Abstract
Various monoclonal antibodies (MAbs) that recognize cell surface proteins on bovine cells were previously shown to efficiently block infection with bovine viral diarrhea virus (BVDV) (C. Schelp, I. Greiser-Wilke, G. Wolf, M. Beer, V. Moennig, and B. Liess, Arch. Virol. 140:1997-2009, 1995). With one of these MAbs, a 50- to 58-kDa protein was purified from calf thymus by immunoaffinity chromatography. Microchemical analysis of two internal peptides revealed significant sequence homology to porcine and human CD46. The cDNA of bovine CD46 (CD46bov) was cloned and further characterized. Heterologously expressed CD46bov was detected by the MAb used for purification. A putative function of CD46bov as a BVDV receptor was studied with respect to virus binding and susceptibility of nonpermissive cells. While the expression of CD46bov correlated well with the binding of [3H]uridine-labeled BVDV, the susceptibility of cells nonpermissive for BVDV was not observed. However, the expression of CD46bov resulted in a significant increase in the susceptibility of porcine cells to BVDV. These results provide strong evidence that CD46bov serves as a cellular receptor for BVDV.
Bovine viral diarrhea virus (BVDV) is a small enveloped RNA virus which belongs to the genus Pestivirus within the family Flaviviridae. Pestiviruses are widespread among cloven-hoofed animals (Artiodactyla), causing disease in ruminants (Bos, Ovis, and Camelidae) and nonruminants (Suidae). BVDV is an important pathogen of cattle and accounts for syndromes of the intestinal, respiratory, and reproductive tracts. While the economic impact of ruminant pestiviruses is mainly due to reduced herd fertility and loss of calves (and lambs), classical swine fever virus (CSFV) causes epidemic hemorrhagic fever in pigs (33). The pathway by which pestiviruses find and enter their host cells has not been studied in detail. It is considered likely that the viruses enter the cells by receptor-mediated endocytosis, but conclusive data with respect to penetration are not available.
Pestiviruses encode three structural glycoproteins, namely, Erns, E1, and E2, which form extensive inter- and intramolecular disulfide bonds (34). Recent data have shown that the glycoprotein Erns can bind to heparan sulfate and other glycosaminoglycans (GAGs) (10, 11). Several proteinaceous structures were proposed to serve as cellular receptors for pestiviruses. A 50-kDa protein was identified by using anti-idiotypic antibodies raised against neutralizing monoclonal antibodies (MAbs) binding to BVDV E2 (35, 36). Two other proteins, of 60 and 93 kDa, were identified by using MAbs directed against cell surface molecules of bovine cells. These MAbs inhibited the infection of bovine lymphocytes by various BVDV strains efficiently in a dose-dependent manner (28). Moreover, the low-density lipoprotein receptor (LDL-R) was proposed to be involved in the entry of BVDV, since infection of Madin-Darby bovine kidney (MDBK) cells with BVDV was blocked by an antibody directed against LDL-R (2).
We describe here the identification and functional characterization of bovine CD46 (CD46bov, or membrane cofactor protein) as a cellular receptor for BVDV. CD46 serves as a cofactor for plasma serine protease factor I to cleave complement factors C3b and C4b deposited on host tissues. Thereby, autologous cells are protected from complement-mediated injury (3, 17, 29). CD46 belongs to the family of regulators of complement activation which includes other cell surface proteins, i.e., CD35, CD21, and CD55 (17). The amino-terminal regions of these proteins consist of variable numbers of tandemly linked cysteine-rich modules of approximately 60 amino acids, termed complement control protein (CCP) repeats (16). CD46 consists of four CCP domains (CCP1 to CCP4) which serve different functions. Binding of complement factors C3b and C4b occurs at CCP2, CCP3, and CCP4 (1, 13). In addition, CD46 has been extensively characterized as a receptor for measles virus (22), human herpesvirus 6 (27), and bacteria (14, 23). Measles virus binds to CCP1 and CCP2 (5, 13, 19, 20) at the tip of an antenna-like structure. The structures of these two domains have been solved by X-ray crystallography to a resolution of 3.1 Å (6).
MATERIALS AND METHODS
Cells and viruses.
MDBK cells (ATCC CCL-22), baby hamster kidney (BHK) cells (ATCC CCL-10), porcine kidney cells (PK15) (ATCC CCL-33), pig lymphoma cells (38A1D) (31), HeLa cells (ATCC CCL-2), mouse L cells (ATCC CCL-1), and Vero cells (ATCC CCL-81) were grown in Dulbecco modified Eagle medium (DMEM) containing nonessential amino acids and 10% fetal calf serum (FCS) at 37°C in 5% CO2. BVDV strain NADL (ATCC VR-534) was propagated on MDBK cells and stored at −70°C.
Antibodies and immunoblotting.
Hybridomas BVD/CA 17, BVD/CA 26, and BVD/CA 27 were grown in DMEM containing nonessential amino acids and 15% FCS. The immunoglobulin G (IgG) subclasses of the MAbs were determined to be IgG2a for MAb BVD/CA 17 and BVD/CA 27. BVD/CA 26 was typed as IgG1. For immunoblotting, a cell culture supernatant of hybridoma BVD/CA 26 was used at a dilution of 1:10, and peroxidase-conjugated anti-mouse IgG (Dianova) was used at a 1:10,000 dilution. Western blot membranes were blocked after semidry transfer with a synthetic blocking agent (Roti-Block) as recommended by the supplier (Carl Roth, Karlsruhe, Germany). The use of milk powder as a blocking reagent resulted in a high background. Signals were revealed by chemiluminescence.
For inhibition experiments, IgG was purified by using immobilized protein A-Sepharose (for MAbs BVD/CA 17 and BVD/CA 27) or protein G-Sepharose (for MAb BVD/CA 26) (Amersham Bioscience) according to the manufacturer's instructions.
Purification and sequencing of CD46.
Fresh calf thymus (750 g) was homogenized in ice-cold phosphate-buffered saline (PBS) in a Varix blender. The homogenate was extracted by the addition of Triton X-114 (Sigma) to a final concentration of 2% (vol/vol) in a total volume of 13 liters. After removal of the insoluble matter by centrifugation at 2,000 × g for 15 min, the solution was heated slowly to 37°C in batches of 5 liters and incubated for 14 h. The lower, detergent-rich phase was extracted twice with ice-cold PBS followed by heating to 37°C. Proteins enriched in the detergent phase were precipitated by the addition of 2.5 volumes of ice-cold acetone and sedimented by centrifugation at 3,000 × g for 30 min. The precipitate was washed with acetone, dried, and dissolved in 50 mM Tris-8 M urea-1% Triton X-100 (pH 8.0).
For preparation of the immunoaffinity matrix, 2 mg of purified IgG from hybridoma BVD/CA 17 (IgG2a) was bound to 1 ml of protein A-Sepharose and immobilized by cross-linking with dimethylpimelimidate (Pierce) in 200 mM sodium borate buffer. The Triton X-114 extract was diluted 1:8 in 50 mM Tris (pH 8.0)-1% Triton X-100-0.1% phenylmethylsulfonyl fluoride and applied to the immobilized-antibody column at a flow rate of 100 μl/min at 4°C. After the column was washed with 100 volumes of 50 mM Tris-HCl-1 M urea-1 M NaCl- 1% Triton X-100-0.1% sodium dodecyl sulfate (SDS) (pH 8.0), the bound material was eluted with 5 ml of 50 mM Tris-HCl-1 M urea-150 mM NaCl- 1% Triton X-100-5% SDS at 37°C. The eluate was dialyzed against 20 mM Tris (pH 8.0)-0.1% SDS-0.5% Triton X-100 for 24 h and concentrated by ultrafiltration. The total amount of eluted protein was 600 μg. SDS-polyacrylamide gel electrophoresis (PAGE) analysis revealed that the 50- to 58-kDa protein was approximately 80% pure.
Microchemical analysis.
For microchemical analysis, the eluate was further purified by preparative SDS-PAGE to remove IgG molecules which had leaked from the column. A gel slice containing 50 μg of the 50- to 58-kDa protein was excised and subjected to a commercial microchemical analyis (Eurosequence BV, Groningen, The Netherlands). The sequences obtained were analyzed by using the FASTA3 and BLAST programs available at the European Molecular Biology Laboratory and the National Center for Biotechnology Information.
cDNA cloning of the CD46 gene.
RNA for Northern blot analysis was extracted with an RNeasy kit as described by the manufacturer (Qiagen), and denaturing agarose gel electrophoresis and Northern blot analysis were performed essentially as described earlier (26). A 0.723-kb fragment from porcine CD46 was amplified from porcine RNA with oligonucleotides KM1 (5′-TTTGAATTCAAAATGATGGCGTTTTGCGCGCTGC) and KM5 (5′-ACATTTGACCACTTTACACTC) by reverse transcription (RT)-PCR (derived from GenBank locus D70897.1; GI 1018988). This fragment served as a hybridization probe after labeling with 32P by nick translation. This probe also was used to screen a bovine spleen cDNA library in lambda ZAPII (Stratagene). Positive lambda clones were plaque purified, and the inserts containing the plasmids were rescued by in vivo excision by following the manufacturer's protocol. Plasmids were sequenced by using a LiCor 800 system and an Amersham PCR sequencing kit. IR800-labeled and unlabeled oligonucleotides were obtained from MWG (Ebersberg, Germany).
Clone pTK12 was obtained by RT-PCR from total MDBK cell RNA with oligonucleotides BVR5 (5′-AAGAATTCACCATGAGGGCGTCTTGCACCC) and BVTK3 (5′-GAATTCATCTGTTCTGTTGCAGTGGTTGC).
Construction of expression plasmids.
From lamdba ZAP clone pKM2, a 2.5-kb BamHI/EcoRI fragment which encompassed the entire coding region for CD46 as well as 87 nucleotides of the 5 nontranslated region was isolated and ligated into expression plasmid pcDNA3 (Invitrogen), resulting in pKM6. From cDNA clone pTK12, a 1.3-kb EcoRI fragment encompassing the coding region was ligated into expression plasmid pcDNA3, resulting in pTK13.
Transient expression and generation of cell lines.
PK15, 38A1D, HeLa, and L cells were transfected with 2 μg of pKM6 and pTK13 DNAs by using Superfect reagent (Qiagen) according to the manufacturer's recommendations. At 24 to 48 h after transfection, the expression of CD46bov was detected by immunohistochemical analysis. Briefly, stable CD46bov-expressing cell lines were selected by the addition of 100 μg of G418 (Calbiochem)/ml to the culture medium at 24 h after transfection. CD46bov-expressing cell colonies were identified by immunohistochemical analysis with a mixture of anti-CD46 MAbs.
Preparation of [3H]uridine-labeled BVDV.
A total of 2 × 107 MDBK cells were infected with BVDV NADL at a multiplicity of infection (MOI) of 0.1. Upon appearance of cytopathic effects (at 23 to 27 h postinfection [p.i.]), the medium was replaced with DMEM containing 10% FCS and 300 μCi of [3H] uridine (Amersham).
The culture medium was harvested at 72 h p.i., cellular debris was removed by centrifugation at 3,900 × g for 30 min, and the virus was concentrated by ultracentrifugation in an SW41 rotor (Beckman) at 125,000 × g for 4 h. For flotation purification, the viral pellet was resuspended in 350 μl of PBS on ice; the suspension was mixed with an equal volume of a 60% Optiprep solution (Sigma). A step gradient with the virus-containing 30% Optiprep solution at the bottom and 25, 20, 15, 10, and 5% Optiprep in PBS (700 μl each) layered on top was centrifuged in an SW60 rotor (Beckman) at 168,000 × g for 6 h at 4°C. Fractions (200 μl) were taken with a pipette, and the radioactivity was determined by liquid scintillation counting (LSC).
For binding, 104 cpm of virus was added to 5 × 105 monolayer cells in PBS- 2% horse serum for 2 h at 0°C. The virus inoculum was removed, and the cells were washed twice with ice-cold PBS. Cells were lysed in 300 μl of PBS- 1% Triton X-100 for 10 min at 25°C, the lysate as well as the inoculum and washes were mixed with scintillation cocktail, and counts were determined by using a liquid scintillation counter. The amount of radioactivity contained in the cell lysate was correlated to the counts per minute in the inoculum and washes.
For inhibition experiments, 2 μg of a mixture of MAbs BVD/CA 17, BVD/CA 26, and BVD/CA 27 was added to cells for 1 h at 0°C. The MAbs were removed before 3H-labeled BVDV was added as described above. All binding experiments were repeated at least three times.
Determination of susceptibility of porcine cells.
A total of 106 PK15, CD46-expressing PK15 (PK15CD46), and MDBK cells were infected with a serial dilution (1:5) of BVDV NADL (2 × 107 PFU/ml) for 1 h at 0°C. For inhibition experiments, cells were preincubated with 2 μg of a mixture of equal volumes of anti-CD46 MAbs for 1 h at 0°C and washed twice with PBS before infection with the virus for 1 h. Cells were fixed at 22 h p.i., and the number of infectious centers was determined by immunohistochemical analysis with anti-BVDV E2 MAb D5. The number of infectious centers was multiplied by the virus dilution to determine the number of focus-forming units per milliliter for each cell line. This value determined for MDBK cells was taken to be 100%. All experiments were performed at least in triplicate.
Nucleotide sequence accession number.
The sequence of the CD46bov cDNA (3,658 nucleotides) has been submitted to GenBank under accession number AY342429.
RESULTS
Purification and microchemical analysis of a 50- to 58-kDa protein from the surface of bovine cells.
Three MAbs, BVD/CA 17, BVD/CA 26, and BVD/CA 27, which efficiently inhibit the infection of bovine cells with BVDV, have been described. The MAbs were described to recognize identical patterns of proteins with apparent molecular masses of 60 and 93 kDa and proposed to be cellular receptors for BVDV (28). Treatment of these proteins with peptide:N-glycosidase F revealed both species to be modified by N glycosylation (data not shown). Although the actual apparent molecular masses were 50 to 58 kDa and 80 to 90 kDa, we termed these proteins gp60 and gp93, according to the first description by Schelp et al. (28).
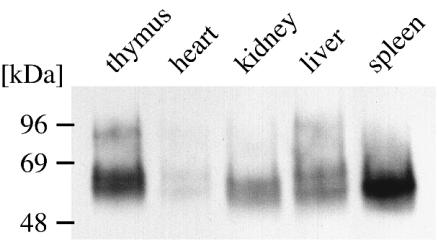
In order to purify and further characterize the identified antigens, immunoaffinity chromatography was attempted. Pilot experiments with immobilized MAbs allowed enrichment of the proteins from lysates of large numbers of cultured cells (>109 MDBK cells), but the amounts of protein recovered were insufficient for further analysis. In a search for alternative sources of antigen, different bovine cell lines (data not shown) and bovine tissues (thymus, heart, kidney, liver, and spleen) were screened for the expression of gp60 and gp93. The respective immunoblots were incubated with MAb BVD/CA 26, and a protein of 50 to 58 kDa was detected in all of the tested tissues. A larger, 80- to 90-kDa molecule representing gp93 was clearly apparent in lysates from thymus tissue and MDBK cells (Fig. 1; see Fig. 4, lane 3). gp93 also was detectable in small amounts in tissues from heart, kidney, liver, and spleen but required extended exposure times (data not shown). Calf thymus was chosen as a convenient source of antigen, especially because of its low content of connective tissue.
FIG. 1.
Detection of 50- to 58-kDa and 80- to 90-kDa proteins in various bovine tissues. Similar amounts of proteins from lysates of bovine thymus, heart muscle, kidney, liver, and spleen tissues were subjected to SDS-PAGE and blotted onto nitrocellulose. The blot was probed with MAb BVD/CA 26.
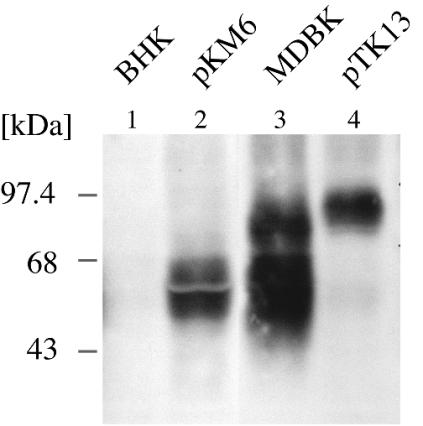
FIG. 4.
CD46bov is recognized by MAb BVD/CA 26. The open reading frames of CD46bov cDNA clones with short and extended STP domains were cloned downstream of a human cytomegalovirus promoter, yielding pKM6 and pTK13. BHK cells were transfected with pKM6 (lane 2) and pTK13 (lane 4), and immunoblot analysis with MAb BVD/CA 26 was performed 24 h later. MDBK cells (lane 3) and mock-transfected BHK cells (lane 1) served as positive and negative controls, respectively.
A total of 750 g of fresh calf thymus tissue was homogenized and extracted with Triton X-114. Triton X-114 is soluble in water at low temperatures but becomes insoluble at 30°C, resulting in the segregation of hydrophobic (membrane) proteins into the detergent phase. Proteins contained in the detergent phase were precipitated with acetone, solubilized, and subjected to immunoaffinity chromatography with MAb BVD/CA 17 (2 mg) immobilized on protein A-Sepharose. Eluted proteins were purified further by preparative SDS-PAGE, and a gel slice containing a 50- to 58-kDa protein was subjected to microchemical analysis. N-terminal sequencing of trypsin-generated internal peptides revealed two unambiguous peptide sequences, NH2-CXYPAIEHGTIVSGFGPK-COOH (peptide 1) and NH2-FVSMKPQGTLKPYSPGEQIV-COOH (peptide 2). Databank searches (BLAST and FASTA3) revealed significant identities with two members of the family of regulators of complement activation. With either peptide, the highest scores were observed for porcine membrane cofactor protein (CD46), with identities of 61% (peptide 1) and 57% (peptide 2). The identities of the peptide sequences to human CD46 and murine CD46 scored at 50% or less. Bovine C4 binding protein was 52.9% identical to peptide 1. Bovine C4 binding protein is a soluble protein of 500 to 570 kDa and consists of disulfide-linked homo- or heterooligomers. Therefore, it was highly unlikely to be identical to the purified 50- to 58-kDa protein. Human CD46 is N and O glycosylated and is located in the plasma membrane, and different splice variants which appear simultaneously in the same cell account for variable apparent molecular masses of 57 to 78 kDa. These data are more or less in agreement with the apparent molecular masses observed for the bovine proteins and led to the working hypothesis that gp60 and gp93 actually represented CD46bov.
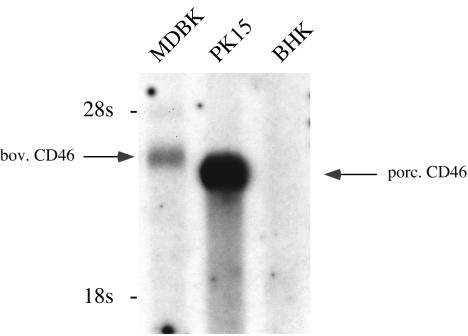
To verify our hypothesis, the CD46bov gene had to be cloned and expressed. CD46 sequences are available for a number of species, such as humans, nonhuman primates, laboratory animals, and pigs; recently, the sequence for cattle also was published (30). At the time of this investigation, however, no sequence data for CD46bov were available. Because the synthesis of oligonucleotides deduced from the determined peptide sequences would have led to highly degenerate primer sequences, we chose a different approach for cloning. Pig and cattle belong to the same zoological order (Artiodactyla), and we therefore expected reasonable homology of the CD46 genes. This idea was tested by using Northern blot analysis with a 0.7-kb fragment from porcine CD46 cDNA as a probe. This fragment, cloned by RT-PCR from PK15 cells, was labeled with 32P and hybridized to poly(A)+ RNA from PK15, MDBK, and BHK cells. When low-stringency conditions were used for hybridization, RNA molecules corresponding to 3.8 and 3.6 kb were detected in bovine and porcine cells, respectively (Fig. 2). No signal was observed with mRNA prepared from BHK cells.
FIG. 2.
Detection of CD46bov (bov. CD46) mRNA by Northern blot analysis. A total of 2 μg of glyoxylated poly(A)+ RNAs from MDBK, PK15, and BHK cells was separated in a denaturing 0.8% agarose gel and blotted onto a nylon membrane. For hybridization, a 700-bp fragment from the 5′ end of porcine CD46 (porc. CD46) cDNA was labeled with [32P]dATP by nick translation. Low-stringency hybridization was performed at 48°C. The positions of 28S (4.2-kb) and 18S (2.4-kb) rRNAs are indicated on the left.
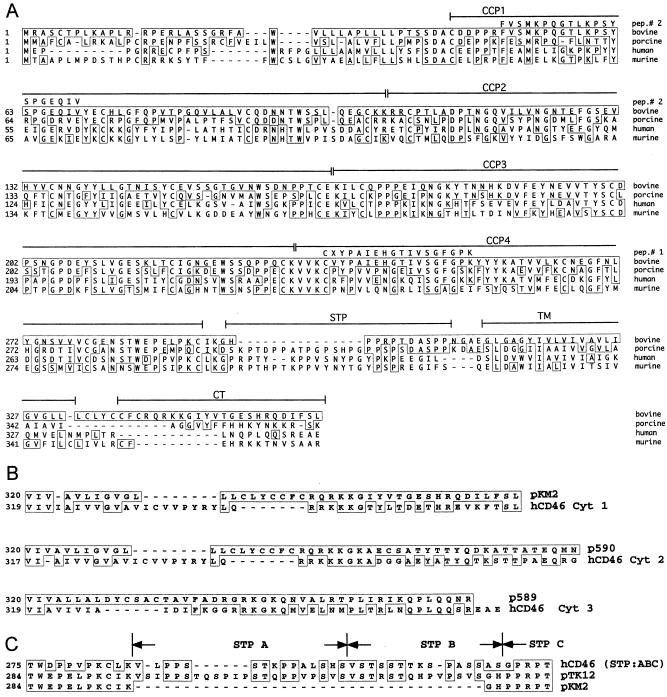
As the next step, a commercially available bovine lymph node cDNA library in lambda ZAPII was screened with the porcine cDNA probe. Two cDNA clones with insert sizes of 3.1 kbp (p589) and 3.3 kbp (p590) were identified. The sequences from the 5′ ends of these cDNA clones showed clear homology to porcine and human CD46 sequences. The amino acid sequence of peptide 1 was found in the translated cDNA sequences of both cDNA clones. Thus, it was likely that we had cloned the correct gene. Using a 300-bp fragment of cDNA clone p590 as a homologous probe, we screened the cDNA library again at a higher stringency. This process led to the isolation of a number of cDNA clones; the largest of these, pKM2, consisted of 3,658 nucleotides (GenBank accession number AY342429) and contained a complete open reading frame (361 codons) of CD46bov (Fig. 3A). This sequence allowed the identification of the structural elements which are typical for CD46, namely, CCP1 to CCP4 (ectodomain); a stretch rich in serine, threonine, and proline residues (STP domain); a transmembrane domain; and a cytosolic tail. A comparison of the complete amino acid sequences revealed an identity to porcine CD46 of 50%, while human CD46 and murine CD46 were more distantly related (39.8 and 34.4%, respectively). When the sequence comparison was restricted to CCP1 to CCP4, the identities increased to 59.5% with porcine CD46, 46.2% with human CD46, and 40.6% with murine CD46.
FIG. 3.
Analysis of the CD46bov sequence. (A) Comparison of the CD46 peptide sequences from different species. The peptide sequence deduced from CD46bov cDNA was aligned to pig (GenBank locus E12732.1), mouse (translated from GI 31981632), and human (translated from GI 27502404) CD46 sequences. The locations of the two internal peptides (pep.# 2 and pep.# 1) and the individual domains of CD46 (CCP1 to CCP4), the STP domain, the transmembrane (TM) region, and the cytosolic tail (CT) are indicated above the sequence. Amino acid residues identical to those in CD46bov are boxed. (B) Comparison of C termini encoded by various CD46bov cDNA clones with splice variants of human CD46 (hCD46). cDNA clones pKM2, p590, and p589 were obtained from a bovine lymph node cDNA library. The sequences corresponding to the C termini of CD46bov were compared to GenBank entries by BLAST analysis, and the best matches are shown. hCD46 Cyt 1 corresponds to GenBank GI 27502406, Cyt 2 corresponds to GI 27502422, and Cyt 3 corresponds to GI 27502420. The numbers on the left indicate the positions in the amino acid sequences of CD46bov and hCD46. (C) Comparison of two CD46bov peptide sequences differing in the STP region and the corresponding human sequence. cDNA clone pTK12 was obtained by RT-PCR from MDBK cell RNA, and the peptide sequence was compared to that of pKM2 and the corresponding human splice variant with an STP ABC domain (GI 27502401). The numbers on the left indicate the positions in the CD46bov amino acid sequence. The organization of STP segments A, B, and C is indicated above the sequence.
Analysis of other cDNA clones from the cDNA library and RT-PCR products from MDBK cells revealed variable sequences close to the 3′ end of the CD46 open reading frame. Variable C termini, which result from alternative splicing, have been described for human CD46. The C termini encoded by CD46bov cDNA clones pKM2, p590, and p589 were analogous to the three known cytosolic tails, Cyt 1, Cyt 2, and Cyt 3, of human CD46 (18, 24, 25, 30) (Fig. 3B). An interesting splice variant which contained an insertion of 108 nucleotides downstream of CCP4 (pTK12) was identified by RT-PCR from MDBK cells (Fig. 3C). This stretch of 36 amino acids forms an extensive STP region (64% of this stretch consists of serine, threonine, and proline residues). By analogy to the terminology of the three known segments of human CD46, the STP domain of pTK12 comprises segments A, B, and C (ABC), while the other cDNAs of CD46 from the bovine lymph node cDNA library and MDBK cells contain only segment C (Fig. 3C) (24, 30). CD46bov from a bovine brain capillary cDNA library (30) is homologous to p589 with respect to STP segment C and Cyt 3 at the C terminus. Compared to the amino acid sequence of p589, there are differences at nine positions, namely, L23R, C39S, R104K, Q106R, N109T, S124N, S185N, K228Q, and Y244H (the first letter refers to the amino acid residue in the CD46bov sequence from reference 30). L23R and C39S are within the N-terminal leader sequence, which is removed from CD46 by a signal peptidase. R104K and Q106R are located at the junction of CCP1 and CCP2. Due to the heterogeneity at S124N, CD46bov from bovine lymph nodes has two potential N-glycosylation sites within CCP2, while CD46bov from bovine brain capillaries has three.
CD46bov is recognized by MAbs BVD/CA 17, BVD/CA 26, and BVD/CA 27.
Evidence from N-terminal sequencing and cDNA cloning suggested that the proteins under investigation likely represent CD46bov. To support this assumption, the reactivity of heterologously expressed CD46bov with MAbs BVD/CA 17, BVD/CA 26, and BVD/CA 27 was investigated. For this purpose, the complete open reading frames of the two CD46bov genes which differed in the STP region were cloned downstream of a human cytomegalovirus promoter into mammalian expression vectors (pKM6 and pTK13). In transient expression experiments, transfected BHK cells clearly were detected by MAbs BVD/CA 17 and BVD/CA 27 in an immunohistochemical analysis, while mock-transfected BHK cells were negative (data not shown). Western blot analysis with MAb BVD/CA 26 revealed the presence of a 50- to 58-kDa protein in BHK cells transfected with pKM6 (Fig. 4, lane 2). Transfection of pTK13 resulted in the detection of an 80- to 90-kDa protein (Fig. 4, lane 4). The apparent molecular masses of these proteins compared well to the typical fuzzy appearances of gp60 and gp93 from MDBK cells (Fig. 4, lane 3). Therefore, it can be concluded that CD46bov is the target of the BVDV-inhibiting MAbs BVD/CA 17, BVD/CA 26, and BVD/CA 27. In addition, gp60 and gp93 are splice isoforms of the same polypeptide which differ by 36 amino acids in the STP domain. The large difference in molecular masses between gp60 and gp93 is probably due to O glycosylation within the STP domain. The appearance of the bipartite broad band (50- to 58-kDa protein in Fig. 4, lane 2; also apparent in liver in Fig. 1) likely is due to posttranslational modifications.
The expression of CD46bov does not confer susceptibility to nonpermissive cells.
CD46bov is the target of BVDV-inhibiting MAbs. Therefore, it is likely that CD46bov is involved in BVDV invasion, possibly as a cellular receptor. It was straightforward to investigate whether the expression of CD46bov correlated with susceptibility to BVDV infection. BHK, HeLa, mouse L, and Vero cells, which are not permissive to BVDV, were transfected with pKM6, pTK13, or the empty plasmid pcDNA3 and incubated with BVDV NADL 24 h later. The cells were fixed at 48 h p.i. and examined for the presence of viral antigen by immunohistochemical analysis. Viral antigen indicative of BVDV replication was detected neither in transfected nor in nontransfected cells (data not shown). To exclude the possibility that transient expression might not be sensitive enough to demonstrate a low level of susceptibility, stable cell lines were selected from pKM6-transfected HeLa, mouse L, and Vero cells by using G418. Vero, HeLa, and mouse L cell lines constitutively expressing CD46bov were incubated with BVDV NADL at an MOI of 10. Again, infection with BVDV did not occur (data not shown).
BVDV binds to CD46bov-expressing cells.
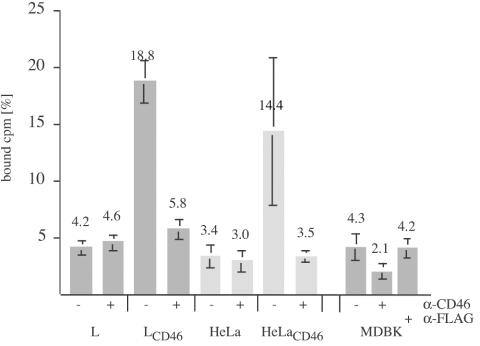
While the presence of CD46bov in HeLa, mouse L, and Vero cells did not result in susceptibility to BVDV, it was important to investigate whether CD46bov was able to bind BVDV. For this purpose, a binding assay based on 3H-labeled BVDV NADL was developed. For the production of labeled virions, 3 × 107 MDBK cells were infected with prototypic BVDV strain NADL at an MOI of 0.1 and metabolically labeled with [3H]uridine for 48 h. Virions were concentrated from the culture medium by ultracentrifugation and then purified by flotation in a 5 to 30% Optiprep- PBS step gradient. BVDV infectivity peaked at a density of 1.093 ± 0.008 g/ml (mean and standard deviation), corresponding to 16.5% (wt/vol) Optiprep. For the binding assay, 104 cpm (5 × 105 focus-forming units) of labeled BVDV was incubated with 5 × 105 cells on ice for 2 h. The inoculum was removed, and the cells were washed twice with PBS. To determine the amount of adsorbed virus, the cells were solubilized, and counts for these samples along with the pooled inoculum and washing solutions were determined. From these numbers, the ratio of adsorbed BVDV to input BVDV was calculated as percent counts per minute. While nontransfected HeLa and mouseL cells bound 4.2 and 3.4% of the radioactive inoculum, respectively, CD46-expressing HeLa (HeLaCD46) cells and CD46-expressing L (LCD46) cells bound 18.8 and 14.4% of 3H-labeled BVDV, respectively (Fig. 5). To examine whether the anti-CD46bov MAbs affected the binding of BVDV, HeLaCD46 and LCD46 cells were preincubated with saturating amounts of anti-CD46bov antibodies before 3H-labeled BVDV was added. Preincubation with anti-CD46bov MAbs specifically reduced the binding of BVDV to LCD46 cells (5.8%) and HeLaCD46 cells (3.5%). The same degrees of inhibition were obtained when the individual MAbs were used (data not shown). Preincubation of LCD46 and HeLaCD46 cells with the same amounts of anti-FLAG MAb M2 had no effect (data not shown).
FIG. 5.
Binding of 3H-labeled BVDV to cells expressing CD46bov. Monolayers with 2 × 105 mouse L, LCD46, HeLa, HeLaCD46, and MDBK cells were incubated with 5 × 103 cpm of 3H-labeled BVDV NADL for 1 h at 0°C without (−) or with (+) preincubation with a mixture of anti-CD46bov MAbs or anti-FLAG MAb M2, respectively. The inoculum was removed, and the cells were washed twice with PBS before being lysed with Triton X-100 (1%) and counted by LSC. The ratio of bound to total input radioactivity is indicated as a percentage. The columns represent mean values of triplicate experiments, and the bars indicate maximum and minimum values.
MDBK cells, which are efficient host cells for BVDV, bound surprisingly small amounts of 3H-labeled BVDV (4.3%), and binding was specifically reduced to 2.1% by preincubation with anti-CD46bov MAbs. The same amount of control IgG (anti-FLAG MAb M2) led to 4.2% binding (Fig. 5).
The expression of CD46bov confers elevated susceptibility to porcine cells.
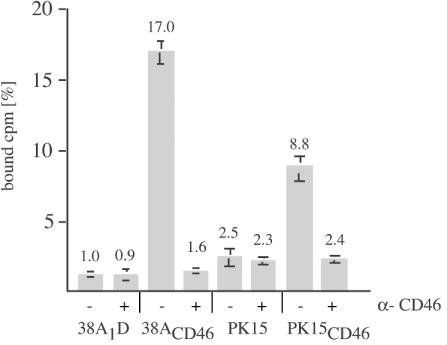
Pestiviruses are restricted to cloven-hoofed animals, but cross-species infections are common among cattle and sheep and also occur between ruminants and pigs. In tissue cultures, bovine pestiviruses can be propagated on porcine cells. However, compared to that on MDBK cells, the plaquing efficiency of, e.g., BVDV NADL on PK15 cells is >100-fold lower. We therefore analyzed whether the expression of CD46bov by porcine cells had an effect on virus binding and plaquing efficiency. CD46bov-expressing cell lines were established from PK15 and 38A1D cell lines by transfection with pKM6 and were selected with G418. In these cells, the expression of CD46bov led to increased binding of 3H-labeled BVDV; PK15CD46 cells bound 8.8% of the labeled inoculum (PK15 cells bound 2.5%), and CD46-expressing 38A1D (38A1DCD46) cells bound 17% (38A1D cells bound 1%). As expected from the previous experiments, preincubation with anti-CD46bov MAbs reduced binding to the level seen in nontransfected cells (Fig. 6), while preincubation with 2 μg of anti-FLAG IgG had no effect (data not shown).
FIG. 6.
Binding of 3H-labeled BVDV to CD46bov-expressing porcine cells. Equal numbers of PK15, PK15CD46, 38A1D, and 38A1DCD46 cells were incubated without (−) or with (+) a mixture of anti-CD46bov MAbs for 1 h at 0°C before incubation with 5 × 103 cpm of 3H-labeled BVDV NADL for 1 h. The inoculum was removed, and the cells were washed twice with PBS before being lysed with Triton X-100 and counted by LSC. The columns represent mean values of triplicate experiments, and the bars indicate maximum and minimum values.
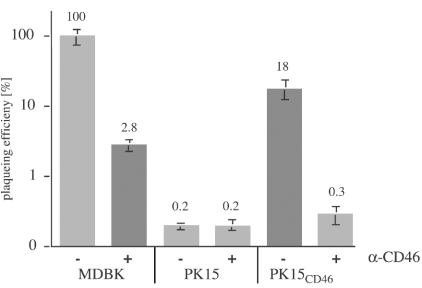
The susceptibilities of PK15, PK15CD46, and MDBK cells were compared by infection of cells with serial dilutions of BVDV NADL. The cells were fixed at 24 h p.i., and infectious centers were revealed by immunohistochemical analysis. Compared to the plaquing efficiency of BVDV NADL on MDBK cells (100%), only 0.2% infectious centers were detected on PK15 cells. Strikingly, PK15CD46 cells (18%) showed a 90-fold increase in susceptibility over nontransfected PK15 cells. Preincubation of PK15CD46 cells with anti-CD46bov antibodies prior to infection with BVDV completely abolished this increase (Fig. 7). The increased susceptibility of porcine cells to BVDV NADL induced by the expression of CD46bov also was evident for porcine lymphoma cell line 38A1D. 38A1DCD46 cells were completely lysed by infection with cytopathic BVDV NADL within 3 days, while parental 38A1D cells exhibited only mild signs of cytopathic effects (data not shown).
FIG. 7.
The expression of CD46bov increases the susceptibility of PK15 cells. Equal numbers of MDBK, PK15, and PK15CD46 cells were infected with serial dilutions of BVDV NADL for 1 h at 0°C with (+) or without (−) preincubation with 2 μg of a mixture of anti-CD46bov MAbs. At 24 h p.i., the cells were fixed, and the number of infectious centers was determined after immunohistochemical detection of intracellular BVDV E2 antigen. Susceptibility was calculated as the number of infectious centers determined for PK15 and PK15CD46 cells divided by the number of infectious centers determined for MDBK cells (100%). The columns represent mean values of triplicate experiments, and the bars indicate maximum and minimum values.
DISCUSSION
The most important result of the present study is the identification of CD46bov as the target of MAbs which were shown previously to inhibit the infection of bovine cells by several BVDV strains (28). CD46bov was unambiguously determined to be the 50- to 58-kDa and 80- to 90-kDa target proteins for these MAbs on the basis of the expression of CD46bov cDNAs in heterologous cells. The identification of splice variants of the CD46bov gene which lead to proteins with three different C termini (analogous to human Cyt 1, Cyt 2, and Cyt 3) and two different STP segments (analogous to human STP segments C and ABC) (24, 25) along with posttranslational modifications explains the variability of the observed molecular masses.
A cellular receptor for a virus must fulfill two claims, namely, binding to virus and mediating virus invasion (32). To demonstrate the adsorption of BVDV to CD46bov, a binding assay had to be established. Attempts to incorporate radioactively labeled amino acids into virions failed, especially because of insufficient purification of BVDV from host cellular components. This problem was overcome by metabolic labeling of the BVDV genome with [3H]uridine. Contaminating cellular RNAs and residual unincorporated label were removed efficiently by flotation in an Optiprep gradient. The binding of 3H-labeled BVDV was evident on all CD46bov-expressing cells, and there was a clear correlation between bound BVDV and the amount of expressed CD46bov. These results were especially evident for MDBK cells, which express far less CD46bov than do CD46bov-overexpressing cell lines (data not shown). Specific binding was inhibited by the presence of anti-CD46 antibodies in a concentration-dependent manner and also by competition with unlabeled BVD virions (data not shown). While whole BVDV virions attach to CD46bov, which of the viral glycoproteins acts as the receptor ligand needs to be determined in future experiments.
A direct link between CD46bov expression and susceptibility to BVDV could be demonstrated only with porcine cells. Porcine cells are susceptible to BVDV, but the plaquing efficiency of BVDV NADL on, e.g., PK15 cells was only 0.2% compared to that observed on MDBK cells. Thus, a 500-fold higher MOI was required to infect the same number of PK15 cells. Upon expression of CD46bov in these cells, the susceptibility increased 90-fold from 0.2 to 18%. This increase was fully reversed by the presence of anti-CD46bov antibodies, but the basal susceptibility of the PK15 cells was unchanged. In contrast, HeLaCD46 and mouse LCD46 cells adsorbed to BVDV in significant amounts, but a productive infection did not occur. It was recently shown that subviral genomes of BVDV replicate well in HeLa cells (4), and the onset of a productive infection after electroporation of genomic BVDV RNA into these cells also was observed (data not shown). These findings suggest that the pathway of entry of BVDV into CD46bov-expressing human and murine cells is blocked somewhere between virus adsorption and the onset of replication.
It is believed that pestiviruses enter the host cell by endocytosis, although only preliminary data support this assumption (D. Boulanger, personal communication; T. Krey, unpublished data). It is unlikely, however, that CD46 directly induces an endocytotic process because human CD46 is excluded from incorporation into endosomes due to a sorting signal (Phe-Thr-Ser-Leu) in the C-terminal cytosolic tail (18). A similar C terminus (Leu-Phe-Ser-Leu) was encoded by a cDNA clone of CD46bov (pKM6) and was sufficient to increase the susceptibility of porcine cells. However, CD46bov molecules with C termini analogous to Cyt 2 and Cyt 3 as well as with STP segments ABC (pTK13) increased the susceptibility of PK15 cells but failed altogether to render HeLa and mouse L cells permissive for BVDV in transient expression experiments. In conclusion, it is reasonable to assume that BVDV interacts with one or more additional cellular molecule(s) which facilitates entry. This factor(s) is specifically present on cells of the Artiodactyla and probably represent a coreceptor(s) which supplements the function of the binding receptor.
According to the literature, other molecules have been claimed to act as pestivirus receptors. The binding of CSFV strain Brescia (9, 10) and BVDV strain PE515 (12) was demonstrated for GAGs, e.g., heparin or heparan sulfate (9). Glycoprotein Erns was identified as a primary ligand. In either case, a single point mutation (CSFV: Ser476Arg) in Erns was the reason for the high affinity for GAGs. Very likely this point mutation is a result of tissue culture adaptation of the particular virus isolates (9, 10, 12), but GAGs also may serve with a low affinity as alternative receptors. An alternative receptor(s) may account for the residual susceptibility (≈1%) of MDBK cells to BVDV NADL, since its entry was not inhibited even by high concentrations of anti-CD46bov antibodies (data not shown). Also, the basal susceptibility of porcine cells to BVDV NADL likely is caused by an alternative receptor molecule(s). The function of two proteinaceous receptor candidates is less clear. A 50-kDa protein was identified by using anti-idiotypic antibodies raised against neutralizing MAbs binding to BVDV E2 (35, 36). The anti-idiotypic antibodies as well as the enriched 50-kDa protein, which is also present on cells of nonsusceptible species, were able to block BVDV infection of bovine cells (37). To further define the function of this protein in BVDV entry, the respective cloned gene would be required. Another receptor candidate is LDL-R; it was reported that anti-LDL-R antibodies interfered with BVDV infection (2). The absence of LDL-R from CRIB cells, which are resistent to BVDV infection (7, 8), was taken as further evidence. Interestingly, LDL-R also has been postulated to act as a cellular receptor for hepatitis C virus (HCV), which is also a member of the Flaviviridae (2, 21).
Taken together, these results provide strong evidence that CD46bov acts as a cellular receptor for BVDV strain NADL and that other BVDV strains most likely use CD46bov as well. These conclusions are based on the reported inhibition of five additional BVDV strains by anti-CD46 MAbs (28) as well as on preliminary experiments which confirmed this result for a larger number of BVDV strains, including clinical isolates not previously passaged in cultured cells (A. Himmelreich, unpublished data). Thus, the use of CD46bov as a receptor is not a result of tissue culture adaptation, as has been observed for a few measles virus isolates which utilize CD46 for entry.
CD46 is present on all nucleated cells and protects cells from autologous complement attack. Is the distribution of a ubiquitous protein like CD46 compatible with the pathogenesis of BVDV? Initial infection with BVDV occurs in the tonsils, and virus spreads to other lymphatic tissues before viremia occurs (15). Thus, tropism to lymphatic tissues is not explained by the distribution of CD46. In the late stage of infection, however, BVDV is found in essentially all organs and tissues (15), indicating the usage of a ubiquitous receptor like CD46.
The identification of CD46 as a BVDV receptor raises a number of questions with regard to the pathway of entry of BVDV, the requirement for a further cellular factor(s), and the role of CD46 in the pathogenesis of disease. It will be interesting to determine whether pig CD46 and sheep CD46 are involved in invasion by CSFV and border disease virus.
Acknowledgments
This work was funded by Deutsche Forschungsgemeinschaft grant SFB 535-A1.
T.K. is a fellow of the Graduiertenkolleg Biochemistry of Nucleoprotein Complexes.
REFERENCES
- 1.Adams, E. M., M. C. Brown, M. Nunge, M. Krych, and J. P. Atkinson. 1991. Contribution of the repeating domains of membrane cofactor protein (CD46) of the complement system to ligand binding and cofactor activity. J. Immunol. 147:3005-3011. [PubMed] [Google Scholar]
- 2.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q.-X. Zhang. 1999. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barilla-LaBarca, M. L., M. K. Liszewski, J. D. Lambris, D. Hourcade, and J. P. Atkinson. 2002. Role of membrane cofactor protein (CD46) in regulation of C4b and C3b deposited on cells. J. Immunol. 168:6298-6304. [DOI] [PubMed] [Google Scholar]
- 4.Behrens, S. E., C. W. Grassmann, H. J. Thiel, G. Meyers, and N. Tautz. 1998. Characterization of an autonomous subgenomic pestivirus RNA replicon. J. Virol. 72:2364-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchholz, C. J., D. Koller, P. Devaux, C. Mumenthaler, J. Schneider-Schaulies, W. Braun, D. Gerlier, and R. Cattaneo. 1997. Mapping of the primary binding site of measles virus to its receptor CD46. J. Biol. Chem. 272:22072-22079. [DOI] [PubMed] [Google Scholar]
- 6.Casasnovas, J. M., M. Larvie, and T. Stehle. 1999. Crystal structure of two CD46 domains reveals an extended measles virus-binding surface. EMBO J. 18:2911-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores, E. F., and R. O. Donis. 1995. Isolation and characterization of a mutant MDBK cell line resistant to bovine viral diarrhea virus infection due to a block in viral entry. Virology 208:365-375. [DOI] [PubMed] [Google Scholar]
- 8.Flores, E. F., L. C. Kreutz, and R. O. Donis. 1996. Swine and ruminant pestiviruses require the same cellular factor to enter bovine cells. J. Gen. Virol. 77:1295-1303. [DOI] [PubMed] [Google Scholar]
- 9.Hulst, M. M., H. G. van Gennip, and R. J. Moormann. 2000. Passage of classical swine fever virus in cultured swine kidney cells selects virus variants that bind to heparan sulfate due to a single amino acid change in envelope protein E(rns). J. Virol. 74:9553-9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hulst, M. M., H. G. van Gennip, A. C. Vlot, E. Schooten, A. J. de Smit, and R. J. Moormann. 2001. Interaction of classical swine fever virus with membrane-associated heparan sulfate: role for virus replication in vivo and virulence. J. Virol. 75:9585-9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iqbal, M., H. Flick-Smith, and J. W. McCauley. 2000. Interactions of bovine viral diarrhoea virus glycoprotein E(rns) with cell surface glycosaminoglycans. J. Gen. Virol. 81:451-459. [DOI] [PubMed] [Google Scholar]
- 12.Iqbal, M., and J. W. McCauley. 2002. Identification of the glycosaminoglycan-binding site on the glycoprotein E(rns) of bovine viral diarrhoea virus by site-directed mutagenesis. J. Gen. Virol. 83:2153-2159. [DOI] [PubMed] [Google Scholar]
- 13.Iwata, K., T. Seya, Y. Yanagi, J. M. Pesando, P. M. Johnson, M. Okabe, S. Ueda, H. Ariga, and S. Nagasawa. 1995. Diversity of sites for measles virus binding and for inactivation of complement C3b and C4b on membrane cofactor protein CD46. J. Biol. Chem. 270:15148-15152. [DOI] [PubMed] [Google Scholar]
- 14.Kallstrom, H., M. K. Liszewski, J. P. Atkinson, and A. B. Jonsson. 1997. Membrane cofactor protein (MCP or CD46) is a cellular pilus receptor for pathogenic Neisseria. Mol. Microbiol. 25:639-647. [DOI] [PubMed] [Google Scholar]
- 15.Liebler-Tenorio, E. M., I. Greiser-Wilke, and J. F. Pohlenz. 1997. Organ and tissue distribution of the antigen of the cytopathogenic bovine viral diarrhea virus in the early and advanced phase of experimental mucosal disease. Arch. Virol. 142:1613-1634. [DOI] [PubMed] [Google Scholar]
- 16.Liszewski, M. K., M. Leung, W. Cui, V. B. Subramanian, J. Parkinson, P. N. Barlow, M. M., and J. P. Atkinson. 2000. Dissecting sites important for complement regulatory activity in membrane cofactor protein. J. Biol. Chem. 275:37692-37701. [DOI] [PubMed] [Google Scholar]
- 17.Liszewski, M. K., T. W. Post, and J. P. Atkinson. 1991. Membrane cofactor protein (MCP or CD46): newest member of the regulators of complement activation gene cluster. Annu. Rev. Immunol. 9:431-455. [DOI] [PubMed] [Google Scholar]
- 18.Maisner, A., G. Zimmer, M. K. Liszewski, D. M. Lublin, J. P. Atkinson, and G. Herrler. 1997. Membrane cofactor protein (CD46) is a basolateral protein that is not endocytosed. Importance of the tetrapeptide FTSL at the carboxyl terminus. J. Biol. Chem. 272:20793-20799. [DOI] [PubMed] [Google Scholar]
- 19.Manchester, M., J. E. Gairin, J. B. Patterson, J. Alvarez, M. K. Liszewski, D. S. Eto, J. P. Atkinson, and M. B. Oldstone. 1997. Measles virus recognizes its receptor, CD46, via two distinct binding domains within SCR1-2. Virology 233:174-184. [DOI] [PubMed] [Google Scholar]
- 20.Manchester, M., A. Valsamakis, R. Kaufman, M. K. Liszewski, J. Alvarez, J. P. Atkinson, D. M. Lublin, and M. B. Oldstone. 1995. Measles virus and C3 binding sites are distinct on membrane cofactor protein (CD46). Proc. Natl. Acad. Sci. USA 92:2303-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monazahian, M., I. Bohme, S. Bonk, A. Koch, C. Scholz, S. Grethe, and R. Thomssen. 1999. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J. Med. Virol. 57:223-229. [DOI] [PubMed] [Google Scholar]
- 22.Naniche, D., G. Varior-Krishnan, F. Cervoni, T. F. Wild, B. Rossi, C. Rabourdin-Combe, and D. Gerlier. 1993. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 67:6025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada, N., M. K. Liszewski, J. P. Atkinson, and M. Caparon. 1995. Membrane cofactor protein (CD46) is a keratinocyte receptor for the M protein of the group A streptococcus. Proc. Natl. Acad. Sci. USA 92:2489-2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Post, T. W., M. K. Liszewski, E. M. Adams, I. Tedja, E. A. Miller, and J. P. Atkinson. 1991. Membrane cofactor protein of the complement system: alternative splicing of serin/threonine/ proline-rich exons and cytoplasmic tails produce multiple isoforms which correlate with protein phenotype. J. Exp. Med. 174:93-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riley, R. C., C. Kemper, M. Leung, and J. P. Atkinson. 2002. Characterization of human membrane cofactor protein (MCP; CD46) on spermatozoa. Mol. Reprod. Dev. 62:534-546. [DOI] [PubMed] [Google Scholar]
- 26.Rümenapf, T., E. G. Strauss, and J. H. Strauss. 1994. Subgenomic mRNA of Aura alphavirus is packaged into virions. J. Virol. 68:56-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santoro, F., P. E. Kennedy, G. Locatelli, M. S. Malnati, E. A. Berger, and P. Lusso. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817-827. [DOI] [PubMed] [Google Scholar]
- 28.Schelp, C., I. Greiser-Wilke, G. Wolf, M. Beer, V. Moennig, and B. Liess. 1995. Identification of cell membrane proteins linked to susceptibility to bovine viral diarrhea virus infection. Arch. Virol. 140:1997-2009. [DOI] [PubMed] [Google Scholar]
- 29.Seya, T., J. Turner, and J. P. Atkinson. 1986. Purification and characterization of a membrane protein (gp45-70) that is a cofactor for cleavage of C3b and C4b. J. Exp. Med. 163:837-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shusta, E. V., C. Zhu, R. J. Boado, and W. M. Pardridge. 2002. Subtractive expression cloning reveals high expression of CD46 at the blood-brain barrier. J. Neuropathol. Exp. Neurol. 61:597-604. [DOI] [PubMed] [Google Scholar]
- 31.Strandström, H., P. Veijalainen, V. Moennig, G. Hunsmann, H. Schwarz, and W. Schäfer. 1974. C-type particles produced by permanent cell line from a leukemic pig. Virology 57:175-188. [DOI] [PubMed] [Google Scholar]
- 32.Tardieu, M., R. L. Epstein, and H. L. Weiner. 1982. Interaction of viruses with cell surface receptors. Int. Rev. Cytol. 80:27-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thiel, H.-J., P. G. W. Plagemann, and V. Moennig. 1996. Pestiviruses, p. 1059-1073. In B. N. Fields et al. (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 34.Thiel, H. J., R. Stark, E. Weiland, T. Rümenapf, and G. Meyers. 1991. Hog cholera virus: molecular composition of virions from a pestivirus. J. Virol. 65:4705-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue, W., and H. C. Minocha. 1993. Identification of the cell surface receptor for bovine viral diarrhea virus by using anti-idiotypic antibodies. J. Gen. Virol. 74:73-79. [DOI] [PubMed] [Google Scholar]
- 36.Xue, W., D. J. Orten, O. Y. Abdelmagid, M. Rider, F. Blecha, and H. C. Minocha. 1991. Anti-idiotypic antibodies mimic viral diarrhea virus antigen. Vet. Microbiol. 29:201-212. [DOI] [PubMed] [Google Scholar]
- 37.Xue, W., S. Zhang, and H. C. Minocha. 1997. Characterization of a putative receptor protein for bovine viral diarrhea virus. Vet. Microbiol. 57:105-118. [DOI] [PubMed] [Google Scholar]