Abstract
Acute myeloid leukemia (AML) is a heterogeneous group of diseases in which chromosomal aberrations, small insertions or deletions, or point mutations in certain genes have profound consequences for prognosis. However, the majority of AML patients present without currently known genetic defects. Retroviral insertion mutagenesis in mice has become a powerful tool for identifying new disease genes involved in the pathogenesis of leukemia and lymphoma. Here we have used the Graffi-1.4 strain of murine leukemia virus, which causes predominantly AML, in a screen to identify novel genes involved in the pathogenesis of this disease. We report 79 candidate disease genes in common integration sites (CISs) and 15 genes whose family members previously were found to be affected in other studies. The majority of the identified sequences (60%) were not found in lymphomas and monocytic leukemias in previous screens, suggesting a specific involvement in AML. Although most of the virus integrations occurred in or near the 5′ or 3′ ends of the genes, suggesting deregulation of gene expression as a consequence of virus integration, 18 CISs were located exclusively within the genes, conceivably causing gene disruption.
Acute myeloid leukemia (AML) is characterized by a block in myeloid differentiation that results in the accumulation of leukemic myeloid cells in the bone marrow and peripheral blood. For AML cases with specific chromosomal translocations, the identification and functional characterization of fusion genes located at translocation breakpoints have resulted in the discovery of pathways involved in leukemic transformation (34). However, it also has become clear that these defects themselves are not sufficient to cause acute leukemia, supporting the theory that the disruption of multiple regulatory mechanisms is required to fully transform hematopoietic stem and progenitor cells toward AML. At present, cytogenetic parameters are used successfully in clinics for risk stratification of leukemia. For instance, AML cases with t(8;21), t(15;17), and inv(16) chromosomal abnormalities are classified as low risk, whereas cases with 3q26, 5q, and 7q abnormalities generally are classified as high risk, with unfavorable treatment outcomes resulting in early relapse and decreased overall survival (34, 47). Importantly, for the majority of AML cases (>60%), tentatively classified as intermediate risk, the genetic parameters predictive for therapy outcome have not been identified (14, 15). Furthermore, in the approximately 20 to 40% of AML patients without chromosomal abnormalities, the molecular pathogenesis remains entirely unknown (33).
Retroviral insertion mutagenesis in mice has become a powerful and rapid method for the identification of new genes involved in cancer (23). This approach has benefitted greatly from both human and mouse genome programs and the recently developed genome database search programs (20, 29). Studies aimed at finding novel genes involved in leukemia thus far have been carried out with virus strains that have a propensity to induce lymphoid malignancies or myelomonocytic tumors (21, 26, 32, 37, 44, 54). To focus this strategy on myeloid leukemia, we have used the Graffi-1.4 (Gr-1.4) virus, which has been demonstrated to induce leukemia with predominantly myeloid or mixed-lineage early hematopoietic phenotypes (9, 48). As with the Moloney and Cas-Br-M viruses used in previous studies, the Gr-1.4 virus does not contain oncogenic sequences but can deregulate or disrupt gene expression due to proviral integration.
Recently, Erkeland et al. reported on a new Gr-1.4 virus common integration site (CIS) that is located in the Yin Yang 1 (YY1) promoter and that causes the deregulation of YY1 gene expression, resulting in defective myeloid differentiation (9). Here we report 94 candidate leukemia disease genes that are targeted by the Gr-1.4 virus. While some of the affected genes in the Gr-1.4 virus-induced leukemia overlapped with those identified in lymphoma screens, suggesting a more general involvement in leukemogenesis and lymphogenesis, the majority of integrations appeared to affect novel genes that may be more specifically involved in the development of myeloid malignancies.
MATERIALS AND METHODS
MuLV Gr-1.4-induced leukemia.
Newborn mice were injected subcutaneously with 100 μl of cell culture supernatant from murine leukemia virus (MuLV) Gr-1.4-producing NIH 3T3 cells (a gift from E. Rassart, Department des Sciences Biologiques, Universite du Quebec à Montreal, Montreal, Quebec, Canada). Mice were treated and analyzed for the development of leukemia as previously described (9). Chromosomal DNA was isolated from leukemic cells for PCR-based screening (9).
Cytological analysis and immunophenotyping of leukemic cells.
For morphological analysis, blood smears and cytospin preparations were fixed in methanol, stained with May-Grünwald-Giemsa stained, and examined by using an Axioscope microscope (Carl Zeiss BV, Weesp, The Netherlands). Single-cell suspensions of different organs were analyzed by flow cytometry with a FACScan flow cytometer (Becton Dickinson and Co., Mountain View, Calif.). Cells were labeled as described previously (25) with the following rat monoclonal antibodies: ER-MP54, ER-MP58, M1/70 (Mac-1), F4/80, RB68C5 (GR-1), ER-MP21 (transferrin receptor), TER119 (glycophorin A), 59-AD2.2 (Thy-1), KT3 (CD3), RA3 6B2 (B220), and E13 161-7 (Sca1). Immunodetection was performed with goat anti-rat antibodies coupled to fluorescein isothiocyanate (Nordic, Tilburg, The Netherlands).
Inverse PCR of MuLV Gr-1.4-induced leukemia.
The inverse PCR strategy was recently described in detail (9). Briefly, genomic DNA from primary tumors was digested with HhaI (CGCG). After circularization by ligation (rapid ligation kit; Roche Diagnostics, Mannheim, Germany), an initial PCR was performed with MuLV Gr-1.4 (long terminal repeat [LTR])-specific primers L1 (5′-TGCAAGATGGCGTTACTGTAGCTAG-3′) and L2 (5′-CCAGGTTGCCCCAAAGACCTG-3′). Cycling conditions were 1 min at 94°C, 1 min at 62°C, and 3 min at 72°C for 30 cycles. For the second, nested PCR, primers L1N (5′-AGCCTTATGGTGGGGTCTTTC-3′) and L2N (5′-AAAGACCTGAAACGACCTTGC-3′) (15 cycles) were used. The PCR mixture contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, 10 pmol of each primer, and 2.5 U of Taq polymerase (Pharmacia, Uppsala, Sweden). The PCR fragments were analyzed on a 1% agarose gel.
Detection of virus integration by a specific nested PCR.
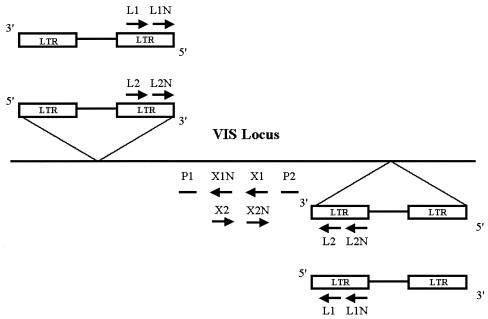
To determine the localization of the Gr-1.4 provirus in specific virus-targeted genes in an extended panel of leukemias, a nested PCR was performed with DNA from primary tumors. For the first PCR, virus integration site locus-specific primers X1 and X2 were used in combination with MuLV Gr-1.4 LTR-specific primers L1 and L2 (Fig. 1). Cycling conditions were 1 min at 94°C, 1 min at 62°C, and 3 min at 72°C for 30 cycles. For the second PCR, nested virus integration site-specific primers X1N and X2N were used in combination with nested LTR-specific primers L1N and L2N under the same conditions (Fig. 1). The PCR products obtained were analyzed by Southern blotting. To verify the correct nature of the amplified bands, the blots were hybridized with radiolabeled gene-specific probes P1 and P2 (Fig. 1) at 45°C in Church buffer (0.5 M phosphate buffer [pH 7.2], 7% [wt/vol] sodium dodecyl sulfate, 10 mM EDTA) overnight. Signals were visualized by autoradiography according to standard procedures (34a).
FIG. 1.
Directed PCR of chromosomal DNA to determine the commonality of virus integration sites (VIS) identified by inverse PCR. Primers X1, X1N, X2, and X2N were designed from a locus that was identified as a virus integration site by inverse PCR. To amplify flanking genomic sequences and to determine the localization and orientation of the integrated provirus, four different nested PCRs were performed. First, primers X1 and X2 were combined with MuLV Gr-1.4 LTR primers L1 and L2. The products were amplified by a nested PCR with primers X1N and X2N in combination with L1N and L2N. The specificity of the amplified bands was checked by Southern blot analysis with probes P1 and P2.
Nucleotide sequence analysis.
PCR products were sequenced by using an ABI 3100 sequencer (Applied Biosystems, Neuwerkerk aan den Ijssel, The Netherlands) with MuLV Gr-1.4-specific forward primer L1. Virus flanking genomic sequences were analyzed by using GenBank (National Center for Biotechnology Information), a Celera discovery system (Celera Genomics, Rockville, Md.) (19, 29), and Ensembl (Wellcome Trust Genome Campus, Hinxton, Cambridgeshire, United Kingdom) (20).
RESULTS
Gr-1.4-induced leukemias.
Eighty-nine newborn FVB/N mice were inoculated with Gr-1.4. When moribund, mice were sacrificed, and hematopoietic organs were isolated. Six mice died without signs of leukemia and were excluded from further investigation. Standard blood cell analysis was performed, and values were compared with the mean value for 10 normal FVB/N mice (24). Most of the leukemic mice had increased numbers of peripheral white blood cells and decreased numbers of platelets and red blood cells compared to normal controls (data not shown). Blast percentages in the bone marrow ranged from 24 to 90%, with an average of 48%. Leukemic cells from 76 mice were immunophenotyped. The major immunophenotypic features of these leukemias are given in Table 1. Based on these criteria, the leukemias were classified as T-lymphoid; mixed lymphoid, erythroid, and myeloid; myeloid; myelomonocytic; or erythroid. Fifty-nine leukemias were analyzed morphologically. Representative examples are shown in Fig. 2. Almost all of the mice showed splenomegaly, with 25% showing thymus enlargement, 20% showing lymph node enlargement, and 55% showing liver involvement. In 5% of the mice, leukemic cells were present in the bone marrow and blood, without overt peripheral organ involvement.
TABLE 1.
Gr-1.4-induced leukemiasa
| Subtype | Leukemia type | Immunophenotype (markers) | No. of leukemias |
|---|---|---|---|
| I | T-lymphoid | MP21+ CD3+ Thy1+ | 2 |
| II | Mixed lymphoid, erythroid, and myeloid differentiation | Gr1+ F4/80+ Mac1+ Imm+ CD3+ B220+ (gcsfr+ Ter119+) | 12 |
| III | Myeloid differentiation | 1mm+ MP21+ (F4/80+ Gr1+ B220+ Mac1+gcsfr+) | 43 |
| IV | Myelocytic or monocytic | 1mm+ Gr1−gcsfr− (F4/80+ Mac1+ B220+) | 15 |
| V | Erythroid | Ter119+, MP21+ (Sca1+) | 4 |
| Total | 76 |
All tumors were tested with the markers MP21, MP58, CD3, Thy1, Gr1, F4/80, Mac1, Sca1, B220, Ter119, and gcsfr. Markers that were consistently negative are not shown, unless they were informative for discrimination between myeloid subtypes III and IV. Imm+ indicates positive staining for immature hematopoietic cell markers Sca1+, MP58+, and MP54+ (Thy1+). Markers in parentheses were not present in all individual tumors.
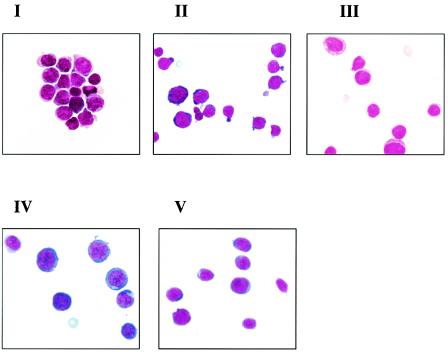
FIG. 2.
Morphological features of MuLV Gr-1.4-induced leukemia. Micrographs were taken with a Zeiss Axioscope microscope (magnification, ×1,000). Morphological subtypes were lymphoblasts (I); mixed erythroblasts, myeloblasts, and lymphoblasts (II); myeloblasts (III); myelo-monoblasts (IV); and erythroblasts (V).
Virus integration sites in Gr-1.4-induced leukemias.
PCR analysis in conjunction with database searches identified a total of 94 different virus integration sites out of 69 tumors, 38 of which were found in earlier screens. Examples of this latter group of CISs are p53 (39, 41), Notch-1 (12, 13, 31), Evi-1 (38), NF1 (Evi-2) (5), Lck-1 (1, 11, 35), Pim-1 (50), HoxA9 (Evi-6) (43), Fli-1 (4, 8), and N-Myc (18, 51). Notably, 79 of the 94 integrations were CISs, directly implicating the affected genes in leukemic transformation. The remaining 15 were included in this report because family members closely related to these genes were found in other studies. Fifty-six of the identified sequences were mapped near or in novel candidate leukemia genes. The products of the affected genes have been classified as receptors and signaling molecules (Table 2), as regulators of transcription (Table 3), and as having regulatory roles in other pathways (e.g., DNA stability and proteasomal targeting) or unknown functions (Table 4).
TABLE 2.
MuLV Gr-1.4 integration sites in receptors and signaling-related molecules
| Genea | Virus integration site localization | Databaseb | Tumorsc | Subtype of (leukemia)d | Chromosome
|
Gr-1.4 virus unique CIS (x) or sourcee | Family member identified in other studies | Unique database identifier | |
|---|---|---|---|---|---|---|---|---|---|
| Mouse | Human | ||||||||
| II-2Rα | 7 kb upstream of gene | C, E | 1# | IV | 2A1 | 10p15.1 | RTCGD | NM_008367 | |
| II-2Rβ | 500 bp upstream of gene | C, E | 1 | III | 15E2 | 22q13.1 | RTCGD | NM_008368 | |
| Notch-1 | Exon 12, intron 14 | C, E, N | 2 | II | 2A3 | 9q34.3 | RTCGD (31) | Notch-2 | NM_008714 |
| Flt3 | 1.5-2 kb upstream of gene | E | 2 | III | 5G2 | 13q12.3 | RTCGD | NM_010229 | |
| Tie-1 | 6-15 kb downstream of gene | C, E | 2 | III, V | 4D1 | 1p34.2 | x | NM_011587 | |
| c-Mpl | 0.25-10 kb upstream of gene | C, E | 2 | III, V | 4D1 | 1p34 | x | NM_010823 | |
| Edg3 (G protein coupled) | 18 kb upstream of gene | C, E | 7/12§ | II, III, IV | 13B1 | 9q22.1 | x | Edg1 (RTCGD) | NM_010101 |
| Lck-1 | Intron 1 (upstream of ATG) | C, E | 1 | III | 4D2.3 | 1p34.3 | RTCGD | XM_110515 | |
| Pim-1 | 9 kb downstream of gene | C, E | 1 | VI | 17B1 | 6p21.2 | RTCGD (50) | Pim-2, Pim-3 | NM_008842 |
| Akt-2 | 11-14 kb upstream of gene | C, E | 4/13 | II, IV | 7A3 | 19q13.1 | RTCGD | Akt-1 | NM_007434 |
| PTPN5 | Exon 1 (upstream of ATG) | C, E, N | 1/24 | III | 7B3 | 11p15.1 | x | PTPN18, PTPNS1 (RTCGD) | NM_013643 |
| PTPN1 (PTP1B) | Intron 1 (downstream of ATG) | C, E, N | 9/13 | II, III, IV | 2H3 | 20q13.13 | RTCGD | PTPN18, PTPNS1 | NM_011201 |
| DUSP10 (MKP-5) | 1 kb upstream of gene | C, E | 1 | III | 1H4 | 1q41 | RTCGD | DUSP2 | NM_022019 |
| Inpp5b | 0.3 kb upstream of gene | C, E | 6/13 | I, III, V | 4D1 | 1p34.3 | x | NM_008385 | |
| Inpp4a | Intron 14 | C, E, N | 6/12 | III, IV, V | 1B | 2q11.2 | RTCGD | NM_030266 | |
| Dok-1 (p62) | 26-28 kb from gene | C, E, N | 10/10 | I, II, III | 6D1 | 2p13 | x | Dok1 (pending) (RTCGD) | NM_001381 |
| Phospholipase D3 (Pld3) | 25 kb upstream of gene | E | 5/13 | II, III, IV | 7A3 | 19q13.1 | RTCGD | Plaa, 2310004B05Rik | NM_011116 |
| Dapp-1 | 0.8 kb upstream of gene to intron 1 (downstream of ATG) | C, E | 11/13 | II, III, IV | 3H2 | 4q24 | RTCGD | NM_011932 | |
| Edaradd | 17 kb downstream of gene | C, E | 1 | III | 13A2 | 1q43 | RTCGD | NM_133643 | |
| Semaphorin 4b | 3 kb upstream of gene | C, E | 1 | V | 7D2 | 15q26.1 | RTCGD | Semaphorin 7a | XM_133534 |
| Insulin receptor substrate 2 | 2.5 kb downstream of gene | C, E | 2 | III, IV | 8A2 | 13q34 | RTCGD | XM_146235 | |
| NF1 (Evi-2) | Intron 35 | C, E | 2 | III | 11B5 | 17q11.2 | RTCGD (5) | NM_010161 | |
| PKI-gamma | 66 kb upstream of gene | C, E | 1 | IV | 2H3 | 20q13.12 | RTCGD | U97170 | |
| Tao-2 | 3-4 kb upstream of gene | C, E | 5/13 | II, III, IV | 7F4 | 16p13.11 | RTCGD | NM_004783 | |
| Shc3 | 41 kb downstream of gene | C, E | 7/12 | II, III, IV | 13B1 | 9q22.1 | x | NM_009167 | |
| Calcyphosine | Intron 4 (downstream of ATG) | C, E | 2/13 | II | 15A2 | 5p13.3 | x | NM_029341 | |
| TNF-related protein 6 | 2.5 kb downstream of gene | C, E | 1 | III | 15E2 | 22q13.1 | RTCGD | Many | AF329842 |
| Lymphotoxin beta (TNF-C) | 8.3 kb upstream of gene | C, E | 1 | III | 17B2 | 6p21.3 | RTCGD | Many | NM_008518 |
| Phosphodiesterase 1 | Intron 3, intron 4 (downstream of ATG) | C, E | 6/13 | I, II, III, IV | 10A3 | 6q22.31 | x | Phosphodiesterase 4b (RTCGD) | NM_006208 |
| Socs-2 | 0.75 kb upstream of gene to intron 1 (downstream of ATG) | C | 8/39 | II, III, IV, V | 10C3 | 12q22 | x | Socs-1, Socs-7, Asb-2 (RTCGD) | NM_003877 |
| ARF-related protein | 1.5 kb upstream of gene, intron 1 (upstream of ATG) | C, E | 8/13 | II, III, IV | 2H4 | 20q13.13 | x | ARF6, ARF2 (RTCGD) | AF217796 |
| RAP-1 | 18 kb upstream of gene | C, E | 1 | III | 15F2 | 12q12 | x | RAP1a (RTCGD) | AF103905 |
IL-2Rα, interleukin 2 receptor alpha; PKI-gamma, phosphokinase gamma; TNF, tumor necrosis factor; ARF, ADP-ribosylation factor.
Sequences from inverse PCR were analyzed with Celera (C), Ensembl (E), and National Center for Biotechnology Information (N) BLAST search programs, and the exact integration site was localized.
Number of tumors found positive for integration by inverse PCR only (out of 69 analyzed) # or by inverse PCR followed by directed PCR§ (positive out of total analyzed).
Source (reference) for other studies in which the Gr-1.4 virus integration site was previously found.
TABLE 3.
MuLV Gr-1.4 integration sites in transcriptional regulators
| Genea | Virus integration site localization | Databaseb | Tumorsc | Subtype of tumord | Chromosome
|
Gr-1.4 virus unique CIS (x) or sourcee | Family member identified in other studies | Unique database identifier | |
|---|---|---|---|---|---|---|---|---|---|
| Mouse | Human | ||||||||
| JunB | 10 kb upstream of gene | C | 14/14 | I, II, III, IV | 8C3 | 19p13.2 | RTCGD | NM_008416 | |
| Fli-1 | Between exon 1a and exon 1 (upstream of ATG) | C, E, N | 7 | III, V | 9A5.2 | 11q24 | RTCGD (8) | NM_008026 | |
| p53 | Introns 1, 5, and 6; exons 5, 6, and 7; 1.4 kb upstream of gene | C, E, N | 7 | III, V | 11B4 | 17p13.1 | RTCGD (39) | NM_011640 | |
| Evi-1 | 2-14 kb upstream of gene | C, E | 2 | III, IV | 3A3 | 3q26 | RTCGD (38) | NM_007963 | |
| n-Myc | Intron 1 upstream of gene | C, E, N | 1 | II | 12A2 | 2p25.1 | RTCGD (51) | Pvt1, c-Myc (RTCGD) (51) | NM_008709 |
| COE (OLF-1/EBF) | 14 kb upstream of gene | C, E | 4/48 | II, III, IV | 11B1.1 | 5q34 | RTCGD | AF208502 | |
| Elf-4 (MEF) | 0.25-5.5 kb upstream of gene, intron 1 (downstream of ATG) | C, E | 11/12 | II, III, IV | XA3.2 | Xq26.1 | RTCGD | NM_019680 | |
| YY1 | 0.5-1.5 kb upstream of gene | E, N | 14/20 | I, III | 12F2 | 14q32.2 | RTCGD (9) | NM_009537 | |
| Sax-1 (NKX-1.1) | 0.3 kb upstream of gene, intron 1, exon 2 (upstream of and downstream of ATG) | C, E, N | 1 | III | 7F4 | 10q26.12 | x | NKX-1.2 (RTCGD) | NM_009123 |
| NK-2.3 homeobox | 6 kb upstream of gene | C, E | 2 | III, IV | 19D1 | 10q24.32 | x | NKX-1.2 (RTCGD) | AF155583 |
| Evi-6 (Hoxa9) | 2 kb upstream of gene | C, E | 1 | IV | 6B3 | 7p15.2 | RTCGD (43) | Hoxa7, Hoxb4 (RTCGD) | NM_010456 |
| mPLZF | 1.5 kb upstream of gene to intron 1 (upstream of ATG) | C, E | 3/59 | III | 9B | 11q23 | x | NM_010823 | |
| ELL | Exon 1, intron 1 (downstream of ATG) | C, E | 11/15 | II, III, IV | 8C1 | 19p13.11 | x | NM_007924 | |
| Cesanne | 0.8 kb, 16 kb upstream of gene | C, E | 2/13 | III, IV | 3F2 | 1q23.1 | x | NM_020205 | |
| Nuclear hormone receptor NR1D1 | 8-10 kb upstream of gene | C, E | 11/12 | II, III, IV | 11D | 17q21.1 | x | BC008989 | |
| Retinoid X receptor alpha | 84 kb upstream of exon 1, 2 kb upstream of gene, intron 1 (upstream of ATG) | C | 3/12 | II, III, IV | 2A3 | 9q34.2 | x | NM_011305 | |
| n-Cor-1 | Intron 1 (downstream of ATG) | E | 3/13 | III | 11B2 | 17p11.2 | x | n-Cor-2 (RTCGD) | NM_011308 |
| HDAC1 | 14 kb upstream of gene | C, E | 1/13 | III | 4D2.3 | 1p34.3 | x | HDAC5, HDAC9 (RTCGD) | NM_008228 |
| HDAC7A | 0.1-8 kb upstream of gene, introns 3 and 4 (downstream of ATG) | C, E | 3/13 | II, III | 15F2 | 12q12 | x | HDAC5, HDAC9 (RTCGD) | NM_019572 |
| Smarcc2 | 0.55 kb upstream of gene | C, E | 1 | III | 10D3 | 12q13.3 | x | Smarcf1 (RTCGD) | NM_003075 |
| HMG-17 | 28 kb upstream of gene, 1-2 kb downstream of gene | C, E | 3/13 | II, III | 8C1 | 19p13.11 | x | HMG-a1, HMG-a2, HMG-b1, HMG-c1 HMG-Cr (RTCGD) | AL590390 |
TABLE 4.
Other MuLV Gr-1.4 integration sites
| Genea | Virus integration site localization | Databaseb | Tumorsc | Subtype of tumord | Chromosome
|
Gr-1.4 virus unique CIS (x) or sourcee | Family member identified in other studies | Unique database identifier | |
|---|---|---|---|---|---|---|---|---|---|
| Mouse | Human | ||||||||
| Protein-arginine deiminase type 1 (Pdi-1, PADI1) | 0.5 kb upstream of gene, intron 1 (upstream of and downstream of ATG) | C, E | 5/13 | II, III, IV | 4E1 | 1p36.1 | RTCGD | Pdi-2 (RTCGD) | NM_011059 |
| Myosin IC synaptic vesicle | Intron 1 (upstream of ATG) | C, E | 12/39 | II, III, IV | 9D | 15q22.2 | RTCGD | Myosin 1B, myosin IIIA (RTCGD) | XM_207764 |
| Glycoprotein 2A (Sv2A) | 27 kb upstream of gene | E | 1 | III | 3F2 | 1q23.1 | RTCGD | NM_022030 | |
| Swap70 | Intron 3 (downstream of ATG) | C, E | 1 | IV | 8C3 | 19p13.2 | RTCGD | NM_009302 | |
| PAFAH1B2 | 11 kb downstream of gene | E, N | 1 | III | 9B | 11q23.3 | RTCGD | NM_008775 | |
| Netrin 1 | Intron 1 (downstream of ATG) | C, E | 1 | ND | 11B3 | 17p11.2 | x | XM_193630 | |
| Multispanning membrane protein (TM9SF2) | 1 kb upstream of gene | C, E | 1 | III | 14E5 | 13q32.3 | RTCGD | NM_080556 | |
| Histone H3.3A (H3F3A) | Exon 1 (downstream of ATG) | C, N | 1 | IV | 1H4 | 1q41 | x | H1b, H1c, H2 (RTCGD) | Z85979 |
| Gene trap locus 13 (GTL1-13) | 17 kb upstream of gene | C, E | 2 | III | 2E1 | 11q12 | x | NM_021512 | |
| Tumor differentially expressed Tde1 | 15 kb upstream of gene | C, E | 1 | IV | 2H3 | 20q13.12 | RTCGD | NM_012032 | |
| O-Methyltransferase | 4.5-25 kb upstream of gene | E | 2 | III | 14B | 10q22.2 | x | AK007659 | |
| CHD2 | 17.3-18.8 kb upstream of gene | C, E | 11/13 | I, II, III, IV | 7D1 | 15q26.1 | x | NM_001271 | |
| Blm | 0.8-3 kb upstream of gene | C, E | 4/30 | II | 7D2 | 15q26.1 | x | NM_007550 | |
| VDUP1 | 1.6 kb upstream of gene to intron 1 (downstream of ATG), exon 8 (downstream of stop) to 3 kb downstream of gene | C, E, N | 14/14 | I, III, IV | 3F2 | 1q21 | x | AF282826 | |
| PrdxII | 2.4 kb upstream of gene to intron 1 (upstream of ATG) | C, E | 14/14 | I, II, III, IV | 8C3 | 19p13.2 | x | NM_011563 | |
| Lymphocyte antigen (Y-6M, LY-6A.2/LY-6E.1; TAP) | 6 kb downstream of gene | C, E | 8/39 | II, III, IV, V | 10C3 | 12q22 | x | Lymphocyte antigens 108, 74, and 6E (RTCGD) | NM_010738 |
| Periaxin | 57 kb upstream of gene | E | 5/13 | II, III, IV | 7A3 | 19q13.1 | x | AJ222968 | |
| RNase H1 subunit | 3 kb upstream of gene | C, E | 14/14 | I, II, III, IV | 8C3 | 19p13.2 | x | AK010292 | |
| Apoptosis inhibitory 5 (Api-5), fibroblast growth factor 2-interacting factor 2 | 6 kb downstream of gene, intron 1 (downstream of ATG) | C, E | 5/25 | I, III | 2E1 | 11p11.2 | x | NM_007466 | |
| Hematopoietic progenitor (CDCA4) | Intron 1 (upstream of ATG) | C, E | 2/13 | IV | 14E5 | 13q23.3 | x | AK010535 | |
| U1 snRNP binding protein | 1 kb upstream of gene | C, E | 2 | II, III | 5F | ? | x | AK018232 | |
| Unnamed | Intron 1 (downstream of ATG) | C, E | 3/13 | II, III | 15F2 | 12q12 | x | AK002729 | |
| Unnamed | 1.5 kb downstream of gene | C, E | 5/13 | II, III, IV | 7F4 | 16p13.11 | x | AK004043 | |
| Unnamed | 0.1-1.5 kb upstream of exon 1 | C, E | 9/12 | II, III, IV | 8C1 | 19p13.11 | x | AK013135 | |
| Unnamed | Intron 1 (downstream of ATG) | E | 5/25 | I, III | 2E1 | 11p11.2 | x | AK001752 | |
| Lim domain protein Ril | Intron 1 (downstream of ATG) | C, E | 2/13 | I, II | 11B1.3 | 5q31.1 | x | NM_019417 | |
| D6MM5e | Intron 4 (downstream of ATG) | C, E, N | 10/10 | I, II, III | 6D1 | 2p13 | x | AF084364 | |
| Psmb-1 | 1 kb upstream of gene | C, E | 1 | ND | 17A2 | 6q27 | x | Psmb-8 (RTCGD) | NM_011185 |
| Ubiquitin | 13 kb upstream of gene | C, E | 9/12 | II, III, IV | 8C1 | 19p13.11 | x | Many (RTCGD) | NM_019639 |
| Psme-1 | 0.34 kb upstream of gene | C, E | 1 | IV | 14C1 | 14q11.2 | x | Psmb-8 (RTCGD) | AB007136 |
| Ddx-21 | 2 kb downstream of gene | C, E | 1 | IV | 10B4 | 10q22.1 | x | Ddx-3, Ddx-6, Ddx-26 (RTCGD) | NM_019553 |
| Actin-like | 0.2 kb upstream of gene | C, E | 1 | III | 10C2 | 12q23.1 | x | β-Actin (RTCGD) | AK008409 |
| Alpha-1 catenin | 83 kb upstream of gene | C, E | 1 | III | 18B3 | 5q31.2 | NM_009818 | ||
| VDAC-2 | 7.5 kb downstream of gene | C | 1 | III | 14B | 10q22.2 | x | VDAC-1 (RTCGD) | NM_011695 |
| Proteoglycan, secretory granule (PRG1) | 1.1 kb upstream of gene | C, E | 1 | I | 10B4 | 10q22.1 | x | PRG4 (RTCGD) | NM_011157 |
| Potassium channel Kcnk5 | Intron 1 (downstream of ATG) | C, E | 1 | II | 14A3-B | 6p21.2 | RTCGD | NM_021542 | |
| FK506BP8 | 0.2-6.5 kb upstream of gene | C, E | 9/12 | II, III, IV | 8C1 | 19p13.11 | x | FK506BP3, FK506BP10 (RTCGD) | NM_010223 |
| MCT related | 10 kb upstream of gene | C, E | 1 | III | 11B4 | 17p13.2 | x | MCT4 (RTCGD) | AB041591 |
| RNA binding motif 5 | 2.2-3 kb upstream of gene | E, N | 3/11 | I, III | 9F2 | 3p21.31 | x | AJ309168 | |
| RNA binding motif 6 | 3-4.8 kb downstream of gene | E, N | 3/11 | I, III | 9F2 | 3p21.31 | x | NM_011251 | |
Approximately 15% of the virus integrations identified by this inverse PCR-based screen were determined to be common (more than two integrations in a particular genomic locus) by the initial screen. To determine the frequency of common integrations more sensitively, we performed integration-specific PCRs with 49 Gr-1.4-targeted genes in multiple tumors (Tables 2 to 4); most appeared to be common. We sequenced the integrations in 19 genes derived from multiple (2 to 10) tumors; these included VDUP1, PrdxII, 11- to 19-lysine-rich leukemia (ELL), promyelocytic leukemia zinc finger (PLZF), and Edg3. In all instances, the integrations were located in the expected gene but at different positions, confirming the specificity of the Southern analysis and ruling out the possibility of cross-contamination by previously amplified PCR products. The genes flanking the virus integration sites were compared with data from the National Cancer Institute retrovirus-tagged cancer gene database (RTCGD) at http://genome2.ncifcrf.gov and other sources (Tables 2 to 4). The Gr-1.4 integrations that are included in this database or that were reported in other studies are indicated by the database name or references in Tables 2 to 4. Family members of Gr-1.4-targeted genes found in other studies are also indicated in those tables. Most of the virus integrations occurred in or near the 5′ or 3′ ends of the genes, suggesting that the levels of expression of these genes are deregulated as a consequence of virus integration. Eighteen CISs were located exclusively within the gene, conceivably causing gene disruption. This group comprises the previously reported virus targets Notch-1, NF1, PTPN1, Inpp4a, p53, SWAP70, and Kcnk5 and 11 new virus targets: calcyphosine, phosphodiesterase 1, ELL, NCOR-1, HDAC-7A, histone H3.3A, Api-5, Ril, D6Mm5e, and two unknown genes.
DISCUSSION
In this study, we have used in vivo retroviral mutagenesis with the Gr-1.4 virus complex to identify novel disease genes specifically involved in the pathogenesis of AML. In comparable studies with other virus strains, e.g., Moloney, AKXD, or Cas-Br-M, the most prominently appearing tumor types are T- and B-cell lymphomas; in the case of BXH2, myelomonocytic tumors are prominent (21, 30, 32, 37, 54). In contrast, more than 80% of the leukemias induced by Gr-1.4 unequivocally exhibited immunophenotypic characteristics of myeloid cells, with immature morphological features (mainly myeloblasts), emphasizing the unique features of the Gr-1.4 virus complex as a tool for characterizing pathogenetic mechanisms in myeloid disease. Indeed, the majority of the CISs described here have thus far not been reported in extensive screens in lymphoma models, although some overlap was observed. The latter observation is not surprising in view of the fact that cell type-specific events usually impinge on downstream common regulatory pathways that can be affected in multiple tumor types.
Although most of the Gr-1.4 CISs have been linked to candidate disease genes based on their proximity to these genes, proviral integrations have been reported to influence gene expression over distances of more than 100 kb. Thus, we cannot exclude the possibilities that genes located more distantly from the CISs also are deregulated and that multiple genes are affected by a single CIS (44). In addition, three other aspects of retroviral screens as they are currently being performed must be emphasized. First, malignancies induced by replication-competent retroviruses are usually oligo- or polyclonal rather than monoclonal (9, 32). Although this characteristic gives rise to high frequencies of CISs in relatively small cohorts of mice, it complicates the search for cooperating events within one leukemic clone. Second, the sensitive PCR-based techniques used to identify virus integrations do not allow distinction between CISs present in a majority of the leukemic cells, initiating an early pathogenic event, and CISs present in only a minor population of the cells, probably affecting leukemia progression genes. Finally, a recent study emphasized that MuLV strains have a preference for integration near transcriptional start sites (62). Our data obtained with Gr-1.4 corroborate these conclusions and indicate that retroviral mutagenesis with MuLV preferentially, although not exclusively, identifies gene deregulation rather than gene disruption. Another important conclusion drawn by Wu and colleagues is that there appear to be no integration hot spots (preferred integration sites in certain loci) for MuLV (62). Therefore, CISs found in viral screens are likely to play a role in leukemia.
Many of the Gr-1.4-affected genes appeared to be related to signaling; some have been linked to human leukemia already. For instance, the Tie-1 gene, which encodes a tyrosine kinase receptor that is normally expressed in vascular endothelial cells and hematopoietic stem cells, is overexpressed in chronic myeloid leukemia (59). Importantly, high Tie-1 levels inversely correlate with the survival of chronic myeloid leukemia patients in the early chronic phase (59). Markedly increased levels of Tie-1 also were detected in bone marrow samples from myelodysplastic syndrome and AML patients (58). Notch-1 is another example of a CIS associated with human disease. The human homologue of the Notch (Tan-1) gene is involved in chromosomal translocation t(7;9)(q34;34.3), which results in a truncated receptor in human T-lymphoblastic neoplasms. Notch-1 is a proviral integration site in mouse lymphoid leukemias (12, 31). Our data suggest that aberrant Notch signaling also may be involved in myeloid leukemia. Notably, the integrations in the Notch gene result in the constitutive formation of the truncated active form of Notch (31), which has been demonstrated to interfere with granulocyte colony-stimulating factor-induced myeloid differentiation in the 32D cell model (55). Edg3 (endothelial differentiation gene 3) is a G-protein-coupled receptor (GPCR) involved in cell proliferation and survival (2). The expression of Edg3 is downregulated during the differentiation of HL60 human leukemia cells and is assumed to mediate sphingosine 1-phosphate-induced Ca2+ responses (49). Although the role of Edg receptors in normal hematopoiesis and malignancies is not yet clear, a closely related GPCR, cannabinoid receptor type 2, was found in a CIS in Cas-Br-M-induced leukemia (57). Notably, perturbed expression of cannabinoid receptor type 2 recently was found to interfere with myeloid differentiation, supporting the notion that this type of GPCR may contribute to the pathogenesis of AML by inducing a maturation block (27).
Both protein and lipid phosphatases are crucial, mostly negative, regulators in growth factor signaling pathways (7, 64). Two members of the protein tyrosine phosphatase nonreceptor (PTPN) family affected by Gr-1.4 are PTPN1 and PTPN5. The virus integrations in the PTPN1 gene are localized in intron 1 and conceivably disrupt its function. The PTPN1 gene was found in a CIS in BXH2-induced leukemia (32). Because TYK2, JAK2, and STAT5—JAK/STAT signaling intermediates involved in growth control by hematopoietic growth factors—are all inactivated by PTPN1 (also known as PTP1B) (3, 16, 42), the disruption of PTPN1 is predicted to contribute to uncontrolled proliferation and survival of leukemic cells. Integrations in the PTPN5 gene occur in exon 1, upstream of the ATG translation start site. At present, little is known regarding the signaling function of PTPN5, which is alternatively termed STEP. Interestingly, recent data suggest that PTPN5/STEP shows a preferential affinity for p38 mitogen-activated protein kinase in a redox-sensitive manner, leaving the Erk kinases largely unaffected (40). Inhibition of p38 mitogen-activated protein kinase activity by pharmacological inhibitors has been shown to interfere with terminal neutrophilic differentiation (G.-J. van de Geijn et al., unpublished data) and to prevent apoptosis in neutrophils (10). Conceivably, dephosphorylation of p38 by PTPN5/STEP may have similar effects and may promote the survival of immature myeloid cells. Other phosphatases that are targeted by Gr-1.4 are the inositol polyphosphate phosphatases Inpp4a and Inpp5b, which catalyze the hydrolysis of phosphatidylinositol polyphosphates at positions 4 and 5, respectively. Intriguingly, Inpp4a is a major target for transcription factor GATA-1 and inhibits the growth of megakaryocytes and NIH 3T3 fibroblasts (60). Inpp5b is required for normal sperm development and function (17), but how this enzyme affects normal or leukemic blood cell development is not known.
Reactive oxidant species (ROS) are generated by multiple cellular mechanisms, including metabolic processes, phagocytic reponses to pathogens (oxidative burst), and membrane receptor-mediated signaling. The tight regulation of ROS concentrations in the cell is controlled by both catabolic and redox mechanisms and is important for signal transduction, as it influences protein kinase and phosphatase reactions as well as the activities of transcription factors (46). When ROS levels in a cell are too high, the resulting oxidative stress will affect normal protein function and genomic stability (46). Genes involved in the redox-controlled regulation of ROS levels that are commonly targeted by Gr-1.4 are those for vitamin D3-upregulated protein 1 (VDUP1) and peroxiredoxin 2 (PrdxII). VDUP1 interacts with thioredoxin (TRX) and thereby counteracts the ameliorating effects of TRX on stress-induced apoptosis via apoptosis signal-regulating kinase 1 (ASK-1) and the function of TRX as an antioxidant (28). The consequence of upregulation of PrdxII is as yet unclear, but it may interfere with the function of peroxiredoxin complexes and therefore inhibit the antistress activities of TRX (6). Importantly, PrdxII is a TRX reductase and has been found to be overexpressed in human breast cancer (45).
The ELL and PLZF genes are two examples of genes that are part of fusion genes in human AML as a result of chromosomal translocations. In translocation t(11;19)(q23;p13.1), the ELL gene is fused to the trithorax-like mixed lineage leukemia (MLL) gene (56). The ELL protein is an RNA polymerase II elongation factor (53). Recently, it was demonstrated that both MLL-ELL and ELL inhibit p53 activity via binding of the C-terminal part of ELL to the transactivating domain of p53 (61). The PLZF gene, involved in translocation t(11;17)(q23;q21), is fused in frame with RARα (22, 36). PLZF is expressed in hematopoietic stem cells and normally is downregulated during differentiation. The overexpression of PLZF plays a role in the maintenance of the immature state of hematopoietic progenitor cells and promotes cell survival (52, 63). The fact that PLZF is found as a common Gr-1.4-targeted gene indicates that the deregulation of PLZF expression can contribute to leukemic development independent of its fusion to RARα. One of the major features of PLZF is its tight binding to transcriptional corepressors. Interestingly, several genes involved in transcriptional repression by chromatin modification, such as those for N-Cor-1 and histone deacetylases HDAC1 and HDAC7, are affected by Gr-1.4. Notably, family members of these proteins (N-Cor-2 and HDAC5) also were found in previous screens (54)
In conclusion, using high-throughput retroviral screens with the Gr-1.4 virus complex, we have identified novel pathways involved in myeloid leukemia. Currently, we are studying the consequences of aberrant gene expression or function by using gene transfer methodology with 32D cells and primary bone marrow cultures (9). In addition, we are analyzing the significance of these pathways for human AML by screening expression levels in a panel of clinical samples with a gene expression array and quantitative reverse transcription-PCR technology. By following this combinatorial approach, we hope to develop new prognostic indicators that will discriminate further between low and high responses to therapy in different risk groups. Furthermore, the results obtained may open new avenues for developing specific and sensitive therapies based on the heterogeneous pathogenetic features of human AML.
Acknowledgments
We thank E. Rassart for providing the cell line that produced MuLV Gr-1.4.
This work was supported by grants from the Dutch Cancer Society Koningin Wilhelminafonds.
REFERENCES
- 1.Adler, H. T., P. J. Reynolds, C. M. Kelley, and B. M. Sefton. 1988. Transcriptional activation of lck by retrovirus promoter insertion between two lymphoid-specific promoters. J. Virol. 62:4113-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, S., Y. Zheng, and T. Bleu. 2000. Sphingosine 1-phosphate-induced cell proliferation, survival, and related signaling events mediated by G protein-coupled receptors Edg3 and Edg5. J. Biol. Chem. 275:288-296. [DOI] [PubMed] [Google Scholar]
- 3.Aoki, N., and T. Matsuda. 2000. A cytosolic protein-tyrosine phosphatase PTP1B specifically dephosphorylates and deactivates prolactin-activated STAT5a and STAT5b. J. Biol. Chem. 275:39718-39726. [DOI] [PubMed] [Google Scholar]
- 4.Ben David, Y., E. B. Giddens, and A. Bernstein. 1990. Identification and mapping of a common proviral integration site Fli-1 in erythroleukemia cells induced by Friend murine leukemia virus. Proc. Natl. Acad. Sci. USA 87:1332-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchberg, A. M., H. G. Bedigian, N. A. Jenkins, and N. G. Copeland. 1990. Evi-2, a common integration site involved in murine myeloid leukemogenesis. Mol. Cell. Biol. 10:4658-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butterfield, L. H., A. Merino, S. H. Golub, and H. Shau. 1999. From cytoprotection to tumor suppression: the multifactorial role of peroxiredoxins. Antioxid. Redox Signal. 1:385-402. [DOI] [PubMed] [Google Scholar]
- 7.Coggeshall, K. M., K. Nakamura, and H. Phee. 2002. How do inhibitory phosphatases work? Mol. Immunol. 39:521-529. [DOI] [PubMed] [Google Scholar]
- 8.Denicourt, C., E. Edouard, and E. Rassart. 1999. Oncogene activation in myeloid leukemias by Graffi murine leukemia virus proviral integration. J. Virol. 73:4439-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erkeland, S. J., M. Valkhof, C. Heijmans-Antonissen, R. Delwel, P. J. Valk, M. H. Hermans, and I. P. Touw. 2003. The gene encoding the transcriptional regulator Yin Yang 1 (YY1) is a myeloid transforming gene interfering with neutrophilic differentiation. Blood 101:1111-1117. [DOI] [PubMed] [Google Scholar]
- 10.Frasch, S. C., J. A. Nick, V. A. Fadok, D. L. Bratton, G. S. Worthen, and P. M. Henson. 1998. p38 mitogen-activated protein kinase-dependent and -independent intracellular signal transduction pathways leading to apoptosis in human neutrophils. J. Biol. Chem. 273:8389-8397. [DOI] [PubMed] [Google Scholar]
- 11.Garvin, A. M., S. Pawar, J. D. Marth, and R. M. Perlmutter. 1988. Structure of the murine lck gene and its rearrangement in a murine lymphoma cell line. Mol. Cell. Biol. 8:3058-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girard, L., Z. Hanna, N. Beaulieu, C. D. Hoemann, C. Simard, C. A. Kozak, and P. Jolicoeur. 1996. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 10:1930-1944. [DOI] [PubMed] [Google Scholar]
- 13.Girard, L., and P. Jolicoeur. 1998. A full-length Notch1 allele is dispensable for transformation associated with a provirally activated truncated Notch1 allele in Moloney MuLV-infected MMTV(D)/myc transgenic mice. Oncogene 16:517-522. [DOI] [PubMed] [Google Scholar]
- 14.Grimwade, D., H. Walker, G. Harrison, F. Oliver, S. Chatters, C. J. Harrison, K. Wheatley, A. K. Burnett, and A. H. Goldstone. 2001. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood 98:1312-1320. [DOI] [PubMed] [Google Scholar]
- 15.Grimwade, D., H. Walker, F. Oliver, K. Wheatley, C. Harrison, G. Harrison, J. Rees, I. Hann, R. Stevens, A. Burnett, A. Goldstone, et al. 1998. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. Blood 92:2322-2333. [PubMed] [Google Scholar]
- 16.Haj, F. G., B. Markova, L. D. Klaman, F. D. Bohmer, and B. G. Neel. 2003. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatase-1B. J. Biol. Chem. 278:739-744. [DOI] [PubMed] [Google Scholar]
- 17.Hellsten, E., J. P. Evans, D. J. Bernard, P. A. Janne, and R. L. Nussbaum. 2001. Disrupted sperm function and fertilin beta processing in mice deficient in the inositol polyphosphate 5-phosphatase Inpp5b. Dev. Biol. 240:641-653. [DOI] [PubMed] [Google Scholar]
- 18.Hirvonen, H., V. Hukkanen, T. T. Salmi, T. T. Pelliniemi, and R. Alitalo. 1993. L-myc and N-myc in hematopoietic malignancies. Leuk. Lymphoma 11:197-205. [DOI] [PubMed] [Google Scholar]
- 19.Hogenesch, J. B., K. A. Ching, S. Batalov, A. I. Su, J. R. Walker, Y. Zhou, S. A. Kay, P. G. Schultz, and M. P. Cooke. 2001. A comparison of the Celera and Ensembl predicted gene sets reveals little overlap in novel genes. Cell 106:413-415. [DOI] [PubMed] [Google Scholar]
- 20.Hubbard, T., D. Barker, E. Birney, G. Cameron, Y. Chen, L. Clark, T. Cox, J. Cuff, V. Curwen, T. Down, R. Durbin, E. Eyras, J. Gilbert, M. Hammond, L. Huminiecki, A. Kasprzyk, H. Lehvaslaiho, P. Lijnzaad, C. Melsopp, E. Mongin, R. Pettett, M. Pocock, S. Potter, A. Rust, E. Schmidt, S. Searle, G. Slater, J. Smith, W. Spooner, A. Stabenau, J. Stalker, E. Stupka, A. Ureta-Vidal, I. Vastrik, and M. Clamp. 2002. The Ensembl genome database project. Nucleic Acids Res. 30:38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang, H. C., C. P. Martins, Y. Bronkhorst, E. Randel, A. Berns, M. Fero, and B. E. Clurman. 2002. Identification of oncogenes collaborating with p27Kip1 loss by insertional mutagenesis and high-throughput insertion site analysis. Proc. Natl. Acad. Sci. USA 99:11293-11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jansen, J. H., M. C. de Ridder, W. M. Geertsma, C. A. Erpelinck, K. van Lom, E. M. Smit, R. Slater, B. A. van der Reijden, G. E. de Greef, P. Sonneveld, and B. Lowenberg. 1999. Complete remission of t(11;17) positive acute promyelocytic leukemia induced by all-trans retinoic acid and granulocyte colony-stimulating factor. Blood 94:39-45. [PubMed] [Google Scholar]
- 23.Jonkers, J., and A. Berns. 1996. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim. Biophys. Acta 1287:29-57. [DOI] [PubMed] [Google Scholar]
- 24.Joosten, M., P. J. Valk, M. A. Jorda, Y. Vankan-Berkhoudt, S. Verbakel, B. M. van den, A. Beijen, B. Lowenberg, and R. Delwel. 2002. Leukemic predisposition of pSca-1/Cb2 transgenic mice. Exp. Hematol. 30:142-149. [DOI] [PubMed] [Google Scholar]
- 25.Joosten, M., P. J. Valk, Y. Vankan, N. de Both, B. Lowenberg, and R. Delwel. 2000. Phenotyping of Evi1, Evi11/Cb2, and Evi12 transformed leukemias isolated from a novel panel of cas-Br-M murine leukemia virus-infected mice. Virology 268:308-318. [DOI] [PubMed] [Google Scholar]
- 26.Joosten, M., Y. Vankan-Berkhoudt, M. Tas, M. Lunghi, Y. Jenniskens, E. Parganas, P. J. Valk, B. Lowenberg, A. E. van den Akker, and R. Delwel. 2002. Large-scale identification of novel potential disease loci in mouse leukemia applying an improved strategy for cloning common virus integration sites. Oncogene 21:7247-7255. [DOI] [PubMed] [Google Scholar]
- 27.Jorda, M. A., B. Lowenberg, and R. Delwel. 2003. The peripheral cannabinoid receptor Cb2, a novel oncoprotein, induces a reversible block in neutrophilic differentiation. Blood 101:1336-1343. [DOI] [PubMed] [Google Scholar]
- 28.Junn, E., S. H. Han, J. Y. Im, Y. Yang, E. W. Cho, H. D. Um, D. K. Kim, K. W. Lee, P. L. Han, S. G. Rhee, and I. Choi. 2000. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J. Immunol. 164:6287-6295. [DOI] [PubMed] [Google Scholar]
- 29.Kerlavage, A., V. Bonazzi, M. di Tommaso, C. Lawrence, P. Li, F. Mayberry, R. Mural, M. Nodell, M. Yandell, J. Zhang, and P. Thomas. 2002. The Celera discovery system. Nucleic Acids Res. 30:129-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Largaespada, D. A. 2000. Genetic heterogeneity in acute myeloid leukemia: maximizing information flow from MuLV mutagenesis studies. Leukemia 14:1174-1184. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J. S., A. Ishimoto, T. Honjo, and S. Yanagawa. 1999. Murine leukemia provirus-mediated activation of the Notch1 gene leads to induction of HES-1 in a mouse T lymphoma cell line, DL-3. FEBS Lett. 455:276-280. [DOI] [PubMed] [Google Scholar]
- 32.Li, J., H. Shen, K. L. Himmel, A. J. Dupuy, D. A. Largaespada, T. Nakamura, J. D. Shaughnessy, Jr., N. A. Jenkins, and N. G. Copeland. 1999. Leukaemia disease genes: large-scale cloning and pathway predictions. Nat. Genet. 23:348-353. [DOI] [PubMed] [Google Scholar]
- 33.Look, A. T. 1997. Oncogenic transcription factors in the human acute leukemias. Science 278:1059-1064. [DOI] [PubMed] [Google Scholar]
- 34.Lowenberg, B., J. R. Downing, and A. Burnett. 1999. Acute myeloid leukemia. N. Engl. J. Med. 341:1051-1062. [DOI] [PubMed] [Google Scholar]
- 34a.Maniatis, T., E.F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Marth, J. D., R. Peet, E. G. Krebs, and R. M. Perlmutter. 1985. A lymphocyte-specific protein-tyrosine kinase gene is rearranged and overexpressed in the murine T cell lymphoma LSTRA. Cell 43:393-404. [DOI] [PubMed] [Google Scholar]
- 36.Melnick, A. M., J. J. Westendorf, A. Polinger, G. W. Carlile, S. Arai, H. J. Ball, B. Lutterbach, S. W. Hiebert, and J. D. Licht. 2000. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol. Cell. Biol. 20:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikkers, H., and A. Berns. 2003. Retroviral insertional mutagenesis: tagging cancer pathways. Adv. Cancer Res. 88:53-99. [DOI] [PubMed] [Google Scholar]
- 38.Morishita, K., D. S. Parker, M. L. Mucenski, N. A. Jenkins, N. G. Copeland, and J. N. Ihle. 1988. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell 54:831-840. [DOI] [PubMed] [Google Scholar]
- 39.Mowat, M., A. Cheng, N. Kimura, A. Bernstein, and S. Benchimol. 1985. Rearrangements of the cellular p53 gene in erythroleukaemic cells transformed by Friend virus. Nature 314:633-636. [DOI] [PubMed] [Google Scholar]
- 40.Munoz, J. J., C. Tarrega, C. Blanco-Aparicio, and R. Pulido. 2003. Differential interaction of the tyrosine phosphatases PTP-SL, STEP and HePTP with the mitogen-activated protein kinases ERK1/2 and p38alpha is determined by a kinase specificity sequence and influenced by reducing agents. Biochem. J. 372:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munroe, D. G., J. W. Peacock, and S. Benchimol. 1990. Inactivation of the cellular p53 gene is a common feature of Friend virus-induced erythroleukemia: relationship of inactivation to dominant transforming alleles. Mol. Cell. Biol. 10:3307-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myers, M. P., J. N. Andersen, A. Cheng, M. L. Tremblay, C. M. Horvath, J. P. Parisien, A. Salmeen, D. Barford, and N. K. Tonks. 2001. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J. Biol. Chem. 276:47771-47774. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura, T., D. A. Largaespada, J. D. Shaughnessy, Jr., N. A. Jenkins, and N. G. Copeland. 1996. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat. Genet. 12:149-153. [DOI] [PubMed] [Google Scholar]
- 44.Neil, J. C., and E. R. Cameron. 2002. Retroviral insertion sites and cancer: fountain of all knowledge? Cancer Cell 2:253-255. [DOI] [PubMed] [Google Scholar]
- 45.Noh, D. Y., S. J. Ahn, R. A. Lee, S. W. Kim, I. A. Park, and H. Z. Chae. 2001. Overexpression of peroxiredoxin in human breast cancer. Anticancer Res. 21:2085-2090. [PubMed] [Google Scholar]
- 46.Nordberg, J., and E. S. Arner. 2001. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 31:1287-1312. [DOI] [PubMed] [Google Scholar]
- 47.Pedersen-Bjergaard, J., D. H. Christiansen, M. K. Andersen, and F. Skovby. 2002. Causality of myelodysplasia and acute myeloid leukemia and their genetic abnormalities. Leukemia 16:2177-2184. [DOI] [PubMed] [Google Scholar]
- 48.Ru, M., C. Shustik, and E. Rassart. 1993. Graffi murine leukemia virus: molecular cloning and characterization of the myeloid leukemia-inducing agent. J. Virol. 67:4722-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato, K., N. Murata, J. Kon, H. Tomura, H. Nochi, K. Tamoto, M. Osada, H. Ohta, Y. Tokumitsu, M. Ui, and F. Okajima. 1998. Downregulation of mRNA expression of Edg-3, a putative sphingosine 1-phosphate receptor coupled to Ca2+ signaling, during differentiation of HL-60 leukemia cells. Biochem. Biophys. Res. Commun. 253:253-256. [DOI] [PubMed] [Google Scholar]
- 50.Selten, G., H. T. Cuypers, and A. Berns. 1985. Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J. 4:1793-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Setoguchi, M., Y. Higuchi, S. Yoshida, N. Nasu, Y. Miyazaki, S. Akizuki, and S. Yamamoto. 1989. Insertional activation of N-myc by endogenous Moloney-like murine retrovirus sequences in macrophage cell lines derived from myeloma cell line-macrophage hybrids. Mol. Cell. Biol. 9:4515-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaknovich, R., P. L. Yeyati, S. Ivins, A. Melnick, C. Lempert, S. Waxman, A. Zelent, and J. D. Licht. 1998. The promyelocytic leukemia zinc finger protein affects myeloid cell growth, differentiation, and apoptosis. Mol. Cell. Biol. 18:5533-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shilatifard, A., W. S. Lane, K. W. Jackson, R. C. Conaway, and J. W. Conaway. 1996. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271:1873-1876. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki, T., H. Shen, K. Akagi, H. C. Morse, J. D. Malley, D. Q. Naiman, N. A. Jenkins, and N. G. Copeland. 2002. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 32:166-174. (Erratum, 32:331.) [DOI] [PubMed] [Google Scholar]
- 55.Tan-Pertel, H. T., L. Walker, D. Browning, A. Miyamoto, G. Weinmaster, and J. C. Gasson. 2000. Notch signaling enhances survival and alters differentiation of 32D myeloblasts. J. Immunol. 165:4428-4436. [DOI] [PubMed] [Google Scholar]
- 56.Thirman, M. J., D. A. Levitan, H. Kobayashi, M. C. Simon, and J. D. Rowley. 1994. Cloning of ELL, a gene that fuses to MLL in t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 91:12110-12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valk, P. J., and R. Delwel. 1998. The peripheral cannabinoid receptor, Cb2, in retrovirally-induced leukemic transformation and normal hematopoiesis. Leuk. Lymphoma 32:29-43. [DOI] [PubMed] [Google Scholar]
- 58.Verstovsek, S., E. Estey, T. Manshouri, M. Keating, H. Kantarjian, F. J. Giles, and M. Albitar. 2001. High expression of the receptor tyrosine kinase Tie-1 in acute myeloid leukemia and myelodysplastic syndrome. Leuk. Lymphoma 42:511-516. [DOI] [PubMed] [Google Scholar]
- 59.Verstovsek, S., H. Kantarjian, T. Manshouri, S. O'Brien, S. Faderl, M. Talpaz, J. Cortes, and M. Albitar. 2002. Prognostic significance of Tie-1 protein expression in patients with early chronic phase chronic myeloid leukemia. Cancer 94:1517-1521. [DOI] [PubMed] [Google Scholar]
- 60.Vyas, P., F. A. Norris, R. Joseph, P. W. Majerus, and S. H. Orkin. 2000. Inositol polyphosphate 4-phosphatase type I regulates cell growth downstream of transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 97:13696-13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiederschain, D., H. Kawai, J. Gu, A. Shilatifard, and Z. M. Yuan. 2003. Molecular basis of p53 functional inactivation by the leukemic protein MLL-ELL. Mol. Cell. Biol. 23:4230-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 30:1749-1751. [DOI] [PubMed] [Google Scholar]
- 63.Zelent, A., F. Guidez, A. Melnick, S. Waxman, and J. D. Licht. 2001. Translocations of the RARalpha gene in acute promyelocytic leukemia. Oncogene 20:7186-7203. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, Z. Y., B. Zhou, and L. Xie. 2002. Modulation of protein kinase signaling by protein phosphatases and inhibitors. Pharmacol. Ther. 93:307-317. [DOI] [PubMed] [Google Scholar]