Abstract
Distamycin A has been described as an inhibitor of the cellular pathogenesis of vaccinia virus in culture. Distamycin is an antibiotic that specifically targets the minor groove of DNA. We show here that distamycin is a potent inhibitor of vaccinia virus replication. Pulse-labeling experiments showed that most major late proteins failed to accumulate in the presence of the antibiotic. We characterized the effect of distamycin on vaccinia virus nucleic acid biosynthesis with the goal of determining the inhibitor's target. Early gene transcription was unaffected. DNA synthesis proceeded at normal rates, but DNA accumulated in large masses in the cytoplasm with no evidence of virion assembly. Transcription from the intermediate class promoter for the I1L gene was partially reduced by distamycin; however, transcription from the intermediate promoters for the three late transcription factor genes was severely inhibited. The accumulation of the late transcripts for the viral F17R and A10L genes also was severely impaired and was shown to be a direct inhibition of late promoter activity. These results indicate that inhibition of postreplicative intermediate and late transcription is the basis for inhibition of vaccinia virus by distamycin and indicate that DNA minor-groove ligands hold promise for effective anti-poxvirus drugs.
Distamycin A is an antibiotic that reduces the pathogenic effects of vaccinia virus. Distamycin blocks virus-induced cell syncytium formation (22) and inhibits the visually observed cytopathic effects of the virus in cultured cells (16). Distamycin targets the minor groove of DNA (10), preferentially binding DNA sequences that are five consecutive A-T pairs, with affinity that varies with the particular sequence (1). The base composition of the DNA genome of vaccinia virus is 66% A-T, with transcriptional promoter regions being close to 90% A-T, making them ideal targets for the antibiotic.
Vaccinia virus replicates its genome and assembles progeny virions exclusively in the cytoplasm of the infected cell. The processes of DNA synthesis and transcription of the three different classes of genes are coordinated in a sequential manner (reviewed in reference 4). Upon entry of the cell, viral early gene transcription begins immediately. The early gene products include factors participating in DNA synthesis that begins about an hour after infection. As DNA synthesis initiates, early gene transcription ceases and intermediate gene transcription ensues. Shortly thereafter, late gene transcription begins and intermediate gene transcription wanes. The virus-encoded proteins required for the transcription of each gene class are products of the preceding class in a gene expression cascade. The promoters for vaccinia virus early, intermediate, and late genes have A-T-rich motifs whose interaction with transcription factors potentially could be affected by a DNA minor-groove ligand. The early promoter element is targeted by the early transcription factor (6). We have shown that distamycin impairs DNA binding by the early transcription factor in vitro (S. S. Broyles, submitted for publication), thus predicting one basis for inhibition of virus replication. Intermediate promoters consist of an upstream element and an initiator-like element at the transcription start site (2). The initiator element of the intermediate I1L promoter may be a target for the cellular transcription factor YY1 (5). YY1 is a zinc finger transcription factor that does not contact the minor groove of DNA (12). The protein that targets the upstream intermediate element is unknown. Late promoters also have upstream and initiator elements. Heterogeneous nuclear riboproteins A2/B1 and RBM3 have been shown to stimulate vaccinia virus late transcription in vitro and target oligo(T) tracts in DNA (23) previously shown to behave as an upstream element in a late promoter (9). The role of the late promoter initiator element is not known. DNA synthesis is another potential target of distamycin. Whether vaccinia virus has an origin of replication that is targeted by origin-binding proteins is unclear.
In this study, we characterized the effect of distamycin on vaccinia virus nucleic acid biosynthesis. We established that distamycin is an inhibitor of virus multiplication and followed the synthesis of viral DNA and the three classes of mRNA to determine the target of the drug.
Antiviral effects of distamycin A against vaccinia virus.
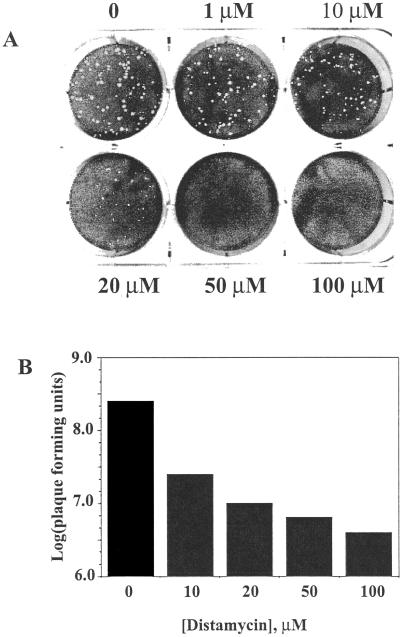
The effect of distamycin A on the replication of vaccinia virus was assessed initially in a virus plaque assay. Monolayer cultures of BSC40 cells were infected with approximately 50 PFU of virus for 48 h in the presence of increasing concentrations of the drug and subsequently stained with crystal violet to visualize remaining adherent cells. Distamycin was applied to the cells immediately after the virus, as it was in all subsequent experiments. High concentrations of the drug (50 and 100 μM) resulted in complete disappearance of plaques (Fig. 1A). Intermediate concentrations of the drug (1 to 20 μM) reduced the diameter of plaques but did not significantly affect the number of visible plaques. We followed the course of infection for up to 6 days, when the antibiotic begins to show cytotoxic effects, and observed no sign of virus plaques.
FIG. 1.
Inhibition of vaccinia virus by distamycin. (A) Effect of distamycin on vaccinia virus plaque formation. Monolayers of BSC40 cells were infected with 60 PFU of vaccinia virus for 48 h in the presence of the indicated concentrations of distamycin A (Sigma). Plaques were visualized by staining with crystal violet. (B) Effect of distamycin on virus yield in a single-step infection. BSC40 cells were infected with 10 PFU of vaccinia virus per cell for 24 h in the presence of the indicated concentrations of distamycin A. Cells were harvested and lysed, and virus yield was quantitated by plaque assay in the absence of the drug.
The effects of distamycin on vaccinia virus multiplication were evaluated further in a single-step growth experiment. BSC40 cells were infected with virus at a multiplicity of 10 PFU per cell for 24 h in the presence of various drug concentrations, and the virus yield was determined in a subsequent plaque assay. High concentrations of distamycin (>50 μM) reduced the virus yield by approximately 2 log units relative to infection in the absence of the drug (Fig. 1B). Virtually identical results were obtained when infection was allowed to proceed for 48 h (data not shown). For all further experiments, the distamycin concentration was 100 μM.
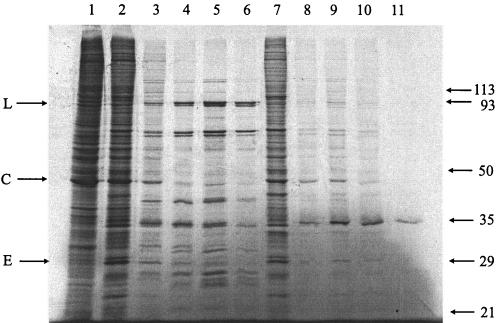
The effect of distamycin on the global vaccinia virus gene expression profile was assessed by monitoring protein synthesis during the course of infection. BSC40 cells were infected with 10 PFU of virus per cell and pulse-labeled for 30 min with [35S]methionine at intervals from 2 to 24 h after infection. In the absence of the drug, shutoff of host protein synthesis was evident within a few hours, early proteins were made within the first few hours and declined, and major proteins species appeared at 4 h after infection (Fig. 2). In the presence of distamycin, host protein shutoff was also evident and early viral proteins also appeared in the first few hours of infection. In the distamycin-treated cells, the early proteins appeared to persist during the course of the experiment and the major late proteins failed to appear. This experiment suggests that early events in the infectious cycle are unaffected; however, late protein synthesis appears to be defective. Interestingly, one major protein with the mobility of a 35-kDa protein appeared late in the presence of distamycin.
FIG. 2.
Protein synthesis in cells infected with vaccinia virus in the absence and presence of distamycin. Cells were infected with vaccinia virus for 2 (lanes 2 and 7), 4 (lanes 3 and 8), 8 (lanes 4 and 9), 12 (lanes 5 and 10), or 24 (lanes 6 and 11) h in the absence (lanes 2 to 6) or presence (lanes 7 to 11) of distamycin and pulsed with 50 mCi of [35S]methionine for 30 min. Lane 1 is protein from uninfected cells. Protein extracts were electrophoresed on a sodium dodecyl sulfate-10% polyacrylamide gel, dried, and autoradiographed. The mobilities of molecular mass standards are given on the right (molecular masses in kilodaltons are shown). On the left, arrows indicate a major late protein (L), a major cellular protein (C), and a major early protein (E).
Effect of distamycin on viral nucleic acid metabolism.
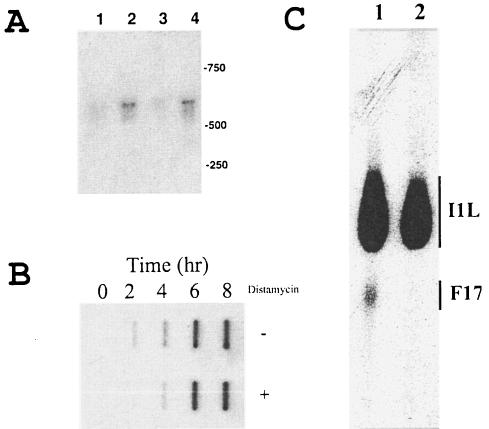
Because distamycin is a DNA minor-groove ligand, it is reasonable to expect it to exert inhibitory effects on DNA replication and/or viral gene transcription. To determine the effect of distamycin on early gene transcription, BSC40 cells were infected with vaccinia virus for 1 and 2 h. Total RNA was isolated and analyzed by Northern blotting with a probe for the C11L (growth factor) gene, which is an early class gene (21). The C11L gene probe detected the expected 600-base transcript in RNA from cells infected for 1 h that increased further at 2 h (Fig. 3A). Inclusion of distamycin in the culture medium did not affect transcript accumulation at either of the two time points. We concluded that distamycin does not affect vaccinia virus early gene transcription significantly in vivo.
FIG. 3.
Effect of distamycin A on vaccinia virus nucleic acid biosynthesis. (A) Effect of distamycin A on vaccinia virus early mRNA accumulation. RNA was isolated 1 (lanes 1 and 3) and 2 (lanes 2 and 4) h after infection by vaccinia virus in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of distamycin with a mini-spin column kit (Qiagen) and analyzed by Northern blotting (19). Membrane blots were probed with a 32P-labeled DNA probe from the vaccinia virus C11L gene. The mobilities of DNA molecular size markers are shown on the right (molecular masses in kilodaltons are shown). (B) Effect of distamycin on vaccinia virus DNA synthesis. Total DNA was isolated from vaccinia virus-infected cells at the indicated times in the absence (upper row) or presence (lower row) of distamycin. DNA was immobilized on nitrocellulose by slot blotting and probed with 32P-labeled genomic vaccinia virus DNA (7). (C) Effect of distamycin on accumulation of an intermediate and a late mRNA. Total mRNA was isolated from cells infected with vaccinia virus for 6 h in the absence (lane 1) or presence (lane 2) of distamycin. RNA was analyzed by primer extension (3) with avian myeloblastosis virus reverse transcriptase (Roche) and 32P-labeled oligonucleotide primers designed to hybridize 120 and 150 bases downstream of the F17R and intermediate I1L transcriptional start sites, respectively. DNA products were resolved by denaturing sequencing polyacrylamide gel electrophoresis. Bars indicate the range of products produced for each transcript. The mobilities of DNA markers are shown on the right.
We next determined the effect of distamycin on replication of the vaccinia virus genome. Total DNA was extracted from cells at various times after infection, and viral DNA was identified by slot blot hybridization with radiolabeled vaccinia virus genomic DNA as the probe. As expected, viral DNA was detectable by 2 h after infection and continued to accumulate through 8 h (Fig. 3B). Inclusion of distamycin in the culture medium affected neither the onset of DNA replication nor the level of accumulation of viral DNA through 8 h postinfection. We concluded that vaccinia virus DNA replication is not affected by distamycin. Because DNA replication requires the expression of multiple early gene products, the ability to synthesize DNA supports the conclusion that distamycin does not affect early gene transcription.
We next examined the effect of distamycin on intermediate and late gene transcription. Intermediate and late transcripts are moderately heterogeneous at their 5′ ends because of apparent slippage of the RNA polymerase on the TTT in the initiator motif on the template DNA, producing mRNAs with 15 to 35 non-template-encoded A residues (3, 20). They are also extremely heterogeneous at their 3′ ends because the RNA polymerase terminates randomly (for example, see reference 24). We therefore used primer extension to detect the 5′ ends of intermediate and late mRNAs from virus-infected cells. We have demonstrated that the I1L promoter belongs to the intermediate class and confirmed that the F17R promoter is a late class gene (X. Liu and S. S. Broyles, unpublished observations). Primers were designed to hybridize 120 nucleotides downstream of the F17R transcription initiation site and 150 nucleotides downstream of the I1L transcription initiation site, respectively. Simultaneous analysis of the primer extension products from both mRNAs showed that the I1L message was reduced by 22% in RNA derived from cells infected with virus in the presence of distamycin relative to its absence, as determined with a phosphorimager (Fig. 2C). The level of F17R mRNA appeared to be more dramatically affected. Primer extension products corresponding to the F17R mRNA appeared to be absent in RNA from distamycin-treated cells.
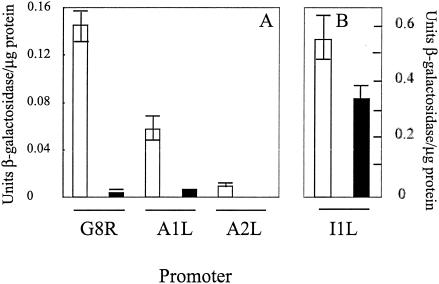
The loss of late transcription coupled with a partial inhibition of transcription from an intermediate promoter could be explained by inhibition of transcription of late factor genes that are intermediate genes. We therefore tested the effect of distamycin on the transcription of the intermediate class A1L, A2L, and G8R genes that encode late transcription factors (14). The activity of these promoters is only a fraction of that of the I1L intermediate promoter (Liu and Broyles, unpublished), making detection of the heterogeneous primer extension products problematic. As an alternative, a reporter gene approach was used to monitor the activity of the late transcription factor promoters. We linked each of the three promoters to the lacZ reporter gene and transfected each into HeLa cells previously infected with vaccinia virus. Reporter gene expression from each of the three promoters was inhibited by distamycin. Reporter activity of the A1L, A2L, and G8R promoters was reduced by greater than 90% in the presence of distamycin relative to its absence (Fig. 4). We conclude that the transcription of some intermediate genes, as typified by the late factor genes, is highly sensitive to the effects of distamycin. The reporter activity of the I1L reporter construct retained almost 60% of its activity in the presence of distamycin, in agreement with the primer extension results.
FIG. 4.
Effect of distamycin A on transcription from intermediate class promoters for the late transcription factor genes. The promoters for the A1L, A2L, G8R, and I1L genes were linked to the Escherichia coli lacZ gene. HeLa cells were infected with 10 PFU of vaccinia virus per cell in the absence (open bars) or presence (closed bars) of 100 μM distamycin A, and plasmid constructs were transfected into HeLa cells (2 μg/35-cm2 dish) with Superfect transfection reagent (Qiagen) in accordance with the manufacturer's recommendations. After 16 h, cells were harvested and permeabilized and β-galactosidase assays were performed as previously described (18). Units of enzyme activity were normalized to the total protein. Error bars indicate standard deviations of assays performed in triplicate. There was no detectable activity from the A2L promoter in the presence of distamycin. Note the different scales in panels A and B.
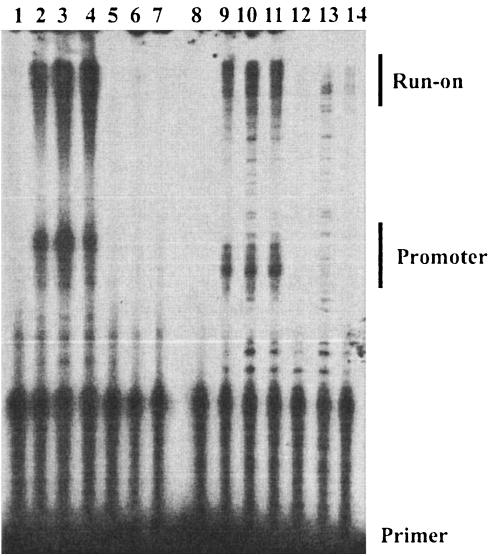
The effect of distamycin on late gene transcription was assessed in more detail by primer extension with primers designed to hybridize to the F17R and A10L (p4a gene) mRNAs. We have confirmed that the promoters for both genes belong to the late class (Liu and Broyles, unpublished). RNA was isolated at 6, 8, and 24 h after infection with vaccinia virus and analyzed by primer extension with 30-mer primers designed to initiate extension 50 nucleotides upstream of the initiation site in each promoter. Primer extension on RNA from cells infected with virus in the absence of the drug produced heterogeneous products of the expected length for both mRNAs (Fig. 5). The range of primer extension product lengths for both mRNAs was about 60 to 85 nucleotides, consistent with 50 nucleotides of mRNA plus the length of the non-template-encoded oligo(A) tract previously described for late mRNAs (3, 20). Primer extension products also included longer products that we call readthrough products, although we do not know whether they reflect transcripts originating upstream of each promoter or are the result of primer hybridization to other transcripts. Primer extension on RNA from cells infected with virus in the presence of distamycin showed virtually no products corresponding to either promoter, nor did they produce appreciable signal from the readthrough RNA (Fig. 5). These results demonstrate that late transcription is extremely sensitive to distamycin.
FIG. 5.
Effect of distamycin on accumulation of the F17R and A10L transcripts. Total RNA was isolated from cells infected with vaccinia virus in the absence (lanes 2 to 4 and 9 to 11) or presence (lanes 5 to 7 and 12 to 14) of distamycin for 6 (lanes 2, 5, 9, and 12), 8 (lanes 3, 6, 10, and 13), or 24 (lanes 4, 7, 11, and 14) h. Primer extension reactions were performed as described in the legend to Fig. 1C, except that primers were designed to hybridize 50 nucleotides downstream of transcription initiation sites. The mobilities of primers, cDNA products expected from mRNA originating from promoters, and cDNA from run-on RNA that would have originated upstream of the promoter are indicated on the right.
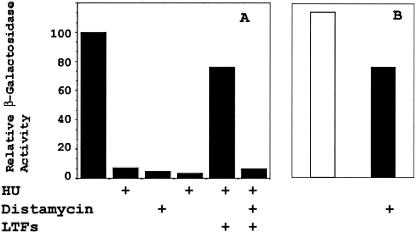
The results presented above raised the question of whether distamycin has a direct effect on late gene transcription or only an indirect effect of impaired expression of the late transcription factors. To address this issue, we circumvented the late transcription requirement for intermediate transcription by expression of the three late transcription factors with a T7 RNA polymerase-directed system (14, 15). HeLa cells were infected with a vaccinia virus expressing the bacteriophage T7 RNA polymerase and transfected with a β-galactosidase reporter plasmid driven by the A10L late vaccinia virus promoter together with plasmids encoding the A1L, A2L, and G8R genes driven by a T7 promoter. Expression of the T7 RNA polymerase is driven by a tandem early-late promoter and therefore proceeds in the absence of DNA synthesis. This protocol allows late promoters to be activated in the absence of intermediate transcription. In the reporter gene assays, distamycin alone was a potent inhibitor of the A10L late promoter, as was hydroxyurea (HU), an inhibitor of DNA synthesis (Fig. 6). The activity of the A10L promoter in the presence of HU was restored by coexpression of the three late transcription factors, which also was inhibited by distamycin. A reporter construct driven by a T7 promoter retained more than 60% of its activity in the presence of distamycin, indicating that inhibition of the T7 RNA polymerase was not the basis for inhibition of late transcription. These results indicate that late transcription is inhibited directly by distamycin, similar to that observed for the intermediate transcription described above.
FIG. 6.
Effect of distamycin on the late A10L promoter. (A) A β-galactosidase reporter gene plasmid (5 μg), driven by the vaccinia virus A10L promoter, was transfected into HeLa cells (previously infected with vaccinia virus strain vTF7-3, which expresses the T7 RNA polymerase. Where indicated, 100 μM distamycin and/or 5 mM HU was included at the time of infection. Also, where indicated, 5 μg each of plasmids encoding the late transcription factors (LTFs) A1L, A2L, and G8R driven by a T7 promoter was cotransfected with the reporter gene plasmid. After 18 h, cells were harvested and β-galactosidase activity was determined. Activity was normalized to protein content. Activities are expressed as that relative to the activity of the reporter gene in the absence of inhibitors. (B) Activity of a T7 promoter β-galactosidase reporter construct in the absence (open bar) and presence (filled bar) of distamycin.
To further characterize the defects in viral replication induced by distamycin, we examined virus-infected cells by transmission electron microscopy. Vaccinia virus morphogenesis proceeds through a series of discrete structures that are readily distinguished by electron microscopy (8). Membrane crescents initially form to envelop nascent genomes to assemble spherical immature particles that later condense to form oblate mature virions. Cells infected with vaccinia virus in the absence of the drug for 24 h displayed all of these morphogenic species (data not shown). Cells infected with vaccinia virus in the presence of distamycin showed no evidence of the virion assembly intermediates normally found in virus-infected cells. Instead, these cells contained large cytoplasmic structures referred to as viroplasm bodies. These viroplasm bodies appear to exclude cytoplasmic organelles, being bordered by a membrane at its periphery in some areas but not in others. We inferred that these structures contain DNA because fluorescence microscopy of cells infected with vaccinia virus in the presence of distamycin detected cytoplasmic staining with the DNA dye 4′,6′-diamidino-2-phenylindole (data not shown). We concluded that distamycin allows the accumulation of high levels of viral DNA but has a profound effect on assembly of progeny virions.
Distamycin A is shown here to be a potent inhibitor of vaccinia virus replication. Infection at the level of a plaque assay was essentially completely negated by high concentrations of distamycin. Virus yield was reduced by about 2 orders of magnitude. Electron microscopy showed no evidence of virus assembly intermediates beyond large masses of DNA in the cytoplasm of antibiotic-treated cells.
Our characterization of viral nucleic acid synthesis in the presence of distamycin indicated that vaccinia virus intermediate and late gene transcription is inhibited by the antibiotic. Early gene transcription and DNA synthesis appeared to proceed normally in the presence of the antibiotic. Transcription from the intermediate I1L promoter was partially inhibited, whereas transcription from the intermediate A1L, A2L, and G8R promoters was almost totally inactivated by distamycin. The reason for differential sensitivity to distamycin among intermediate promoters is unclear. We noted that an abundant 35-kDa protein appeared to be resistant to the effects of distamycin. The predicted mass of the I1L protein is 35 kDa (13). The protein that targets the upstream element in intermediate promoters has not been identified, and no protein interacting with the minor groove of DNA has been implicated in vaccinia virus intermediate transcription. The phenotype of vaccinia virus in the presence of distamycin is one that closely recapitulates the phenotype of some conditional lethal mutants with lesions in RNA polymerase subunit genes (11). RNA polymerase mutants are defective for late gene transcription but normal for early gene transcription. While the characterization of these mutants was reported prior to the discovery of the intermediate class of vaccinia virus genes, RNA polymerase mutants are likely defective for intermediate transcription as well. The RNA polymerase mutants also produce large viroplasm masses of DNA in the cytoplasm and fail to produce virion assembly intermediates. These mutants also are defective for processing of genome DNA concatemers into unit lengths (17), which probably explains why the DNA remains associated in large aggregates.
The combined effects of inhibition of late transcription factor synthesis and inhibition of late transcription by distamycin no doubt constitute a particularly potent means by which to block vaccinia virus late gene expression. The results presented here indicate that DNA minor-groove ligands may hold promise for the development of new anti-poxvirus drugs. While distamycin itself is too toxic for therapeutic purposes, other minor-groove ligands with specificity for vaccinia virus promoters may be developed. DNA minor-groove ligands may also be useful for investigations into which inhibiting postreplicative mRNA synthesis is desired without disturbing DNA replication.
Acknowledgments
We are grateful to Debra Sherman for expert electron microscopy.
B.A.K. was supported by NIH predoctoral training grant GM08737.
REFERENCES
- 1.Abu-Daya, A., P. M. Brown, and K. R. Fox. 1995. DNA sequence specificity of several AT-selective minor groove binding ligands. Nucleic Acids Res. 23:3385-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick, C. J., Jr., J. G. Keck, and B. Moss. 1992. Mutational analysis of the core, spacer and initiator regions of vaccinia virus intermediate class promoters. J. Virol. 66:4710-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertholet, C., E. Van Meir, B. ten Heggeler-Bordier, and R. Wittek. 1987. Vaccinia virus produces late mRNAs by discontinuous synthesis. Cell 50:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broyles, S. S. 2003. Vaccinia virus transcription. J. Gen. Virol. 84:2293-2303. [DOI] [PubMed] [Google Scholar]
- 5.Broyles, S. S., X. Liu, M. Zhu, and M. Kremer. 1999. Transcription factor YY1 is an activator of vaccinia virus late promoters. J. Biol. Chem. 274:35662-35667. [DOI] [PubMed] [Google Scholar]
- 6.Broyles, S. S., L. Yuen, S. Shuman, and B. Moss. 1988. Purification of a factor required for transcription of vaccinia virus early genes. J. Biol. Chem. 263:10754-10760. [PubMed] [Google Scholar]
- 7.Condit, R. C., and A. Motyczka. 1981. Isolation and preliminary characterization of temperature sensitive mutants of vaccinia virus. Virology 113:224-241. [DOI] [PubMed] [Google Scholar]
- 8.Dales, S. 1963. The uptake and development of vaccinia virus in strain L cells followed with labeled viral deoxyribonucleic acid. J. Cell Biol. 18:51-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davison, A. J., and B. Moss. 1989. The structure of vaccinia virus late promoters. J. Mol. Biol. 210:771-784. [DOI] [PubMed] [Google Scholar]
- 10.Geierstanger, B. H., and D. E. Wemmer. 1995. Complexes of the minor groove of DNA. Annu. Rev. Biophys. Biomol. Struct. 24:463-493. [DOI] [PubMed] [Google Scholar]
- 11.Hooda-Dhingra, U., C. L. Thompson, and R. C. Condit. 1989. Detailed phenotypic characterization of five temperature-sensitive mutants in the 22- and 147-kilodalton subunits of vaccinia virus DNA-dependent RNA polymerase. J. Virol. 63:714-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houbaviy, H. B., A. Usheva, T. Shenk, and S. K. Burley. 1996. Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc. Natl. Acad. Sci. USA 93:13577-13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, G. P., S. J. Goebel, and E. Paoletti. 1993. An update on the vaccinia virus genome. Virology 196:381-401. [DOI] [PubMed] [Google Scholar]
- 14.Keck, J. G., C. J. Baldick, Jr., and B. Moss. 1990. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late transactivator genes. Cell 61:801-809. [DOI] [PubMed] [Google Scholar]
- 15.Kovacs, G. R., N. Vasilakis, and B. Moss. 2001. Regulation of viral intermediate gene expression by the vaccinia virus B1 protein kinase. J. Virol. 75:4048-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lown, J. W., K. Krowicki, J. Balzarini, R. A. Newman, and E. De Clercq. 1989. Novel linked antiviral and antitumor agents related to netropsin and distamycin: synthesis and biological evaluation. J. Med. Chem. 32:2368-2375. [DOI] [PubMed] [Google Scholar]
- 17.Merchlinsky, M., and B. Moss. 1989. Resolution of vaccinia virus DNA concatemer junctions requires late gene expression. J. Virol. 63:1595-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Schwer, B., P. Visca, J. C. Vos, and H. G. Stunnenberg. 1987. Discontinuous transcription or RNA processing of vaccinia virus later messengers results in a 5′ poly(A) leader. Cell 50:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatesan, S., B. M. Baroudy, and B. Moss. 1981. Distinctive nucleotide sequences adjacent to multiple initiation and termination sites of an early vaccinia virus gene. Cell 25:805-813. [DOI] [PubMed] [Google Scholar]
- 22.Verini, M. A., and M. Ghione. 1964. Activity of distamycin A on vaccinia virus infection in culture. Chemotherapia 9:144-157. [DOI] [PubMed] [Google Scholar]
- 23.Wright, C. F., B. W. Oswald, and S. Dellis. 2001. Vaccinia virus late transcription is activated in vitro by cellular heterogeneous nuclear ribonucleoproteins. J. Biol. Chem. 276:40680-40686. [DOI] [PubMed] [Google Scholar]
- 24.Xiang, Y., D. A. Simpson, J. Spiegel, A. Zhou, R. H. Silverman, and R. C. Condit. 1998. The vaccinia virus A18R DNA helicase is a postreplicative negative transcription elongation factor. J. Virol. 72:7012-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]