Abstract
Small interfering RNAs (siRNAs) are applied for post-transcriptional gene silencing by binding target mRNA. A target coding region is usually chosen, although the 3′-untranslated region (3′-UTR) can also be a target. This study elucidates whether the coding region or 3′-UTR elicits higher repression. pFLuc and pRLuc are two reporter plasmids. A segment of FLuc gene was PCR-amplified and inserted behind the stop codon of the RLuc gene of the pRLuc. Similarly, a segment of RLuc gene was inserted behind the stop codon of FLuc. Two siFLuc and two siRLuc were siRNAs designed to target the central portions of these segments. Therefore, the siRNA encountered the same targets and flanking sequences. Results showed that the two siFLuc elicited higher repression when the FLuc segment resided in the coding region. Conversely, the two siRLuc showed higher repression when the RLuc segment was in the 3′-UTR. These results indicate that both the coding region and the 3′-UTR can be more effective targets. The thermodynamic stability of the secondary structures was analyzed. The siRNA elicited higher repression in the coding region when the target configuration was stable, and needed to be solved by translation. A siRNA may otherwise favor the target at 3′-UTR.
1. Introduction
Two major classes of small regulatory RNAs, small interfering RNAs (siRNAs) and microRNA (miRNAs), are involved in posttranscriptional gene silencing [1]. miRNAs are noncoding endogenous RNAs that direct posttranscriptional repression by binding to partially complementary sites in the 3′-untranslated region (3′-UTR) of the target mRNAs [2]. siRNAs are exogenous RNA designed to bind the target sequence in a perfect match. siRNAs are loaded into the RNA-induced silencing complex (RISC) [3, 4], in which one of the strands is preferentially selected by an Argonaute protein [5], and are guided toward the perfectly paired target. The siRNA/RISC then mediates the endonucleolytic cleavage of the complementary target RNAs and/or causes translational repression [6, 7]. siRNA has become one of the most powerful tools for suppression [8]. However, the efficacy of siRNA varies dramatically [9, 10].
Many studies in genomics and sequence analysis approaches, such as in identifying nucleosomes [11], predicting cysteine S-nitrosylation sites [12], identifying recombination spots [13], identifying antimicrobial peptides and their functional types [14], predicting signal propagation during colorectal cancer progression [15], and predicting HIV protease cleavage sites [16], can timely provide very useful information and insights for drug development and hence are widely welcomed by science community. The present study attempted to propose a novel approach for comparing the repression potency of siRNA targeting the coding region and the 3′-untranslated region of mRNA in hopes that the new method can become a useful tool for both basic research and drug development.
2. Materials and Methods
2.1. Reporter Vectors Used
pFLuc (pGL3-Control Vector, Promega, Madison, WI, USA), is a firefly luciferase (FLuc, coding region 1 nt to 1653 nt) reporter vector. A segment of firefly luciferase gene (91 nt to 454 nt) was PCR-amplified by primers 5′-TATCTAGAGATACGCCCTGGTTCCTG-3′ and 5′-TATCTAGAGATGATAATAATTTTTTGGATG-3′. The introduced Xba I restriction cutting sites are underlined. The amplicon was cut with Xba I and inserted into the Xba I site of the pRLuc (i.e., pRL-TK Vector, Promega) vector. This cutting site is right behind the stop codon of the Renilla luciferase gene. The recombinant vector obtained was designated as pRLuc-f. Similarly, pRLuc is a Renilla luciferase (RLuc, coding region 1 nt to 935 nt) reporter. A segment of Renilla luciferase gene (334 nt to 715 nt) was PCR-amplified by primers 5′-ATATCTAGAAGATCATTTTTGTCGGCCA-3′ and 5′-ATATCTAGATTCCTAACAATTTGTACAAC-3′. After cutting by Xba I, the amplicon was inserted into the Xba I site of the pFLuc vector. This cutting site is right behind the stop codon ofthe firefly luciferase gene. The recombinant vector obtained was designated as pFLuc-r. PSEAP2-Control (Clotech, Inc., Palo Alto, CA, USA), which is a vector encoding secretable alkaline phosphatase (SEAP), was used as an internal control for transfection efficiency.
2.2. siRNA
siFLuc and siRLuc are siRNAs targeting the FLuc and RLuc gene segments described previously. The siFLuc and siRLuc sequences are shown in Figure 1. These siRNAs were obtained from MDBio, Inc. (Taiwan). The siRNAs were purified by HPLC, and their molecular weights were verified by LC/Mass. siRNA negative control (siNC) used as a negative control siRNA was purchased from MDBio, Inc.
Figure 1.

Sequences of siRNA used in this report. The double-stranded sequences of siRNA and their locations in the segments and genes of FLuc and RLuc are illustrated. The lower strands represent the antisense strands. The numbering starts from nucleotide A of the start codon.
2.3. Transfection of Reporter Vectors and siRNA
H1299 is a non-small-cell lung carcinoma cell line purchased from the Bioresource Collection and Research Center, Taiwan. The cells grow fast and are easily transfected. H1299 cells were cultured in DMEM containing 10% fetal bovine serum (bought from Gibco BRL, Gaithersburg, MD, USA) in a humidified CO2 incubator at 37°C. For transfection, the cells were grown in 6 cm Petri dishes at 70% confluence. One microgram (1 μg) pSEAP2-Control, 1 μg pFLuc (pRLuc, pFLuc-r, or pRLuc-f), and siRNA or siNC (20 nM final concentration) were added to DMEM, bringing it to a final volume of 500 μL. Lipofectamine 2000 (Invitrogen, 10 μL) was added to 490 μL DMEM, and the reagents were well mixed and left undisturbed for 2 min. The two DMEM solutions were mixed and incubated for 20 min. The medium was aspirated from each dish. The mixed DMEM solution and 1 mL DMEM were added to each dish. The cells were incubated in a CO2 incubator at 37°C for 4.5 h prior to the addition of complete medium to replace the transfection medium.
2.4. Assays of SEAP, FLuc, and RLuc Activities
The culture medium was collected 24 h post-transfection and subjected for SEAP assay as described previously [17, 18]. After the withdrawal of the medium, the cells were washed once with 2 mL of 1x PBS and then lysed with 0.5 mL Glo Lysis Buffer (Promega) at room temperature for 5 min, followed by centrifugation to remove the debris. The supernatant was then subjected to FLuc and RLuc activity assays using Bright-Glo Luciferase Assay System (Promega) and Ready-To-Glow Reporter Assay (Clontech), respectively. Luminescence was quantified by a Tecan Infinite M1000 (Tecan Group Ltd., Switzerland) with the luminescence mode.
2.5. Measurement of Fold Repression Caused by siFLuc or siRluc
Fold repression is defined as B/T, where “B” is the FLuc (or RLuc) activity divided by its SEAP activity (internal control) for the siNC-treated groups, and “T” is the FLuc (or RLuc) activity divided by SEAP activity for the siFLuc- (or siRLuc-) treated groups.
3. Results and Discussion
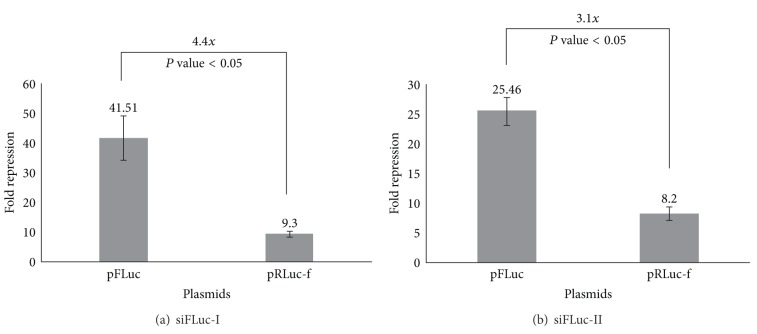
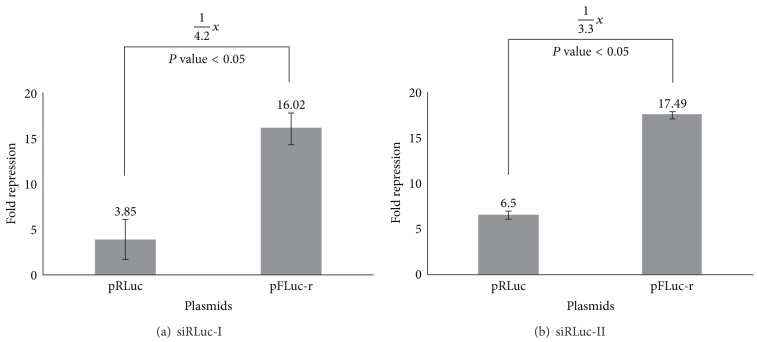
The cloned segment DNAs in pRLuc-f and pFLuc-r were about 0.4 kb in length. We designed four siRNAs against the middle portion of these segments (see Figure 1). Therefore, the siFLuc binding sites had the same binding and surrounding sequences for both pFLuc and pRLuc-f, except that one resided in the coding region and the other in the 3′-UTR. The same situation was set for siRLuc against the targets on the pRLuc and pFLuc-r vectors. The fold repressions of siFLuc-I on the coding region (pFLuc) and the 3′-UTR (pRLuc-f) are shown in Figure 2(a). The siFLuc-I against the coding region showed significantly higher repression than that against the 3′-UTR. We further evaluated the repression activity of siFLuc-II (Figure 2(b)). siFLuc-II also showed significantly higher repression at the coding region of the FLuc segment. These results indicate that the two siFLuc both rendered 3-fold to 4-fold higher repression when targeting the coding region compared when targeting the 3′-UTR. We subsequently tested siRLuc-I and siRLuc-II on their suppression of pRLuc and pFLuc-r (Figures 3(a) and 3(b)). Conversely, the two siRLuc showed higher repression when the RLuc segment was in the 3′-UTR. Therefore, both the coding region and the 3′-UTR can be more effective targets, and other factors may affect the outcome. One possible factor is the thermodynamic stabilities of the secondary structures of these two mRNA segments. We analyzed them by using an RNAfold program (http://bibiserv.techfak.uni-bielefeld.de/rnafold/). RNAfold server predicts minimum free energy structures and base pair probabilities from single RNA sequence [18–20]. Fold algorithms and basic options we used are minimum free energy and partition function, with avoiding isolated base pairs. Figures 4(a) and 4(b) show the 2D structures of these two mRNA segments. The entropy assigned to each nucleotide is also shown in these figures. Higher value of positive entropy indicates that these nucleotides are more stably fitted in this structure. Therefore, the 2D structure around the two siFLuc binding sites was more stable than that of the binding site of siRLuc. We speculate that the on-going translational process helped expose the binding sites; therefore, the binding sites at the coding region resulted in higher repression. On the contrary, the configurations of the siRLuc binding sites were thermodynamically less stable and were accessible to siRLuc. The binding sites located in the 3′-UTR, where the interruption by working ribosomes is avoided, rendered higher repression. Most binding sites of endogenous miRNA resided in the 3′-UTR of their target mRNAs [21, 22]. The advantage may be that miRNA-guided translational repression can uncouple the ongoing translation. Consequently, the translational repression of an mRNA can be reflected and fine-tuned by the number of miRNA binding to its 3′-UTR in a cooperative manner [2, 7].
Figure 2.
Fold repression caused by (a) siFLuc-I and (b) siFLuc-II on the reporter vectors pFluc and pRLuc-f. For transfection, the H1299 cells were grown in 6 cm Petri dishes at 70% confluence. One microgram (1 μg) pSEAP2-Control, 1 μg pFLuc or pRLuc-f, and siNC or siFLuc (20 nM final concentration) were mixed and transfected into the cells (see Section 2 for details). Measurement of fold repression is mentioned in Section 2. The data presented are the mean ± SD of triplicates. P values are also indicated.
Figure 3.
Fold repression caused by (a) siRLuc-I and (b) siRLuc-II on the reporter vectors pRluc and pFLuc-r. For transfection, the H1299 cells were grown in 6 cm Petri dishes at 70% confluence. One microgram (1 μg) pSEAP2-Control, 1 μg pRLuc or pFLuc-r, and siNC or siRLuc (20 nM final concentration) were mixed and transfected into the cells. Measurement of fold repression is mentioned in Section 2. The data presented are the mean ± SD of triplicates. P values are also indicated.
Figure 4.
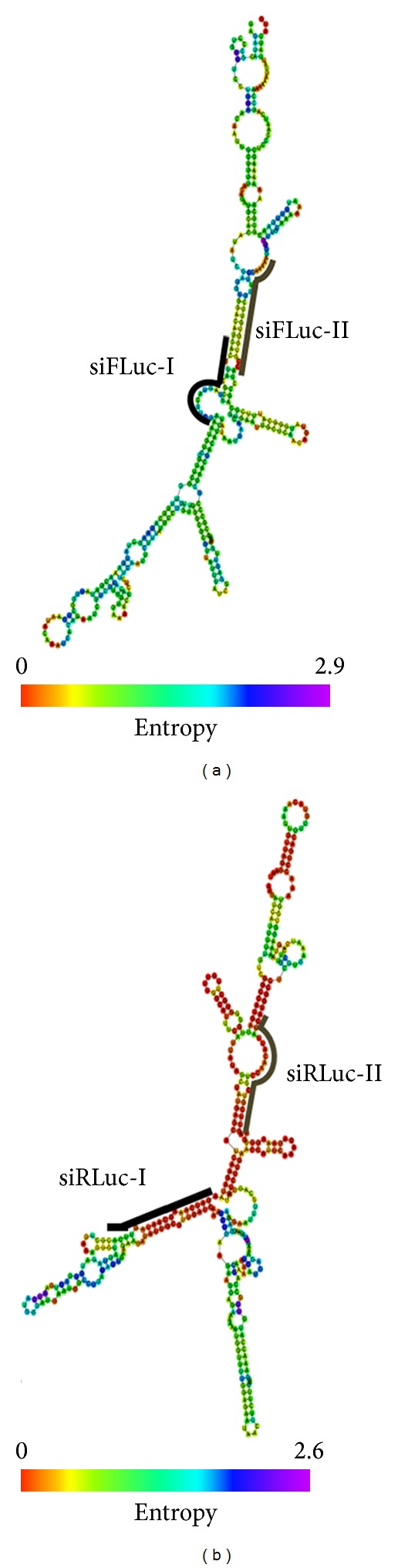
Secondary structures and thermodynamic stabilities of the two mRNA segments. The mRNA segments of the (a) Fluc gene (91 nt to 454 nt) and (b) RLuc gene (334 nt to 715 nt) were analyzed using an RNAfold program (http://bibiserv.techfak.uni-bielefeld.de/rnafold/). The 2D structures and the entropy assigned to each nucleotide are shown. The siRNA binding sites are indicated by heavy lines.
Since user-friendly and publicly accessible web servers represent the future direction for developing practically more useful models, simulated methods, or predictors [23], we shall make efforts in our future work to provide a web server for the method presented in this paper.
4. Conclusion
In the present report, we illustrate that both the coding region and the 3′-UTR can be highly repressive siRNA targets. The effectiveness of the coding region or the 3′-UTR as a binding site may depend on the thermodynamic stabilities of the secondary structures of the target and the flanking sequence.
Conflict of Interests
The authors declare that they have no conflict of interests.
Acknowledgments
This work is supported by the National Science Council, Taiwan, Grant NSC102-2325-B-075-002, and Taipei Veterans General Hospital, Grant V102C-116.
References
- 1.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nature Reviews Drug Discovery. 2007;6(6):443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 3.Sakurai K, Amarzguioui M, Kim DH, et al. A role for human Dicer in pre-RISC loading of siRNAs. Nucleic Acids Research. 2011;39(4):1510–1525. doi: 10.1093/nar/gkq846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123(4):607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 5.Caplen NJ, Fleenor J, Fire A, Morgan RA. dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene. 2000;252(1-2):95–105. doi: 10.1016/s0378-1119(00)00224-9. [DOI] [PubMed] [Google Scholar]
- 6.Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends in Biotechnology. 2010;28(11):570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee HC, Chen CY, Au LC. Systemic comparison of repression activity for miRNA and siRNA associated with different types of target sequences. Biochemical and Biophysical Research Communications. 2011;411(2):393–396. doi: 10.1016/j.bbrc.2011.06.159. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 9.Kurreck J. siRNA efficiency: structure or sequence—that is the question. Journal of Biomedicine and Biotechnology. 2006;2006:7 pages. doi: 10.1155/JBB/2006/83757.83757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel R, T’Wallant NC, Herbert MH, White D, Murison JG, Reid G. The potency of siRNA-mediated growth inhibition following silencing of essential genes is dependent on siRNA design and varies with target sequence. Oligonucleotides. 2009;19(4):317–328. doi: 10.1089/oli.2009.0207. [DOI] [PubMed] [Google Scholar]
- 11.Yigit E, Bischof JM, Zhang Z, et al. Nucleosome mapping across the CFTR locus identifies novel regulatory factors. Nucleic Acids Research. 2013;41(5):2857–2868. doi: 10.1093/nar/gks1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li BQ, Hu LL, Niu S, Cai YD, Chou KC. Predict and analyze S-nitrosylation modification sites with the mRMR and IFS approaches. Journal of Proteomics. 2012;75(5):1654–1665. doi: 10.1016/j.jprot.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Chen W, Feng PM, Lin H, Chou KC. iRSpot-PseDNC: identify recombination spots with pseudo dinucleotide composition. Nucleic Acids Research. 2013;41(6):p. e68. doi: 10.1093/nar/gks1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao X, Wang P, Lin WZ, Jia JH, Chou KC. iAMP-2L: a two-level multi-label classifier for identifying antimicrobial peptides and their functional types. Analytical Biochemistry. 2013;436(2):168–177. doi: 10.1016/j.ab.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 15.Jiang Y, Huang T, Chen L, Gao YF, Cai Y, Chou KC. Signal propagation in protein interaction network during colorectal cancer progression. BioMed Research International. 2013;2013:9 pages. doi: 10.1155/2013/287019.287019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai YD, Liu XJ, Xu EB, Chou KC. Support vector machines for predicting HIV protease cleavage sites in protein. Journal of Computational Chemistry. 2002;23(2):267–274. doi: 10.1002/jcc.10017. [DOI] [PubMed] [Google Scholar]
- 17.Huang PC, Chen CY, Yang FY, Au LC. A multisampling reporter system for monitoring microRNA activity in the same population of cells. Journal of Biomedicine and Biotechnology. 2009;2009:5 pages. doi: 10.1155/2009/104716.104716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Research. 1981;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. Journal of Molecular Biology. 1999;288(5):911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 20.McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29(6-7):1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 21.Meister G. miRNAs get an early start on translational silencing. Cell. 2007;131(1):25–28. doi: 10.1016/j.cell.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nature Structural & Molecular Biology. 2010;17(1):5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou KC, Shen HB. Recent advances in developing web-serves for predicting protein attributes. Natural Science. 2009;1(2):63–92. [Google Scholar]