Abstract
Both human and mouse cytomegaloviruses (HCMV and MCMV) avoid peptide presentation through the major histocompatibility complex (MHC) class I pathway to CD8+ T cells. Within the MHC class I pathway, the vast majority of antigenic peptides are generated by the proteasome system, a multicatalytic protease complex consisting of constitutive subunits, three of which can be replaced by enzymatically active gamma interferon (IFN-γ)-inducible subunits, i.e., LMP2, LMP7, and MECL1, to form the so-called immunoproteasomes. Here, we show that steady-state levels of immunoproteasomes are readily formed in response to MCMV infection in the liver. In contrast, the incorporation of immunoproteasome subunits was prevented in MCMV-infected, as well as HCMV-infected, fibroblasts in vitro. Likewise, the expression of the IFN-γ-inducible proteasome regulator PA28αβ was also impaired in MCMV-infected cells. Both MCMV and HCMV did not alter the constitutive-subunit composition of proteasomes in infected cells. Quantitative assessment of LMP2, MECL1, and LMP7 transcripts revealed that the inhibition of immunoproteasome formation occurred at a pretranscriptional level. Remarkably, a targeted deletion of the MCMV gene M27, encoding an inhibitor of STAT2 that disrupts IFN-γ receptor signaling, largely restored transcription and protein expression of immunoproteasome subunits in infected cells. While CMV block peptide transport and MHC class I assembly by posttranslational strategies, immunoproteasome assembly, and thus the repertoire of proteasomal peptides, is controlled by pretranscriptional mechanisms. We hypothesize that the blockade of immunoproteasome formation has considerable consequences for shaping the CD8+-T-cell repertoire during the effector phase of the immune response.
Human cytomegalovirus (HCMV), a prototype member of the betaherpesvirus subfamily, is an important pathogen and can cause a wide range of disease manifestations. Primary infection in the immunocompetent host is usually asymptomatic, whereas in the immunocompromised host infection or virus reactivation from latency can cause severe and even fatal disease. Mouse cytomegalovirus (MCMV) shows a similar pathobiology and has a collinear genome (47). Studies of humans and of the mouse have revealed that virus-specific CD8+ T lymphocytes represent the dominant effector arm of protective immunity. For immune recognition, the infected cells present virus-derived peptides on their major histocompatibility complex (MHC) class I molecules to CD8+ cytotoxic T lymphocytes. The processing and presentation of viral peptides is therefore the basis for immune recognition of infected cells, and a change in or lack of peptide supply can undermine the efficiency of T-cell recognition and result in immune evasion of the virus. Both HCMV and MCMV avoid peptide presentation by the expression of several viral glycoproteins (gps) controlling distinct checkpoints of the MHC class I presentation pathway. Specifically, the HCMV US6-encoded gp shuts off the translocation of peptides across the endoplasmic reticulum membrane by the transporter associated with antigen processing (TAP) (2, 31). MHC class I complex formation and transport to the cell surface is blocked by the HCMV gps US2, US11, and US3. MCMV affects peptide presentation by the gps m152/gp40 (70), m06/gp48 (57), and m04/gp34 (37, 40).
The majority of endogenous proteins are degraded via the ubiquitin-proteasome pathway. In this pathway, the proteolytic core is the 20S proteasome. Although it is freely accessible in the cytoplasm and nucleoplasm, its geometry prevents the uncontrolled access of substrates. The 20S proteasome is shaped like a closed cylinder constituted from four stacked rings. Each of the two outer rings is composed of seven different subunits of the α type, whereas each of the two inner rings is constituted from seven different β-type subunits. Both β-rings contain a copy of the subunits delta (β1), MB1 (β5), and Z (β2), which bear the active centers facing the luminal side of the 20S proteasome. Upon stimulation of cells with gamma interferon (IFN-γ), these three subunits are replaced by the IFN-γ-inducible subunits LMP2, LMP7, and MECL-1, respectively (8, 11, 13, 24), to form so-called immunoproteasomes. These subunit replacements have been shown to alter the different catalytic activities of the proteasome (42) and to promote the production of peptide ligands for MHC class I molecules (12, 63, 69). Overexpression of the immunoproteasome subunits LMP2, LMP7, and MECL1 in cell lines enhanced the presentation of different viral epitopes, such as the NP118 epitope of the lymphocytic choriomeningitis virus (LCMV) nucleoprotein (62) or an epitope derived from the GagL protein of Moloney murine leukemia virus (68). Further evidence for the significance of the proteasome in antigen processing comes from studies using proteasome inhibitors, which have been found to abolish MHC class I antigen presentation (59). In addition, mice deficient in LMP2 and LMP7 showed some deficiencies in peptide processing and presentation (12, 69).
26S proteasome holoenzymes consist of catalytic 20S proteasomes and one or two copies of the 19S regulatory complex. Moreover, so-called hybrid proteasomes, which consist of 20S proteasomes bound by the 19S regulator (PA700), as well as an 11S regulator (PA28α/β), have been described (28). PA700 and PA28 serve to control the hydrolytic activity of the 20S core particle. PA28αβ consists of two subunits, α and β, which form hexa- or heptameric rings that can bind to one or both sides of the 20S proteasome (18). Both subunits of PA28 are inducible by IFN-γ (1), and PA28 has been shown to play a role in antigen presentation and the generation of peptide ligands for MHC class I molecules (10, 23, 50).
Since the proteasome is the key protease generating peptides for the MHC class I antigen presentation pathway, we analyzed the impact of HCMV and MCMV infection on the proteasome subunit composition. We hypothesized that CMVs may alter the makeup of the proteasome and its regulatory complex, PA28αβ, to affect the cleavage and presentation of antigenic peptides. The formation of immunoproteasomes was dramatically induced during acute MCMV replication in the liver. In clear contrast, MCMV, as well as HCMV, prevented the incorporation of the IFN-γ-inducible proteasome subunits LMP2, MECL1, and LMP7 in infected cells in vitro. Moreover, the expression of the proteasome regulator PA28αβ was also affected by MCMV infection. The blockade of immunoproteasome assembly was due to pretranscriptional inhibition and required the expression of the MCMV gene M27, encoding an inhibitor of IFN-γ receptor signaling (A. Zimmermann, M. Trilling, M. Wagner, M. Wilborn, I. Bubic, T. Ziade, S. Jonjic, U. H. Koszinowski, and H. Hengel, submitted for publication). The data identify different cellular compartments and opposing principles of proteasome regulation by CMV infection in infected cells versus neighboring cells in infected tissues. The findings predict differences between the repertoires of viral peptides generated by immunoproteasomes in professional antigen-presenting cells (APC) on one hand and by housekeeping proteasomes in productively infected cells on the other.
MATERIALS AND METHODS
Mice.
Female BALB/cJ (H-2d) mice were purchased from the University of Konstanz (Konstanz, Germany). The mice were kept in a conventional pathogen-free environment and used at 8 to 9 weeks of age.
Viruses, cells, and infection conditions.
The Smith strain of MCMV (ATCC VR-194) and the MCMV mutant ΔM27 (Zimmermann et al., submitted) were propagated in third-passage mouse embryo fibroblasts (MEF) and purified by being pelleted through a sucrose cushion before the virus titers were determined by a standard plaque assay. Tissue cultures were infected with MCMV at a multiplicity of infection of 10 and harvested at the appropriate times postinfection (p.i.). The animals were infected by intraperitoneal injection with 106 PFU of MCMV before they were sacrificed on day 6 p.i. and their organs were removed. Stocks of HCMV strain AD 169 were prepared using human MRC5 cells (30). Infectious supernatants were harvested when 100% of the cells showed cytopathic effects. Virus titers were determined by a standard plaque assay. HCMV infection was enhanced by centrifugation at 800 × g for 30 min. In all experiments, MRC5 cells were infected with HCMV at a multiplicity of infection of 5 to 10. The cells were processed further as indicated below.
Purification of 26S proteasome from mouse livers.
The 26S proteasome was purified from uninfected and MCMV-infected BALB/c mouse livers according to a protocol originally designed for the purification of 26S proteasomes from rabbit muscle, which we adopted for mouse liver tissue (9). The livers were homogenized in TSDG buffer (10 mM Tris-HCl, 1 mM dithiothreitol [DTT], 1 mM NaN3, 25 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 2 mM ATP, 50 mM NaF, 0.1 mM Na8VO4, pH 7.5) using a Dounce 40-ml glass homogenizer. The homogenate was centrifuged for 20 min at 20,000 × g, and the supernatant was filtered through a layer of glass wool; the supernatant was centrifuged once more, this time for 45 min at 100,000 × g, and recovered after filtering it over the glass wool. The clarified crude lysate was adsorbed onto the DEAE-TSK 650S resin (Tosoh Biosep, Stuttgart, Germany) that had been equilibrated in TSDG buffer (2 g of liver tissue per g of DEAE matrix) by being tumbled end over end for at least 1 h at 4°C. Subsequently, unbound proteins were removed by washing the DEAE gel with TSDG buffer. The DEAE resin was suspended in 50 ml of TSDG and poured into the column (C10/20; 1 by 15 cm; Pharmacia, Zürich, Switzerland). After the column was packed, 20 ml of TSDG was pumped over the column at 1 ml/min. The column was washed with 20 ml of 75 mM KCl in TSDG, bound proteins were eluted with a linear gradient (190 ml) of 75 to 400 mM KCl in TSDG buffer, and 2-ml fractions were collected. After elution, peptidase activity was measured using the fluorogenic peptide substrate N-succinyl-LLVY-7-amido-4-methylcoumarin (Suc-LLVY-MCA) at a final concentration of 200 μM. The fluorogenic-peptide assay was performed exactly as previously described (22). Fractions with peak activity were pooled, and the 26S proteasome sample was concentrated by ultracentrifugation at 100,000 × g for 21 h at 4°C. The 26S proteasome precipitated by ultracentrifugation was subjected to Sepharose 6B using a column with the dimensions 1.6 by 98 cm (Pharmacia C16/100; Amersham Bioscience, Zürich, Switzerland) in TSDG buffer, proteins were eluted with a flow rate of 0.4 ml/min, and 2-ml fractions were collected and tested for proteasome activity. The pooled active fractions were then applied to an arginine-Sepharose column (Amersham Bioscience) with the dimensions 1.5 by 6 cm and equilibrated in TSDG, and unbound proteins were washed away with 20 ml of TSDG. The proteins were eluted with a linear gradient (300 ml) from 25 to 400 mM KCl in TSDG buffer at a flow rate of 1 ml/min, and 3-ml fractions were collected. The fractions containing peak activity were pooled and subjected to another ultracentrifugation at 100,000 × g for 21 h at 4°C. After the concentration step, the precipitated proteasome was loaded onto glycerol gradients of 20 to 40% glycerol in Kopp buffer (20 mM Tris, 1.2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 1 mM NaN3, pH 7.5) and centrifuged for 20 h at 100,000 × g. After the run, 0.5-ml fractions were recovered and peptidase activity was measured. Fractions with peak activity were pooled and processed further for nondenaturing polyacrylamide gel electrophoresis (PAGE) and two-dimensional gel electrophoresis. The purified 26S proteasome was separated by isoelectric focusing (IEF) using the Immobiline DryStrip pH gradient strips from Amersham BioScience, and the second dimension was performed on a vertical 12.5% sodium dodecyl sulfate (SDS) gel. Subsequently, the gels were silver stained. For quantification, the gels were scanned and analyzed using AIDA software (Fuji, Tokyo, Japan).
Nondenaturing PAGE and substrate overlay.
Nondenaturing polyacrylamide gels (2.5% [wt/vol] stacking gels and 4% [wt/vol] separating gel) were prepared using Bio-Rad Mini Protean electrophoresis units and gels (10 by 8 cm by 0.75 mm). The gels were run at 10 mA per gel for 2 h at 4°C. Thereafter, the gel was covered with substrate buffer (30 mM Tris, pH 7.5, 10 mM KCl, 1 mM DTT, 5 mM MgCl2, 2 mM ATP, 100 μg of creatine kinase/ml, 100 mM creatine phosphate) containing 200 μM Suc-LLVY-MCA and incubated in a humid chamber for 10 min at 37°C. Activity was then visualized by exposing the gel to UV light (365-nm wavelength). Subsequently, the gels were stained with Coomassie blue.
IFN-γ treatment and metabolic labeling.
MEF were stimulated with 100 U of mouse IFN-γ/ml (Alexis Biochemicals, Grünberg, Germany), and MRC5 cells were stimulated with 500 U of human IFN-γ/ml (Alexis Biochemicals) for periods as described in the figure legends. Thereafter, subconfluent monolayers of cells were either harvested for real-time reverse transcription (RT)-PCR and Western blot analysis or cells were labeled with [35S]methionine and [35S]cysteine (1,200 Ci/mmol; Amersham-Pharmacia, Freiburg, Germany) at a concentration of 500 μCi/ml for 6 h in methionine-free medium before overnight chase. After being washed with 2% phosphate-buffered saline (PBS), the cells were lysed in lysis buffer for immunoprecipitation.
Immunoprecipitation and NEPHGE.
The labeled cells were lysed in buffer A (25 mM Tris-HCl, pH 7.5, 1 mM DTT, 2 mM ATP, 2 mM MgCl2) containing 0.2 mg of creatine kinase/ml and 40 mM creatine phosphate as an ATP-regenerating system (28). After sonication, the lysates were clarified by centrifugation and precleared for 60 min using protein G-Sepharose beads. The supernatants were incubated overnight with protein G-Sepharose, precoated with the monoclonal antibody MCP444 (mouse anti-human HN3; generously provided by Klavs Hendil, Copenhagen, Denmark) (29) or a polyclonal anti-proteasome serum generated in rabbits against purified 20S proteasome (61). The beads were washed five times in 1 ml of buffer A and once in 25 mM ammonium acetate, pH 7.5, and the immunoprecipitated proteasome was then separated by nonequilibrium pH gradient electrophoresis (NEPHGE)-SDS-PAGE as described previously (28). The dried gels were exposed to Kodak BioMaxMR films for 7 days. For quantification, the films were scanned and analyzed using AIDA software.
Real-time RT-PCR.
Extraction of total RNA from cells and real-time RT-PCR were conducted as described previously (39). The primers used for the LMP2, LMP7, delta, and hypoxanthine phosphoribosyltransferase PCR amplifications were as described previously (39), whereas the primers for PA28α PCR amplification were 5′-AAGAGAAGAAGAAAGGGGACG-3′ and 5′-AGCTTGGTGTGAAGGTTGG-3′, with an annealing temperature of 60°C. The sense and antisense primers used for MECL1 were 5′-CGTCTGCCCTTTACTGC-3′ and 5′-CCACTTCATTCCACCTCC-3′, with an annealing temperature of 60°C. In each run, the mock samples (uninfected and unstimulated cells) were compared to IFN-γ-stimulated and MCMV-infected samples. The value calculated by the quantification analysis was always within the range covered by three concentrations of the mock sample, which was taken as arbitrary units to construct the standard curve for linear regression with r equal to 1.0 and a P value of <0.0001. The amounts of template cDNAs were normalized to those of hypoxanthine phosphoribosyltransferase mRNA. The amplification was checked by melting-curve analysis of the products.
Western blotting.
Cells were lysed in buffer B (50 mM Tris, pH 7.5, 5 mM MgCl2, 1 mM EDTA, 0.5% Triton) followed by sonication, and the lysates were clarified by centrifugation. Thereafter, the lysates were boiled for 5 min in Laemmli sample buffer (10 mM Tris, pH 6.8, 10.4 mM SDS, 38 nM bromophenol blue, 2.5% glycerol) and separated by SDS-PAGE. The proteins were blotted onto nitrocellulose (Schleicher & Schüll, Dassel, Germany), which was blocked with PBS-5% (wt/vol) low-fat dry milk-0.1% Tween 20 for 1 h and agitated overnight at 4°C with the appropriate antibodies in PBS-5% low-fat dry milk. Immunoblots were performed with the following antibodies: anti-MCMV pp89 monoclonal antibody Croma 101, anti-β-actin (Sigma, Munich, Germany), anti-proteasome subunit C7 (α1; a kind contribution of Klaus Scherrer, Paris, France), anti-LMP2, anti-LMP7, anti-MECL-1, and anti-PA28α (39). The blots were washed and incubated for 1 h with a horseradish peroxidase-conjugated secondary antibody, goat anti-rabbit immunoglobulin G (Jackson ImmunoResearch, West Grove, Pa.). After extensive washing with PBS-0.1% Tween 20, the proteins were visualized on X-ray films by chemiluminescence. For quantification, the films were scanned and analyzed using AIDA software.
RESULTS
Induction of immunoproteasomes during MCMV infection in vivo.
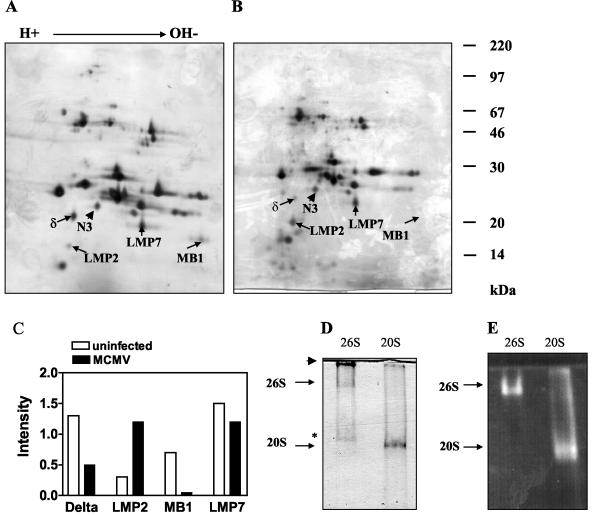
Recently, it was shown that LCMV infection results in a rapid and dramatic induction of immunoproteasome formation in vivo that is mediated by cytokines, particularly IFN-γ (39). In MCMV-infected mice, IFN-γ has been demonstrated to govern the yield of processing, as well as the presentation of the MCMV ie1/pp89-derived, H-2 Ld-restricted peptide YPHFMPTNL, in infected organs (15, 32). On the other hand, MCMV has been demonstrated to block IFN-γ-induced antiviral responses (43), the MHC class I presentation function in infected fibroblasts (32), and IFN-γ-induced MHC class II expression in infected macrophages (27). To determine whether immunoproteasomes are formed in MCMV-infected tissues in vivo, BALB/c mice were infected intraperitoneally with 106 PFU of MCMV, and 26S proteasomes were purified from the livers (see Materials and Methods) of MCMV-infected and control animals on day 6 p.i. At this point, MCMV replication reaches high titers of ∼4 to 5 log10 PFU per g of tissue (67; Zimmermann et al., submitted). The purified 26S proteasomes from uninfected and infected mice were separated on IEF-SDS-PAGE two-dimensional gels before the gels were silver stained (Fig. 1A and B) and quantified using the invariant subunit MN3 (β7) as a standard (Fig. 1C). The assignment of proteasome subunits was performed according to their migratory positions in two-dimensional gels, which were previously identified by protein microsequencing (21). The immunoproteasome subunit LMP2 was markedly upregulated during MCMV infection, while LMP7 was already quite prominent in uninfected mice. However, since both of the corresponding constitutive subunits delta and MB1 disappear from the MCMV-infected liver, a further replacement of MB1 by LMP7 must also have occurred. Changes in the subunit pattern of the 19S regulator or among enzymatically inactive subunits of the 20S proteasome were not observed. To verify that the purified 26S proteasome was indeed intact and enzymatically active, the preparation was subjected to a native acrylamide gel electrophoresis. 26S proteasomes were visualized by overlaying the gel with the fluorogenic peptide substrate Suc-Leu-Leu-Val-Tyr methyl-coumaryl-7-amide (Fig. 1E). Proteolytic activity was found at one distinct band of the 26S proteasome preparation. This band migrated significantly more slowly than the proteolytic activity of a purified 20S proteasome preparation. In summary, this finding indicated that under steady-state conditions, functional immunoproteasomes are readily generated in response to acute MCMV infection in vivo.
FIG. 1.
(A and B) Two-dimensional IEF-SDS-PAGE of 26S proteasome purified from the livers of uninfected and MCMV-infected mice. Purified 26S proteasomes (100 μg) from uninfected (A) or MCMV-infected (B) BALB/c mice were separated by IEF-SDS-PAGE, and the gels were silver stained. The constitutive subunits N3, delta, and MB1 and the immunoproteasome subunits LMP2 and LMP7 are indicated. (C) Densitometric evaluation of the indicated proteasome subunits from the two-dimensional gels shown in panels A and B. The intensity values are standardized on the expression of the constitutive and invariant β-type subunit N3, setting the intensity of N3 to 1. (D and E) Nondenaturing PAGE of the purified 26S proteasome. Purified proteasomes were electrophoresed for 2 h at 10 mA on a 4.5% native polyacrylamide gel, and 30 μg of proteins was loaded in each lane. Lanes: 26S, 26S proteasomes; 20S, purified 20S proteasome. Enzyme activity was localized by a peptide overlay using the fluorogenic peptide Suc-LLVY-MCA (E), and the proteins were subsequently stained with Coomassie blue (D). The arrowhead in panel D indicates the top of the gel, whereas the asterisk indicates a band corresponding to the PA700 complex, as determined by Western blot analysis (data not shown). The gels shown were reproduced three times each.
HCMV infection prevents the formation of immunoproteasomes.
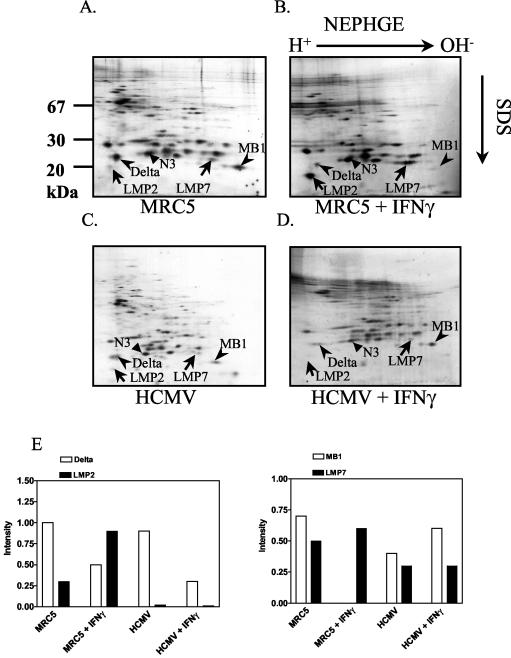
Like MCMV, HCMV was reported to prevent IFN-γ receptor-mediated cellular responses, like an elevation of MHC class II expression, by interfering with JAK/STAT signaling (45, 46). In view of the dramatic upregulation of immunoproteasome formation during MCMV replication in the liver, we wanted to settle the question of whether immunoproteasomes are formed in IFN-γ-exposed infected cells. To study proteasome assembly, human MRC5 fibroblasts were metabolically labeled for 6 h using [35S]methionine and [35S]cysteine before overnight chase. After the lysis of cells, the proteasomes were immunoprecipitated using the monoclonal antibody MCP444, which recognizes the beta subunit HN3 (β7) (29) and does not interfere with the binding of the regulatory complexes to the 20S proteasome. The precipitation of immune complexes was conducted under conditions maintaining the interaction between the 20S proteasome and the PA700 complex (28). The immunoprecipitated 26S proteasome was subsequently separated by NEPHGE and SDS-PAGE (Fig. 2A to D). Quantification of the induction of immunoproteasomes is shown in Fig. 2E. Treatment of MRC5 cells with IFN-γ resulted in the incorporation of the immunoproteasome subunits LMP2 and LMP7 into the proteasome by replacing the subunits delta and MB1 (Fig. 2B), as reported previously with other cell lines (3, 14). Infection of MRC5 cells with HCMV for 72 h did not significantly change the relative amount of incorporated LMP7 in relation to the MB1 subunits compared to uninfected cells (Fig. 2C versus A and E). In contrast, LMP2 expression was decreased three- to fourfold after HCMV infection. Next, we assessed the IFN-γ-induced increase of immunoproteasomes in HCMV-infected MRC5 cells. Remarkably, IFN-γ treatment of HCMV-infected cells did not enhance the levels of incorporated immunoproteasome subunits (Fig. 2D), whereas an almost-complete exchange of LMP7 for MB1 occurred in uninfected MRC5 cells after stimulation with IFN-γ. Again, no changes in the subunit pattern of the 19S regulator or among enzymatically inactive subunits of the 20S proteasome were apparent. In conclusion, the data demonstrated an impaired generation of immunoproteasomes in HCMV-infected cells.
FIG. 2.
HCMV infection reduces the IFN-γ-dependent induction of immunoproteasomes. MRC5 cells were left uninfected (A and B) or were infected with the HCMV strain AD169 for 72 h (C and D). IFN-γ (500 U/ml) was added for 24 h after 48 h of infection (D) or for 24 h to uninfected cells (B) prior to pulse-chase labeling and immunoprecipitation of the proteasome. Proteasome subunits were separated by NEPHGE-SDS-PAGE. The invariant β subunit N3 and the IFN-γ-inducible subunits LMP2 and LMP7, as well as their constitutively expressed homologues delta and MB1, are indicated. (E) Quantification of radioactivity in the two-dimensional gels. Intensity values are standardized on the expression of the constitutive and invariant β-type subunit N3. The gels shown are from one experiment out of three.
Inhibition of IFN-γ-mediated immunoproteasome induction in MCMV-infected cells.
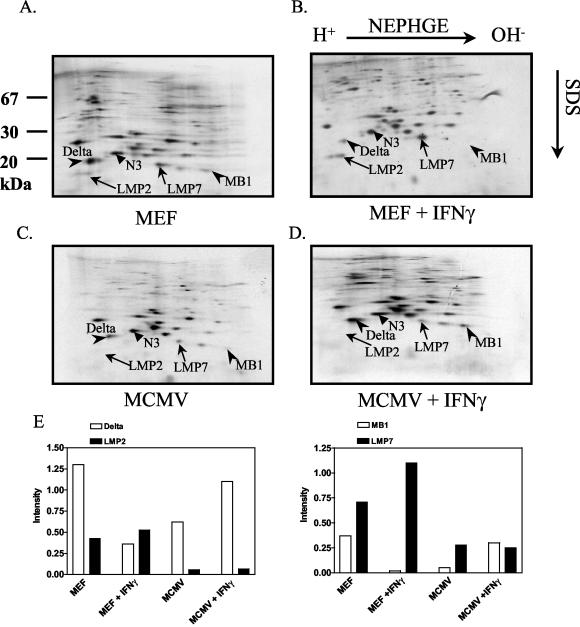
To find out whether the induction of immunoproteasomes in MCMV-infected fibroblasts is blocked, as observed in HCMV infection, we analyzed MEF 30 h p.i. As shown in Fig. 3, MCMV infection led to an inhibition of the IFN-γ-mediated induction and incorporation of LMP2 and LMP7 into proteasomes (compare Fig. 3B with D), while mock-infected MEF exposed to IFN-γ responded with an increase of LMP2- and LMP7-containing proteasomes (compare Fig. 3A with B). A quantification of the expression of the respective subunits revealed that MCMV infection led to a four- to fivefold inhibition of LMP2 and LMP7 induction in IFN-γ-stimulated infected MEF compared to uninfected MEF. The expression of the immunoproteasome subunits in MCMV-infected cells decreased by two- to fourfold compared to uninfected MEF cells. Taken together, the findings indicated that the generation of immunoproteasomes is inhibited to similar degrees in HCMV- and MCMV-infected cells in vitro. This effect could be due to either decreased synthesis of immunoproteasome subunits or inhibition of immunoproteasome subunit incorporation during 20S proteasome assembly.
FIG. 3.
MCMV suppresses the IFN-γ-dependent induction of the immunoproteasome subunits LMP2 and LMP7. MEF were left uninfected (A and B) or infected with MCMV for 30 h (C and D). After 6 h of infection, 100 U of mouse IFN-γ/ml was added (D), or it was added to uninfected cells (B) prior to pulse-chase labeling. Proteasomes were immunoprecipitated and separated by NEPHGE-SDS-PAGE. The IFN-γ-inducible subunits LMP2 and LMP7 and their constitutively expressed homologues delta and MB1 are indicated, along with the constitutive β-type subunit N3. (E) Quantitative evaluation of the two-dimensional gels, as indicated in the legend to Fig. 2. The gels shown are from one experiment out of three independent experiments.
Changes in steady-state levels of immunoproteasome subunits LMP2, LMP7, MECL-1, and PA28.
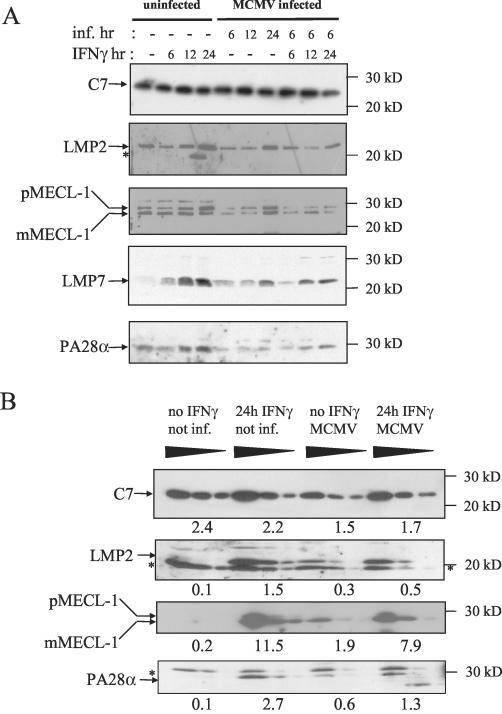
To test whether MCMV infection decreases the steady-state levels of the immunoproteasome subunits LMP2, LMP7, MECL-1, and PA28, Western blot analysis was performed. As shown in Fig. 4A, the total proteasome content remained unchanged following MCMV infection, as indicated by the same levels of the constitutive subunit C7 (α1). Upon stimulation of MEF cells with IFN-γ for 24 h, the abundances of the immunoproteasome subunits LMP2, LMP7, and MECL-1 were strongly elevated. MCMV infection alone resulted in a modest but significant induction of these subunits in vitro. Interestingly, MCMV infection inhibited the induction of LMP2, LMP7, and MECL-1 when the cells were infected for 6 h with MCMV before they were treated with IFN-γ for 6, 12, or 24 h. In order to achieve a more quantitative evaluation of steady-state immunoproteasome expression levels, we analyzed titrated amounts of lysates on Western blots probed with C7-, LMP2-, and MECL-1-specific antibodies (Fig. 4B). Quantification by densitometry in the linear range of detection yielded 15- and 57-fold induction of LMP2 and MECL-1, respectively, after stimulation with IFN-γ for 24 h, which was inhibited 3- and 1.5-fold, respectively, when the cells were infected with MCMV. The proteasome regulator PA28αβ, which enhances the processing of several viral epitopes (61, 68), was enhanced 27-fold by IFN-γ treatment in uninfected cells and 13-fold in MCMV-infected cells (Fig. 4A). In summary, these findings suggested that MCMV impedes the synthesis, assembly, or stability of immunoproteasome components.
FIG. 4.
Steady-state levels of the proteasome subunits LMP2, LMP7, and MECL-1 and the proteasome regulator subunit PA28α. MEF were left uninfected (−) or were infected (inf.) with MCMV for 6, 12, or 24 h as indicated before they were stimulated with 100 U of mouse IFN-γ/ml for 6, 12, or 24 h as shown at the top of each panel. (A) Crude cell lysates were separated by SDS-PAGE and blotted. The Western blots were probed with antibodies specific for the constitutive proteasome subunit C7 (α1); the immunoproteasome subunits LMP2, LMP7, and MECL-1; and the α subunit of the proteasome regulator PA28. For MECL-1 both the precursor (pMECL-1) and the mature subunit (mMECL-1) are visible. The asterisks indicate cross-reactive irrelevant bands. (B) For a semiquantitative assessment of steady-state protein expression, threefold dilutions of lysates were analyzed on Western blots specific for C7, LMP2, MECL-1, and PA28α. IFN-γ treatment was performed for 24 h with or without MCMV infection 6 h before the onset of IFN-γ stimulation. The bands were analyzed by densitometry in the linear range of detection, and the raw density data of volume integration minus background density are indicated below the samples.
MCMV inhibits gene transcription of immunoproteasomes and the PA28αβ regulator.
To determine whether the MCMV-mediated inhibition of the synthesis of immunoproteasome subunits and the PA28α activator occurred at a pretranscriptional or posttranscriptional level, real-time RT-PCR was conducted to assess the mRNA levels of LMP2, MECL1, LMP7, and PA28α gene transcription. The results from one of two independent experiments are listed in Table 1. As expected, incubation of MEF with IFN-γ resulted in up to a 100-fold increase in the mRNA levels of the immunoproteasome subunits LMP2, LMP7, MECL1, and PA28α. Plateau levels were reached 12 to 24 h after the exposure of cells to IFN-γ for LMP2 and LMP7. MCMV infection alone induced LMP2 and LMP7 transcription ∼2- to 4-fold compared to uninfected cells. The mRNA level for the constitutive β subunit delta remained virtually unchanged, indicating that the inhibition of the IFN-γ-inducible expression of immunoproteasome and PA28α genes was not due to a general effect on proteasome gene transcription during MCMV infection. The data demonstrated that IFN-γ induction of immunoproteasome gene transcription is inhibited in MCMV-infected cells.
TABLE 1.
Real-time RT-PCR analysis of mRNA in MEF after MCMV infection and/or IFN-γ treatmenta
| Time (h) | Level
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LMP2
|
LMP7
|
δ
|
MECL1
|
PA28α
|
|||||||||||||||||||||
| Mock + IFN-γ | MCMV (wt) | MCMV (wt) + IFN-γ | ΔM27 | ΔM27 + IFN-γ | Mock + IFN-γ | MCMV (wt) | MCMV (wt) + IFN-γ | ΔM27 | ΔM27 + IFN-γ | Mock + IFN-γ | MCMV (wt) | MCMV (wt) + IFN-γ | ΔM27 | ΔM27 + IFN-γ | Mock + IFN-γ | MCMV (wt) | MCMV (wt) + IFN-γ | ΔM27 | ΔM27 + IFN-γ | Mock + IFN-γ | MCMV (wt) | MCMV (wt) + IFN-γ | ΔM27 | ΔM27 + IFN-γ | |
| 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ND | ND | 1 | 1 | 1 | ND | ND | 1 | 1 | 1 | ND | ND |
| 6 | 31 | 4.2 | 5 | 4.5 | 13.8 | 14 | 3.1 | 5.2 | 5.3 | 30 | 1.6 | 1.5 | 1.8 | ND | ND | 6.9 | 1.9 | 1.4 | ND | ND | 6.3 | 1.5 | 2.4 | ND | ND |
| 12 | 126 | 4.0 | 21 | 10.8 | 94 | 127 | 2.5 | 11 | 17.9 | 32 | 1.5 | 1.4 | 1.5 | ND | ND | 14.2 | 1.8 | 4.1 | ND | ND | 9.2 | 1.9 | 3.2 | ND | ND |
| 24 | 90 | 4.6 | 2.7 | 14.8 | 108 | 35 | 2.3 | 0.8 | 9.3 | 51 | 1.3 | 1.0 | 0.8 | ND | ND | 89 | 3.2 | 21 | ND | ND | 21 | 4.8 | 4.8 | ND | ND |
Indicated as relative levels (LMP2/HPRT, LMP7/HPRT, δ/HPRT, MECL1/HPRT, and PA28α/HPRT). wt, wild type; Mock, mock infected; ND, not done.
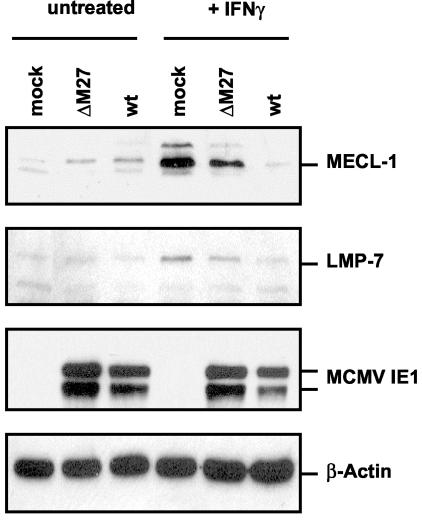
The MCMV gene M27 is essential for the full suppression of immunoproteasome subunits.
The MCMV early gene M27 has been shown to downregulate STAT2 and thereby confers resistance in viral replication to IFN-γ in vitro and in vivo (Zimmermann et al., submitted). As a consequence, the replication of a ΔM27 mutant is completely blocked in the presence of IFN-γ. We surmised from this finding that IFN-γ-induced transcription of immunoproteasome genes might be restored in ΔM27-infected cells. When the levels of LMP2 and LMP7 transcripts were quantitated by real-time PCR, a significant induction by IFN-γ was found, which reached a little lower values than in mock-infected IFN-γ-treated MEF (Table 1). Western blot analysis revealed that levels of MECL-1 and LMP7 (Fig. 5) were also significantly higher in ΔM27-infected cells than in wild-type MCMV-infected cells. However, the expression levels in ΔM27-infected cells were not as high as in uninfected controls, leaving open the possibility that other MCMV-encoded genes may contribute to immunoproteasome suppression. In conclusion, the data indicated that MCMV already prevents immunoproteasome formation at the stage of IFN-γ receptor signal transduction.
FIG. 5.
Steady-state levels of MECL-1 and LMP7 in wild-type (wt) MCMV- and ΔM27-infected MEF. Equivalent amounts of cell lysates of mock- and MCMV-infected MEF, either unstimulated or stimulated with IFN-γ (+ IFN-γ), were separated by SDS-PAGE and blotted. Western blot analysis was performed using antibodies specific for MECL-1, LMP7, MCMV IE1 (pp89 and pp76), and β-actin.
DISCUSSION
Here, we demonstrate that proteasomes are subject to CMV regulation and identify how CMV infection modulates proteasome subunit composition. In response to acute MCMV infection, mice generate strong induction of immunoproteasomes in the liver, i.e., the constitutive proteolytically active subunits delta, MB1, and Z are rapidly replaced by the IFN-γ-inducible subunits LMP2, LMP7, and MECL-1 within the 20S core proteasome complex. In sharp contrast, the formation of immunoproteasomes in vitro is blocked in MCMV-infected, as well as HCMV-infected, fibroblasts treated with high doses of IFN-γ. Further analysis of the viral blockade of immunoproteasome formation revealed a decrease in protein expression and gene transcription, suggesting that CMV infection results in a state of unresponsiveness to IFN-γ-induced immunoproteasome gene induction. The diminished incorporation of LMP2 and LMP7 into the proteasome after IFN-γ treatment of HCMV-infected cells is in agreement with a recent study by Miller and colleagues reporting that HCMV leads to a transcriptional downregulation of IFN-γ-sensitive genes within the MHC class II locus (including TAP1, TAP2, LMP2, and LMP7 genes) (46). Taking advantage of an MCMV mutant, ΔM27 (Zimmermann et al., submitted), which lacks an inhibitor of STAT2, we identified IFN-γ receptor signaling as a critical step for CMV interference with immunoproteasome gene expression and function.
Immunoproteasome induction in vivo.
At first glance, our in vitro and in vivo findings appear paradoxical. The likely explanation of the strong increase in the steady-state levels of immunoproteasomes in response to MCMV infection in the liver is a scenario in which (i) uninfected bystander cells in infected tissue become exposed to inflammatory cytokines, like IFN-γ and tumor necrosis factor alpha, known to induce immunoproteasome synthesis (3, 11, 13) and (ii) immune cells which constitutively express immunoproteasome subunits immigrate in great numbers into sites of virus replication in the liver (48). Hepatic 20S proteasomes have an extraordinarily long half-life of 12 to 15 days, as demonstrated in metabolically labeled rats (66), indicating that the replacement of constitutive proteasomes by immunoproteasomes in MCMV-infected livers occurs surprisingly fast. A strong increase in immunoproteasome formation has also been documented in the livers of mice during viral, bacterial, and fungal infections with LCMV, Listeria monocytogenes, and Histoplasma capsulatum (6, 39). This effect was markedly reduced in IFN-γ-deficient mice, indicating that this cytokine is indeed a leading factor driving proteasome replacement in vivo (39). On the organ level and under steady-state conditions, the inflammatory increase far surpasses the direct inhibition of immunoproteasome formation in infected cells. The fact that only a small fraction of liver parenchyma is productively infected with MCMV (26, 49) resolves the seeming paradox between the in vitro and in vivo findings.
Prevention of immunoproteasome assembly: restriction of peptide antigen complexity?
A pertinent question concerns what consequences the downregulation of immunoproteasomes may have for antigen presentation and clearance of CMV-infected cells. There is ample evidence that immunoproteasomes have altered peptidolytic properties. This is based on the fact that of the 14 different proteasomal subunits, only the three enzymatically active components, i.e., delta, Z, and MB1, have IFN-γ-inducible counterparts. Incorporation of LMP2, MECL1, and LMP7 is carried out interdependently, favoring the assembly of homogeneous immunoproteasomes (19, 25). As a result, two different types of proteasomes are built, those with constitutively expressed catalytic subunits and immunoproteasomes. Proteasomal peptides can be subdivided into three distinct subsets, those exclusively cleaved by constitutive proteasomes, those exclusively generated by immunoproteasomes, and common peptides produced by both proteasome types. Thus, immunoproteasomes generate a distinct and different repertoire of MHC class I ligands in inflamed tissues than in uninflamed tissues, which express very low levels of LMP2, LMP7, and MECL-1 (64). The immunoproteasome-dependent change in epitope production may serve to focus the specificity of the CD8+-T-cell response but also to avoid autoimmune assaults, since different peptides are processed from normal tissues and from sites of inflammation (20).
Given the fact that proteasome assembly is regulated by opposing principles in CMV-infected cells on the one hand and professional APC on the other, direct consequences for the processing of viral epitopes and the specificity of responding antiviral CD8+ T cells are likely. During the initial priming phase of the immune response CD8+ T cells are activated by dendritic cells (DC) constitutively expressing immunoproteasomes (44). Irrespective of whether the DC is cross-presenting CMV peptides from exogenous antigens (5, 65) or processing peptides from proteins synthesized within the cell, a repertoire of viral peptides divergent from that in CMV-infected cells lacking immunoproteasomes should be expected. The deviation in epitope production during the priming versus the effector phase of the immune response should lead to a reduction in the overall antigenicity of CMV. The CMV inhibition of immunoproteasome formation restricts the diversity of epitopes presented in productively infected nonhematopoietic stromal and parenchymal tissue. Additionally, in these cell types, the MHC class I function is most efficiently downregulated by a multitude of CMV inhibitors (7). Taking these data together, it is tempting to speculate that CMV misleads the CD8+-T-cell response when primed by peptides derived from infected or cross-presenting professional APC that are not generated by CMV-infected nonhematopoietic cells lacking immunoproteasomes.
Potential implications of the specificity of the antiviral CD8+-cytotoxic-T-lymphocyte response.
The mechanism of immune deviation may contribute to the relatively low number of MHC class I-restricted immunodominant CMV epitopes compared with the very large MCMV and HCMV proteome encompassing ∼200 antigenic proteins (for a review, see references 51 and 52). In this context, it is worth mentioning that the CD8+-T-effector-memory cell population in latently MCMV-infected mice is surprisingly focused on very few peptides in both the H-2d (34, 36) and the H-2b haplotypes (17). In relation to humans, the expansion of a similar CD8+-effector-memory population has been demonstrated which is characterized by relatively few peptides (16, 38). Interestingly, the MCMV- and HCMV-specific T-cell populations remain small in CD62L and may therefore effectively patrol and protect nonlymphoid tissue (4, 35).
Moreover, one should predict that naturally processed CMV peptides exhibiting a privileged immunogenicity and able to induce dominant and protective CD8+-T-cell reponses in vivo belong to a set of shared epitopes which is generated by both housekeeping proteasomes and immunoproteasomes. Although there are so far few experimental data on the proteasome-dependent processing of CMV peptides, this assumption holds true for the H-2Ld-restricted immunodominant MCMV pp89-derived epitope YPHFMPTNL (35, 53-55). In previous studies, this epitope was found to be efficiently processed by both types of 20S proteasomes from a precursor peptide in vitro (8, 22), as well as naturally processed in vivo in IFN-γ-treated and untreated fibroblasts and in bone marrow-derived hematopoietic cells and parenchymal cells in mice, respectively (32, 33). The degradation studies of a 25-mer peptide derived from the MCMV IE-pp89 protein using purified proteasomes and PA28αβ showed that, compared to the 20S proteasome alone, an enhancement in the generation of the 11-mer precursor peptide DMYPHFMPTNL occurred when PA28αβ was present (10). This 11-mer precursor was recently demonstrated to be efficiently transported into the endoplasmic reticulum by TAPs for final processing of its N-terminal end (41). Neither overexpression of LMP2 and LMP7 in ie1/pp89-transfected fibroblasts nor treatment with IFN-γ affected pp89 antigen presentation (23). Based on these data, we make the tacit assumption that these features also apply to other immunodominant CMV epitopes and that the processing of such peptides remains efficient in the absence of LMP2, LMP7, and MECL-1. Furthermore, we conjecture that T-cell epitopes of HCMV and MCMV which rely on immunoproteasomes will not significantly contribute to the cellular immune response against these viruses. This would be a potential mechanism of CMV immune escape which is also applicable to the oncogenic adenovirus strain 12, which shuts off the expression of immunoproteasomes on a transcriptional level (60).
The MCMV mutant ΔM27 is not able to counteract IFN-γ-mediated gene expression and inhibition of viral replication due to downregulation of STAT2 (Zimmermann et al., submitted). This let us conclude that the viral interference with IFN-γ signaling is responsible for the prevention of immunopreatosome expression. This mutant, which still expresses the whole “machinery” of MHC class I inhibitors, is dramatically attenuated in vivo (Zimmermann et al., submitted). It is conceivable that the IFN-γ-induced expression of cellular immune response genes, including immunoproteasome components, is responsible for the attenuated phenotype by increasing antigenicity to CD8+ T cells. The analysis of the CD8+-T-cell response induced by this mutant could allow us to test the hypothesis that inhibition of immunoproteasome gene expression shapes the protective T-cell response and the accumulation of memory T cells. A better understanding of the biochemical basis of the processing of protective epitopes with privileged immunogenicity and salient antigenicity will promote the design of CMV vaccines and the selection of optimal peptides used to expand CD8+ T cells in vitro for adoptive immunotherapy against CMV disease (56, 58).
Acknowledgments
We are grateful to Katja Wichmann and Rita de Giuli for expert technical assistance. We warmly acknowledge the technical help and advice of Lother Kuehn for the purification of 26S proteasomes. We thank Klavs Hendil for providing the antibody MCP444, Klaus Scherrer for the C7 antibody, and Stipan Jonjic for pp89 monoclonal antibody Croma 101.
This study was supported by the Deutsche Forschungsgemeinschaft through SFB 421 project A8 and EU QLRT-2001-01112 and by grant 31-52284 from the Swiss National Science Foundation.
REFERENCES
- 1.Ahn, J. Y., N. Tanahashi, K. Akiyama, H. Hisamatsu, C. Noda, K. Tanaka, C. H. Chung, N. Shibmara, P. J. Willy, J. D. Mott, et al. 1995. Primary structures of two homologous subunits of PA28, a gamma-interferon-inducible protein activator of the 20S proteasome. FEBS Lett. 366:37-42. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, K., A. Gruhler, B. Galocha, T. R. Jones, E. J. Wiertz, H. L. Ploegh, P. A. Peterson, Y. Yang, and K. Fruh. 1997. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6:613-621. [DOI] [PubMed] [Google Scholar]
- 3.Aki, M., N. Shimbara, M. Takashina, K. Akiyama, S. Kagawa, T. Tamura, N. Tanahashi, T. Yoshimura, K. Tanaka, and A. Ichihara. 1994. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J. Biochem. (Tokyo) 115:257-269. [DOI] [PubMed] [Google Scholar]
- 4.Appay, V., P. R. Dunbar, M. Callan, P. Klenerman, G. M. Gillespie, L. Papagno, G. S. Ogg, A. King, F. Lechner, C. A. Spina, S. Little, D. V. Havlir, D. D. Richman, N. Gruener, G. Pape, A. Waters, P. Easterbrook, M. Salio, V. Cerundolo, A. J. McMichael, and S. L. Rowland-Jones. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8:379-385. [DOI] [PubMed] [Google Scholar]
- 5.Arrode, G., C. Boccaccio, J. Lule, S. Allart, N. Moinard, J. P. Abastado, A. Alam, and C. Davrinche. 2000. Incoming human cytomegalovirus pp65 (UL83) contained in apoptotic infected fibroblasts is cross-presented to CD8+ T cells by dendritic cells. J. Virol. 74:10018-10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton, L. F., M. Cruz, R. Rangwala, G. S. Deepe, Jr., and J. J. Monaco. 2002. Regulation of immunoproteasome subunit expression in vivo following pathogenic fungal infection. J. Immunol. 169:3046-3052. [DOI] [PubMed] [Google Scholar]
- 7.Benz, C., U. Reusch, W. Muranyi, W. Brune, R. Atalay, and H. Hengel. 2001. Efficient downregulation of major histocompatibility complex class I molecules in human epithelial cells infected with cytomegalovirus. J. Gen. Virol. 82:2061-2070. [DOI] [PubMed] [Google Scholar]
- 8.Boes, B., H. Hengel, T. Ruppert, G. Multhaup, U. H. Koszinowski, and P. M. Kloetzel. 1994. Interferon gamma stimulation modulates the proteolytic activity and cleavage site preference of 20S mouse proteasomes. J. Exp. Med. 179:901-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahlmann, B., L. Kuehn, and H. Reinauer. 1995. Studies on the activation by ATP of the 26 S proteasome complex from rat skeletal muscle. Biochem. J. 309:195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dick, T. P., T. Ruppert, M. Groettrup, P. M. Kloetzel, L. Kuehn, U. H. Koszinowski, S. Stevanovic, H. Schild, and H. G. Rammensee. 1996. Coordinated dual cleavages induced by the proteasome regulator PA28 lead to dominant MHC ligands. Cell 86:253-262. [DOI] [PubMed] [Google Scholar]
- 11.Driscoll, J., M. G. Brown, D. Finley, and J. J. Monaco. 1993. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature 365:262-264. [DOI] [PubMed] [Google Scholar]
- 12.Fehling, H. J., W. Swat, C. Laplace, R. Kuhn, K. Rajewsky, U. Muller, and H. von Boehmer. 1994. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science 265:1234-1237. [DOI] [PubMed] [Google Scholar]
- 13.Gaczynska, M., K. L. Rock, and A. L. Goldberg. 1993. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature 365:264-267. [DOI] [PubMed] [Google Scholar]
- 14.Gaczynska, M., K. L. Rock, T. Spies, and A. L. Goldberg. 1994. Peptidase activities of proteasomes are differentially regulated by the major histocompatibility complex-encoded genes for LMP2 and LMP7. Proc. Natl. Acad. Sci. USA 91:9213-9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geginat, G., T. Ruppert, H. Hengel, R. Holtappels, and U. H. Koszinowski. 1997. IFN-gamma is a prerequisite for optimal antigen processing of viral peptides in vivo. J. Immunol. 158:3303-3310. [PubMed] [Google Scholar]
- 16.Gillespie, G. M., M. R. Wills, V. Appay, C. O'Callaghan, M. Murphy, N. Smith, P. Sissons, S. Rowland-Jones, J. I. Bell, and P. A. Moss. 2000. Functional heterogeneity and high frequencies of cytomegalovirus-specific CD8+ T lymphocytes in healthy seropositive donors. J. Virol. 74:8140-8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gold, M. C., M. W. Munks, M. Wagner, U. H. Koszinowski, A. B. Hill, and S. P. Fling. 2002. The murine cytomegalovirus immunomodulatory gene m152 prevents recognition of infected cells by M45-specific CTL but does not alter the immunodominance of the M45-specific CD8 T cell response in vivo. J. Immunol. 169:359-365. [DOI] [PubMed] [Google Scholar]
- 18.Gray, C. W., C. A. Slaughter, and G. N. DeMartino. 1994. PA28 activator protein forms regulatory caps on proteasome stacked rings. J. Mol. Biol. 236:7-15. [DOI] [PubMed] [Google Scholar]
- 19.Griffin, T. A., D. Nandi, M. Cruz, H. J. Fehling, L. V. Kaer, J. J. Monaco, and R. A. Colbert. 1998. Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-γ)-inducible subunits. J. Exp. Med. 187:97-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groettrup, M., S. Khan, K. Schwarz, and G. Schmidtke. 2001. Interferon-gamma inducible exchanges of 20S proteasome active site subunits: why? Biochimie 83:367-372. [DOI] [PubMed] [Google Scholar]
- 21.Groettrup, M., R. Kraft, S. Kostka, S. Standera, R. Stohwasser, and P. M. Kloetzel. 1996. A third interferon-gamma-induced subunit exchange in the 20S proteasome. Eur. J. Immunol. 26:863-869. [DOI] [PubMed] [Google Scholar]
- 22.Groettrup, M., T. Ruppert, L. Kuehn, M. Seeger, S. Standera, U. Koszinowski, and P. M. Kloetzel. 1995. The interferon-gamma-inducible 11 S regulator (PA28) and the LMP2/LMP7 subunits govern the peptide production by the 20 S proteasome in vitro. J. Biol. Chem. 270:23808-23815. [DOI] [PubMed] [Google Scholar]
- 23.Groettrup, M., A. Soza, M. Eggers, L. Kuehn, T. P. Dick, H. Schild, H. G. Rammensee, U. H. Koszinowski, and P. M. Kloetzel. 1996. A role for the proteasome regulator PA28α in antigen presentation. Nature 381:166-168. [DOI] [PubMed] [Google Scholar]
- 24.Groettrup, M., A. Soza, U. Kuckelkorn, and P. M. Kloetzel. 1996. Peptide antigen production by the proteasome: complexity provides efficiency. Immunol. Today 17:429-435. [DOI] [PubMed] [Google Scholar]
- 25.Groettrup, M., S. Standera, R. Stohwasser, and P. M. Kloetzel. 1997. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc. Natl. Acad. Sci. USA 94:8970-8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grzimek, N. K., J. Podlech, H. P. Steffens, R. Holtappels, S. Schmalz, and M. J. Reddehase. 1999. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J. Virol. 73:5043-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heise, M. T., M. Connick, and H. W. T. Virgin. 1998. Murine cytomegalovirus inhibits interferon gamma-induced antigen presentation to CD4 T cells by macrophages via regulation of expression of major histocompatibility complex class II-associated genes. J. Exp. Med. 187:1037-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendil, K. B., S. Khan, and K. Tanaka. 1998. Simultaneous binding of PA28 and PA700 activators to 20 S proteasomes. Biochem. J. 332:749-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendil, K. B., P. Kristensen, and W. Uerkvitz. 1995. Human proteasomes analysed with monoclonal antibodies. Biochem. J. 305:245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hengel, H., C. Esslinger, J. Pool, E. Goulmy, and U. H. Koszinowski. 1995. Cytokines restore MHC class I complex formation and control antigen presentation in human cytomegalovirus-infected cells. J. Gen. Virol. 76:2987-2997. [DOI] [PubMed] [Google Scholar]
- 31.Hengel, H., J. O. Koopmann, T. Flohr, W. Muranyi, E. Goulmy, G. J. Hammerling, U. H. Koszinowski, and F. Momburg. 1997. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity 6:623-632. [DOI] [PubMed] [Google Scholar]
- 32.Hengel, H., P. Lucin, S. Jonjic, T. Ruppert, and U. H. Koszinowski. 1994. Restoration of cytomegalovirus antigen presentation by gamma interferon combats viral escape. J. Virol. 68:289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hengel, H., U. Reusch, G. Geginat, R. Holtappels, T. Ruppert, E. Hellebrand, and U. H. Koszinowski. 2000. Macrophages escape inhibition of major histocompatibility complex class I-dependent antigen presentation by cytomegalovirus. J. Virol. 74:7861-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holtappels, R., N. K. Grzimek, C. O. Simon, D. Thomas, D. Dreis, and M. J. Reddehase. 2002. Processing and presentation of murine cytomegalovirus pORFm164-derived peptide in fibroblasts in the face of all viral immunosubversive early gene functions. J. Virol. 76:6044-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holtappels, R., M. F. Pahl-Seibert, D. Thomas, and M. J. Reddehase. 2000. Enrichment of immediate-early 1 (m123/pp89) peptide-specific CD8 T cells in a pulmonary CD62L(lo) memory-effector cell pool during latent murine cytomegalovirus infection of the lungs. J. Virol. 74:11495-11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karrer, U., S. Sierro, M. Wagner, A. Oxenius, H. Hengel, U. H. Koszinowski, R. E. Phillips, and P. Klenerman. 2003. Memory inflation: continuous accumulation of antiviral CD8+ T cells over time. J. Immunol. 170:2022-2029. [DOI] [PubMed] [Google Scholar]
- 37.Kavanagh, D. G., M. C. Gold, M. Wagner, U. H. Koszinowski, and A. B. Hill. 2001. The multiple immune-evasion genes of murine cytomegalovirus are not redundant: m4 and m152 inhibit antigen presentation in a complementary and cooperative fashion. J. Exp. Med. 194:967-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan, N., M. Cobbold, R. Keenan, and P. A. Moss. 2002. Comparative analysis of CD8+ T cell responses against human cytomegalovirus proteins pp65 and immediate early 1 shows similarities in precursor frequency, oligoclonality, and phenotype. J. Infect. Dis. 185:1025-1034. [DOI] [PubMed] [Google Scholar]
- 39.Khan, S., M. van den Broek, K. Schwarz, R. de Giuli, P. A. Diener, and M. Groettrup. 2001. Immunoproteasomes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J. Immunol. 167:6859-6868. [DOI] [PubMed] [Google Scholar]
- 40.Kleijnen, M. F., J. B. Huppa, P. Lucin, S. Mukherjee, H. Farrell, A. E. Campbell, U. H. Koszinowski, A. B. Hill, and H. L. Ploegh. 1997. A mouse cytomegalovirus glycoprotein, gp34, forms a complex with folded class I MHC molecules in the ER which is not retained but is transported to the cell surface. EMBO J. 16:685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knuehl, C., P. Spee, T. Ruppert, U. Kuckelkorn, P. Henklein, J. Neefjes, and P. M. Kloetzel. 2001. The murine cytomegalovirus pp89 immunodominant H-2Ld epitope is generated and translocated into the endoplasmic reticulum as an 11-mer precursor peptide. J. Immunol. 167:1515-1521. [DOI] [PubMed] [Google Scholar]
- 42.Kuckelkorn, U., S. Frentzel, R. Kraft, S. Kostka, M. Groettrup, and P. M. Kloetzel. 1995. Incorporation of major histocompatibility complex-encoded subunits LMP2 and LMP7 changes the quality of the 20S proteasome polypeptide processing products independent of interferon-gamma. Eur. J. Immunol. 25:2605-2611. [DOI] [PubMed] [Google Scholar]
- 43.Lucin, P., S. Jonjic, M. Messerle, B. Polic, H. Hengel, and U. H. Koszinowski. 1994. Late phase inhibition of murine cytomegalovirus replication by synergistic action of interferon-gamma and tumour necrosis factor. J. Gen. Virol. 75:101-110. [DOI] [PubMed] [Google Scholar]
- 44.Macagno, A., M. Gilliet, F. Sallusto, A. Lanzavecchia, F. O. Nestle, and M. Groettrup. 1999. Dendritic cells up-regulate immunoproteasomes and the proteasome regulator PA28 during maturation. Eur. J. Immunol. 29:4037-4042. [DOI] [PubMed] [Google Scholar]
- 45.Miller, D. M., B. M. Rahill, J. M. Boss, M. D. Lairmore, J. E. Durbin, J. W. Waldman, and D. D. Sedmak. 1998. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J. Exp. Med. 187:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, D. M., Y. Zhang, B. M. Rahill, K. Kazor, S. Rofagha, J. J. Eckel, and D. D. Sedmak. 2000. Human cytomegalovirus blocks interferon-gamma stimulated up-regulation of major histocompatibility complex class I expression and the class I antigen processing machinery. Transplantation 69:687-690. [DOI] [PubMed] [Google Scholar]
- 47.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 48.Olver, S. D., P. Price, and G. R. Shellam. 1994. Cytomegalovirus hepatitis: characterization of the inflammatory infiltrate in resistant and susceptible mice. Clin. Exp. Immunol. 98:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Podlech, J., R. Holtappels, N. Wirtz, H. P. Steffens, and M. J. Reddehase. 1998. Reconstitution of CD8 T cells is essential for the prevention of multiple-organ cytomegalovirus histopathology after bone marrow transplantation. J. Gen. Virol. 79:2099-2104. [DOI] [PubMed] [Google Scholar]
- 50.Preckel, T., W. P. Fung-Leung, Z. Cai, A. Vitiello, L. Salter-Cid, O. Winqvist, T. G. Wolfe, M. Von Herrath, A. Angulo, P. Ghazal, J. D. Lee, A. M. Fourie, Y. Wu, J. Pang, K. Ngo, P. A. Peterson, K. Fruh, and Y. Yang. 1999. Impaired immunoproteasome assembly and immune responses in PA28−/− mice. Science 286:2162-2165. [DOI] [PubMed] [Google Scholar]
- 51.Reddehase, M. J. 2002. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol. 2:831-844. [DOI] [PubMed] [Google Scholar]
- 52.Reddehase, M. J. 2000. The immunogenicity of human and murine cytomegaloviruses. Curr. Opin. Immunol. 12:390-396. [DOI] [PubMed] [Google Scholar]
- 53.Reddehase, M. J., and U. H. Koszinowski. 1984. Significance of herpesvirus immediate early gene expression in cellular immunity to cytomegalovirus infection. Nature 312:369-371. [DOI] [PubMed] [Google Scholar]
- 54.Reddehase, M. J., W. Mutter, K. Munch, H. J. Buhring, and U. H. Koszinowski. 1987. CD8-positive T lymphocytes specific for murine cytomegalovirus immediate-early antigens mediate protective immunity. J. Virol. 61:3102-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reddehase, M. J., J. B. Rothbard, and U. H. Koszinowski. 1989. A pentapeptide as minimal antigenic determinant for MHC class I-restricted T lymphocytes. Nature 337:651-653. [DOI] [PubMed] [Google Scholar]
- 56.Reddehase, M. J., F. Weiland, K. Munch, S. Jonjic, A. Luske, and U. H. Koszinowski. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reusch, U., W. Muranyi, P. Lucin, H. G. Burgert, H. Hengel, and U. H. Koszinowski. 1999. A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J. 18:1081-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Riddell, S. R., K. S. Watanabe, J. M. Goodrich, C. R. Li, M. E. Agha, and P. D. Greenberg. 1992. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science 257:238-241. [DOI] [PubMed] [Google Scholar]
- 59.Rock, K. L., C. Gramm, L. Rothstein, K. Clark, R. Stein, L. Dick, D. Hwang, and A. L. Goldberg. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78:761-771. [DOI] [PubMed] [Google Scholar]
- 60.Rotem-Yehudar, R., M. Groettrup, A. Soza, P. M. Kloetzel, and R. Ehrlich. 1996. LMP-associated proteolytic activities and TAP-dependent peptide transport for class 1 MHC molecules are suppressed in cell lines transformed by the highly oncogenic adenovirus 12. J. Exp. Med. 183:499-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwarz, K., M. Eggers, A. Soza, U. H. Koszinowski, P. M. Kloetzel, and M. Groettrup. 2000. The proteasome regulator PA28α/β can enhance antigen presentation without affecting 20S proteasome subunit composition. Eur. J. Immunol. 30:3672-3679. [DOI] [PubMed] [Google Scholar]
- 62.Schwarz, K., M. van Den Broek, S. Kostka, R. Kraft, A. Soza, G. Schmidtke, P. M. Kloetzel, and M. Groettrup. 2000. Overexpression of the proteasome subunits LMP2, LMP7, and MECL-1, but not PA28 alpha/beta, enhances the presentation of an immunodominant lymphocytic choriomeningitis virus T cell epitope. J. Immunol. 165:768-778. [DOI] [PubMed] [Google Scholar]
- 63.Sibille, C., K. G. Gould, K. Willard-Gallo, S. Thomson, A. J. Rivett, S. Powis, G. W. Butcher, and P. De Baetselier. 1995. LMP2+ proteasomes are required for the presentation of specific antigens to cytotoxic T lymphocytes. Curr. Biol. 5:923-930. [DOI] [PubMed] [Google Scholar]
- 64.Stohwasser, R., S. Standera, I. Peters, P. M. Kloetzel, and M. Groettrup. 1997. Molecular cloning of the mouse proteasome subunits MC14 and MECL-1: reciprocally regulated tissue expression of interferon-gamma-modulated proteasome subunits. Eur. J. Immunol. 27:1182-1187. [DOI] [PubMed] [Google Scholar]
- 65.Tabi, Z., M. Moutaftsi, and L. K. Borysiewicz. 2001. Human cytomegalovirus pp65- and immediate early 1 antigen-specific HLA class I-restricted cytotoxic T cell responses induced by cross-presentation of viral antigens. J. Immunol. 166:5695-5703. [DOI] [PubMed] [Google Scholar]
- 66.Tanaka, K., and A. Ichihara. 1989. Half-life of proteasomes (multiprotease complexes) in rat liver. Biochem. Biophys. Res. Commun. 159:1309-1315. [DOI] [PubMed] [Google Scholar]
- 67.Trgovcich, J., D. Stimac, B. Polic, A. Krmpotic, E. Pernjak-Pugel, J. Tomac, M. Hasan, B. Wraber, and S. Jonjic. 2000. Immune responses and cytokine induction in the development of severe hepatitis during acute infections with murine cytomegalovirus. Arch. Virol. 145:2601-2618. [DOI] [PubMed] [Google Scholar]
- 68.van Hall, T., A. Sijts, M. Camps, R. Offringa, C. Melief, P. M. Kloetzel, and F. Ossendorp. 2000. Differential influence on cytotoxic T lymphocyte epitope presentation by controlled expression of either proteasome immunosubunits or PA28. J. Exp. Med. 192:483-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Kaer, L., P. G. Ashton-Rickardt, M. Eichelberger, M. Gaczynska, K. Nagashima, K. L. Rock, A. L. Goldberg, P. C. Doherty, and S. Tonegawa. 1994. Altered peptidase and viral-specific T cell response in LMP2 mutant mice. Immunity 1:533-541. [DOI] [PubMed] [Google Scholar]
- 70.Ziegler, H., R. Thale, P. Lucin, W. Muranyi, T. Flohr, H. Hengel, H. Farrell, W. Rawlinson, and U. H. Koszinowski. 1997. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity 6:57-66. [DOI] [PubMed] [Google Scholar]