tyr | Arg | Gln | 0.91 | *0701 |
| Ser | Ser or Gly | Arg | Ala | 0.88 | *1101, *1104, *1201 |
| Ser | Ser | Arg | Glu | 0.84 | *1401 |
| Leu | Phe | Glu | Ala | 0.73 | *0103 |
| Ser | Gly | Arg | Leu | 0.71 | *0801, *0804 |
| Ser | Ser | Lys | Arg | 0.63 | *0301 |
| Ser | Ser | Glu | Ala | 0.59 | *1102, *1103, *1301, *1302 |
The most common haplotype (ProArgAlaAla, corresponding to HLA-DRB1*1501 and *1502) has been used as reference to calculate odds ratios.
The classical shared epitope alleles are indicated. Reproduced from Raychaudhuri et al. Nat. Genet. 44, 291–296 (2012).38 © NPG.
Linking HLA alleles to function
The fine-mapping of MHC polymorphisms that we describe above confirms that HLA-DRB1 modulates susceptibility to RA, and defines a few amino acids, including positions 71 and 74 originally highlighted in the shared epitope hypothesis, as determining the effect.38 Further, the data extend associations with RA to HLA-B and HLA-DPB1. Most interestingly in biological terms, sidechains of amino acids in the positions that alter susceptibility to RA all point towards the peptide-binding groove, revitalizing the ‘arthritogenic peptide hypothesis’. Lack of identification of an ‘RA antigen’ to date might, therefore, be related more to technical challenges than to its non-existence. Indeed, the well-established effect-size hierarchy of classical shared epitope alleles (Table 1 and reviewed elsewhere)51,55 might correlate with the HLA-binding affinity of an antigenic peptide61 and, ultimately, with its immunogenicity.62,63 The next step in characterizing this potential pathogenic mechanism consists of identifying T-cell autoantigens in ACPA-positive RA. New structural information regarding the peptide-binding groove38 and the importance of citrullination with regard to binding affinity will help in the selection of peptides from putative target proteins for reverse engineering experiments.64
Non-HLA SNPs
High-throughput SNP genotyping
Since 2000, high-throughput SNP genotyping has successfully facilitated case–control association studies in RA to test putative links with genetic variants outside the MHC.24,65–67 In 2003, Suzuki et al.68 identified a SNP in the third intron of PADI4 that contributed to the risk of RA in Japanese populations (Figure 2). A year later, Begovich et al.16 identified a non-synonymous SNP in PTPN22 as a risk variant in white individuals in the USA; this variant remains the most strongly RA-associated SNP identified to date with an odds ratio of 1.8 for ACPA-positive RA. Subsequent case–control studies investigating other candidate gene associations have suggested only a handful of additional susceptibility loci (among them CTLA-4,18 TRAF169 and FCRL370); most loci identified in candidate-gene studies were not reproducible in independent studies.18 In 2007, investigators in RA genetics published three separate GWAS in RA,71–73 including one within the multi-disease Wellcome Trust Case–Control Consortium study.73 Several of the many new RA susceptibility SNPs (reviewed elsewhere)37 identified in these GWAS71–73 and subsequent studies are described in this section. GWAS have now been used to identify risk factors for RA in populations of European and Asian descent.21,22,68–71,74–77
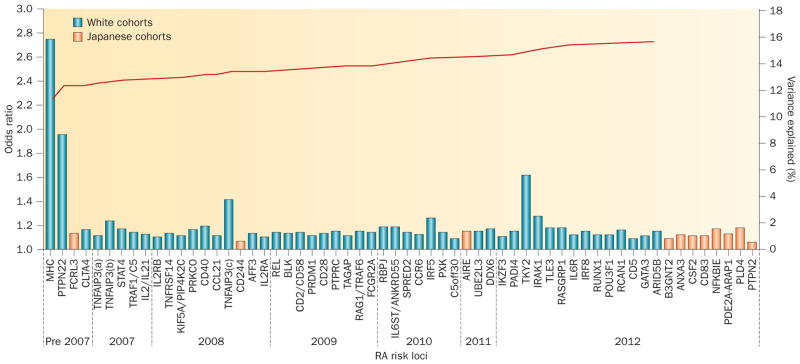
Figure 2.
RA genetic susceptibility loci identified to date, and cumulative proportion of observed variance in disease susceptibility explained thus far. As of 2012, approximately 16% of phenotypic variance has been accounted for genetically. Odds ratios10,21,22,79,80 for RA genetic susceptibility loci are presented in the approximate chronological order of discovery (blue and orange bars). Only susceptibility loci in white cohorts (blue) were included in calculating the proportion of phenotypic variance explained. Loci validated only in east-Asian populations are shown in orange; although the loci identified by Okada et al.21 were published a few months before the Immunochip study,10 they are presented at the far right of the plot for graphical convenience. Shared loci across different populations are shown at the time of their discovery in white cohorts. A 0.5% disease prevalence was assumed for calculations. For simplification, every locus is represented once, even if multiple independent effects were identified (except for TNFAIP3). The cumulative percent variance explained by the loci is indicated by the red line. Abbreviation: RA, rheumatoid arthritis.
Meta-analyses of data from GWAS
Imputation techniques, which have become standard tools to determine the genotype of ungenotyped SNPs,78 facilitate powerful meta-analyses of GWAS data originating from different genotyping platforms. Two large RA GWAS meta-analyses have independently examined different populations: Stahl et al.22 analysed data from people of European descent in 2010 (initially 5,539 patients with ACPA-positive RA and 20,169 controls, replicated using data from a further 6,768 patients and 8,806 controls), whereas Okada et al.21 used data from Japanese individuals (initially 4,074 with RA and 16,891 controls, then a further 5,277 patients and 21,684 controls) in 2012—these analyses identified 7 and 9 novel RA risk alleles, respectively.
Another approach to investigating the genetic basis of susceptibility to RA is to examine shared genetic bases between it and other autoimmune diseases, or across different ethnicities; genes, including ubiquitin-conjugating enzyme E2 L3 (UBE2L3)79, DEAD (Asp-Glu-Ala-Asp) box helicase 6 (DDX6; encoding probable ATP-dependent RNA helicase DDX6)80, and IKAROS family zinc finger 3 (Aiolos) (IKZF3, encoding zinc finger protein Aiolos),81 have been thus implicated. Okada et al.21 conducted a multi-ancestry comparative analysis of 46 risk loci between the Japanese data we have mentioned and data from individuals of European descent—5,539 patients with RA and 20,169 controls. Six of these sites were monomorphic in Japanese people (that is, all Japanese individuals have the same genotype at that locus), but all were polymorphic in individuals of European descent. Significant associations with RA (false discovery rate <0.05, P <0.0030) were found at 22 loci in Japanese people and at 36 loci in those of European descent; 14 of these signals were shared. Indeed, a comparison of all tested SNPs across the two populations showed a positive correlation of odds ratios for of a large proportion of SNPs between cohorts of individuals of European descent and Japanese cohorts, indicating shared genetic susceptibility alleles.21 Ethnogenetic heterogeneity in RA has been reviewed previously in this journal.82
Of note, the focus of meta-analyses to date has been almost exclusively on ACPA-positive RA. Genetic architecture differs between ACPA-positive and ACPA-negative RA,48 and RA susceptibility loci are only partially shared between the two serotypes.10,28
Immunochip
Immunochip, a custom SNP array, facilitates dense genotyping and fine-mapping at 186 genetic loci, including confirmed autoimmune loci and other alleles with nominal GWAS-based evidence of an association with an autoimmune disease. Collaborating with investigators worldwide, our group genotyped 11,475 patients of European descent with RA and 15,870 controls at 130,000 markers using Immunochip, identifying 14 novel RA risk loci.10 Furthermore, we refined to single genes the association signals of 19 previously identified loci. Secondary independent effects (defined as a remaining association at P <5 × 10−4 after conditioning on the most associated SNP of the region) were identified at 6 loci, and non-synonymous exonic SNPs or SNPs located within an essential splice site suggested putative causality at 7 loci. Interestingly, PADI4 polymorphisms, unequivocally associated with RA in Asian populations in previous studies, were significantly associated (genome-wide; P <5 × 10−8) with RA in patients of European descent in this study.10 Although a PADI4 variant was historically the first RA-associated polymorphism to be identified outside the HLA, its association has been controversial in populations of European descent.
Aetiopathogenetic mechanisms
Connecting genotype to phenotype in RA
Few genetic markers of RA susceptibility have been experimentally linked to functions, and many different molecular mechanisms are implicated. Indeed, the first molecular steps by which a SNP influences phenotype might involve alterations in transcriptional activity, epigenetic modifications, microRNA regulation, splicing, mRNA or protein stability, translation, protein activity or post-translational modifications. In this section, we review RA susceptibility loci for which roles have been investigated experimentally after their discovery in GWAS.
PTPN22
The most studied polymorphism in RA to date is the PTPN22 non-synonymous Arg620Trp SNP rs2476601. Tyrosine-protein phosphatase non-receptor type 22 (known as PTPN22 and encoded by PTPN22), down-regulates TCR signalling by dephosphorylating Src family kinases, such as Lck or Fyn (Figure 3). Although evidence indicates that the PTPN22 risk allele affects the enzymatic activity of the encoded phosphatase,83 the influence of the Arg620Trp variant on the immune response has been controversial—in 2005, a gain-of-function consequence was reported,84 but further functional studies have been inconsistent. In 2011, Zhang et al.85 showed that rs2476601 is a loss-of-function allele that mediates its effect by destabilizing PTPN22 (or its mouse homolog). The variant phosphatase is targeted for degradation both by calpain proteases and through ubiquitin-mediated proteasomal degradation. Reduced levels of the protein correlate with increased number, activation and thymic positive selection of T cells, and with dendritic-cell and B-cell activation. In 2012, the function of PTPN22 was linked to the thymic development of regulatory T cells,86,87 and alternative molecular mechanisms as heterogeneous as imbalance in the expression of PTPN22 splice variants88 and differential allelic expression89 have been suggested.
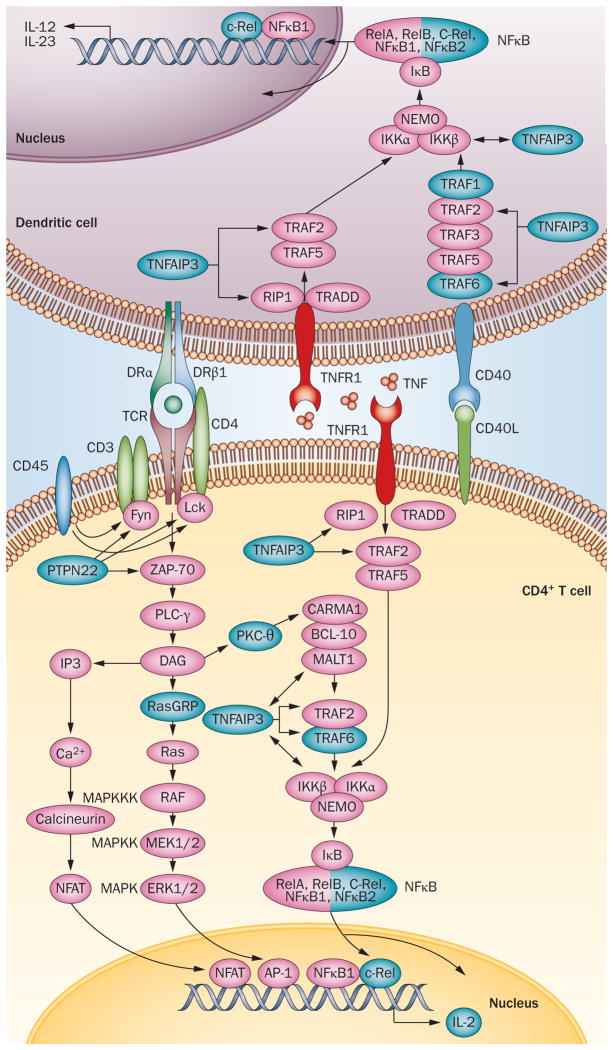
Figure 3.
Mapping of 11 RA susceptibility loci to pathways involved in the ‘T-cell–dendritic-cell dialogue’. The gene products (blue) of several RA susceptibility loci are implicated in the three pathways represented here—the TCR, TNF and CD40 signalling pathways. To avoid overloading the picture and to represent pathways as linear and chronological successions of molecular interactions, several membrane-associated factors are schematically represented in the cytosol and many key molecules or interactions have been omitted (for example, the p38 and JNK pathways, CD28 co-stimulation, and adhesion mediated by CD2 and CD58). NF κB is a homodimer or heterodimer containing 2 of the following subunits: RelA, RelB, c-Rel, NF κB1, NF κB2. Only c-Rel is associated with RA. CD45 and PKC-θ are encoded by PTPRC and PRKCQ respectively; both of these genes are within RA susceptibility loci. Abbreviations: NF κB, nuclear factor κB; PTPN22, tyrosine-protein phosphatase non-receptor type 22; TCR, T-cell receptor; TNFR1: TNF receptor 1; TNFAIP3, TNF-induced protein 3.
PADI4
PADI4 mediates post-translational conversion of arginine residues to citrulline. Originally,68 an RA susceptibility haplotype was shown to increase the stability of PADI4 mRNA transcripts and was associated with ACPA positivity in patients with RA. Citrullinated peptides bind with higher affinity to HLA-DRβ1 shared epitope molecules,61,90 are naturally processed,91 and are immunogenic.62 Thus, it seems that increased translation of variant PADI4 mRNA boosts production of citrullinated peptides, which act as autoantigens and elicit profound adaptive immune responses. Whereas many other risk loci seem to be connected to multiple autoimmune diseases, the PADI4 locus is specific to RA.
CCR6
CCR6 encodes a chemokine receptor expressed by CD4+ type 17 T helper (TH17) cells. A polymorphism in CCR6 correlated with expression level of CCR6 mRNA and with the presence of IL-17 in the sera of patients with RA, highlighting the importance of the TH17 pathway in RA pathogenesis.74
Inferring function from data outside RA
Other than PTPN22, PADI4 and CCR6, few other risk loci have been investigated functionally in RA. Nevertheless, knowledge has been gained from studies in healthy individuals or in the context of other auto-immune diseases.
IL2RA
Autoimmunity-associated SNPs located in non-coding genomic regions in the vicinity of IL2RA (encoding IL-2 receptor subunit α) have been shown to correlate with IL2RA mRNA and surface protein expression levels in monocytes, CD4+ naive T cells and memory T cells, but not in other cell types tested.94 According to the quantal theory of immunity, T-cell responses depend on a critical number of stimuli mediated by TCR and IL-2R,95 which could explain different activation thresholds in the T-cell compartment of individuals polymorphic at the IL2RA locus.
TNFAIP3
TNFAIP3 encodes TNF-induced protein 3 (TNFAIP3), a ubiquitin-modifying enzyme that is a key regulator of nuclear factor κB activity (Figure 3). Three SNPs within the TNFAIP3 locus are independently associated with RA susceptibility.96 In patients with systemic lupus erythematosus (SLE), a polymorphism located in a highly conserved region of TNFAIP3 has been shown to reduce mRNA and protein expression of the gene, seemingly by reducing the avidity with which a nuclear protein complex of NF κB subunits binds to it.97 Indicating the importance of cell-type specific expression, mice with conditional knockout of Tnfaip3 expression in dendritic cells develop an SLE-like phenotype,98 whereas mice lacking Tnfaip3 in myeloid cells develop an RA-like phenotype.99
Bioinformatic analysis approaches
A biologically pragmatic way to define a pathway is to consider it as a chronological succession of molecular interactions occurring between cells or within a cell, starting with a signal (input) and ultimately resulting in a response (output). This linear definition of pathways conveniently allows experimental testing, as responses to signals can be measured. Several bioinformatic techniques have been proposed to analyse post-GWAS data as a whole and to identify RA-specific mechanisms of disease progression.98 In this section, we describe how bioinformatic techniques, such as pathways and networks analyses and integrative systematic approaches, are applied to functional analysis of putative RA risk alleles. Importantly, bioinformatic definitions of pathways are often non-linear and do not lend themsleves to experimental validation; an important challenge for the future will be how to biologically validate integrated bioinformatic analysis approaches.
Representing the TCR intracellular signalling pathway as a schematic linear series of molecular events provides an example of how RA susceptibility loci can be matched with potential roles in a biological model of pathology, and yet also illustrates how complex such efforts are. Indeed, many of the known loci associated with RA are involved in the TCR signalling pathway, but intricate interactions link these components with other signalling pathways in even a simplified depiction (Figure 3). Thus, although plausible functional explanations for how genetic variants confer RA risk can be generated, it remains to be experimentally demonstrated that RA risk alleles in aggregate alter, for example, the efficiency of TCR engagement (input), subsequent signal transduction events, and consequent production of IL-2 (output).
Pathway analysis
The generic term ‘pathway analysis’ is loosely defined in the literature and has been used to refer broadly to systematic analyses examining sets of genes for common functional properties. In some instances, this broad definition, instead of a clear linear one, can be misleading; for example, the ‘cellular compartment’ ontology (or pathway) in the Gene Ontology (GO) classification does not describe a biological pathway, rather it describes the specific cellular locations where a protein localizes.99
Database-driven pathway analysis
Several studies have analysed GWAS data for enrichment in genes belonging to specific biological pathways, as defined by pathway classification tools such as GO, Kyoto Encyclopedia of Genes and Genomes (KEGG), Reactome, PantherDB and BioCarta.100–103 Biological notions of pathways, as well as detail and quality of annotation, differ between pathway databases.104–106 As a result, outcomes of database-driven pathway analyses depend on the pathway ontologies used.107 Nevertheless, such analyses have confirmed broad statements about the aetiology of RA by showing enrichment of RA susceptibility loci in pathways related to immune functions,100,101 T-cell activation and/or differentiation,101,102 JAK-STAT pathway signalling,102 and TNF signalling.101 Novel approaches are required to gain a more differentiated picture of causal molecular events.
Network analysis
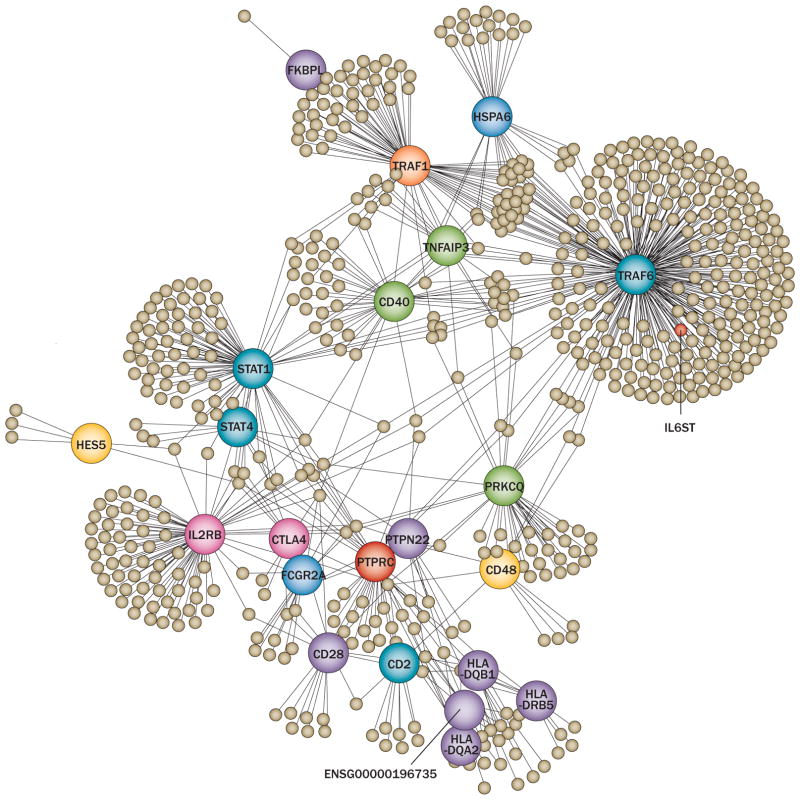
Another way to investigate the function of RA susceptibility loci in biological pathways is through bioinformatic identification of proteins that their products physically interact with. Such approaches commonly include protein–protein interaction network analyses. Protein–protein interaction databases such as the Human Protein Reference Database108 or text mining techniques such as GRAIL35 can be used to construct networks. Other emerging techniques integrate pathway and network-oriented analysis.109,110 Several proteins encoded by RA susceptibility genes are consequently thought to interact or bind with each other (Figure 4).108,109
Figure 4.
Network analysis to infer functional characteristics of genetic variants implicated in RA susceptibility. Analysis of RA susceptibility genes using protein–protein interaction databases shows the extent of potential physical interaction between their protein products. This network has been built using RA candidate genes and comprises major and minor network nodes (large and small circles respectively), both representing proteins. Several confirmed RA suceptibiltiy loci can be recognized among major nodes. The lines indicate physical interactions. Reproduced from Rossin, E. J. et al. PLoS Genet. 2011 Jan; 7(1): e1001273,109 which is published under an open-access license under the terms of the Creative Commons Attribution License.
Inferring pathways from expression data
Cell-specific expression analysis can, as a proxy for gene function, be particularly useful in identifying pathways, cell types, and regulatory programmes relevant to RA. Our group recently mapped RA susceptibility markers to specific cell types.33 As a comprehensive and unbiased catalogue of gene functions is not available, we used a compendium of gene expression data as an objective proxy for tissue-specific gene function. We observed that CD4+ effector memory T cells were highly enriched for the specific expression of genes within RA risk loci.
Integrating data—the SLE example
A 2012 systematic review of putative pathogenic mechanisms in SLE illustrates how experimental data from various sources can be integrated into a single disease model: SLE pathology is hypothesized to result from type I interferon (IFN) misregulation.111 Data from single-gene disorders, GWAS, gene expression micro-arrays, and serologic studies tend to converge towards a linear disease model: immune complexes bind to Toll-like receptors (TLRs) on plasmacytoid dendritic cells; type I IFN production by dendritic cells is triggered; IFN binds to its receptor on target cells; JAK-STAT signalling is activated and the expression of hundreds of genes—the ‘IFN signature’—is altered, leading to disease manifestation. Type I IFN levels and its signature can both be directly measured in peripheral blood. Of 47 loci associated with SLE susceptibility, 27 (57%) are involved in type I IFN production or signalling. Evidence supporting the direct involvement of type I IFN in SLE pathogenesis has paved the way for new therapeutic approaches targeting type I IFN.111 No such evidence of a clear disease pathway is available yet for RA.
Epigenetics of RA
Results from twin studies support a substantial role for environmental triggers in determining RA risk, as evidenced by high discordance rates between monozygotic twins.6 However, the identities of non-shared environmental exposures remain largely elusive. One of the ways in which individuals may respond to an environmental exposure is through changes in their epigenome. The best-understood epigenetic phenomena include post-translational histone modifications and DNA methylation, both of which have a profound influence on gene expression.112
Epigenetic mechanisms
Methylation of DNA cytosine residues at the carbon 5 position, generating 5-methylcytosine, occurs primarily in the context of cytosine-guanine dinucleotides (CpGs). An unexpected feature of the human genome is the relative paucity of CpGs due to the frequent mutation of 5-methylcytosine to thymine.113 Regions of the genome with high CpG content, termed CpG islands (CGI),114 are often hypomethylated and are associated with the promoter regions of actively transcribed genes.115 Methylation in regions up to 2kb away from CGIs (termed GpG island shores) can also strongly influence gene expression.116
N-terminal tails of histone proteins are subject to a wide range of different modifications including acetylation, methylation, phosphorylation and ubiquitylation. More than 60 different histone modification sites have been described.117 A mechanistic connection clearly exists between histone modifications and DNA methylation.118 For example, the presence of DNA methylation reportedly promotes deacetylation of histone 4 and dimethylation of histone 3 at lysine 9, as well as inhibiting methylation of histone 3 at lysine 4, all of which are important modifications that inhibit gene repression.119
Environmental influences
The epigenome has sufficient plasticity to react to the internal and external environment; a range of environmental exposures (such as xenobiotic chemicals and behavioural cues)120 can alter the epigenome. For example, DNA methylation levels at the F2RL3 locus (the gene for proteinase-activated receptor 4) are significantly lower in individuals exposed to cigarette smoke.121 In a related study, F2RL3 methylation status was reported to mediate smoking-associated mortality in patients with stable coronary heart disease.122 Induced epigenetic changes can be inherited during cell division, thereby maintaining the acquired phenotype in daughter cells.120 In addition, stochastic epigenetic instability may accumulate over time in multiple cell types in the absence of obvious environmental stimuli. For example, methylation patterns are more poorly conserved than DNA sequences during mitosis. The error rate for the maintenance of methylation is approximately 10−3 per base pair, whereas the error rate for DNA sequence is approximately 10−6 per base pair.123 Phenotypic differences in genetically identical siblings could conceivably be determined more by this stochastic variation in the epigenome than by epigenetic differences due to non-shared environmental effects.124
Cell-type considerations
The pluripotency of cells decreases during cellular differentiation as gene expression programmes become more restricted.125 This process, which results in the acquisition of cell-type specific features, is controlled epigenetically and is characterized by a specific set of histone modifications and DNA methylation patterns. For example, histone acetylation at the IFN-γ promoter occurs during differentiation of naive T helper (TH0) cells into cells with the TH1 phenotype. 126 This modification reduces the affinity between the histone and DNA, increasing access for transcription factors. When investigating epigenetic alterations in the context of disease pathogenesis it is therefore essential to focus on a pure or enriched cell type that is relevant to the disease under investigation. This requirement is particularly challenging in RA, wherein the most relevant cell subsets are not immediately obvious.
Epigenetic studies in RA
Data for epigenetic phenomena in RA are currently limited, especially in terms of study scale and power.127 However, some interesting observations from studies of DNA methylation patterns are beginning to emerge. For example, analysis of DNA methylation in T cells has revealed global hypomethylation in cells derived from patients with RA compared with those from healthy controls.128 DNA hypomethylation has also been observed in RA fibroblast-like synoviocytes (FLS), as compared with normal FLS, derived from small joint post-trauma biopsy samples.129 In 2013, Nakano et al.130 published a genome-wide evaluation of FLS derived from patients with RA or osteoarthritis (OA), reporting that as many as 1,859 loci, relevant to cell movement, adhesion and trafficking, were differentially methylated in RA (732 hypomethylated and 1,127 hypermethylated). This study was performed using the latest Illumina HumanMethylation450 BeadChip, which provides comprehensive gene region (for example, promoter, exon 1, gene body, 5′ and 3′ untranslated regions) coverage of over 96% of NCBI Reference Sequence genes.
In a gene-targeted approach Nile et al.131 investigated DNA methylation patterns in the promoter region of IL6 in peripheral blood mononuclear cells (PBMC) derived from patients with RA (n = 8) and healthy controls (n = 5). This study identified a single CpG motif 1,099 base pairs upstream of the IL6 transcription start site that was less methylated in patients with RA than in controls. Electrophoretic mobility-shift assay (EMSA) experiments supported this finding in that reduced methylation at the –1,099 locus was reported to correlate with increased binding of nuclear proteins to the genomic DNA.131 However, further experiments in isolated B cells will be needed to further support these interesting data.
Representing a new class of modulators of gene expression, miRNAs base-pair with the 3′-untranslated region of target mRNAs leading to mRNA degradation or inhibition of translation.132 Increased expression of miRNA-115133 and miRNA-203134 has been observed in RA FLS (compared with OA FLS) and this increase correlates with elevated levels of matrix metalloproteinase-1 (MMP-1) and IL-6. It is important to note that expression of miRNA-115132 and of miRNA-203134 are inversely correlated with levels of DNA methylation.
Improving epigenetic knowledge in RA
Correct study design will be critical if our understanding of the role of epigenetic alterations in RA is to expand. Retrospective case–control studies are possible and may also include GWAS information, but care must be taken to ensure that observed differences reflect true epigenetic differences and not variance in, for example, cell-type composition. Retrospective studies are limited in that they cannot determine whether an observed epigenetic mark is causal or consequential (secondary, for example, to therapeutic intervention or the inflammatory response). Investigations including disease discordant monozygotic twins are useful as they control for differences due to germline sequence variation and gender; however, unless samples are collected longitudinally, which would be very difficult in a late onset disease such as RA, cause will not be distinguished from consequence. Longitudinal cohorts of people initially free from disease (for example, the 1958 birth cohort in the UK)135 would avoid confounding due to differences in recruitment of cases and controls, and avoid bias due to case-control differences in the measurement of non-genetic risk factors. Longitudinal cohorts would be essential for establishing the temporal origin of deleterious events and distinguishing causal from consequential effects.136 Important considerations in designing epigenetic studies include sample throughput methods and genome coverage and resolution. For studies of DNA methylation, array-based approaches involving bisulphite conversion are currently the most powerful, but a shift towards whole-genome bisulphite sequencing is likely in the future.137
Epigenetic alterations might prove useful in the clinical setting as markers of disease progression or response to treatment. Furthermore, epigenetic alterations provide new and important targets for the development of therapeutics in RA. Histone deacetylase inhibitors (HDACIs) are currently the best-studied epigenetic therapeutic agents.138 The anti-inflammatory properties of HDACIs include reductions in the levels of cytokines such as TNF, IL-6 and INF-γ.139,140 HDACIs could represent, in the future, a suitable therapeutic option for the treatment of autoimmune diseases such as RA as they are well tolerated at low doses and orally active.141
Future directions
Future challenges in understanding and leveraging RA genetics and epigenetics include further identification of causal genetic variants and their functional characterization, investigation of the role of epigenetic modifications in RA pathogenesis, and translation of fundamental discoveries into clinical practice. Accurate risk prediction in susceptible individuals142 will allow preventive intervention; in patients, individual predictions of disease outcome143 and treatment response144 will pave the way to personalized medicine and allow more efficient patient care. A better understanding of RA molecular pathogenesis will enable the development of new intervention strategies.
Those future tasks are likely to be achieved by the use of new technologies and innovative research strategies. Next-generation sequencing will facilitate whole-exome and whole-genome investigations, in particular studies of the role of rare (<0.5%) genetic variants in large cohorts. Rare variants might explain a certain proportion of the missing heritability of RA, and growing evidence indicates that such alleles are functionally important, penetrant, and harbour larger effect sizes than common variations.145,146 Deep re-sequencing, which identified new and independent effects in other autoimmune diseases,147,148 could be applied to RA.
New technologies will also drive epigenetic studies.137 Epigenetic modifications are potentially influenced by genetic factors as well as by environmental signals including those, such as cigarette smoke, that are known to influence RA risk.
Conclusions
Despite the large number of RA susceptibility loci identified in recent years, genetic risk prediction of RA cannot be performed with sufficient accuracy to enter clinical practice.149,142 Nevertheless, as a result of GWAS and related studies, new pathogenic pathways have been revealed, and mechanisms of some existing drugs are becoming clearer. Genetic sequence variants are unlikely to explain all of the variation in gene function that underpins RA. Correct gene function also depends on appropriate epigenetic programming, which differs between cell and tissue types, and between different stages of cellular development.
Little is currently known about the extent of epigenetic burden in RA; however, epigenetic data in this disease are beginning to accumulate. It will be important for future epigenetic studies in RA to focus on the correct cell types and, in targeted approaches, the correct biological pathways.
Genetic testing has already entered clinical practice in oncology and predicting drug response is currently part of everyday practice in some oncologic subspecialities. Although the genetic architecture of disease susceptibility, severity and treatment response differs significantly between cancers and autoimmune diseases, genetic testing is likely to enter clinical practice in rheumatology in the next decade.
Key points.
Nearly 60 loci associated with susceptibility to rheumatoid arthritis (RA) have been identified across multiple populations, and are at least partially shared between ethnicities
Five amino acid positions, all located in peptide-binding grooves, almost completely explain the association between MHC polymorphisms and RA risk, revitalizing the ‘arthritogenic peptide hypothesis’
Cumulatively, genetic markers identified to date explain only 50% of RA heritability
Using genetics to identify causative disease pathways represents a major challenge for the future
Epigenetic changes in RA remain underexplored and represent a promising new area to link genetics and gene expression with disease risk
Although genetics can be used to stratify disease risk, clinical predictions for the development and progression of RA cannot yet be performed with sufficient accuracy in individual patients
Review criteria.
The PubMed database was searched using the following terms: “genetics AND (rheumatoid OR arthritis)”, “epigenetics AND (rheumatoid OR arthritis)” for full papers and abstracts published online and/or in print in English up to June 2012. References to be included were selected by the authors according to their opinion of their relevance to the scope of this Review, and further papers were identified from the reference lists of relevant publications. Some reports published after June 2012 and identified during revisions to this manuscript have also been included. Pathways presented in Figure 3 were curated manually from the literature; only well established interactions were considered.
Acknowledgments
S. Viatte’s research activities are supported by a grant from the Swiss Foundation for Medical-Biological Scholarships (SSMBS), managed by the Swiss National Science Foundation and financed by a donation from Novartis (PASMP3 134380). The work of S. Raychaudhuri is supported by grants from the National Institutes of Health (5K08AR055688 and 1R01AR062886) and an Arthritis Foundation Innovator Award. This manuscript was also funded by a core programme grant from Arthritis Research UK (17552).
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
All authors contributed equally to researching data for the article, writing the article, discussions of the content, and review and/or editing of the article before submission.
References
- 1.Arend WP, Firestein GS. Pre-rheumatoid arthritis: predisposition and transition to clinical synovitis. Nat Rev Rheumatol. 2012;8:573–586. doi: 10.1038/nrrheum.2012.134. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence JS. Heberden Oration, 1969. Rheumatoid arthritis—nature or nurture? Ann Rheum Dis. 1970;29:357–379. doi: 10.1136/ard.29.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.del Junco D, Luthra HS, Annegers JF, Worthington JW, Kurland LT. The familial aggregation of rheumatoid arthritis and its relationship to the HLA-DR4 association. Am J Epidemiol. 1984;119:813–829. doi: 10.1093/oxfordjournals.aje.a113802. [DOI] [PubMed] [Google Scholar]
- 4.Hemminki K, Li X, Sundquist J, Sundquist K. Familial associations of rheumatoid arthritis with autoimmune diseases and related conditions. Arthritis Rheum. 2009;60:661–668. doi: 10.1002/art.24328. [DOI] [PubMed] [Google Scholar]
- 5.Jones MA, Silman AJ, Whiting S, Barrett EM, Symmons DP. Occurrence of rheumatoid arthritis is not increased in the first degree relatives of a population based inception cohort of inflammatory polyarthritis. Ann Rheum Dis. 1996;55:89–93. doi: 10.1136/ard.55.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacGregor AJ, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43:30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 7.Silman AJ, Newman J, MacGregor AJ. Cigarette smoking increases the risk of rheumatoid arthritis. Results from a nationwide study of disease-discordant twins. Arthritis Rheum. 1996;39:732–735. doi: 10.1002/art.1780390504. [DOI] [PubMed] [Google Scholar]
- 8.Cutolo M. Sex and rheumatoid arthritis: mouse model versus human disease. Arthritis Rheum. 2007;56:1–3. doi: 10.1002/art.22322. [DOI] [PubMed] [Google Scholar]
- 9.Luckey D, Medina K, Taneja V. B cells as effectors and regulators of sex-biased arthritis. Autoimmunity. 2012;45:364–376. doi: 10.3109/08916934.2012.665528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eyre S, et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44:1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 12.Astorga GP, Williams RC., Jr Altered reactivity in mixed lymphocyte culture of lymphocytes from patients with rheumatoid arthritis. Arthritis Rheum. 1969;12:547–554. doi: 10.1002/art.1780120602. [DOI] [PubMed] [Google Scholar]
- 13.Stastny P. Mixed lymphocyte cultures in rheumatoid arthritis. J Clin Invest. 1976;57:1148–1157. doi: 10.1172/JCI108382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stastny P. Association of the B-cell alloantigen DRw4 with rheumatoid arthritis. N Engl J Med. 1978;298:869–871. doi: 10.1056/NEJM197804202981602. [DOI] [PubMed] [Google Scholar]
- 15.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 16.Begovich AB, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki A, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 18.Plenge RM, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77:1044–1060. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 20.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 21.Okada Y, et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet. 2012;44:511–516. doi: 10.1038/ng.2231. [DOI] [PubMed] [Google Scholar]
- 22.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raychaudhuri S, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raychaudhuri S. Recent advances in the genetics of rheumatoid arthritis. Curr Opin Rheumatol. 2010;22:109–118. doi: 10.1097/BOR.0b013e328336474d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stahl EA, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet. 2012;44:483–489. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willemze A, Trouw LA, Toes RE, Huizinga TW. The influence of ACPA status and characteristics on the course of RA. Nat Rev Rheumatol. 2012;8:144–152. doi: 10.1038/nrrheum.2011.204. [DOI] [PubMed] [Google Scholar]
- 27.Huizinga TW, et al. Refining the complex rheumatoid arthritis phenotype based on specificity of the HLA-DRB1 shared epitope for antibodies to citrullinated proteins. Arthritis Rheum. 2005;52:3433–3438. doi: 10.1002/art.21385. [DOI] [PubMed] [Google Scholar]
- 28.Viatte S, et al. Genetic markers of rheumatoid arthritis susceptibility in anti-citrullinated peptide antibody negative patients. Ann Rheum Dis. 2012;71:1984–1990. doi: 10.1136/annrheumdis-2011-201225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daha NA, Toes RE. Rheumatoid arthritis: Are ACPA-positive and ACPA-negative RA the same disease? Nat Rev Rheumatol. 2011;7:202–203. doi: 10.1038/nrrheum.2011.28. [DOI] [PubMed] [Google Scholar]
- 30.Arnett FC, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;3:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 31.van der Woude D, et al. Quantitative heritability of anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum. 2009;60:916–923. doi: 10.1002/art.24385. [DOI] [PubMed] [Google Scholar]
- 32.Shi J, et al. Autoantibodies recognizing carbamylated proteins are present in sera of patients with rheumatoid arthritis and predict joint damage. Proc Natl Acad Sci USA. 2011;108:17372–17377. doi: 10.1073/pnas.1114465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu X, et al. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am J Hum Genet. 2011;89:496–506. doi: 10.1016/j.ajhg.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gregersen PK, et al. REL, encoding a member of the NF-κB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41:820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raychaudhuri S, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Vries R. Genetics of rheumatoid arthritis: time for a change! Curr Opin Rheumatol. 2011;23:227–232. doi: 10.1097/BOR.0b013e3283457524. [DOI] [PubMed] [Google Scholar]
- 37.McAllister K, Eyre S, Orozco G. Genetics of rheumatoid arthritis: GWAS and beyond. OA Rheumatol Res Rev. 2011;3:31–46. doi: 10.2147/OARRR.S14725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raychaudhuri S, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.EMBL-EBI. IMGT/HLA database. 2012 [online], http://www.ebi.ac.uk/imgt/hla/
- 40.Thomas R, Lipsky PE. Could endogenous self-peptides presented by dendritic cells initiate rheumatoid arthritis? Immunol Today. 1996;17:559–564. doi: 10.1016/s0167-5699(96)20030-1. [DOI] [PubMed] [Google Scholar]
- 41.Jenkins MK, Mueller D. On the trail of arthritogenic T cells. Arthritis Rheum. 2011;63:2851–2853. doi: 10.1002/art.30440. [DOI] [PubMed] [Google Scholar]
- 42.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423:356–361. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 43.Cornelis F, et al. New susceptibility locus for rheumatoid arthritis suggested by a genome-wide linkage study. Proc Natl Acad Sci USA. 1998;95:10746–10750. doi: 10.1073/pnas.95.18.10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jawaheer D, et al. A genomewide screen in multiplex rheumatoid arthritis families suggests genetic overlap with other autoimmune diseases. Am J Hum Genet. 2001;68:927–936. doi: 10.1086/319518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jawaheer D, et al. Screening the genome for rheumatoid arthritis susceptibility genes: a replication study and combined analysis of 512 multicase families. Arthritis Rheum. 2003;48:906–916. doi: 10.1002/art.10989. [DOI] [PubMed] [Google Scholar]
- 46.MacKay K, et al. Whole-genome linkage analysis of rheumatoid arthritis susceptibility loci in 252 affected sibling pairs in the United Kingdom. Arthritis Rheum. 2002;46:632–639. doi: 10.1002/art.10147. [DOI] [PubMed] [Google Scholar]
- 47.Shiozawa S, et al. Identification of the gene loci that predispose to rheumatoid arthritis. Int Immunol. 1998;10:1891–1895. doi: 10.1093/intimm/10.12.1891. [DOI] [PubMed] [Google Scholar]
- 48.Ding B, et al. Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum. 2009;60:30–38. doi: 10.1002/art.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee HS, et al. Several regions in the major histocompatibility complex confer risk for anti-CCP-antibody positive rheumatoid arthritis, independent of the DRB1 locus. Mol Med. 2008;14:293–300. doi: 10.2119/2007-00123.Lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vignal C, et al. Genetic association of the major histocompatibility complex with rheumatoid arthritis implicates two non-DRB1 loci. Arthritis Rheum. 2009;60:53–62. doi: 10.1002/art.24138. [DOI] [PubMed] [Google Scholar]
- 51.Mackie SL, et al. A spectrum of susceptibility to rheumatoid arthritis within HLA-DRB1: stratification by autoantibody status in a large UK population. Genes Immun. 2012;13:120–128. doi: 10.1038/gene.2011.60. [DOI] [PubMed] [Google Scholar]
- 52.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pereyra F, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Woude D, et al. Protection against anti-citrullinated protein antibody-positive rheumatoid arthritis is predominantly associated with HLA-DRB1*1301: a meta-analysis of HLA-DRB1 associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in four European populations. Arthritis Rheum. 2010;62:1236–1245. doi: 10.1002/art.27366. [DOI] [PubMed] [Google Scholar]
- 55.Holoshitz J. The rheumatoid arthritis HLA-DRB1 shared epitope. Curr Opin Rheumatol. 2010;22:293–298. doi: 10.1097/BOR.0b013e328336ba63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.du Montcel ST, et al. New classification of HLA-DRB1 alleles supports the shared epitope hypothesis of rheumatoid arthritis susceptibility. Arthritis Rheum. 2005;52:1063–1068. doi: 10.1002/art.20989. [DOI] [PubMed] [Google Scholar]
- 57.Michou L, et al. Validation of the reshaped shared epitope HLA-DRB1 classification in rheumatoid arthritis. Arthritis Res Ther. 2006;8:R79. doi: 10.1186/ar1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morgan AW, et al. The shared epitope hypothesis in rheumatoid arthritis: evaluation of alternative classification criteria in a large UK Caucasian cohort. Arthritis Rheum. 2008;58:1275–1283. doi: 10.1002/art.23432. [DOI] [PubMed] [Google Scholar]
- 59.van der Helm-van Mil AH, et al. An independent role of protective HLA class II alleles in rheumatoid arthritis severity and susceptibility. Arthritis Rheum. 2005;52:2637–2644. doi: 10.1002/art.21272. [DOI] [PubMed] [Google Scholar]
- 60.Shadick NA, et al. Opposing effects of the D70 mutation and the shared epitope in HLA-DR4 on disease activity and certain disease phenotypes in rheumatoid arthritis. Ann Rheum Dis. 2007;66:1497–1502. doi: 10.1136/ard.2006.067603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hill JA, et al. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J Immunol. 2003;171:538–541. doi: 10.4049/jimmunol.171.2.538. [DOI] [PubMed] [Google Scholar]
- 62.Snir O, et al. Identification and functional characterization of T cells reactive to citrullinated vimentin in HLA-DRB1*0401-positive humanized mice and rheumatoid arthritis patients. Arthritis Rheum. 2011;63:2873–2883. doi: 10.1002/art.30445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Law SC, et al. T-cell autoreactivity to citrullinated autoantigenic peptides in rheumatoid arthritis patients carrying HLA-DRB1 shared epitope alleles. Arthritis Res Ther. 2012;14:R118. doi: 10.1186/ar3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Viatte S, Alves PM, Romero P. Reverse immunology approach for the identification of CD8 T-cell-defined antigens: advantages and hurdles. Immunol Cell Biol. 2006;84:318–330. doi: 10.1111/j.1440-1711.2006.01447.x. [DOI] [PubMed] [Google Scholar]
- 65.Bax M, van HJ, Huizinga TW, Toes RE. Genetics of rheumatoid arthritis: what have we learned? Immunogenetics. 2011;63:459–466. doi: 10.1007/s00251-011-0528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kunz M, Ibrahim SM. Non-major histocompatibility complex rheumatoid arthritis susceptibility genes. Crit Rev Immunol. 2011;31:99–114. doi: 10.1615/critrevimmunol.v31.i2.20. [DOI] [PubMed] [Google Scholar]
- 67.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suzuki A, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 69.Kurreeman FA, et al. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLoS Med. 2007;4:e278. doi: 10.1371/journal.pmed.0040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kochi Y, et al. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat Genet. 2005;37:478–485. doi: 10.1038/ng1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Plenge RM, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Plenge RM, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis—a genome-wide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kochi Y, et al. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat Genet. 2010;42:515–519. doi: 10.1038/ng.583. [DOI] [PubMed] [Google Scholar]
- 75.Thomson W, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39:1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barton A, et al. Rheumatoid arthritis susceptibility loci at chromosomes 10p15, 12q13 and 22q13. Nat Genet. 2008;40:1156–1159. doi: 10.1038/ng.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Freudenberg J, et al. Genome-wide association study of rheumatoid arthritis in Koreans: population-specific loci as well as overlap with European susceptibility loci. Arthritis Rheum. 2011;63:884–893. doi: 10.1002/art.30235. [DOI] [PubMed] [Google Scholar]
- 78.de Bakker PI, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Orozco G, et al. Study of the common genetic background for rheumatoid arthritis and systemic lupus erythematosus. Ann Rheum Dis. 2011;70:463–468. doi: 10.1136/ard.2010.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhernakova A, et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 2011;7:e1002004. doi: 10.1371/journal.pgen.1002004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kurreeman FA, et al. Use of a multiethnic approach to identify rheumatoid-arthritis-susceptibility loci, 1p36 and 17q12. Am J Hum Genet. 2012;90:524–532. doi: 10.1016/j.ajhg.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kochi Y, Suzuki A, Yamada R, Yamamoto K. Ethnogenetic heterogeneity of rheumatoid arthritis-implications for pathogenesis. Nat Rev Rheumatol. 2010;6:290–295. doi: 10.1038/nrrheum.2010.23. [DOI] [PubMed] [Google Scholar]
- 83.Rhee I, Veillette A. Protein tyrosine phosphatases in lymphocyte activation and autoimmunity. Nat Immunol. 2012;13:439–447. doi: 10.1038/ni.2246. [DOI] [PubMed] [Google Scholar]
- 84.Vang T, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37:1317–1319. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 85.Zhang J, et al. The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat Genet. 2011;43:902–907. doi: 10.1038/ng.904. [DOI] [PubMed] [Google Scholar]
- 86.Maine CJ, et al. PTPN22 alters the development of regulatory T cells in the thymus. J Immunol. 2012;188:5267–5275. doi: 10.4049/jimmunol.1200150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brownlie RJ, et al. Lack of the phosphatase PTPN22 increases adhesion of murine regulatory T cells to improve their immunosuppressive function. Sci Signal. 2012;5:ra87. doi: 10.1126/scisignal.2003365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ronninger M, et al. The balance of expression of PTPN22 splice forms is significantly different in rheumatoid arthritis patients compared with controls. Genome Med. 2012;4:2. doi: 10.1186/gm301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harrison P, et al. Evidence of cis-acting regulatory variation in PTPN22 in patients with rheumatoid arthritis. Scand J Rheumatol. 2012;41:249–252. doi: 10.3109/03009742.2012.658859. [DOI] [PubMed] [Google Scholar]
- 90.James EA, et al. HLA-DR1001 presents “altered-self” peptides derived from joint-associated proteins by accepting citrulline in three of its binding pockets. Arthritis Rheum. 2010;62:2909–2918. doi: 10.1002/art.27594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Feitsma AL, et al. Identification of citrullinated vimentin peptides as T cell epitopes in HLA-DR4-positive patients with rheumatoid arthritis. Arthritis Rheum. 2010;62:117–125. doi: 10.1002/art.25059. [DOI] [PubMed] [Google Scholar]
- 92.Dendrou CA, et al. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat Genet. 2009;41:1011–1015. doi: 10.1038/ng.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith KA, Popmihajlov Z. The quantal theory of immunity and the interleukin-2-dependent negative feedback regulation of the immune response. Immunol Rev. 2008;224:124–140. doi: 10.1111/j.1600-065X.2008.00654.x. [DOI] [PubMed] [Google Scholar]
- 94.Orozco G, et al. Combined effects of three independent SNPs greatly increase the risk estimate for RA at 6q23. Hum Mol Genet. 2009;18:2693–2699. doi: 10.1093/hmg/ddp193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Adrianto I, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kool M, et al. The ubiquitin-editing protein A20 prevents dendritic cell activation, recognition of apoptotic cells, and systemic autoimmunity. Immunity. 2011;35:82–96. doi: 10.1016/j.immuni.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 97.Matmati M, et al. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat Genet. 2011;43:908–912. doi: 10.1038/ng.874. [DOI] [PubMed] [Google Scholar]
- 98.Raychaudhuri S. Mapping rare and common causal alleles for complex human diseases. Cell. 2011;147:57–69. doi: 10.1016/j.cell.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265–272. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 102.Eleftherohorinou H, et al. Pathway analysis of GWAS provides new insights into genetic susceptibility to 3 inflammatory diseases. PLoS ONE. 2009;4:e8068. doi: 10.1371/journal.pone.0008068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Beyene J, et al. Pathway-based analysis of a genome-wide case-control association study of rheumatoid arthritis. BMC Proc. 2009;3 (Suppl 7):S128. doi: 10.1186/1753-6561-3-s7-s128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8:e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kraft P, Raychaudhuri S. Complex diseases, complex genes: keeping pathways on the right track. Epidemiology. 2009;20:508–511. doi: 10.1097/EDE.0b013e3181a93b98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 107.Green ML, Karp PD. The outcomes of pathway database computations depend on pathway ontology. Nucleic Acids Res. 2006;34:3687–3697. doi: 10.1093/nar/gkl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nakaoka H, et al. A systems genetics approach provides a bridge from discovered genetic variants to biological pathways in rheumatoid arthritis. PLoS ONE. 2011;6:e25389. doi: 10.1371/journal.pone.0025389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bakir-Gungor B, Sezerman OU. A new methodology to associate SNPs with human diseases according to their pathway related context. PLoS ONE. 2011;6:e26277. doi: 10.1371/journal.pone.0026277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bronson PG, Chaivorapol C, Ortmann W, Behrens TW, Graham RR. The genetics of type I interferon in systemic lupus erythematosus. Curr Opin Immunol. 2012;24:530–537. doi: 10.1016/j.coi.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 112.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 113.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 114.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 115.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 118.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 119.Hashimshony T, Zhang J, Keshet I, Bustin M, Cedar H. The role of DNA methylation in setting up chromatin structure during development. Nat Genet. 2003;34:187–192. doi: 10.1038/ng1158. [DOI] [PubMed] [Google Scholar]
- 120.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Breitling LP, Salzmann K, Rothenbacher D, Burwinkel B, Brenner H. Smoking, F2RL3 methylation, and prognosis in stable coronary heart disease. Eur Heart J. 2012;33:2841–2848. doi: 10.1093/eurheartj/ehs091. [DOI] [PubMed] [Google Scholar]
- 123.Ushijima T, et al. Fidelity of the methylation pattern and its variation in the genome. Genome Res. 2003;13:868–874. doi: 10.1101/gr.969603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bouchard TJ, Jr, Lykken DT, McGue M, Segal NL, Tellegen A. Sources of human psychological differences: the Minnesota Study of Twins Reared Apart. Science. 1990;250:223–228. doi: 10.1126/science.2218526. [DOI] [PubMed] [Google Scholar]
- 125.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 126.Avni O, et al. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 127.Ballestar E. Epigenetic alterations in autoimmune rheumatic diseases. Nat Rev Rheumatol. 2011;7:263–271. doi: 10.1038/nrrheum.2011.16. [DOI] [PubMed] [Google Scholar]
- 128.Richardson B, et al. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 129.Karouzakis E, Gay RE, Michel BA, Gay S, Neidhart M. DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2009;60:3613–3622. doi: 10.1002/art.25018. [DOI] [PubMed] [Google Scholar]
- 130.Nakano K, Whitaker JW, Boyle DL, Wang W, Firestein GS. DNA methylome signature in rheumatoid arthritis. Ann Rheum Dis. 2013;72:110–117. doi: 10.1136/annrheumdis-2012-201526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008;58:2686–2693. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- 132.Baer C, et al. Extensive promoter DNA hypermethylation and hypomethylation is associated with aberrant microRNA expression in chronic lymphocytic leukemia. Cancer Res. 2012;72:3775–3785. doi: 10.1158/0008-5472.CAN-12-0803. [DOI] [PubMed] [Google Scholar]
- 133.Stanczyk J, et al. Altered expression of microRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 134.Stanczyk J, et al. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 2011;63:373–381. doi: 10.1002/art.30115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study) Int J Epidemiol. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- 136.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 138.Khan N, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 139.Leoni F, et al. The histone deacetylase inhibitor ITF2357 reduces production of pro-inflammatory cytokines in vitro and systemic inflammation in vivo. Mol Med. 2005;11:1–15. doi: 10.2119/2006-00005.Dinarello. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Leoni F, et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proc Natl Acad Sci USA. 2002;99:2995–3000. doi: 10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.DeSantis M, Selmi C. The therapeutic potential of epigenetics in autoimmune diseases. Clin Rev Allergy Immunol. 2012;42:92–101. doi: 10.1007/s12016-011-8293-8. [DOI] [PubMed] [Google Scholar]
- 142.van der Helm-van Mil AH, Toes RE, Huizinga TW. Genetic variants in the prediction of rheumatoid arthritis. Ann Rheum Dis. 2010;69:1694–1696. doi: 10.1136/ard.2009.123828. [DOI] [PubMed] [Google Scholar]
- 143.Viatte S, Barton A. The role of rheumatoid arthritis genetic susceptibility markers in the prediction of erosive disease. European Musculoskeletal Rev. 2012;7:102–107. [Google Scholar]
- 144.Plant D, et al. Genome-wide association study of genetic predictors of anti-tumor necrosis factor treatment efficacy in rheumatoid arthritis identifies associations with polymorphisms at seven loci. Arthritis Rheum. 2011;63:645–653. doi: 10.1002/art.30130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Raychaudhuri S, et al. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet. 2011;43:1232–1236. doi: 10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rivas MA, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Aschard H, et al. Inclusion of gene–gene and gene–environment interactions unlikely to dramatically improve risk prediction for complex diseases. Am J Hum Genet. 2012;90:962–972. doi: 10.1016/j.ajhg.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]