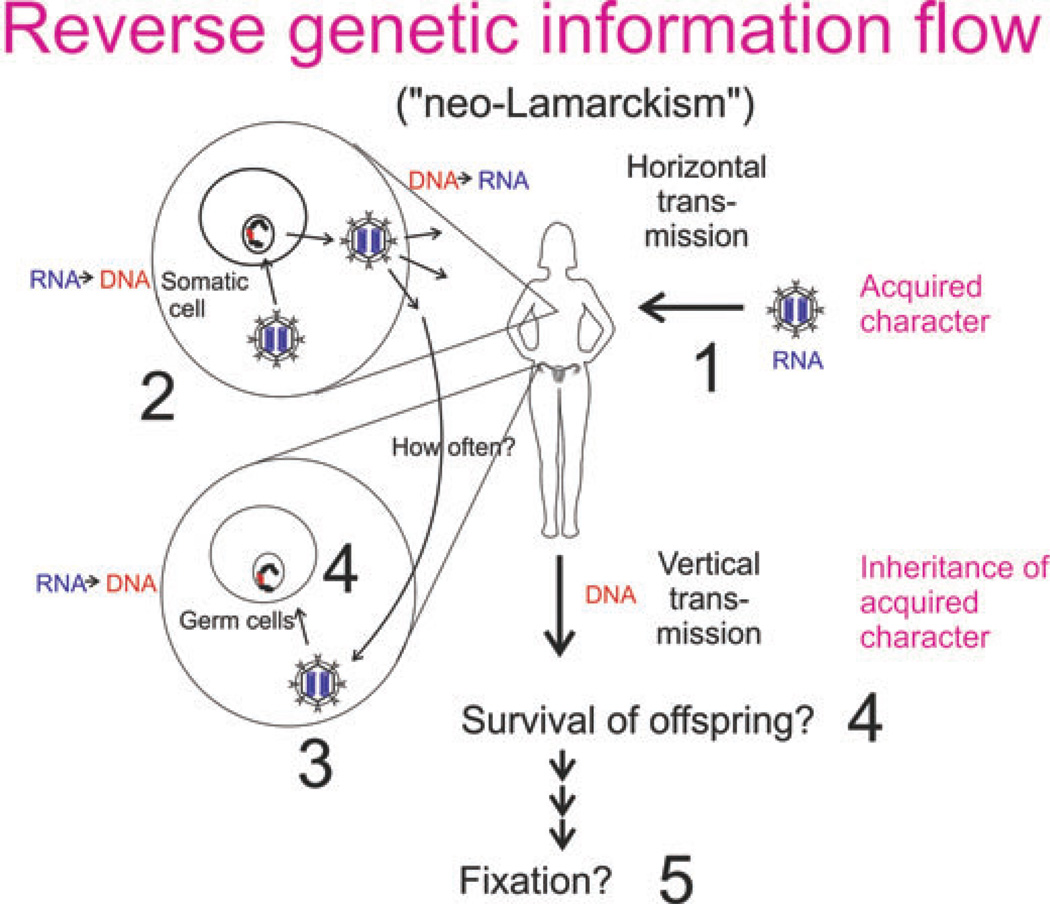
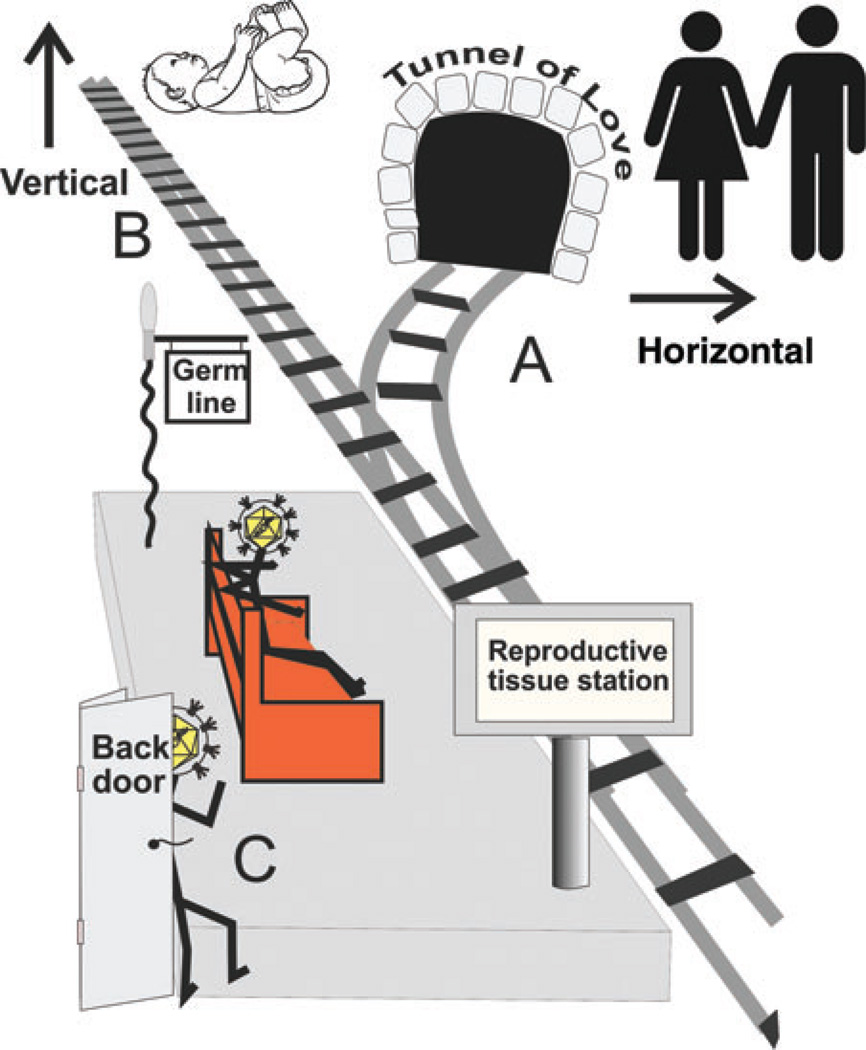
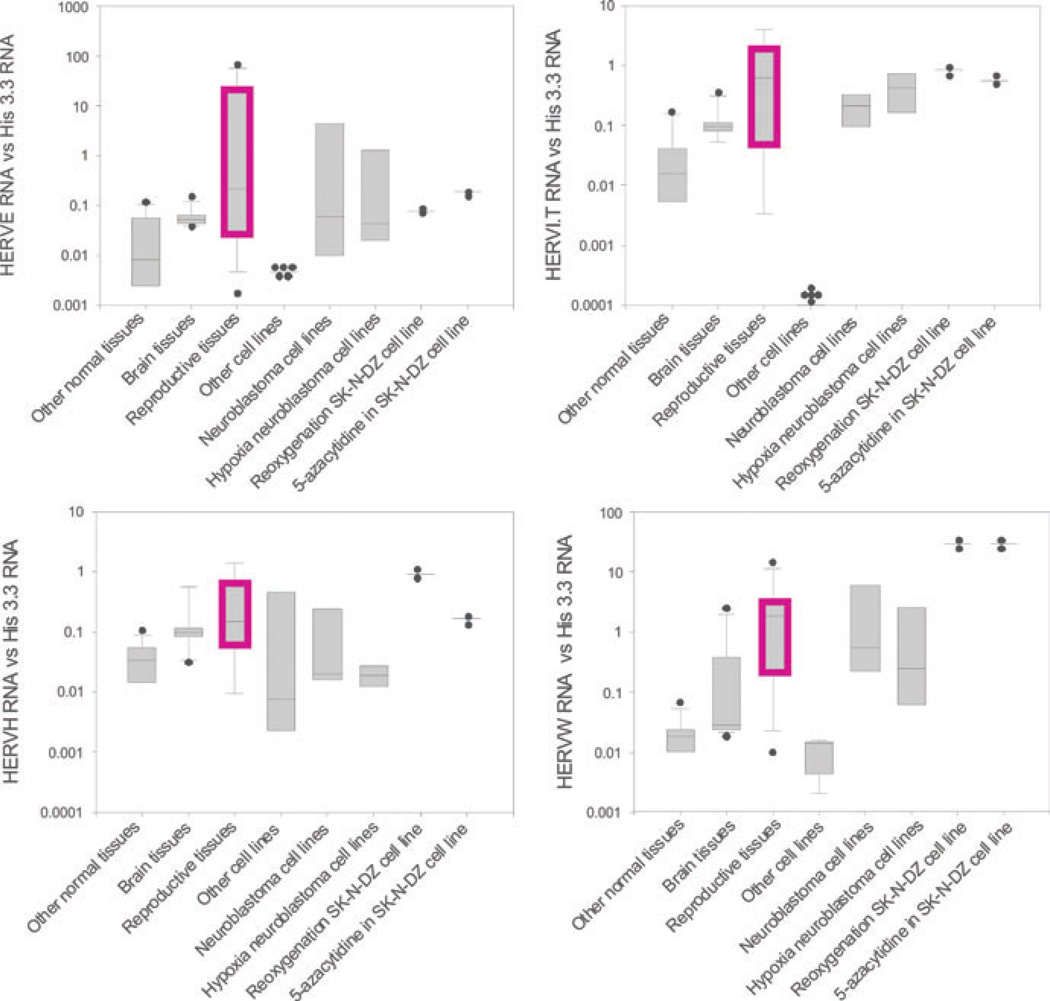
Abstract
The following series of concise summaries addresses the evolution of infectious agents in relation to sex in animals and humans from the perspective of three specific questions: (1) what have we learned about the likely origin and phylogeny, up to the establishment of the infectious agent in the genital econiche, including the relative frequency of its sexual transmission; (2) what further research is needed to provide additional knowledge on some of these evolutionary aspects; and (3) what evolutionary considerations might aid in providing novel approaches to the more practical clinical and public health issues facing us currently and in the future?
Keywords: evolution, infectious agents, sexual transmission, econiche
Animal agents
Lice as both troublesome parasites and informative markers of host evolutionary history
David L. Reed
dlreed@ufl.edu
At present there are approximately 3,000 described species of lice.1,2 There is conflicting evidence surrounding the monophyly of the Order Phthiraptera. Recent molecular data suggests that lice are paraphyletic with the Psocoptera (book and bark lice), which would mean that obligate parasitism of birds and mammals evolved more than once within what is now known as Phthiraptera.3 Morphological evidence suggests that the Order Phthiraptera is a monophyletic group, but opponents contend that the characters that unite them are merely convergent, owing to the constraints of an obligate parasitic lifestyle. Within the Phthiraptera resides the Suborder Anoplura, which contains the obligate blood feeding lice of mammals. This clade of approximately 530 species is monophyletic based on morphological and molecular data. Within the Anoplura are the species of lice that infest humans, which are of interest here.
There are three types of lice that infest humans, clothing lice, head lice, and pubic lice. Clothing lice (also called body lice) and head lice are two morphotypes of a single species, Pediculus humanus.4 Head and clothing lice are modestly distinct in terms of their morphology, but have pronounced differences in their feeding behavior, egg deposition, and their ability to transmit disease.5 Clothing lice feed only a few times per day, lay eggs in clothing, and transmit three deadly microbial pathogens, whereas head lice feed frequently throughout the day, attach eggs to individual hair shafts, and are not known to transmit disease to human hosts in natural environments. Humans are also infested with the pubic louse (Pthirus pubis), which is the family Pthiridae. Humans likely acquired this parasite from a gorilla-like ancestor about three million years ago.6 Pubic lice are morphologically very different from clothing and head lice, and the two genera (Pthirus and Pediculus) have not shared a common ancestor for 13 million years.6 Pubic lice are unique among human lice in that they are primarily transmitted sexually. The presence of pubic lice is correlated with the presence of other sexually transmitted infections, but the lice themselves are not known to be vectors of sexually transmitted diseases.
The evolutionary events that led pubic lice to their current ecological niche are only partially known. Reed et al. showed that the evolutionary origins of pubic lice can be traced to a gorilla-associated ancestor, but the conditions under which that might have happened are speculative.6 The successful establishment of Pthirus on hominid hosts is interesting because hominids already played host to lice in the genus Pediculus when Pthirus invaded three million years ago. It is possible that humans had already lost much of their body hair, which caused lice of the genus Pediculus to retreat to head hair, leaving the pubic niche available for colonization. However, this also presumes that humans had already developed pubic hair, which is absent in other great apes, and develops at sexual maturity. The evolutionary loss of body hair, the development of pubic hair, and the later development of clothing in humans are watershed events in the evolutionary history of lice, and likely spawned the birth of new louse lineages.
Although pubic lice are estimated to infest about 2% of the world’s population, the exact number is exceedingly hard to quantify. Infestations are easy to acquire and sometimes difficult to eradicate. Permethrin-based products are often used to treat phthiriasis; and unlike head lice, pubic lice do not appear to be widely resistant to its use. The prevalence of pubic lice is declining in certain developed countries among men and women that remove their pubic hair.7
The lice of the genus Pediculus have provided great insight into primate and human evolutionary history—they have told us that humans began wearing clothing 170,000 years ago, prior to leaving Africa,8 and that modern humans had direct physical contact with archaic hominids perhaps around 25,000 years ago.9 Because pubic lice also parasitized humans during this time they represent ecological replicates of Pediculus that can be used to test the predictions gleaned from Pediculus about human evolution.
Furthermore, because Pthirus is a sexually transmitted parasite, its coevolutionary history with humans may differ from that of the more casually transmitted lice of the genus Pediculus. For example, there is strong evidence that head lice from Neanderthals made it onto modern humans, where traces of this host switch can still be seen in louse DNA. Reed et al. (2004) concluded that the host switch from Neanderthals to modern humans must have been caused by direct physical contact between the two species of host.9 If the pubic lice of living humans also show traces of Neanderthal louse DNA, then we might propose that interbreeding occurred supporting the findings of recent genome studies hinting at introgression between modern and archaic hominins.
We now have a complete genome of the human clothing louse (Pediculus humanus), which will facilitate much new research in the treatment of pediculosis and in studies of human/louse evolution. Similar efforts are needed to sequence the genome of the pubic louse to better understand its similarities and differences to Pediculus. The genome of Pediculus was incredibly small, owing to the reduced and simple environment that the louse lives within. A comparison to another louse living in roughly the same environment would be very useful. Whether the two different types of lice have similar gene composition will help us to understand how the external environment shapes genome evolution.
The treatment of pediculosis is a billion dollar industry, and new treatments are constantly being sought. Now that louse populations are resistant to insecticidal shampoos in most developed countries, new “evolution proof” ways of dealing with lice are being investigated. One such method uses hot air to kill lice and their eggs with no discomfort to the patient.10 It is likely that resistance to hot air would be difficult to evolve in louse populations. Similar “evolution-proof” treatments are not required for pubic lice, as this species has not yet evolved widespread resistance to the insecticides used to treat them.
Scabies in animals and humans
Russell W. Currier and Shelley F. Walton
ruscurrier@yahoo.com
Scabies is one of the great epidemic diseases of man. Not only are humans affected by the common itch mite Sarcoptes scabiei, but also numerous species of animals. Host species are infested with different variants of the same genus and species of mite, as is true in a number of other animal hosts—reflecting the overall observation that ectoparasites tend to be very host specific. Sarcoptic mange in animals and infestations in humans date back to antiquity, with some historians noting that Aristotle described these as lice-like creatures of the skin. However, most historians credit Avenzoar, the Moorish physician, and his student Averroes of Seville in the 12th century, with accurately describing the mites. Successive discoveries and descriptions were detailed by St. Hildegarde (12th century), August Hauptmann in 1654, and Giovanni Bonomo in 1687, who described and drew the mite.11 The Swedish physician and botanist Carolus Linnaeus established the parasitic nature of scabies in 1746, but eleven years later confused the mite with the common flour mite.12 These developments, however, were ignored and forgotten in the field of medicine. Finally a senior medical student in Paris, France, Simon Francois Renucci, extracted a mite from a female patient in August, 1834 with a needle probe—a technique he learned from peasant women in his native Corsica—to usher in the modern era of scabies diagnosis. This “rediscovery” was expanded in subsequent decades by Ferdinand von Hebra, the father of modern dermatology in Vienna, who established our basic knowledge of scabies.13
Scabies mites are transmitted by skin-to-skin con-tact, including sexual contact; after varying periods of time, usually three to six weeks, hypersensitivity develops to metabolites and secretions of the mites that result in pruritis, especially in the evening, and prompts patients to seek treatment. Diagnosis can be determined most commonly by skin scrapings examined microscopically with mineral oil (author RWC prefers microscopic immersion oil). Observing adult and larval mites, eggs, and scybala (discrete golden brown fecal pellets) are confirmatory. Since most otherwise healthy patients have fewer than 15 adult mites, it is important to collect sufficient scrapings to ensure a reasonable chance of recovering a mite. A slightly less traumatic and very specific method to identify scabies is needle extraction with the aid of a loupe or hand lens.14
Affected patients should be treated, and a variety of products are available including topicals: Lindane lotion, 5% permethrin lotion, crotamiton lotion or cream, and 6–10% precipitated sulfur in petrolatum. For the past 10 years, oral administration of ivermectin has produced good results, especially with a heavy mite burden, that is, patients profoundly immunosuppressed and for patients with “crusted” or keratotic scabies. In the UK and Commonwealth, benzyl benzoate preparations are licensed for treatment of scabies as well.15,16 Prophylactic treatment of close physical contacts, for example, family members, romantic acquaintances, and preschool siblings who are not yet exhibiting signs/symptoms should be considered on a case by case basis.
How did humans become colonized with S. scabiei? The scientific consensus is that humans, and protohumans before them, were the principal hosts for Sarcoptes mites.11,13 When humans domesticated various species of animals over the past 10,000 years, Sarcoptes mites jumped species and established new colonizations among recently domesticated animals. These species were more susceptible, likely because domestication per se tended to reduce immunocompetence and, thus, increased the probability of mites adapting to new hosts.17–19 Domesticated animals that escaped, in turn, infected several wild nondomesticated species.
Transmission of Sarcoptes in both humans and animals is by direct contact in most situations, but indirect contact may serve a role depending on crowding and individual host mite populations. In modern humans, sexual transmission is important especially among sexually active young people, who may further transmit the mite during care of young children. Scabies is, or should be, routinely considered in the framework of screening patients at risk for other sexually transmitted infections (STIs). In evolutionary terms, humans may have become partially protected through the advent of clothing, with social trends towards monogamous relationships also reducing risk of exposure.
Walton et al.20,21 have demonstrated that canine and human scabies are phylogenetically distinct variants of S. scabiei with minimal gene exchange. (Heretofore there have been no definitive means to distinguish taxonomically the various strains that manifest a fairly high degree of host specificity for their respective host species.) As such, even if humans are infested, as well as their dogs, it would not require elaborate coordinated efforts to treat both groups simultaneously. Interestingly, these evolutionary studies in aboriginal indigenous communities in northern Australia with endemic scabies have led to changes in health policies at the regional level, with the development of cost effective public health intervention programs that are directed at humans only.20
Future research would best be directed at defining parasitic secretions and hidden antigens and their role in stimulating and modulating host immune responses. Better definition of S. scabiei gene families and their products will provide better insights into the molecular biology and host immune evasion strategies of the mite. Clearly, one of the major difficulties of managing scabies infestations in humans, as well as in animals, is recognizing infested hosts. An immunological test for diagnosis based on mite proteins would be most welcome.20
Another issue is the evidence for an inverse relation between parasites and allergy, as noted by Walton et al., observing that asthma and hay fever are rare in parts of the world where the population is heavily parasitized.21 “The relationship between susceptibility to, and severity of, symptoms with scabies and asthma are as yet unknown and conceivably may be interdependent. Those who suffer from house dust mite (HDM) allergy may have reduced (or possibly increased) symptoms of scabies and presumably the reverse—thosewith scabies could have a reduced (or increased) susceptibility to (HDM) antigens.”21
Investigating the evolutionary relationship of house dust and scabies mites, as well as their cross-antigenicity, may lead to improved diagnostics for both scabies and HDM allergy and potentially protective immunoprophylaxis for asthma.
Trichomonas infections of humans and animals
Melissa Conrad, Steven A. Sullivan, and Jane M. Carlton
Melissa.Conrad@nyumc.org
Trichomonads are protozoa of the class Parabasalia that inhabit the alimentary canals or urogenital tracts of vertebrates and insects. Members are characterized by having three to five anterior flagella, and many species have highly diverged mitochondrion-like organelles called hydrogenosomes. Trichomonads inhabit vertebrate hosts ranging from reptiles and amphibians to pigs and humans. Most are considered commensal, but even these can cause diarrhea and abdominal pain, as is the case for Pentatrichomonas hominis, a commensal found in the gastrointestinal tracts of several mammals. Three species of trichomonads are generally recognized as pathogens: Trichomonas gallinae, Trichomonas vaginalis, and Tritrichomonas foetus. T. gallinae is a parasite of the upper alimentary tracts of bird species and can cause severe necrotic ingluvitis. The remaining two species are sexually transmitted parasites that reside in the urogenital tracts of humans and cattle, respectively.
T.vaginalis is the causative agent of trichomoniasis, the most prevalent nonviral sexually transmitted disease in humans (reviewed in Ref. 22). The parasite adheres to the lining of the urogenital tracts of both men and women and can result in reproductive health sequelae, including pelvic inflammatory disease and adverse pregnancy outcomes, and has been associated with an increased risk in HIV transmission. About half of infected women manifest symptoms, including a frothy vaginal discharge, odor, itching, and punctate microhemorrhages on the cervix known as “strawberry cervix.” Men generally remain asymptomatic. T. foetus causes bovine trichomoniasis. Comparable to T. vaginais, T. foetus inhabits the reproductive tract and typically causes few symptoms in bulls, while cows frequently suffer from urogenital inflammation. Bovine trichomoniasis can also lead to severe consequences, such as infertility and abortion. Both T. vaginais and T. foetus appear to have a simple life cycle consisting of a trophozoite stage and a putative pseudocyst stage, and both divide asexually through mitosis.
With these similarities in mind, it is tempting to speculate that trichomoniasis is shared between cows and humans. However, phylogenetic analyses using genetic markers (SSU rRNA, GAPDH, enolase, (β-tubulin, α-tubulin, and Rpb1), and several morphological distinctions indicate that exploitation of the urogenital niche by the two species is the result of convergent evolution. T. vaginais and T. foetus are not sister taxa, being separated on a phylogenetic tree by many species that inhabit the gastrointestinal tract.23 In addition, they are morphologically distinct; T. vaginalis is characterized by four flagella, while T. foetus only has three. Both Trichomonas and Tritrichomonas are genera composed of a large number of gastrointestinal Parabasalids, suggesting that T. vaginais and T. foetus share a common gastrointestinal ancestor but evolved independently to colonize the urogenital niche. Indeed, T. foetus has been isolated from the feces of cats and dogs with diarrhea, suggesting that the parasite retains ancestral characteristics that allow it to continue to colonize and survive in the intestine.24
Instances of inferred convergent evolution provide an opportunity to compare derived similarities, in order to identify common traits that coincide with the shared niche. In the case of T. vaginais and T. foetus, we may be able to learn what characteristics provided these two trichomonad species with the appropriate “fitness” to transition from the gastrointestinal tract (the ancestral state) to the urogenital tract. Genomics will be a powerful tool in determining this evolutionary history.
The T. vaginalis genome was published in 2007.25 At 160 Mb, it is one of the largest parasite genomes known, dwarfing those of Entamoeba (20 Mb), Plasmodium (25 Mb), and Toxopasma (63 Mb). The large genome is thought to be the result of a sudden expansion of “repeats” that occurred after T. vaginalis and Trichomonas tenax, its sister taxon, split from their most recent common ancestor. At least 65% of T. vaginalis DNA consists of hundreds of families of transposable elements and multigene families that have undergone massive amplification. These gene expansions may have occurred after some evolutionary event, such as the reduction in effective population size,26 which could have followed the transition of the parasite from an enteric to a urogenital environment. However, a drastic increase in genome size over a short period of evolutionary time is typically detrimental,27 making it unlikely that such a trait would have been fixed by genetic drift alone; rather, it must have provided some selective advantage to the organism in its new environment.28 One possible selective advantage relates to the positive correlation between genome size and cell volume in trichomonads.29 An increase in cell volume, resulting from the increase in genome size due to fixation of expanded gene families, could have resulted in the parasite being able to augment phagocytosis of bacteria, reduce its vulnerability to phagocytosis by macrophages, and increase its cell surface to allow better adhesion to the vaginal mucosa. These advantages may have allowed T. vaginalis to thrive in its new vaginal environment.
One way to test this hypothesis would be to look for similar evolutionary tendencies in the T. foetus genome. It has been suggested that gene family expansions are common among Parabasalids,30 which we have confirmed through Roche 454 sequencing of the T. foetus and P. hominis genomes (unpublished). The genomes of these species are composed of a minimum of 28% and 20% “repeats,” respectively. Consistent with the difference in percentage of genome repeats, T. foetus has a 166–188 Mb genome (comparable to that of T. vaginalis ), while the P. ho-minis genome is 86–102 Mb.29 Perhaps here, too, a sudden expansion of gene families has contributed to a larger cell volume, which may have provided T. foetus with the necessary characteristics to diverge from its enteric ancestors and successfully colonize the bovine urogenital system.
Admittedly, our hypothesis is far from being proven. Unfortunately, there is also a lack of sufficient data to disprove it. If we ever hope to understand the mechanism by which trichomonads successfully moved from an enteric environment to the urogenital tract, it will be necessary to expand the inventory of whole genomes available from diverse Parabasalid species. Among the top priorities should be gastrointestinal species closely related to T. vaginalis and T. foetus , such as Dientamoeba fragilis and Trichomonastenax. Only by elucidating the state of ancestral genomes will we be able to determine how these species have evolved to adapt to new environments, and ultimately cause sexually transmitted diseases of significant veterinary and public health importance.
Chlamydia animal sexually transmitted infections
Timothy D. Read
tread@emory.edu
The Chlamydiaceae are an ancient group of Gram-negative obligate intracellular pathogens, currently divided into two genera, Chlamydia and Chlamydophila. All Chlamydiaceae share a distinctive biphasic lifecycle, alternating between a dormant infectious elementary body (EB) and a cell-bound vegetative reticulate body (RB). Complete sequencing projects of bacteria representing seven species have revealed relatively small genomes (1.1–1.2 Mb) with about 90% of genes conserved within the genus. Despite their limited genetic repertoire, Chlamydiaceae causea very diverse array of diseases in a broad range of hosts. These infections include sexually transmitted infections (STIs), pneumonia, abortions, gastritis, and sepsis. The range of hosts includes mollusks, amphibians, reptiles, and mammals. Genomic studies have revealed ongoing adaptive evolution in the Chlamydiaceae, including acquisition of genes by Horizontal Gene Transmission (HGT) and frequent gene conversion.31 We know from evolutionary reconstructions that Chlamydiaceae can make dramatic host and disease shifts. The common human STI, Chlamydia trachomatis, is very similar to strains causing trachoma,32 and genomic evidence from koala-infecting strains indicate that the common human respiratory pathogen Chlamydia pneumoniae evolved from a zoonotic ancestor.33
Interest has been particularly focused on Chlamydiaceae in economically important animals, including C.abortus and C.pecorum in cattle, sheep, and/or goats, and mixed infections are also of interest.33–37 In the wild, Australian koalas have been found to be infected by C. pneumoniae and C. pecorum— the latter being the more pathogenic of the two, causing urogenital and ocular infections, and occasional outbreaks.38
There are two fundamental questions about Chlamydiaceae evolution that we are some way from answering: how do Chlamydia adapt to new host species, and how do Chlamydiaceae evolve new modalities of infection.
Genome sequencing is an increasingly cost-effective way to investigate these questions, and it will be soon possible to sequence thousands of Chlamydiaceae strains per week. We will soon be limited by well-sourced isolates. The well-maintained existing collections of Chlamydiaceae need to be made available for sequencing, and a major effort to acquire new, diverse strains from animals and the environment needs to be undertaken. Epidemiologic models for natural chlamydial sexually transmitted infection in wild animal species, for example, koalas and perhaps primates, may be useful to parallel information about these infections in humans.
Several recent examples, e.g., HIV, serve to warn us of the importance of tracking emerging zoonoses. Comparing the genomes of human- and non-human–infecting Chlamydiaceae on a large scale may allow us to define the genes acquired by HGT, or even genetic variants in core genes (single nucleotide polymorphisms, insertions, deletions, and inversion) that are associated specifically with human infection. This may lead us to better understand the traits that make a zoonotic pathogen more likely to cross over to infecting humans [e.g., Refs. 31 and 32]. Comparative genome analysis also provides information about epitopes on surface and secreted proteins that may be evolutionary constrained and hence viable targets for vaccine or immunologically targeted therapies. In the same vein, comparative genomics can help improve selecting targets for genetic- and immunologic-based diagnostics and inform where diagnostics are likely to fail.
Comparative sequencing will warn of the dangerous possibility for acquisition of antibiotic resistance genes into human Chlamydia STIs from animal-infecting strains. A recent paper described the uptake via HGT of a tetracycline resistance cassette into the genome of a pig pathogen—C. suis— that probably originated from Enterococcus.39 As we learn more about the pathways for gene exchange in Chlamydiaceae, we could anticipate the potential for losing drug efficacy for human infections based on patterns emerging in animal strains. Also, the possible effectiveness of a C. pecorum vaccine for animals, now in development,40 may provide a model for human chlamydia vaccines, providing information of their possible epidemiological and evolutionary impact.
Molecular evolution of human and simian simplexviruses
Alberto Severini, Shaun Tyler, and R. Eberle
alberto.severini@phac-aspc.gc.ca
Simplexviruses (or α-1 herpesviruses) belong to a genus within the subfamily Alphaherpesvirinae comprising closely related viruses that have mostly primates as natural hosts. Bovine herpesvirus 2, however, infects cattle, and the poorly characterized macropodid herpesviruses 1 and 2 have been isolated from species of wallaby, a marsupial.41 Well characterized simplexviruses of primates are the human HSV-1 and HSV-2, B virus in Asian macaques, SA8 in vervets, herpesvirus papio 2 (HVP2) in African baboons, herpesvirus saimiri 1 (HVS) in squirrel monkeys, and the herpesvirus ateles 1 (HVA1) in spider monkeys.
Phylogenetic analyses based on nucleotide and amino acid sequencing have shown that simplexviruses from humans, old world monkeys, and new world monkeys each cluster in separate branches, indicating that, in general, they coevolved with their hosts.42,43 Despite the relative diversity of natural hosts, primate simplexviruses cause remarkably similar clinical manifestations, that is, orogenital vesicular lesions closely resembling HSV infections in humans including a generally benign course, establishment of latency with the possibility of reactivation, and occasionally severe neurological or other disseminated infections in newborns.44 Thus coevolution spanning about 50 million years seem to have preserved intact the sexual/oral tropism of this group of simplexviruses. However, infections of other primate species other than the natural host often results in more severe pathology, such as deadly B virus encephalomyelitis in humans44 or disseminated HSV infections in some non-human primate species.45,46
Complete genome sequencing of HSV-1,47–49 HSV-2,50 B virus,51 HVP2,52 SA8,53 and chimpanzee herpesvirus (ChHV; manuscript in preparation) has shown that their genomes are overall highly homologous (from 60% to 90% at the DNA evel) and almost all genes and features are collinear. Finer comparison of these genomes leads to the formulation of some hypothesis on the mechanisms of simplexvirus evolution and pathogenesis. Old World monkey simplexviruses (B virus, HVP2, and SA8) lack a homolog of the HSV RL1 open reading frame, the product of which (the γ1 34.5 protein) inhibits the cell interferon-activated, PKR-dependent system that shuts down protein synthesis in response to viral infection.54 As this is considered to be a factor of virulence in mice and humans for HSV, it will be interesting to study how Old World monkey simplexviruses circumvent the PKR system to infect their hosts and to cause severe diseases in humans (B virus) and mice (B virus and HVP2).
Another interesting comparison is the glycoprotein G (gG). This envelope glycoprotein is involved in virus entry in polarized cells and it is the only antigen that differs substantially between HSV-1 and HSV-2, and therefore it has been used for HSV type-specific serological assays.55 HSV-2 gG is a 699 amino acid protein which is cleaved roughly in half into a secreted amino terminus fragment and a transmembrane portion (mgG2) that is incorporated into the viral envelope.56 In contrast, the HSV-1 gG consists only of a 238 amino acid transmembrane protein homologous to mgG2. All of the other Old World simian simplexviruses and, surprisingly, bovine herpesvirus 2, possess an HSV-2-like glycoprotein gene. In contrast, the New World virus HVS1 has a putative gG gene that is similar in structure (but not in sequence) to the HSV-1 gG.
We have recently sequenced the entire genome of chimpanzee herpesvirus, a simplexvirus isolated from a captive animal with oral lesions.57 The genome sequence is remarkably similar to that of HSV-2 (almost 90% homology at the DNA level), but still distinct enough from all known variants of HSV-2 to be considered a distinct virus. The level of DNA sequence divergence is consistent with the human/chimpanzee divergence estimated to have occurred 4 to 6 million years ago.57 The more divergent HSV-1 was estimated to have separated from HSV-2 about 8–9 million years ago,58 and, if these estimates are correct, we have to consider the possibility that HSV-1 did not coevolve with the human species, but rather that its progenitor “colonized” us at a later time in evolution, jumping from an as yet unidentified primate host. Interestingly the aforementioned RL1 virulence gene is the only gene that is most similar between HSV-1 and HSV-2. A comparison of the mechanism of the virulence of the RL1 genes of HVS-1, HSV-2, and ChHV may shed light on the species tropism of these viruses.
The recent whole genome sequence of HSV-1 has confirmed the more distant relationship between New World monkey simplexviruses and the Old World monkey simplexviruses.59 Most HSV-1 genes are homologous to those of other simian simplexviruses, but their arrangement and the overall structure of the genome is similar to that of VZV, α varicellovirus. We can hypothesize that HSV-1 is closer to the common progenitor of all α-herpesviruses, a hypothesis that may be tested by sequencing other α-herpesviruses and by comparative analysis of the function of their gene products.
Whole genome analysis of simplexviruses is a powerful tool aiding in understanding the evolution of this genus in relation to their hosts and to other groups of herpesviruses. Comparative analysis in vitro and in animal models of the function of the few divergent genes may provide insight on the pathogenesis mechanisms of HSV-1 and HSV-2 in humans. All of the primate simplexviruses produce in their natural hosts the same, generally benign, often recurrent vesicular disease and are transmitted by oral or genital contact. Yet, their virulence differs dramatically when these same viruses are transmitted to other host species. A comparative analysis of the virus/natural host interaction maybe a good approach in defining the still elusive pathogenic mechanisms of HSV and the evolution of herpesvirus sexual transmission.
Primate lentiviruses and their hosts
Welkin E. Johnson and Guido Silvestri
welkin_johnson@hms.harvard.edu
The primate lentiviruses include the human pathogens, HIV-1 andHIV-2, and the closely related simian immunodeficiency viruses (SIVs), which are endemic among many species of African apes and monkeys.60,62 The phylogenetic relationships among the primate lentiviruses do not consistently mirror host phylogeny, an indication that the distribution of SIVs among modern primates is the result of numerous cross-species jumps followed by adaptation and emergence of new host–virus combinations (the relatively recent emergence of HIV-1 and HIV-2 in humans being notable examples). In addition to extant SIVs, which have been detected in more than 40 species of African primate, ancient SIV-like proviral sequences have been found embedded in the genomes of lemurs.61 These defective relics of an ancient ancestor of the modern SIVs are direct evidence that lentiviruses were already present in ancestral primate populations millions of years ago.
A distinctive trait of all primate lentiviruses is their capacity for replication in the face of what is, at least initially, a robust virus-specific immune response.63 Infections are characterized by continuous, uninterrupted viral replication and rapid turnover of the virus population. A consequence of high-titer virion production is killing of CD4+ T cells, a central player in the adaptive immune response. The capacity to generate and maintain very large population sizes within the infected individual (coupled with an error-prone viral polymerase) permits these viruses to adapt rapidly to the slightest shifts in selective pressure, including the onset of virus-specific immune responses in a newly infected individual and changes in genetic landscape encountered when jumping between individuals or populations of individuals.
Among many primate species harboring endemic SIVs, there is a notable absence of pathogenesis. Most intriguing to AIDS researchers, the non-pathogenic condition does not correlate with fundamental differences in virulence.64 For example, viral replication kinetics and T cell killing are strikingly similar in natural hosts of SIVs, where pathogenesis is the exception, and in animal models of AIDS (e.g., the SIV-infected rhesus macaque) or humans with HIV, in which pathogenesis is the rule. The lack of pathogenic outcome in many natural hosts of SIVs is viewed as a strong indication that these particular virus–host relationships are the result of long periods of coevolution, possibly spanning hundreds or thousands of host generations. Nonpathogenic infection, which may have evolved independently on multiple occasions, suggests that these natural hosts represent genetic “uncoupling” of overt pathogenesis from viral replication.
While the major focus of HIV/AIDS research is on humans, comparative approaches encompassing the primate lentiviruses and their hosts as a whole are desirable. In particular, two areas deserve closer examination of SIV infections in natural hosts.
Why does infection with HIV result in AIDS?
Identifying the specific adaptive mechanism(s) by which the natural hosts of SIVs uncouple pathogenesis from viremia may suggest previously unimagined avenues for treatment of patients with established HIV infection and should be the target of more intensive research efforts. Toward the same end, efforts to identify and investigate rare cases of HIV-infected individuals with evidence of long-term high-level viremia but notable absence of disease should also be pursued.
How do host genes influence cross-species transmission and emergence of viral pathogens?
Given the close evolutionary relationships between humans and other primates, a key question in virology is whether viruses of other primates are more likely to spread and adapt in human populations than viruses from more distantly related species (anecdotally, the worldwide spread of HIV-1, the result of transmission of SIVcpz from chimpanzees to humans, compared to the more limited success of HIV-2, which arose by transmission of SIVsm from sooty mangabey monkeys, is consistent with this notion). Along these lines, comparative studies of humans and nonhuman primates and their respective viruses could prove useful in identifying host genes that have the strongest influence on limiting the spread of viruses between species; such studies should include efforts to identify the corresponding viral mechanisms for overcoming genetic barriers to interspecies transmission.
Human agents
Evolution of Chlamydia trachomatis
Ian N. Clarke
inc@soton.ac.uk
We know surprisingly little about the recent evolutionary origins of chlamydia (C. trachomatis) as a sexually transmitted agent of infection. Research has been severely restricted because these bacteria are obligate intracellular pathogens, hence clinical samples taken for isolation of C. trachomatis have to be handled in the same way as swabs taken for virus isolation. Live C. trachomatis was first cultivated in hens’ eggs, but isolation in tissue culture took a long time to perfect. While the isolation of chlamydial strains requires expertise in mammalian cell culture systems, the antibiotics used to suppress the contaminating bacteria for virus isolation, such as streptomycin and penicillin, also inhibit C. trachomatis, and this confounded early attempts to develop cell culture systems that supported C. trachomatis. Furthermore, genital tract strains of C. trachomatis have low infectivity, and thus centrifugation is required to ensure these bacteria can infect susceptible, cultured mammalian host cells. C. trachomatis retains viability in long-term storage only if kept at ultra low temperatures and with stabilizing cryopreservatives; consequently, there is a paucity of historical isolates.
Since the advent of rapid and sensitive nucleic acid amplification tests over the past 15 years, most diagnostic laboratories have stopped attempting to diagnose C. trachomatis infections by isolation and these skills are rapidly becoming a “lost art.” The American Type Culture Collection (ATCC) holds a small collection of isolates that cover all of the genital tract serotypes, and it is mainly these strains, chosen for their ability to grow well in the laboratory, that have been the focus of laboratory-based studies. From limited phylogenetic studies, using either fragments of the chromosome or using whole genomes, it is clear that C. trachomatis can be divided into two distinctive groupings. These also reflect the groupings of isolates on the basis of their biological properties. Thus there are the highly invasive and rapidly growing strains that make up the “lymphogranuloma venereum” biovar that appear to be the more ancient lineage. Branching from this comes the trachoma biovar, which comprises two different sets of strains: those from the eye and those from the genital tract.
From the first few genomes analyzed, it appeared that the C. trachomatis genome was monomorphic. These first genomes possessed high synteny and extremely high sequence conservation with as little as 20 single nucleotide polymorphisms (SNPs) accounting for differences between some strains.65 A comprehensive analysis of the nature and extent of genome diversity is now required by collecting as many different isolates/genomes from around the world as possible. This will define the chlamydial pan-genome. Recent indications from partial genome analyses suggest that recombination is a mechanism for generating diversity,66 and a comprehensive genome-sequencing program will allow evaluation of this process and elucidate possible mechanisms and the forces that drive the generation of diversity. While sexually transmitted C. trachomatis appears to be an exclusively human disease, the evolutionary origins of the infection are unclear.67 There is no accurate molecular clock by which to measure the evolution of C. trachomatis. It has been speculated that chlamydial infections are an ancient disease and their evolution has been closely linked to that of their human host. However, quite closely related chlamydia have been isolated from other species (e.g., pigs and mice) but there is only very little knowledge of the presence of chlamydia in higher primates.68 A detailed investigation screening wild primates for chlamydia might be profitable in illuminating the evolutionary origins of this bacterial species.
The recent emergence of the Swedish new variant illuminates the importance of understanding how strain diversity is generated and focusing attention on the plasmid as well as the genome. In the case of the Swedish new variant “selection” was by failure to detect the strain because of mutations within the plasmid and the subsequent absence of treatment and contact tracing follow-up.69 Attempts to identify precisely when the strain appeared in the Swedish population suggest that the primary event (emergence or introduction) occurred very recently. The way the Swedish new variant was able to spread rapidly in the population is a cause for alarm. Certainly some significant lessons have been learned, the most important that the chlamydial population is not merely a static isolated backwater with limited potential for adapting rapidly to a changing world. The second is that at least two targets should be used for detection, thereby insuring that the new generation of nucleic acid amplifications tests should all conform to this criterion. Recent studies suggest that the plasmid is a virulence factor and critical for the spread of C. trachomatis in the population.
The widespread use of antibiotics has created additional concerns that antibiotic-resistant strains of C. trachomatis might be created in the future, and these could be disseminated as apidly as the Swedish new variant.
Haemophilus ducreyi and Klebsiella granulomatis
Teresa Lagerård
teresa.lagergard@microbio.gu.se
Haemophilus ducreyi is a sexually transmitted, obligate human pathogen that primarily infects skin as well as epithelial surfaces in the genital tract and causes local, persistent ulcers (chancroid). This disease is most commonly seen in developing countries and is recognized as a cofactor in the transmission of HIV-1.70
This Gram-negative bacterium was classified to family Pasteurellaceae, genus Haemophilus based on morphology of cells and nutrition requirements. Later studies based on nucleic acid methodology showed that the phylogenic structure of the family is more complex. Based on sequence studies of 16SrRNA and different housekeeping genes, H. ducreyi was shown to differ from the majority of family members and is less related to the typical Haemophilus cluster. Based on studies of 50 highly conserved housekeeping gene sequences, it was shown that H. ducreyi together with Mannheimia haemolytica and Actinobacillus pleuropenaumoniae forms a group that is divergent from the other Pasteurellaceae.71 Relationship between species in this group are, however, less obvious, because the chancroid bacterium is a human genital pathogen and the two other species are both animal commensals/pathogens in respiratory tract. Taken together, the phylogenic position of H. ducreyi in relation to other Pasteurellaceae is still not clear.
It is known that bacterial genomic fluidity and adaptation to the environment play an important role in diversity and reflects ability in colonization or in development to a successful pathogen. Different species acquired their virulence genes from another species through, for example, horizontal gene transfer, to better adapt to grow and survive. H. ducreyi has a rather small genome (1.8 Mb). The bacterium does not show pronounced genetic variation concerning antigens and virulence factors of different isolates. This may indicate a good adaptation to the genital tract in humans according to the rule that the level of genetic variability that maximizes the fitness of the population varies with the degree of its adaptation to the environment: low when the environment is stable and high when the environment is variable. The genetic bases for adaptation of H. ducreyi to its ecological niche are unknown. The bacterium, however, developed the strategies to adhere to each other and to attach to the epithelial cells, to compete with skin/genital bacteria for nutrition, to produce toxins that destroy host cells, and to survive, despite generating vigorous innate and adaptive immune responses. It can persist in lesions probably by killing cells involved in immune response as well as cells involved in epithelialization.72 Moreover, the bacterium harbored some resistance plasmids from other bacteria, helping it to survive antibiotic treatment. Since H. ducreyi is not known to colonize ecological niches other than the genital tract; sexual contact seems to be the only way to transfer pathogen to other hosts. This successful sexually transmitted infection (STI) pathogen probably evolved the ability to persist in the genital tract lesions, causing a lingering type of disease in order to maximize its transmission fitness as many other STI pathogens do.
Another strictly human pathogen and sexually transmitted Gram-negative bacterium causing genital ulcers is Calymmatobacterium granulomatis— the etiologic agent of donovannosis (granuloma inquinale). This disease is uncommon, with a curious geographical distribution of “hotspots” in Papua New Guinea, South Africa, India, and Australia. The lesions are presented on the genitals/perianal area and start as raised nodules (granulomas), and subsequently erode to form granulomatous, red ulcers that gradually increase in size and can persist for a long time. Two studies contributed to the evaluation of the taxonomic position of C. granulomatis using 16rRNA and phoE gene sequences.73,74 Both studies showed a close relationship with Klebsiella pneumoniae and K. rhinoscleromatis. Carter et al. proposed that C. granuomatis should be reclassified as K. granulomatis comb. nov.73
The high degree of similarity of K. granulomatis to K. pneumoniae on the basis of DNA is interesting because the bacteria differ profoundly in their ecological niches and also probably their pathogenic mechanisms. K. pneumoniae bacteria are found in soil, water, cereal grains, and the intestinal tract of humans and other animals. They are extracellular microorganisms causing pneumonia, wound, soft tissue, and urinary tract infections, and are easy to cultivate on artificial media, while K. granulomatis is known as an intracellular pathogen found in monocytes and is difficult to cultivate.
There are many unsolved questions concerning the evolution of H. ducreyi, such as (i) the origin, development, regulation, and expression of H. ducreyi virulence factors; (ii) what environmental stimuli may influence genome plasticity and what mechanisms do the bacterium use to respond to environmental changes? and (iii) what communication takes place among incoming pathogens, host cells, and resident microflora in genital tract epithelium/skin?
More information is definitely needed to understand the genetic and host factors related to the pathogenesis, immunology, and evolution of K. granulomatis. Such studies could help to ex-plain the possible selective factors responsible for the adaptation to the genital econiche of the Klebsiella gut organisms, and their particular sexual transmissibility in certain geographical “hot spot” populations of the world.
Understanding bacterial genome dynamics, including that of H. ducrey and K. granulomatis, has important consequences for the development of antibiotic resistance and clinical management of the disease, development of diagnostics tools, and new molecular epidemiological methods. Genome instability and variable antigenic repertoire may have a serious impact on the development of therapeutic interventions against infection disease (e.g., vaccines) and the protective efficacy of vaccines and impact in selection of new pathogenic variants.
Genetic diversity of Treponema pallidum
Sheila A. Lukehart
lukehart@u.washington.edu
Four very closely related human pathogenic Treponema species have been identified as Treponemapallidum subspecies pallidum (venereal syphilis), pertenue (yaws), endemicum (bejel) and Treponemacarateum (pinta). Limited isolates of the T. pallidum subspecies are available for study, and no known isolates of T. carateum exist. Pathogenic treponemes, including those causing genital lesions, also exist in nonhuman primates;75,76 only one isolate is known to exist.77 A decades-old “nature” versus “nurture” argument exists about whether the disease manifestations of treponemal infections have a genetic basis or are due to environmental factors (e.g., heat, aridity).78,79 Analysis of existing strains shows that genetic diversity is limited primarily to the tpr paralog family and a few other genes.80–82 Phylogenetic analysis of the tpr gene family in these strains supports division of this species into three human sub-species.81,83 Despite claims to the contrary,84 it is not possible to tell which subspecies is ancestral.83,85
It is doubtful that there is a biological basis for the sexual transmission of T. p . pallidum , compared to the nonvenereal transmission of the pertenue and endemicum subspecies. It is well recognized that T. p. pallidum can cause non-genital and non-mucosal primary infections (e.g., chancres on the shaft of the penis [nonmucosal], digital chancres in dentists prior to universal precautions, and breast ulcers in wet-nurses). Conversely, treponemes associated with severe genital lesions in baboons are more closely related to the pertenue subspecies than to pallidum.76 Careful analysis of the genes comprising the tpr family reveals genetic diversity even within the pallidum subspecies, with five distinct groups identified within the pallidum isolates (unpublished results). There are no data on the respective infectiousness among the species, subspecies, or groups within subspecies.
The two related issues that most impede our knowledge of the evolution of the pathogenic Treponema are the inability to culture this organism in vitro and, consequently, the very limited number of human and animal isolates available for study. It is only recently that investigators have appreciated the genetic diversity represented by these organisms and the wealth of information that can be gained by examining isolates other than the standard Nichols type strain.
A new enhanced molecular typing method,86 based upon genetic diversity among strains, can divide T. p .pallidum isolates into molecular strain types, with discriminatory capability far surpassing that of previous typing schemas.87 This will permit public health officials and epidemiologists to map sexual networks and to identify specific population groups for more effective syphilis control interventions. Studies of genetic diversity may also lead to the ability to predict which patients are infected with strains that are more likely to cause serious disease. The first association of genotype to clinical outcome in syphilis has been reported, in which molecular strain type 14d/f was significantly associated with neurosyphilis than were seven other strain types circulating in Seattle, Washington between 1999 and 2008.86 If future studies confirm associations between type and outcome, clinicians maybe better able to determine which patients could benefit from cerebrospinal fluid (CSF) examination for neurosyphilis or from more intensive treatment.
Evolution and diversity of Neisseria gonorrhoeae
Magnus Unemo and William M. Shafer
magnus.unemo@orebroll.se
In this paper, current knowledge of the evolution and diversity of Neisseria gonorrhoeae, and remaining research questions in the field as well as public health importance of solving these questions are discussed.
Gonorrhea is the second most prevalent bacterial STI globally and a major public health concern that, in the near future, may become untreatable in certain circumstances, that is, due to the absence of a vaccine and the rapid development and spread of resistance to all antimicrobials introduced for treatment.88 Gonorrhea has been recognized for thousands of years, for example, it is referred to in the Bible (Leviticus). However, the phylogenetic origin of the etiological agent, N. gonorrhoeae, is not completely elucidated, but taxonomically it belongs to the Phylum Protobacteria, Taxa (3-proteobacteria, Order Neisseriales, Family Neisseriaceae, and Genus Neisseria. The Neisseria genus contains many human commensal species but only two human pathogenic species—N. gonorrhoeae (obligate pathogen) and N. meningitidis. N. meningitidis causes meningitis and/or septicemia, however, the bacterium is also frequently found in the (naso)pharynx of healthy carriers. N. gonorrhoeae has instead been adapted to effectively adhere to and invade urogenital nonciliated columnar epithelial cells, but also conjunctival, anorectal, and pharyngeal epithelium. By using mechanisms such as antigenic and phase variation of outer membrane structures, blocking antibodies, molecular mimicry (e.g., lipooligosaccharides [LOS]), sialylation of LOS, inhibition and/or induction of apoptosis, and rapid development of resistance to all antimicrobials introduced for gonorrhea treatment, this “master of survival” has effectively adapted to new ecological niches and evaded host immune systems, new antimicrobial treatments, and other interventions.89 N. gonorrhoeae displays an extensive and frequent horizontal gene transfer (partial or whole genes) within the species and with closely related species, using transformation (natural competence during its entire life cycle) including usage of a specific DNA uptake sequence (DUS)90 and conjugation (plasmid DNA). This, combined with a high mutational frequency in many genes and a substantial number of phase-variable genes, causes an exceedingly high heterogeneity in the N. gonorrhoeae population. In fact, N. gonorrhoeae has a nonclonal, sexual, and panmictic population structure, which is rare for bacterial species. Relatively few N. gonorrhoeae genomes have been published, and genome studies to elucidate the origin and phylogeny of all human Neisseria species have been rare. The size of the N. gonorrhoeae genome is ∼2.1–2.2 Mbp (∼2,000 predicted genes), and it shares nearly 900 core genes, mostly with housekeeping functions but also many virulence genes, with all other humanNeisseria species.4 The apparent widespread exchange of virulence, metabolic, adaptation, and antimicrobial resistance genes (whole or partial genes) among human Neisseria species and the commensal Neisseria species seem to represent an extensive reservoir for these genes (“shareable gene pool”). The prevalence of several Neisseria species in the same ecological niches in the human body provides an excellent opportunity for a high frequency horizontal gene transfer, which can increase biological fitness, enhance host adaptation, and affect bacterial pathogenicity/virulence.91 Most human Neisseria species share the majority of the core genome as well as the “pathogenome,”91,92 and the Neisseria genus seems to comprise an open pan-genome. Accordingly, relatively few gene complements (and gene combinations) combined with smaller genetic differences (single nucleotide polymorphisms [SNPs] and insertions/deletions [indels]), gene expression, and regulation (e.g., ON/OFF phase switching of genes under certain circumstances) seem to differentiate the divergent Neisseria species. The genomes of the two pathogenic Neisseria species, N. meningitidis and N. gonorrhoeae, form a distinct monophyletic clade that is derived from the commensal Neisseria genomes. Compared to all Neisseria commensal species, the two pathogenic Neisseria species have some gene differences, such as additional silent pilS loci (increased antigenic variation of Type IV Pilus) and opa genes (enhanced antigenic variation of the outer membrane Opa proteins), which can be important for enhanced adaptation, antigenic diversity, and attachment to and invasion of host cells. Genomic (as well as biochemical, morphological, and antigenic) differences between the two pathogenic Neisseria species, which result in the widely different preferred ecological niche, infection spectrum, and epidemiology, are relatively few.91 One plausible theory regarding the origin and evolution of pathogenic Neisseria species may be that the pathogenic Neisseria species, when evolving from the human commensal Neisseria species, obtained additional genes such as porA (encoding a second porin [PorA]), iga (IgA proteases cleaving human IgA), lamp1 (lysosome-associated membrane protein 1), and additional opa and pilS loci. For the speciation of N. gonorrhoeae, the porA evolved into a pseudogene (possibly for enhanced preference for urogenital and a wider ecological niche), and outer membrane protein A (OmpA), additional opa, and pilS loci (possibly for enhanced adaptation, antigenic diversity, evasion of host immune response, and adherence to and invasion of epithelial cells in different ecological niches) were acquired.
Many questions regarding the origin, evolution, and entire population structure of N. gonorrhoeae remain. It would be valuable to have a substantially higher number of finished and closed (with high confidence) N. gonorrhoeae genomes, and appropriate comparisons of these genomes in many different aspects involving population structure, genome (pan, core, dispensable genome, and possible pathogenome), evolution (in vitro and in vivo, i.e., in different hosts, anatomical sites, and sexual networks), and also preference of ecological niche, pathogenesis, virulence, and biological fitness. Furthermore, adequate and well-designed studies combining genomics (including investigations of small gene variations such as SNPs and indels, and highlighting the importance of retained pseudogenes); transcriptomics (gene expression and regulation); proteomics; immunobiological–physiological experiments; and, ideally, also involving host factors would be exceedingly valuable. Finally, new models using modern methodologies for more precise studies, for example, regarding single cell genomics and expression, interplay between different cells and mechanisms, and genomics on non-cultured, primary samples (reflecting the entire population and possible mixed infection) would be the way forward.
The importance and future benefits for management of bacterial STIs (both case and population level) and the public health purposes of using new technologies, epi- and/or metagenomics, expression analysis, and evolution are large and numerous. For instance, the complete phyologenetic origin, evolution, and even predictions of future evolution of N. gonorrhoeae might be possible to elucidate. This would result in enhanced knowledge regarding mechanisms for emergence, evolution, and transmission of single bacterial strains in human and bacterial population (locally and globally), greater understanding of altered biological fitness, and the basis for more successful N. gonorrhoeae clones. We would also obtain an enhanced understanding regarding pathogenesis, virulence, and immunobiology (natural course of infection, asymptomatic infection, and complications). Furthermore, the data provided would improve diagnostics (new technologies, new targets [essential and multicopy], understanding of evolution of targets [inter- and intrapatient in different sexual networks], and appropriate point-of-care (POC) tests) as well as enhance the molecular epidemiological typing used to describe the N. gonorrhoeae population that circulates globally, including its dynamics (new technologies and thinking [possibly using genomic SNP-based typing], new targets, and evolution of targets [inter-and intrapatient]. Finally, we would be able to develop more effective treatment regimens (new drugs and drug targets including evolutionary stability of and selective pressure on these, tailor-made antimicrobials [+/– adjuvants/immunomodulators], antibacterial peptides, microbiocides, prediction of antimicrobial resistance emergence, effects on biological fitness, and mechanisms for spread of more successful resistant strains worldwide) and enhanced prevention of gonorrhea (contact tracing, treatment failures, sexual networks, new vaccine candidates [possibly using reverse vaccinology to identify genome-derived immunogens], and new ways to administer vaccines.
Neisseria gonorrhoeae and drug resistance
William M. Shafer and Magnus Unemo
wshafer@emory.edu
In the absence of vaccines, we rely on the efficacy of diagnostics and subsequent antibiotic treatment to stop the spread of bacterial agents that cause sexually transmitted infections (STIs). Clinically, of the main bacterial agents of STIs, only Neisseria gonorrhoeae (the gonococcus) has developed resistance to multiple antibiotics, which has necessitated major changes in drug treatment regimens over the past decades.93,94 Hence, gonococci serve as a good model system for studies on the evolution of STI bacteria and development of antibiotic resistance.
It is important to remember that gonococci have spent thousands of years infecting humans, and over the millennia the bacterium has adapted itself for survival; hence, its capacity to develop resistance to antimicrobials used clinically to treat infections should not be surprising. In relationship to the evolution of antibiotic resistance in N. gonorrhoeae, several important issues need to be considered. First, both gene (whole or parts of a gene) acquisition and spontaneous mutation events (high mutational rate in some resistance genes), which are effectively selected due to antibiotic pressure in the society, are responsible for the development and spread of antibiotic resistance in gonococci. Gonococci have a very efficient transformation system (natural competence for transformation during the entire life cycle) that employs the ubiquitous Type IV pili and a sequence-specific uptake system. The bacteria also have a conjugation system, but this is less efficient and the conjugal plasmid, which can carry the tetM determinant (causing tetracycline resistance), fails to mobilize chromosomal genes. Often, gonococci initially acquire DNA sequences involved in antibiotic resistance by transformation from commensal neisserial species, and these resistance determinants can then spread among gonococcal strains. Pharyngeal gonorrhea, where gonococci frequently coincide with commensal neisserial species, may be an effective, asymptomatic reservoir both for infection and initial emergence of antibiotic resistance, by transformation, in gonococci. For instance, commensal neisserial species can harbor a penA allele that encodes a penicillin binding protein (PBP) termed PBP2, which has reduced affinity for beta-lactam antibiotics compared to the wild-type PBP2 produced by gonococci sensitive to beta-lactams. Since PBP2 is the lethal target for beta-lactam antibiotics, remodeling of PBP2 decreases gonococcal susceptibility to beta-lactams and further genomic changes due to spontaneous mutation or acquisition of alleles of other genes (e.g., those encoding the MtrC–MtrD–MtrE efflux pump, porin, secretin, or PBP1), can further reduce beta-lactam susceptibility.93,94 The demise of penicillin as a first-line antibiotic was the result of at least five such events in a 1983 clinical isolate from Durham, NC.95 The importance of spontaneous mutations in the evolution of antibiotic resistance can be learned from how gonococci developed resistance to quinolones as this involved spontaneous mutations in gyrA and parC.93,94 Once these mutations developed, they were rapidly transferred to sensitive gonococci by the highly efficient transformation system.
A second important issue to consider is why resistant strains persist in the community despite removal of the antibiotic from therapy, that is, causing a loss of selective pressure. Do these strains have a fitness advantage (or at least lack a fitness disadvantage) over wild-type strains in the community? Results from experimental murine vaginal infection studies showed96 that strains possessing an mtrR mutation (causing enhanced production of the MtrCDE efflux pump and most probably altered regulation of other chromosomal genes94), which is needed for high level penicillin resistance, are more fit in vivo than strains with the wild type allele. Certainly, however, antibiotics used to treat other bacterial infections, inappropriate use of the antibiotic, or antigonococcal agents used topically to prevent STIs and HIV transmission or pregnancy (e.g., the spermicide nonoxynol-9) could inadvertently maintain selective pressure in the community for resistant strains.
The recent emergence of strains in the Far East, especially in Japan, with decreased susceptibility and resistance (one strain reported97) to third generation cephalosporins including ceftriaxone, the internationally recommended and main class of antibiotic used today, and the global spread over the past 25 years of strains resistant to relatively cheap antibiotics (e.g., penicillin, ciprofloxacin, and tetracycline) highlight the critical and immediate need for new antibiotics to treat gonorrhea. In fact, gonorrhea may become untreatable in certain circumstances and settings.93 Strains expressing decreased susceptibility (and now one single strain displaying high-level resistance97) to ceftriaxone should remind us of those gonococci in late 1950s and 1960s94 that expressed decreased susceptibility to penicillin and the subsequent emergence of strains expressing higher levels (i.e., chromosomal and not caused by beta-lactamase production) of resistance. Clearly, history is repeating itself and lessons learned from how gonococci developed resistance to penicillin are applicable to how they will most likely develop clinical resistance to ceftriaxone that rapidly spread globally.
For bacteria in general, and for gonococci in particular, unfortunately, no “rediscovery” of effective older antibiotics or new antibiotics with unique targets are in the immediate future. However, recent advances in research laboratories give some hope and may have applications for treating gonorrhea when ceftriaxone-resistant strains spread globally. Of these new antimicrobials, which specifically targets multidrug resistant bacteria, are efflux pump inhibitors (EPIs). These EPIs block the energy-dependent export action of efflux pumps rendering the target bacteria susceptible to antibiotics that are pump substrates. These EPIs could poison the gonococcal MtrCDE efflux pump, which based on earlier studies (reviewed in Ref. 94) would make the target gonococcus susceptible to penicillin. Moreover, extrapolation of the results from the murine vaginal infection model in which MtrCDE pump deficient mutants were unable to cause a sustained vaginal infection and were more rapidly cleared than the wild type strain (also reviewed in Ref. 94), predicts that poisoning the pump would diminish bacterial fitness in vivo. Additional antimicrobials currently in the developmental phase deserve mention. One class of antimicrobials is the host defense, cationic pep-tides, such as the human cathelicidin LL-37 that can have both direct and indirect antibacterial action. LL-37, for instance, is antigonococcal directly, but is also a substrate for the MtrCDE pump (reviewed in Ref. 94); again, poisoning the pump with EPIs could render gonococci more susceptible to the antibiotic-like action of LL-37. Finally, novel inhibitors of enzymes involved in lipid A biosynthesis, for example, LpcC inhibitors, are being studied by R. Nicholas and coworkers (personal communication, 2011).
In the absence of new antimicrobials on the immediate horizon, we need to consider use of increased doses or multiple doses of third generation cephalosporins or possibly other antibiotics, which, however, only offer limited hope for long-term effective treatment. Furthermore, combination therapy (several antibiotics administered at the same time point) needs to be strongly considered for use and further evaluated in appropriate studies. In this respect, a combination of gentamicin and the new formula of azithromycin (with enhanced gastrointestinal tolerability) have been suggested as an alternative therapy; however, possible undesirable side effects will need to be considered and monitored. Finally, it may be crucial to maximize local surveillance efforts to identify those strains that have remained susceptible to antibiotics, including those used previously, but removed from treatment regimens due to high prevalence of resistance. Thus, if in a community there are strains that remain susceptible to such antibiotics, which are no longer recommended by national healthcare agencies, then such antibiotics should be used anyway if warranted (microbiologically directed treatment). Certainly, this will be a point of contention, but curing the patient and stopping local spread of gonorrhea are the objectives, and if this means culturing and doing antimicrobial susceptibility testing, which will be expensive, to accomplish this, then this option must be seriously considered and used by local health care providers. Rapid sequencing technologies can help in identifying resistant and sensitive strains since mutations needed for resistance can be detected. Nevertheless, in regards to the last remaining options for treatment, that is, the third generation cephalosporins, knowledge remains lacking and better correlates between different genetic polymorphisms, in vitro determined antimicrobial susceptibility, and outcome of clinical therapy are essential. For enhanced knowledge regarding these issues, genome sequencing of additional isolates displaying decreased susceptibility and resistance to third generation cephalosporins combined with biological studies would be exceedingly valuable. A concerted effort to develop appropriate point-of-care sequencing technologies for this purpose is required to bring such technology to the clinic.
We need to ask: how (and when) will clinically significant levels of gonococcal resistance to third generation cephalosporins develop, will such resistance provide a fitness advantage or disadvantage to these strains, and how quickly will the resistant strains spread globally? These issues can be addressed by more detailed studies on the genetics and molecular biology of resistance, fitness studies, and studies regarding mechanisms of national and international spread (including modeling in different populations) of ceftriaxone-resistance as resistant strains emerge and spread. We can then use this information with DNA sequencing studies to rapidly identify such strains and catalog mutations in different genes or new genes that contribute to such resistance. In relationship to new antimicrobials that come into clinical practice, investments into antibiotic development and basic studies on resistance, including in vitro development and selection of resistance, must continue and be enhanced. How will resistance to current and future antibiotics impact fitness in the community? To address this issue, we must maximize our surveillance efforts to learn if resistant strains, based on different antimicrobial resistance mechanisms, have a fitness advantage (or disadvantage) in the community.
Epidemiology and evolution of hepatitis viruses
R. Palmer Beasley
r.p.beasley@uth.tmc.edu
Introduction and characteristics of the five hepatitis viruses
Substantial research has resulted in a rapidly expanding body of knowledge about the hepatitis viruses, their epidemiology, transmission modes and, more recently, their possible origins and evolutionary progression. The five hepatitis viruses, grouped because of their hepatotropism and inflammation of the hepatic parenchyma, have little else in common and almost certainly have no common evolutionary pathway. This paper summarizes current knowledge on classification, transmission, origin and evolution of the hepatitis viruses. Several schemata are used to classify viruses including clinical, epidemiological (transmission), nucleic acid type, and replication cycle. The classification, developed by David Baltimore, uses genome composition and structure, and method of replication all possibly helpful for understanding evolution. The morphology and classification of hepatitis viruses A through E are summarized in Table A1. Hepatitis A and C (and GVB) are Class IV RNA viruses, HBV is a Class VII DNA virus, HDV and HEV are as yet unclassified RNA viruses. The lettered names now used for the hepatitis viruses was adopted following the recommendation of Robert MacCallum in the 1970s as it was becoming apparent that the descriptive names then in use were inaccurate and misleading. Thus, infectious hepatitis became hepatitis A and serum hepatitis became hepatitis B. The next agent was called non-A non-B for several years before the agent was identified and named hepatitis C. The next agent, initially called the delta agent, fit well into the alphabetic sequence when its viral nature was established and was renamed HDV. Hepatitis E was initially called the New Delhi hepatitis agent, then non-A, non-B, non-C before becoming HEV. There was a brief claim to HFV, which was not substantiated, and then HGV, which exits but doesn’t seem to cause hepatitis and is now called GVB. Irrespective of its classification, the GVB virus remains of evolutionary interest because it is most closely related to HCV.
Table A1.
Hepatitis virus classification and characteristics
| Morphology |
Classification |
|||||||
|---|---|---|---|---|---|---|---|---|
| Name | Nucleic Acid |
Structure | Baltimore class |
Diameter (nm) |
Nucleotides Base pairs |
Family | Genus | Genotypes |
| A HAV | RNA | ss+ strand | IV | 25 | 7,500 | Picorna | Hepato | 3 |
| B HBV | DNA | ds circle (RT) | VII | 42 | 3,200 | Hepadna | Orthohepadna | 8 |
| C HCV Delta | RNA | ss+ strand | IV | 55–65 | 9,500 | Flavi | Hepaci | 6 |
| D HDV | RNA | ss circular | ? | 36 | 1,700 | Not assign | Delta | 3 |
| E HEV | RNA | ss+ strand | ? | 32–34 | 7,200 | Not assign | Hepe | 4 |
Incubation period and clinical spectrum
Long-incubation periods are generally characteristic of the hepatitis viruses. While the incubation periods for HBV and HCV are generally longer than for HAV or HEV, high variability of the incubation periods is characteristic of all of the hepatitis viruses, probably relating to dose, route of infection, a variety of poorly characterized host factors, and possibly viral strain differences (Table A2).
Table A2.
Clinical characteristics of hepatitis virus infection
| Clinical characteristics |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Virus | Incubation period (weeks) |
Acute hepatitis |
Chronic hep |
Chronic outcomes |
|||||
| Asymp | Moderate | Fulminant | Viral persistence |
Cirrhosis | HCC | ||||
| HAV | 2–6 | Common | Usual | Rare | No | No | No | No | |
| HBV | 4–20 | Common | Common | Occasional | Yes | Yes | Yes | Yes | |
| HCV | 2–26 | Common | Common | Rare | Yes | Yes | Yes | Yes | |
| HDV | 3–7 | Common | Common | Yes | Yes | Yes | Yes | Yesa | |
| HEV | 2–10 | Common | Common | Rare | No | No | No | No | |
HDV increases the risk associated with HBV.
The clinical spectrum and natural history of infection is also highly variable.98–100 The acute response to all five viruses ranges in severity from asymptomatic to severe acute illnesses with hepatic failure with occasional deaths. Persistent infections may occur with HBV, HCV, and HDV with infections of any severity, but more commonly when the initial infection is mild or asymptomatic, and are probably determined by immunological factors. Hepatitis infections are usually asymptomatic in infants and children. HEV in pregnancy is well known for its severity and high mortality.101 Infection with more than one hepatitis virus may result in more severe illness and higher mortality rates. HDV infection markedly aggravates the course of HBV.
Chronicity and carriers
Chronic carrier states, often lifelong, sometimes follow infections with HBV and HCV. HDV may persist with chronic HBV infections. The probability of chronicity for HBV is inversely related to age of the person when infected. There is no evidence for this age effect with HCV.
Immunology
Resolved hepatitis infections result in long-lasting protection from reinfection, so that recurrent episodes of acute hepatitis are due either to infections with different virus types or acute/subacute relapses of chronic infections (HBV and HCV). First infection immunity appears to preclude recurrent new infections with the same virus. Periodic clinical relapses in carriers can appear as recurrent new infections with HBV and HCV, or even HDV can resemble reinfections.
Highly immunogenic commercial vaccines, which induce long-term protection exist for HAV and HBV. Experimental vaccines for HDV and HEV have also shown protection, while a satisfactory vaccine against HCV is still to be developed.
Human hepatitis viruses in animals
HAV, HCV, and HDV appear to be confined to humans in nature (Table A3). A number of wild mammals and birds are infected with hepadna viruses, similar to HBV, which provides interesting clues regarding the origins and evolution of this group, as discussed below.102,103 Pigs and chickens are frequently infected with HEV- or HEV-like agents. Strains from pigs, but not chickens, appear to be infectious to man, and there is increasing data suggesting that swine are the origin, if not the current reservoir, for human infections.7
Table A3.
Occurrence of human hepatitis viruses in animals
| Human virus infections in animals |
Similar animal viruses in nature |
||||||
|---|---|---|---|---|---|---|---|
| Name | Lab | Nature | Primates | Other mammals |
Birds | Transmission to humans |
Viral persistence |
| A | Yes | No | No | No | No | N/A | No |
| B | Yes | No | Yes | Yes | Yes | No | Yes |
| C | Chimps | No | No | No | No | No | Yes |
| D | Yes | No | No | No | No | N/A | Yes |
| E | Yes | No | Unknown | Yes | Yes | Yes | Unknown |
Geographic distribution and transmission
All hepatitis viruses have been found in all parts of the world but with strikingly varying distributions. Epidemics of jaundice associated with poverty and war, presumably due to HAV, have been reported since ancient times. Both HAV and HEV are correlated with fecal contamination of food and water. Markers of HBV infection were identified in every population in the world as soon as tests became available in the early 1970s. High HBV carrier rates are found in East Asia, the Pacific, and Africa, while the lowest rates are found in the UK and Scandinavia. Most or all countries probably had low to very low prevalence of HCV infections before the widespread medical use of blood and needles by medicine and illicit drug users. Currently, Egypt has the highest HCV prevalence in the world following a national program from the 1950s to the 1980s for using intravenous tartar emetic for the treatment and prevention of schistosomiasis.105 HDV is characterized by striking geographic variation, which oddly doesn’t follow the distribution of HBV. Concentrations of HDV infections have been found among Indians of the Amazon and Orinoco basins, central Africa and among pockets of drug users in areas with generally infrequent HDV prevalence; but infection rates are rare in Asia where HBV infections are common.106
Transmission patterns of the hepatitis virus are variable, complex, and not fully understood (Table A4). HAV and HEV are primarily transmitted following fecal contamination of food or water. Transmission of HBV, HBV/HDV, and HCV is usually through infected blood from chronic carriers by transfusion of unscreened blood or blood products, perinatal transmission from mother to infant during labor and delivery from microhematologic leaks across the placenta;107 or reuse of contaminated injection equipment. There is no evidence of arthropod borne transmission of any of the hepatitis viruses. HBV, and probably HDV, are also commonly transmitted by contact with saliva of infected persons. HAV, HBV, and HDV infections are also correlated with sexual activity with infected persons all of which may related to saliva or HBV and HDV or oral–genital contact for HAV. Because kissing simultaneously occurs, it is essentially impossible to establish whether transmission also occurs from genital–genital contact. Carefully designed epidemiological studies suggest that virtually all HCV infections are transmitted by inoculation with blood from HCV carriers, occasionally including perinatal transmission. Mother-to-infant transmission occurs in about 5% of HCV carrier pregnancies.108 Convincing evidence of sexual transmission of HCV, either from saliva or genital contact is lacking. While traumatic intercourse with bleeding cannot be ruled out, the widespread belief that some HCV infections are due to sexual contact with carriers is more likely wishful thinking among those trying to obscure a history of illicit drug use.109 HDV infections can only be transmitted as co-infections or super-infections with HBV because HDV replication depends on the presence of the hepatitis B virus.110 Thus, HDV can be transmitted with HBV by any of the myriad of routes used by HBV, but the modes of HDV super-infection are poorly understood. In addition, to contaminated water, HEV transmission between man and swine appears to occur frequently, but much is still to be learned about the relative importance of cross species transmission.101,104 HEV appears to be the only hepatitis virus for which there is good evidence for of frequent transmission between man and animals.
Table A4.
Transmission modes of hepatitis viruses
| Transmission |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Name | Chronicity viral persistence |
Blood |
Vector (Insect) |
Airborne (Distance) |
Vehicle (Fec–oral) |
Person to person |
||||
| Vertical | Parenteral | Resp | Saliva | Sexual | Genital/genital | |||||
| A | No | No | Rare | No | No | ++++ | No | No | + | Unknown |
| B | Yes | +++ | +++ | No | No | No | No | + | + | Unknown |
| C | Yes | + | ++++ | No | No | No | No | No | No | Unknown |
| D | Yes | + | +++ | No | No | No | No | + | + | Unknown |
| E | No | No | No | No | ++++ | No | No | ? | Unknown | |
Note: HBV and HDV can be found in saliva of carriers of those viruses, and epidemiological data suggest that some infections occur through contact with saliva such as kissing. Transmission of HAV probably also occurs from oral– genital contact, particularly among MSM. None of the hepatitis viruses have been established to be transmitted by genital-to-genital contact.
Origins and evolution genetics
The hepatitis viruses have remarkably different mechanisms of replication and transmission, but it is noteworthy, in view of the title of the conference, The Evolution of Infectious Agents in Relation to Sex–focus on sexually transmitted and transmissible diseases in animals and humans, that none of the hepatitis viruses are known to be transmitted by genital-to-genital contact. On the other hand, sexual activity may be responsible for transmission of HBV and HDV through kissing, and HAV and HEV through enhanced exposure to fecal material during sex. Very little is known about the origins of the hepatitis viruses, but fragments of information have stimulated interesting speculation regarding several of these agents.
Tools to evaluate the possible origins of viruses are limited. Molecular divergence studies of populations over time suggest relatively recent divergence from common ancestors that seems too recent to plausibly fit with the distribution of the agents in their host agents. Thus, so called molecular clock studies have shown annual nucleotide substitutions rates for each of the hepatitis viruses as: HAV, 1.99 × 10−4 (Ref. 111); HBV, 2.1 × 10−5 (Ref. 112); HCV, 4 × 10−4 (Ref. 113); HDV, unstudied; and GBV-C, 3.9 × 10−4 (Ref. 114).
Hepatitis A is a common disease and one of the first distinct etiologic disease entities recognized. Published reports of epidemic jaundice exist in the literature as far back as Hippocrates in the 5th century B.C.115 and have long been associated with war, social disruption, and poor sanitary conditions. It has gone by many names such as campaign jaundice, infectious jaundice, epidemic jaundice, acute catarrhal jaundice, and infectious hepatitis (IH) before becoming A as it is now called. It was not cultured in tissue culture until after aggregates of the virus were demonstrated by electron microscopy when convalescent serum was mixed with acute phase stools R. It is a Group IV virus, is a member of the Picornaviridae family of RNA viruses, and has only been found in humans. The Picornaviridae is a large family, which includes the enteroviruses, found in humans and animals. HAV is classified by itself in the genus Hepatoviradae. A single serotype and 3 genotypes of HAV are recognized based on variability of the VP1 region.116,117
Hepatitis B virus is a member of the Hepadna Virus family, which constitutes the unique Group VII viruses created to accommodate these small circular DNA viruses now known to occur in a number of avian and mammalian species.103 Infected birds include domestic ducks, a number of species of wild ducks, including the mallard from which the domestic duck is thought to arise, and some species of geese, cranes, herons, and storks. Hepadna viruses have also been found in one wild species of marmot (the American woodchuck), several species of ground squirrel, and five species of higher apes, in addition to man. Particularly interesting is hepadana viruses found in the Woolley monkey of South America, which may imply a very early origin of this group. These agents are distinct from HBV, but resemble them quite closely, and in several species have been found to be transmitted vertically and to cause liver cancer.
Hepatitis C virus is a Class IV virus belonging to the large Flaviridae family of RNA viruses. It is most closely related to the GBV viruses, which replicate in the liver without causing disease. GBV, also a Flavi virus, has four genotypes and is noteworthy here because it possibly provides insights regarding the origins of HCV, because animal hosts have been found for GBV, even though none have been found for HCV. GBV-genotype A and B have been found in tamarins, a primate species from the New World, but not humans. GBV-C has been found in humans and chimps, while GBV-D has been found in wild-caught fruit bats (Pteropus giganteus) from Bangladesh, but not in humans. These findings suggest an ancient origin of the GBV group of viruses and possibly a common root that will tie them to HCV.
Hepatitis D virus, the delta agent, is a tiny (1,700 bp) RNA virus that requires HBV to replicate. It is classified in a unique genus, but is yet to be assigned to a virus family. Similarities between HDV and viroids have stimulated speculation that the HDV might be of plant origin. Similarities include size, common RNA features, their modes of replication, and the protein interactions stimulated by their RNA genomes. Taylor has speculated that HDV is of plant origin.118
Hepatitis E virus is an RNA virus of the Hepe genus, but is yet to be assigned to a Baltimore Class or a family. Four genotypes have been identified. It is not clear whether pigs are the source of HEV infections in man or vice versa. There is archeological evidence that pigs were domesticated as early as 13,000–12,700 BC in the Tigris Basin,119 so this could be another example of an animal virus acquired through domestic animals. On the other hand, pigs have long been used as “human toilets” to clean up feces, so perhaps HEV had a different origin and later went from man to pigs.
Genital molluscum contagiosum
Tomas Bergström and Peter Norberg
tomas.bergstrom@gu.se
Poxviruses form an extensive family of large brick-shaped DNA viruses, with a genome size of approximately 200 kb, whose members infect a variety of animals from insects to vertebrates. From studies of 26 sequenced members of the divergent poxvirus family, a central core of conserved genes has been observed flanked by more diverse genes that may be fragmented in some viruses. Phylogenetic analysis also revealed that interspecies recombination has contributed to the evolution of these viruses.121 Cross-species transfer of poxviruses seems to occur relatively often compared to other virus families, and hijacking of host genes may contribute to the viral evolution.121,122 At present, since the eradication of smallpox and its causative agent variola (VAR) virus declared by the World Health Organization in 1980, molluscum contagiosum (MC) virus is the only known poxvirus with humans as reservoir host. This virus commonly causes benign umbilicated skin lesions in children. In adolescents and young adults, these lesions are often localized to the genital region. Some other poxviruses, such as monkeypox virus and cowpox virus, may occasionally infect humans after zoonotic transmission, but have hitherto not succeeded to establish themselves as a human infection. MC, on the other hand, appears to be an exclusive human pathogen, and infection of other species has not been reported. Although transmission mainly occurs after skin contact, MC can be spread through intercourse and may manifest itself as a sexually transmitted infection (STI). This development has accelerated in conjunction with the HIV pandemic, and genital MC lesion may be one of the most common localizations on the body.
Four distinct genotypes of MC, obtained after cleavage with restriction enzymes, have been reported, of which, one subtype is much more prevalent than the others and has been fully sequenced.123 The 190 kb genome potentially encodes for 163 proteins, of which, 59 genes lack homology to previously known proteins. Although functional data on MC viral proteins are lacking, several of these gene products were predicted to be homologs of human proteins involved in the inflammatory response such as chemokines, MHCI, and gluthatione peroxidase.122 On the other hand, of the 83 genes that were present in the VAR virus but absent in MC virus, most of these were also involved in evasion of the immune response of the host. Thus, although little is known about the evolution of the MC virus in general, and in the genital niche in particular, it may be speculated that the interaction/escape from various immune mechanisms122 is an important driving force for this process.
The typical skin lesion caused by the MC virus is restricted to the epidermis and has a characteristic nodular appearance with a size of less than 5 mm in most cases. MC is therefore easily diagnosed without virological/histological confirmation. The MC virus is most likely transmitted through skin-to-skin contact or indirectly by fomites. In children, the MC lesions are often localized to the trunk and the limbs, and multiple MC lesions can be clustered together in skin regions where the virus has been spread by autoinoculation. Individual lesions may disappear within two months, but may also last for years in patients with immunosuppressive conditions.
During the last decades, MC has been increasingly recognized as a venereal disease.124 It may be debated whether MC is a “true” STI or if the genital lesions are just an ordinary form of contact transmission by rubbing an infected skin surface against an un-infected one. Regardless of this, patients with MC lesions in the genital tract, especially in the age of 15–29 years, are increasingly common in STI clinics. Moreover, a large proportion of HIV-infected patients present with MC.124 Another intriguing presentation of the MC virus is as a possible congenital or perinatal infection in which the skin lesions may be presented on the scalp early after birth, compatible with a cervical location of the virus in the mother.120 This clinical presentation of MC virus infection clearly associates it to similar properties of other STIs. However, no studies on cervical persistence of the MC virus have been reported.
To increase understanding of genital MC virus infection and evolution, several molecular virology studies should be undertaken addressing the following questions: (1) does a phase of MC virus persistence exist in the genital tract; (2) how genetically diverged are the four types of the MC virus family based on nucleotide differences, what are their phylogenetic relationships and, how closely are the genitally derived viruses related to pediatric strains; (3) does homologous recombination continuously occur between MC strains as described for other STIs such as HIV and HSV; and (4) how does the evolution of MC compare with that of other viral STI’s such as HIV and HSV-2? On a clinical basis, an extended disease spectrum of the hitherto un-cultivable MC virus is now possible to search for based on PCR diagnostics. The outcome of such studies will aid in future discussions on possibilities of prevention and therapy.
Human herpesviruses
Andrew J. Davison
andrew.davison@glasgow.ac.uk
Herpesviruses are large, spherical viruses of animals that share a distinct virion structure: an icosahedral capsid embedded in a proteinaceous matrix (tegument) surrounded by a glycoprotein-studded lipid envelope. Their large, linear, double-stranded DNA genomes are 125–295 kb in size and contain 70–170 genes. Herpesviruses are numerous—over 300 examples have been identified—and intensively studied animals host several herpesvirus species. Research into the genetic content and evolution of herpesviruses has proceded for over three decades, and the field is in a mature state.43,125 Taxonomically, herpesviruses belong to the order Herpesvirales, which consists of the families Herpesviridae (infecting reptiles, birds, and mammals), Alloherpesviridae (fish and frogs), and Malacoherpesviridae (bivalves and gastropods). Since a pervasive degree of coevolution with the host appears to have occurred, it is considered that herpesviruses predate vertebrates. Indeed, there is evidence that they may have very ancient evolutionary links with bacteriophages in the order Caudovirales.
The eight recognized human herpesviruses represent all three branches of the family Herpesviridae: herpes simplex virus types 1 and 2 (HSV1, HSV2) and varicellazoster virus (VZV) in the subfamily Alphaherpesvirinae, human cytomegalovirus (HCMV) and human herpesviruses6 and 7(HHV6, HHV7) in the subfamily Betaherpesvirinae, and Epstein–Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV) in the subfamily Gammaherpesvirinae. The marked genetic relationships among these viruses constitute strong evidence for their evolution from a common ancestor, which has been proposed as having existed 400 million years ago. This ancestor has passed to its modern descendents about 44 genes, which are involved in basic aspects of the lytic life cycle (DNA replication, DNA packaging, virion structure). The rest of the genes have accumulated at later stages, and provide econiche functions. Some of these functions are involved in aspects of the latent life cycle, which differ among the subfamilies, and have evidently been replaced at various stages to resource novel biological capabilities. Thus, HSV1, HSV2, and VZV are latent in sensory neurons, HCMV in monocytes, HHV6 and HHV7 in T lymphocytes, and EBV and KSHV in B lymphocytes; the latency site utilized by the common ancestor cannot be determined. Among the evolutionary mechanisms that have molded herpesvirus genomes are gradual mutation (accelerated in hypervariable genes), capture (lateral transmission, usually from the host), duplication (a form of recycling that gives rise to gene families), de novo generation of genes, recombination, and gene loss.
Four of the eight human herpesviruses are transmitted sexually. HSV2 is most commonly transmitted thus (as genital herpes) and is classed as a sexually transmitted (STI) agent. EBV, KSHV, HCMV, and HSV1 are most commonly transmitted by other means and are classed as sexually transmissible (StxI) agents. Each of these viruses has a number of notable evolutionary features. HCMV has the largest genome among known human viruses, encodes many captured genes and gene families, and exhibits a remarkable degree of strain variation. Its hypervariable genes probably arose as a result of immune exposure and population isolation early during human evolution, and are now distributed worldwide. EBV and KSHV encode many captured genes and a few gene families. Versions of the single hypervariable gene (K1) in KSHV track geographically with early human migrations out of Africa. KSHV also encodes two highly diverged versions of another gene (K15), which are the legacy of an ancient recombination event involving a related virus whose other descendants have not been found. HSV1 and HSV2 encode few captured genes and only a single gene family. They are each other’s closest relative, sharing equivalent gene sets (74 genes, three duplicated in inverted repeats) whose coding sequences are approximately 83% identical. The greatest difference between the two viruses exists in gene US4, which encodes a virion glycoprotein and is much shorter in HSV1. HSV1 strain distribution is to some extent geographical, with greater variation among African strains. HSV1 and HSV2 have been estimated to have diverged about 8 million years ago, and this has prompted speculation that divergence in their respective econiches (orofacial for HSV1 and genital for HSV2) may have been promoted by steps in the evolution of human sexual behavior.126 In addition, growing recognition of the involvement of HSV1 in genital herpes is exciting interest in both the evolutionary and clinical fields.
There are several research areas that interconnect with the perspective on herpesvirus evolution. It has become clear that herpesviruses manipulate host immune responses by many mechanisms— often using captured cellular genes—and it is likely that the full panoply of such functions is yet to be revealed. Recombination is becoming more prominent as a major mechanism by which herpesviruses have generated (and may in future generate) diversity; including within, but not between, HSV1 and HSV2.127 The involvement of genetic variation in the history, geography, epidemiology, and pathogenesis of herpesvirus infections is an area of increasing activity. Finally, the mechanisms involved in latency128 must remain a crucial focus, since this part of the herpesvirus life cycle is essential to survival and is yet so flexible evolutionarily. The areas of research described above entail obvious practical considerations in terms of prognosis of the likely outcome of, and intervention into, herpesvirus infections.
Evolution and genital infection of herpes simplex viruses Type1and 2
Peter Norberg and Tomas Bergstro¨m
peter.norberg@gu.se
Herpesviruses constitute a large family of over 100 slowly evolving DNA viruses that are highly disseminated among animals, and have coevolved with their hosts for hundreds of millions of years. Eight herpesviruses have been identified in humans, of which herpes simplex virus HSV-1 and HSV-2 belongs to the alphaherpesvirus subfamily. These are closely related viruses with approximately 80% nucleotide identity, and were suggested to have diverged from a common ancestor 8.5 to 8.4 million years before present (BP).130 HSV-1 and HSV-2 thus existed prior to the emergence of Homo sapiens, and have probably coevolved with their human hosts, and earlier Homo-species, for over eight million years. It has been debated whether the common HSV ancestor had its natural econiche in the oral or genital tract.
HSV-2 is one of the most common human genital pathogens globally. Although the primary infections on rare occasions may affect the oral region and reactivations may occur over most parts of the body, HSV-2 has a clear tropism for the genital tract to which it readily reactivates. HSV-1 is typically associated with oral lesions but is also considered a major and increasing cause of primary genital herpes in several western world countries, including Sweden.129 Genital HSV-1 infection is most common in young women and most often contracted by the oral–genital route, but genital–genital transmission cannot be excluded. The genital econiche may well harbor HSV-1, because the primary genital infection is as clinically severe as compared with the corresponding infection of HSV-2. In addition, both viruses replicates equally well during such conditions. However, reactivation to the genital tract of HSV-1 is much less common than that of HSV-2, suggesting a lesser degree of adaptation to the genital econiche by this virus. This question involves the important mechanisms of latency and reactivation, which are intensely studied for HSV-1 (for a review, see Ref. 133) but much less known for HSV-2.
An important challenge for further HSV research will be to follow the genetics and evolution of HSV-1 and 2 with focus on the genital econiche and the possible adaptation of HSV-1 to genital–genital transmission. No complete genome evolutionary studies have, however, been presented for either HSV-1 or 2, but phylogenetic analyses on subgenomic regions have been presented for clinical strains of both viruses. Analysis based on the genes US4, US7, and US8 of clinical HSV-1 strains isolated in the western world has demonstrated a divergence into three distinct phylogenetic clades, A, B, and C.131 The strain DNA diversity in this genomic region was less than 2%, and it was demonstrated that homologous recombination is common among HSV-1 strains in natural populations.131 HSV-2 is more conserved than HSV-1 with a DNA diversity of less than 0.4% between strains in the genes US4, US7, and US8, and analysis of clinical strains collected in Sweden, Norway, and Tanzania demonstrated a divergence into two straggling phylogenetic clades A and B. Homologous recombination was even more frequently detected in HSV-2 than in HSV-1.132 Consequently, clade diversity and homologous recombination has been demonstrated for both HSV-1 and HSV-2. Although the absolute frequency of naturally occurring recombinant HSV strains is unknown, it has been suggested that all globally circulating HSV-1 and HSV-2 strains may be recombinants if their complete genomes were analyzed.129,131–132
Complete genome sequencing and phylogeny of genital and nongenital HSV-1 and HSV-2 strains would not only increase our understanding of HSV evolution, the extent of divergence and recombination, and geographical spread of different genotypes, but could also be used in comparative studies of the evolution of these two closely related viruses. Despite their similar biological properties, some questions about their evolutionary differences remain unanswered. For example, why is the recombination frequency (at least in the US4, US7, and US8 genes) significantly higher for HSV-2 than for HSV-1? And why is HSV-2 more conserved than HSV-1 considering single nucleotide substitution? Is it possible that differences in natural sites of infection, for example, genital versus oral, and routes of transmission have influences on the evolutionary clock and recombination rate? Can HSV-1/HSV-2 recombinant emerge when both viruses compete for the genital econiche? Furthermore, to what extent are the different clinical traits correlated with the genetic setups of the viruses? Future studies including analysis of complete genome DNA from a large number of clinical strains in patients with genital herpes followed prospectively may shed light on these questions. These studies should preferably focus on geographic distribution of genotypes, the long-term viral evolutionary process, evolutionary mechanisms, and genotype–phenotype associations. Furthermore, although infections with HSV-1 and 2 are typically asymptomatic or associated with mucocutaneous lesions, both viruses can also cause severe neonatal infections(neonatal transmission may be genital but not sexual), encephalitis, meningitis, keratitis, and facial palsy. The underlying reasons for these devastating infections are poorly understood, but are likely to be linked, at least partially, to viral genetic factors.
Another question of interest would be to analyze the extent of within-host heterogeneity of different strains. Deep sequencing of genital and non-genital HSV-1 and HSV-2 strains might reveal differences in minority sequences. Information about, for example, the existence of acyclovir-resistant minority strains would increase our understanding of the emergence and spread of such strains. The existence of such resistant HSV-1 and 2 mutants is well described, but little is known about how these variants emerge and spread through the viral population. From the studies of other microorganisms, it is well known that resistant strains might spread and evolve rapidly. Furthermore, although no vaccine is currently available for HSV-1 or 2, a live vaccine strain of another alphaherpesvirus, the varicellazoster virus (VZV), was recently introduced on a global scale to prevent the spread of VZV and reactivating zoster. Several cases of rashes caused by this vaccine have subsequently been reported, and the monitoring of the emergence of possible vaccine-wildtype recombinants may provide an interesting avenue for further study that may be of importance for future development of a HSV vaccine. Taken together, a thorough knowledge of the evolutionary and genetic characteristics of HSV-1 and 2 is important to better understand the impact of antiviral-resistant mutants and how such strains should be treated and their transmission and spread prevented. Furthermore, such knowledge might help in the design of an HSV vaccine and in the prediction of possible long-term consequences of its introduction into the population. To increase understanding of the evolutionary process and its impact for public health care issues, such studies of HSV genetics should be performed on viral samples from clinically well characterized and prospectively followed patients.
AIDS: the origins and evolution of HIV-1
Paul M. Sharp and Beatrice H. Hahn
paul.sharp@ed.ac.uk
Acquired immune deficiency syndrome (AIDS) is primarily a sexually transmitted disease, although it can also be spread by percutaneous and perinatal routes. AIDS in humans is caused by two related but distinct retroviruses, human immunodeficiency virus types 1 and 2 (HIV-1 and HIV-2), each of which can be divided into a number of phylogenetically distinct groups. However, one particular form of HIV-1, termed group M, is responsible for the vast majority (more than 95%) of AIDS cases globally. AIDS was first described in 1981, but it has become clear that HIV-1 group M must have been spreading in the human population for at least 50 years prior to that. It is estimated that about 25 million people have died because of the AIDS pandemic and that more than 30 million people are currently infected with HIV-1. While the apparently sudden emergence of AIDS prompted numerous conspiracy theories about the creation of HIV-1, over the past 25 years a more and more detailed understanding of the evolutionary origins of HIV-1 has emerged.133
The closest relatives of HIV-1 and HIV-2 are lentiviruses infecting primates from Africa. These have been termed simian immunodeficiency viruses (SIVs), although in many cases they appear to be nonpathogenic. Host-specific forms of SIV have so far been found naturally infecting about 40 of the 70 or so different species of primates in sub-Saharan Africa. Phylogenetic analyses have revealed that the two types of human virus arose from cross-species transmissions of SIVs from different primates. HIV-2 was first, and is still most commonly, found infecting individuals from west Africa, and was derived from SIVsmm infecting sooty mangabeys (Cercocebus atys), a monkey species indigenous to that region. The precursor of HIV-1 was SIVcpz, a virus infecting our closest relative, the chimpanzee (Pan troglodytes). On the basis of mitochondrial DNA sequences, chimpanzees have been divided into four subspecies: SIVcpz was first found in blood samples from a small number of captive apes belonging to two of these subspecies, P. t. schweinfurthii in east central Africa and P. t. troglodytes in west central Africa. The development of noninvasive techniques, identifying SIV antibodies, as well as host and viral nucleotide sequences in fecal samples, has allowed the sampling of much larger numbers of wild individuals from numerous locations across west and central Africa.134 This has confirmed that both subspecies are infected by SIV in the wild, with prevalences around 30% in some communities. All of these SIVcpz strains fall into one of two clades, depending on the subspecies from which they were obtained. In contrast, SIV has not been found in P. t. verus from west Africa or P. t. ellioti from northern Cameroon, despite extensive sampling.
Testing of fecal samples has also led to the discovery that western gorillas (Gorilla gorilla) are infected with an SIVcpz-like virus, although only a few strains of this virus have been found. Both HIV-1 and these gorilla viruses (SIVgor) are more closely related to SIVcpz from P. t. troglodytes than to SIVcpz from P. t. schweinfurthii. The SIVgor strains form a single lineage, probably reflecting a single chimpanzee-to-gorilla transmission. In contrast, HIV-1 strains fall into four clades, termed groups M, N, O, and P, which are interspersed among the SIV lineages, and so must reflect four independent ape-to-human transmissions. Groups M and N originated in chimpanzees, while group P was probably acquired from gorillas; it is not yet clear which ape was the immediate source of group O. The genetic diversity among SIVcpz strains is geographically structured. The SIVcpz strains most closely related to HIV-1 group M have been found in the south east corner of Cameroon,134 suggesting that the transmission from a chimpanzee to a human that ultimately gave rise to the AIDS pandemic took place in that region. We cannot know how this transmission occurred, but it was most likely through contact with contaminated blood during the hunting and butchering of a chimpanzee.
Since then,HIV-1 group M has diversified rapidly. The rate of evolution of the HIV-1 genome is sufficiently fast, on the order of 10−3 nucleotide substitutions per site per year, that it can be estimated from comparisons of viruses isolated in different years. Use of this molecular clock leads to an estimate that the common ancestor of HIV-1 group M existed around 1910–1930, and that the jump from chimpanzees to humans occurred at some time prior to that. Surveys of the geographical distribution of diversity among HIV-1 group M strains suggest that Kinshasa, in the Democratic Republic of Congo, was likely the location where the early diversification of these viruses occurred. Both this timescale and the location of the early evolution of HIV-1 group M are supported by sequences of two viruses obtained from tissue samples first collected in 1959 and 1960, respectively. These two “ancient” strains were highly divergent from one another, indicating that HIV-1 group M had, even at that time, already been spreading in humans for many years, and both samples were obtained from subjects in Kinshasa (then called Leopoldville). Although Leopoldville was located some 800 km from the region where the chimpanzees with group M-like strains of SIVcpz live, it is notable that this area in southeast Cameroon is surrounded by rivers, which flow south, eventually joining the Congo River and flowing to Kinshasa. At the time when HIV-1 group M emerged, these rivers were the main routes of travel. Humans have likely been hunting chimpanzees for thousands of years, and so it is probable that many more SIVcpz transmissions occurred in the past. However, it was not until the 20th century that substantial cities grew up in central Africa, providing an environment in which the virus could spread extensively enough to start an epidemic. And although it might, at first, seem surprising that HIV-1 group M could have been present in humans for more than 50 years before AIDS was recognized, with hindsight it is not, because for much of this time rather few individuals would have been infected.135
The observation that HIV-1 originated from our closest relatives raises a number of interesting questions about the virus in chimpanzees. First, since most forms of SIV infect monkeys, what was the origin of SIVcpz? Extensive phylogenetic analyses have revealed that SIVcpz has a mosaic genome, derived by recombination between members of SIV lineages infecting two different monkey species.133 The implication is that chimpanzees acquired two viruses from monkeys on which they prey, that these two viruses recombined, and that this hybrid virus then spread through two of the four subspecies of chimpanzee. This also suggests that SIV infection of apes is rather younger than SIV in monkeys.
Second, given the close relationship of chimpanzees and humans, and the suggestion that SIVcpz may have originated comparatively recently, is it true (as generally assumed) that SIV is non-pathogenic in chimpanzees? It has been possible to address this question through 10 years of observation of chimpanzees of known SIV-infection status in the habituated communities at Gombe National Park, Tanzania.136 These studies revealed significantly (10–16 fold) higher age-corrected risk of death, as well as reduced fertility and lower CD4+ T cell counts, among SIV-positive chimpanzees. These data strongly suggest that SIVcpz infection can cause AIDS in chimpanzees.
Finally, did prior adaptation of SIV to infecting chimpanzees predispose the virus to successfully infect humans? This is difficult to answer, but it has become clear that SIVcpz still had considerable adaptive hurdles to overcome. Most interestingly, differences between the chimpanzee and human forms of tetherin, an antiviral protein that “tethers” budding virions to the cell membrane, suggest that SIVcpz would not be able to replicate and spread efficiently in humans. It is intriguing to note that, among the different groups of HIV-1, only group M has fully adapted to counteract human tetherin.137 Thus, it is tempting to speculate that this is why group M has spread so much more successfully than the other forms of HIV-1 and that this adaptation gave rise to the AIDS pandemic.
Retroviruses in reproductive tissue: waiting for sexual transmission or the germ line?
Jonas Blomberg
jonas.blomberg@medsci.uu.se
Retroviruses frequently infect reproductive tissue. From this site, they have access to two transmission modes, horizontal and vertical. Sexual transmission is horizontal. Vertical transmission (to offspring) of retrovirus from reproductive tissue can be via breast milk, transplacentally, or via gametes. The latter may give rise to an endogenous retrovirus (ERV). Although each ERV has its own expression pattern, ERV expression is generally higher in testes, placenta, and ovaries compared to other tissues. This indicates that the retroviruses that endogenized have a predilection for replication in reproductive tissue. I here discuss the possibility that some retroviruses may stay in reproductive tissue without giving a symptomatic systemic infection (a “stealth” infection). In that case, sexual transmission and endogenization may take place silently and perhaps more frequently than previously known.
Ubiquity of ERVs
ERVs exist in all vertebrate genomes. Generally speaking, a vertebrate genome may contain 100–10,000 more or less complete integrated retroviral genomes (proviruses). Thus, they are ubiquitous genetic components and fundamental parts of the formation of a complex multicellular organism like a human.
The endogenization process
As shown in Figure A1, there are many imperfectly known steps in the endogenization process. The purpose of bringing this up is to emphasize their fundamental biological importance, as acquired characters, and their intimate relation to the germ line. One issue is the degree of systemic infection that preceeds endogenization. It may be more or less symptomatic. For example, murine leukemia virus, feline leukemia virus, and koala retrovirus can give severe a systemic infection with immune disturbance and opportunistic infections. However, retroviral infections may also be very asymptomatic. The most extreme cases are human T lymphotropic virus infection, bovine leukosis, and spumaretroviral infections. Viruses which are asymptomatic may thus occasionally reach the germ line without causing disease. In fact, such an apathogenic retrovirus would have a higher chance of endogenization than a pathogenic one, because the fitness of the offspring will not be affected. Thus, apathogenic retroviruses that reside in reproductive tissue have a particularly high chance to transmit in two different fashions, either sexually (horizontally), or genetically (vertically; Fig. A2).
Figure A1.
Germ line transmission of ERVs is a form of Lamarckian inheritance of acquired characters, creating a reverse flow of genetic information. Before reaching germ cells, the virus must give a systemic infection of varying intensity, then infect reproductive tissue, and finally reach germ line cells, presumably spermatogonia or oogonia. There are several bottlenecks in this process; (1) A systemic infection must be established, then (2) reproductive tissue must be infected, and (3) finally the germ cell precursor must be infected, (4) the offspring must be fit enough to pass on the new acquired character, and (5) in a few cases the new ERV is “fixed” in the germ line, i.e., is present in all individuals in a species. The many restrictions and the ubiquity of fixed ERVs indicate that stages 1–4 are common, but surprisingly little is known about this process.
Figure A2.
Retroviral replication in some reproductive tissues may lead to transmission through two routes: sexually (A) and via the germ line (B). Such transmission may or may not be associated with significant systemic infection. The possibility that some retroviruses are apathogenic and home to reproductive tissue with little engagement of the rest of the body (sneaking in through the “back door”), (C) should be considered. In this paper it is discussed whether some ERVs are a distinct, apathogenic, subset of retroviruses specialized for germ line transmission.
Why, then, are not all sexually transmitted retroviruses apathogenic? HIV-1 and HIV-2 are pathogenic sexually transmitted retroviruses. HTLV-1 and (especially) HTLV-2 are much less pathogenic. None of them are endogenous. A possible explanation is that the manipulations of these complex retroviruses with cellular physiology through their transactivating proteins impair the fitness of the offspring. One reason why we do not know of as many asymptomatic retroviruses is because we do not see them. Modern detection methods like PCR, and high throughput sequencing, might reveal them.
Do ERVs have a special predilection for reproductive tissue?
Several observations indicate that reproductive tissue is unique in its high expression of ERVs. We studied ERV RNA expression in a panel of human tissues (Fig. A3). An especially high expression was found in tissues like testes and placenta;138 this is in line with the reasoning presented here.
Figure A3.
ERVs are often highly expressed in reproductive tissue. Results from real-time PCRs specific for HERV-E, HERV-I and –T, HERV-H and HERV-W are shown. Reproductive tissue (testes, ovaries, prostate, and placenta) have a tendency toward high ERV expression. Brain expression is also relatively high. Many cell lines have a low ERV expression (“other cell lines”), but after hypoxia and demethylation some ERVs may be highly expressed in certain neuroblastoma cell lines. RNA expression is given as a ratio between HERV and Histone 3.3 target equivalents per PCR reaction. Histone 3.3 RNA is a reference that is relatively evenly expressed in many tissues. From Ref. 138.
Conclusions
The actual transmission route leading to eventual endogenization is nearly always obscure. However, expression in reproductive tissue is central for endogenous retroviruses. Especially, expression close to sperm production, for example, in testes, is typical. This indicates that these viruses often have been transmitted sexually.
The transmission of retroviruses that can be both endogenous and exogenous, like the murine and feline leukemia viruses, can, however, be studied. In these cases, secretion in body fluids like semen, saliva, and breast milk is common. Another way of studying the pathobiology of ERVs could be to recreate them as infectious forms,139,140 although this would have ethical aspects to consider.
What further research is needed to provide additional knowledge on some of these evolutionary aspects? Better classification141 is needed to understand the pathobiology of the exogenous ancestors of ERVs. Only by doing this they can be defined as host-independent entities. Then we can follow how they spread between species. It will also clarify other aspects of the evolution of ERVs. Data from more vertebrate genomes are also needed. Bioinformatic tools for detecting ERVs must be improved and further evaluated. Only by a multipronged attack can we avoid idiosyncratic biases of individual ERV detection approaches.142,143
What evolutionary considerations might aid in providing novel approaches to the more practical clinical and public health issues facing us currently and in the future? I think that we should search for ERV-related extant exogenous retroviruses in humans and in other animals. This can be done with RetroBank asa base.144 Potentially, these viruses may develop into the next HIV. Searching for infectious retroviruses related to ERVs (like XMRV/HMRV) in humans necessitates a phylogeny-directed approach.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Durden LA, Musser GG. The sucking lice (Insecta, Anoplura) of the world - a taxonomic checklist with records of mammalian hosts and geographical distributions. Bull. Amer. Mus. Nat. Hist. 1994;218:1–90. [Google Scholar]
- 2.Price RD, Hellethal RA, Palma RL, et al. The Chewing Lice: World Checklist and Biological Overview. Champaign, IL: Special Publication 24, Illinois Natural History Survey; 2003. [Google Scholar]
- 3.Johnson KP, Yoshizawa K, Smith VS. Multiple origins of parasitism in lice. Proc. R. Soc. Lond., Ser. B. 2004;271:1771–1776. doi: 10.1098/rspb.2004.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Light JE, Toups MA, Reed DL. What’sin a name: the taxonomic status of human head and body lice. Mol. Phylogen. Evol. 2008;47:1203–1216. doi: 10.1016/j.ympev.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Kim KC, Pratt HD, Stojanovich CJ. The Sucking Lice of North America: An Illustrated Manual for Identification. University Park, PA: Pennsylvania State University Press; 1986. [Google Scholar]
- 6.Reed DL, Light JE, Allen JM, Kirchman JJ. Pair of lice lost or parasites regained: The evolutionary history of Anthropoid primate lice. BMC Biol. 2007;5:7. doi: 10.1186/1741-7007-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong NR, Wilson JD. Did the “Brazilian” kill the pubic louse? Sex Transm Inf. 2006;82:265–266. doi: 10.1136/sti.2005.018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toups MA, Kitchen A, Light JE, Reed DL. Origin of clothing lice indicates early clothing use by anatomically modern humans in Africal. Mol. Biol. Evol. 2011;28:265–266. doi: 10.1093/molbev/msq234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed DL, Smith VS, Rogers AR, et al. Molecular genetic analysis of human lice supports direct contact between modern and archaic humans. PLoS Biol. 2004;2:e304. doi: 10.1371/journal.pbio.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goates BM, Atkin JS, Wilding KG, et al. An effective non-chemical treatment for head lice: a lot of hot air. Pediatrics. 2006;118:1962–1970. doi: 10.1542/peds.2005-1847. [DOI] [PubMed] [Google Scholar]
- 11.Green MS. Epidemiology of Scabies. Epidemiol. Rev. 1989;11:126–150. doi: 10.1093/oxfordjournals.epirev.a036033. [DOI] [PubMed] [Google Scholar]
- 12.Heilesen B. Studies on Acarus Scabiei and Scabies. Acta Dermato-venerologica. 1946;XXVI(Suppl XIV):1–370. [Google Scholar]
- 13.Friedman F. The story of Scabies - written for the Centenary of Renucci’s re-discovery of the Acarus Scabiei. Med. Life. 1934;41:381–476. [Google Scholar]
- 14.Orkin M, Maibach HI. Modern Aspects of Scabies. Curr. Probl. Dermatol. 1985;31:109–127. doi: 10.1159/000410146. [DOI] [PubMed] [Google Scholar]
- 15.Leone PA. Chapter 46. In: Holmes KK, editor. Pubic lice and Scabies Sexually Transmitted Diseases. 4th ed. McGraw Hill Medical; pp. 839–851. [Google Scholar]
- 16.Currier RW, Ceilley RI. Scabies. In: Rabinowitz PM, Conti LA, editors. Human - Animal Medicine. Saunders Elsevier; 2010. [Google Scholar]
- 17.Fain A. Epidemiological problems of Scabies. Int. J. Dermatol. 1978;17:20–30. doi: 10.1111/j.1365-4362.1978.tb06040.x. [DOI] [PubMed] [Google Scholar]
- 18.Fain A. Speciation and evolution in. Acari. Acarology VI. 1984;1:10–18. [Google Scholar]
- 19.Andrews JRH. The origin and evolution of host associations of Sarcoptes scabiei and the subfamily Sarcoptinae Murray. Acarologia Vol. 1983;24:85–94. [PubMed] [Google Scholar]
- 20.Walton SF, Currie BJ. Problems in diagnosing scabies, a global disease in human and animal populations. Clin. Microbiol. Rev. 2007;20:268–279. doi: 10.1128/CMR.00042-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walton SF, Holt DC, Currie BJ, Kemp DJ. Scabies: new future for a neglected disease. Adv. Parasitol. 2004;57:309–327. doi: 10.1016/S0065-308X(04)57005-7. [DOI] [PubMed] [Google Scholar]
- 22.Cudmore SL, Garber GE. Prevention or treatment: the benefits of Trichomonas vaginalis vaccine. J. Infect Public Health. 2010;3:47–53. doi: 10.1016/j.jiph.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Malik SB, Brochu CD, Bilic I, et al. Phylogeny of parasitic Parabasalia and free-living relatives inferred from conventional markers vs. Rpb1, a single-copy gene. PLoS One. 2011;6(6):e20774. doi: 10.1371/journal.pone.0020774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slapeta J, Craig S, McDonell D, Emery D. Tritrichomonas foetus from domestic cats and cattle are genetically distinct. Exper. Parasitol. 2010;126:209–213. doi: 10.1016/j.exppara.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 25.Carlton JM, Hirt RP, Silva JC, et al. Draft genome sequence of the sexually transmitted pathogen Tri-chomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 27.Knight CA, Molinari NA, Petrov DA. The large genome constraint hypothesis: evolution, ecology and phenotype. Ann. Bot. (Lond) 2005;1:177–190. doi: 10.1093/aob/mci011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory TR. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. Camb. Philos. Soc. 2001;1:65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- 29.Zubacova Z, Cimburek Z, Tachezy J. Comparative analysis of trichomonad genome sizes and karyotypes. Mol. Biochem. Parasitol. 2008;161:49–54. doi: 10.1016/j.molbiopara.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Gerbod D, Sanders E, Moriya S, et al. Molecular phylogenies of Parabasalia inferred from four protein genes and comparison with rRNA trees. Mol. Phylogenet Evol. 2004;2:572–580. doi: 10.1016/j.ympev.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 31.Read TD, Myers GS, Brunham RC, et al. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 2003;31:2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dean D, Bruno WJ, Wan R, et al. Predicting phenotype and emerging strains among Chlamydia trachomatis infections. Emerging Infect. Diseases. 2009;15:1385–1394. doi: 10.3201/eid1509.090272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers GS, Mathews SA, Eppinger M, et al. Evidence that human Chlamydia pneumoniae was zoonotically acquired. J. Bacteriol. 2009;191:7225–7233. doi: 10.1128/JB.00746-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nietfeld JC. Chlamydial infections in small ruminants. Vet. Clin. North Am. Food Anim. Pract. 2001;17:301–314. doi: 10.1016/s0749-0720(15)30030-x. vi. [DOI] [PubMed] [Google Scholar]
- 35.DeGraves FJ, Gao D, Hehnen HR, et al. Quantitative detection of Chlamydia psittaci and C. pecorum by high-sensitivity real-time PCR reveals high prevalence of vaginal infection in cattle. J. Clin. Microbiol. 2003;41:1726–1729. doi: 10.1128/JCM.41.4.1726-1729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clarkson MJ, Philips HL. Isolation of faecal chlamydia from sheep in Britain and their characterization by cultural properties. Vet J. 1997;153:307–310. doi: 10.1016/s1090-0233(97)80064-0. [DOI] [PubMed] [Google Scholar]
- 37.Borel N, Dumrese C, Ziegler U, et al. Mixed infections with Chlamydia and porcine epidemic diarrhea virus - a new in vitro model of chlamydial persistence. BMC Microbiol. 2010;10:201. doi: 10.1186/1471-2180-10-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson MN, White P, Giffard P, Timms P. Epizootiology of Chlamydia infections in two free-range koala populations. Vet. Microbiol. 1999;65:255–264. doi: 10.1016/s0378-1135(98)00302-2. [DOI] [PubMed] [Google Scholar]
- 39.Dugan J, Rockey DD, Jones L, Andersen AA. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob. Agents Chemother. 2004;48:3989–3995. doi: 10.1128/AAC.48.10.3989-3995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carey AJ, Timms P, Rawlinson G, et al. A multi-subunit chlamydial vaccine induces antibody and cell-mediated immunity in immunized koalas (Phascolarctos cinereus): comparison of three different adjuvants. Am. J. Reprod. Immunol. 2010;63:161–172. doi: 10.1111/j.1600-0897.2009.00776.x. [DOI] [PubMed] [Google Scholar]
- 41.Davinson AJ, Eberle R, Hayward GS, et al. Family herpesviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger E, Ball LA, editors. Virus Taxonomy: Classification and Nomenclature of Viruses. San Diego: Academic Press; 2005. pp. 193–212. [Google Scholar]
- 42.Eberle R, Black D. Sequence analysis of herpes simplex virus gB gene homologs of twoplatyrrhine monkey alpha-herpesviruses. Arch. Virol. 1993;129:167–182. doi: 10.1007/BF01316893. [DOI] [PubMed] [Google Scholar]
- 43.McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117:90–104. doi: 10.1016/j.virusres.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Weigler BJ. Biology of B virus in macaque and human hosts: a review. Clin. Infect. Dis. 1992;14:555–567. doi: 10.1093/clinids/14.2.555. [DOI] [PubMed] [Google Scholar]
- 45.Loomis MR, O’Neill T, Bush M, Montali RJ. Fatal herpesvirus infection in patas monkeys and a black and white colobus monkey. J. Am. Vet. Med. Assoc. 1981;179:1236–1239. [PubMed] [Google Scholar]
- 46.Ramsay E, Stair EL, Castro AE, Marks MI. Fatal Herpesvirus hominis encephalitis in a white-handed gibbon. J. Am. Vet. Med. Assoc. 1982;181:1429–1430. [PubMed] [Google Scholar]
- 47.McGeoch DJ, Dolan A, Donald S, Brauer DH. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 1986;4:1727–1745. doi: 10.1093/nar/14.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGeoch DJ, Dolan A, Donald S, Rixon FJ. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 1985;1:1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- 49.McGeoch DJ, Dalrymple MA, Davison AJ, et al. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 50.Dolan A, Jamieson FE, Cunningham C, et al. The genome sequence of herpes simplex virus type 2. J. Virol. 1998;3:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perelygina L, Zhu L, Zurkuhlen H, et al. Complete sequence and comparative analysis of the genome of herpes B virus (Cercopithecine herpesvirus 1) from a rhesus monkey. J. Virol. 2003;11:6167–6177. doi: 10.1128/JVI.77.11.6167-6177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tyler SD, Peters GA, Severini A. Complete genome sequence of cercopithecine herpesvirus 2 (SA8) and comparison with other simplexviruses. Virology. 2005;331:429–440. doi: 10.1016/j.virol.2004.09.042. [DOI] [PubMed] [Google Scholar]
- 53.Tyler SD, Severini A. The complete genome sequence of herpesvirus papio 2 (Cercopithecine herpesvirus 16) shows evidence of recombination events among various progenitor herpesviruses. J. Virol. 2006;80:1214–1221. doi: 10.1128/JVI.80.3.1214-1221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chou J, Roizman B. The gamma 1(34.5) gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programed cell death in neuronal cells. Proc. Natl. Acad. Sci. USA. 1992;89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashley RL, Militoni J, Lee F, et al. Comparison of Western blot (immunoblot) and glycoprotein G-specific immunodot enzyme assay for detecting antibodies to herpes simplex virus types 1 and 2 in human sera. J. Clin. Microbiol. 1988;26:662–667. doi: 10.1128/jcm.26.4.662-667.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Su HK, Fetherston JD, Smith ME, Courtney RJ. Orientation of the cleavage site of the herpes simplex virus glycoprotein G-2. J. Virol. 1993;67:2954–2959. doi: 10.1128/jvi.67.5.2954-2959.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luebcke E, Dubovi E, Black D, et al. Isolation and characterization of a chimpanzee alphaherpesvirus. J. Gen. Virol. 2006;87:11–19. doi: 10.1099/vir.0.81606-0. [DOI] [PubMed] [Google Scholar]
- 58.McGeoch DJ, Cook S, Dolan A, et al. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- 59.Tyler S, Severini A, Black D, et al. Structure and sequence of the saimiriine herpesvirus 1 genome. Virology. 2011;410:181–191. doi: 10.1016/j.virol.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Apetrei C, Robertson DL, Marx PA. The history of SIVS and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa. Front Biosci. 2004;9:225–254. doi: 10.2741/1154. [DOI] [PubMed] [Google Scholar]
- 61.Gifford RJ, Katzourakis A, Tristem M, et al. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. USA. 2008;105:20362–20367. doi: 10.1073/pnas.0807873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 63.Desrosiers RC. In: Nonhuman Lentiviruses Fields Virology. Knipe D, Howley P, editors. Philadelphia, PA: Lippincott Williams & Wilkins; 2001. pp. 2095–2121. [Google Scholar]
- 64.Paiardini M, Pandrea I, Apetrei C, Silvestri G. Lessons learned from the natural hosts of HIV-related viruses. Annu. Rev. Med. 2009;60:485–495. doi: 10.1146/annurev.med.60.041807.123753. [DOI] [PubMed] [Google Scholar]
- 65.Thomson NR, Clarke IN. Chlamydia trachomatis: small genome, big challenges. Future Microbiol. 2010;5:555–561. doi: 10.2217/fmb.10.31. [DOI] [PubMed] [Google Scholar]
- 66.Gomes JP, Bruno WJ, Nunes A, et al. Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hotspots. Genome Res. 2007;17:50–60. doi: 10.1101/gr.5674706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stephens RS. Chlamydial evolution: a billion years and counting. In: Schachter J, Christiansen G, Clarke IN, Hammerschlag MR, Kaltenboeck B, Rank RG, Ridgway GL, Saikku P, Stamm WE, Stephens RS, Summersgill JT, Timms P, Wyrick PB, editors. Proceedings of the Tenth International Symposium on Human Chlamydial Infections; International Chlamydia Symposium; 2002. pp. 3–12. [Google Scholar]
- 68.Ochman H, Worobey M, Kuo CH, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Unemo M, Clarke IN. The Swedish new variant of Chlamydia trachomatis. Curr. Opin. Infect. Dis. 2011;24:62–69. doi: 10.1097/QCO.0b013e32834204d5. [DOI] [PubMed] [Google Scholar]
- 70.Lagergård T. In: Haemophilus ducreyi. Chapter 86, in Molecular Medical Microbiology. Sussman M, editor. vol 3. London UK, San Diego, USA: Academic Press. A Harcourt Science and Technology Company; 2002. pp. 1809–1824. [Google Scholar]
- 71.Gioia J, Qin X, Jiang H, et al. The genome sequence of Mannheimia haemolytica A1: insights into virulence, natural competence, and Pasteurellaceae phylogeny. J. Bacteriol. 2006;188:7257–7266. doi: 10.1128/JB.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Janowicz DM, Li W, Bauer ME. Host-pathogen interplay of Haemophilus ducreyi. Curr. Opin. Infect Dis. 2010;23:64–69. doi: 10.1097/QCO.0b013e328334c0cb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carter JS, Bowden FJ, Bastian I, et al. Phylogenetic evidence for reclassification of Calymmatobacterium granulomatis as Klebsiella granulomatis comb. nov. Int. J. Syst. Bacteriol. 1999;49(Pt 4):1695–1700. doi: 10.1099/00207713-49-4-1695. [DOI] [PubMed] [Google Scholar]
- 74.Kharsany AB, Hoosen AA, Kiepiela P, et al. Phylogenetic analysis of Calymmatobacterium granulomatis based on 16S rRNA gene sequences. J. Med. Microbiol. 1999;48:841–847. doi: 10.1099/00222615-48-9-841. [DOI] [PubMed] [Google Scholar]
- 75.Mlengeya TDK. Distribution pattern of a sexually transmitted diseast (STD) of Olive Baboon in Lake Manyara National Park, Tanzania. Moshi: College of Wildlife Management; 2004. [Google Scholar]
- 76.Knauf S, Batamusi E, Mlengeya TDK, et al. Treponema infection associated with Genital Ulceration in Wild Baboons. Vet. Pahtol. 2011 Mar 16; doi: 10.1177/0300985811402839. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 77.Fribourg-Blanc A, Mollaret HH. Natural treponematosis of the African primate. Primates Med. 1969;3:113–121. [PubMed] [Google Scholar]
- 78.Hackett CJ. On the origin of the human treponematoses. Bull. World Health Org. 1963;29:7–41. [PMC free article] [PubMed] [Google Scholar]
- 79.Hudson EH. Treponematosis in perspective. Bull. World Health Org. 1965;32:735–748. [PMC free article] [PubMed] [Google Scholar]
- 80.Weinstock GM, Smajs D, Hardham JM, Norris SJ. From microbial genome sequence to applications. Res. Microbiol. 2000;151:151–158. doi: 10.1016/s0923-2508(00)00115-7. [DOI] [PubMed] [Google Scholar]
- 81.Mikalova´ L, Strouhal M, Ĉejková D, et al. Genome analysis of Treponema pallidum subsp., pallidum and subsp., pertenue strains: most of the genetic differences are localized in six regions. PLoS One. 2010;5:15713. doi: 10.1371/journal.pone.0015713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Centurion-Lara A, Molini BJ, Godornes C, et al. Molecular differentiation of Treponema pallidum subspecies. J. Clin. Microbiol. 2006;44:3377–3380. doi: 10.1128/JCM.00784-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gray R, Mulligan C, Molini B, et al. Molecular evolution of the tprC, D, I, K, G, and J genes in the pathogenic genus Treponema. Mol. Biol. Evol. 2006;23:2220–2233. doi: 10.1093/molbev/msl092. [DOI] [PubMed] [Google Scholar]
- 84.Harper KN, Ocampo PS, Steiner BM, et al. On the origin of the Treponematoses: a phylogenetic approach. PLoS Negl. Trop. Dis. 2008;2(1):e148. doi: 10.1371/journal.pntd.0000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Melo FL, de Mello JC, Fraga AM, et al. Syphilis at the crossroads of Phylogenetics and Paleopathology. PLoS Negl. Trop. Dis. 2010;4:e575. doi: 10.1371/journal.pntd.0000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marra CM, Sahi SK, Tantalo L, et al. Enhanced molecular typing of Treponema pallidum: geographical distribution of strain types and association with neurosyphilis. J. Infect. Dis. 2010;202:1380–1388. doi: 10.1086/656533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pillay A, Liu H, Chen CY, et al. Molecular subtyping of Treponema pallidum subspecies pallidum. Sex Transm. Dis. 1998;25:408–414. doi: 10.1097/00007435-199809000-00004. [DOI] [PubMed] [Google Scholar]
- 88.Tapsall JW, Ndowa F, Lewis DA, Unemo M. Meeting the public health challenge of multidrug- and extensively drug-resistant Neisseria gonorrhoeae. Expert Rev. Anti. Infect. Ther. 2009;7:821–834. doi: 10.1586/eri.09.63. [DOI] [PubMed] [Google Scholar]
- 89.Sparling PF. Biology of Neisseria gonorrhoeae . In: Holmes KK, Sparling PF, Stamm WE, Piot P, Wasserheit JN, Corey L, Cohen MS, Watts DH, editors. Sexually Transmitted Diseases. 4th ed. New York, USA: McGraw-Hill Professional; 2007. pp. 607–626. [Google Scholar]
- 90.Hamilton HL, Dillard JP. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 2006;59:376–385. doi: 10.1111/j.1365-2958.2005.04964.x. [DOI] [PubMed] [Google Scholar]
- 91.Marri PR, Paniscus M, Weyand NJ, et al. Genome sequencing reveals widespread virulence gene exchange among human Neisseria species. PLoS One. 2010;5:e11835. doi: 10.1371/journal.pone.0011835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dunning Hotopp JC, Grifantini R, Kumar N, et al. Comparative genomics of Neisseria meningitidis: core genome, islands of horizontal transfer and pathogen-specific genes. Microbiology. 2006;152(Pt 12):3733–3749. doi: 10.1099/mic.0.29261-0. [DOI] [PubMed] [Google Scholar]
- 93.Tapsall JW, Ndowa F, Lewis DA, Unemo M. Meeting the public health challenge of multidrug- and extensively drug-resistant Neisseria gonorrhoeae. Expert Rev. Anti. Infect. Ther. 2009;7:821–834. doi: 10.1586/eri.09.63. [DOI] [PubMed] [Google Scholar]
- 94.Shafer WM, Folster JP, Nicholas RA. In Neisseria: Molecular Mechanisms of Pathogenesis. Norfolk, UK: Caister Academic Press; 2010. Molecular mechanisms of antibiotic resistance expressed by the pathogenic Neisseriae. [Google Scholar]
- 95.Faruki H, Kohmescher RN, McKinney WP, Sparling PF. A community-based outbreak of infection with penicillin-resistant Neisseria gonorrhoeae not producing penicillinase (chromosomally-mediated resistance) N. Engl. J. Med. 1985;313:607–611. doi: 10.1056/NEJM198509053131004. [DOI] [PubMed] [Google Scholar]
- 96.Warner DM, Shafer WM, Jerse AE. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE Efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol. Microbiol. 2008;70:462–478. doi: 10.1111/j.1365-2958.2008.06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ohnishi M, Golparian D, Shimuta K, et al. Is Neisseria gonorrhoeae initiating a future era of untreatable gonorrhea? Detailed characterization of the first strain with high-level resistance to ceftriaxone. Antimicrob Agents Chemother. 2011;55:3538–3545. doi: 10.1128/AAC.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hoofnagle JH. Hepatitis C: the clinical spectrum of disease. Hepatology. 1997;26(Suppl 1):158–205. doi: 10.1002/hep.510260703. [DOI] [PubMed] [Google Scholar]
- 99.Promrat K, Wands JR. Aetiology of fulminant hepatitis. In: Thomas HC, Lemon HCS, Zuckerman A, editors. Viral Hepatitis. 3rd ed. Blackwell; 2005. pp. 651–665. [Google Scholar]
- 100.Acharya SK, Dasarathy S, Kumer TL, et al. Fulminant Hepatitis in a tropical population: clinical course, cause, and early predictors of outcome. Hepatology. 1996;23:1448–1455. doi: 10.1002/hep.510230622. [DOI] [PubMed] [Google Scholar]
- 101.Anderson DA, Cheng RH. Hepatitis E Virus Epidemiology, clinical and pathologic features, diagnosis, and experimental models. In: Thomas HC, Lemon HCS, Zuckerman A, editors. Viral Hepatitis. 3rd ed. Blackwell; 2005. pp. 603–610. [Google Scholar]
- 102.Simmonds P. Reconstructing the origins of human hepatitis viruses. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2001;356:1013–1026. doi: 10.1098/rstb.2001.0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Simmonds P. The origin and evolution of hepatitis viruses in humans. J. Gen. Virol. 2001;82:693–712. doi: 10.1099/0022-1317-82-4-693. [DOI] [PubMed] [Google Scholar]
- 104.Meng XJ. From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 2011 Feb 21; doi: 10.1016/j.virusres.2011.01.016. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Strickland GT. Liver disease in Egypt: hepatitis C superseded schistosomiasis as a result of iatrogenic and biological factors. Hepatology. 2006;43:915–922. doi: 10.1002/hep.21173. [DOI] [PubMed] [Google Scholar]
- 106.Nordenfelt E. Epidemiology of hepatitis delta virus and non-A, non-B hepatitis. Scand J. Infect. Dis. Suppl. 1990;69:49–53. [PubMed] [Google Scholar]
- 107.Stevens CE, Beasley RP, Tsui JJ, Lee WC. Vertical transmission of hepatitis B antigen in Taiwan. N. Eng. J. Med. 1975;292:771–774. doi: 10.1056/NEJM197504102921503. [DOI] [PubMed] [Google Scholar]
- 108.Mast EE, Hwang LY, Seto DS, et al. Risk factors for perinatal transmission of hepatitis C virus (HCV) and the natural history of HCV infection acquired in infancy. J. Infect. Dis. 2005;192:1880–1889. doi: 10.1086/497701. [DOI] [PubMed] [Google Scholar]
- 109.Quer J, Muir JIE. Hepatitis C Virus Epidemiology. In: Thomas HC, Lemon S, Zuckerman A, editors. Viral Hepatitis. 3rd ed. Blackwell; 2005. pp. 407–425. [Google Scholar]
- 110.Rosina F, Rizzetto M. Hepatitis D Virus: Epidemiology and natural history. In: Thomas HC, Lemon S, Zuckerman A, editors. Viral Hepatitis. 3rd ed. Blackwell; 2005. pp. 583–592. [Google Scholar]
- 111.Kulkarni MA, Walimbe AM, Cherian S, Arankalle VA. Full length genomes of genotype IIIA Hepatitis A Virus strains (1995–2008) from India and estimates of the evolutionary rates and ages. Infect. Genet. Evol. 2009;9:1287–1294. doi: 10.1016/j.meegid.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 112.Hannoun CP, Horal P, Lindh M. Long-term mutation rates in the hepatitis B virus genome. J. Gen. Virol. 2000;81:75–83. doi: 10.1099/0022-1317-81-1-75. [DOI] [PubMed] [Google Scholar]
- 113.Smith DB, Pathirana S, Davidson F, et al. The origin of hepatitis C virus genotypes. J. Gen. Virol. 1997;78:321–328. doi: 10.1099/0022-1317-78-2-321. [DOI] [PubMed] [Google Scholar]
- 114.Nakao H, Okamoto H, Fukuda M, et al. Mutation rate of GB virus C/hepatitis G virus over the entire genome and in subgenomic regions. Virology. 1997;233:43–50. doi: 10.1006/viro.1997.8615. [DOI] [PubMed] [Google Scholar]
- 115.Krugman S. The ABC’S of Viral Hepatitis - The Gordon Lecture. Trans. Am. Clin. Climatol. Assoc. 1992;101:145–156. [PMC free article] [PubMed] [Google Scholar]
- 116.Robertson BH, Jansen RW, Khanna B, et al. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J. Gen. Virol. 1992;73:1365–1377. doi: 10.1099/0022-1317-73-6-1365. [DOI] [PubMed] [Google Scholar]
- 117.Costa-Mattioli M, DiNapoli A, Ferre V, et al. Genetic variability of hepatitis A virus. J. Gen. Virol. 2003;84:3191–3201. doi: 10.1099/vir.0.19532-0. [DOI] [PubMed] [Google Scholar]
- 118.Taylor J, Pelchat M. Origin of hepatitis delta virus. Future Microbiol. 2010;5:393–402. doi: 10.2217/fmb.10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nelson SM. Pigs in prehistory. University of Pennsylvania Museum of Archaeology and Anthropology; 1998. Ancestors for the Pigs. ISBN 978–1–931707–09–1. [Google Scholar]
- 120.Connell CO’, Oranje A, Van Gysel D, Silverberg NB. Congenital Molluscum Contagiosum: report of four cases and review of the literature. Pediatric Dermatol. 2008;25:553–556. doi: 10.1111/j.1525-1470.2008.00730.x. [DOI] [PubMed] [Google Scholar]
- 121.Gubser C, Kellam SP, Smith GL. Poxvirus genomes: a phylogenetic analysis. J. Gen Virol. 2004;85:105–117. doi: 10.1099/vir.0.19565-0. [DOI] [PubMed] [Google Scholar]
- 122.Moss B, Shisler JL, Xiang Y, Senkevich TG. Immune-defense molecules of Molluscum contagiosum virus, a human poxvirus. Trends Microbiol. 2000;8:473–477. doi: 10.1016/s0966-842x(00)01838-2. [DOI] [PubMed] [Google Scholar]
- 123.Senkevich TG, Sisler JR, Koonin EV, et al. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 124.Tyring SK. Molluscum contagiosum: the importance of early diagnosis and treatment. Am. J. Obst. Gynecol. 2003;189:S12–S16. doi: 10.1067/s0002-9378(03)00793-2. [DOI] [PubMed] [Google Scholar]
- 125.McGeoch DJ, Davison AJ, Dolan A, et al. Molecular evolution of the Herpesvirales . In: Domingo E, Parrish CR, Holland JJ, editors. Origin and Evolution of Viruses. 2nd ed. London: Elsevier; 2008. pp. 447–475. [Google Scholar]
- 126.Gentry GA, Lowe M, Alford G, Nevins R. Sequence analyses of herpesviral enzymes suggest an ancient origin for human sexual behavior. Proc. Natl. Acad. Sci. USA. 1988;85:2658–2661. doi: 10.1073/pnas.85.8.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Norberg P. Divergence and genotyping of human alpha-herpesviruses: an overview. Infect. Genet. Evol. 2010;10:14–25. doi: 10.1016/j.meegid.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 128.Preston CM, Efstathiou S. Molecular basis of HSV latency and reactivation. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K, editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge: Cambridge University Press; 2007. pp. 602–615. [Google Scholar]
- 129.Lowhagen GB, Tunback P, Andersson K, et al. First episodes of genital herpes in a Swedish STD population: a study of epidemiology and transmission by the use of herpes simplex virus (HSV) typing and specific serology. Sex Transm. Infect. 2000;76:179–182. doi: 10.1136/sti.76.3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McGeoch DJ, Cook S. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J. Mol. Biol. 1994;238:9–22. doi: 10.1006/jmbi.1994.1264. [DOI] [PubMed] [Google Scholar]
- 131.Norberg P, Bergstrom T, Rekabdar E, et al. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses. J. Virol. 2004;78:10755–10764. doi: 10.1128/JVI.78.19.10755-10764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Norberg P, Kasubi MJ, Haarr L, et al. Divergence and recombination of clinical herpes simplex virus type 2 isolates. J. Virol. 2007;81:13158–13167. doi: 10.1128/JVI.01310-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sharp PM, Hahn BH. The evolution of HIV-1 and the origin of AIDS. Phil. Trans. Roy. Soc. London, Series B. 2010;365:2487–2494. doi: 10.1098/rstb.2010.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Keele BF, et al. Chimpanzee reservoirs of pandemic and non-pandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sharp PM, Hahn BH, et al. Prehistory of HIV-1. Nature (London) 2008;455:605–606. doi: 10.1038/455605a. [DOI] [PubMed] [Google Scholar]
- 136.Keele BF, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sauter D, et al. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and non-pandemic HIV-1 strains. Cell Host Microbe. 2009;6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hu L. Expression of Endogenous Retroviruses in Humans. Thesis. Uppsala: Uppsala University; 2007. [Google Scholar]
- 139.Dewannieux M, Harper F, Richaud A, et al. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee YN, Bieniasz PD. Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog. 2007;3:e10. doi: 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Blomberg J, Benachenhou F, Blikstad V, et al. Classification and nomenclature of endogenous retroviral sequences (ERVs) Problems and recommendations. Gene. 2009;448(2):115123. doi: 10.1016/j.gene.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 142.Sperber G, Lovgren A, Eriksson NE, et al. Retro-Tector online, a rational tool for analysis of retroviral elements in small and medium size vertebrate genomic sequences. BMC bioinformatics. 2009;10(Suppl 6):S4. doi: 10.1186/1471-2105-10-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sperber GO, Airola T, Jern P, Blomberg J. Automated recognition of retroviral sequences in genomic data–RetroTector. Nucleic Acids Res. 2007;35:49644976. doi: 10.1093/nar/gkm515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Blomberg J, Sperber G, Jern P, Benachenhou F. Towards a retrovirus database, RetroBank. In: Hejnar RDJ, Skalka A, Svoboda J, editors. Proceedings of the Centennial Retrovirus Meeting; April 29–May 4; Prague: Medimond International Proceedings; 2010. pp. 19–22. [Google Scholar]