Abstract
CRISPR interference confers adaptive, sequence-based immunity against viruses and plasmids and is specified by CRISPR RNAs (crRNAs) that are transcribed and processed from spacer-repeat units. Pre-crRNA processing is essential for CRISPR interference in all systems studied thus far. Here, our studies of crRNA biogenesis and CRISPR interference in naturally competent Neisseria spp. reveal a unique crRNA maturation pathway in which crRNAs are transcribed from promoters that are embedded within each repeat, yielding crRNA 5’ ends formed by transcription and not by processing. Although crRNA 3’ end formation involves RNase III and trans-encoded tracrRNA, as in other Type II CRISPR systems, this processing is dispensable for interference. The meningococcal pathway is the most streamlined CRISPR/cas system characterized to date. Endogenous CRISPR spacers limit natural transformation, which is the primary source of genetic variation that contributes to immune evasion, antibiotic resistance, and virulence in the human pathogen N. meningitidis.
Keywords: Neisseria, CRISPR/Cas, dRNA-seq, RNA surveillance system, pathogen, RNA processing, crRNA, Cas9, Type II-C
INTRODUCTION
Clustered, regularly interspaced, short palindromic repeat (CRISPR) loci confer sequencebased, adaptive immunity against virus infection and plasmid conjugation in bacteria and archaea (Haft et al., 2005; Makarova et al., 2006; Barrangou et al., 2007; Terns and Terns, 2011; Wiedenheft et al., 2012). CRISPRs consist of short repeats separated by similarly sized, non-repetitive sequences called spacers, which are derived from previously encountered invasive sequences such as viral genomes or plasmids (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005). CRISPR loci are transcribed as long CRISPR RNA (crRNA) precursors that are processed into small crRNAs (Brouns et al., 2008; Hale et al., 2008). Pre-crRNA transcription is generally driven by promoters within “leader” sequences outside of the CRISPR array. The crRNAs assemble with CRISPR-associated (Cas) proteins into complexes that cleave complementary “protospacer” sequences within invasive nucleic acids, a phenomenon known as CRISPR interference (Karginov and Hannon, 2010; Marraffini and Sontheimer, 2010; Terns and Terns, 2011; Wiedenheft et al., 2012). The sequence information in crRNAs is used to guide Cas complexes to their targets on viruses and plasmids, leading to their destruction (Barrangou et al., 2007; Brouns et al., 2008; Marraffini and Sontheimer, 2008; Hale et al., 2009; Garneau et al., 2010; Westra et al., 2012). Most CRISPR/Cas systems cleave incoming DNAs (Marraffini and Sontheimer, 2008; Garneau et al., 2010; Westra et al., 2012), though RNA-cleaving systems have also been identified (Hale et al., 2009, 2012; Zhang et al., 2012).
CRISPR/Cas systems have been classified into types I, II and III based primarily on their cas gene complement (Makarova et al., 2011a). Common to all of these three types is that the CRISPR array is transcribed as a multimeric pre-crRNA that is processed into crRNAs that each contain an individual spacer flanked on one or both sides by partial repeat sequences (Bhaya et al., 2011). However, the molecular events underlying processing dramatically differ. Whereas in Type I and III systems the processing enzymes are encoded within the CRISPR/cas locus, Type II systems use the host enzyme RNase III (encoded by the rnc gene) and a noncoding RNA called tracrRNA (Deltcheva et al., 2011). In Streptococcus pyogenes SF370, an rnc mutant abolishes the function of a Type II CRISPR/cas locus, indicating that pre-crRNA processing is essential (Deltcheva et al., 2011).
The importance of the Type II CRISPR/Cas pathway has been dramatically enhanced by its development into a system for RNA-guided DNA cleavage in vitro (Jinek et al., 2012) and genome editing in vivo (Jinek et al., 2013; Cho et al., 2013; Cong et al., 2013; DiCarlo et al., 2013; Hwang et al., 2013; Jiang et al., 2013; Mali et al., 2013). Our ability to exploit this new technology further will depend on a deeper understanding of the underlying molecular mechanisms, and will be increased by the characterization of systems that are as simplified and streamlined as possible. Type II CRISPR/Cas loci, which are found in bacteria but not archaea, usually contain four cas genes: cas1, cas2, cas9, and either csn2 (subtype II-A) or cas4 (subtype II-B) (Makarova et al., 2011b). Cas9 is the effector protein for the interference function of existing spacer sequences in the CRISPR array (Sapranauskas et al., 2011; Gasiunas et al., 2012; Jinek et al., 2012), whereas the other proteins are thought to be involved in the acquisition of new CRISPR spacers. The tracrRNA is essential for crRNA-directed DNA recognition and Cas9-catalyzed DNA cleavage in vitro, even with crRNAs that bypass processing (Jinek et al., 2012). DNA targeting in both Type I and Type II systems requires not only crRNA/target complementarity but also a protospacer adjacent motif (PAM), which is a short (2–5 nt), conserved sequence just outside of the protospacer (Deveau et al., 2008; Horvath et al., 2008; Mojica et al., 2009; Semenova et al., 2011; Sashital et al., 2012).
Although CRISPR interference was originally defined as a phage defense pathway, CRISPR/Cas systems are now understood to play a broader role in limiting horizontal gene transfer (HGT) (Marraffini and Sontheimer, 2008). The three primary routes of HGT are transformation, conjugation, and phage transduction, and the latter two are well established as being subject to interference by naturally occurring spacers. Artificial means of transformation (e.g. electroporation) can also be blocked by CRISPR interference (Marraffini and Sontheimer, 2008; Deltcheva et al., 2011; Sapranauskas et al., 2011; Semenova et al., 2011), though natural transformation uses a very different DNA uptake process (Chen et al., 2005). An engineered spacer can prevent natural transformation specified by an S. pyogenes CRISPR/cas locus transplanted into Streptococcus pneumonia (Bikard et al., 2012). However, although this artificial system suggests that natural CRISPR/Cas contexts may do likewise, the fundamental question of whether native CRISPR/Cas systems play a role in natural transformation remains to be addressed.
Strains from the genus Neisseria serve as paradigms for natural transformation, as they are competent during all phases of their life cycle and use this process for frequent genetic exchange (Hamilton and Dillard, 2006). Although functional CRISPR/cas systems have not been identified in Neisseria gonorrhoeae, some strains of Neisseria lactamica and Neisseria meningitidis carry apparent Type II CRISPR/cas loci (Grissa et al., 2007). Meningococci are obligate human commensals that can invade the bloodstream and cerebrospinal fluid (Bratcher et al., 2012), and meningococcal disease is responsible for tens of thousands of deaths per year (Stephens et al., 2007).
Here we characterize the CRISPR pathway in neisseriae and find that it exhibits several unique features, most notably a streamlined functional architecture that includes a previously unknown, processing-independent mode of crRNA biogenesis. Furthermore, naturally occurring spacers match sequences from other Neisseria genomes, including a prophage-like meningococcal disease-associated (MDA) island that correlates with invasiveness and pathogenicity (Bille et al., 2005, 2008). We find that a native meningococcal CRISPR/cas locus prevents natural transformation of spacer-matched sequences, suggesting that it can limit the horizontal spread of virulence genes.
RESULTS
dRNA-seq reveals that each repeat in the Neisseria CRISPR carries its own promoter
We analysed all 19 sequenced Neisseria genomes available in the NCBI database (fifteen from N. meningitidis, three from N. gonorrhoeae, and one from N. lactamica) using CRISPRFinder (Grissa et al., 2007) or CRISPRi (Rousseau et al., 2009). We identified seven putative Type II CRISPR/cas loci: six in N. meningitidis strains, and one in N. lactamica 020-06. All were highly similar, and unlike other Type II loci characterized previously (Barrangou et al., 2007; Deltcheva et al., 2011; Magadán et al., 2012), included a set of only three predicted protein-coding genes (cas9, cas1 and cas2) but neither csn2 nor cas4. To examine the expression status of a representative locus, we performed our recently developed dRNA-seq approach (Sharma et al., 2010) on N. lactamica 020-06. We prepared two cDNA libraries from total RNA using a strategy that allows us to distinguish between transcripts with either primary or processed 5’ ends: one library is generated from untreated RNA, whereas the other is treated with terminator exonuclease (TEX), which specifically degrades RNAs with 5’-monophosphate ends (including the abundant rRNAs and tRNAs) that are formed by processing. Primary transcripts with 5’-triphosphate ends survive TEX treatment, resulting in their relative enrichment in the TEX+ library.
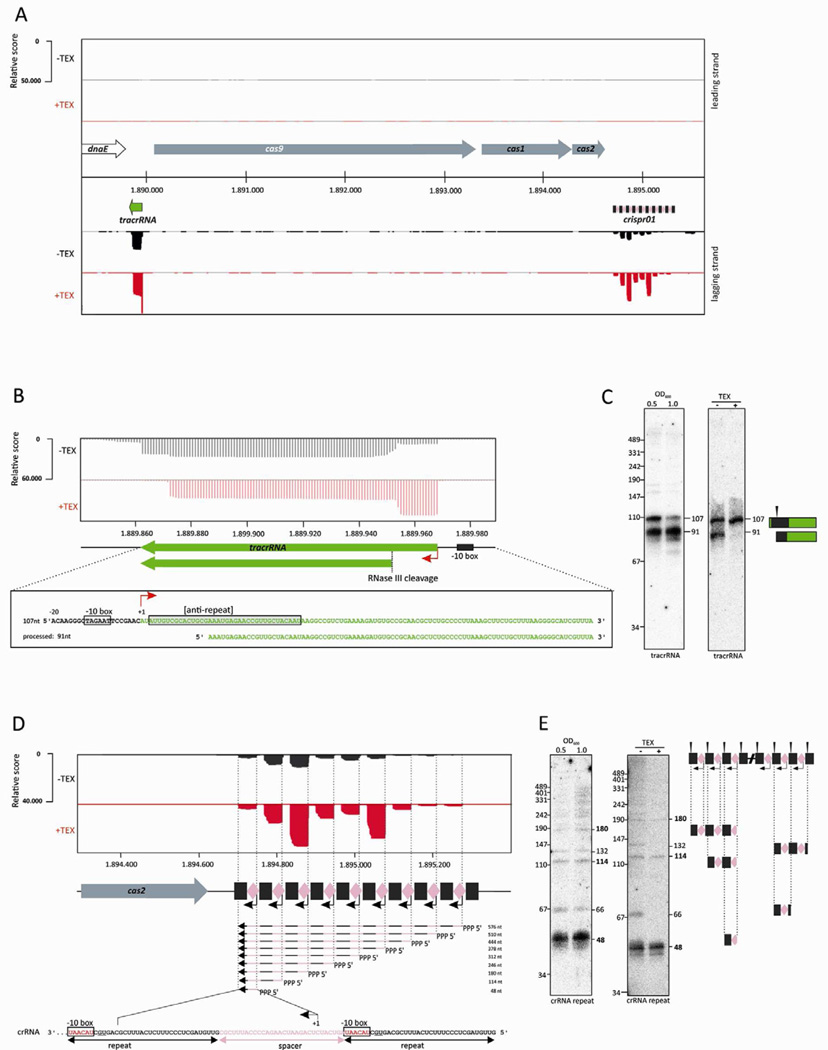
Our global mapping of cDNA reads identified a tracrRNA and small crRNAs as highly abundant classes of transcripts (Figure 1A), which suggests a highly active CRISPR/Cas system. Reads obtained from the tracrRNA locus reveal the existence of two tracrRNA forms [107 nucleotides (nt) and 91 nt] (Figure 1B). The treatment with TEX eliminated the shorter tracrRNA reads, which indicates that they are products of processing as observed in a Type II-A CRISPR system (Deltcheva et al., 2011). These sequencing results were confirmed by Northern analyses (Figure 1C). In striking contrast, crRNAs were enriched rather than depleted in the TEX-treated libraries (Figure 1D), suggesting that formation of many if not most crRNA 5’ ends is processing-independent. TEX treatment did affect the relative amounts of individual crRNAs; for example, crRNAs from spacers 4, 6 and 9 exhibited very similar read counts in the untreated sample but varied over a 5-fold range in the TEX-treated sample. These results suggest that the crRNA pool contains some 5’-monophosphorylated crRNAs, the fraction of which varies from spacer to spacer. Northern blots confirmed the resistance of a population of mature, ~48-nt crRNAs, as well as a subset of longer crRNA precursors, to TEX digestion (Figure 1E). This crRNA profile indicates a different mode of crRNA biogenesis from that reported for other Type II systems (Deltcheva et al., 2011). Intriguingly, we noted that the terminal nine nucleotides of each CRISPR repeat exhibit sequence hallmarks of an extended −10 box promoter element (Hook-Barnard and Hinton, 2007) and that the TEX-resistant crRNA 5’ ends map 9–10 nt downstream from the center of each such element (Figure 1D).
Figure 1. A newly identified mode of crRNA biogenesis in Neisseria lactamica.
(A) Differential RNA-seq (dRNA-seq)-based analysis of the minimal CRISPR/Cas system in N. lactamica 020-06 reveals expression of tracrRNA and crRNAs. Approximately three million cDNAs from untreated and TEX-treated RNA were sequenced and mapped to the genome. Read counts are plotted here for the CRISPR/cas locus. Both strands of all libraries were adjusted to the same scale (maximum of 50,000 for leading strand; minimum of −50,000 for lagging strand) that reflects a relative expression score. The horizontal black bar encompasses the genomic organization of the CRISPR/cas locus. Black rectangles, CRISPR repeats; pink diamonds, CRISPR spacers; green arrow, tracrRNA; gray arrows, cas genes. The number of reads obtained for tracrRNA and crRNAs are in the range of 40,000 to 50,000 each, which is comparable to the range we observe for other high-abundance classes of RNAs.
(B) Top: dRNA-seq data were mapped onto the genomic region corresponding to the tracrRNA gene. Expression scores at each position were adjusted to the same relative scale. Bottom: sequence of the full-length 107-nt form and processed 91-nt form of tracrRNA. The red arrow indicates the primary tracrRNA transcription start site (TSS) based on enrichment in the TEX+ libraries. Boxes denote the extended −10 promoter element and the anti-CRISPR-repeat region of the tracrRNA.
(C) Northern analysis of N. lactamica 020-06 tracrRNA. OD600 (left panel) and TEX treatment (right panel) is denoted at the top of each lane. Processed and unprocessed tracrRNAs are schematized on the right, with the anti-repeat region in black and the RNase III processing site indicated with an arrowhead.
(D) Top: dRNA-seq data were mapped onto the genomic region corresponding to the CRISPR array. Expression scores at each position were adjusted to the same relative scale. Middle: the primary TSS of each CRISPR spacer-repeat unit (based on enrichment in the TEX+ libraries) is indicated by a black arrow. Primary CRISPR transcripts of different lengths with likely 5’ triphosphates are indicated by black/pink arrows. Bottom: The sequence of spacer 4 and its flanking CRISPR repeats, with the putative extended −10 box (consensus sequence 5’-tgnTACAAT-3’) in each single repeat highlighted in red.
(E) Northern analysis of N. lactamica 020-06 crRNA, using a probe complementary to the CRISPR repeat. OD600 (left panel) and TEX treatment (right panel) is denoted at the top of each lane. Candidate monomeric, dimeric and trimeric crRNAs are schematized on the right, each of a predicted size consistent with bands observed on the blots.
crRNA biogenesis in Neisseria lactamica depends on single promoter elements in each CRISPR repeat
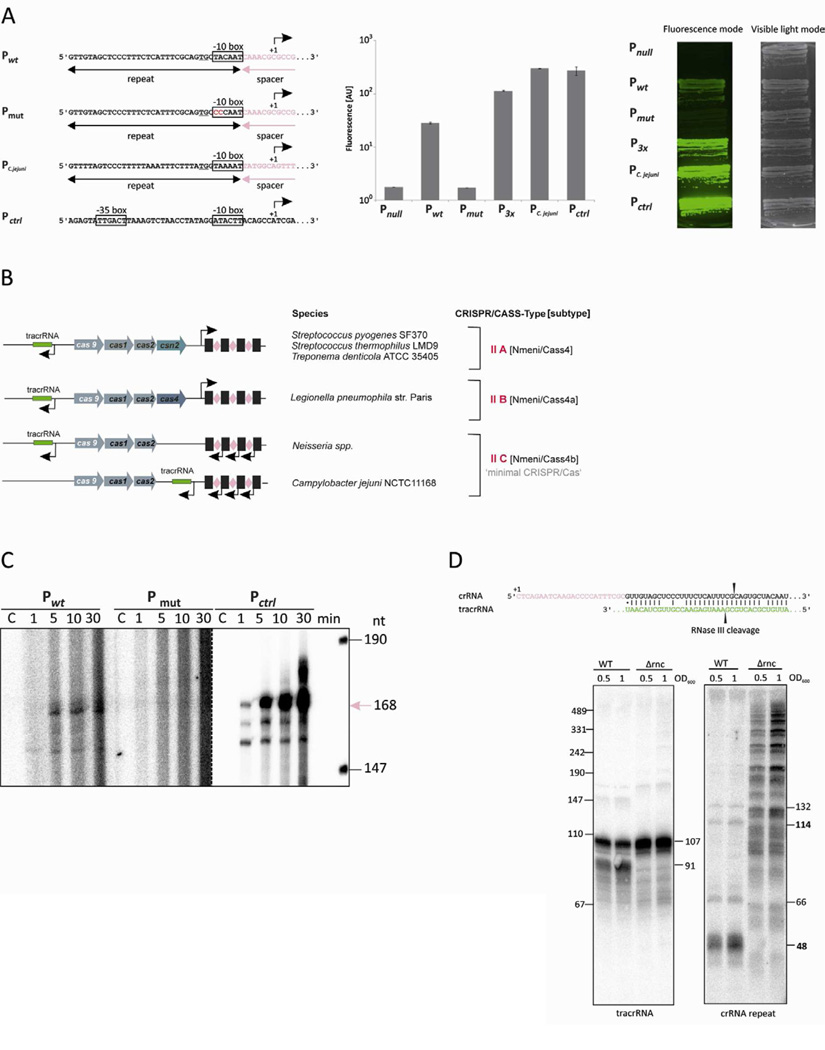
The dRNA-seq results and −10 box similarity suggest that in N. Iactamica 020-06, each CRISPR repeat carries its own minimal promoter, and that pre-crRNA transcription initiates independently within each spacer. As an initial test of this hypothesis, we designed a series of transcriptional green fluorescent protein (gfp) fusion constructs containing either single or multiple CRISPR repeats, introduced these constructs into E. coli, and analysed cellular GFP fluorescence. As shown in Figure 2A, the gfp fusion construct with a wild-type CRISPR repeat led to robust cellular fluorescence, whereas a two-nt substitution in the extended −10 promoter TG motif (Hook-Barnard and Hinton, 2007) reduced gfp expression to background levels. Constructs with gfp fused to three CRISPR repeat-spacer units increased the fluorescence signal almost two-fold, consistent with the possibility of increased transcription from the multiple repeats. We obtained similar results using repeat sequences from a Type II CRISPR/cas locus in Campylobacter jejunii NCTC11168 (Figure 2A); this locus, like that of N. lactamica 020-06, also contains only cas9, cas1 and cas2 and has CRISPR repeats that include an extended −10 box consensus. Thus, our gfp reporter assays prove that the putative promoter elements in each N. lactamica repeat are indeed active in bacterial cells, and are likely also present in some other Type II CRISPR/cas loci with similar, minimized cas gene content (Figure 2B and Table S1). The Neisseria and Campylobacter systems are also unusual in that the crRNAs and cas genes are transcribed in the opposite direction, and the CRISPR arrays lack recognizable “leader” sequences with external promoters. Based on these considerations as well as independent phylogenetic analyses (Koonin and Makarova, 2013; Chylinski et al., 2013), we now consider these variant Type II CRISPR/Cas loci as members of a distinct and newly defined subtype, Type II-C (Figure 2B).
Figure 2. CRISPR repeats contain active promoters that form the 5’ ends of mature crRNAs, whereas RNase III processing forms crRNA 3’ ends.
(A) Left panel: Promoter-element-containing sequences used for gfp fusions. Plasmids included a wild-type (wt) and mutated (mut) N. lactamica 020-06 CRISPR repeat (pNH13 and pNH14, respectively), a C. jejunii NCTC11168 CRISPR repeat (pNH18), and a positive control promoter from T7 phage A1. The promoterless control gfp construct (pAS00046) and a construct with three wildtype N. lactamica 020-06 CRISPR repeats (pNH17) are not shown. Promoter elements are indicated by boxes, with mutated sequences colored in red. Middle panel: Flow-cytometric fluorescence intensity measurements of cells containing the transcriptional gfp fusions described in the left panel. Fluorescence values are expressed in arbitrary units (AU). Error bars indicate standard deviation for three independent biological replicates. Right panel: Fluorescence images of transcriptional gfp fusion strains grown on agar plates. The right image was captured in the visible light mode; the left image shows the same plate in fluorescence mode (460 nm excitation, 510 nm emission).
(B) Classification of Type II CRISPR/cas loci. The genomic organization of representative Type II-A or Type II-B loci, as defined previously, is given on top. Below are two CRISPR/cas loci (including from neisseriae) from the newly defined Subtype II-C.
(C) Gel electrophoresis of radiolabeled RNAs from in vitro transcription reactions using linear DNA templates from a subset of the CRISPR repeat-containing sequences given in (A). Full- length run-off transcripts (168 nt) are denoted by the pink arrow. The area to the right of the dotted line was imaged at lower contrast to avoid overexposure. See also Figure S1.
(D) Top: Base pairing between a primary crRNA and tracrRNA. RNase III cleavage sites inferred from dRNA-seq are indicated by black arrows. Bottom: Northern analysis of total RNA from N. meningitidis WUE2594 and its Δrnc derivative during mid-log and early stationary phase, probed for tracrRNA (left) and crRNAs (right).
To obtain additional proof that each N. lactamica CRISPR repeat carries its own promoter element, we used purified E. coli σ70 RNA polymerase (RNAP) holoenzyme in in vitro transcription assays with linear DNA templates containing either a wild-type or a mutant repeat. A transcript of the expected length (168 nt) was obtained with the wild-type CRISPR repeat template (and with a control −10/−35 promoter construct), but not with the mutated repeat (Figure 2C and Figure S1). This result demonstrated that the extended −10 motif in the N. lactamica CRISPR repeat was recognized even by the heterologous E. coli σ70 RNAP holoenzyme. These data show that crRNAs in N. lactamica exhibit a unique mode of biogenesis involving transcription from extended −10 class promoters located within CRISPR repeats.
RNase III is involved in 3’ end formation of Neisseria crRNAs
The observation that crRNA 5’ ends correspond to sites of transcription initiation in N. lactamica suggests a reduced dependence on processing relative to other CRISPR systems. To determine whether this reduced dependence extends to crRNA 3’ end formation as well or if 3’ processing still occurs, and to extend our studies to other Neisseria strains, we deleted the rnc gene (which encodes RNase III) in N. meningitidis WUE2594, and then compared the tracrRNA and crRNA populations from this Δrnc mutant with wild-type bacteria by northern analysis. As shown in Figure 2D, the Δrnc strain exhibited a complete loss of the shorter (91 nt), TEX-sensitive tracrRNA, indicating that the 5’ end of this RNA is generated by RNase III-dependent processing as observed previously in a Type II-A system (Deltcheva et al., 2011). We also observed dramatic differences in the crRNA population in the Δrnc mutant: the 48-nt population is virtually abolished, and longer pre-crRNAs accumulate. This result strongly suggests that Neisseria spp. crRNA 3’ end formation depends upon RNase III rather than direct transcription termination.
Repeat/spacer organization and potential targets of Neisseria Type II-C CRISPR loci
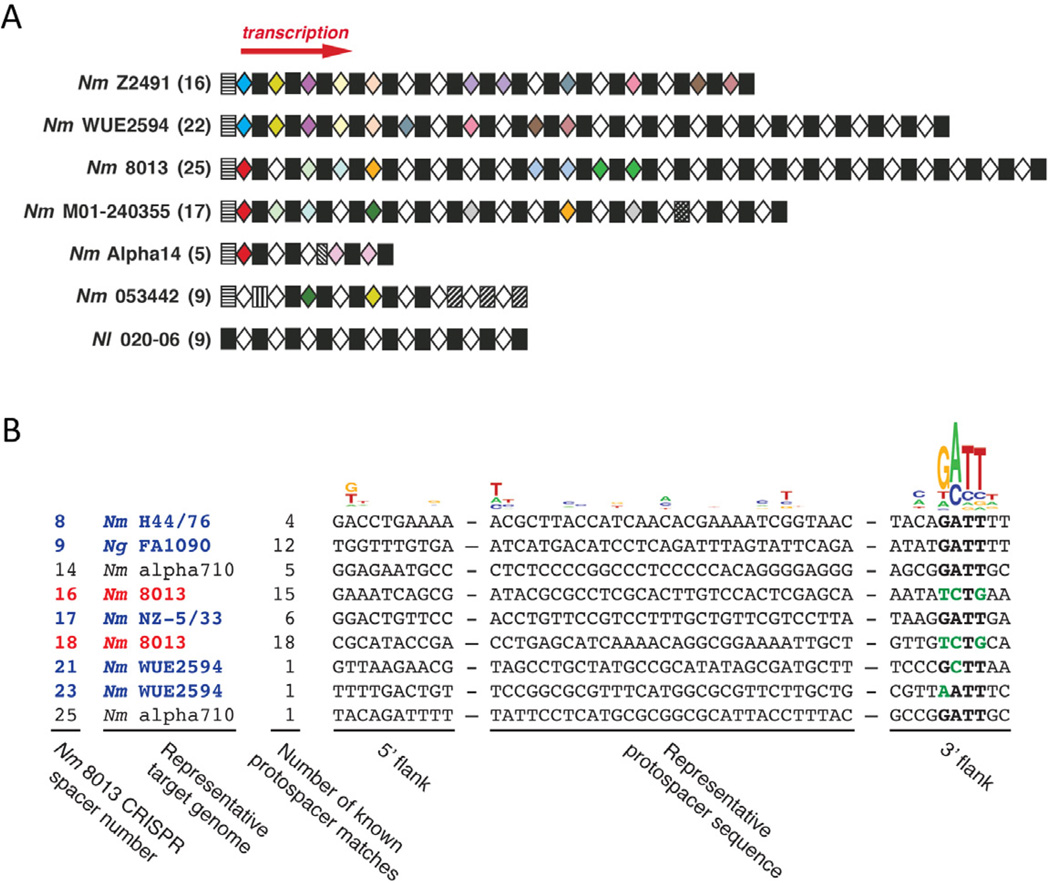
Having defined unique features of CRISPR/Cas systems in neisseriae, we turned our attention towards functional analyses, beginning with an examination of CRISPR organization and spacer content. Of the 103 spacers found in the seven CRISPR-positive genomes (Figure 3A and Table S2), one is 29 nt long while all others are 30 nt. All seven CRISPRs have the same 36-nt repeat consensus, with only a few repeats that deviate from this consensus (Figure 3A and Table S3). Intriguingly, the polarity of spacer conservation is opposite to that generally observed in other CRISPR loci. Conserved spacers that are shared among multiple strains in Neisseria spp. (color-coded in Figure 3A) are enriched at the upstream end of the array (relative to the direction of transcription) but are far less common at the downstream end. In contrast, other CRISPRs described thus far have the most recently derived and therefore least conserved spacers at the upstream end, i.e., proximal to the promoter (Makarova et al., 2011b). This observation suggests that new spacer acquisition in neisseriae, and perhaps in other Type II-C loci, occurs at the downstream end. In addition, deviations from the repeat consensus are most common at the upstream end in neisseriae (Table S3), whereas other CRISPRs most frequently exhibit the opposite tendency. Although technical limitations have thus far precluded direct tests of spacer acquisition in neisseriae, several considerations make it likely that these CRISPRs are competent for the adaptive component of the pathway. First, Type II-C loci are relatively common, with many more apparent examples among sequenced bacterial genomes than Type II-B loci (Chylinski et al., 2013). It is unlikely that type II-C systems would be so widespread if they were unable to provide adaptive protection to their hosts. Second, despite the conservation of the Type II-C loci in closely related Neisseria strains, there are many differences in spacer content (Figure 3A). This implies that these CRISPR loci are fluid, i.e. can adapt by adding and deleting spacers. Third, reports from other systems indicate that cas1 and cas2 can suffice for spacer acquisition (Yosef et al., 2012). It is therefore plausible that cas1 and cas2 likewise suffice for adaptation in Type II-C systems.
Figure 3. CRISPR organization in Neisseria.
(A) Schematic representation of CRISPR loci from seven different Neisseria strains. Strain names are indicated (Nm N. meningitidis; Nl N. lactamica) with the total number of spacers in each strain shown in parentheses. The red arrow indicates the direction of crRNA transcription. Repeats and spacers are shown as rectangles and diamonds, respectively. Unique spacers are in white, and non-unique spacers are color-coded. Black rectangles indicate repeats that match the consensus, whereas patterned rectangles are variant repeats (see Tables S2 and S3).
(B) Potential natural targets for N. meningitidis 8013 spacers. 9 out of 25 spacers match varying numbers of Neisseria genomes. For each spacer, its number in the 8013 array, the quantity of known protospacer matches, and a representative target genome (Nm N. meningitidis; Ng N. gonorrhoea) are listed. Protospacers and 10 flanking nts (on both sides) from the representative target genomes are aligned. Sequence similarities are indicated at the top, revealing the 5’-GATT-3’ PAM consensus 5–8 nts 3’ of the protospacer. The WebLogo is derived from the alignment of all Neisseria-matched protospacers, not just those shown here that match spacers from the N. meningitidis 8013 CRISPR. The PAM regions in the putative targets are in bold, with non-consensus nucleotides in green. Potential self-targeting spacers are in red, and spacers with possible prophage-like targets are in blue. See also Figure S2.
BLASTN searches with the 83 unique spacer sequences for similar sequences in the NCBI database allowed us to identify at least one potential target for 35 (~42%) of them. For simplicity we required either a perfect match, or at most a single mismatch in the 10 protospacer nucleotides furthest from the PAM (i.e., well outside of the presumptive “seed” sequence that requires perfect complementarity for interference) (Sapranauskas et al., 2011; Semenova et al., 2011; Wiedenheft et al., 2011; Jinek et al., 2012). Figure 3B shows representative protospacer sequences that match CRISPR spacers from N. meningitidis strain 8013; representative protospacers that match spacers from all strains are shown in Figure S2. Protospacer alignments reveal an apparent PAM of 5’-NNNNGATT-3’ (Figure 3B and Figure S2). Of the 325 distinct candidate protospacers that match these 35 CRISPR spacers, all are in Neisseria sequences: 248 (76%) in N. meningitidis genomes, 69 (21%) in N. gonorrhoeae genomes, 6 (2%) in N. lactamica plasmids, and 1 each in N. meningitidis and N. flavescens plasmids (Table S4; also see Supplementary Text). In some cases (shown in red in Figure 3B and Figure S2), potential protospacers are present in the same genome as the targeting CRISPR spacer, suggesting the potential for autoimmunity (Stern et al., 2010). However, these protospacers all include significant deviations from the PAM consensus, perhaps explaining the apparent lack of self-targeting that is implied by the persistence of the matching protospacer. Intriguingly, 22 out of 35 CRISPR spacers (shown in blue in Figure 3B and Figure S2) with identifiable targets match at least one potential prophage sequence (Table S5), including the meningococcal disease-associated (MDA) island that is associated with invasiveness and pathogenicity in young adult patients (Bille et al., 2005, 2008).
CRISPR interference blocks natural transformation in N. meningitidis
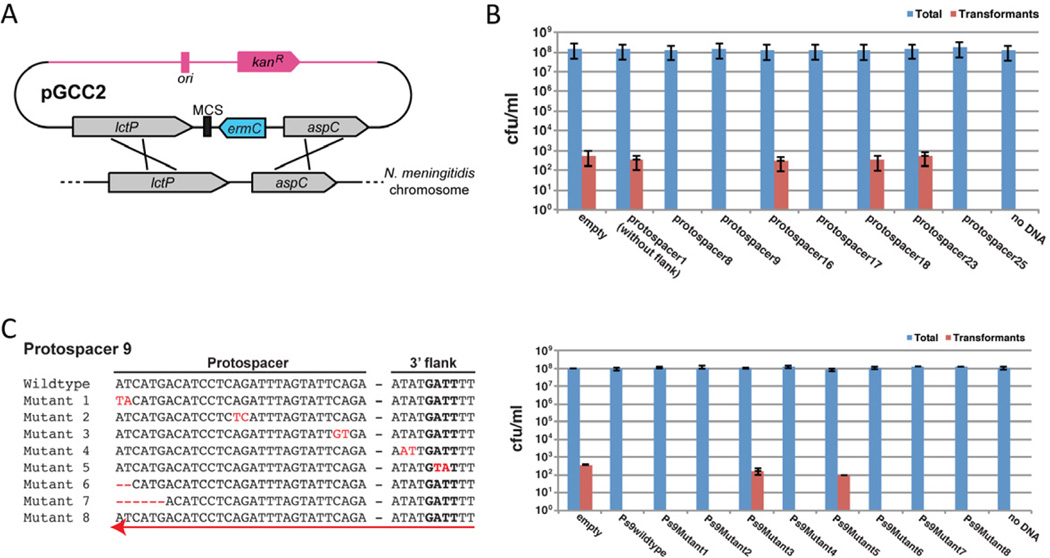
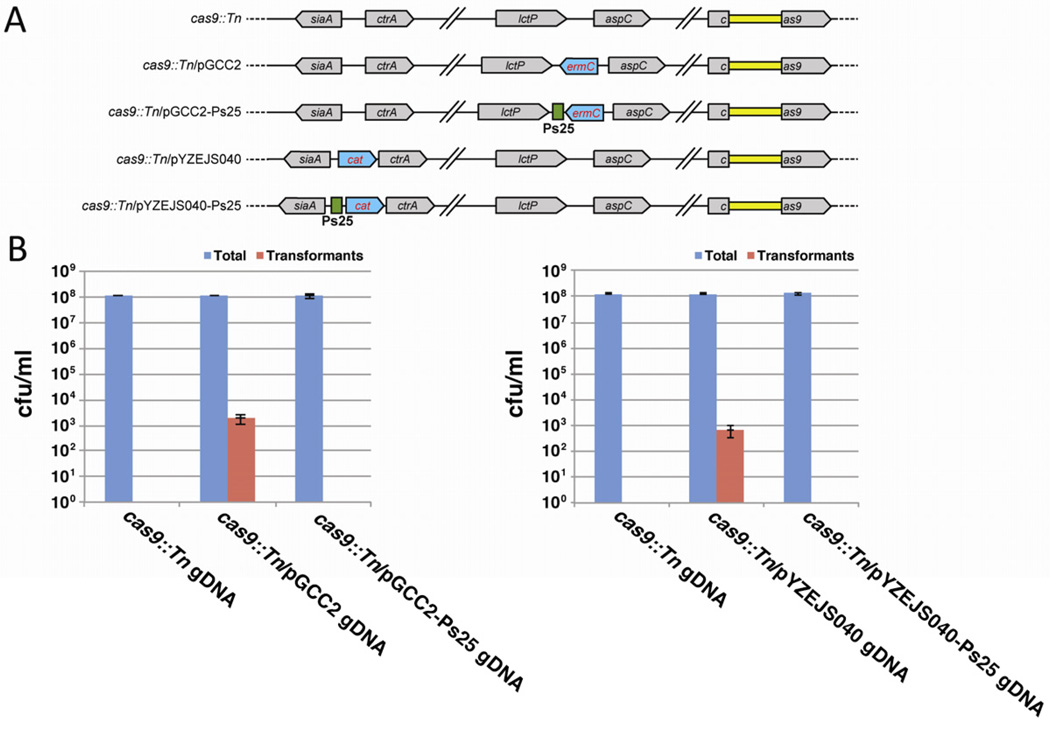
The preponderance of protospacers in Neisseria spp. genomes suggests that the CRISPR/cas loci could interfere with natural transformation. For our functional analyses addressing this possibility, we focused on N. meningitidis 8013, primarily because it exhibits the most robust transformation competence in our hands. For transformation assays we used the vector pGCC2, which contains an erythromycin resistance gene (ermC) and polylinker inserted into sequences from the gonococcal lctP/aspC locus (Figure 4A). Upon transformation into N. meningitidis, homologous recombination into the meningococcal lctP/aspC locus leads to ermC insertion and erythromycin-resistant (ErmR) transformants. We selected potential natural target sequences, including ten or more nucleotides on both sides of the protospacer, for seven of the spacers of N. meningitidis 8013, namely spacers 8, 9, 16, 17, 18, 23, and 25 (Figure 3B and Figure S3). Upon cloning them into pGCC2, the resulting vectors were used in liquid-medium natural transformation assays into wild-type 8013 cells, and transformation frequencies (antibiotic-resistant cfu ml−1/total cfu ml−1) were determined. For comparison we also cloned a spacer 1 target without any flanking sequences. The results are shown in Figure 4B. Empty pGCC2 exhibited a transformation frequency of 3.9 × 10−6 (Table S6), consistent with previous reports (Rusniok et al., 2009). Plasmids carrying targets for spacers 1, 16, 18 and 23 all exhibited transformation frequencies of 2–4 × 10−6 (Table S6), i.e. comparable to that of the empty vector. The cloned protospacers in these plasmids either lack flanking Neisseria sequences or have flanking sequences that deviate significantly from the PAM consensus (Figure 3B and Figure S3). In contrast, protospacers 8, 9, 17 and 25, all four of which have flanking sequences that conform to the PAM consensus, consistently failed to yield transformants, indicating that they likely elicited CRISPR interference. Protospacers matching CRISPR spacers 9 and 25 (Figure S3) cloned into a different transformation vector, pYZEJS040 (which confers chloramphenicol resistance), for targeted integration into the distinct capsule locus also yielded no transformants (Figure S4), demonstrating that the observed effect was independent of the vector, the integration locus, and the selectable marker.
Figure 4. Natural transformation is limited by the native Neissiera CRISPR/Cas system.
(A) Schematic representation of integrational vector pGCC2, and recombination between pGCC2 and the meningococcal chromosome (Tobiason and Seifert, 2010). Grey arrows, lctP and aspC genes; blue arrow, ermC gene; pink, regions required for maintenance in E. coli; black box, multiple cloning site. Individual elements are not drawn to scale.
(B) pGCC2 derivatives with potential targets for different N. meningitidis 8013 spacers (1, 8, 9, 16, 17, 18, 23, and 25) were tested by natural transformation assays using wild-type 8013 as the recipient strain. The data show log-scale plots of colony-forming units (cfu) per ml [mean ± s.e.m. (standard error of the mean) for three independent biological replicates] for total cells (blue bars) and ErmR transformants (red bars) from three independent experiments. See also Figure S3.
(C) Left panel, sequences of a series of mutations in the pGCC2 derivative carrying the 350 nt target for spacer 9. Sequence alterations are in red. The red arrow indicates reversed orientation of the target sequence in the plasmid. Right panel, pGCC2 constructs containing the spacer 9 target mutations are tested by natural transformation into wild-type N. meningitidis 8013. Data are presented as in (B). Error bars represent s.e.m. for three independent biological replicates.
To examine targeting requirements further, we generated a series of mutations in the protospacer or flanking sequences of the pGCC2-derived plasmid targeted by spacer 9 (Figure 4C). Substitutions of two consecutive nucleotides within the PAM or the seed sequence of the protospacer yielded plasmids with transformation frequencies comparable to that of the empty vector (mutants 3 and 5; Figure 4C and Table S6), indicating that interference was abolished. In contrast, two-nucleotide substitutions in a non-PAM flanking region, substitutions in non-seed protospacer positions, small deletions at the PAM-distal end of the spacer, or a wild-type protospacer cloned in the opposite orientation all had no effect on interference (mutants 1, 2, 4, 6, 7 and 8). All of these observations are consistent with previously defined characteristics of functional Type II CRISPR/Cas systems (Deveau et al., 2008; Sapranauskas et al., 2011; Gasiunas et al., 2012; Jinek et al., 2012), indicating that CRISPR interference is indeed responsible for the observed effects on transformation frequencies. The effects of single-nucleotide mutations in this apparent 4-nt PAM remain to be defined.
Genetic analysis of the N. meningitidis CRISPR/cas locus
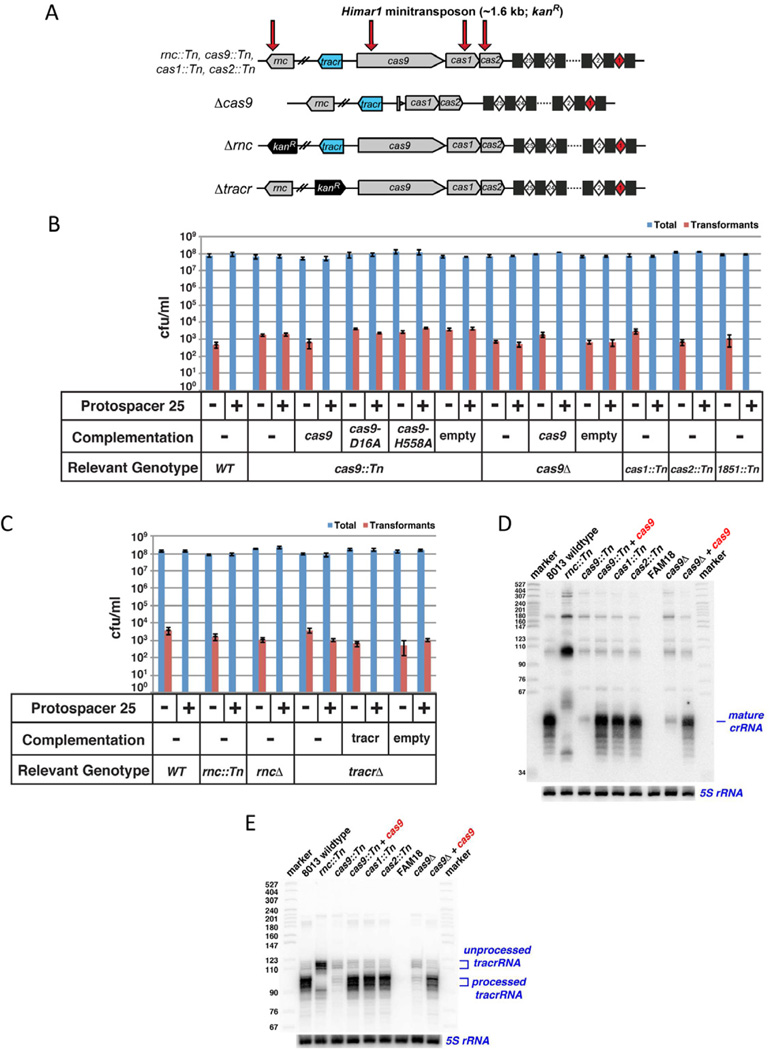
In other Type II CRISPR/Cas systems, Cas9 is the only Cas protein that is necessary for interference specified by existing spacers (Barrangou et al., 2007; Deltcheva et al., 2011; Sapranauskas et al., 2011; Jinek et al., 2012). To investigate if Type II-C CRISPR/Cas systems exhibit the same Cas protein requirements, we introduced transposon insertion mutations in the three cas genes – cas1, cas2, and cas9 – in N. meningitidis 8013 (Figure 5A). We also generated an unmarked, in-frame cas9 deletion strain to avoid potential polar effects and to generate a guaranteed null allele (Figure 5A). We transformed wild-type and mutant strains with a pYZEJS040 construct carrying protospacer 25 (as in Figure S4B), and compared their transformation frequencies with those of empty pYZEJS040. As expected, the empty vector readily transformed all strains, with transformation frequencies in the range of ~0.5–7 × 10−5 (Figure 5B and Table S6). No transformants were observed when the protospacer 25 construct was used with wild-type cells, indicating effective CRISPR interference. Similarly, transposon insertion mutants in cas1, cas2, or a control irrelevant gene (cas1::Tn, cas2::Tn and 1851::Tn, respectively) exhibited complete interference, consistent with previous results in Type II-A systems (Sapranauskas et al., 2011). In contrast, CRISPR function is abolished in both the transposon-induced (cas9::Tn) and deletion (Δcas9) mutations in cas9. The CRISPR interference defect of both alleles could be complemented with wild-type cas9 under the control of its native promoter (integrated chromosomally via pGCC2), whereas empty pGCC2 had no effect (Figure 5B).
Figure 5. Type II-C CRISPR interference requires Cas9 and tracrRNA but is independent of RNase III-mediated processing.
(A) Schematic representations of N. meningitidis 8013 mutant strains. Grey arrows, cas1 cas2 cas9 and rnc genes; blue arrow, tracrRNA gene; black arrow, kanamycin resistance gene; squares and diamonds, CRISPR repeats and spacers (respectively), colored as in Figure 3A. Red arrows indicate transposon insertions in the rnc::Tn cas9::Tn cas1::Tn, and cas2::Tn mutants.
(B, C) pYZEJS040 (−) and its protospacer 25-containing derivative (+) were tested by natural transformation assays using N. meningitidis 8013 and its mutant derivatives as recipients. Relevant genotypes as well as the presence or absence of pGCC2-mediated cas9 complementation are given at the bottom. The data show cfu/ml (log scale, mean ± s.e.m. for three independent biological replicates) for total cells (blue bars) and chloramphenicol-resistant transformants (red bars). See also Figure S4.
(D) Total RNA from the indicated strains were subjected to northern analysis using a probe complementary to spacer 25 (top). In the lower panel, the same blot was probed for 5S RNA as a loading control. See also Figure S5.
(E) As in (D), except that a probe specific for tracrRNA was used.
Previous studies of cas9 orthologs from S. thermophilus and S. pyogenes identified two distinct active sites in RuvC-like and HNH nuclease motifs that are important for Cas9 function in vivo and in vitro (Sapranauskas et al., 2011; Gasiunas et al., 2012; Jinek et al., 2012). We engineered alanine mutants in corresponding catalytic residues (D16 in the RuvC domain and H588 in the HNH domain) and tested the abilities of both single mutants to complement the interference defect of the cas9::Tn mutant. Both alanine mutants failed to restore interference (Figure 5B). Altogether these analyses demonstrate that the Neisseria Type II-C CRISPR/Cas system requires cas9 but not cas1 or cas2 for interference of natural transformation, and that the presence of intact RuvC-like and HNH motifs are essential for cas9 function.
RNase III-catalysed pre-crRNA processing is dispensable for Type II-C CRISPR interference
Two additional loci – tracrRNA and rnc (the gene encoding RNase III) – have been shown to be essential for crRNA processing and interference in the Type II-A system of S. pyogenes SF370 (Deltcheva et al., 2011). The unique Neisseria biogenesis pathway described above, in which repeat-driven promoters yield crRNAs with unprocessed 5’ ends, raises questions about the roles of tracrRNA and RNase III in this Type II-C system. To examine this issue, we generated N. meningitidis 8013 derivatives carrying a transposon-induced allele of rnc, or a complete deletion of either rnc or tracrRNA (Figure 5A). These strains were tested in liquid transformation assays as described above, using pYZEJS040 and its protospacer 25- bearing derivative. The results are shown in Figure 5C. The Δ tracrRNA strain was completely defective in CRISPR interference, but the defect was restored upon integration of a tracrRNA gene with its native promoter in a distinct chromosomal locus. These results are consistent with the strict requirement for S. pyogenes tracrRNA for pre-crRNA processing and interference in vivo (Deltcheva et al., 2011), as well as crRNA-directed, Cas9-catalyzed DNA cleavage in vitro (Jinek et al., 2012) and in eukaryotes (Cong et al., 2013).
Intriguingly, despite the previously demonstrated importance of the tracrRNA as a guide for RNase III-mediated processing, we detected no interference defect in either the rnc::Tn or Δrnc mutants (Figure 5C). This result is in stark contrast to that observed in the Type II-A system in S. pyogenes SF370 (Deltcheva et al., 2011). The lack of an interference defect was observed with a vector that is targeted by an internal spacer (spacer 9; Table S6) as well as a terminal spacer (spacer 25; Figure 5C and Table S6). Northern analyses revealed clear processing defects in both crRNA (Figure 5D) and tracrRNA (Figure 5E) for the N. meningitidis 8013 rnc::Tn mutant, consistent with the results described above with a Δrnc mutant in the WUE2594 strain background (Figure 2D). Mature, 48 nt crRNAs are virtually absent in the rnc::Tn mutant, and longer precursors accumulate (Figure 5D and Figure S5). CrRNAs are also strongly depleted in cas9 mutants, but unlike in the rnc::Tn mutant, pre-crRNAs do not accumulate. The latter observation suggests that Cas9 is important for crRNA stability but not processing, or that Cas9 functions in processing and also stabilizes unprocessed precursors. We conclude that the N. meningitidis Type II-C CRISPR/Cas system, unlike all other CRISPR/Cas systems characterized to date, does not require pre-crRNA processing for interference activity. RNase III-catalyzed pre-crRNA processing occurs within the bacterial cell but is dispensable for interference. The tracrRNA requirement likely reflects its involvement in target DNA binding and cleavage by crRNA-programmed Cas9, as observed in vitro (Jinek et al., 2012).
CRISPR interference limits transformation by Neisseria genomic DNA
Plasmids are rare in N. meningitidis (van Passel et al., 2006), and Neisseria genomic DNA (gDNA) is thought to be the most frequent substrate for natural transformation (Hamilton and Dillard, 2006). To test whether our results with E. coli-isolated plasmids extend to Neisseria gDNA, we generated strains carrying a selectable marker tightly linked to a validated target (protospacer 25). We used the cas9::Tn strain to enable transformation and integration of both empty pGCC2 (Figure 4A) and protospacer 25-containing pGCC2 into the meningococcal chromosome (Figure 6A). We then isolated gDNA from these strains and used them as donors in liquid-medium transformation assays with wildtype N. meningitidis 8013 cells. Transformants (transformation frequency of 1.6 × 10−5; Table S6) were readily obtained using DNA that lacked protospacer 25 adjacent to the ermC marker, whereas no transformants were observed when the protospacer was present (Figure 6B, left panel). Similar results were obtained with gDNA strains carrying the CAT marker at the capsule locus with and without tightly-linked protospacer 25 (Figure 6 and Figure S4A, and Table S6), indicating that the interference effect was not marker- or locus-specific. We conclude that CRISPR interference is effective against the most common natural substrate for transformation in N. meningitidis.
Figure 6. Neisseria type II-C CRISPR/Cas limits natural transformation of meningococcal chromosomal DNA.
(A) Schematic representation of the siaA/ctrA and lctP/aspC chromosomal loci in the cas9::Tn strain (top; the transposon insertion is in yellow). Below are derivatives of the same cas9::Tn strain following transformation with the ermC-marked vector pGCC2 with or without a protospacer that matches CRISPR spacer 25, or with the cat-marked vector pYZEJS040 with or without a protospacer that matches CRISPR spacer 25.
(B) For the left panel, gDNA from the ermC-marked strains shown in (A), as well as the unmarked control strain, was used in transformation assays with wild-type N. meningitidis 8013. Natural transformations were performed and presented as in Figure 4B. The right panel shows an analogous experiment using gDNA from the cat-marked strains shown in (A), as well as the unmarked control strain. Error bars represent s.e.m. for three independent biological replicates.
DISCUSSION
CRISPR interference and the third major pillar of horizontal gene transfer
CRISPR/Cas pathways have been revealed as RNA-directed immune systems that protect bacteria and archaea from phage infection and HGT (Karginov and Hannon, 2010; Marraffini and Sontheimer, 2010; Terns and Terns, 2011; Wiedenheft et al., 2012). Several dozen bacterial species are known to be competent for HGT via natural transformation. Of this subset of bacteria, Neisseria spp. are unusual in that their transformation competence is constitutive (Hamilton and Dillard, 2006). Only a few phages are known to infect N. meningitidis, and although conjugative plasmids are present in some meningococcal isolates (van Passel et al., 2006), transformation is the major mechanism for mobilization of meningococcal chromosomal loci (Moxon and Jansen, 2005). Neisseria genomic sequences are preferred substrates for natural transformation, given that DNA uptake is strongly promoted by a short DNA uptake sequence (DUS) that is highly overrepresented in Neisseria spp. chromosomes (Budroni et al., 2011). DNA exchange is so frequent that the population structures of most neisseriae are effectively panmictic, with little propensity for the emergence of clonal subpopulations (Smith et al., 1993). Frequent HGT in N. meningitidis is thought to promote capsule switching and other forms of antigenic variation, homology-based DNA repair, and other functions (Hamilton and Dillard, 2006). Native CRISPR/Cas systems have previously been shown to prevent phage infection (and, by inference, phage transduction) and conjugation, which constitute two of the primary routes of HGT. Our results reveal a role for a native CRISPR/Cas system in preventing the third main route of HGT, natural transformation. This is consistent with recent reports that CRISPR/Cas systems can target loci that are already established in bacterial or archaeal chromosomes (Edgar and Qimron, 2010; Gudbergsdottir et al., 2011; Jiang et al., 2013), indicating that interference does not depend on the invasive DNA’s route of entry. Similarly, an engineered, heterologous CRISPR/Cas system introduced into Streptococcus pneumoniae can block natural transformation during active infection in mice (Bikard et al., 2012). We find that a native CRISPR/Cas system in N. meningitidis can block the transformation events that can be so important for immune evasion and other critical aspects of invasiveness and pathogenicity. Intriguingly, the ability of native CRISPR systems to block natural transformation would be expected to enable the selection of spacers that discriminate against specific chromosomal loci that negatively affect the fitness of certain strains or under certain conditions.
Although relatively few phages are known to infect N. meningitidis, they are not unknown (Kawai et al., 2005). Several genomic islands have been identified that resemble phages and could therefore represent prophage sequences (Bille et al., 2005, 2008; Joseph et al., 2011). One such sequence, the MDA island, correlates with invasiveness and pathogenicity in young adults (Bille et al., 2005, 2008). The existence of numerous CRISPR spacers with the potential to target these sequences suggests that CRISPR interference plays a role in shaping prophage content and serves phage defense functions in N. meningitidis, as elsewhere. CRISPR interference could limit the spread of prophages via either transformation or infection. Accordingly, CRISPR interference could negatively correlate with meningococcal pathogenicity, as suggested in enterococci (Palmer and Gilmore, 2010) and streptococci (Bikard et al., 2012). Alternatively, meningococcal Cas9 could participate in other regulatory events that contribute to pathogenicity, as suggested very recently (Sampson et al., 2013).
It is noteworthy that many N. meningitidis and N. lactamica strains encode CRISPR systems, while strains of the closely related N. gonorrhoeae with clearly functional CRISPR systems have not been identified. It is believed that these organisms split in relatively recent times (<100,000 years ago), evolutionarily speaking, but exact estimates have been stymied by the large recombination frequencies in these species (Bennett et al., 2010). It is equally possible that the nasopharyngeal-localized species gained the system after the split, or that N. gonorrhoeae lost the CRISPR system after the split. Both pathogens have been suggested not to establish long-lasting clones and tend towards linkage equilibrium (Buckee et al., 2008). It may not be coincidental that N. meningitidis carries a CRISPR system and can develop semi-clonal lineages (Bart et al., 2001), given that the CRISPR system could provide a short-term barrier to HGT. It is also possible that the co-existence of commensal Neisseria species such as N. lactamica and N. meningitidis in the nasal pharynx (Feil and Spratt, 2001) selects for a system that limits genetic exchange, and intriguingly, some N. lactamica and N. meningitidis isolates show large amounts of exchange while others show lower signatures of exchange (Hanage et al., 2005; Corander et al., 2012). In contrast, N. gonorrhoeae inhabits a niche that is devoid of other closely related bacteria that encode the DUS to allow efficient transformation of their sequences (Vazques et al., 1993; Cehovin et al., 2013).
Towards a minimal CRISPR/Cas system
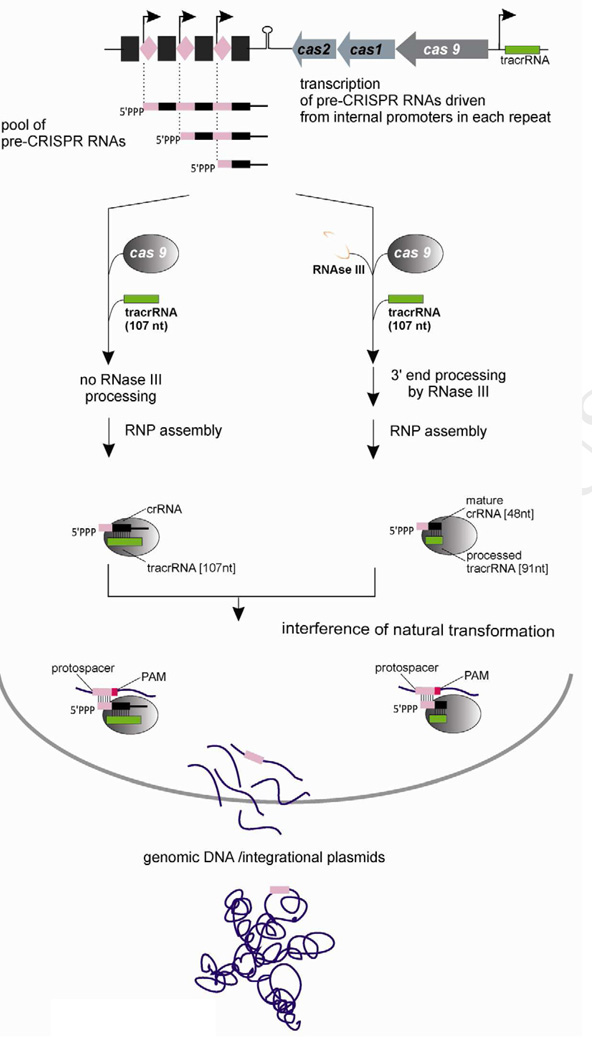
In CRISPR/Cas systems investigated to date, crRNAs are transcribed from an external promoter, generating a multimeric precursor. The pre-crRNA is processed by endonucleolytic cleavage to generate mature crRNAs (Carte et al., 2008; Haurwitz et al., 2010; Gesner et al., 2011), and processing is essential for interference in vivo (Brouns et al., 2008; Deltcheva et al., 2011; Hale et al., 2012; Westra et al., 2012). The potential presence of minimal and apparently fortuitous promoter elements has been noted within certain CRISPRs of Sulfolobus solfataricus P2, though they are not thought to account for the functional expression of crRNA and in fact appear to be suppressed by the repeat-binding protein Cbp1 (Deng et al., 2012). The results presented here reveal that streamlined CRISPR/Cas systems exist in which pre-crRNA processing is not essential (Figure 7). In CRISPR-containing strains of N. meningitidis and N. lactamica, as well as other species such as C. jejuni, the CRISPR repeats each contain an extended −10 box that drives transcription initiation within the downstream spacer. Thus, many crRNAs contain 5’-triphosphate ends that are not subject to further 5’-processing.
Figure 7. CrRNA biogenesis and CRISPR interference in Neisseria.
Type II-C CRISPR/cas loci in Neisseria spp. initiate transcription within each spacer, driven by promoter elements embedded within each repeat. The resulting crRNAs and pre-crRNAs carry 5’-terminal triphosphates. Following tracrRNA annealing, RNase III can cleave both strands of the tracrRNA/pre-crRNA duplex (right pathway). Unexpectedly, pre-crRNA processing is not required: when RNase III is unavailable or fails to act, Cas9 can still form functional complexes with tracrRNA and crRNA (left pathway). The naturally-encoded crRNAs target sequences present in other Neisseria spp. chromosomes, consistent with the high frequency of genetic exchange by natural transformation. Because Type II-C have only three protein-coding genes, lack leader sequences upstream of the CRISPR array, and do not require the host factor RNase III, they are the most streamlined natural CRISPR/Cas systems known.
Like other Type II CRISPR/Cas systems, neisseriae produce a tracrRNA that apparently anneals to pre-crRNA and enables binding and cleavage by a RNase III. This reaction generates crRNA 3’ ends, and rnc mutants accumulate multimeric crRNA precursors. However, these rnc mutants exhibit no interference defect, indicating that processing is not essential. In addition, while the tracrRNA is essential for interference, its role in directing processing is not, since processing is itself dispensable. This provides the first clear indication that the tracrRNA is required for post-processing events such as target DNA binding and cleavage in bacterial cells, as it is in vitro (Jinek et al., 2012).
Among the three main types of CRISPR/Cas pathways, the Type II systems are the simplest ones characterized thus far, as judged by the number of components and essential steps. Both Type II-A and Type II-B systems include the CRISPR array itself, a tracrRNA, four protein-coding genes encoded within the cas locus, and the host factor RNase III (Deltcheva et al., 2011; Makarova et al., 2011b; Magadán et al., 2012; Chylinski et al., 2013). The Neisseria systems that we have characterized are even more streamlined: they do not require a separate leader sequence to drive crRNA transcription, they lack one of the four cas/csn genes present in Type II-A or II-B systems, and they do not require RNase III or crRNA processing. The Neisseria systems are among the founding members of a new CRISPR/Cas subtype (Type II-C) that is characterized by a smaller number of cas/csn proteins (Koonin and Makarova, 2013; Chylinski et al., 2013), and in at least some cases by repeat-embedded promoters and processing independence.
Importantly, recent reports have shown that Type II CRISPR/Cas systems can be ported into eukaryotic cells and employed for RNA-directed genome editing and genome binding, including multiplexed applications specified by multiple spacers (Jinek et al., 2012, 2013; Cho et al., 2013; Cong et al., 2013; DiCarlo et al., 2013; Hwang et al., 2013; Mali et al., 2013; Qi et al., 2013). The Cas9 effector proteins from neisseriae share the conserved features observed in the S. pyogenes and S. thermophilus Cas9 enzymes used in these studies (Chylinski et al., 2013). The fewer the functional requirements for the operation of such systems, the greater their versatility and applicability will be. Separately encoded crRNAs and tracrRNAs are more efficient in vivo than single-guide RNAs that combine essential crRNA and tracrRNA domains in the same transcript (Cong et al., 2013). Although endogenous eukaryotic activities can substitute for bacterial RNase III to process tracrRNA/pre-crRNA hybrids in human and mouse cells (Cong et al., 2013), it is not known whether RNase III will be dispensable in other eukaryotic contexts, or indeed in all mammalian cell types. Accordingly, the identification of processing-independent CRISPR/Cas systems could increase the range of applications in eukaryotic genome editing, especially in light of the potential toxicity of bacterial RNase III expression (Pines et al., 1988). Such applications will benefit from further analysis of meningococcal Cas9 activity, including the definition of the presumptive cleavage sites relative to the PAM.
The dispensability of pre-crRNA processing in neisseriae also raises intriguing mechanistic questions that are unique to the II-C subtype. First, can any spacer in a multimeric pre-crRNA engage Cas9 and target DNA cleavage, or is the 5’-terminal spacer the only one that can function in interference? If any spacer can function in target cleavage, can a single multimeric pre-crRNA engage multiple copies of the Cas9 protein and simultaneously direct the interference of multiple targets? Answers to these questions will be valuable in applying the CRISPR machinery as a versatile, adaptable and scalable tool for multiplex genome editing and genome binding.
EXPERIMENTAL PROCEDURES
Detailed experimental and analytical procedures can be found in the Supplemental information.
Bacterial Strains, Plasmids, and Oligonucleotides
N. lactamica 020-06, N. meningitidis WUE2594 and 8013, and mutant derivatives thereof that were used in this study are listed in Supplemental Experimental Procedures, as are complete lists of all plasmids and DNA oligonucleotides.
Mutant Strain Construction
All mutants were confirmed by PCR and DNA sequencing. Most mutant strains were generated by transformation with appropriately constructed plasmids. For generation of the cas9, rnc, and control NMV_1851 transposon-induced alleles in the 8013 strain background, we used gDNA from the corresponding mutant in the NeMeSys collection (Rusniok et al., 2009) to transform 8013. For generation of the Δrnc derivative of 8013, we used gDNA from the WUE2594 Δrnc derivative that was initially made by a plasmid-based approach.
For complementation of cas9::Tn, Δcas9, and ΔtracrRNA mutants, we cloned wildtype copies of the relevant gene into plasmid pGCC2 and transformed the resulting plasmids into the parental mutant strain.
RNA Extraction and Depletion of Processed RNAs
For 020-06, WUE2594 and its mutant derivatives, total RNA was extracted from frozen cell pellet lysates using the hot-phenol method described previously (Blomberg et al., 1990). For depletion of processed transcripts, equal amounts of total RNA were incubated with Terminator™ exonuclease (TEX) (Epicentre) or in buffer alone as described (Sharma et al., 2010). For 8013 and its mutant derivatives, total RNAs were extracted from frozen cell pellets using miRNeasy Mini Kit (Qiagen) with two additional steps: a ten minute initial cell lysis with lysozyme and Proteinase K, and a later on-column DNase digestion step (the RNase-Free DNase Set, Qiagen).
dRNA-seq
Libraries for Solexa sequencing (HiSeq) of cDNA were constructed by vertis Biotechnology AG, Germany (http://www.vertis-biotech.com/), as described previously for eukaryotic microRNA (Berezikov et al., 2006) but omitting the RNA size-fractionation step prior to cDNA synthesis. cDNA libraries were sequenced using a HiSeq 2000 machine (Illumina) in single read mode at the Max Planck Genome Centre Cologne (Cologne, Germany). Data analysis was done as described (Chao et al., 2012), with the only exception being that the minimal read length after trimming and clipping was 12 nt instead of 20 nt.
Transcriptional gfp Fusions
The inserts used for the construction of the transcriptional gfp fusion plasmids were generated with synthetic DNA oligonucleotides. E. coli cells were transformed with these plasmids and grown on agar plates for fluorescence imaging. To measure GFP fluorescence, single colonies were grown in LB broth, fixed, and analysed by flow cytometry.
In vitro Transcription
Templates for in vitro transcription assays were PCR-generated, gel-purified 210 bp DNA fragments amplified from pNH13, pNH14, or pNH15. Transcription reactions with σ-saturated E. coli RNA Polymerase holoenzyme (Epicentre) included α-[32P]-ATP.
Natural Transformation
Natural transformation assays were performed as described (Duffin and Seifert, 2012). Transformation frequencies were reported as antibiotic-resistant cfu/ml divided by total cfu/ml from at least three independent experiments (mean ±s.e.m.).
Supplementary Material
Highlights.
Unlike previously described CRISPRs, each Neisseria repeat carries its own promoter
Pre-crRNA processing is dispensable for CRISPR interference in Neisseria spp.
CRISPR interference blocks natural transformation in the pathogen N. meningitidis
Neisseria CRISPR/Cas systems are the most streamlined observed to date
ACKNOWLEDGMENTS
We thank V. Pelicic for genomic DNAs from meningococcal transposon insertion strains; Y. Chao, G. Dugar and C. M. Sharma for discussions; and R. Reinhardt and K. Förstner for dRNA-seq and data mapping, respectively. This work was supported by DFG grants (875/4-2 and 875/7-1) to J.V., and by National Institutes of Health grants R37 AI033493 and R01 AI044239 to H.S.S. and R01 GM093769 to E.J.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The Gene Expression Ominbus (GEO) accession number for the dRNA-Seq data reported in this paper is GSE44582.
REFERENCES
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bart A, Barnabé C, Achtman M, Dankert J, van der Ende A, Tibayrenc M. The population structure of Neisseria meningitidis serogroup A fits the predictions for clonality. Infect. Genet. Evol. 2001;1:117–122. doi: 10.1016/s1567-1348(01)00011-9. [DOI] [PubMed] [Google Scholar]
- Bennett JS, Bentley SD, Vernikos GS, Quail MA, Cherevach I, White B, Parkhill J, Maiden MCJ. Independent evolution of the core and accessory gene sets in the genus Neisseria: insights gained from the genome of Neisseria lactamica isolate 020-06. BMC Genomics. 2010;11:652. doi: 10.1186/1471-2164-11-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host & Microbe. 2012;12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Bille E, Ure R, Gray SJ, Kaczmarski EB, McCarthy ND, Nassif X, Maiden MCJ, Tinsley CR. Association of a bacteriophage with meningococcal disease in young adults. PloS One. 2008;3:e3885. doi: 10.1371/journal.pone.0003885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bille E, Zahar J-R, Perrin A, Morelle S, Kriz P, Jolley KA, Maiden MCJ, Dervin C, Nassif X, Tinsley CR. A chromosomally integrated bacteriophage in invasive meningococci. J. Exp. Med. 2005;201:1905–1913. doi: 10.1084/jem.20050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Bratcher HB, Bennett JS, Maiden MCJ. Evolutionary and genomic insights into meningococcal biology. Future Microbiol. 2012;7:873–885. doi: 10.2217/fmb.12.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, Van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckee CO, Jolley KA, Recker M, Penman B, Kriz P, Gupta S, Maiden MCJ. Role of selection in the emergence of lineages and the evolution of virulence in Neisseria meningitidis. Proc. Natl. Acad. Sci. USA. 2008;105:15082–15087. doi: 10.1073/pnas.0712019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budroni S, Siena E, Dunning Hotopp JC, Seib KL, Serruto D, Nofroni C, Comanducci M, Riley DR, Daugherty SC, Angiuoli SV, et al. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc. Natl. Acad. Sci. USA. 2011;108:4494–4499. doi: 10.1073/pnas.1019751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, Brady J, Baldwin GS, Lea SM, Matthews SJ, Pelicic V. Specific DNA recognition mediated by a type IV pilin. Proc. Natl. Acad. Sci. USA. 2013;110:3065–3070. doi: 10.1073/pnas.1218832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3’ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Christie PJ, Dubnau D. The ins and outs of DNA transfer in bacteria. Science. 2005;310:1456–1460. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature Biotech. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013 doi: 10.4161/rna.24321. in press [ePub ahead of print (doi:10.4161/rna24321)] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J, Connor TR, O'Dwyer CA, Kroll JS, Hanage WP. Population structure in the Neisseria, and the biological significance of fuzzy species. J. Royal Soc. Interface. 2012;9:1208–1215. doi: 10.1098/rsif.2011.0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Kenchappa CS, Peng X, She Q, Garrett RA. Modulation of CRISPR locus transcription by the repeat-binding protein Cbp1 in Sulfolobus. Nucleic Acids Res. 2012;40:2470–2480. doi: 10.1093/nar/gkr1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bact. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffin PM, Seifert HS. Genetic transformation of Neisseria gonorrhoeae shows a strand preference. FEMS Microbiol. Letters. 2012;334:44–48. doi: 10.1111/j.1574-6968.2012.02612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Qimron U. The Escherichia coli CRISPR system protects from l lysogenization, lysogens, and prophage induction. J. Bact. 2010;192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Spratt BG. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 2001;55:561–590. doi: 10.1146/annurev.micro.55.1.561. [DOI] [PubMed] [Google Scholar]
- Garneau JE, Dupuis M-È, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA. 2012;109:2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nature Struct. Mol. Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbergsdottir S, Deng L, Chen Z, Jensen JVK, Jensen LR, She Q, Garrett RA. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol. Microbiol. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comp. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CVC, Graveley BR, Terns RM, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton HL, Dillard JP. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 2006;59:376–385. doi: 10.1111/j.1365-2958.2005.04964.x. [DOI] [PubMed] [Google Scholar]
- Hanage WP, Fraser C, Spratt BG. Fuzzy species among recombinogenic bacteria. BMC Biol. 2005;3:6. doi: 10.1186/1741-7007-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence-and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook-Barnard IG, Hinton DM. Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters, Gene Reg. Systems Biol. 2007;1:275–293. [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coûté-Monvoisin A-C, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bact. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh J-RJ, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature Biotech. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature Biotech. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2 doi: 10.7554/eLife.00471. e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Schwarz RF, Linke B, Blom J, Becker A, Claus H, Goesmann A, Frosch M, Müller T, Vogel U, et al. Virulence evolution of the human pathogen Neisseria meningitidis by recombination in the core and accessory genome. PloS One. 2011;6:e18441. doi: 10.1371/journal.pone.0018441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Uchiyama I, Kobayashi I. Genome comparison in silico in Neisseria suggests integration of filamentous bacteriophages by their own transposase. DNA Research. 2005;12:389–401. doi: 10.1093/dnares/dsi021. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS. CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol. 2013 doi: 10.4161/rna.24022. in press [ePub ahead of print (doi:10.4161/rna24022)] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magadán AH, Dupuis M-È, Villion M, Moineau S. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS ONE. 2012;7:e40913. doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct. 2011a;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nature Rev. Microbiol. 2011b;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nature Rev. Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Moxon ER, Jansen VAA. Phage variation: understanding the behaviour of an accidental pathogen. Trends Microbiol. 2005;13:563–565. doi: 10.1016/j.tim.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Palmer K, Gilmore M. Multidrug-resistant enterococci lack CRISPR-cas. mBio. 2010;1 doi: 10.1128/mBio.00227-10. e00227–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines O, Yoon H, Inouye M. Expression of double-stranded-RNA-specific RNase III of Escherichia coli is lethal to Saccharomyces cerevisiae. J. Bact. 1988;170:2989–2993. doi: 10.1128/jb.170.7.2989-2993.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau C, Gonnet M, Le Romancer M, Nicolas J. CRISPRi: a CRISPR interactive database. Bioinformatics. 2009;25:3317–3318. doi: 10.1093/bioinformatics/btp586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusniok C, Vallenet D, Floquet S, Ewles H, Mouzé-Soulama C, Brown D, Lajus A, Buchrieser C, Médigue C, Glaser P, et al. NeMeSys: a biological resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol. 2009;10:R110. doi: 10.1186/gb-2009-10-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Saroj SD, Llewellyn AC, Tzeng Y-L, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013 doi: 10.1038/nature12048. in press [ePub ahead of print (doi:10.1038/nature12048)] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Smith JM, Smith NH, O’Rourke M, Spratt BG. How clonal are bacteria? Proc. Natl. Acad. Sci. USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DS, Greenwood B, Brandtzaeg P. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet. 2007;369:2196–2210. doi: 10.1016/S0140-6736(07)61016-2. [DOI] [PubMed] [Google Scholar]
- Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobiason DM, Seifert HS. Genomic content of Neisseria species. J. Bact. 2010;192:2160–2168. doi: 10.1128/JB.01593-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Passel MWJ, Van der Ende A, Bart A. Plasmid diversity in neisseriae. Infect. Immun. 2006;74:4892–4899. doi: 10.1128/IAI.02087-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez JA, de la Fuente L, Berron S, O'Rourke M, Smith NH, Zhou J, Spratt BG. Ecological separation and genetic isolation of Neisseria gonorrhoeae and Neisseria meningitidis. Curr. Biol. 1993;3:567–572. doi: 10.1016/0960-9822(93)90001-5. [DOI] [PubMed] [Google Scholar]
- Westra ER, Van Erp PBG, Künne T, Wong SP, Staals RHJ, Seegers CLC, Bollen S, Jore MM, Semenova E, Severinov K, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Van Duijn E, Bultema JB, Bultema J, Waghmare SP, Waghmare S, Zhou K, Barendregt A, Westphal W, Heck AJR, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. USA. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DANN elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers S-V, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.