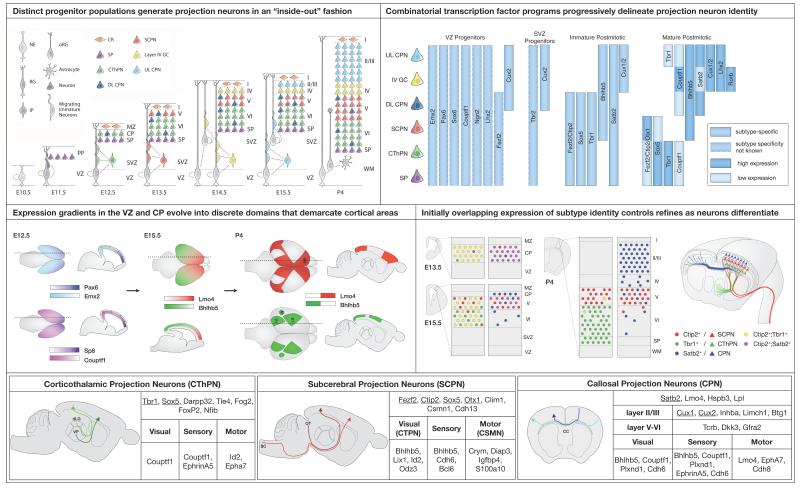
This SnapShot summarizes current knowledge of mammalian cortical development, with a particular focus on the molecular controls that orchestrate the stepwise decisions leading from multiple types of undifferentiated forebrain progenitors to fully mature projection neurons with correctly-targeted axons and carefully-elaborated dendritic trees, as well as appropriate electrophysiology and gene expression, reflective of precise subtype and area identity.
Neocortical Progenitors
Early in development, the telencephalic wall is composed of undifferentiated neuroepithelial (NE) cells, which give rise to diverse progenitor populations. Radial glial cells (RG) divide asymmetrically to self-renew and generate intermediate progenitor (IP) cells or neurons. IP cells divide symmetrically to produce two neurons. In the mouse, small numbers of neurons are produced by radial glia-like (oRG) cells, but oRG cells are abundant in the outer SVZ of human fetal cortex where they generate transit amplifying cells that in turn produce most cortical neurons.
Projection Neuron Diversity
Specific subtypes of neocortical projection neurons are generated by neural progenitors during distinct temporal windows, beginning in mice at approximately E11.5, and continuing through late embryonic development. These young postmitotic neurons migrate away from the ventricular zone to populate progressively more superficial positions in the cortical plate. Projection neurons can be classified on the basis of their mature axonal projections: corticothalamic projection neurons (CThPN) are located in layer VI and send axons to thalamus; subcerebral projection neurons (SCPN) are located in layer V and send axons to optic tectum, brainstem, or spinal cord; and callosal projection neurons (CPN) are located in layers II/III, V, and VI and send axons to contralateral cortex. Importantly, neurons of each subtype are further specialized based on their positions in specific cortical areas. For example, CThPN establish area-specific connections with thalamic nuclei (motor cortex CThPN with VL; sensory cortex CThPN with VP; visual cortex CThPN with dLG).
Molecular Controls over Subtype and Area Identity
Both subtype and area identity are specified in a stepwise fashion, with early overlapping expression of critical controls resolving over the course of development to specific subtypes and areas. Area identity begins to be imparted embryonically by smooth gradients of transcription factors in progenitors and postmitotic neurons, but during the first postnatal week, expression of critical controls, such as Lmo4 and Bhlhb5, becomes restricted to domains that sharply delineate cortical areas. Similarly, subtype identity is progressively specified, as molecular controls that are initially co-expressed by newly-generated postmitotic neurons later refine to a single subtype, or to high levels in some subtypes and low levels in others. Several central identified controls over subtype development, including Fezf2, Ctip2, Satb2, and Tbr1, interact combinatorially (although not linearly) as part of a broader molecular network and nested molecular logic that directs subtype identity acquisition.
SnapShot: Cortical Development.
Abbreviations
- A1
primary auditory cortex
- Bhlhb5
basic helix-loop-helix domain-containing, class B5
- Btg1
B cell translocation gene 1, anti-proliferative
- Cdh6
cadherin 6
- Cdh8
cadherin 8
- Cdh13
cadherin 13
- Clim1
carboxyl-terminal LIM domain-binding protein 1
- Couptf1
chicken ovalbumin upstream transcription factor I
- CC
corpus callosum
- CP
cortical plate
- CPN
callosal projection neuron(s)
- CR
Cajal-Retzius cell(s)
- Crym
mu crystallin
- CSMN
corticospinal motor neuron(s)
- Csmn1
zinc finger protein 703
- CThPN
corticothalamic projection neuron(s)
- CTPN
corticotectal projection neuron(s)
- Ctip2
Couptf-interacting protein 2
- Cux1
cut-like homeobox 1
- Cux2
cut-like homeobox 2
- Darpp32
dopamine- and cAMP-regulated neuronal phosphoprotein
- Diap3
diaphanous homolog 3
- Dkk3
dickkopf homolog 3
- DL
deep-layer (layers V and VI)
- dLG
dorsal lateral geniculate nucleus of thalamus
- E
embryonic day
- Emx2
empty spiracles homeobox 2
- Epha7
Eph receptor A7
- Fezf2
Fez family zinc finger 2
- Fog2
friend of GATA 2
- FoxP2
forkhead box P2
- GC
granule cell(s)
- Gfra2
glial cell line derived neurotrophic factor family receptor alpha 2
- Hspb3
heat shock protein 3
- Id2
inhibitor of DNA binding 2
- Igfbp4
insulin-like growth factor binding protein 4
- Inhba
inhibin beta-A
- IP
intermediate progenitor
- Lhx2
LIM homeobox protein 2
- Limch1
LIM and calponin homology domains 1
- Lix1
limb expression homolog 1
- Lmo4
LIM domain only 4
- Lpl
lipoprotein lipase
- M1
primary motor cortex
- MZ
marginal zone
- NE
neuroepithelial cell
- Nfib
nuclear factor IB
- Ngn2
neurogenin 2
- Odz3
odd Oz/ten-m homolog 3
- oRG
outer radial glia
- OT
optic tectum (superior colliculus)
- Otx1
orthodenticle homolog 1
- P
postnatal day
- Pax6
paired box gene 6
- Plxnd1
plexin D1
- PP
preplate
- RG
radial glia
- Rorb
RAR-related orphan receptor beta
- S1
primary sensory cortex
- S100a10
S100 calcium binding protein A10
- Satb2
special AT-rich sequence binding protein 2
- SC
spinal cord
- SCPN
subcerebral projection neuron(s)
- Sox5
SRY box-containing gene 5
- Sox6
SRY box-containing gene 6
- SP
subplate neuron(s)
- Sp8
trans-acting transcription factor 8
- SVZ
subventricular zone
- Tbr1
T-box brain gene 1
- Tbr2
T-box brain gene 2
- Tcrb
T cell receptor beta chain
- Tle4
transducin-like enhancer of split 4
- UL
upper-layer (layers II/III and IV)
- V1
primary visual cortex
- VL
ventral lateral nucleus of thalamus
- VP
ventral posterior nucleus of thalamus
- VZ
ventricular zone
- WM
white matter
REFERENCES
- Custo Greig LF*, Woodworth MB*, Galazo MJ#, Padmanabhan H#, Macklis JD. Molecular logic of neocortical projection neuron development. Nat. Rev. Neurosci. 2012 doi: 10.1038/nrn3586. in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Rubenstein JLR, Kriegstein AR. Deriving excitatory neurons of the neocortex from pluripotent stem cells. Neuron. 2011;70:645–660. doi: 10.1016/j.neuron.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr. Opin. Neurobiol. 2008;18:1–8. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ*, Arlotta P*, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- O’Leary DDM, Chou S-J, Sahara S. Area patterning of the mammalian cortex. Neuron. 2007;56:252–269. doi: 10.1016/j.neuron.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Shoemaker LD, Arlotta P. Untangling the cortex: advances in understanding specification and differentiation of corticospinal motor neurons. BioEssays. 2010;9999:1–10. doi: 10.1002/bies.200900114. [DOI] [PMC free article] [PubMed] [Google Scholar]