Summary
A growing class of proteins regulates transcription through interaction with DNA-dependent RNA polymerase. Here we report that a recently identified, highly conserved sporulation gene ylyA encodes a novel RNA polymerase-binding protein that influences the expression of genes under the control of the late-acting, sporulation sigma factor σG in Bacillus subtilis. Spores from a ylyA mutant exhibited defects in germination corresponding to changes in the levels of membrane receptors for spore germinants and a protein channel governing the release of dipicolinic acid and hydration of the spore core during germination. Purified YlyA interacted with RNA polymerase and stimulated transcription from promoters dependent on σG but not promoters dependent on the house-keeping sigma factor σA. YlyA is a previously unrecognized RNA polymerase-binding protein that is dedicated to modulating the expression of genes involved in spore germination.
Keywords: Bacillus subtilis, spore germination, gene regulation, RNA polymerase
Introduction
Bacillus subtilis undergoes a complex process of cellular differentiation in response to changes in the environment that culminates in the formation of a dormant cell type known as the endospore (or simply spore). Spore formation takes place in a two-chamber sporangium consisting of a forespore and a mother-cell compartment. The forespore, which is nurtured by the mother cell, matures into the spore and is ultimately released by lysis of the mother cell. Through membrane-embedded receptors, mature spores monitor the environment, and are capable of rapidly germinating and resuming growth in response to small molecule germinants (Setlow, 2003). Spores of B. subtilis contain three germinant receptors (i.e. GerA, GerB and GerK), each produced from a single tricistronic mRNA. Germinant receptors are present at moderate abundance in spores, and their levels are a major determinant of the rate of germination (Paidhungat & Setlow, 2000, Cabrera-Martinez et al., 2003). Binding of a germinant to one of the germinant receptors triggers the release of the large depot of pyridine-2, 6-dicarboxylic acid (dipicolinic acid (DPA)) from the spore, which results in initiation of the process of full hydration of the spore core, ultimately enabling the spore to resume enzymatic activity (Setlow, 2006). Many of the proteins encoded by the spoVA operon are required for the initial uptake of DPA during sporulation and are also involved in the release of DPA during germination (Li et al., 2012, Vepachedu & Setlow, 2007, Tovar-Rojo et al., 2002). Here we report the identification of a novel regulatory protein involved in the expression of genes governing germination.
The gene regulatory program of sporulation is governed in part by the successive appearance of four sporulation-specific sigma factors, appearing in the order σF, σE, σG, and σK. The activities of these sigma factors are confined to the forespore (σF and σG) or mother cell (σE and σK) compartments (Losick & Stragier, 1992). The late-acting forespore-specific sigma factor σG activates the expression of approximately 100 genes, including the gene for σG itself (sigG) and SpoVT (Wang et al., 2006). SpoVT is a DNA-binding protein that can act as both a positive and a negative regulator of σG-dependent transcription (Bagyan et al., 1996, Wang et al., 2006). Genes under the control of σG and SpoVT include the operons coding for all three of the germinant receptors and the spoVA operon.
We and others recently used gene conservation as an alternative approach to identify previously unrecognized sporulation genes (Traag et al., 2013, Abecasis et al., 2013). One gene identified using this approach, ylyA, is expressed under the control of the forespore-specific transcription factors σG and SpoVT, and deletion of ylyA resulted in a defect in the efficiency of spore germination (Traag et al., 2013). ylyA was previously suggested to have a low level of sequence similarity to E. coli dksA (Krasny & Gourse, 2004). Members of the DksA family transcriptional regulators, which are highly conserved among Gram-negative bacteria, are thought to interact directly with the secondary channel of RNA polymerase and modulate transcription in conjunction with the nucleotide alarmones guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) (Srivatsan & Wang, 2008). This prompted us to investigate a potential role for ylyA in modulating sporulation-specific gene expression. Here we show that YlyA influences the expression of certain σG-controlled genes, including genes involved in germination and that a mutant lacking YlyA produces spores that exhibit specific defects in spore germination. We also show that YlyA binds to RNA polymerase and stimulates transcription by σG-containing RNA polymerase, but not RNA polymerase containing the housekeeping sigma factor σA. Thus, YlyA is a conserved sporulation-specific transcription factor that modulates the expression of a specialized subset of genes involved in spore germination through interaction with RNA polymerase.
Results
YlyA shares similarity with members of a family of RNA polymerase-binding proteins
ylyA, together with yocK and yteA, is one of the three closest homologs of E. coli dksA in B. subtilis (Krasny & Gourse, 2004, Traag et al., 2013). Although the sequence similarity of YlyA to DksA is weak and mostly limited to the C-terminal part of YlyA (Fig 1), structure prediction using the Homology Detection & Structure prediction by HMM-HMM comparison open-software program (HHPred; (Soding et al., 2005)) suggests that the structure of YlyA is highly related (probability score >99.9) to the solved three-dimensional structures of DksA from E.coli (Perederina et al., 2004) and Agrobacterium tumefaciens (DOI:10.2210/pdb2kq9/pdb). DksA consists of two characteristic domains: (i) an N-terminal coiled-coil region with two invariable acidic residues at its tip known as the DxxDxA motif and (ii) N-terminal and C-terminal globular regions including a conserved four cysteine (Cys-4) zinc finger motif (IPR012783) (Perederina et al., 2004). The coiled-coil prediction program COILS/PCOILS (Lupas et al., 1991) predicts that the N-terminal region of YlyA indeed forms a coiled-coil. YlyA, however, lacks an N-terminal globular extension upstream of the coiled-coil, and has no acidic residues (and hence lacks the DxxDxA motif) near the predicted tip of the coiled-coil (Fig 1; note that YocK and YteA each have one of the conserved aspartate residues). In a phylogenetic study of 965 DksA-like proteins, 677 homologs had an intact DxxDxA motif. The remaining sequences, however, contained variations in one or more of the conserved residues, suggesting a possible function distinct from the canonical motif (Furman et al., 2013). Activity has previously only been demonstrated for DksA homologs with this motif intact.
Fig. 1. Alignment of the amino acid sequences of DksA-like proteins.
ClustalW was used to align the amino acid sequence of E. coli and A. tumefaciens DksA, and B. subtilis YlyA, YocK and YteA. Identical residues (*), conserved substitutions of residues with similar properties (:), and semi-conserved substitutions of residues with similar steric confirmations (.) are indicated below the alignment. The N-terminal α-helix coiled-coil (light grey) and the C-terminal Cys-4 zinc finger motif (dark grey) are indicated above the alignment. The conserved residues of the DxxDxA motif, and the conserved first cysteine residue of the Cys-4 zinc finger motif are shaded in grey.
An intact Cys-4 zinc finger motif was found in 561 out of 965 DksA homologs (Furman et al., 2013). The C-terminal motif of YlyA has only a single conserved cysteine residue (Fig 1), which is insufficient for a monomer of YlyA to coordinate zinc. One of the two DksA paralogs from Pseudomonas aeruginosa (named DksA2) lacks an intact Cys-4 zinc finger motif and zinc was not found in association with this protein. This variant, however, could functionally substitute for the canonical DksA in vivo and in vitro (Blaby-Haas et al., 2011). Note that YocK, YteA, and A. tumefaciens DksA, to which YlyA was found to be structurally similar, also only contain the first of the four conserved cysteine residues in the motif (Fig 1). We conclude that ylyA encodes a divergent homolog of the DksA family, lacking some of the features characteristic of canonical DksA proteins.
YlyA influences σG-directed transcription
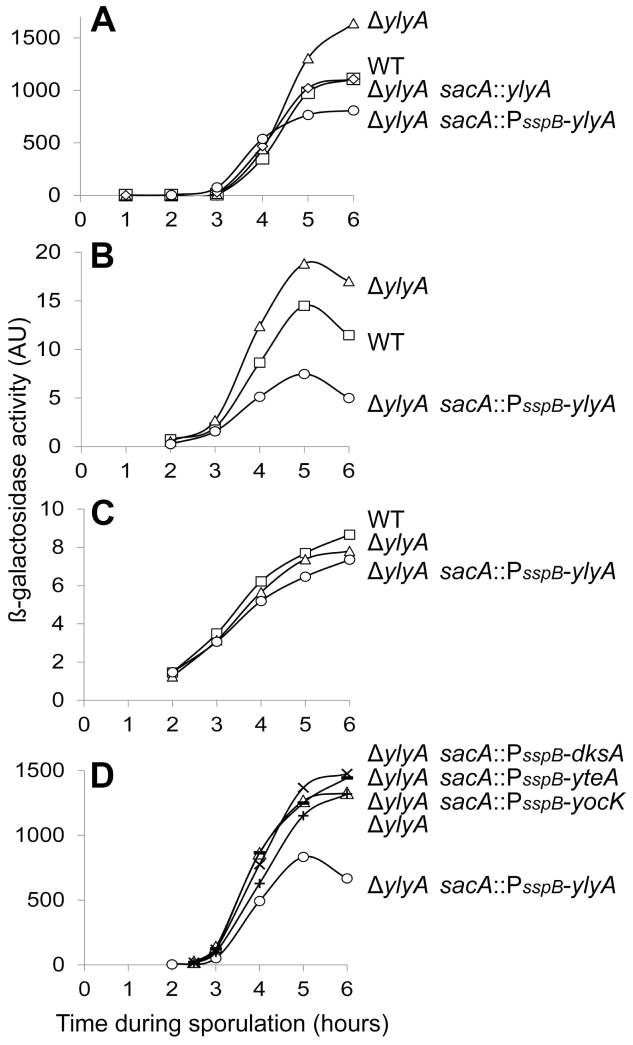
The forespore line of gene expression is a hierarchical cascade in which σF, the earliest-acting regulatory protein in the forespore, turns on the synthesis of σG, which in turn activates the gene for the DNA-binding protein SpoVT. The ylyA gene is turned on at the end of this cascade by σG in conjunction with SpoVT (Traag et al., 2013). Since YlyA resembles a transcription factor, we tested the effects of deleting and overexpressing ylyA on late gene expression in the forespore using fusions of lacZ to the promoters for spoVT (PspoVT) (Bagyan et al., 1996) and another σG-controlled gene sspB (PsspB) (Sun et al., 1991). Deleting ylyA increased β-galactosidase production from the PsspB-lacZ and PspoVT-lacZ reporters (Fig 2A, B). To overexpress ylyA, we placed the gene under the control of PsspB itself, which is a strong, σG-controlled promoter (Mason et al., 1988). Using the PsspB-ylyA overexpression construct, we found that β-galactosidase production from both PsspB-lacZ and PspoVT-lacZ was significantly decreased (Fig 2A, B). As a test of whether the observed effects were due to an effect on host cell RNA polymerase, we used cells engineered to produce phage T7 RNA polymerase in the forespore (Camp & Losick, 2009). Deleting or overexpressing ylyA in cells producing the phage RNA polymerase had little effect on β-galactosidase production from a fusion of lacZ to a phage T7 promoter (Fig 2C).
Fig. 2. YlyA exhibits a specific effect on σG-dependent transcription.
β-galactosidase activity (arbitrary units; AU) was monitored for samples taken at the indicated time points after sporulation was induced by resuspension. (A) PsspB-directed lacZ activity was determined in wild type cells (squares), ylyA mutant cells (triangles), ylyA mutant cells carrying a copy of ylyA at the ectopic sacA locus (diamonds), and ylyA mutant cells carrying a copy of ylyA expressed from the sspB promoter at the ectopic sacA locus (circles). All strains carry the PsspB-lacZ reporter construct at the amyE locus. (B) PspoVT-directed lacZ activity was determined in wild type cells (squares), ylyA mutant cells (triangles), and ylyA mutant cells carrying a copy of ylyA expressed from the sspB promoter at the ectopic sacA locus (circles). All strains carry the PspoVT-lacZ reporter construct at the amyE locus. (C) Forespore-specific T7 RNA polymerase-directed PT7-lacZ activity was determined in wild type cells (squares), ylyA mutant cells (triangles), and ylyA mutant cells carrying a copy of ylyA expressed from the sspB promoter at the ectopic sacA locus (circles). All strains carry the construct expressing T7 RNA polymerase (PspoIIQ-T7 RNAP) at the ylnF locus, whereas the PT7-lacZ reporter gene was integrated at the ywrK locus. (D) PsspB-directed lacZ activity was determined in ylyA mutant cells carrying a copy of ylyA (circles), yocK (plus sign), yteA (minus sign) and E. coli dksA (cross) expressed from the sspB promoter at the ectopic sacA locus (circles).
To determine whether the observed effects were specific to YlyA, we tested the effect of overexpressing the two other B. subtilis homologs of ylyA (i.e. yocK and yteA), and E. coli dksA on β-galactosidase production from the PsspB-lacZ reporter. Whereas overexpression of ylyA decreased β-galactosidase production approximately two-fold as compared to the ylyA null mutant, constructs in which yocK, yteA or dksA were fused to PsspB did not significantly affect PsspB-directed β-galactosidase production (Fig 2D). We conclude that the inhibitory effect of YlyA on σG-directed transcription is specific to YlyA itself.
YlyA influences germination protein levels
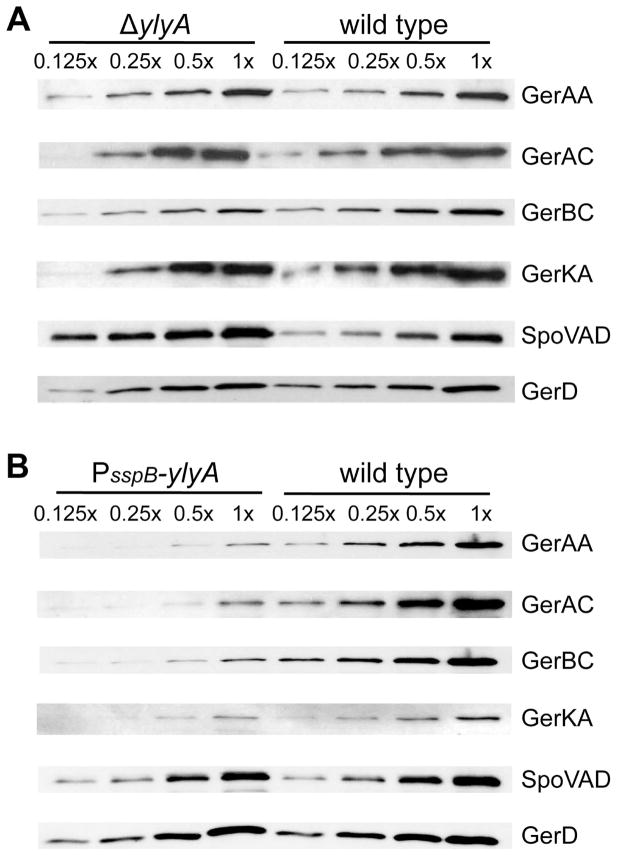
We previously reported that spores mutant for ylyA are defective in germination in LB medium (Traag et al., 2013). To investigate the basis for this germination defect, we examined the relative levels of several germination proteins in wild type spores and spores from a ylyA mutant and from a ylyA overexpression strain (PsspB-ylyA). B. subtilis spores contain three germinant receptors (i.e. GerA, GerB and GerK), which are present at moderate abundance in the inner membrane of the dormant spore (Paidhungat & Setlow, 2001, Hudson et al., 2001, Moir, 2006). To determine the relative levels of these proteins we used antibodies against the germinant receptor subunits GerAA, GerAC, GerBC and GerKA (Ramirez-Peralta et al., 2012b). We also determined the levels of SpoVAD, one of the seven proteins encoded by the spoVA operon, which is required for the accumulation of DPA in the spore core and is involved in its release during germination (Tovar-Rojo et al., 2002, Vepachedu & Setlow, 2007), and GerD, which plays a role in rapid response to nutrient germinants (Pelczar et al., 2007). GerAA, GerAC, GerKA, and GerD levels were relatively unaffected by the ylyA deletion. Interestingly, GerBC levels were reduced 4-fold in the mutant compared to the wild type, while the levels of SpoVAD were approximately 2.5 times higher (Fig 3A, Table S2). Overexpression of ylyA resulted in decreased levels of all germinant receptor proteins, but did not affect SpoVAD and GerD levels (Fig 3B, Table S2).
Fig. 3. YlyA influences the levels of germination proteins.
Equal amounts of inner membrane protein fractions prepared from purified spores of the wild type, ylyA mutant, and ylyA overexpression strains were subjected to SDS-PAGE and Western blot analysis. Western membranes were probed with antibodies against GerAA, GerAC, GerBC, GerKA, SpoVAD and GerD. For direct comparison, dilutions of samples prepared from wild type and ylyA mutant (A) or wild type and ylyA overexpression (B) spores were run on the same gel.
YlyA influences the response of spores to specific germinants
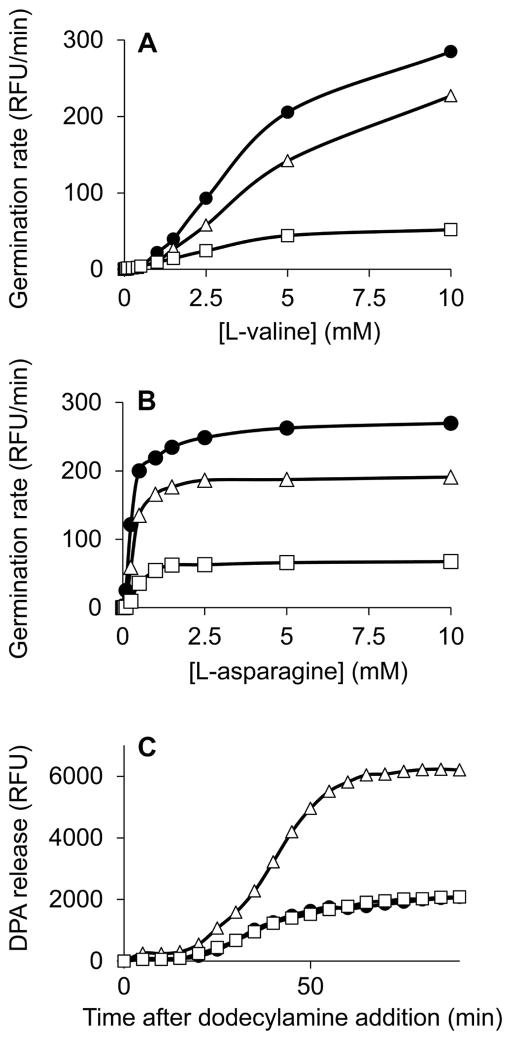
Germinant receptor levels are a major determinant of the rate of spore germination with particular nutrient germinants (Cabrera-Martinez et al., 2003, Paidhungat & Setlow, 2000). Our findings predict that ylyA mutant spores would be impaired when germination is induced through the GerB receptor, and that spores from the ylyA overexpression strain would be impaired in nutrient germination through any of the three germinant receptors. We induced germination of purified spores using germinants specific to the GerA receptor (i.e. L-valine) or the GerB and GerK receptors (i.e. a mixture of L-asparagine, glucose, fructose, potassium chloride (AGFK)), and measured the rate of germination as the release of DPA. ylyA mutant spores and spores from a ylyA overexpression strain showed reduced germination rates with both nutrient germinations (Fig 4A, B). Spores from the ylyA overexpression strain in particular showed severe impairment in germination, consistent with the reduced levels of all germinant receptors in these spores. The reduced rate observed for ylyA mutant spores with AGFK is in agreement with the lower GerBC levels in these spores. GerAA and GerAC levels were not significantly changed in mutant spores (Fig 3A, Table S2). The modest germination defect observed for mutant spores in the presence of L-valine, which acts through the GerA receptor (Atluri et al., 2006, Moir & Smith, 1990), therefore possibly reflects changes in additional factors involved in nutrient germination.
Fig. 4. YlyA affects the efficiency of spore germination.
Germination kinetics of purified spores were determined by measuring the formation of the fluorescent complex between terbium (Tb3+) and the DPA released from spores. (A) Germination was induced with different millimolar concentrations of L-valine, which is specific for the GerA receptor. Results are plotted as the velocity of DPA release (relative fluorescence units per minute (RFU/min)). (B) Germination was induced with different millimolar concentrations of L-asparagine in the presence of 10 mM each of glucose, fructose and potassium chloride (AGFK), which together are specific for the GerB and GerK receptors. Results are plotted as the velocity of DPA release (RFU/min). (C) Germination was induced with 0.8 mM dodecylamine, which acts on the SpoVA protein channel, and DPA release was measured over time. Results are plotted as the total release of DPA (RFU). The symbols used are: (●), wild-type; (△), ylyA mutant; and (□) ylyA overexpression.
Spores can also be induced to germinate through direct activation of the SpoVA protein channel for DPA by non-nutrient germinants such as dodecylamine, triggering the release of DPA, and bypassing the necessity for the germinant receptors. Elevated levels of SpoVA results in faster release of DPA during non-nutrient germination (Vepachedu & Setlow, 2007). We induced germination by addition of the cationic surfactant dodecylamine and monitored the release of DPA over time. Spores from the ylyA overexpression strains germinated as efficiently as wild type spores, while the release of DPA by ylyA mutant spores was significantly enhanced (Fig 4C). These results are in agreement with those expected from the observed effects of YlyA on the levels of SpoVAD (Fig 3B). Taken together, these results indicate that YlyA modulates the levels of proteins involved in nutrient sensing and DPA release and ensures that spores can germinate efficiently.
Purified YlyA binds to RNA polymerase
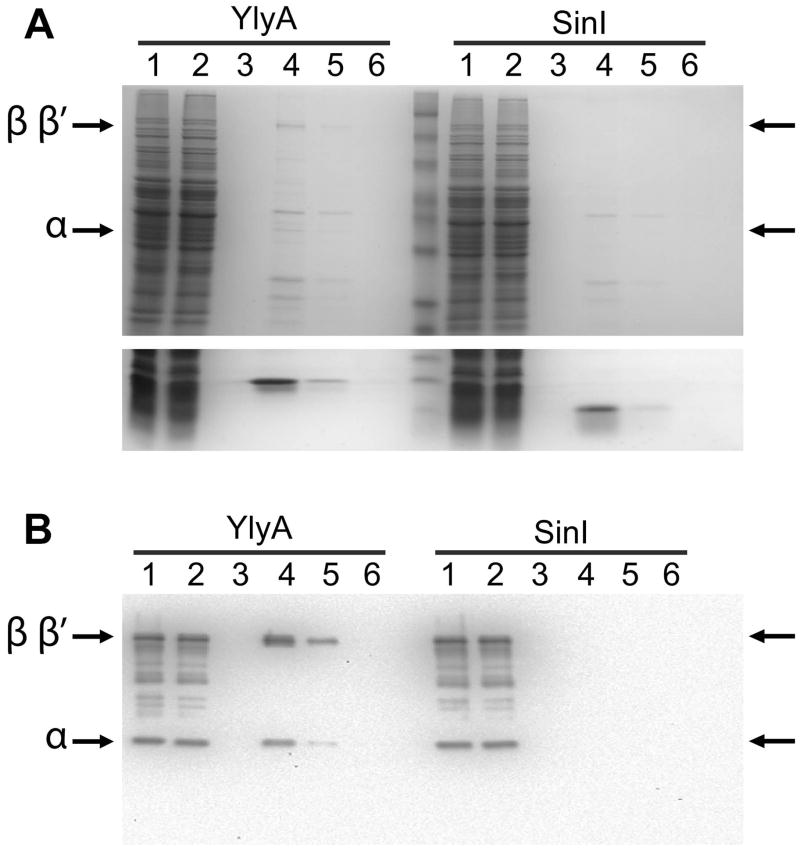
Our results indicate that YlyA is a regulator of gene expression and that its activity depends on the host cell RNA polymerase. The similarity of YlyA to DksA, which interacts with RNA polymerase (Perederina et al., 2004, Paul et al., 2004), motivated us ask if YlyA too interacts with RNA polymerase. To this end, we created a C-terminal, hexa-histidine-tagged YlyA and then passed it over a metal affinity resin. We then applied a soluble protein lysate from sporulating cells of a ylyA mutant to the column. After the column was washed, YlyA was eluted from the column with imidazole. Bands seemingly corresponding to the molecular weight of the β, β′ and αsubunits eluted together with YlyA (Fig 5A). Western blot analysis confirmed the identity of these bands as RNA polymerase subunits (Fig 5B). In contrast, the RNA polymerase subunits were not present in the elution fractions when a control protein unrelated to YlyA, namely SinI, was bound to the column (Fig 5A, B). RNA polymerase from a lysate from vegetatively grown cells was similarly co-immobilized with hexa-histidine-tagged YlyA, indicating that the RNA polymerase-YlyA interaction was not dependent on a sporulation-specific factor (Fig S1).
Fig. 5. YlyA interacts with RNA polymerase from a lysate.
Recombinant hexahistidine-tagged YlyA or SinI was passed over a metal affinity resin. Next a soluble protein lysate from sporulating cells of a ylyA mutant was applied to the column. The column was washed and then eluted with imidazole. Lanes: soluble protein lysate (1), flow through (2), wash (3), elution fractions (4–6). (A) Top panel: 10% SDS/PAGE gel showing bands corresponding to the β/β′ and αsubunits of RNA polymerase (indicated by arrows) which co-eluted with YlyA (left half of the gel) but not SinI (right half of the gel); Bottom panel: 15% SDS/PAGE gel showing the bands for YlyA (left) and SinI (right) eluted from the column. (B) Western blot analysis. Bands were transferred to a membrane, and the membrane was probed with antibodies raised against purified Bacillus RNA polymerase. Arrows indicate bands corresponding to the β/β′ and α subunits of RNA polymerase which co-eluted with YlyA (left half of the gel) but not SinI (right half of the gel).
YlyA stimulates transcription from σG-dependent promoters
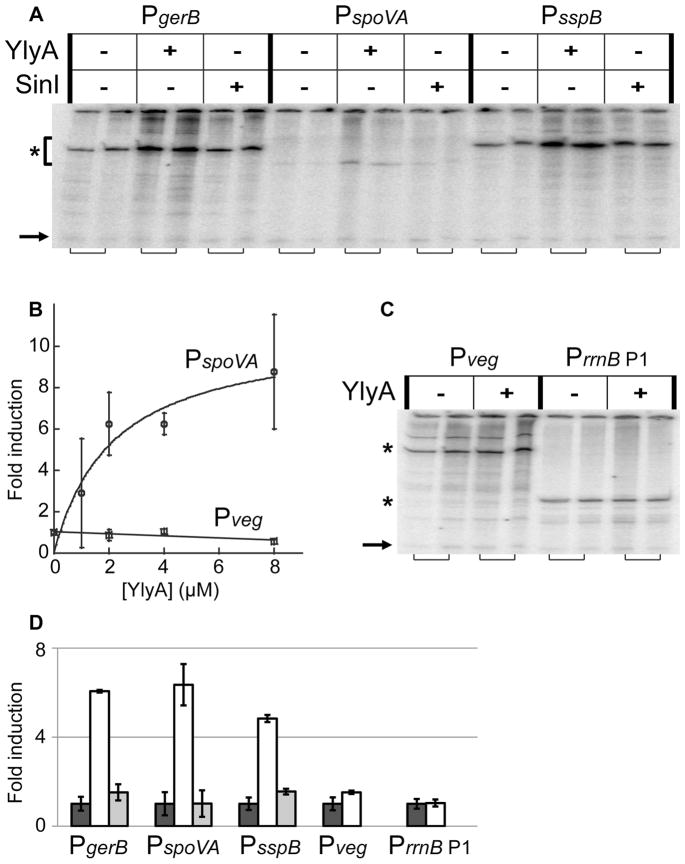
The effects of ylyA deletion on σG-dependent expression in vivo and the interaction of YlyA with RNA polymerase in vitro, prompted us to investigate the effect of YlyA on transcription in multiple round, in vitro transcription assays. First, we tested the effect of YlyA on σG-dependent transcription using recombinant σG and three supercoiled plasmids carrying promoters that were found to be differentially affected by ylyA in vivo, namely PgerB, PspoVA and PsspB. At 8 μM, YlyA stimulated the activity from all three σG-dependent promoters approximately 6-fold (Fig 6A, D). This effect on transcription was dose-dependent and non-linear, seemingly reaching near saturation at 8 μM YlyA (approximately 800-fold molar excess over RNA polymerase) under these conditions (Fig 6B). In contrast, recombinant SinI did not have an effect on transcription from any of these promoters (Fig 6A, D). As a further test of specificity, we investigated the effect of YlyA on transcription from promoters that are dependent on the house-keeping sigma factor σA. We tested this using recombinant σA and two plasmids carrying σA-promoters, namely Pveg and PrrnB P1 (the E. coli rrnB P1 promoter was previously found to be inhibited by DksA (Paul et al., 2004)). Interestingly, YlyA had no effect on transcription using either template (Fig 6B–D). We conclude that YlyA is a regulator of transcription, and that its activity appears to be specific to the alternative sigma factor σG.
Fig. 6. YlyA stimulates σG-dependent, but not σA-dependent, promoters in vitro.
Multiple round in vitro transcription assays using 10 nM purified B. subtilis RNA polymerase, recombinant sigma factors, and hexahistidine-tagged YlyA or SinI. (A) Experiments with purified σG and plasmid templates carrying the σG-dependent PgerB, PspoVA or PsspB promoters, showing stimulation by YlyA. Dilution buffer, 8 μM YlyA, or 8 μM SinI were included in the reactions. Specific signals are indicated by an asterisk to the left of the gel. Adjacent lanes (indicated by brackets below the gel) are from duplicate reactions. Arrow to the bottom left of the gel indicates the end-labeled probe included during phenol/chloroform extraction and ethanol precipitation (see Experimental Procedures). (B) The effect of YlyA on transcription with σG-dependent PspoVA is dose-dependent. Transcription with σA-dependent Pveg is unaffected by YlyA. Results are averages from three separate experiments. Error bars indicate standard errors (SEM). (C) Experiments with purified σA and plasmid templates carrying the σA-dependent Pveg or PrrnB P1 promoters, showing these promoters are unaffected by YlyA. Dilution buffer, 8 μM YlyA, or 8 μM SinI were included in the reactions. Specific signals are indicated by asterisks to the left of the gel. Adjacent lanes (indicated by brackets below the gel) are from duplicate reactions. Arrow to the left of the gel indicates end-labeled probe included during phenol/chloroform extraction and ethanol precipitation. (D) Quantification of the specific signals from gels from (A) and (B). Values are averages from the two duplicate experiments. Average signal from the lanes without YlyA or SinI added were set to one, and fold induction with protein added is given relative to that. Error bars indicate standard errors (SEM).
Discussion
We describe an RNA polymerase-binding protein, YlyA, that is produced at a late stage of spore formation in B. subtilis and that modulates the expression of genes involved in spore germination. Our results suggest that YlyA fine-tunes forespore-specific transcription and indirectly prepares the spore for a future germination event.
YlyA shares some similarity with members of the DksA family of regulators, which are conserved among Gram-negative bacteria. YlyA, however, lacks two features characteristic of studied DksA homologs, namely the N-terminal DxxDxA motif at the coiled-coil tip and the C-terminal Cys-4 zinc finger motif. The zinc finger motif has previously been found to be dispensable for the function of other DksA homologs (Blaby-Haas et al., 2011). Two residues in the DxxDxA motif, namely D74 and A76, are oriented towards the trigger loop in the β′ subunit of RNA polymerase (Lennon et al., 2012). Deletion of the β′ trigger loop or substitution of these essential DksA residues eliminated DksA function in negative and positive control of transcription, without affecting the affinity of DksA for RNA polymerase (Lennon et al., 2012, Lee et al., 2012, Rutherford et al., 2009). YlyA lacks both aspartate residues, and the region around the predicted coiled-coil tip of YlyA orthologs in different Bacillus species shows little sequence conservation (Fig S2), strongly suggesting that these residues are not important for YlyA function, and that YlyA likely acts through a mechanism distinct from that of DksA. Further studies are required to identify residues or motifs important for YlyA activity. Preliminary work with truncated ylyA constructs indicates that the C-terminal region including the zinc finger motif is at least partially dispensable in vivo, highlighting the importance of the N-terminus for YlyA function (Fig S3).
The similarity between YlyA and DksA, albeit weak, led us to the finding that recombinant YlyA co-immobilizes RNA polymerase from a crude cell lysate. In addition, YlyA stimulated transcription from three different σG-dependent promoters in a reconstituted in vitro system using purified RNA polymerase, suggesting that YlyA indeed interacts with RNA polymerase. One possibility is that YlyA, like DksA, interacts with the secondary channel of RNA polymerase. It is entirely conceivable, however, that the weak similarity of YlyA to DksA is adventitious and that it interacts with RNA polymerase at a site distinct from the DksA-binding site. Interestingly, YlyA does not act in conjunction with the house-keeping sigma factor σA. Instead, YlyA seems to be specific to promoters under the control of the sporulation-specific sigma factor σG. (We cannot distinguish whether YlyA is truly specific to σG-containing RNA polymerase or whether the three promoters tested have some distinctive feature other than sigma factor recognition elements that distinguishes them from the σA–controlled promoters tested). Importantly, then, YlyA is dedicated to the transcription of a specialized set of genes involved in spore germination. These include the operons coding for the GerB germinant receptor and the SpoVA channel for the uptake and release of DPA.
A complication in our analysis is that YlyA stimulated transcription from all σG-controlled promoters tested in vitro, even promoters of genes that were upregulated in ylyA mutant cells. Thus, and as expected, YlyA stimulated transcription from the gerB promoter, which correlates to the decrease in GerBC levels in the absence of the RNA-polymerase-binding protein in vivo. On the other hand, YlyA also stimulated transcription from PsspB and PspoVA, whose levels of expression in vivo were higher in the absence of YlyA than in its presence. Conceivably, the precise biochemical conditions we used to carry out our in vitro transcription experiments did not faithfully mimic the conditions for RNA synthesis in the late-stage, forespore. Alternatively, however, the discrepancy between our in vitro and in vivo results may be apparent and reflect the complicated, multi-component, feedback circuit operating at the terminal stage of gene expression in the forespore (Fig 7). This circuit involves σG, which stimulates the transcription of the gene (sigG) for σG itself, and the DNA-binding protein SpoVT (Wang et al., 2006), which, in turn stimulates or represses the expression of other σG-controlled genes, including activating ylyA and repressing spoVT and sigG (Traag et al., 2013, Bagyan et al., 1996, Wang et al., 2006). Further complicating this picture, deletion of ylyA results in elevated spoVT expression. This suggests that, in the absence of YlyA, SpoVT becomes the dominant modulator of σG-directed gene expression, resulting in upregulation of genes activated by SpoVT (e.g. sspB and spoVA) and downregulation of genes repressed by SpoVT (e.g. gerB). We therefore speculate that YlyA is a stimulator of σG-directed transcription (as we observed biochemically) but that the net in vivo output of individual promoters depends on the complex interplay among σG, SpoVT and YlyA.
Fig. 7. The multi-component feedback circuit regulating the σG regulon.
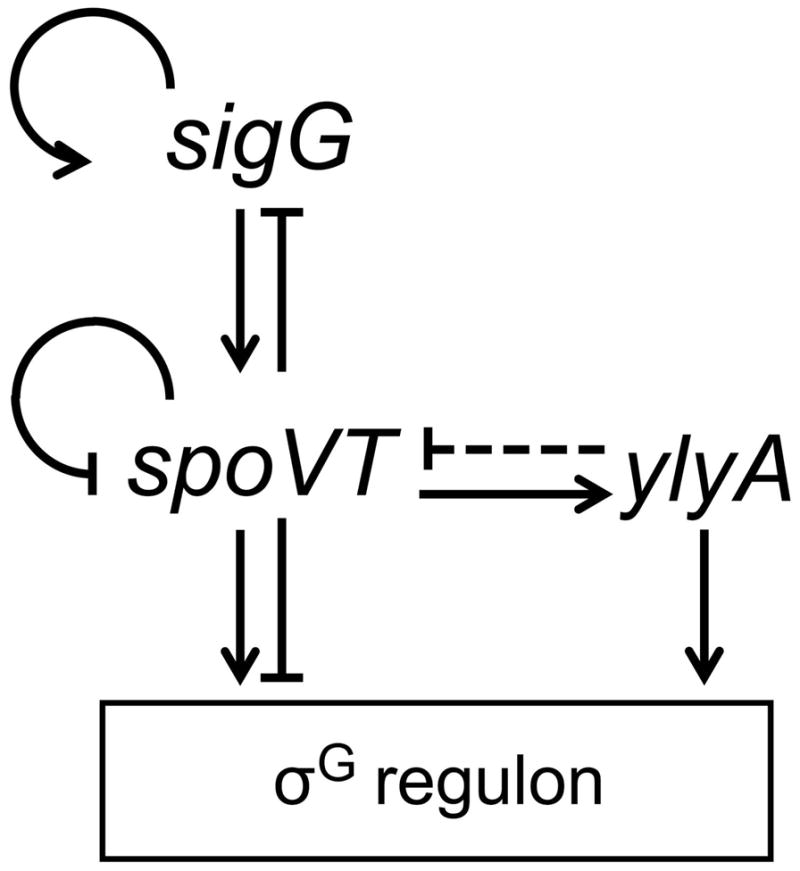
σG, encoded by the sigG gene, stimulates the transcription of sigG and spoVT, encoding the DNA-binding protein SpoVT. SpoVT in turn stimulates or represses the expression of other σG-controlled genes, including activating ylyA and repressing spoVT and sigG. YlyA interacts with RNA polymerase and stimulates σG-dependent gene expression. Through an unknown mechanism, spoVT gene expression is upregulated in cells lacking YlyA (indicated by the dashed repressor symbol from ylyA to spoVT), resulting in the upregulation of genes activated by SpoVT (e.g. sspB and spoVA) and downregulation of genes repressed by SpoVT (e.g. gerB).
In conclusion, YlyA is a novel RNA polymerase-binding protein that is conserved among endospore-forming bacteria and dedicated to the transcription of late-activated sporulation genes involved in germination of the mature spore.
Experimental Procedures
General methods
E. coli strain DH5α was used for propagating plasmids, and grown and transformed using standard procedures (Sambrook et al., 1989). E. coli BL21 (DE3) was used for the expression and purification of recombinant proteins. Details on plasmid sources and construction can be found in SI Experimental Procedures. B. subtilis strains used in this work are listed in Table S1. Transformation of Bacillus was done as previously described (Wilson & Bott, 1968). Spores were purified from liquid Difco sporulation (DS) medium as previously described (Nicholson & Setlow, 1990).
β-galactosidase activity assays
Sporulation of strains carrying lacZ reporter constructs was induced by resuspension in Sterlini-Mandelstam (SM) medium (Nicholson & Setlow, 1990), and samples were collected at various time points after induction. Activity was measured in a Synergy 2 plate reader (BioTek) as previously described (Camp & Losick, 2009). β-galactosidase activity is reported in arbitrary (AU) units as the rate of 2-nitrophenyl β-D-galactopyranoside (ONPG) hydrolysis (i.e. Vmax, with units of OD420 per minute) divided by the optical density at 600 nm (OD600) of the culture at the time of collection.
Determination of the levels of spore germination proteins
Levels of germinant receptor subunits, GerD and SpoVAD were determined in the inner membrane fraction of spores of different strains by Western blot analyses using rabbit antibodies against these proteins and a secondary antibody as described previously (Ramirez-Peralta et al., 2012b, Ramirez-Peralta et al., 2012a, Paidhungat & Setlow, 2001). Quantitative comparisons of relative levels of various proteins were made using the ImageJ program, and the overall analysis was carried out with replicate blots on two independent spore preparations.
Spore preparation, purification and germination
Spores of various strains were prepared at 37°C on 2x Schaeffer’s-glucose agar plates, harvested, purified and stored as described previously (Nicholson & Setlow, 1990). All spores used in this work were free (>98%) of growing or sporulating cells, germinated spores and cell debris as determined by phase contrast microscopy. Spores at an OD600 of 5–10 were heat-activated at 70°C for 30 min, cooled on ice for at least 15 min, and germinated at an OD600 of 0.5 in the presence of various concentrations of L-valine or L-asparagine, or 0.8 mM dodecylamine, at 37°C (nutrient) or 45°C (dodecylamine), as previously described (Yi & Setlow, 2010). The incubations with L-asparagine additionally contained 10 mM each of D-glucose, D-fructose and KCl. The kinetics of spore germination in these incubations was followed by measurement of DPA release by Tb-DPA fluorescence in a multi-well fluorescence plate reader as described previously (Yi & Setlow, 2010). All germination experiments were repeated at least twice with two independent spore preparations with essentially identical results.
Protein expression and purification
E. coli BL21 (DE3) derivative strains were used for the overexpression and purification of recombinant YlyA, SinI, σG and σA. The tagged variant of YlyA could functionally replace native YlyA in vivo. B. subtilis strain RL5493, which encodes a hexahistidine-tagged β′ subunit as the only copy in the cell, was constructed, and B. subtilis RNA polymerase was purified from crude lysates similar to previously described (Anthony et al., 2000). Protein purification was done using one-step Ni-NTA (Qiagen) affinity chromatography purification protocols. Details on the construction of RL5493 and the purifications can be found in SI Experimental Procedures.
Multiple-round in vitro transcription
Supercoiled plasmids carrying promoter fragments of gerB (−120/+150 relative to the transcription start site), spoVA (−71/+92), sspB (−220/+151), veg (−334/+185), rrnB P1 (−58/+1), used as templates for in vitro transcription, are derivatives of pRLG770 (Ross et al., 1990). Multiple-round in vitro transcription was done similar to previously described (Lee et al., 2012), with some differences. In brief, 10 nM reconstituted RNA polymerase holoenzyme (B. subtilis RNA polymerase core and appropriate sigma factor in a 1:10 ratio) was incubated with buffer or micromolar concentrations of YlyA or SinI for 30 minutes at room temperature. Transcription was initiated by the addition of template (0.5 nM) and NTP (500 μM ATP, 200 μM GTP and CTP, 10 μM UTP/1 μCi [α-32P]UTP), and incubated for 15 minutes at 30°C. For rrnB P1 the initiating nucleotide is GTP, and therefore the concentrations of ATP and GTP were reversed. Reactions were terminated after 15 min by extraction with phenol/chloroform. At this stage a single-stranded end-labeled probe was included to ensure that the extraction and precipitation of all samples was similar. The aqueous phase was ethanol precipitated in the presence of glycogen, and the pellet was washed with 70% ethanol, dried, and resuspended in buffer containing 95% formamide and 20 mM EDTA. Samples were run on 7 M urea-6% polyacrylamide gels, and visualized and quantified by phosphorimaging. Multiple-round in vitro transcription experiments were repeated at least twice in duplicate for all tested promoter fragments.
Supplementary Material
Acknowledgments
We thank R. Gourse, W. Ross and J. Winkelman and members of the Losick lab for helpful discussions, and J. Winkelman for construction of plasmid pRLG10611. This work was supported in part by a Netherlands Organization for Scientific Research (NWO) Rubicon grant to B.A.T., a National Science Foundation graduate research fellowship to A.F.W.E., and National Institutes of Health grant GM18568 to R.L. Work in the Setlow lab was supported by a Department of Defense Multi-disciplinary University Research Initiative through the U.S. Army Research Laboratory and the U.S. Army Research Office under contract number W911NF-09-1-0286.
References
- Abecasis AB, Serrano M, Alves R, Quintais L, Pereira-Leal JB, Henriques AO. A genomic signature and the identification of new sporulation genes. J Bacteriol. 2013;195:2101–2115. doi: 10.1128/JB.02110-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony LC, Artsimovitch I, Svetlov V, Landick R, Burgess RR. Rapid purification of His(6)- tagged Bacillus subtilis core RNA polymerase. Protein Expr Purif. 2000;19:350–354. doi: 10.1006/prep.2000.1272. [DOI] [PubMed] [Google Scholar]
- Atluri S, Ragkousi K, Cortezzo DE, Setlow P. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J Bacteriol. 2006;188:28–36. doi: 10.1128/JB.188.1.28-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagyan I, Hobot J, Cutting S. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J Bacteriol. 1996;178:4500–4507. doi: 10.1128/jb.178.15.4500-4507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby-Haas CE, Furman R, Rodionov DA, Artsimovitch I, de Crecy-Lagard V. Role of a Zn- independent DksA in Zn homeostasis and stringent response. Mol Microbiol. 2011;79:700–715. doi: 10.1111/j.1365-2958.2010.07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Martinez RM, Tovar-Rojo F, Vepachedu VR, Setlow P. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J Bacteriol. 2003;185:2457–2464. doi: 10.1128/JB.185.8.2457-2464.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 2009;23:1014–1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman R, Tsodikov OV, Wolf YI, Artsimovitch I. An insertion in the catalytic trigger loop gates the secondary channel of RNA polymerase. J Mol Biol. 2013;425:82–93. doi: 10.1016/j.jmb.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson KD, Corfe BM, Kemp EH, Feavers IM, Coote PJ, Moir A. Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J Bacteriol. 2001;183:4317–4322. doi: 10.1128/JB.183.14.4317-4322.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasny L, Gourse RL. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004;23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lennon CW, Ross W, Gourse RL. Role of the coiled-coil tip of Escherichia coli DksA in promoter control. J Mol Biol. 2012;416:503–517. doi: 10.1016/j.jmb.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon CW, Ross W, Martin-Tumasz S, Toulokhonov I, Vrentas CE, Rutherford ST, Lee JH, Butcher SE, Gourse RL. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev. 2012;26:2634–2646. doi: 10.1101/gad.204693.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Davis A, Korza G, Zhang P, Li YQ, Setlow B, Setlow P, Hao B. Role of a SpoVA protein in dipicolinic acid uptake into developing spores of Bacillus subtilis. J Bacteriol. 2012;194:1875–1884. doi: 10.1128/JB.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mason JM, Hackett RH, Setlow P. Regulation of expression of genes coding for small, acid- soluble proteins of Bacillus subtilis spores: studies using lacZ gene fusions. J Bacteriol. 1988;170:239–244. doi: 10.1128/jb.170.1.239-244.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir A. How do spores germinate? J Appl Microbiol. 2006;101:526–530. doi: 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- Moir A, Smith DA. The genetics of bacterial spore germination. Annu Rev Microbiol. 1990;44:531–553. doi: 10.1146/annurev.mi.44.100190.002531. [DOI] [PubMed] [Google Scholar]
- Nicholson WL, Setlow P. Sporulation, Germination and Outgrowth. In: Harwood CR, Cutting SM, editors. Molecular Biological Methods for Bacillus. Wiley-Interscience Publication; 1990. pp. 391–450. [Google Scholar]
- Paidhungat M, Setlow P. Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J Bacteriol. 2000;182:2513–2519. doi: 10.1128/jb.182.9.2513-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paidhungat M, Setlow P. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J Bacteriol. 2001;183:3982–3990. doi: 10.1128/JB.183.13.3982-3990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Pelczar PL, Igarashi T, Setlow B, Setlow P. Role of GerD in germination of Bacillus subtilis spores. J Bacteriol. 2007;189:1090–1098. doi: 10.1128/JB.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Ramirez-Peralta A, Stewart KA, Thomas SK, Setlow B, Chen Z, Li YQ, Setlow P. Effects of the SpoVT regulatory protein on the germination and germination protein levels of spores ofBacillus subtilis. J Bacteriol. 2012a;194:3417–3425. doi: 10.1128/JB.00504-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Peralta A, Zhang P, Li YQ, Setlow P. Effects of sporulation conditions on the germination and germination protein levels of Bacillus subtilis spores. Appl Environ Microbiol. 2012b;78:2689–2697. doi: 10.1128/AEM.07908-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Thompson JF, Newlands JT, Gourse RL. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1989. [Google Scholar]
- Setlow P. Spore germination. Curr Opin Microbiol. 2003;6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Sun D, Fajardo-Cavazos P, Sussman MD, Tovar-Rojo F, Cabrera-Martinez RM, Setlow P. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by E sigma F: identification of features of good E sigma F-dependent promoters. J Bacteriol. 1991;173:7867–7874. doi: 10.1128/jb.173.24.7867-7874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar-Rojo F, Chander M, Setlow B, Setlow P. The products of the spoVA operon are involved in dipicolinic acid uptake into developing spores of Bacillus subtilis. J Bacteriol. 2002;184:584–587. doi: 10.1128/JB.184.2.584-587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag BA, Pugliese A, Eisen JA, Losick R. Gene conservation among endospore-forming bacteria reveals additional sporulation genes in Bacillus subtilis. J Bacteriol. 2013;195:253–260. doi: 10.1128/JB.01778-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vepachedu VR, Setlow P. Role of SpoVA proteins in release of dipicolinic acid during germination of Bacillus subtilis spores triggered by dodecylamine or lysozyme. J Bacteriol. 2007;189:1565–1572. doi: 10.1128/JB.01613-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. The forespore line of gene expression in Bacillus subtilis. J Mol Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- Wilson GA, Bott KF. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1968;95:1439–1449. doi: 10.1128/jb.95.4.1439-1449.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Setlow P. Studies of the commitment step in the germination of spores of Bacillus species. J Bacteriol. 2010;192:3424–3433. doi: 10.1128/JB.00326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.