Abstract
Cigarette smoke creates a molecular field of injury in epithelial cells that line the respiratory tract. We hypothesized that transcriptome sequencing (RNA-Seq) will enhance our understanding of the field of molecular injury in response to tobacco smoke exposure and lung cancer pathogenesis by identifying gene expression differences not interrogated or accurately measured by microarrays. We sequenced the high-molecular-weight fraction of total RNA (>200 nt) from pooled bronchial airway epithelial cell brushings (n = 3 patients per pool) obtained during bronchoscopy from healthy never smoker (NS) and current smoker (S) volunteers and smokers with (C) and without (NC) lung cancer undergoing lung nodule resection surgery. RNA-Seq libraries were prepared using 2 distinct approaches, one capable of capturing non-polyadenylated RNA (the prototype NuGEN Ovation RNA-Seq protocol) and the other designed to measure only polyadenylated RNA (the standard Illumina mRNA-Seq protocol) followed by sequencing generating approximately 29 million 36 nt reads per pool and approximately 22 million 75 nt paired-end reads per pool, respectively. The NuGEN protocol captured additional transcripts not detected by the Illumina protocol at the expense of reduced coverage of polyadenylated transcripts, while longer read lengths and a paired-end sequencing strategy significantly improved the number of reads that could be aligned to the genome. The aligned reads derived from the two complementary protocols were used to define the compendium of genes expressed in the airway epithelium (n = 20,573 genes). Pathways related to the metabolism of xenobiotics by cytochrome P450, retinol metabolism, and oxidoreductase activity were enriched among genes differentially expressed in smokers, whereas chemokine signaling pathways, cytokine–cytokine receptor interactions, and cell adhesion molecules were enriched among genes differentially expressed in smokers with lung cancer. There was a significant correlation between the RNA-Seq gene expression data and Affymetrix microarray data generated from the same samples (P < 0.001); however, the RNA-Seq data detected additional smoking- and cancer-related transcripts whose expression was were either not interrogated by or was not found to be significantly altered when using microarrays, including smoking-related changes in the inflammatory genes S100A8 and S100A9 and cancer-related changes in MUC5AC and secretoglobin (SCGB3A1). Quantitative real-time PCR confirmed differential expression of select genes and non-coding RNAs within individual samples. These results demonstrate that transcriptome sequencing has the potential to provide new insights into the biology of the airway field of injury associated with smoking and lung cancer. The measurement of both coding and non-coding transcripts by RNA-Seq has the potential to help elucidate mechanisms of response to tobacco smoke and to identify additional biomarkers of lung cancer risk and novel targets for chemoprevention.
Introduction
Cigarette smoking is a causative factor for chronic obstructive pulmonary disease (COPD) and lung cancer, with 10% to 20% of smokers developing these diseases (1). Cigarette smoke creates a field of injury in the epithelial cells of the respiratory tract (2–8). Our group and others have shown smoking-related gene and miRNA expression alterations in the cytologically normal large and small airway epithelium by using microarray technology (2, 9–14). These expression alterations have been categorized by their degree of reversibility upon smoking cessation, providing insights into genomic changes that may account for persistent lung cancer risk (12, 14). Similar gene expression alterations have been found in the epithelia of the nose and mouth of smokers (15–17). We have shown that lung cancer also significantly alters the airway transcriptome and have developed a gene expression–based biomarker for the detection of lung cancer by using cells collected from the mainstem bronchus during bronchoscopy that are cytologically normal and distant from the primary tumor (11, 18, 19).
For the past decade, microarrays have been the most comprehensive approach to measure gene expression and have led to significant advances in our knowledge of the airway field of injury (20). Microarrays, however, have several limitations, including probe hybridization kinetics, probe selection (genomic loci and features interrogated), background hybridization that may limit ability to accurately estimate low-level transcripts, and cross-platform comparability. Transcriptome sequencing has been shown to be comparable to microarrays (21, 22) and has potential advantages, such as a larger dynamic range, the ability to detect all expressed transcripts as a function of depth of read coverage, and the ability to detect transcript structure. Transcriptome sequencing, for example, has been used to identify long noncoding RNAs (lincRNA; 23) that have important transcriptional and posttranslational gene regulatory roles (24).
In this study, we have used whole transcriptome sequencing (RNA-Seq) to characterize the airway transcriptome and gene expression alterations associated with cigarette smoke exposure and lung cancer. To our knowledge, this is the first study to apply this emerging technology to profile RNA in airway epithelial cells. We compared and contrasted 2 approaches for RNA-Seq of these airway samples and compared RNA-Seq data to microarray data generated from these same samples. Our data suggest that transcriptome sequencing of both polyadenylated and nonpolyadenylated RNA from airway epithelium provides novel biological insights into the airway field of injury induced by tobacco smoke and additional candidate biomarkers for lung cancer detection.
Methods
Patient population
We recruited healthy never (NS; n = 3) and current (S; n = 3) smokers without cancer to undergo flexible bronchoscopy as volunteers at Boston University Medical Center. We also recruited current and former smokers with cancer (C; n = 8) and without cancer (NC; n = 5) undergoing flexible bronchoscopy in the operating room for lung nodule resection at Boston University Medical Center. Patients were classified as having lung cancer or an alternative benign disease of the chest (e.g. organizing pneumonitis, sarcoidosis, or chronic inflammation due to foreign body material) on the basis of pathologic results from the lung biopsy.
Airway epithelial cell collection
In both the healthy volunteer cohort and the clinical cohort undergoing bronchoscopy pre-nodule resection, we obtained bronchial airway epithelial cells from the uninvolved right mainstem bronchus with an endoscopic cytobrush (Cellebrity Endoscopic Cytobrush; Boston Scientific). If a suspicious lesion (endobronchial or submucosal) was seen in the right mainstem bronchus, brushings were obtained from the uninvolved left mainstem bronchus. The brushes were immediately stored in TRIzol reagent (Invitrogen) at −80°C. RNA was extracted from the brushes as previously described (11).
RNA-Seq library preparation and sequencing
High-molecular-weight (>200 nt) RNA (300 ng) was pooled from 3 individuals within each phenotype: never smokers (NS), healthy current smokers (S), smokers with lung cancer (C), and smokers with benign diseases of the chest (NC), for a total of 4 samples. For each phenotype, 2 pools of 900 ng of RNA were created and libraries were prepared using 2 distinct approaches. The rationale for pooling the RNA from individual samples was to obtain sufficient quantities of RNA for library preparation using samples (with the exception of one) that had previously been processed and hybridized to microarrays.
First, a prototype NuGEN Ovation RNA-Seq protocol was carried out on 10 ng from each pool. In brief, total RNA is reverse transcribed using oligo-d(T) and random primers to generate cDNA, followed by fragmentation and SPIA (NuGEN) linear amplification. Amplified cDNA then underwent end repair and adapter ligation, as described in the Illumina mRNA-Seq sample preparation protocol. The ligation products were run on a 2% TAE (Tris-acetate EDTA) gel to isolate 200 nt fragments. DNA from the gel bands was purified using the QIAquick Gel Extraction Kit. The purified, size-selected cDNA was then PCR amplified (12 cycles) by using an input amount of 5 µL. The purified PCR products were then quantified using an Agilent bioanalyzer (DNA-1000 kit). Each sample was sequenced using Illumina GAII sequencer on 3 lanes of the flow cell generating 36 nt single-end (SE) reads.
For the second library preparation, the standard Illumina mRNA-Seq protocol was carried out on 900 ng from each RNA pool. In brief, mRNA was purified using Sera-Mag Magnetic Oligo(dT) beads and fragmented, followed by cDNA synthesis with random hexamers. This product then underwent end repair, adapter ligation, and gel purification (2% TAE) to isolate 300 nt fragments. DNA from the gel bands was purified using the QIAquick Gel Extraction Kit, PCR amplified (15 cycles), and libraries were quantified using an Agilent bioanalyzer (DNA-1000 kit). Each library was sequenced using Illumina GAIIX sequencer on 1 lane of the flow cell, generating 75 nt paired-end (PE) reads.
Microarray data analysis
For each microarray data set, CEL files were analyzed using Robust Multichip Average (RMA; ref. 25) and the Ensembl v58 CDF file (26) using R statistical software (27). Differential expression analyses were conducted using the R package limma (28) to generate empirical Bayes moderated t-statistics and P values for each Ensembl Gene ID interrogated on the microarray.
Total RNA from the 3 NS and 3 S patients used in this study was processed and hybridized to Affymetrix Exon 1.0 ST microarrays as described in the work of Schembri and colleagues (13). These microarray profiles were previously deposited in the Gene Expression Omnibus (GEO) series GSE14633 (NS samples were GSM365356, GSM365360, and GSM365366 and S samples were GSM365345, GSM365353, and GSM365355). The entire set of microarray profiles in GSE14633 was used when deriving RMA expression values, but differential expression analysis was done across only the 3 NS and 3 S samples from the same individuals profiled by RNA-Seq. Bronchial airway epithelium obtained from smokers with (n = 8) and without (n = 5) cancer was processed, RNA was hybridized to Affymetrix HGU133A 2.0 microarrays as described in the work of Spira and colleagues (11), the CEL files were analyzed as described earlier, and differential expression analysis was done between 8 NC and 5 C samples. The microarray data for the surgical cohorts have been deposited in GEO Series GSE28835, part of superSeries GSE29007. The 3 samples without cancer and 2 of the 3 samples with cancer used in this study were a subset of the 13 samples (NC samples were GSM714147, GSM714148, and GSM714150 and C samples were GSM714157 and GSM714159).
RNA-Seq data analysis
For each sample processed using the NuGEN protocol, the sequencing reads obtained from 3 lanes were combined and aligned to human genome build 19 (hg19) by using Tophat v1.0.14 (29). The Illumina libraries were also aligned to hg19 by using Tophat in 3 separate ways: (i) using the entire data set (PE reads), (ii) aligning each read of the pair separately, and (iii) truncating each read of the pair to 36 nt and aligning each end of the pair separately. The alignments were conducted using Tophat mammalian default parameters, but the maximum number of multi-reads was limited to 10. Gene expression measurements were calculated on the basis of alignments of the NuGEN libraries and the PE Illumina libraries by using the score function of the Scripture software package (23) or Cufflinks software (30). The annotation file used by Scripture was based on Ensembl v59 (n = 49,702 genes), wherein the union set of transcripts mapping to a single gene was used to summarize gene-level expression. Gene-level expression measurements are reported in reads per kilobase per million reads (RPKM) by Scripture and in fragments per kilobase per million reads (FPKM) by Cufflinks. The family-wide error rate (FWER)-corrected P-value for the observed read count across the transcript (Scripture) was used to establish the confidence of each gene being expressed in a given sample (genes with an FWER for a value of P < 0.05 were designated as present). If a gene was detected as expressed in at least 1 of the 4 samples in each sequencing experiment, the gene was included in the airway transcriptome. Genes detected only in samples processed using the NuGEN library preparation protocol were designated as differentially expressed if the gene measurement was greater than zero in both samples in the comparison (Sv. NS or Cvs. NC), if the | log2 (fold change)| >log2(1.5), S/NS or C/NC, using both Scripture and Cufflinks, and if the direction of change was consistent between the 2 software packages. Genes detected in samples processed using the Illumina library preparation protocol were designated as differentially expressed using the same criteria as earlier, with the addition that the genes also had to have a false discovery rate (FDR)-corrected P < 0.05 for differential expression by Cuffdiff (part of Cufflinks suite of analysis tools). The R package goseq (31) was used to find KEGG pathways and Gene Ontology molecular function categories enriched among differentially expressed genes. The background set of genes used in the functional analyses was the 20,573 airway transcriptome genes. The GTF file used by Scripture, Scripture- and Cufflinks-derived gene expression measurements, FWER-corrected P-values for the observed read counts across the genes, alignment wig files, and FASTQ files for each sample processed either using the Illumina or NuGEN protocols have been deposited in GEO Series GSE29006, part of superSeries GSE29007.
Quantitative real-time PCR validation
Quantitative real-time PCR (qRT-PCR) was used to confirm differential expression of select genes and transcripts that were differentially expressed [|log2(fold change)| > log2(1.5), S/NS or C/NC] by RNA-Seq but not by microarray analysis. This validation was done using RNA from each individual from the pooled samples, with the exception of 2 NC samples for which insufficient RNA remained. These samples were replaced with RNA samples from 2 other NC patients with similar demographics (Supplementary Table S8). The gene, ALDH3A1, which was differentially expressed by both microarrays and RNA-Seq, was used as a positive control (data not shown).
qRT-PCRs were carried out as follows: high-molecular-weight RNA (650 ng) from each individual was treated with TURBO DNA-free (Ambion), according to the manufacturer’s instructions to remove contaminating genomic DNA. RNA was reverse-transcribed using random hexamers (Applied Biosystems) and Superscript II reverse transcriptase (Invitrogen) to produce cDNA. Primer sequences for candidate genes/transcripts and a housekeeping gene (GADPH) were designed using PRIMER3 v.0.4.0 (ref. 32; Supplementary Table S10). SYBR Green (Applied Biosystems) PCR reactions (25 µL) containing 20 ng of cDNA and 300 nmol/L of forward and reverse primers were carried out in triplicate for each sample. Forty cycles of amplification and data acquisition were carried out on a StepOnePlus real-time PCR system (Applied Biosystems). Data were analyzed using the comparative threshold (Ct) method, and all samples were normalized to GAPDH. Fold changes (S/NS or C/NC) were calculated from the average expression value across the 3 individuals in each phenotype (NS, S, NC, or C).
Results
Study design and patient population
Total RNA prepared from bronchial airway epithelial cells obtained via brushings during bronchoscopy of healthy never smoker (NS) and current smoker (S) volunteers or smokers with (C) and without (NC) lung cancer undergoing surgery for lung nodule resection (n = 3 patients per group) was pooled and sequenced using 2 different library preparation protocols (Fig. 1A). Demographics of the subjects recruited into our study are reported in Supplementary Table S1. The demographics of the 2 comparative groups, NS versus S and NC versus C, were well matched. The only variable that differed significantly between experiment and control was age (P < 0.05, S vs. NS). The NC and C samples each had 1 current smoker and 2 former smokers and thus were matched for smoking status, and there was no significant difference between the smoking histories of the cancer patient and non-cancer patient pools.
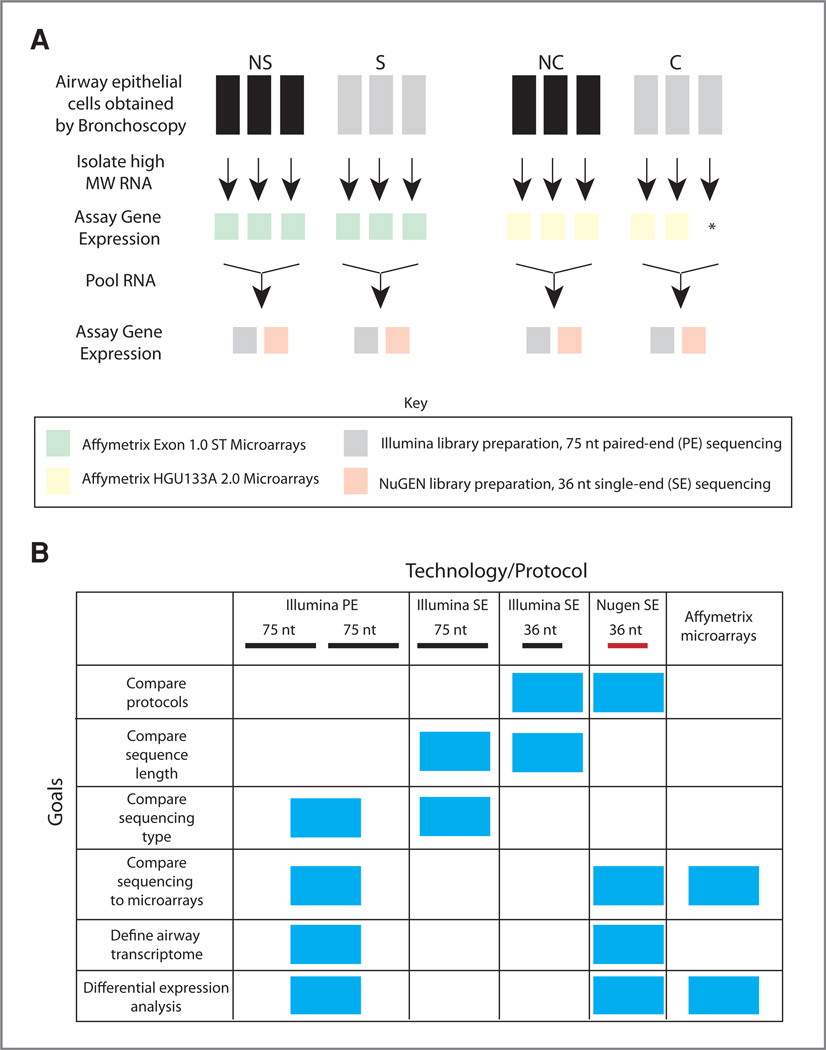
Figure 1.
Study design and goals. A, airway epithelial cells were obtained from 3 never smoker (NS) and 3 current smoker (S) volunteers. The high molecular weight (MW) RNA fraction was isolated from each sample and processed and hybridized to Affymetrix Exon 1.0 ST microarrays (green). Equal amounts of RNA from each sample were then pooled within the NS and S groups. Gene expression was assayed using the standard Illumina RNA-Seq protocol on the Illumina GAIIX sequencer generating 75 nt PE reads (gray) or the prototype NuGEN Ovation RNA-Seq protocol on the Illumina GAII sequencer generating 36 nt SE reads (orange). The same study design was used for the smokers without (NC) and with (C) lung cancer with the exception that RNA from only 2 of the 3 C samples was processed and hybridized to Affymetrix HGU133A 2.0 microarrays (yellow). B, chart displaying the various study goals (y-axis) and the technology and protocol used to accomplish each goal (x-axis), blue boxes indicate which technology and protocol were used to accomplish each goal. Samples were processed and hybridized to microarrays or sequenced using either the NuGEN library preparation protocol (36 nt SE reads) or the Illumina library preparation protocol (75 nt PE reads). The samples processed using the Illumina protocol were analyzed in 3 different ways: to compare library preparation protocols, the 75 nt reads were trimmed to 36 nt and each read of the pair was aligned separately, to compare sequencing length the 75 nt reads were trimmed to 36 nt and each read of the pair was aligned separately, and to compare sequencing type the 75 nt reads aligned separately were compared with the 75 nt reads aligned as a pair.
Alignment differences between library preparation protocols, sequencing lengths, and sequencing types (SE vs. PE sequencing)
Samples prepared using the prototype NuGEN protocol were each sequenced on 3 lanes of a flow cell using an Illumina GAII sequencer. The reads from the 3 lanes were pooled resulting in 28.98, 30.34, 26.94, and 27.8 million 36 nt SE reads for the NS, S, NC, and C samples, respectively. The samples prepared using the standard Illumina mRNA-Seq protocol were sequenced on 1 lane of a flow cell using an Illumina GAIIX sequencer yielding 28.22, 17.24, 22.26, and 20.93 million 75 nt PE reads for the NS, S, NC, and C samples, respectively. The latter experiment provided more reads per lane as a result of advances in sequencer technology and software. We then compared differences in read alignment between protocols, read lengths, and sequencing types (PE vs. SE; Fig. 1B).
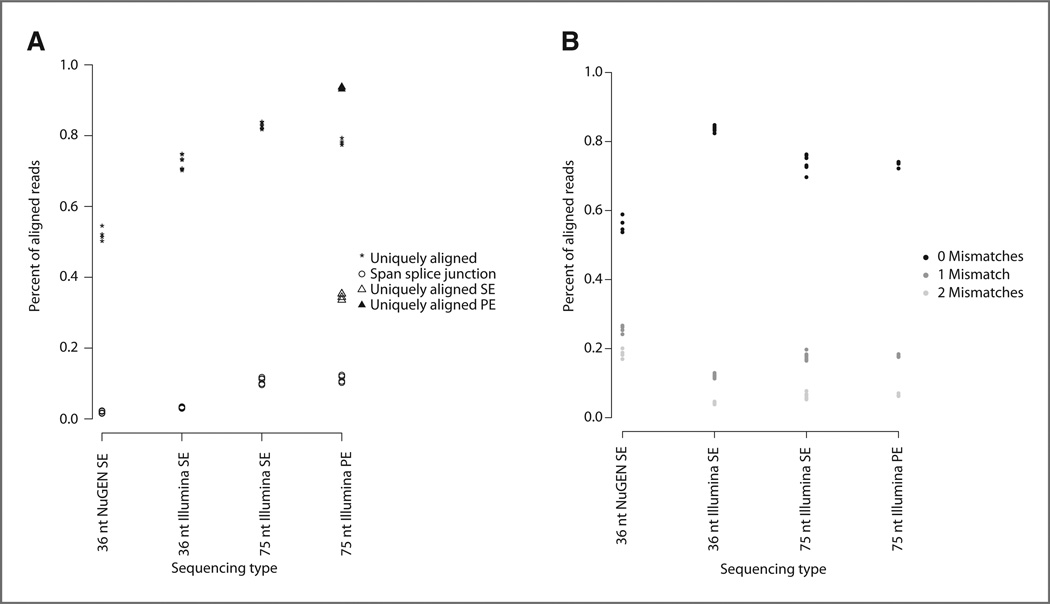
To properly compare the 2 protocols, the 75 nt PE reads from the Illumina protocol were trimmed to 36 nt and each read in the pair was aligned separately. An average of 52% of total aligned reads aligned to a unique location in the genome in the samples prepared using the NuGEN protocol versus 72% for the samples prepared using the Illumina protocol (Fig. 2A, 36 nt Illumina SE). A higher percentage of reads from samples prepared using the NuGEN protocol aligned to the mitochondrial chromosome (an average of 37% vs. 12% using Illumina) and to rRNA (2% vs. 0.1% using Illumina, calculated by summing reads that aligned to rRNA and rRNA pseudogenes as designated in Ensembl v59 annotation). In addition, the reads from samples processed using the Illumina library preparation protocol were of higher quality and aligned with fewer mismatches (Fig. 2B). Next, we compared the alignment of 75 nt reads from the Illumina protocol to the alignment of the same reads trimmed to 36 nt; in both cases, each read from the pair was aligned separately. As expected, increased read length resulted in a greater number of uniquely aligning reads (an average of 82% vs. 72%) and reads aligned to splice junctions (an average of 11% vs. 3%; Fig. 2A, 36 nt Illumina SE vs. 75 nt Illumina SE). Finally, using the 75 nt reads from the Illumina protocol, we compared the alignment of reads as a pair versus alignment of each read of the pair separately. PE versus SE sequencing increases the number of uniquely aligned reads from an average of 82% to an average of 88% (Fig. 2A, 75 nt Illumina SE vs. 75 nt Illumina PE). A summary of the statistics for all alignments is shown in Supplementary Table S2.
Figure 2.
Read alignment statistics. A, the percentage of reads that align to a unique genomic location (asterisk) and the percentage of reads that span splice junctions (open circle; y-axis) versus the sequencing type (x-axis). The sequencing types are as follows: 36 nt NuGEN SE, 36 nt SE reads generated using the NuGEN protocol (n = 4); 36 nt IIIumina SE, 75 nt PE reads generated using the Illumina protocol were trimmed to 36 nt and each read of the pair was aligned separately (n = 8); 75 nt Illumina SE, each pair of the 75 nt PE reads generated using the Illumina protocol were aligned separately (n = 8); 75 nt Illumina PE, 75 nt PE reads generated using the Illumina protocol and aligned as pairs. For the 75 nt Illumina PE sequencing type, the percentage of uniquely aligned reads (asterisk) contains both reads that align as a pair (black triangle) and reads for which only one read in the pair aligned (open triangle). B, the percentage of reads aligning with zero mismatches (black), 1 mismatch (dark gray), or 2 mismatches (light gray) on y-axis versus sequencing type on x-axis.
Defining genes expressed in the airway transcriptome
Given that the 2 library preparation protocols are complementary in their abilities to detect polyadenylated and non-polyadenylated transcripts, genes whose expression was detected in at least 1 sample in either protocol (n = 20,573 genes) were used to define the airway transcriptome (Fig. 3A). This definition of the airway transcriptome is inclusive and numbers of genes in the airway transcriptome using alternative definitions are shown in Supplementary Table S3. Despite the technical differences between the 2 protocols, 93% of the genes detected using the NuGEN protocol were also detected as being expressed by the Illumina protocol. Furthermore, read counts for genes detected by both protocols were significantly correlated (r = 0.59, P < 0.001, Fig. 3B), although the Illumina protocol yielded higher coverage (slope of line is >1). The higher coverage may explain the additional 9,647 genes whose expression was detected using only the Illumina protocol. However, a group of non-protein-coding transcripts had markedly higher read counts by using the NuGEN protocol versus the Illumina protocol (Fig. 3B). In addition, the set of 787 genes detected only when the samples were processed using the NuGEN protocol was composed of higher percentages of non-coding RNAs [lincRNAs, small nucleolar RNAs (snoRNA), small nuclear RNAs (snRNA)], pseudogenes, and processed transcripts (Fig. 3C). Only 32% of the NuGEN protocol–specific genes are classified as "known" in Ensembl (genes that match a human sequence in a public, scientific database such as NCBI RefSeq or UniProtKB) versus 69% or 86% of the genes detected only in samples processed using the Illumina protocol or by both protocols, respectively. Together, the complementary protocols define a compendium of protein-coding and non-coding genes that are expressed in the bronchial epithelium across the pheno-types in this study.
Figure 3.
The airway transcriptome. A, pie chart of genes detected as present in the airway when the samples were processed using the NuGEN (NG) protocol only (blue), using the Illumina (IL) protocol only (red), or by both protocols (green). B, the correlation between read counts (fifth-root transformed) for genes detected as present by both protocols (Illumina y-axis; NuGEN x-axis) across all 4 samples (NS, S, NC, and C), r = 0.59 and P < 0.001. Light green dots represent protein-coding genes and dark green dots represent nonprotein-coding genes as designated in Ensembl annotation. The least squares ine is shown in black (y = 1.57×). C, the distribution of Ensembl-designated gene biotypes for each of the 3 airway transcriptome categories defined in A.
Smoking- and lung cancer–associated gene expression alterations
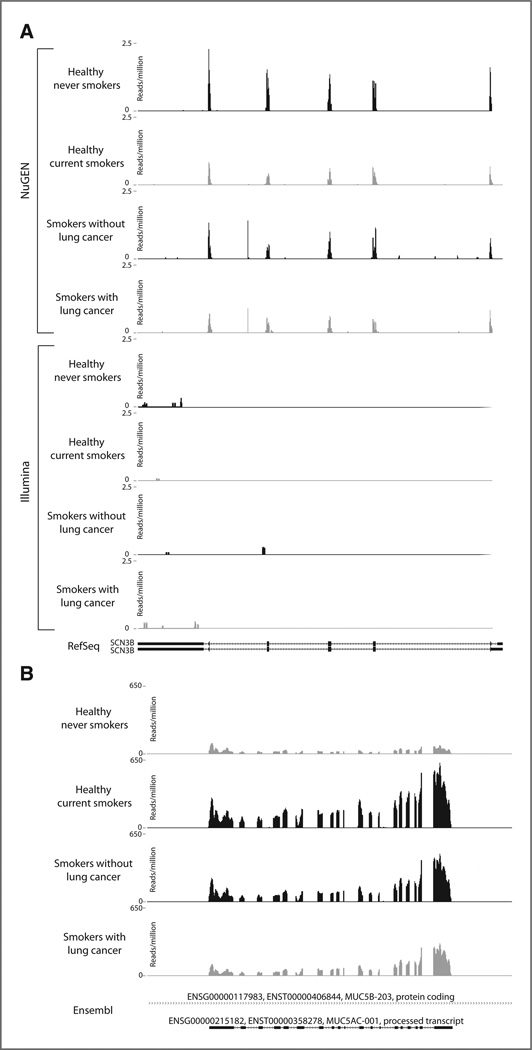
The airway transcriptome defined earlier includes 787 genes detected only in samples prepared using the NuGEN library preparation protocol. These genes are potentially non-polyadenylated transcripts that are not captured using the Illumina protocol. There were 156 genes differentially expressed between S and NS samples and 100 genes differentially expressed between C and NC samples among the 787 genes [differentially expressed genes had a non-zero RPKM value in both samples in each comparison and log2(fold change)| > log2(1.5) by both Cufflinks and Scripture software]. These gene sets were checked for enrichment of Gene Ontology molecular function categories by using goseq (ref. 31; the background gene set was all genes in the airway transcriptome). Categories related to ion channel activity were enriched among genes differentially expressed between NS and S, and a category related to oxidoreductase activity was enriched among genes differentially expressed between NC and C samples (goseq FDR-corrected P < 0.05, Supplementary Table S4). The smoking-associated differentially expressed genes enriched in voltage-gated ion channel activity (GO:005244) were KNCJ5, KNCJ8, KNCJ3, KNCJ12, KNCJ11, KCND3, SCN1B, SCN2B, and SCN3B. An example of one of these ion channel genes detected only using the NuGEN library preparation protocol, SCN3B (sodium channel, voltage-gated, type III, beta), that is down-regulated in smoking and in lung cancer is shown in Figure 4A.
Figure 4.
Read coverage plots of selected genes. For each plot, the reads normalized by the total number of reads (reads per million) are displayed on they-axis and the genomic coordinates are displayed on the x-axis. Within each group (S vs. NS and C vs. NC), the sample with higher expression is shown in black and the sample with lower expression is shown in gray. A, SCN3B (sodium channel, voltage-gated, type III, beta), reads from samples processed using the NuGEN protocol are represented in the top 4 panels and reads from samples processed using the Illumina protocol are represented in the bottom 4 panels. B, MUC5AC (mucin 5AC), reads from samples processed using the Illumina protocol are shown.
We next focused on gene expression detected in samples processed with the Illumina protocol, as the increased number of sequencing reads mapping to mRNA-encoding genes with this protocol suggested that it might potentially be better able to accurately quantitate gene expression levels. Smoking- and cancer-associated differentially expressed genes detected by both library preparation protocols or just the Illumina protocol were identified by choosing genes with a non-zero RPKM for both samples in each comparison, an |log2(fold change)| > log2(1.5) by Cufflinks and Scripture, and an FDR-corrected P < 0.05 by Cuffdiff. There were 517 genes differentially expressed between S and NS samples and 192 genes differentially expressed between C and NC samples by these criteria. These gene sets were checked for statistical enrichment of Gene Ontology molecular function categories and KEGG pathways using goseq (31). Pathways and molecular functions such as oxidoreductase activity, metabolism of xenobiotics by cytochrome P450 and retinol metabolism were enriched among genes differentially expressed between current and never smokers. Cytokine–cytokine receptor interaction, chemokine signaling pathway, and cell adhesion molecules were enriched among gene differentially expressed between smokers with and without cancer (goseq FDR-corrected P < 0.05, Supplementary Table S5). The smoking- and lung cancer–associated pathways and molecular functions uncovered earlier are consistent with gene expression alterations in previous microarray studies (2, 11, 12).
RNA-Seq and microarray gene expression measurements are correlated
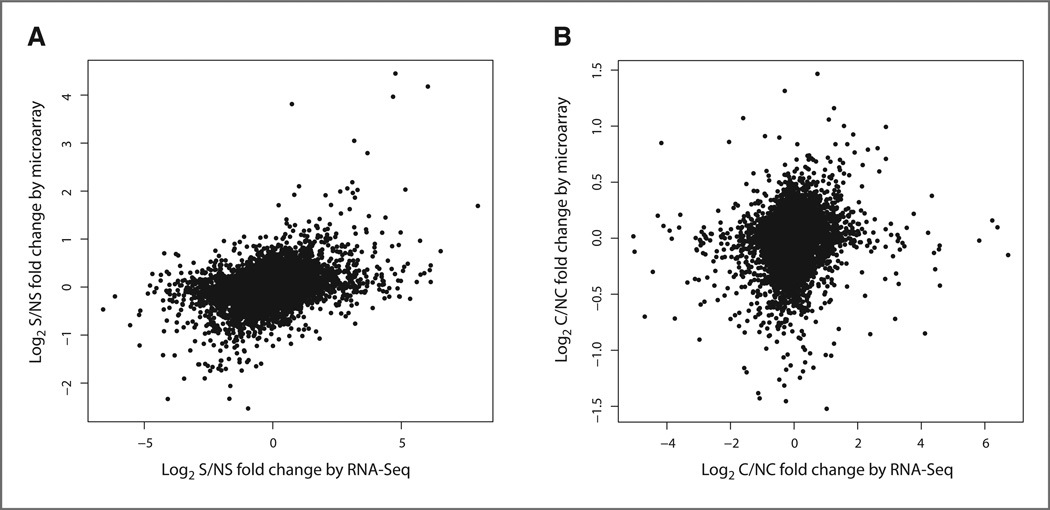
The logarithmically transformed fold changes of Scripture-derived RPKM values between the S and NS samples were computed across the genes defined in the airway transcriptome with non-zero RPKM values in both samples. Gene expression measurements from samples processed using the NuGEN protocol were used to determine the fold changes of genes detected only using the NuGEN protocol, whereas gene expression measurements from samples processed using the Illumina protocol were used to compute the fold changes for all other genes. The RNA-Seq fold changes between S and NS samples were compared with those obtained using microarray profiles on the individual samples in the pool across the 17,005 airway transcriptome genes that were also interrogated by the Affymetrix Exon 1.0 ST microarray. The fold changes measured by sequencing and arrays were significantly correlated (r = 0.36, P < 0.001, Fig. 5A) and the RNA-Seq fold changes were also correlated with the microarray t-statistics between the S and NS samples (r = 0.33, P < 0.001). The correlation between RNA-Seq- and microarray-derived data was slightly lower than the correlation between RNA-Seq fold changes computed on the basis of data derived using the NuGEN protocol versus the Illumina protocol (r = 0.42, P < 0.001, for the genes with non-zero RPKM values detected by both protocols).
Figure 5.
Correlation between RNA-Seq and microarray-detected expression differences between the NS and S samples. A, the log2 fold change between the S and NS samples on the Affymetrix Exon 1.0 ST microarray (y-axis) versus the log2 fold change between the RPKM values for the S sample divided by the RPKM values for the NS sample (y-axis). The fold changes between the platforms are significantly correlated (r = 0.36, P < 0.001) across genes measured by both platforms and genes with a non-zero RPKM in the NS and S samples (n = 17,005). B, the log2 fold change between the C and NC samples on the Affymetrix HGU133A 2.0 microarray (y-axis) versus the log2 fold change between the RPKM values for the C sample divided by the RPKM values for the NC sample (y-axis). The fold changes between the platforms are significantly correlated (r = 0.16, P < 0.001) across genes measured by both platforms and genes with a non-zero RPKM in the NC and C samples (n = 9,308).
The 517 differentially expressed genes between NS and S samples identified earlier (Supplementary Table S6) were examined for differential expression in the microarray data. A total of 109 of the 517 genes differentially expressed by sequencing had either |log2(fold change)| > log2(1.5) or a P < 0.05 (based on a t test) between S and NS samples on the Affymetrix Exon 1.0 ST microarray. A total of 319 genes of the 517 genes were interrogated by but were not differentially expressed on the microarray. Many of these genes are non-coding RNAs and potentially important genes in the response to tobacco smoke exposure and tumorgenesis. Finally, 89 of the 517 genes with smoking-associated expression levels as determined by RNA-Seq are not interrogated on the microarray (see Table 1 for the top most highly expressed subset of the 89 genes).
Table 1.
Genes not interrogated by microarray and significantly differentially expressed by RNA-Seq.
| Ensembl ID | Direction | Gene biotype |
Gene name | Description | Associated gene database |
Source | Gene status |
|---|---|---|---|---|---|---|---|
| ENSG00000215182 | Up S | Pt | MUC5AC | mucin 5AC, oligomeric mucus/gel-forming | HGNC c | H | K |
| ENSG00000225972 | Up S | ps | RP5-857K21.9 | cb V | H | K | |
| ENSG00000160932 | Up S | pc | LY6E | lymphocyte antigen 6 complex, locus E | HGNC a | E | K |
| ENSG00000250477 | Up S | pc | AL161445.1 | BAG family molecular chaperone regulator 1 (BAG-1)(Bcl-2-associated athanogene 1) |
cb E | E | K |
| ENSG00000235058 | Up S | pt | AC002481.5 | cb V | H | P | |
| ENSG00000225410 | Up S | ps | AC107016.2 | cb E | E | N | |
| ENSG00000233458 | Up S | pc | AC123789.1 | Putative uncharacterized protein ENSP00000403148 | cb E | E | K |
| ENSG00000237411 | Up S | pt | AC003043.2 | cb V | H | N | |
| ENSG00000214274 | Up S | pc | ANG | angiogenin, ribonuclease, RNase A family, 5 | HGNC c | E | K |
| ENSG00000234045 | Up S | pc | AC069356.1 | cb E | E | K | |
| ENSG00000232581 | Up S | pt | AC079742.4 | cb V | H | P | |
| ENSG00000215089 | Up S | ps | RP13-138P15.3 | cb V | E | K | |
| ENSG00000183336 | Up S | pc | BOLA2 | bolA homolog 2 (E. coli) | HGNC a | E | K |
| ENSG00000243135 | Up S | pc | UGT1A3 | UDP glucuronosyltransferase 1 family | HGNC c | E | K |
| ENSG00000241119 | Up S | pc | UGT1A9 | UDP glucuronosyltransferase 1 family, polypeptide A9 | HGNC c | E | K |
| ENSG00000181625 | Up S | pc | GIYD1 | GIY-YIG domain containing 1 | HGNC c | E | K |
| ENSG00000235867 | Up S | pc | AC013356.2 | Cell growth-inhibiting protein 48c | cb E | E | K |
| ENSG00000248581 | Up S | lincRNA | AC005618.1 | cb E | E | N | |
| ENSG00000240831 | Dn S | rRNA ps | AC112777.1 | cb E | E | N | |
| ENSG00000242604 | Dn S | rRNA ps | AL512503.1 | cb E | E | N | |
| ENSG00000249529 | Dn S | pc | AC046176.2 | Tyrosine-protein kinase Lyn | cb E | E | K |
| ENSG00000232150 | Dn S | ps | RP11-480P3.1 | cb V | H | K | |
| ENSG00000165807 | Dn S | pc | C14orf50 | Uncharacterized protein C14orf50 | HGNC c | E | K |
| ENSG00000249037 | Dn S | pc | AL121852.1 | sed family domain-containing protein 1 isoform b | cb E | E | K |
| ENSG00000235878 | Dn S | pc | AP001468.1 | cDNA FLJ46393 fis, clone THYMU3002887 | cb E | E | K |
| ENSG00000161055 | Up C | pc | SCGB3A1 | secretoglobin, family 3A, member 1 | HGNC c | E | K |
| ENSG00000057149 | Up C | pc | SERPINB3 | serpin peptidase inhibitor, clade B (ovalbumin), member 3 | HGNC c | E | K |
| ENSG00000206072 | Up C | pc | SERPINB11 | serpin peptidase inhibitor, clade B (ovalbumin), member 11 (gene/pseudogene) |
HGNC c | H | K |
| ENSG00000129824 | Up C | pc | RPS4Y1 | ribosomal protein S4, Y-linked 1 | HGNC c | E | K |
| ENSG00000240831 | Up C | rRNA ps | AC112777.1 | cb E | E | N | |
| ENSG00000244067 | Up C | pc | GSTA2 | glutathione S-transferase alpha 2 | HGNC c | E | K |
| ENSG00000182853 | Up C | pc | VM01 | vitelline membrane outer layer 1 homolog (chicken) | HGNC a | E | K |
| ENSG00000186261 | Up C | pc | AC027277.1 | MSTP160 | cb E | E | K |
| ENSG00000234964 | Up C | ps | AP000640.1 | cb E | E | N | |
| ENSG00000232150 | Up C | ps | RP11–480P3.1 | cb V | H | K | |
| ENSG00000232220 | Up C | Pt | AC008440.5 | cb V | H | P | |
| ENSG00000247017 | Up C | lincRNA | AC009520.1 | cb E | E | N | |
| ENSG00000214265 | Up C | pc | SNURF | SNRPN upstream reading frame | HGNC c | E | K |
| ENSG00000233458 | Up C | pc | AC123789.1 | Putative uncharacterized protein ENSP00000403148 | cb E | E | K |
| ENSG00000224543 | Up C | ps | AC012318.1 | cb E | E | N | |
| ENSG00000170509 | Up C | pc | HSD17B13 | hydroxysteroid (17-beta) dehydrogenase 13 | HGNC c | E | K |
| ENSG00000188643 | Up C | pc | S100A16 | S100 calcium binding protein A16 | HGNC c | E | K |
| ENSG00000006075 | Up C | pc | CCL3 | chemokine (C-C motif) ligand 3 | HGNC c | E | K |
| ENSG00000215182 | DnC | Pt | MUC5AC | mucin 5AC, oligomeric mucus/gel-forming | HGNC c | H | K |
| ENSG00000206172 | DnC | pc | HBA1 | hemoglobin, alpha 1 | HGNC c | E | K |
| ENSG00000204950 | Dn C | pc | LRRC10B | leucine rich repeat containing 10B | HGNC a | E | K |
| ENSG00000226284 | Dn C | ps | RP3-470L14.1 | cb V | H | K | |
| ENSG00000239930 | DnC | Pt | AP001625.4 | cb V | H | N | |
| ENSG00000227053 | DnC | Pt | RP11-395B7.4 | cb V | H | N | |
| ENSG00000248411 | DnC | lincRNA | AC084125.1 | cb E | E | N |
NOTE: The top 25 most highly expressed genes are listed for each group (see Supplementary Tables S6 and S7 for the complete list). The following information is given for each gene based on Ensembl v59 annotation: Ensembl Gene ID, direction of change, gene biotype (pc = protein coding, ps = pseudogene, pt = processed transcript, rRNA ps = rRNA pseudogene), gene name, gene description, associated gene database (cb E = Clone-based (Ensembl), cb V = Clone-based (Vega), HGNC c = HGNC (curated), HGNC a = HGNC automatic), source (E = Ensembl, H = Havana), and gene status (K = known, N = novel, P = putative).
We then carried out a similar analysis using the C and NC samples. The logarithmically transformed fold changes of RPKM values between the C and NC samples were computed across the airway transcriptome genes as described earlier for the S and NS samples. The C versus NC RNA-Seq fold change results were significantly correlated with the fold change computed using the 5 NC and 8 C samples processed on the HGU133A 2.0 microarray (r = 0.16, P < 0.001, Fig. 5B) across 9,308 genes measured on both sequencing and microarrays. The RNA-Seq fold change was also significantly correlated to the t-statistics computed on the basis of microarray data (r = 0.14, P < 0.001). The correlation between RNA-Seq- and microarray-derived data was slightly lower than the correlation between RNA-Seq fold changes computed on the basis of data derived using the NuGEN protocol versus the Illumina protocol across the same C and NC samples (r = 0.24, P < 0.001, for genes with non-zero RPKM values and detected by both protocols).
Of the 192 genes found to be differentially expressed between C and NC samples (Supplementary Table S7), 20 genes had either a |log2(fold change)| > log2(1.5) or a P < 0.05 between C and NC samples on the Affymetrix HGU133A 2.0 microarray. A total of 66 of the 192 genes were interrogated on the microarray but did not have absolute log2 fold change > 1.5 or P < 0.05, including interesting genes related to inflammation and tumorigenesis. Finally, 106 the 192 genes with cancer-associated expression levels as determined by RNA-Seq are not interrogated on the microarray (see Table 1 for the top most highly expressed subset of these 106 genes). Relatively little is known about many of the genes in Table 1; however, one interesting example of a gene with potentially important expression difference in smoking and lung cancer is a processed transcript of MUC5AC. The gene (MUC5AC; Ensembl ID ENSG00000215182) is upregulated in current smokers compared with never smokers but is downregulated in smokers with lung cancer compared with smokers without lung cancer, and there were no probes on the Affymetrix Exon 1.0 ST microarray designed to interrogate the transcript. A plot of the sequencing reads mapping to this gene is shown in Figure 4B.
Quantitative RT-PCR confirms RNA-Seq changes
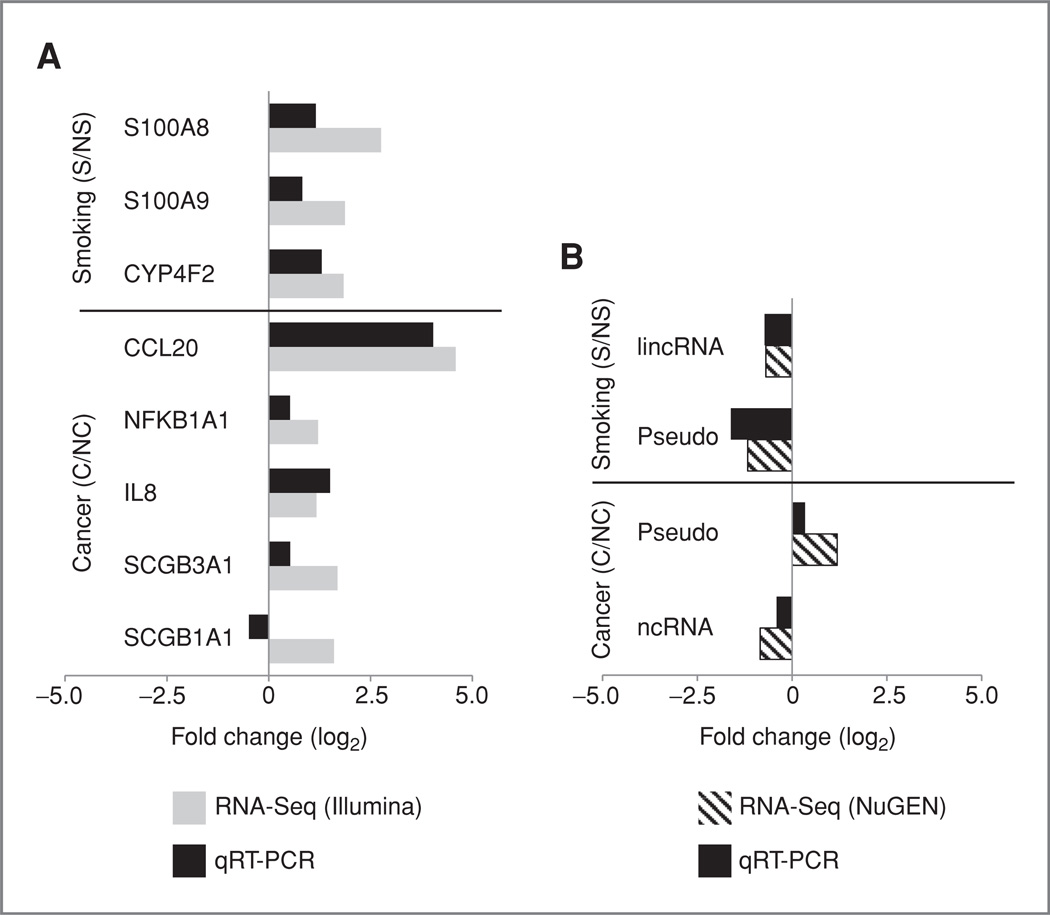
To confirm differential expression by RNA-Seq, we used qRT-PCR to validate a subset of genes that were either not differentially expressed [|log2(fold change)| <log2(1.5)] or were not interrogated when using microarrays (Supplementary Table S9) within NS, S, NC, and C individuals (Supplementary Table S8). Figure 6A provides a comparison of fold changes between Illumina RNA-Seq and qRT-PCR for each gene within smoking or cancer comparisons. All genes except SCGB1A1 show concordant direction of fold change between RNA-Seq and qRT-PCR. In addition to protein-coding genes, we selected a subset of non-protein-coding transcripts represented as differentially expressed in the NuGEN RNA-Seq method for qRT-PCR validation. Smokers showed downregulation of a lincRNA (AC004968.2) and a pseudogene (CTD-2325P2.2) in RNA-Seq data, and this was confirmed with qRT-PCR (Fig. 6B). Similarly, PCR data confirmed the upregulation of the pseudogene (CTD-2325P2.2) and the downregulation of a noncoding RNA (RP11-295J3.2) in individuals with cancer (Fig. 6B).
Figure 6.
Correlation of differential expression between RNA-Seq and qRT-PCR. Genes and transcripts were selected as differentially expressed by RNA-Seq. A, log2 fold change (S/NS or C/NC; x-axis) derived on the basis of samples processed using the Illumina protocol (gray) versus log2 fold change derived on the basis of qRT-PCR results wherein expression values for each phenotype (NS, S, NC, or C) are averaged from 3 samples (black). B. Same as A, except the log2 fold change is derived on the basis of samples processed using the NuGEN protocol (diagonal lines).
Discussion
In this study, we carried out transcriptome sequencing on airway epithelial cells from healthy, never and current smoker volunteers and from smokers with a diagnosis of lung cancer or an alternative benign disease of the chest. The goals of this study were to compare several methods for conducting airway transcriptome sequencing and to evaluate the potential of this technology to provide new biological insights into the altered gene expression in the airway epithelium in response to tobacco smoke and lung cancer.
The design of this study (Fig. 1) made it possible to compare different RNA-Seq protocols, read lengths, and sequencing types (SE or PE) across the same set of pooled samples. The samples were processed using 2 different, but complementary, protocols: the standard Illumina protocol, which selects polyadenylated RNA from total RNA prior to cDNA synthesis, and a prototype NuGEN Ovation RNA-Seq protocol, which uses a combination of oligo-d(T) primers and random hexamers to synthesize cDNA from total RNA. The samples processed using the NuGEN protocol had a lower percentage of uniquely aligned reads than samples processed using the Illumina protocol. This difference may be partly due to higher-quality reads from the samples prepared with Illumina protocol using a new Illumina sequencer (greater number of reads aligning with zero mismatches, Fig. 2B) and to a higher content of repetitive RNA in the samples prepared using the NuGEN protocol. The NuGEN protocol had a much higher percentage of reads aligning to mitochondrial RNA and rRNA (39% vs. 12%) due to the fact that it captures both polyadenylated and non-polyadenylated RNAs and that the protocol may not have been fully optimized (a prototype version of the protocol was used). We were able to assess the effects of read length and sequencing type, using the Illumina-protocol processed samples, by trimming the reads and considering each read of each pair separately. Increased read length (36 to 75 nt) results in 10% more reads aligning to a unique location in the genome and 8% more reads that span splice junctions. PE versus SE sequencing gives an additional 6% increase in the number of reads that align uniquely as a pair. The results show that there are clear advantages to longer sequencing length and PE sequencing versus SE sequencing. In addition, the standard Illumina protocol results in higher coverage of polyadenylated genes (mostly protein coding), but it fails to capture a subset of non-polyadenylated transcripts. In this study, the subset of genes detected only in samples processed using the NuGEN protocol is small; however, it is likely that an optimized protocol (incorporating methods to reduce repetitive and highly abundant RNA) combined with higher-quality PE 75 nt (or greater) reads would yield additional transcripts. Our data suggest that the optimal approach for sequencing the airway transcriptome would be to use an optimized protocol that captures both poly-adenylated and non-polyadenylated transcripts or a combination of protocols followed by 75 nt (or longer) PE sequencing at a depth of coverage greater than 30 million reads.
The union set of genes measured by both the Illumina and NuGEN protocols was used to define the compendium of genes expressed in the airway transcriptome. We believe this is the first comprehensive catalogue of genes expressed in the bronchial airway epithelium. Despite the small sample size, the numbers of genes (even the conservative airway transcriptome definition, Supplementary Table S3) exceeds the number of genes determined to be expressed in the airway using microarrays (2). The majority of genes were detected when the samples were processed using the Illumina protocol, resulting in higher read coverage of annotated genes (19,786 vs. 10,926 genes for samples processed using NuGEN, Fig. 3A), because less reads were lost to alignments to mitochondrial RNA, rRNA, and non-polyadenylated transcripts. The fact that the expression of some genes could only be detected when the samples were processed using the NuGEN protocol was therefore probably not the result of coverage differences but rather because of differences between the protocols. The genes whose expression is detected only by the samples processed using the NuGEN protocol (n = 787) are more likely to belong to gene biotypes other than protein coding (Fig. 3C) and to the set of Ensembl genes not categorized as "known." Despite the differences between the 2 protocols, the read counts obtained are highly correlated (r = 0.59, P < 0.001) within the set of genes detected by both protocols (n = 10,139). There is a group of genes (among the 10,139 genes) with markedly higher counts when using the NuGEN protocol versus the Illumina protocol (Fig. 3B) that predominantly belong to gene biotypes other than protein coding. One interesting example is the lincRNA MALAT1, which has a mean RPKM of 14,915 (NuGEN) versus 2352 (Illumina). Reads aligning to MALAT1 from samples processed using the Illumina protocol are distributed along the entire length of the gene, whereas reads from samples processed using the NuGEN protocol are concentrated in a shorter, approximately 300-bp region of the transcript, which may represent an additional non-polyadenylated processed RNA transcribed from this locus (Supplementary Fig. S1). Defining the airway transcriptome by combining the strengths of both RNA-Seq library preparation protocols is an important step in fully understanding the biology of the airway field of injury.
Genes in the airway transcriptome were classified as differentially expressed between the S and NS samples or the C and NC samples to find enriched biological pathways and functions. Surprisingly, genes involved in ion channel activity were enriched among the differentially expressed genes detected only in samples processed using the NuGEN protocol (Supplementary Table S4). It is unclear if this finding is because of biases in the library preparation protocols, polyadenylated tail length, or the presence of non-polyadenylated isoforms. The presence of non-polyadenylated isoforms of transcripts measured in samples processed using the NuGEN protocol, which may have important regulatory functions, needs to be confirmed with further studies. Among genes detected in samples processed using the Illumina protocol (n = 19,786), there were several smoking-related pathways such as oxidoreductase activity, retinol metabolism, and metabolism of xenobiotics by cytochrome P450 that were enriched among genes differentially expressed between the NS and S samples. Cell adhesion molecules, cytokine–cytokine receptor interaction, and chemokine activity were enriched among genes differentially expressed between the NC and C samples (Supplementary Table S5). The RNA-Seq data appear to find gene expression alterations that are smoking and cancer related despite the small sample size.
RNA-Seq–derived fold changes between the NS and S samples and between the NC and C samples are significantly correlated with changes measured by microarrays among genes interrogated by both platforms. There is, however, a weaker correlation among the samples with and without cancer (Fig. 5B) that can be partially explained by the fact that the an older microarray platform was used and that only a subset of the cancer samples used in the pool had microarray data. In addition, the cancer signal is weaker than the smoking signal [less genes have an absolute log2 fold change > log2(1.5)], and therefore, the correlations of C/NC fold change between data generated using the Illumina and the NuGEN protocols or between RNA-Seq and microarray are weaker.
An advantage of RNA-Seq over microarray technology lies in the number of genes that are significantly differentially expressed by RNA-Seq but are not interrogated on the microarray as exemplified by the genes listed in Table 1. One example is a MUC5AC (mucin 5AC)-processed transcript located within an intron of MUC5B (Fig. 4B) that is upregulated in current smokers compared with never smokers and downregulated in smokers with lung cancer compared with smokers without lung cancer. MUC5AC is a mucin gene expressed in the respiratory tract and is found in patients with asthma, cystic fibrosis, and COPD (33). We also found that ANG (angiogenin, ribonuclease, RNase A family, 5), a gene that is not measured by these microarrays but which is involved in lung adenocarcinoma cell proliferation and angiogenesis (34), was up-regulated in smokers. Another gene, SCGB3A1 (secretoglobin, family 3A, member 1), which is not interrogated by the microarrays used in this study but was found to be upregulated in the normal airway of lung cancer patients using RNA-Seq, has been linked to poor prognosis in non-small-cell lung cancer (35). Table 1 also includes non-coding RNAs (lincRNAs, pseudogenes, and processed transcripts) whose biological functions have not been well described but which may have important gene regulatory functions in lung carcinogenesis. Identification of their cancer-associated differential expression by sequencing provides a rationale for studying their expression using targeted assays in larger cohorts of samples.
In addition to examining concordance with microarrays, we also used qRT-PCR to validate the concordance of fold change direction among genes detected only as differentially expressed log2(1.5) by sequencing. The expression of the genes S100A8 and S100A9, which are known to be involved in the inflammatory response in the lung (36–38), and CYP4F2, a member of the cytochrome P450 family of enzymes that play a role in xenobiotic pathways (39), were found to be up-regulated in smokers by both RNA-Seq and qRT-PCR. Similarly, the expression of the genes CCL20, IL8, NFKB1A, and SCGB3A1 was found to be up-regulated in the normal airway of patients with lung cancer versus those with benign disease using both RNA-Seq and qRT-PCR (Fig. 6A). One gene, SCGB1A1, however, was not concordant between RNA-Seq and microarrays. We also validated that the expression of select non-coding RNAs, which may have an important role in gene regulation (41–43), changed in the same directions as measured by either RNA-Seq or qRT-PCR (Fig. 6B). The correlation between qRT-PCR fold change and RNA-Seq fold change was significant (r = 0.888, P< 0.001; data not shown). The concordance between qRT-PCR and RNA-Seq has been confirmed across a small set of genes, suggesting that RNA-Seq is a good method for assaying genes important in epithelial cell response to smoke and lung cancer.
In summary, we have established that transcriptome sequencing has the potential to provide new insights into the biology of the smoking- and cancer-related airway field of injury. While much of the airway transcriptome is captured with RNA-Seq of libraries enriched in polyadenylated transcripts, library preparation protocols that measure the expression of non-polyadenylated RNAs are needed to completely characterize the transcriptome, as are longer read lengths and PE sequencing strategies, both of which yield a higher percentage of mapped reads. Our results suggest that the expression of both protein-coding and non-protein-coding RNAs are impacted by exposure to tobacco smoke and the presence of lung cancer, and that long non-protein-coding RNAs that we have begun to characterize in this study may be important in the response to tobacco smoke in airway epithelial cells. Larger sample sizes are needed to characterize the RNAs uncovered in this study and to confidently assess transcript splicing patterns and the presence of novel transcripts. Novel coding and non-coding transcripts uncovered by RNA-Seq provide amore complete portrait of the smoking- and cancer-related airway field of injury have the potential to help elucidate mechanisms of response to tobacco smoke and to function as additional biomarkers of disease risk or novel targets for chemoprevention.
Supplementary Material
Acknowledgement
We would like to thank Yevgeniy Gindin for his help in creating the Ensembl annotation file used in the manuscript. This work was funded by R01 CA 124640 and U01 CA152751 (Spira and Lenburg) as part of the NCI’s Early Detection Research Network (EDRN).
Footnotes
Note: Supplementary data for this article are available at Cancer Prevention Research Online (http://cancerprevres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
A. Spira and M. Lenburg own equity in and are consultants to Allegro Diagnostics, Inc.
References
- 1.Shields PG. Molecular epidemiology of lung cancer. Ann Oncol. 1999;10(Suppl 5):S7–S11. doi: 10.1093/annonc/10.suppl_5.s7. [DOI] [PubMed] [Google Scholar]
- 2.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, et al. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci U S A. 2004;101:10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miyazu YM, Miyazawa T, Hiyama K, Kurimoto N, Iwamoto Y, Matsuura H, et al. Telomerase expression in noncancerous bronchial epithelia is a possible marker of early development of lung cancer. Cancer Res. 2005;65:9623–9627. doi: 10.1158/0008-5472.CAN-05-0976. [DOI] [PubMed] [Google Scholar]
- 4.Guo M, House MG, Hooker C, Han Y, Heath E, Gabrielson E, et al. Promoter hypermethylation of resected bronchial margins: a field defect of changes? Clin Cancer Res. 2004;10:5131–5136. doi: 10.1158/1078-0432.CCR-03-0763. [DOI] [PubMed] [Google Scholar]
- 5.Franklin WA, Gazdar AF, Haney J, Wistuba II, La Rosa FG, Kennedy T, et al. Widely dispersed p53 mutation in respiratory epithelium. A novel mechanism for field carcinogenesis. J Clin Invest. 1997;100:2133–2137. doi: 10.1172/JCI119748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wistuba II, Lam S, Behrens C, Virmani AK, Fong KM, LeRiche J, et al. Molecular damage in the bronchial epithelium of current and former smokers. J Natl Cancer Inst. 1997;89:1366–1373. doi: 10.1093/jnci/89.18.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powell CA, Klares S, O’Connor G, Brody JS. Loss of heterozygosity in epithelial cells obtained by bronchial brushing: clinical utility in lung cancer. Clin Cancer Res. 1999;5:2025–2034. [PubMed] [Google Scholar]
- 8.Auerbach O, Hammond EC, Kirman D, Garfinkel L. Effects of cigarette smoking on dogs. II. Pulmonary neoplasms. Arch Environ Health. 1970;21:754–768. doi: 10.1080/00039896.1970.10667329. [DOI] [PubMed] [Google Scholar]
- 9.Hackett NR, Heguy A, Harvey BG, O’Connor TP, Luettich K, Flieder DB, et al. Variability of antioxidant-related gene expression in the airway epithelium of cigarette smokers. Am J Respir Cell Mol Biol. 2003;29:331–343. doi: 10.1165/rcmb.2002-0321OC. [DOI] [PubMed] [Google Scholar]
- 10.Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med. 2007;85:39–53. doi: 10.1007/s00109-006-0103-z. [DOI] [PubMed] [Google Scholar]
- 11.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 12.Beane J, Sebastiani P, Liu G, Brody JS, Lenburg ME, Spira A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol. 2007;8:201. doi: 10.1186/gb-2007-8-9-r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci USA. 2009;106:2319–2324. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chari R, Lonergan KM, Ng RT, Macaulay C, Lam WL, Lam S. Effect of active smoking on the human bronchial epithelium transcriptome. BMC Genomics. 2007;8:297. doi: 10.1186/1471-2164-8-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sridhar S, Schembri F, Zeskind J, Shah V, Gustafson AM, Steiling K, et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genomics. 2008;9:259. doi: 10.1186/1471-2164-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyle JO, Gumus ZH, Kacker A, Choksi VL, Bocker JM, Zhou XK, et al. Effects of cigarette smoke on the human oral mucosal transcriptome. Cancer Prev Res. 2010;3:266–278. doi: 10.1158/1940-6207.CAPR-09-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Sebastiani P, Liu G, Schembri F, Zhang X, Dumas YM, et al. Similarities and differences between smoking-related gene expression in nasal and bronchial epithelium. Physiol Genomics. 2010;41:1–8. doi: 10.1152/physiolgenomics.00167.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beane J, Sebastiani P, Whitfield TH, Steiling K, Dumas YM, Lenburg ME, et al. A prediction model for lung cancer diagnosis that integrates genomic and clinical features. Cancer Prev Res. 2008;1:56–64. doi: 10.1158/1940-6207.CAPR-08-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blomquist T, Crawford EL, Mullins D, Yoon Y, Hernandez DA, Khuder S, et al. Pattern of antioxidant and DNA repair gene expression in normal airway epithelium associated with lung cancer diagnosis. Cancer Res. 2009;69:8629–8635. doi: 10.1158/0008-5472.CAN-09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gower A, Steiling K, Brothers J, Lenburg M, Spira A. Transcriptomic studies of the airway "field of injury" associated with smoking-related lung disease. Proc Am Thorac Soc. doi: 10.1513/pats.201011-066MS. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 23.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 25.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 26.Ensembl. Ensembl CDF file (v58) http://brainarray.mbni.medumich.edu/Brainarray/Database/CustomCDF/13.0.0/ensg.asp.
- 27.R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. ISBN 3-900051-07-0. http://www.R-project.org. [Google Scholar]
- 28.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 29.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 33.Voynow JA, Gendler SJ, Rose MC. Regulation of mucin genes in chronic inflammatory airway diseases. Am J Respir Cell Mol Biol. 2006;34:661–665. doi: 10.1165/rcmb.2006-0035SF. [DOI] [PubMed] [Google Scholar]
- 34.Yuan Y, Wang F, Liu XH, Gong DJ, Cheng HZ, Huang SD. Angiogenin is involved in lung adenocarcinoma cell proliferation and angiogen-esis. Lung Cancer. 2009;66:28–36. doi: 10.1016/j.lungcan.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 35.Marchetti A, Barassi F, Martella C, Chella A, Salvatore S, Castrataro A, et al. Down regulation of high in normal-1 (HIN-1) is a frequent event in stage I non-small cell lung cancer and correlates with poor clinical outcome. Clin Cancer Res. 2004;10:1338–1343. doi: 10.1158/1078-0432.ccr-1174-03. [DOI] [PubMed] [Google Scholar]
- 36.Hsu K, Champaiboon C, Guenther BD, Sorenson BS, Khammanivong A, Ross KF, et al. Anti-infective protective properties of S100 calgranulins. Antiinflamm Antiallergy Agents Med Chem. 2009;8:290–305. doi: 10.2174/187152309789838975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim SY, Raftery MJ, Goyette J, Hsu K, Geczy CL. Oxidative modifications of S100 proteins: functional regulation by redox. J Leukoc Biol. 2009;86:577–587. doi: 10.1189/jlb.1008608. [DOI] [PubMed] [Google Scholar]
- 38.Henke MO, Renner A, Rubin BK, Gyves JI, Lorenz E, Koo JS. Up-regulation of S100A8 and S100A9 protein in bronchial epithelial cells by lipopolysaccharide. Exp Lung Res. 2006;32:331–347. doi: 10.1080/01902140600959580. [DOI] [PubMed] [Google Scholar]
- 39.Ding X, Kaminsky LS. Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol. 2003;43:149–173. doi: 10.1146/annurev.pharmtox.43.100901.140251. [DOI] [PubMed] [Google Scholar]
- 40.Sjodin A, Guo D, Sorhaug S, Bjermer L, Henriksson R, Hedman H. Dysregulated secretoglobin expression in human lung cancers. Lung Cancer. 2003;41:49–56. doi: 10.1016/s0169-5002(03)00126-0. [DOI] [PubMed] [Google Scholar]
- 41.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea MD, et al. Many human large intergenic noncoding RNAs associate with chro-matin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.