Abstract
Background
Preterm delivery (PTD) is the leading cause of neonatal morbidity and mortality. Epidemiologic studies indicate recurrence of PTD is maternally inherited creating a strong possibility that mitochondrial variants contribute to its etiology. This study examines the association between mitochondrial genotypes with PTD and related outcomes.
Methods
This study combined, through meta-analysis, two case-control, genome-wide association studies (GWAS); one from the Danish National Birth Cohort (DNBC) Study and one from the Norwegian Mother and Child Cohort Study (MoBa) conducted by the Norwegian Institute of Public Health. The outcomes of PTD (≤36 weeks), very PTD (≤32 weeks) and preterm prelabor rupture of membranes (PPROM) were examined. 135 individual SNP associations were tested using the combined genome from mothers and neonates (case vs. control) in each population and then pooled via meta-analysis.
Results
After meta-analysis there were four SNPs for the outcome of PTD below p≤0.10, and two below p≤0.05. For the additional outcomes of very PTD and PPROM there were three and four SNPs respectively below p≤0.10.
Conclusion
Given the number of tests no single SNP reached study wide significance (p=0.0006). Our study does not support the hypothesis that mitochondrial genetics contributes to the maternal transmission of PTD and related outcomes.
INTRODUCTION
Preterm delivery (PTD; gestational age at delivery <37 weeks) and its effects are the leading causes of neonatal morbidity and mortality, responsible for ~35% of infant deaths in the US1 and ~28% of the 4 million neonatal deaths worldwide2. Neonates who survive are at increased risk for blindness, deafness, cerebral palsy and cognitive delay3,4. These disorders have onset in the neonatal period, and thus cause more disability adjusted life years (DALY) than adult onset diseases. Of concern, is that rates of PTD in the United States have been rising, peaking in 2006 at 12.8 percent overall (11.1% in singleton births) with only slight declines since5.
Contributing factors to PTD are many and varied. Environmental factors including smoking6 and other substance abuse, low socioeconomic status7, maternal stress8, extremes in maternal age9,10 and previous adverse pregnancy outcomes11 contribute to PTD. Also, physiological risk factors such as uterine anomalies12, abnormal cholesterol13, low vitamin D14,15, maternal infection16 and elevated inflammatory states17 put delivering women at risk. The multi-factorial nature of this disease makes it difficult to establish broadly applicable principles about its etiology.
Underlying the established risk factors is the role genetic predisposition plays in PTD18. A familial component has been established with genetic studies showing the heritability of PTD is up to 40%19. In addition, associations of PTD and single nucleotide polymorphisms (SNPs) in the nuclear genome have been reported20-22. An under-explored, yet highly plausible cause of the matrilineal risk for PTD is the role of the mitochondrial genome. Epidemiologic studies in two European populations indicate that recurrence of PTD is maternally associated suggesting that mitochondrial variants may contribute to risk23-25. Specific case-control studies have pointed to association within the mitochondrial genome as well26. These indications of a mitochondrial component to PTD call for a comprehensive, genome-wide approach to identifying associated mitochondrial SNPs implicated in PTD27.
This study examines the link between mitochondrial genome variation, which is maternally inherited, and PTD by examining SNPs from across the mitochondrial genome. This study combines results from two mitochondria-wide scans, one from a Danish population and one from a Norwegian population. The results from each study were meta-analyzed to screen for any potential candidate SNPs related to PTD.
RESULTS
Case and control numbers for each study population, Danish and Norwegian, concerning the outcomes of PTD, very PTD and PPROM are summarized in Table 1. Mitochondrial SNPs were tested using the outcomes defined in these tables for each population and then pooled via random-effects meta-analysis. Figure 1 is a graphical representation of the results for the Denmark and Norway populations and meta-analyzed results.
Table 1.
Demographic Data
| Outcome | Denmark | Norway | ||
|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |
| PTD | 1307 | 988 | 981 | 983 |
| Very PTD | 252 | 988 | 95 | 983 |
| PPROM | 379 | 988 | 287 | 983 |
Cases and controls for both the Danish and Norwegian populations.
Figure 1.
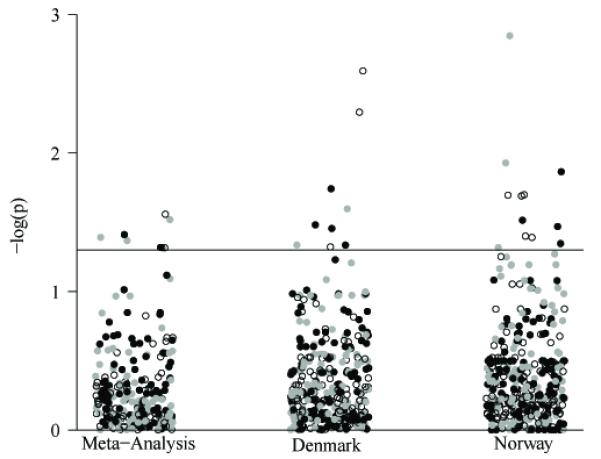
Significance of all tested SNPs. The significance, −log(p), of tests performed is represented graphically. Tests are stratified by the population (Denmark and Norway) and the pooled meta-analysis. Black points indicate PTD, open circles indicate very PTD, and gray points indicate PPROM. The horizontal line on the figure corresponds to p=0.05.
In the Danish population ten SNPs (four for PTD, three for very PTB and two for PPROM) fell below p≤0.05 and in the Norwegian population 12 SNPs were below p≤0.05 (four for PTD, five for very PTD and three for PPROM). These results can be seen in Tables 2 and 3 respectively. None of these SNPs were below p≤0.05 in both populations and none approached study-wide significance threshold (p≤0.0006).
Table 2.
SNPs in the Danish Population with p<0.05
| Location | Random Effects P-value |
Random Effects OR |
P-Value Denmark |
OR Denmark |
P-Value Norway |
OR Norway |
|---|---|---|---|---|---|---|
| PTD | ||||||
| 9541 | 0.529 | 0.56 | 0.018 | 0.19 | 0.465 | 1.24 |
| 8870 | --a | --a | 0.033 | --a | 0.316 | 3.01 |
| 9378 | 0.325 | 0.50 | 0.035 | 0.21 | 0.809 | 0.89 |
| 10045 | --b | --b | 0.046 | --b | 0.678 | 1.19 |
| Very PTD | ||||||
| 13106 | 0.028 | 3.39 | 0.003 | 4.67 | 0.807 | 1.30 |
| 8870 | --a | --a | 0.005 | --a | 0.756 | 0.00 |
| 2485 | --a | --a | 0.048 | --a | --c | --c |
| PPROM | ||||||
| 4337d | 0.980 | 0.98 | 0.025 | 2.24 | 0.095 | 0.37 |
| 3395 | 0.108 | 1.93 | 0.046 | 3.16 | 0.511 | 1.38 |
In the Danish population the controls had a MAF=0 for this SNP.Therefore the OR and meta-analysis could not be evaluated.
In the Danish population the cases had a MAF=0 for this SNP.Therefore the OR and meta-analysis could not be evaluated.
SNP not available in the Norway population.
SNPs indicate those that were not available in the additional 500 Danish samples. The meta-analysis results are based on the original Danish population and the Norwegian population.
Table 3.
SNPs in the Norwegian Population with p<0.05.
| Location | Random Effects P-Value |
Random Effects OR |
P-Value Denmark |
OR Denmark |
P-value Norway |
OR Norway |
|---|---|---|---|---|---|---|
| PTD | ||||||
| 16145 | 0.979 | 0.97 | 0.197 | 0.25 | 0.014 | 2.10 |
| 479 | 0.545 | 0.80 | 0.531 | 1.12 | 0.031 | 0.52 |
| 2160 | 0.142 | 0.67 | 0.846 | 0.93 | 0.034 | 0.54 |
| 10689 | --a | --a | 0.843 | 0.76 | 0.045 | --a |
| Very PTD | ||||||
| 14583 | 0.518 | 1.71 | 0.581 | 0.56 | 0.020 | 3.15 |
| 4025b | 0.048 | 2.53 | 0.973 | 0.96 | 0.020 | 3.16 |
| 5005b | 0.048 | 2.53 | 0.973 | 0.96 | 0.020 | 3.15 |
| 5443 | --c | --c | 0.614 | --c | 0.040 | 10.44 |
| 13966b | 0.652 | 1.86 | 0.360 | 0.62 | 0.041 | 10.37 |
| PPROM | ||||||
| 16145 | --c | --c | 0.283 | --c | 0.001 | 3.10 |
| 1191b | 0.616 | 0.68 | 0.205 | 1.39 | 0.012 | 0.29 |
| 752b | 0.108 | 1.59 | 0.853 | 1.09 | 0.048 | 1.99 |
In the Norwegian population the controls had a MAF=0 for this SNP. Therefore, the OR and meta-analysis could not be evaluated.
SNPs indicate those that were not available in the additional 500 Danish samples. The meta-analysis results are based on the original Danish population and the Norwegian population.
In the Danish population the cases had a MAF=0 for this SNP. Therefore, the OR and meta-analysis could not be evaluated.
In the meta-analyzed results, for the outcome of PTD there were a total of four SNPs that were below p≤0.10 (Table 4). Of these two SNPs were below p≤0.05 at positions 6777 and 5047. None of these SNPs were below p≤0.05 in either the Danish or Norwegian populations alone and none approached study-wide significance (p≤0.0006)
Table 4.
Nominally significant SNPs after meta-analysis for PTD, Very PTD, and PPROM
| Location | Random Effects P-Value |
Random Effects OR |
P-Value Denmark |
OR Denmark |
P-Value Norway |
OR Norway |
|---|---|---|---|---|---|---|
| PTD | ||||||
| 5047 | 0.048 | 1.73 | 0.059 | 1.94 | 0.388 | 1.45 |
| 6777a | 0.039 | 0.69 | 0.143 | 0.67 | 0.139 | 0.71 |
| 3916 | 0.076 | 0.73 | 0.207 | 0.75 | 0.203 | 0.69 |
| 16146 | 0.097 | 0.78 | 0.335 | 0.81 | 0.171 | 0.76 |
| Very PTD | ||||||
| 13106 | 0.028 | 3.39 | 0.003 | 4.67 | 0.807 | 1.30 |
| 4025a | 0.048 | 2.53 | 0.973 | 0.96 | 0.020 | 3.16 |
| 5005a | 0.048 | 2.53 | 0.973 | 0.96 | 0.020 | 3.15 |
| PPROM | ||||||
| 13106 | 0.030 | 2.41 | 0.188 | 2.19 | 0.068 | 2.60 |
| 5047 | 0.041 | 2.06 | 0.082 | 2.16 | 0.239 | 1.92 |
| 5657a | 0.043 | 1.79 | 0.133 | 2.56 | 0.120 | 1.64 |
| 15905 | 0.081 | 1.52 | 0.062 | 1.80 | 0.573 | 1.22 |
SNPs indicate those that were not available in the additional 500 Danish samples. These results are based on a meta-analysis between the original Danish population and the Norwegian population.
Results were similar when using the outcome of very PTD. After meta-analysis, three SNPs had p≤0.10 and all were below p≤0.05. Of these three one was below p≤0.05 in the Danish population and two were below p≤0.05 in the Norwegian population. None approached study-wide significance (p≤0.0006).
For the outcome of PPROM four SNPs were below p≤0.10 and three were below p≤0.05. None of these SNPs were below p≤0.05 in either the Danish or Norwegian populations and none approached study wide significance (p≤0.0006).
No individual SNP, in the Danish cohort, Norwegian cohort or pooled meta-analysis was even on the same order of magnitude of the study-wide significance threshold, p≤0.0006. Complete results for all tested SNPs are available from the authors upon request.
DISCUSSION
Our analysis of mitochondrial SNPs and their relation to PTD and related outcomes did not implicate any particular locus that was significantly associated with any of the defined outcomes. This study does not support the hypothesis that of maternal link to PTD through transmission of the mitochondrial genome.
Assessing the mitochondrial impact on PTD is a necessary step in unraveling its etiology. In addition to epidemiological evidence for a mitochondrial role in PTD, biological factors point to this possibility. Oxidative stress from natural processes, both pregnancy28 and labor29, as well as outside factors like toxins30 and infection, can contribute to adverse pregnancy outcomes and would be worsened in the setting of mitochondrial dysfunction. Similarly, given the central role of the mitochondrion in energy metabolism31, variations in necessary processes, from mother or fetus, could lead to detrimental effects.
Mitochondrial genetics was an obvious explanation for the observed maternal link to PTD risk but this may be explained by other modes of transmission. Genetically, the possibility of imprinting, whereby the maternal or paternal copy of a gene is expressed over the suppressed paternal or maternal copy, at key locations related to pregnancy and parturition may explain the observed inheritance pattern. Imprinting appears to be rare32 and its impact in complex, common diseases is uncertain, but has been shown to possibly contribute to other common conditions33. Alterations in the maternal nuclear genome or fetal-maternal genetic interactions could also explain the witnessed maternal inheritance. Finally, any risk factor for PTD that is preferentially passed from mother to daughter, as opposed to mother to son, could explain this association. Such possibilities include biological factors such as genital tract microflora34, or, conceivably, non-genetic transmission of response to social factors like stress35.
This study using mitochondrial genetics did have some limitations. First, while every effort was made to include only spontaneous PTDs it is possible that our case population included a small portion of PTDs resulting from infections. In the original Danish cohort 12 cases (<1.5%) had documented chorioamnionitis. We believe that the additional Danish samples and the Norwegian population have similarly low infection rates and thus our case population appropriately homogeneous. Also, predefined haplogroups (H, U, J, etc. for Western Europeans) were not tested because the Illumina chip did not contain the necessary SNPs to define these haplogroups in the populations. Meta-analysis of two study populations increased our power to detect any potential associated SNPs but the current study was still limited in its ability to detect the effects of rare variations. A sample size of around 5000 cases and controls would be needed to be to detect an odds ratio of 2.0 for SNPs with MAF around 1%. This is especially true for outcomes of PPROM and very PTD because of the significant decrease in the case population. Studies of the current size can only detect common SNP variations with large disease risk or uncommon SNP variations with extreme impact on disease. Traditionally it has been thought that common SNP variations lead to common diseases but this may not be true for all conditions36. Therefore, only future studies with very large sample sizes and more granular sequencing can completely assess the impact of rare variations in the mitochondrial genome on the risk of PTD.
This study has several strengths that contribute to its validity. The data come from two populations that are genetically homogeneous, reducing population stratification and possible false positives. However, this may also limit the application of these results to other external populations. Definitions of PTD, were clearly established for cases and controls, as they were based on last menstrual period and/or ultrasound. The case and control groups were both clean of many possible confounding conditions including any induction of delivery, multiple births, congenital malformations, pre-eclampsia, or other placental abnormalities.
Previous epidemiological and genetic studies indicate that genetics plays some role in the etiology of PTD. Future studies should include analyses of the effects of imprinting and a full genome-wide approach is currently underway within these same populations. Extending studies of mitochondrial genetics in a larger cohort of this population or in different ethnic populations may yield contrasting results and gene-gene and gene-environment interactions between factors such as inflammation and cholesterol are of interest. Genetic interactions from mother and/or fetus likely play a role in the etiology of PTD. However, no obvious common mitochondrial variant was associated with any of the study outcomes in these populations.
MATERIALS AND METHODS
Study Populations
Danish Population
Mother-neonate pairs were selected as cases and controls from the Danish National Birth Cohort (DNBC) Study37. This cohort consisted of ~100,000 total births occurring between 1997 and 2002. Controls were defined as 39-40 weeks gestational age (GA) and cases as infants between 20-36 GA. (Only three births were <22 weeks GA. As only live-born infants were included in the study, these infants were either unlikely survivors, had underestimated GA or had a data error in their stillborn status. Underestimated GA is most likely thus, they have been kept in the cohort.) All four grandparents of a neonate were born in Denmark except for 24 individuals who had 1 or 2 grandparents born in other Nordic countries. The parent genome wide association study (GWAS) included 1000 case mother-neonate pairs and 1000 control mother-neonate pairs. Biological samples collected from mothers and neonates were stored in the Biobank at Statens Serum Institut. Of the original 2000 mothers chosen, 1937 were successfully genotyped. Of these 41 mothers were removed for having GA outside the case or control definitions, 165 cases for delivery via cesarean section, 24 controls for having a previous PTD and 2 mothers for pre-eclampsia. Additionally, after genotyping some mothers were identified as sisters, thus 5 women were removed via random selection of one sister-neonate pair. This left 1700 mothers for analysis (965 controls 735 cases). Of the original 2000 neonates chosen, 1901 were successfully genotyped. Of these 47 were removed for GA not within the case or control definition, 154 cases for delivery via cesarean section, 23 controls for their mothers having a previous PTD and 2 for pre-eclampsia. Additionally 4 neonates were removed for being born to mothers that were sisters to others in the study. This left 1671 neonates for analysis (958 controls and 713 cases).
To increase the power for SNP detection for the original GWAS described above, an additional set of ~500 mother-baby pairs, mostly cases, were selected from the DNBC BioBank or the Danish Newborn Screening Biobank. Of these 500 mothers, 496 were successfully genotyped and 2 were removed for GA not within the case or control range leaving 490 cases and 4 controls. Of the 500 neonates all were successfully genotyped and 2 were removed for GA not within the case or control range leaving 494 cases and 4 controls.
Norwegian Population
Mother-neonate pairs were selected from the Norwegian Mother and Child Cohort Study (MoBa) conducted by the Norwegian Institute of Public Health38. MoBa is a population-based prospective pregnancy cohort including 108,000 children, 90,700 mothers and 71,500 fathers recruited from all over Norway from 1999 to 2008. Each participant had to give a written informed consent upon recruitment and the participation rate was 38.5% among invited women. Blood samples were collected from both parents at the routine ultrasound examination in week 17-18 of pregnancy and from mothers and children at delivery. Patient samples were stored at the Biobank of the Norwegian Institute of Public Health39. Epidemiological data were obtained through questionnaires given at regular intervals and linkage to the Medical Birth Registry of Norway. The current study is based on version 4 of the quality-assured data files released for research in 2008.
Controls were selected from infants born at 39-40 weeks gestation and cases were born from 22-36 weeks gestation. Only mothers who delivered between 20-34 years of age were eligible for selection. Exclusions were made for women with a medical history of diabetes, chronic hypertension, autoimmune diseases like systemic lupus erythematosus, scleroderma, rheumatoid arthritis, inflammatory bowel disease or immunocompromised diseases. Women with gestational diabetes, hypertension in pregnancy or cervical cerclage, pregnancies conceived by in vitro fertilization, stillbirths and neonates who were small or large for gestational age were also excluded. Before quality control 1921 mothers (951 cases and 970 controls) were selected for genotyping. Of these 81 were removed for low genotyping rate, 6 for unknown sex and 13 for non-relatedness to the assigned child. In addition 19 control mothers were excluded for having a previous PTD, leaving 1802 individuals successfully genotyped (903 cases and 899 controls). There were 1200 children available for genotyping (582 cases and 618 controls). Of these 42 were removed for low genotyping and 6 for non-relatedness to the assigned mother. Furthermore, 15 control children were excluded for having a mother with a previous PTD, leaving 1137 children for further analysis (556 cases and 581 controls). Fewer children than mothers were genotyped due to lack of available DNA.
Common Inclusion–Exclusion Criteria
Exclusion criteria for each study were any obstetrical induction of labor (iatrogenic delivery), placental abnormalities (previa or abruption), pre-eclampsia, congenital malformations, stillbirths and multiple births. All cesarean sections were removed from the case populations as well. Subjects were enrolled with the appropriate consent for future research. In both the Danish and Norwegian cohorts the number of eligible births for this study represented approximately 25% of the total PTD population. Informed consent was obtained from all individuals according to each country’s protocols 37,38 and approval for this study was granted by the University of Iowa Institutional Review Board, study number 200608748.
Genotyping
Genotyping was performed using the Illumina Human660W-Quadv1_A (Illumina Inc, CA, USA) which contained 135 SNPs assumed to have minor allele frequency (MAF) > 1% in Caucasian populations based on public databases (www.mitomap.org). This Illumina chip uses the HapMap Yoruba sample (accession number AF347015) as a reference. The standard reference for mitochondrial SNPs is the revised Cambridge Reference Sequence (rCRS, accession number NC_012920). Positions reported in this paper are according to the AF347015 sequence. The conversion from AF347015 to NC_012920 can be made using the map located at this website: http://www.mitomap.org/Sandbox/YorubanConversion. In total 88 SNPs were meta-analyzed between the two populations. For the Danish samples the call rate was >99% for all included meta-analyzed SNPs and the median MAF was 0.025. For the Norwegian population the call rate was also >99% and the median MAF was 0.024. For the additional 500 Danish samples only 109 SNPs were called during genotyping.
Union of Maternal and Neonatal Genetic information
For mitochondrial data mothers and neonates are assumed to have identical genotypes. The genetic concordance rate between maternal and neonatal pairs was 100% for all SNPs in the Danish population and >99.4% in the Norway population. Thus, maternal and neonatal genetic information was analyzed as one individual, using the mother’s genotype as the default and supplementing the neonates genotype if the mother’s was missing. This increased the power of the individual studies by increasing genotyping rates and increasing the number of cases and controls. Table 1 indicates the number of case and control genotypes available to be analyzed for each of the populations.
Genetic Analysis
Single SNP allelic associations were tested using chi-square tests in both the Danish and Norwegian populations. The outcomes examined were PTD (≤36 weeks GA), very PTD (≤32 weeks GA) and preterm prelabor rupture of membranes (PPROM). PPROM was defined by using the International Classification of Diseases (ICD) code in the Medical Birth Register for each country. After testing within each population, the results were meta-analyzed using a random-effects model. No correction for the number of tests was made to the reported pooled p-values or odds ratios. All analyses were performed in PLINK40.
Of the 135 mitochondrial SNPs on the panel a total of 88 SNPs were meta-analyzed between the two populations. The 47 SNPs removed were either not called in one of the populations or had a minor allele frequency of zero in either the cases or controls for one of the populations. For the additional 500 Danish cases and controls only 109 SNPs were called. Given the number of SNPs tested a conservative threshold for significance in this study would be p=0.0006 (p=0.05 / 88 SNPs).
Acknowledgments
We thank Teri Manolio, who was instrumental in the completion of this GWAS. In addition, we thank Hakon K. Gjessing for the useful comments and discussion.
Statement of Financial Support This work was supported by the National Institutes of Health (NIH) (grants HD52953 and HD57192 to J.M.). The Danish National Birth Cohort (DNBC) was established with the support of a major grant from the Danish National Research Foundation. Additional support for the DNBC has been provided by the Danish Pharmacists’ Fund, the Egmont Foundation, the March of Dimes Birth Defects Foundation, the Augustinus Foundation, and the Health Fund of the Danish Health Insurance Societies.
The generation of GWAS genotype data for the DNBC samples was funded as part of the Gene Environment Association Studies (GENEVA) under the Genes, Environment and Health Initiative (GEI). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research (CIDR) and the Broad Institute of MIT and Harvard, was provided by the NIH GEI (U01HG004438 for CIDR; U01HG04424 for Broad Institute) and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C for CIDR only).
The Norwegian Mother and Child Cohort Study is supported by the Norwegian Ministry of Health and the Ministry of Education and Research, NIH/National Institute of Environmental Health Sciences (NIEHS) (contract NO-ES-75558), NIH/National Institute of Neurological Disorders and Stroke (NINDS) (grant 1 UO1 NS 047537-01), and the Norwegian Research Council/Research in Functional Genomics in Norway (FUGE) (grant 151918/S10). The study was approved by the Regional Committee for Medical Research Ethics in South-Eastern Norway (S-06075) and the Norwegian Data Inspectorate (05/016784). Support came also from the Norwegian Research Council (FUGE 183220/S10, FRIMEDKLI-05 ES236011), Swedish government grants to researchers in the public health service (ALF) (ALFGBG-136431), Sahlgrenska University Hospital and Sahlgrenska Academy (Gothenburg, Sweden), the Swedish Medical Society (Stockholm, Sweden; 2008-21198), and the Jane and Dan Olsson Research Foundation (Gothenburg, Sweden).The MoBa samples were genotyped at the genotyping core facility at Oslo University Hospital (Norway).
Footnotes
Conflicts of Interest: All authors indicate that they have no conflicts of interest to disclose.
References
- 1.Callaghan WM, MacDorman MF, Rasmussen SA, Qin C, Lackritz EM. The contribution of preterm birth to infant mortality rates in the United States. Pediatrics. 2006;118:1566–73. doi: 10.1542/peds.2006-0860. [DOI] [PubMed] [Google Scholar]
- 2.Lawn JE, Cousens S, Zupan J. 4 million neonatal deaths: When? Where? Why? The Lancet. 2005;365:891–900. doi: 10.1016/S0140-6736(05)71048-5. [DOI] [PubMed] [Google Scholar]
- 3.Marlow N, Wolke D, Bracewell MA, Samara M, the EPICure Study Group Neurologic and developmental disability at six years of age after extremely preterm birth. N Engl J Med. 2005;352:9–19. doi: 10.1056/NEJMoa041367. [DOI] [PubMed] [Google Scholar]
- 4.Wood N, Marlow N, Costeloe K, Gibson A, Wilkinson A. Neurologic and developmental disability after extremely preterm birth. N Engl J Med. 2000;343:378–84. doi: 10.1056/NEJM200008103430601. [DOI] [PubMed] [Google Scholar]
- 5.Martin J, Hamilton B, Sutton P, Ventura S, Mathews T, Osterman M. Births: Final Data for 2008. Natl Vital Stat Rep. 2010;59:1–105. [PubMed] [Google Scholar]
- 6.Cnattingius S. The epidemiology of smoking during pregnancy: smoking prevalence, maternal characteristics, and pregnancy outcomes. Nicotine & Tobacco Research. 2004;6:S125–S40. doi: 10.1080/14622200410001669187. [DOI] [PubMed] [Google Scholar]
- 7.Thompson J, Irgens L, Rasmussen S, Daltveit A. Secular trends in socio-economic status and the implications for preterm birth. Paediatr Perinat Epidemiol. 2006;20:182–87. doi: 10.1111/j.1365-3016.2006.00711.x. [DOI] [PubMed] [Google Scholar]
- 8.Dole N, Savitz D, Hertz-Picciotto I, Siega-Riz A, McMahon M, Buekens P. Maternal stress and preterm birth. Am J Epidemiol. 2003;157:14–24. doi: 10.1093/aje/kwf176. [DOI] [PubMed] [Google Scholar]
- 9.Fraser A, Brockert J, Ward R. Association of young maternal age with adverse reproductive outcomes. N Engl J Med. 1995;332:1113–17. doi: 10.1056/NEJM199504273321701. [DOI] [PubMed] [Google Scholar]
- 10.Jacobsson B, Ladfors L, Milsom I. Advanced maternal age and adverse perinatal outcome. Obstet Gynecol. 2004;104:727–33. doi: 10.1097/01.AOG.0000140682.63746.be. [DOI] [PubMed] [Google Scholar]
- 11.Mercer B, Goldenberg R, Moawad A, et al. The preterm prediction study: effect of gestational age and cause of preterm birth on subsequent obstetric outcome. Am J Obstet Gynecol. 1999;181:1216–21. doi: 10.1016/s0002-9378(99)70111-0. [DOI] [PubMed] [Google Scholar]
- 12.Jakobsson M, Gissler M, Sainio S, Paavonen J, Tapper A. Preterm delivery after surgical treatment for cervical intraepithelial neoplasia. Obstet Gynecol. 2007;109:309–13. doi: 10.1097/01.AOG.0000253239.87040.23. [DOI] [PubMed] [Google Scholar]
- 13.Edison RJ, Berg K, Remaley A, et al. Adverse birth outcome among mothers with low serum cholesterol. Pediatrics. 2007;120:723–33. doi: 10.1542/peds.2006-1939. [DOI] [PubMed] [Google Scholar]
- 14.Bodnar L, Catov J, Simhan H, Holick M, Powers R, Roberts J. Maternal vitamin D deficiency increases the risk of preeclampsia. J Clin Endocrinol Metab. 2007;92:3517–22. doi: 10.1210/jc.2007-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodnar LM, Krohn MA, Simhan HN. Maternal Vitamin D Deficiency Is Associated with Bacterial Vaginosis in the First Trimester of Pregnancy. J Nutr. 2009;139:1157–61. doi: 10.3945/jn.108.103168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldenberg R, Hauth J, Andrews W. Intrauterine infection and preterm delivery. N Engl J Med. 2000;342:1500–7. doi: 10.1056/NEJM200005183422007. [DOI] [PubMed] [Google Scholar]
- 17.Romero R, Espinoza J, Gonçalves LF, Kusanovic JP, Friel LA, Nien JK. Inflammation in preterm and term labour and delivery. Seminars in Fetal and Neonatal Medicine. 2006;11:317–26. doi: 10.1016/j.siny.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muglia L, Katz M. The Enigma of Spontaneous Preterm Birth. N Engl J Med. 2010;362:529–35. doi: 10.1056/NEJMra0904308. [DOI] [PubMed] [Google Scholar]
- 19.Kistka Z, DeFranco E, Ligthart L, et al. Heritability of parturition timing: an extended twin design analysis. Am J Obstet Gynecol. 2008;199:43.e1–e5. doi: 10.1016/j.ajog.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 20.Crider K, Whitehead N, Buus R. Genetic variation associated with preterm birth: a HuGE review. Genetics in Medicine. 2005;7:593–604. doi: 10.1097/01.gim.0000187223.69947.db. [DOI] [PubMed] [Google Scholar]
- 21.Engel S, Erichsen H, Savitz D, Thorp J, Chanock S, Olshan A. Risk of spontaneous preterm birth is associated with common proinflammatory cytokine polymorphisms. Epidemiology. 2005;16:469–77. doi: 10.1097/01.ede.0000164539.09250.31. [DOI] [PubMed] [Google Scholar]
- 22.Menon R, Velez D, Simhan H, et al. Multilocus interactions at maternal tumor necrosis factor- , tumor necrosis factor receptors, interleukin-6 and interleukin-6 receptor genes predict spontaneous preterm labor in European-American women. Am J Obstet Gynecol. 2006;194:1616–24. doi: 10.1016/j.ajog.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 23.Boyd HA, Poulsen G, Wohlfahrt J, Murray JC, Feenstra B, Melbye M. Maternal contributions to preterm delivery. Am J Epidemiol. 2009;170:1358–64. doi: 10.1093/aje/kwp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svensson AC, Sandin S, Cnattingius S, et al. Maternal Effects for Preterm Birth: A Genetic Epidemiologic Study of 630,000 Families. Am J Epidemiol. 2009;170:1365–72. doi: 10.1093/aje/kwp328. [DOI] [PubMed] [Google Scholar]
- 25.Lunde A, Melve KK, Gjessing HK, Skjærven R, Irgens LM. Genetic and environmental influences on birth weight, birth length, head circumference, and gestational age by use of population-based parent-offspring data. Am J Epidemiol. 2007;165:734–41. doi: 10.1093/aje/kwk107. [DOI] [PubMed] [Google Scholar]
- 26.Velez DR, Menon R, Simhan H, Fortunato S, Canter JA, Williams SM. Mitochondrial DNA variant A4917G, smoking and spontaneous preterm birth. Mitochondrion. 2008;8:130–5. doi: 10.1016/j.mito.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. The Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myatt L, Cui X. Oxidative stress in the placenta. Histochem Cell Biol. 2004;122:369–82. doi: 10.1007/s00418-004-0677-x. [DOI] [PubMed] [Google Scholar]
- 29.Fainaru O, Almog B, Pinchuk I, Kupferminc MJ, Lichtenberg D, Many A. Active labour is associated with increased oxidisibility of serum lipids ex vivo. BJOG: An International Journal of Obstetrics & Gynaecology. 2002;109:938–41. doi: 10.1111/j.1471-0528.2002.01494.x. [DOI] [PubMed] [Google Scholar]
- 30.Mohorovic L. First two months of pregnancy--critical time for preterm delivery and low birthweight caused by adverse effects of coal combustion toxics. Early Hum Dev. 2004;80:115–23. doi: 10.1016/j.earlhumdev.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, Wang Q, Cook TJ, Knipp GT. Effect of placental fatty acid metabolism and regulation by peroxisome proliferator activated receptor on pregnancy and fetal outcomes. J Pharm Sci. 2007;96:2582–606. doi: 10.1002/jps.20973. [DOI] [PubMed] [Google Scholar]
- 32.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–65. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Cui H, Cruz-Correa M, Giardiello FM, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–5. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 34.Guaschino S, Seta FD, Piccoli M, Maso G, Alberico S. Aetiology of preterm labour: bacterial vaginosis. BJOG: An International Journal of Obstetrics & Gynaecology. 2006;113:46–51. doi: 10.1111/j.1471-0528.2006.01122.x. [DOI] [PubMed] [Google Scholar]
- 35.Caldji C, Diorio J, Meaney MJ. Variations in maternal care in infancy regulate the development of stress reactivity. Biol Psychiatry. 2000;48:1164–74. doi: 10.1016/s0006-3223(00)01084-2. [DOI] [PubMed] [Google Scholar]
- 36.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsen J, Melbye M, Olsen SF, et al. The Danish National Birth Cohort - its background, structure and aim. Scandinavian journal of public health. 2001;29:300–7. doi: 10.1177/14034948010290040201. [DOI] [PubMed] [Google Scholar]
- 38.Magnus P, Irgens LM, Haug K, et al. Cohort profile: The Norwegian Mother and Child Cohort Study (MoBa) Int J Epidemiol. 2006;35:1146–50. doi: 10.1093/ije/dyl170. [DOI] [PubMed] [Google Scholar]
- 39.Rønningen K, Paltiel L, Meltzer H, et al. The biobank of the Norwegian mother and child cohort Study: A resource for the next 100 years. Eur J Epidemiol. 2006;21:619–25. doi: 10.1007/s10654-006-9041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]


