Abstract
Plants respond to pathogens using elaborate networks of genetic interactions. Recently, significant progress has been made in understanding RNA silencing and how viruses counter this apparently ubiquitous antiviral defense. In addition, plants also induce hypersensitive and systemic acquired resistance responses, which together limit the virus to infected cells and impart resistance to the noninfected tissues. Molecular processes such as the ubiquitin proteasome system and DNA methylation are also critical to antiviral defenses. Here, we provide a summary and update of advances in plant antiviral immune responses, beyond RNA silencing mechanisms—advances that went relatively unnoticed in the realm of RNA silencing and nonviral immune responses. We also document the rise of Brachypodium and Setaria species as model grasses to study antiviral responses in Poaceae, aspects that have been relatively understudied, despite grasses being the primary source of our calories, as well as animal feed, forage, recreation, and biofuel needs in the 21st century. Finally, we outline critical gaps, future prospects, and considerations central to studying plant antiviral immunity. To promote an integrated model of plant immunity, we discuss analogous viral and nonviral immune concepts and propose working definitions of viral effectors, effector-triggered immunity, and viral pathogen-triggered immunity.
INTRODUCTION
Plant viruses are superb entities for the elucidation of host–microbe interactions as they encode relatively few proteins and are exclusively dependent on host cellular metabolism for multiplication and movement. Virologists have had many “firsts” in the study of plant immune responses, including the description of the hypersensitive response (HR), systemic acquired resistance (SAR), and elaboration of the gene-for-gene resistance response—contemporary immune response paradigms that were discovered more than 50 years ago studying plant-infecting viruses (Samuel, 1934; Holmes, 1937, 1938, 1954; Ross, 1961a, 1961b). Paradoxically, in the molecular genetics era, critical advances in our mechanistic understanding of innate immunity have been made primarily by studying plant pathogenic bacteria and fungi and primarily using dicotyledonous (dicot) hosts. Although there have been studies with viruses such as Tobacco mosaic virus (TMV), Tomato bushy stunt virus (TBSV), Potato virus X (PVX), potyviruses, cucumoviruses, and bromoviruses, almost without exception dicotyledonous plants, primarily Nicotiana benthamiana and Arabidopsis thaliana, were used as experimental hosts (Whitham et al., 2003, 2006; Ascencio-Ibáñez et al., 2008; García-Marcos et al., 2009; Hanssen et al., 2011; Postnikova and Nemchinov, 2012). Far fewer studies pertain to viruses that infect grasses (the Poaceae). In this review, our intent is fivefold: (1) provide a brief historical precedence to the origins of contemporary immune response concepts in host–pathogen interactions, (2) summarize recent developments and the current state of knowledge of virus–host interactions, (3) present a primer on the emergence of monocot host–virus pathosystems toward understanding antiviral immune responses in grasses, (4) outline areas that may be particularly fruitful for study in the coming years with an overarching goal of unifying plant biology and virology, and (5) discuss contentious semantics (or lack thereof) in describing antiviral immunity in contrast with the nonviral immune responses.
ANTIVIRAL IMMUNE RESPONSES
The initial reports by Francis O. Holmes in 1929, working with TMV infection of Nicotiana glutinosa, that local necrotic lesions were a sign of plant virus infection rapidly opened up the prospects to determine virus titer, isolate viruses, dissect antiviral defenses, and most importantly to quantify viruses using bioassays (Holmes, 1929). This method ultimately led to breakthroughs in understanding the nature of the virus, extending from the crystallization of TMV by Wendell M. Stanley in 1935 to building an infectious cDNA construct (Dawson et al., 1986; Scholthof et al., 1999; Creager, 2002; Scholthof, 2004, 2011, 2013). Both Stanley and Holmes were at the Rockefeller Institute for Medical Research at the Princeton, NJ research facility. Holmes made the interesting discovery that the necrotic local lesions that developed on TMV-inoculated N. glutinosa leaves were due to a resistance response associated with a gene, N (for necrotic lesion response). Within a decade, Holmes had moved the N gene from N. glutinosa to economically important tobacco (Nicotiana tabacum) and had become the first scientist to demonstrate that a dominant gene was associated with the resistance response against TMV infection. As such, Holmes provided the first genetic and physiological evidence for a phenomenon that a virus infection could be limited to the infection sites through the action of host defense machinery. Holmes is also credited with identifying several resistance (R) genes in tomato (Solanum lycopersicum) and pepper (Capsicum annuum) and for moving them into different cultivars for protection of field and greenhouse-grown plants against TMV (Holmes, 1929, 1937, 1938, 1954).
Holmes’ findings were seminal to the field of plant pathology and led to the emergence of core concepts in plant disease resistance. For example, the TMV:N-gene response showed that a gene-for-gene interaction induced a local lesion or HR in the infected plant. Interestingly, Frank Ross was also at the Rockefeller Institute for Medical Research in the late 1930s with Stanley and in the same laboratory where Holmes was working. Ross is credited with discovering SAR. In 1961, Ross found that the zone surrounding TMV-induced local lesions on some tobacco species was completely resistant to subsequent TMV infection, as well as to unrelated viruses, including Tobacco necrosis virus and Tobacco ringspot virus (Ross, 1961a, 1961b). However, in beans (Phaseolus vulgaris), the zone around the TMV-induced lesions protected the plant only from subsequent challenges by TMV, not against infection by heterologous viruses such as Tobacco necrosis virus or Alfalfa mosaic virus. From these results, Ross suggested that the differences in these “local acquired resistance” responses were indicative of differential host responses to virus infections. In subsequent experiments, Ross used Holmes’ N gene–expressing N. tabacum Samsun NN plants and inoculated a half-leaf with TMV. Necrotic local lesions were observed within a few days. Subsequent challenge of the opposite half-leaf or upper leaves of the same plant with TMV resulted in no detectable virus. He described this as “systemic acquired resistance” and determined that it was activated within 2 to 3 d of TMV inoculation (and local lesion formation). This immune response persisted for >20 d with fewer and smaller lesions observed on the upper leaves as well, as summarized by Russell (1978). Ross’s observations led to further studies toward elucidating the nature of such immune response in plants.
As we now know, virus-associated chlorotic lesions or spots, ringspots, and necrotic lesions on leaves, stems, and fruits are various symptomatic manifestations of host immune responses triggered in the infected cells. In the instances of HR and necrosis, virus accumulation is limited to a few hundred infected cells. Classically, HR-mediated resistance is known to be triggered when a pathogen-encoded avirulence factor (Avr) is recognized in plants by a host R gene product (Albar et al., 2006; Moffett, 2009). According to current plant immunity descriptions, there are two layers of plant immune responses against microbial pathogens. First, the recognition of certain conserved pathogen- or microbe-associated molecular patterns (P/MAMPs) by plant pattern recognition receptors (PRRs) initiates the so called P/MAMP-triggered immune (PTI) response, which may occasionally result in HR (Jones and Dangl, 2006; Bent and Mackey, 2007; Boller and Felix, 2009; Dodds and Rathjen, 2010; Schwessinger and Ronald, 2012). As a counter-response to plant PTI defenses, adapted microbes deliver specific effector proteins into plant cells, which compromise PTI defenses and interfere with host defense signaling. To further defend the action of the microbial effectors, plants evolved specific surveillance systems involving receptor-like proteins (R proteins) that directly or indirectly recognize the microbial effectors or monitor their activities in the cell to trigger the so-called effector-triggered immune (ETI) response. Paradoxically, an effector protein can also be the elicitor of ETI defense. Whether the effector or elicitor role of an effector protein prevails is primarily predicated on the presence of the complementing R gene in the plant. The ETI responses, and to a somewhat lesser extent the PTI responses, are closely associated with or even culminate in HR, thus imparting resistance against the invading pathogen (Jones and Dangl, 2006).
Based on current definitions of microbial P/MAMPs and effectors (Jones and Dangl, 2006; Bent and Mackey, 2007; Boller and Felix, 2009; Dodds and Rathjen, 2010; Schwessinger and Ronald, 2012; Spoel and Dong, 2012), viruses are not generally viewed as encoding P/MAMPs or effectors, and antiviral immune responses triggered via the R proteins are not typically classified as ETI responses. In fact, antiviral immune concepts are generally excluded from plant innate immunity models (Jones and Dangl, 2006; Bent and Mackey, 2007; Boller and Felix, 2009; Hogenhout et al., 2009; Dodds and Rathjen, 2010; Schwessinger and Ronald, 2012; Spoel and Dong, 2012). One intent of this review is to present a discussion of such analogous plant antiviral immune responses. Furthermore, we attempt to unify the semantics of plant immune responses through integration of antiviral immune concepts and definitions in the current plant immunity models.
HYPERSENSITIVE AND NECROTIC RESISTANCE RESPONSES TO VIRUS INFECTION
HR and necrotic responses impart resistance against diverse plant pathogenic fungi, bacteria, and viruses, and, to some extent, use similar mechanisms. During a viral infection, in a manner similar to nonviral infections, an HR response is initiated by Avr/R protein interactions that lead to metabolic changes in defense hormone levels, such as salicylic acid (SA), jasmonic acid (JA), and nitric oxide (NO), and the accumulation of reactive oxygen species (ROS), such as O2− and hydrogen peroxide, both in the infected and noninfected tissues (Culver and Padmanabhan, 2007; Carr et al., 2010; Pallas and García, 2011; Mandadi and Scholthof, 2012). At the cellular level, HR affects calcium (Ca2+) ion homeostasis and alters membrane potential and permeability (Mur et al., 2008). For example, TMV and turnip crinkle virus (TCV) infections both induce callose deposition at the plasmodesmata and alter membrane permeability permitting electrolyte leakage in tobacco and Arabidopsis, respectively (Weststeijn, 1978; Ueki and Citovsky, 2005; Culver and Padmanabhan, 2007; Carr et al., 2010; Pallas and García, 2011; Zavaliev et al., 2011). Furthermore, during HR, several caspase-like proteinases, such as the vacuolar processing enzymes, are activated. Vacuolar processing enzymes primarily act as effectors of cell death or necrosis during HR (Mur et al., 2008). Although necrosis is observed concomitantly with HR-mediated resistance, necrosis can be uncoupled from the resistance response. For example, the potato (Solanum tuberosum) resistance protein (Rx1) recognizes the PVX capsid protein (CP) and inhibits PVX replication, thus imparting resistance independent of the HR-associated necrosis triggered subsequently upon PVX CP accumulation (Bendahmane et al., 1999). Another example is the distinct resistance responses exhibited by N. glutinosa and Nicotiana clevelandii against cauliflower mosaic virus (CaMV) strain W260 (Cole et al., 2001). N. clevelandii triggers resistance responses against CaMV W260 by producing discernible necrotic lesions, while N. glutinosa produces only mild non-necrotic chlorotic lesions (Cole et al., 2001). Furthermore, these two manifestations of resistance segregate independently, as evidenced from hybridization experiments between N. glutinosa and N. clevelandii, reminiscent of Holmes’s experiments with TMV and solanaceous plants that showed that symptoms and immunity appeared to be separate phenomena (Holmes, 1932). Similarly, mutations in the TBSV p19 protein, within amino acid residues 43 through 85, abolished the typical HR-associated necrotic lesions on N. tabacum during TBSV infection, instead producing mild chlorotic lesions, suggesting that the cell death/necrosis phenotype can be uncoupled from resistance response (Chu et al., 2000). Finally, the tomato resistance protein (Tm-1) imparts resistance against Tomato mosaic virus (ToMV) by inactivating the ToMV replicase protein, without eliciting HR-associated cell death (Ishibashi et al., 2007, 2009). Taken together, multiple studies with PVX, TBSV, CaMV, and ToMV underscore that HR-associated necrosis and resistance responses, although related, are distinct processes and that the interactions among viral and host proteins could impose differential constraints on the manifestation of resistance phenotypes.
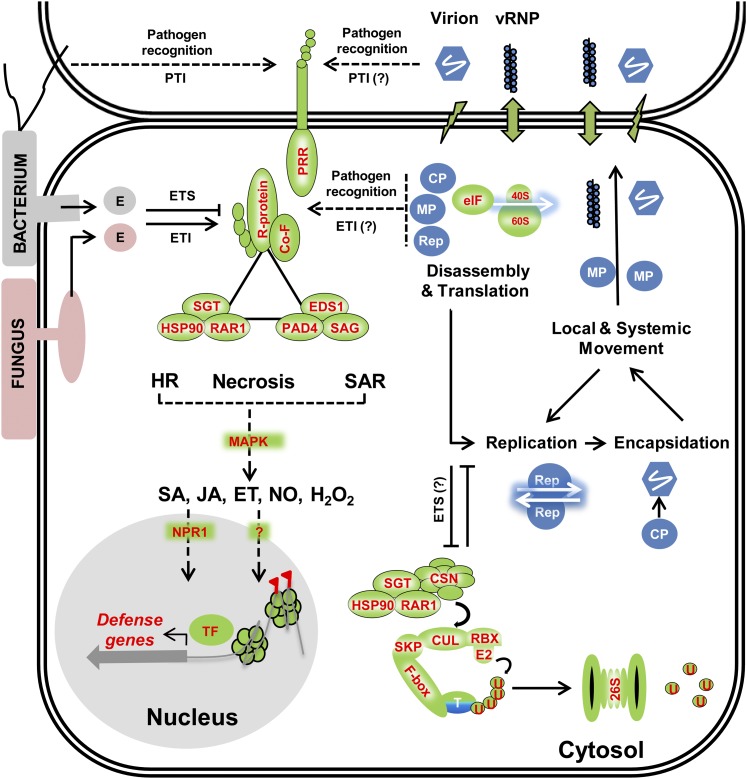
At the molecular and biochemical level, several genetic signaling cascades are activated during HR to induce multiple proteins, including mitogen-activated protein (MAP) kinase proteins, and are reviewed in detail by others (Mur et al., 2008). Downstream of these primary signaling cascades, expression of several defense-related proteins, such as glucanases, chitinases, and defensins, in the pathogenesis-related protein family are upregulated (Mur et al., 2008). Remarkably, the genetic components that mediate HR against diverse viral and nonviral pathogens are conserved across plant genera. For example, of the three major signaling modules that function early in HR signaling (Figure 1), two modules mediate HR against viral and nonviral pathogens alike.
Figure 1.
Antiviral Immune Responses Analogous to Bacterial and Fungal Immune Responses.
Viruses typically enter plant cells through cellular damage (lightning bolt) and move from cell to cell primarily via plasmodesmata (up-down arrow) as viral ribonucleoprotein complexes (vRNP) and/or virions. Virus-encoded proteins such as replicase (Rep), capsid protein (CP), and movement protein (MP) are translated within the host cell cytosol and cooperatively function in translation, replication, encapsidation, and movement of the virus. Similar to events that occur during bacterial and fungal-triggered immune responses, virus-associated factors, such as virion components or virus-encoded proteins, could be perceived by putative cell surface PRRs or cytosolic NB-LRR receptors (e.g., R proteins) to trigger analogous ETI or susceptible (ETS) responses, culminating in HR, SAR, and/or necrosis phenotypes. Bacterial and fungal secreted effector proteins involved in ETI signaling are indicated by “E.” In a manner similar to bacterial and fungal ETI responses, virus-triggered ETI responses also involve functional SGT1/RAR1/HSP90 (Liu et al., 2004) and EDS1/PAD4/SAG101 (Zhu et al., 2011) protein complexes. Combinatorial interactions between viral proteins, R proteins, R cofactors (Co-F), SGT1/RAR1/HSP90, and EDS1/PAD4/SAG101 complexes mediate distinct downstream changes in SA, JA, ethylene (ET), NO, and hydrogen peroxide levels or signaling via MAP kinase signaling cascades. NPR1, a nucleo-cytoplasmic protein critical for nonviral SA defense responses, mediates transcriptional changes in defense gene expression via interactions with specific transcription factors (TFs) (Dong, 2004). The majority of virus-triggered SA responses, however, appear to be NPR1 independent (Whitham et al., 2003). In addition to TF-mediated transcriptional changes, viruses also trigger chromatin modifications including DNA methylation changes (red flag) and increased homologous recombination rates (Kovalchuk et al., 2003; Boyko et al., 2007; Kathiria et al., 2010), epigenetic changes that could be stable inherited by progeny. The SGT1/RAR1/HSP90 complex also interacts with CSN components to mediate degradation of either viral or host proteins (T) via 26S-proteosome complex (Liu et al., 2002b; Shirasu, 2009). The 26S proteasome also functions in the nucleus, aspects that are discussed in detail elsewhere (Padmanabhan and Dinesh-Kumar, 2010). Ubiquitin protein is abbreviated as “U.” Unknown or putative paradigms are indicated as “?.”
The first functional module is comprised of an adaptor protein, SUPPRESSOR OF THE G2 ALLELE OF SKP1 (SGT1), which physically interacts with REQUIRED FOR MLA12 RESISTANCE1 (RAR1), HEAT SHOCK PROTEIN90 (HSP90), and the R proteins (Austin et al., 2002; Takahashi et al., 2003; Bieri et al., 2004). Together, the SGT1/RAR1/HSP90/R protein complex mediates downstream MAP kinase activation and changes in defense gene expression and hormone levels (Dodds and Rathjen, 2010). As a molecular chaperone, the SGT1/RAR1/HSP90 complex also ensures correct folding and stability of R proteins and facilitates R protein recognition of specific pathogen elicitors. Interestingly, SGT1 also interacts with multiple E3-ubiquitin ligase components, such as S PHASE KINASE-ASSOCIATED PROTEIN1 (SKP1) and CULLIN1 in the CULLIN-RING LIGASE (CRL) complex and CSN4 and CSN5 in the COP9 signalosome (CSN) (Azevedo et al., 2002). During TMV infection of tobacco, both SGT1 and RAR1 interact with CSN3 and CSN8 to mediate the N gene resistance against TMV; silencing of either CSN3 or CSN8 compromises the N gene resistance (Liu et al., 2002b; Shirasu, 2009). HSP90 is also a key regulator of N gene resistance, as it both physically interacts with the N protein and is indispensable for N gene resistance (Liu et al., 2004). Together, these interactions suggest a role for ubiquitin-mediated proteolysis in R protein–mediated resistances and are discussed in further detail in subsequent sections of the review. Although the dynamics of interactions between the viral and host proteins needs further elucidation, the functions of the SGT1/RAR1/HSP90 complex appear strikingly conserved to impart resistance against diverse pathogen types.
A second functional module that mediates HR against viral and nonviral pathogens alike requires the interaction of two lipases, ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) (Aarts et al., 1998; Falk et al., 1999) and PHYTOALEXIN DEFICIENT4 (PAD4) (Feys et al., 2001) with SENESCENCE-ASSOCIATED GENE101 (SAG101) (Feys et al., 2005). In Arabidopsis, the EDS1/PAD4/SAG101 complex regulates HRT-mediated resistance against TCV (Zhu et al., 2011). Typically, R proteins with a Toll-interleukin1-like (TIR) domain in their N terminus require EDS1 to mediate the resistance response (Aarts et al., 1998), while R proteins with a coiled coil (CC) domain require NON-RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1), a plasma membrane glycophosphatidyl-inositol–anchored protein (Day et al., 2006), to mediate the R gene resistance response (Aarts et al., 1998). An exception to this is the Arabidopsis HRT protein, which possesses a CC domain and mediates resistance against TCV through interactions with EDS1 and not NDR1 (Chandra-Shekara et al., 2004). HRT-mediated resistance also requires a functional SA-mediated signaling pathway (Chandra-Shekara et al., 2004). Reduction of endogenous SA through mutations in SA biosynthesis genes such as SALICYLIC ACID INDUCTION-DEFICIENT2/ISOCHORISMATE SYNTHASE1 (ICS1) (Wildermuth et al., 2001) compromises HRT-mediated resistance (Chandra-Shekara et al., 2004), without affecting the HRT-mediated HR. When both EDS1 and SA signaling were disrupted, HRT-mediated HR and resistance against TCV were abolished, suggesting that both molecules are required for HRT-mediated resistance (Chandra-Shekara et al., 2004). This finding also supports the aforementioned notion that HR and resistance responses, although closely related, are unique processes.
A third functional module thus far only described for bacterial infection mediates HR and resistance response. It comprises NDR1 and RPM1 INTERACTING PROTEIN4 (RIN4) proteins (Century et al., 1997; Coppinger et al., 2004; Day et al., 2006). RIN4 functions as a broad-spectrum molecular switch for R protein–mediated resistances against bacterial pathogens. For example, in Arabidopsis and Pseudomonas syringae pv tomato DC3000 (PstDC3000) infections, RIN4 physically interacts with R proteins, RESISTANCE TO P. SYRINGAE PV MACULICOLA1 (RPM1) and RESISTANCE TO P. SYRINGAE2 (RPS2), respectively, only in the absence of the bacterial elicitors, AvrRpm1 and AvrRpt2. This interaction maintains the R proteins in an inactive conformation (Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003). However, in the presence of AvrRpm1 and AvrRpt2, RIN4 protein is either phosphorylated or targeted for AvrRpt2-mediated proteolysis, resulting in the activation of the R proteins (Mackey et al., 2002; Coaker et al., 2005). RIN4 also physically interacts with NDR1 (Day et al., 2006). As a surveillance protein, NDR1 interacts with RIN4, thus depleting the total pool of RIN4 available for interaction with RPM1 and RPS2. This, in turn, activates R protein–mediated resistance and HR. NDR1 has topologies similar to integrin proteins and functions in plant stress signaling by promoting plasma membrane–cell wall adhesions (Knepper et al., 2011). Whether NDR1 and RIN4 signaling is critical for virus-triggered immune responses is yet to be determined.
In summary, though diverse plant pathogenic bacteria, fungi, and viruses encode different Avr proteins, possess different elicitors, and use distinct infection strategies, they appear to similarly alter the metabolic, physiological, and cellular states of the hosts. Furthermore, signaling components such as the SGT1/RAR1/HSP90 and EDS1/PAD4/SAG101 complexes, similarly serve to mediate resistance responses against diverse viral and nonviral pathogens, suggesting that plants have evolved such broad-spectrum disease resistances to simultaneously defend against diverse pathogen types.
SYSTEMIC NECROSIS RESPONSES
In contrast with the resistant (or incompatible) host–virus interactions, most susceptible (or compatible) virus infections do not trigger HR and do not produce localized necrotic lesion phenotypes to limit the virus spread in the host plants. However, a similar yet distinct form of necrosis, termed systemic necrosis, is observed in susceptible interactions. For example, systemic necrosis was reported in young Nicotiana rustica plants inoculated with TMV (Holmes, 1936); N. benthamiana with mixed infections of potexviruses, PVX, or Plantago asiatica mosaic virus (PLAMV) isolate Li1 and Potato virus Y (González-Jara et al., 2004; Ozeki et al., 2006); TBSV-infected N. benthamiana and N. clevelandii (Chu et al., 2000); Cucumber mosaic virus (CMV) and satellite RNA-D infected tomato (Xu and Roossinck, 2000); and Panicum mosaic virus (PMV) and its satellite virus (SPMV) infected Brachypodium distachyon and millet species (Panicum miliaceum, Pennisetum glaucum, and Setaria italica; Scholthof, 1999; Mandadi and Scholthof, 2012).
Systemic necrosis resembles necrosis commonly observed in lesion mimic mutants, resulting either from constitutive or uncontrolled cell death (Lorrain et al., 2003; Moeder and Yoshioka, 2008). Unlike HR-associated necrosis, systemic necrosis is manifested much later in the infection and is primarily observed in the upper noninoculated tissues. Another distinction between systemic necrosis and HR-associated necrosis is that systemic necrosis is a lethal response that can kill the host plant, while HR does not. Moreover, systemic necrosis is thought not to preclude virus multiplication or its systemic movement, thereby resulting in a susceptible infection. In contrast with the relatively well understood mechanisms leading to HR and associated necrosis, we are just beginning to understand the molecular processes that underlie systemic necrosis responses and how systemic necrosis responses relate to antiviral immunity. Recent findings revealed that despite the differing roles or outcomes, systemic necrosis and HR-associated necrosis share remarkable similarities at the biochemical and molecular level. For example, both systemic necrosis and HR-associated necrosis involve programmed cell death, alter expression of similar defense-related genes, and trigger ROS accumulation (Xu and Roossinck, 2000; Kim et al., 2008; Komatsu et al., 2010; Xu et al., 2012).
Komatsu et al. (2010) investigated the molecular determinants leading to systemic necrosis elicited by infection with PLAMV, a potexvirus, in N. benthamiana. Using a candidate gene approach, they determined that genes mediating HR in an incompatible interaction also mediate systemic necrosis in compatible infections. For example, systemic necrosis responses depend on a functional SGT1/RAR1 complex and also require MAPKKKα/MEK2 signaling (Komatsu et al., 2010). Unexpectedly, silencing of SGT1 and RAR1 promoted the accumulation of PLAMV (Komatsu et al., 2010) in the infected N. benthamiana plants. This result conflicts with a prevailing assumption that systemic necrosis does not impede viral accumulation and suggests that systemic necrosis may indeed promote antiviral immunity during a compatible plant–virus interaction.
Systemic necrosis is also elicited in N. benthamiana plants infected with a recombinant PVX vector expressing the potyviral helper component-proteinase (HC-Pro) (González-Jara et al., 2004). To decipher systemic necrosis responses, Pacheco et al. (2012) performed global analysis of gene expression of N. benthamiana plants after systemic necrosis was triggered by the recombinant PVX-HC-Pro virus. Comparisons of transcriptional changes associated with HR-associated necrosis during an incompatible interaction revealed striking similarities among the altered gene expression profiles in systemic necrosis and HR-associated necrosis. Together, these studies support an emerging theme that systemic necrosis and HR-associated necrosis involve similar physiological, molecular, and biochemical features. However, the biological relevance of systemic necrosis in compatible infections remains ambiguous. Perhaps systemic necrosis is merely an uncontrolled or incomplete HR-associated necrosis response that is triggered in distal tissues when the localized HR fails to limit virus spread. As a consequence, it may serve as a last attempt of tissue-level suicide to save the remainder of the plant. Further studies are needed to understand the biological relevance of systemic necrosis in antiviral immunity and the molecular constraints that distinguish systemic necrosis from HR-associated necrosis.
SAR
Similar to HR, SAR is triggered during an incompatible interaction involving Avr and R proteins in the primary infected cells. However, the resistance is transduced to the noninfected distant tissues. Although the exact mechanisms of SAR are not defined, it is initiated through a local interaction among Avr and R proteins and results in accumulation of phytohormones such as SA and JA in the distant tissues (reviewed in Vlot et al., 2008a). The NONEXPRESSOR OF PR1 (NPR1) protein functions downstream of SA to mediate changes in expression of defense genes (Dong, 2004). Depending on the pathogen type and stage of infection, NPR1 also interacts with components of JA signaling (Figure 1). Unlike HR, SAR is a long-lasting immune response primed to provide distant tissue resistance against subsequent infections. In the case of TMV-triggered SAR, the response persists up to 3 weeks (Ross, 1961b). How SAR can be sustained for so long is not clear. However, epigenetic modifications, such as DNA methylation and chromatin remodeling, may be critical to maintain a stable SAR signal (reviewed in Spoel and Dong, 2012). Recent studies of Arabidopsis infected with PstDC3000 demonstrated that SAR can be stably inherited to the next generation, even when the progeny was not exposed to the pathogens—possibly via PstDC3000-triggered hypomethylation of host chromatin (Luna et al., 2012). Interestingly, the transgenerational stability of SAR requires NPR1, as progeny of the SA-insensitive npr1-1 mutant plants failed to possess SAR in the next generation (Luna et al., 2012). This induced resistance phenomena is also triggered in the progeny of plants exposed to caterpillar herbivory (Rasmann et al., 2012). In this case, the stable resistance response is dependent on intact JA signaling and requires the biogenesis of short interfering RNA that could mediate the epigenetic chromatin modifications (Rasmann et al., 2012).
With respect to viral responses, tobacco plants infected with TMV or Oilseed rape mosaic virus were shown to display increased frequency of host DNA homologous recombination in both infected and distant noninoculated leaves (Kovalchuk et al., 2003). The increased homologous recombination rate persisted in the progeny of the TMV-infected plants, but not Oilseed rape mosaic virus–infected plants, and resulted in increased DNA rearrangements and hypomethylation of the Leucine-rich repeat (LRR) gene loci homologous to the N gene (Boyko et al., 2007). In addition, progeny of TMV-infected plants with higher recombination rates exhibited broad-spectrum tolerance to TMV, P. syringae, and Phytophthora nicotianae infections (Kathiria et al., 2010). Whether these apparently stable resistance responses mediated by increased homologous recombination frequency are comparable to next-generation SAR is unclear. Additional studies of virus-induced homologous recombination frequency and the molecular determinants of viral SAR are needed to understand if and how SAR and its associated (epi)genetic alterations are established and maintained during a viral infection.
Another intriguing aspect of SAR is the nature of the systemic signal that mediates SAR in noninfected tissues. Although the exact nature of the SAR signal remains unclear, several metabolites have been proposed as putative signals that mediate SAR in viral and nonviral infections. These include glycerol-3-phosphate (Chanda et al., 2011), indole derivatives (Truman et al., 2010), azeleic acid (Jung et al., 2009), methyl salicylate (MeSA) (Park et al., 2007; Vlot et al., 2008b), and glycerolipids (Chaturvedi et al., 2008). SAR likely involves interaction among multiple SAR signals, such as MeSA, lipid-transfer proteins, and glycerolipids (Liu et al., 2011). Significant crosstalk among the SAR signals and environmental factors, such as light, adds yet another layer of complexity to SAR signaling (Vlot et al., 2008a). Since the first discovery of SAR in 1961 by Ross, the molecular responses involved in virus-triggered SAR have been investigated using the TMV pathosystem. Multiple signals including MeSA appear to perpetuate SAR during TMV infection (Park et al., 2007; Vlot et al., 2008a; Dempsey and Klessig, 2012). Whether these signals also perpetuate SAR in other viral pathosystems requires further investigation. Nevertheless, SAR is yet another conserved plant defense response triggered against diverse pathogenic bacteria, fungi, and viruses. Moreover, in contrast with the HR, SAR renders a broader and long-lasting resistance to diverse pathogen types simultaneously.
R GENE–MEDIATED RESPONSES TO VIRUS INFECTION
Over the past decade, several R genes that mediate resistance against viruses have been identified (Collier and Moffett, 2009; Gururani et al., 2012). The majority of the cloned dominant R genes encode the conserved nucleotide binding (NB) and LRR family proteins (Collier and Moffett, 2009; Moffett, 2009; Gururani et al., 2012). NB-LRR proteins also contain additional N-terminal domains such as the TIR homology domain, a CC domain, a Solanaceae domain, or a predicted BED zinc finger domain. Until recently, the LRR domain was thought to be the major domain critical for R protein function. However, growing evidence indicates that both the LRR and the N terminus domains (TIR or CC) are critical for proper resistance responses. The two domains function through intramolecular interactions and interactions with other proteins (R cofactors) to mediate recognition of pathogen elicitors (Collier and Moffett, 2009; Moffett, 2009). R cofactors are also essential for host-mediated HR and SAR responses. For example, the tobacco N protein that recognizes TMV replicase (p50) to trigger the resistance response requires a chloroplast-localized sulfurtransferase, N RECEPTOR-INTERACTING PROTEIN1 (NRIP1) (Caplan et al., 2008). NRIP1 physically interacts with the N-terminal TIR domain of the N protein and TMV p50 protein (Caplan et al., 2008). Interestingly, p50 alters the endogenous localization of NRIP1 from the chloroplast to the cytosol and nucleus where it interacts with the N protein (Caplan et al., 2008).
Similarly, the potato resistance proteins Rx, Rx2, and GREEN PEACH APHID2 interact with RAN GTPASE-ACTIVATING PROTEIN (RanGAP2) (Sacco et al., 2007). The interaction of RanGAP2 with the N-terminal CC domain of Rx and related proteins is required for specific recognition of PVX coat protein and resistance against PVX (Sacco et al., 2007). Recently, it was shown that RanGAP2 directs the Rx protein from the nucleus to the cytosol and contributes to the resistance response (Tameling et al., 2010). Such interactions of R proteins and cofactor proteins are observed for fungal, bacterial, and other viral resistance responses (Moffett, 2009). In summary, a plethora of plant:pathogen resistance scenarios often require combinatorial interactions among multiple host and pathogen factors such as the Avr, R, and R cofactors. In turn, the resistance responses triggered by such interactions (variously described as the guard hypothesis [Dangl and Jones, 2001], the decoy model [van der Hoorn and Kamoun, 2008], or the bait and switch model [Collier and Moffett, 2009]) culminate in HR and SAR responses via the action of hormone and signaling molecules such as SA, JA, ethylene, NO, and ROS, as reviewed recently (Culver and Padmanabhan, 2007; Truniger and Aranda, 2009; Carr et al., 2010).
In viral infections, in addition to the dominant R gene–related resistance responses, another form of recessive resistance exists that is typically derived by a loss of function in host proteins critical for the establishment of disease (Robaglia and Caranta, 2006; Iyer-Pascuzzi and McCouch, 2007; Truniger and Aranda, 2009; Gururani et al., 2012). For example, amino acid mutations in the eukaryotic translation initiation factor, eIF4E, mediates resistance against several viruses in Arabidopsis, tomato, pepper, pea (Pisum sativum), melon (Cucumis melo), and barley (Hordeum vulgare) (Lellis et al., 2002; Ruffel et al., 2002, 2005; Nicaise et al., 2003; Gao et al., 2004; Kanyuka et al., 2005; Piron et al., 2010). In addition, mutations in another subunit of eukaryotic translation initiation factor, eIF4G, imparts recessive resistance against Rice yellow mottle virus (RYMV) in rice (Oryza sativa) (Albar et al., 2006) and to CMV and TCV in Arabidopsis (Yoshii et al., 2004). In these situations, the host cells are challenged in their ability to efficiently translate viral proteins, thus affecting viral replication and/or local movement (Collier and Moffett, 2009; Gururani et al., 2012). Other host proteins also mediate recessive resistance, such as the Arabidopsis tonoplast-localized transmembrane proteins, TOM1 and TOM2A, which interact with each other and with the TMV-encoded 130K and 180K replicase proteins (Ishikawa et al., 1993; Yamanaka et al., 2000; Tsujimoto et al., 2003). The rate of intracellular accumulation of TMV and its associated subgenomic RNAs was significantly decreased in the loss-of-function mutants tom1 and tom2a. Furthermore, the resistance imparted is specific against TMV but not against CMV (a cucumovirus) or TCV (a carmovirus) (Ishikawa et al., 1993; Ohshima et al., 1998; Yamanaka et al., 2000; Hagiwara et al., 2003; Tsujimoto et al., 2003).
In addition to the aforementioned resistance proteins, other host proteins can contribute to viral resistance responses. In this category, lectin proteins are novel and intriguing resistance-imparting proteins. In animals, lectins activate immune responses against diverse pathogens. Certain lectin receptors, such as the C-type lectins, recognize fungal PAMPS (Willment and Brown, 2008). In plants, lectins bind to mono- or oligosaccharide molecules to discriminate self- from non-self-originated carbohydrates, thus rendering them ideal for pathogen perception (Van Damme et al., 2004). For example, the soybean (Glycine max) lectin β-glucan binding protein binds with high affinity to a 1,3-branched heptaglucoside, a PAMP found in the cell walls of Phytophthora sojae (Mithöfer et al., 2000; Fliegmann et al., 2004), and triggers defense reactions. In this context, the discovery of an Arabidopsis jacalin-type lectin, RESTRICTED TEV MOVEMENT1 (RTM1), that mediates resistance against Tobacco etch virus (TEV), was surprising (Chisholm et al., 2000), as the TEV CP is not glycosylated.
More recently, another jacalin-type lectin, JAX1, was shown to confer resistance against multiple potexviruses (PVX, PLAMV, White clover mosaic virus, and Asparagus virus 3; Yamaji et al., 2012). Although JAX1 and RTM1 are closely related lectin proteins, they impart distinct forms of resistance. JAX1 functions at the cellular level, inhibiting replication of PLAMV (Yamaji et al., 2012), and RTM1 inhibits systemic movement of TEV (Chisholm et al., 2000). An intriguing aspect of lectin-mediated resistance (LMR) compared with the NB-LRR resistance is that it does not invoke HR and SAR responses, nor does it alter SA levels, signaling, or other typical defense gene expression changes commonly modulated in immune resistance responses (Yamaji et al., 2012). This suggests that LMR engages an as yet unidentified and distinct mechanism of antiviral immunity. According to the proposed working model of LMR function (Chisholm et al., 2000; Yamaji et al., 2012), lectin proteins such as JAX1 and RTM1 recognize certain viral proteins that are glycosylated within plant cells to trigger resistance. In the case of JAX1, LMR responses could inhibit viral replication by promoting aggregation of replicase-associated proteins, while RTM1 could inhibit viral movement through interference with viral movement–associated proteins. In addition to RTM1, a small heat shock-like protein (RTM2) (Whitham et al., 2000) and a meprin and TRAF homology domain-containing protein (RTM3) that physically interacts with RTM1 (Cosson et al., 2010) also restrict viral movement. RTM2 possesses a transmembrane domain, but unlike other HSPs, its expression is not heat inducible and does not contribute to thermotolerance (Whitham et al., 2000). Notably, both RTM1 and RTM2 are expressed in phloem-associated tissues and sieve elements, correlating with their function in impeding virus movement. Interestingly, the N terminus of the potyvirus coat protein contains features that can overcome RTM resistance, suggesting that interactions of RTM proteins with viral CPs may mediate resistance responses (Decroocq et al., 2009). Further studies on LMR should reveal the mechanism of this intriguing antiviral resistance response.
In another study, complementary proteomic approaches were used to identify a class of endoplasmic reticulum (ER)–residing chaperones that influence antiviral immune responses (Caplan et al., 2009). In N. benthamiana, the ER proteins ERp57, P5, calreticulin2, and CRT3 are strongly expressed within 2 h of TMV infection in plants expressing the N gene (Caplan et al., 2009). Virus-induced gene silencing (VIGS) of the ER chaperones resulted in loss of N-mediated resistance against TMV, although the cell death/necrosis symptoms and TMV movement in the upper noninoculated leaves was only partially abolished, probably due to incomplete knockdown of the respective transcripts by VIGS. Since ER chaperones function in folding and accumulation of other membrane proteins, the authors tested the effects on other membrane proteins. Loss of Nb-CRT3 and Nb-CRT5 resulted in reduced accumulation of a plasma membrane–localized induced receptor-like kinase, IRK (Caplan et al., 2009). Similar to the expression of calreticulins, IRK is strongly upregulated early in TMV infection, and VIGS knockdown of IRK expression resulted in loss of N-mediated resistance against TMV. Although the mechanism of how an ER chaperone alters IRK accumulation is not clear, these results reveal yet another intricate interplay among ER-residing chaperones and plasma membrane receptor proteins in N-mediated immune responses.
Taken together, resistance genes, particularly those encoding the NB-LRR proteins, have well-conserved roles in plants, which are to guard the host cells against diverse viral and nonviral pathogens and to trigger disease resistance. Moreover, the general mechanism of the recognition of R proteins and Avr factors appears to be similar for viral and nonviral pathogens, whereby R cofactors play crucial roles to guide or modulate R/Avr interactions, ultimately activating HR and resistance responses. Finally, while the predominant antiviral resistance responses are mediated by the dominant R genes, other host proteins, such as the elongation initiation factors, TOM proteins, ER chaperones, calreticulins, and lectin proteins, also influence host resistance against diverse viral infections.
UBIQUITIN PROTEASOME SYSTEM
The ubiquitin proteasome system (UPS) has emerged as a prominent player in influencing virus–host interactions at almost every stage of antiviral defense, both in plants and animals (Gao and Luo, 2006; Citovsky et al., 2009). In turn, viruses use a plethora of strategies to modulate UPS processes. UPS regulates cellular activities including cell cycle, transcription, and signal transduction (Hershko and Ciechanover, 1998). The plant UPS primarily involves the activity of ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin-ligase (E3) (Hua and Vierstra, 2011). These three proteins form the E3 ubiquitin ligase complex that specifically polyubiquitinates cellular proteins that are subsequently targeted for degradation by the 26S proteasome. SKP1 is another critical component of the SCF (for SKP1, CULLIN, and F-box) class of E3 ubiquitin ligases and, through interactions with CULLIN and F-box proteins, recruits proteins for polyubiquitination (Hua and Vierstra, 2011).
Whether UPS processes are employed by the plants to defend against virus infections or viruses use the UPS to promote virulence is ambiguous. For example, the tobacco N gene–mediated resistance response against TMV requires the function of the RAR1/SGT1 complex. RAR1/SGT1 physically interacts with SKP1 and the CSN (Liu et al., 2002b), another E3 ubiquitin ligase that targets cellular proteins for turnover in plant growth and development (Nezames and Deng, 2012). VIGS of RAR1, SGT1, SKP1, or the CSN gene products abolishes the N gene–mediated resistance (Liu et al., 2002b) and supports a role for UPS in mediating the N-mediated HR and resistance responses. Movement proteins of TMV and Turnip yellow mosaic virus are also specifically targeted for degradation by the host UPS machinery, resulting in decreased virulence and pathogenicity (Reichel and Beachy, 2000; Drugeon and Jupin, 2002), highlighting the role of the UPS in antiviral immune responses.
Viruses also can use UPS processes to promote virulence. The TBSV replication component, p33, interacts with a host E2 ubiquitin-conjugase, Cdc34 (Li et al., 2008). Although the p33 protein is a substrate for UPS-mediated turnover in vivo, an optimal level of p33 polyubiquitination appears to mediate positive interaction among the TBSV replicase components and a host ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT protein, Vps23p, which is critical for effective replication of TBSV (Li et al., 2008). This result suggests that host UPS processes may also aid the virus by facilitating infection. Similarly, downregulation of a 26S proteasome subunit RPN1 in N. benthamiana inhibits systemic transport of TMV and PVX, two taxonomically distinct viruses (Jin et al., 2006), again supporting the conclusion that UPS can promote virulence. In the case of RPN1 silencing, the virulence-promoting effect occurs in part through modulation of auxin transport and brassinosteroid signaling, both of which are required for efficient vascular development (Jin et al., 2006). As the vasculature is the means by which viruses systemically infect plant tissues, the altered vasculature in RPN1-silenced plants prohibits viral movement. Given these contradictory outcomes of UPS functions in different virus–host responses, it appears that viruses and hosts employ a variety of strategies to enhance virulence or promote host immunity.
Among other examples of viruses that target UPS processes to promote virulence is Beet severe curly top virus (BSCTV), a geminivirus. BSCTV encodes a pathogenicity factor, C2, which interacts and interferes with the UPS-mediated degradation of a host S-adenosyl-methionine decarboxylase 1 (SAMDC1) enzyme (Zhang et al., 2011). SAMDC1 mediates the decarboxylation of S-adenosyl-methionine (SAM) to dcSAM. Because SAM serves as a donor for methyl groups in various methylation reactions, altering the levels of SAM and dcSAM (as a result of modulating SAMDC1 activity) may have an effect on host DNA methylation status. As predicted, loss of function of either SAMDC1 or the BSCTV C2 protein results in enhanced de novo methylation of the dsDNA replicative form of the BSCTV genome, thus reducing BSCTV replication and dampening BSCTV infectivity (Zhang et al., 2011). Together, these results suggest that the BSCTV C2 protein acquired its remarkable function of modulating host DNA methylation-mediated gene silencing via attenuation of UPS-mediated degradation of SAMDC1.
In plants, the CSN complex regulates the activity of another class of ubiquitin ligases, the CRLs, primarily through the addition or removal of a RELATED TO UBIQUITIN modifier to the CRLs, a process dubbed rubylation (Hua and Vierstra, 2011). The appropriate function of CRLs is thus determined by its rubylation status. Similar to BSCTV, Tomato yellow leaf curl Sardinia virus (TYLCSV), another geminivirus, also encodes a C2 protein. TYLCSV C2 protein physically interacts with a CSN subunit, CSN5, in N. benthamiana. This interaction interferes with the derubylation activity of CSN and, consequently, its function (Lozano-Durán et al., 2011). Compromised CSN function also suppresses JA-mediated defense responses and several other host processes in the infected plants. In line with these results, treating plants with JAs reduced the virulence of TYLCSV, suggesting that TYLCSV C2 protein targeting of CSN (and thereby CSNs polyubiquitination activity) has biologically relevant consequences (Lozano-Durán et al., 2011).
Finally, among other strategies viruses employ to modulate host ubiquitin proteasome systems, some viruses encode proteins that resemble the UPS components. For example, P0, an RNA silencing suppressor encoded by poleroviruses, is a biological mimic of an F-box protein (Pazhouhandeh et al., 2006). F-box proteins are also UPS components and associate with CULLIN proteins to mediate protein polyubiquitination. The P0 protein forms a complex with the Arabidopsis SKP1 homologs, ASK1 and ASK2 (Pazhouhandeh et al., 2006), and then assembles into a CULLIN-based ubiquitin ligase to specifically target ARGONAUTE1 (AGO1) for degradation (Baumberger et al., 2007). Intriguingly, P0-mediated AGO1 degradation is insensitive to proteasome inhibitors such as MG132, suggesting that the 26S proteasome may not be involved in the turnover of AGO1. Further studies to understand host UPS components and interactions with viral proteins, as well as the identification of UPS protein targets during a viral infection, will shed more light on this fascinating cellular process associated with antiviral immune responses. Given the diversity of outcomes and several viruses targeting multiple UPS components, a critical role for UPS-mediated resistance or susceptibility in virus–host interactions and antiviral immune responses can be inferred.
MONOCOT ANTIVIRAL RESPONSES: A KNOWLEDGE GAP
As discussed above, studies of virus–host interactions have unequivocally advanced our understanding of plant immune responses and revealed strategies utilized by viruses to target host defense components. However, there is a gap in our knowledge of monocotyledonous plant antiviral responses. For instance, of the 31 cloned plant antiviral resistance genes, only one is from a monocot (Gururani et al., 2012). This is a rice recessive resistance gene, eIF(iso)4G1, that confers resistance against RYMV (Albar et al., 2006) by interacting with the viral genome-linked protein (VPg) (Hébrard et al., 2010). In rice, a mutation in eIF(iso)4G1 disrupts its interaction with VPg, thus imparting resistance against RYMV.
Several economically important monocot-infecting viruses such as Barley stripe mosaic virus (BSMV), Wheat streak mosaic virus, and Brome mosaic virus, which also infect N. benthamiana, have been studied using dicot host plants to dissect viral processes and pathogenicity determinants causing disease (Bragg and Jackson, 2004; French and Stenger, 2004; Mise and Pocsai, 2004). However, little experimental data are available pertaining to the altered host immune responses in their respective grass host species. Moreover, although there have been studies in identification of several resistance gene loci for monocot crops, including rice, barley, maize (Zea mays), and wheat (Triticum aestivum) (Trottet and Gouis, 2004; Redinbaugh and Pratt, 2009), most of these studies were primarily aimed toward breeding for resistance against diverse monocot-infecting viruses. The underlying mechanisms of immunity imparted by the resistance loci remain largely unknown. We also know little of the host molecular processes altered in compatible monocot plant virus infections and whether the antiviral responses are conserved among dicot and monocot host–virus interactions.
The lack of a tractable monocot genetic model system, akin to Arabidopsis or N. benthamiana, has been a major hindrance to research aimed at understanding the antiviral immune responses in monocots. However, emerging monocot model plants such as Brachypodium distachyon (Brachypodium) are bridging this gap. Brachypodium is a temperate grass with all the essential qualities of a genetic model system (Vogel and Bragg, 2009; Brkljacic et al., 2011). Its fully sequenced genome is highly syntenic to the genomes of important monocot crops such as wheat, rice, barley, millet, and maize, thus allowing functional and comparative genomic studies with potential for direct translation to field crops (International Brachypodium Initiative, 2010). Many genomic and functional resources have been developed around Brachypodium, including microarrays, yeast two-hybrid libraries, ∼200 inbred accessions, EMS/TILLING lines, and annotated T-DNA knockout lines (Filiz et al., 2009; Fox et al., 2010; International Brachypodium Initiative, 2010; Brkljacic et al., 2011; Cao et al., 2011; Bragg et al., 2012). Brachypodium has become an accepted model plant for research related to plant–pathogen interactions, as evidenced by the growing list of publications (Routledge et al., 2004; Parker et al., 2008; Peraldi et al., 2011; Cui et al., 2012; Lee et al., 2012; Mandadi and Scholthof, 2012). The reported bacterial, fungal, and viral pathogens that infect Brachypodium species (Table 1) will likely prompt further studies in understanding the molecular basis of disease in monocots.
TABLE 1. Pathogens infecting Brachypodium distachyon and B. sylvaticum. a.
| Viruses |
| Barley stripe mosaic virus (BSMV) |
| Brome mosaic virus (BMV) |
| Panicum mosaic virus (PMV) + SPMV b |
| Maize mild mottle virus (MMMV) |
| Sorghum yellow banding virus (SYBV) |
| Foxtail mosaic virus (FoMV) |
| Fungi: Basidiomycetes |
| Ustilago bromivora - loose smut |
| Tilletia olidea - smut |
| Puccinia brachypodii - leaf rust |
| Puccinia striiformis - stripe rust |
| Puccinia coronata - crown rust |
| Fungi: Ascomycetes |
| Claviceps purpurea - ergot |
| Blumeria graminis - powdery mildew |
| Epichloë sylvatica - endophyte |
| Neotyphodium sp. - endophyte |
| Stagonospora nodorum - glume blotch |
| Setosphaeria turcica - northern leaf blight |
| Pyrenophora teres - net blotch |
| Pyrenophora erythrospila - leaf spot |
| Cochliobolus sativus - root rot/leaf spot |
| Alternaria sp. - leaf spot |
| Ascochyta sp. - leaf spot |
| Sclerotinia homoeocarpa - dollar spot |
| Cladosporium sp. - leaf mold |
| Epicoccum sp. - glume spot |
| Fusarium graminearum - head blight |
| Fusarium culmorum - head blight |
| Magnaporthe grisea - rice blast |
| Fungal-like organisms |
| Pythium ultimum - root rot |
| Bacteria |
| Agrobacterium tumefaciens - crown gall |
Plant viruses infecting Brachypodium distachyon were compiled from Cui et al., 2012; Lee et al., 2012; Mandadi and Scholthof, 2012; and, K.K. Mandadi, J.D. Pyle, and K.-B.G. Scholthof, unpublished data. Fungi, fungal-like organisms, and bacteria (with their common names) that infect Brachpodium species were compiled from Parker et al., 2008; Brkljacic et al., 2011; and, Halbritter et al., 2012.
This reflects the infection of PMV alone or the mixed infection of PMV plus its satellite virus, SPMV (Mandadi and Scholthof, 2012).
THE PANICOVIRUS COMPLEX: NEW FINDINGS IN PLANT VIROLOGY USING BRACHYPODIUM
PMV is a single-stranded positive-sense RNA that is the type member of genus Panicovirus in the family Tombusviridae (Turina et al., 1998, 2000). PMV infects turfgrass, forage grasses, switchgrass, and food crops, such as millets. A peculiar feature of PMV is that it supports the replication of a satellite virus, SPMV, resulting in a viral synergism (Scholthof, 1999; Mandadi and Scholthof, 2012). As a complex, PMV+SPMV causes disease in the host plants characterized by exacerbated symptoms relative to those produced by PMV alone (Scholthof, 1999; Omarov et al., 2005). We are using this unique panicovirus complex as a foundational tool for fundamental advances in host–virus interactions, as well as potential translation to the field due to the distribution of PMV on agriculturally important crop hosts.
The utility of Brachypodium in studying monocot virus–host responses is evident from work done by our group and others. In our laboratory, we used transcriptomic approaches to identify Brachypodium molecular responses to compatible infections of PMV and SPMV. By comparison to the reported dicot virus host responses, we identified both conserved and unique molecular pathways altered in monocot host–virus responses (Mandadi and Scholthof, 2012). Among the conserved responses, the monocot antiviral defenses involve classical defense hormones such as SA, JA, and ethylene. Multiple genes in SA biosynthesis and signaling, such as ICS1, ABERRANT GROWTH AND DEATH2, ALTERNATIVE OXIDASE, pathogenesis-related proteins, and WRKY transcription factors, were upregulated in Brachypodium infected with PMV alone and PMV+SPMV (Mandadi and Scholthof, 2012). By contrast, several genes in JA and ET signaling, such as LIPOXYGENASE2, ALLENE OXIDE SYNTHASE, VEGETATIVE STORAGE PROTEIN1, FATTY ACID DESATURASE7, ETHYLENE RESPONSE FACTOR1 (ERF1), ERF3-like, ERF4, and ERF/Related to AP2.2, were downregulated (Mandadi and Scholthof, 2012). These results also support an existing notion that SA signaling and JA/ET signaling are antagonistic to each other in response to pathogen infection (An and Mou, 2011; Van der Does et al., 2013). In addition, several genes in biological processes, such as carbon metabolism, metabolite transport, cell wall remodeling, protein synthesis and degradation, and photosynthesis, were altered in a manner similar to those described for dicot virus interactions (Whitham et al., 2003, 2006; Ascencio-Ibáñez et al., 2008; García-Marcos et al., 2009; Hanssen et al., 2011; Postnikova and Nemchinov, 2012). These include genes encoding glutathione S-transferases, cytochrome P450s, glucosyl hydrolases, heat shock proteins, ribosomal components, UPS proteins, and chlorophyll a/b binding and photosystem-related proteins (Mandadi and Scholthof, 2012). However, unlike other known dicot antiviral responses, we found ∼30 protein kinases, particularly those in the receptor-like kinase subfamily, which are uniquely altered in both PMV and PMV+SPMV triggered responses in Brachypodium (Mandadi and Scholthof, 2012). This overrepresentation of protein kinases appears to be a unique feature of monocots, as our transcriptome comparisons with the few other reported monocot virus infections, such as maize infected with Rice black-streaked dwarf virus (genus Phytoreovirus) (Jia et al., 2012) and rice infected with Rice stripe virus (genus Tenuivirus) (Satoh et al., 2010), also revealed a higher proportion of upregulated kinases in the respective monocot virus infections. Together, these results suggest that although monocot and dicot plants share some commonalities, they have discrete and unique responses to plant virus infections.
Recently, through a screen for BSMV resistance among diverse Brachypodium accessions using BSMV strain North Dakota (ND18) followed by genetic fine-mapping of a resistance accession Bd3-1, the first dominant R gene (BSR1) for monocot virus resistance was identified (Cui et al., 2012). Similar to known dominant R proteins in dicots, the putative Brachypodium BSR1 gene encodes a CC-NB-LRR protein (Cui et al., 2012; Lee et al., 2012). Interestingly, genome reassortment experiments using the avirulent BSMV ND18 strain and a virulent BSMV Norwich (NW) strain in the Bd3-1 accession revealed that the BSMV NW triple gene block 1 (TGB1NW) movement protein is the virulence trigger. Site-directed mutagenesis assays further revealed that the TGB1NW amino acid residues at position 390 to 392 are critical for elicitation of HR and necrosis-like symptoms, presumably through interactions of this region with BSR1 (Lee et al., 2012). More research to understand these and other monocot antiviral defenses will continue to provide crucial insights into the currently poorly understood monocot antiviral immune responses.
FUTURE PROSPECTS AND CONSIDERATIONS
As mentioned above, one obvious area of future research focus is studying monocot antiviral immune responses. Comparative studies of antiviral responses in dicot and monocots will likely open new areas of investigation owing to the divergent evolutionary relationships of dicots and monocots. Moreover, because monocot and dicot plants have significantly different morphological and anatomical characteristics, including shoot-root architecture and cell wall composition and vasculature, as well as the genome-level differences, research aimed to understand how these differences affect antiviral immunities has intrinsic merit. To this end, Brachypodium and other emerging grass models such as Setaria viridis (Brutnell et al., 2010) are gaining reputations as outstanding experimental plants to study monocot-infecting viruses such as BSMV (Cui et al., 2012; Lee et al., 2012) and PMV (Mandadi and Scholthof, 2012). Brachypodium is also an excellent model for studying nonviral pathogens (Table 1). Brachypodium will continue to play a critical role in addressing questions about monocot pathobiology, as a “working grass hero” (Garvin, 2007).
Much more research is needed toward elaboration of the molecular pathways that lead to SAR in virus–host interactions. For example, the identity of the exact SAR signal in host–virus interactions is elusive. The precise roles of MeSA, azeleic acid, and glycerolipids in antiviral SAR need further investigation. Whether virus-triggered SAR also involves combinatorial interactions among the SAR signaling molecules and environmental factors remains to be tested. Finally, contribution of epigenetic factors in triggering and perpetuating SAR signals and whether virus-triggered SAR can be stably inherited to the next generation need to be determined, particularly in cereals and other grasses.
Although the antiviral HR mechanisms and the downstream defense hormone signaling are known for some viruses, in general, antiviral signal transduction is a black box, especially for monocots. Moreover, relative to the well understood early events in the plant immune responses triggered in fungal and bacterial infections, much work remains before we understand the early signaling events during the perception of virus or virus-encoded factors by the host receptor proteins, particularly at the cell membrane. For example, we do not know if any plasma membrane or cytosolic receptor proteins can directly perceive viral features analogous to PRRs to trigger viral resistance responses (Figure 1) nor do we know if any endogenous danger/damage-associated molecular patterns, which function in eliciting nonviral immune responses (Brutus et al., 2010), have any role in eliciting antiviral immune responses. These questions necessarily transition into an area of semantics that we suggest would likely benefit from some convergence, particularly when describing antiviral immune responses that are analogous to nonviral immune responses. For example, the R gene–mediated virus resistance involves specific recognition of virus-encoded Avr proteins that culminate in HR and SAR responses, as shown for TMV N gene resistance and others (Whitham et al., 1994; Liu et al., 2002a; Schoelz, 2006). However, it is unclear whether they can be classified as ETI responses because of the existing definition (or lack thereof) of what constitutes an effector protein for diverse pathogen types (i.e., viruses versus bacteria). To resolve these irregularities, we propose working definitions of viral effectors, ETI, and PTI responses in viral infections (Table 2).
Table 2. Working Definitions for Plant–Virus Interactions and Immune Responses.
| Term | Working Definition |
|---|---|
| Viral effectors | Virus-encoded proteins that when present in host cells interfere with host defense signaling components to promote virulence |
| Viral ETI | A type of host immune response triggered by R proteins that recognize, directly or indirectly, virus-encoded effectors or their activities within the host cells |
| Viral PTI | A type of basal host immune response triggered upon recognition of conserved viral molecular features by specific membrane-bound receptor-like proteins |
Typical bacterial and fungal effector proteins are encoded by these microbes and delivered into the plant cells, wherein they interfere with PTI or other immune regulators (Jones and Dangl, 2006; Bent and Mackey, 2007; Dodds and Rathjen, 2010; Spoel and Dong, 2012). Plant viruses do not encode effector proteins per se if the definition is limited to proteins that are delivered inside the host cells via microbial secretion systems. Yet, viruses encode proteins that are translated in the host cells and promote virulence by interfering with host defense pathways using a variety of strategies as discussed earlier. For example, the TBSV-encoded P19 protein promotes TBSV accumulation and virulence by suppressing host RNA silencing defenses (Scholthof, 2006). The BSCTV-encoded C2 protein is a silencing suppressor protein that also usurps host UPS processes to promote susceptibility (Zhang et al., 2011). The CaMV-encoded P6 protein suppresses SA-mediated defenses, as well as RNA silencing defenses (Love et al., 2007, 2012). Oftentimes, these viral proteins are recognized by R genes to trigger immune responses. For example, the CaMV P6 protein is an avirulence determinant recognized by R genes in several plants, including Datura stramonium, Nicotiana bigelovii, N. glutinosa, Nicotiana edwardsonii, and Arabidopsis ecotype Tsu-0. Similarly, the TBSV P19 silencing suppressor is also an elicitor of HR in certain Nicotiana species (Scholthof et al., 1995; Hsieh et al., 2009), including Nicotiana sylvestris, N. tabacum, and Nicotiana bonariensis (Angel and Schoelz, 2013). Furthermore, in the case of tobacco N-mediated resistance, the TMV-encoded p50 protein is recognized by the N protein and its cofactor NRIP1 to elicit the N-mediated host immune response (Caplan et al., 2008). These and several other instances reviewed in detail elsewhere (Schoelz, 2006) beg the following question: Should these virulence-promoting factors be referred to as effectors and the immune responses they trigger be classified as ETI responses? Based on the analogous functions of nonviral effector proteins, we propose a working definition for viral effectors: Viral effectors are virus-encoded proteins that when present in host cells interfere with host defense signaling components to promote virulence.
Earlier reviews discussing microbial effectors and their functional definitions noted that several bacterial and fungal avirulence factors are indeed “double agents” that can promote pathogenesis while eliciting ETI responses, suggesting that some avirulence proteins are also effectors (Alfano and Collmer, 2004; Schoelz, 2006). For example, the AvrPtoB protein of PstDC3000 can simultaneously suppress HR, as well as elicit ETI responses in the presence of the appropriate R gene, Pto (Abramovitch et al., 2003). As discussed above, several virus-encoded Avr factors, such as CaMV P6 protein, can interfere with plant defense pathways while eliciting R-mediated resistance responses (Schoelz, 2006). According to our proposed definition for viral effectors, virus Avr factors that interfere with host defenses should thus be referred as effectors. Logically, the immune responses they trigger should be classified as ETI responses. From this, we propose a working definition: ETI in viral infection is a form of host immune response triggered by R proteins that recognize, either directly or indirectly, virus-encoded effectors or their activities within the host cells.
Another example pertains to P/MAMPs and PTI responses. By definition, viruses do not possess conserved P/MAMP-like features such as flagellin or chitin. However, structures such as, but not limited to, the virion (or capsid), viral ribonucleoprotein complexes, and viral-encoded glycoproteins embedded on the host-derived lipid membranes of plant rhabdoviruses (Goldberg et al., 1991; Jackson et al., 2005) are analogous to P/MAMPs. Importantly, these structures are conserved among members of related virus taxa. Thus, we hypothesize that such viral patterns are analogous to P/MAMPs and are accessible for recognition by membrane-bound receptor-like proteins (PRRs or WAKs) to trigger PTI-like responses. As a working definition for viral PTI, we propose the following: PTI in viral infection is a form of basal host immune response triggered upon recognition of conserved viral molecular features by specific membrane-bound receptor-like proteins.
The above descriptions of viral effectors and virus-triggered immune responses are functionally synonymous with plant immune responses triggered in other microbial infections. Consilience of an integrated view of plant–pathogen interactions will be predicated on including viruses that are often overlooked in leading plant innate immunity models (Jones and Dangl, 2006; Bent and Mackey, 2007; Boller and Felix, 2009; Hogenhout et al., 2009; Dodds and Rathjen, 2010; Schwessinger and Ronald, 2012; Spoel and Dong, 2012).
CONCLUSION: MORE QUESTIONS THAN ANSWERS
In 1958, Sam Wildman, in a summary of the state-of-the-art of plant virology noted “our knowledge of what the virus is doing inside the host is not nearly as ‘sophisticated’ as our knowledge of what a plant virus is” (Wildman, 1959). By the early 1980s, technical difficulties associated with studying virus–host molecular interactions coupled with the virologists greater fascination with structural biology, mutagenesis, transgenic crops, and being able to do reverse genetics on plant viruses resulted in virologists taking a very different path from plant biologists and plant pathologists working with bacterial or fungal pathosystems. In recent years, despite having all the tools in place, the focus of many virologists was viral physicochemistry and/or identification of virulence determinants. This has led, in our opinion, to a gap in understanding the mechanisms of infection and host immune responses at the cellular level—especially when compared with the successes of those working on phytopathogenic fungi, bacteria, nematodes, and vector-borne host interactions (Jones and Dangl, 2006; Dodds and Rathjen, 2010; Schwessinger and Ronald, 2012; Spoel and Dong, 2012). As also mentioned above, several questions pertaining to plant viruses and their interactions within the host cells still remain unanswered: What are the major viral elicitors that induce HR and SAR? Which host proteins (e.g., PRRs) mediate viral recognition and by what mechanism? Unlike nonviral pathogens that can be recognized in the extracellular or periplastic spaces, how does an obligate viral pathogen elicit an intracellular host immune response? How is this response reiterated or maintained as the virus moves from cell to cell through the plasmodesmata and subsequently to the noninoculated tissues? Is this response cell autonomous? Wildman in 1958 also asked a very similar question: Is the “behavior” of the virus in the inoculated cell the same as that in a noninoculated systemically infected cell? Unfortunately, the question remains unanswered. Furthermore, despite the direct relevance with production agriculture and our nearly complete reliance on grasses (Poaceae) for food, feed, forage, recreation, and biofuel needs, critical aspects of grass–virus interactions largely remain understudied. In recent years, some work has been performed with laboratory dicotylendous plants as alternate hosts, including Arabidopsis and N. benthamiana; however, we are just beginning to disentangle the fundamental molecular interactions between plant viruses and grasses that result in disease or resistance. As such, by outlining here the status quo and gaps in our knowledge of virus–host interactions, we hope to provide the direction and impetus for a new generation of plant biologists and plant pathologists to explore the mystery of host–virus interactions within the broader context of host-pathogen interactions.
Acknowledgments
This work was supported in part by a Texas Higher Education Coordinating Board’s Norman Hackerman Advanced Research Program Grant (00517-002-2009) and the Texas A&M University Program to Enhance Scholarly and Creative Activities. We thank Veria Alvarado, Christopher Lyons, Jesse Pyle, and Herman Scholthof for helpful comments.
AUTHOR CONTRIBUTIONS
All authors contributed to writing this article.
References
- Aarts N., Metz M., Holub E., Staskawicz B.J., Daniels M.J., Parker J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitch R.B., Kim Y.-J., Chen S., Dickman M.B., Martin G.B. (2003). Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22: 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albar L., Bangratz-Reyser M., Hébrard E., Ndjiondjop M.-N., Jones M., Ghesquière A. (2006). Mutations in the eIF(iso)4G translation initiation factor confer high resistance of rice to Rice yellow mottle virus. Plant J. 47: 417–426 [DOI] [PubMed] [Google Scholar]
- Alfano J.R., Collmer A. (2004). Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42: 385–414 [DOI] [PubMed] [Google Scholar]
- An C., Mou Z. (2011). Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 53: 412–428 [DOI] [PubMed] [Google Scholar]
- Angel C.A., Schoelz J.E. (2013). A survey of resistance to Tomato bushy stunt virus in the genus Nicotiana reveals that the hypersensitive response is triggered by one of three different viral proteins. Mol. Plant Microbe Interact. 26: 240–248 [DOI] [PubMed] [Google Scholar]
- Ascencio-Ibáñez J.T., Sozzani R., Lee T.-J., Chu T.-M., Wolfinger R.D., Cella R., Hanley-Bowdoin L. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148: 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin M.J., Muskett P., Kahn K., Feys B.J., Jones J.D.G., Parker J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Staskawicz B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Azevedo C., Sadanandom A., Kitagawa K., Freialdenhoven A., Shirasu K., Schulze-Lefert P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076 [DOI] [PubMed] [Google Scholar]
- Baumberger N., Tsai C.-H., Lie M., Havecker E., Baulcombe D.C. (2007). The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17: 1609–1614 [DOI] [PubMed] [Google Scholar]
- Bendahmane A., Kanyuka K., Baulcombe D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A.F., Mackey D. (2007). Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Bieri S., Mauch S., Shen Q.-H., Peart J., Devoto A., Casais C., Ceron F., Schulze S., Steinbiss H.H., Shirasu K., Schulze-Lefert P. (2004). RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16: 3480–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Boyko A., Kathiria P., Zemp F.J., Yao Y., Pogribny I., Kovalchuk I. (2007). Transgenerational changes in the genome stability and methylation in pathogen-infected plants: (virus-induced plant genome instability). Nucleic Acids Res. 35: 1714–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg, J.N., and Jackson, A.O. (2004). Barley stripe mosaic. In Viruses and Virus Diseases of Poaceae (Gramineae). H. Lapierre and P. Signoret, eds (Paris: INRA), pp. 456–457. [Google Scholar]
- Bragg J.N., Wu J., Gordon S.P., Guttman M.E., Thilmony R., Lazo G.R., Gu Y.Q., Vogel J.P. (2012). Generation and characterization of the Western Regional Research Center Brachypodium T-DNA insertional mutant collection. PLoS ONE 7: e41916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brkljacic J., et al. (2011). Brachypodium as a model for the grasses: Today and the future. Plant Physiol. 157: 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell T.P., Wang L., Swartwood K., Goldschmidt A., Jackson D., Zhu X.-G., Kellogg E., Van Eck J. (2010). Setaria viridis: A model for C4 photosynthesis. Plant Cell 22: 2537–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A., Sicilia F., Macone A., Cervone F., De Lorenzo G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Siriwardana C.L., Kumimoto R.W., Holt B.F., III (2011). Construction of high quality Gateway™ entry libraries and their application to yeast two-hybrid for the monocot model plant Brachypodium distachyon. BMC Biotechnol. 11: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J.L., Mamillapalli P., Burch-Smith T.M., Czymmek K., Dinesh-Kumar S.P. (2008). Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132: 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J.L., Zhu X., Mamillapalli P., Marathe R., Anandalakshmi R., Dinesh-Kumar S.P. (2009). Induced ER chaperones regulate a receptor-like kinase to mediate antiviral innate immune response in plants. Cell Host Microbe 6: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr J.P., Lewsey M.G., Palukaitis P. (2010). Signaling in induced resistance. Adv. Virus Res. 76: 57–121 [DOI] [PubMed] [Google Scholar]
- Century K.S., Shapiro A.D., Repetti P.P., Dahlbeck D., Holub E., Staskawicz B.J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965 [DOI] [PubMed] [Google Scholar]
- Chanda B., Xia Y., Mandal M.K., Yu K., Sekine K.T., Gao Q.-M., Selote D., Hu Y., Stromberg A., Navarre D., Kachroo A., Kachroo P. (2011). Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 43: 421–427 [DOI] [PubMed] [Google Scholar]
- Chandra-Shekara A.C., Navarre D., Kachroo A., Kang H.-G., Klessig D., Kachroo P. (2004). Signaling requirements and role of salicylic acid in HRT- and rrt-mediated resistance to Turnip crinkle virus in Arabidopsis. Plant J. 40: 647–659 [DOI] [PubMed] [Google Scholar]
- Chaturvedi R., Krothapalli K., Makandar R., Nandi A., Sparks A.A., Roth M.R., Welti R., Shah J. (2008). Plastid ω3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J. 54: 106–117 [DOI] [PubMed] [Google Scholar]
- Chisholm S.T., Mahajan S.K., Whitham S.A., Yamamoto M.L., Carrington J.C. (2000). Cloning of the Arabidopsis RTM1 gene, which controls restriction of long-distance movement of Tobacco etch virus. Proc. Natl. Acad. Sci. USA 97: 489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu M., Desvoyes B., Turina M., Noad R., Scholthof H.B. (2000). Genetic dissection of Tomato bushy stunt virus p19-protein-mediated host-dependent symptom induction and systemic invasion. Virology 266: 79–87 [DOI] [PubMed] [Google Scholar]
- Citovsky V., Zaltsman A., Kozlovsky S.V., Gafni Y., Krichevsky A. (2009). Proteasomal degradation in plant-pathogen interactions. Semin. Cell Dev. Biol. 20: 1048–1054 [DOI] [PubMed] [Google Scholar]
- Coaker G., Falick A., Staskawicz B. (2005). Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 308: 548–550 [DOI] [PubMed] [Google Scholar]
- Cole A.B., Király L., Ross K., Schoelz J.E. (2001). Uncoupling resistance from cell death in the hypersensitive response of Nicotiana species to Cauliflower mosaic virus infection. Mol. Plant Microbe Interact. 14: 31–41 [DOI] [PubMed] [Google Scholar]
- Collier S.M., Moffett P. (2009). NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14: 521–529 [DOI] [PubMed] [Google Scholar]
- Coppinger P., Repetti P.P., Day B., Dahlbeck D., Mehlert A., Staskawicz B.J. (2004). Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J. 40: 225–237 [DOI] [PubMed] [Google Scholar]
- Cosson P., Sofer L., Le Q.H., Léger V., Schurdi-Levraud V., Whitham S.A., Yamamoto M.L., Gopalan S., Le Gall O., Candresse T., Carrington J.C., Revers F. (2010). RTM3, which controls long-distance movement of potyviruses, is a member of a new plant gene family encoding a meprin and TRAF homology domain-containing protein. Plant Physiol. 154: 222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creager, A.N.H. (2002). The Life of a Virus: Tobacco mosaic virus as an Experimental Model, 1930–1965. (Chicago: University of Chicago Press). [Google Scholar]
- Cui Y., et al. (2012). Fine mapping of the Bsr1 Barley stripe mosaic virus resistance gene in the model grass Brachypodium distachyon. PLoS ONE 7: e38333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver J.N., Padmanabhan M.S. (2007). Virus-induced disease: Altering host physiology one interaction at a time. Annu. Rev. Phytopathol. 45: 221–243 [DOI] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Dawson W.O., Beck D.L., Knorr D.A., Grantham G.L. (1986). cDNA cloning of the complete genome of Tobacco mosaic virus and production of infectious transcripts. Proc. Natl. Acad. Sci. USA 83: 1832–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day B., Dahlbeck D., Staskawicz B.J. (2006). NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 18: 2782–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroocq V., Salvador B., Sicard O., Glasa M., Cosson P., Svanella-Dumas L., Revers F., García J.A., Candresse T. (2009). The determinant of potyvirus ability to overcome the RTM resistance of Arabidopsis thaliana maps to the N-terminal region of the coat protein. Mol. Plant Microbe Interact. 22: 1302–1311 [DOI] [PubMed] [Google Scholar]
- Dempsey D.A., Klessig D.F. (2012). SOS - Too many signals for systemic acquired resistance? Trends Plant Sci. 17: 538–545 [DOI] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Dong X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7: 547–552 [DOI] [PubMed] [Google Scholar]
- Drugeon G., Jupin I. (2002). Stability in vitro of the 69K movement protein of Turnip yellow mosaic virus is regulated by the ubiquitin-mediated proteasome pathway. J. Gen. Virol. 83: 3187–3197 [DOI] [PubMed] [Google Scholar]
- Falk A., Feys B.J., Frost L.N., Jones J.D.G., Daniels M.J., Parker J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B.J., Moisan L.J., Newman M.-A., Parker J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B.J., Wiermer M., Bhat R.A., Moisan L.J., Medina-Escobar N., Neu C., Cabral A., Parker J.E. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiz E., Ozdemir B.S., Budak F., Vogel J.P., Tuna M., Budak H. (2009). Molecular, morphological, and cytological analysis of diverse Brachypodium distachyon inbred lines. Genome 52: 876–890 [DOI] [PubMed] [Google Scholar]
- Fliegmann J., Mithöfer A., Wanner G., Ebel J. (2004). An ancient enzyme domain hidden in the putative β-glucan elicitor receptor of soybean may play an active part in the perception of pathogen-associated molecular patterns during broad host resistance. J. Biol. Chem. 279: 1132–1140 [DOI] [PubMed] [Google Scholar]
- Fox, S.E., Priest, H.D., Bryant, D.W., Wilhelm, L.J., Michael, T.P., and Mockler, T.C. (2010). Universal genome array and transcriptome atlas for Brachypodium distachyon (abstract no. W095). In Plant & Animal Genomes XVIII Conference, January 9–13, (San Diego, CA). [Google Scholar]
- French, R., and Stenger, D.C. (2004). Wheat streak mosaic. In Viruses and Virus Diseases of Poaceae (Gramineae). H. Lapierre and P. Signoret, eds (Paris INRA), pp. 602–604. [Google Scholar]
- Gao G., Luo H. (2006). The ubiquitin-proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 84: 5–14 [DOI] [PubMed] [Google Scholar]
- Gao Z., Johansen E., Eyers S., Thomas C.L., Noel Ellis T.H., Maule A.J. (2004). The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 40: 376–385 [DOI] [PubMed] [Google Scholar]
- García-Marcos A., Pacheco R., Martiáñez J., González-Jara P., Díaz-Ruíz J.R., Tenllado F. (2009). Transcriptional changes and oxidative stress associated with the synergistic interaction between Potato virus X and Potato virus Y and their relationship with symptom expression. Mol. Plant Microbe Interact. 22: 1431–1444 [DOI] [PubMed] [Google Scholar]
- Garvin D.F. (2007). Brachypodium: A new monocot model plant system emerges. J. Sci. Food Agric. 87: 1177–1179 [Google Scholar]
- Goldberg K.-B., Modrell B., Hillman B.I., Heaton L.A., Choi T.-J., Jackson A.O. (1991). Structure of the glycoprotein gene of Sonchus yellow net virus, a plant rhabdovirus. Virology 185: 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Jara P., Tenllado F., Martínez-García B., Atencio F.A., Barajas D., Vargas M., Díaz-Ruiz J., Díaz-Ruíz J.R. (2004). Host-dependent differences during synergistic infection by Potyviruses with Potato virus X. Mol. Plant Pathol. 5: 29–35 [DOI] [PubMed] [Google Scholar]
- Gururani M.A., Venkatesh J., Upadhyaya C.P., Nookaraju A., Pandey S.K., Park S.W. (2012). Plant disease resistance genes: Current status and future directions. Physiol. Mol. Plant Pathol. 78: 51–65 [Google Scholar]
- Hagiwara Y., Komoda K., Yamanaka T., Tamai A., Meshi T., Funada R., Tsuchiya T., Naito S., Ishikawa M. (2003). Subcellular localization of host and viral proteins associated with tobamovirus RNA replication. EMBO J. 22: 344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbritter A.H., Carroll G.C., Güsewell S., Roy B.A. (2012). Testing assumptions of the enemy release hypothesis: Generalist versus specialist enemies of the grass Brachypodium sylvaticum. Mycologia 104: 34–44 [DOI] [PubMed] [Google Scholar]
- Hanssen I.M., van Esse H.P., Ballester A.-R., Hogewoning S.W., Parra N.O., Paeleman A., Lievens B., Bovy A.G., Thomma B.P.H.J. (2011). Differential tomato transcriptomic responses induced by Pepino mosaic virus isolates with differential aggressiveness. Plant Physiol. 156: 301–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébrard E., Poulicard N., Gérard C., Traoré O., Wu H.-C., Albar L., Fargette D., Bessin Y., Vignols F. (2010). Direct interaction between the Rice yellow mottle virus (RYMV) VPg and the central domain of the rice eIF(iso)4G1 factor correlates with rice susceptibility and RYMV virulence. Mol. Plant Microbe Interact. 23: 1506–1513 [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hogenhout S.A., Van der Hoorn R.A.L., Terauchi R., Kamoun S. (2009). Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 22: 115–122 [DOI] [PubMed] [Google Scholar]
- Holmes F.O. (1929). Local lesions in tobacco mosaic. Bot. Gaz. 87: 39–55 [Google Scholar]
- Holmes F.O. (1932). Symptoms of tobacco mosaic disease. Contrib. Boyce Thompson Inst. 4: 323–357 [Google Scholar]
- Holmes F.O. (1936). Interspecific transfer of a gene governing type of response to tobacco-mosaic infection. Phytopathology 26: 1007–1014 [Google Scholar]
- Holmes F.O. (1937). Inheritance of resistance to tobacco-mosaic disease in the pepper. Phytopathology 27: 637–642 [Google Scholar]
- Holmes F.O. (1938). Inheritance of resistance to tobacco-mosaic disease in tobacco. Phytopathology 28: 553–561 [Google Scholar]
- Holmes F.O. (1954). Inheritance of resistance to viral diseases in plants. Adv. Virus Res. 2: 1–30 [DOI] [PubMed] [Google Scholar]
- Hsieh Y.-C., Omarov R.T., Scholthof H.B. (2009). Diverse and newly recognized effects associated with short interfering RNA binding site modifications on the Tomato bushy stunt virus p19 silencing suppressor. J. Virol. 83: 2188–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Vierstra R.D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62: 299–334 [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Ishibashi K., Naito S., Meshi T., Ishikawa M. (2009). An inhibitory interaction between viral and cellular proteins underlies the resistance of tomato to nonadapted tobamoviruses. Proc. Natl. Acad. Sci. USA 106: 8778–8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi K., Masuda K., Naito S., Meshi T., Ishikawa M. (2007). An inhibitor of viral RNA replication is encoded by a plant resistance gene. Proc. Natl. Acad. Sci. USA 104: 13833–13838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M., Naito S., Ohno T. (1993). Effects of the tom1 mutation of Arabidopsis thaliana on the multiplication of Tobacco mosaic virus RNA in protoplasts. J. Virol. 67: 5328–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi A.S., McCouch S.R. (2007). Recessive resistance genes and the Oryza sativa-Xanthomonas oryzae pv. oryzae pathosystem. Mol. Plant Microbe Interact. 20: 731–739 [DOI] [PubMed] [Google Scholar]
- Jackson A.O., Dietzgen R.G., Goodin M.M., Bragg J.N., Deng M. (2005). Biology of plant rhabdoviruses. Annu. Rev. Phytopathol. 43: 623–660 [DOI] [PubMed] [Google Scholar]
- Jia M.-A., Li Y., Lei L., Di D., Miao H., Fan Z. (2012). Alteration of gene expression profile in maize infected with a double-stranded RNA fijivirus associated with symptom development. Mol. Plant Pathol. 13: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Li S., Villegas A., Jr (2006). Down-regulation of the 26S proteasome subunit RPN9 inhibits viral systemic transport and alters plant vascular development. Plant Physiol. 142: 651–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jung H.W., Tschaplinski T.J., Wang L., Glazebrook J., Greenberg J.T. (2009). Priming in systemic plant immunity. Science 324: 89–91 [DOI] [PubMed] [Google Scholar]
- Kanyuka K., Druka A., Caldwell D.G., Tymon A., McCallum N., Waugh R., Adams M.J. (2005). Evidence that the recessive bymovirus resistance locus rym4 in barley corresponds to the eukaryotic translation initiation factor 4E gene. Mol. Plant Pathol. 6: 449–458 [DOI] [PubMed] [Google Scholar]
- Kathiria P., Sidler C., Golubov A., Kalischuk M., Kawchuk L.M., Kovalchuk I. (2010). Tobacco mosaic virus infection results in an increase in recombination frequency and resistance to viral, bacterial, and fungal pathogens in the progeny of infected tobacco plants. Plant Physiol. 153: 1859–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Masuta C., Matsuura H., Takahashi H., Inukai T. (2008). Veinal necrosis induced by Turnip mosaic virus infection in Arabidopsis is a form of defense response accompanying HR-like cell death. Mol. Plant Microbe Interact. 21: 260–268 [DOI] [PubMed] [Google Scholar]
- Knepper C., Savory E.A., Day B. (2011). Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol. 156: 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K., Hashimoto M., Ozeki J., Yamaji Y., Maejima K., Senshu H., Himeno M., Okano Y., Kagiwada S., Namba S. (2010). Viral-induced systemic necrosis in plants involves both programmed cell death and the inhibition of viral multiplication, which are regulated by independent pathways. Mol. Plant Microbe Interact. 23: 283–293 [DOI] [PubMed] [Google Scholar]
- Kovalchuk I., Kovalchuk O., Kalck V., Boyko V., Filkowski J., Heinlein M., Hohn B. (2003). Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 423: 760–762 [DOI] [PubMed] [Google Scholar]
- Lee M.Y., et al. (2012). Brachypodium distachyon line Bd3-1 resistance is elicited by the Barley stripe mosaic virus triple gene block 1 movement protein. J. Gen. Virol. 93: 2729–2739 [DOI] [PubMed] [Google Scholar]
- Lellis A.D., Kasschau K.D., Whitham S.A., Carrington J.C. (2002). Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12: 1046–1051 [DOI] [PubMed] [Google Scholar]
- Li Z., Barajas D., Panavas T., Herbst D.A., Nagy P.D. (2008). Cdc34p ubiquitin-conjugating enzyme is a component of the tombusvirus replicase complex and ubiquitinates p33 replication protein. J. Virol. 82: 6911–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Burch-Smith T., Schiff M., Feng S., Dinesh-Kumar S.P. (2004). Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 279: 2101–2108 [DOI] [PubMed] [Google Scholar]
- Liu P.-P., von Dahl C.C., Park S.-W., Klessig D.F. (2011). Interconnection between methyl salicylate and lipid-based long-distance signaling during the development of systemic acquired resistance in Arabidopsis and tobacco. Plant Physiol. 155: 1762–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Marathe R., Dinesh-Kumar S.P. (2002a). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to Tobacco mosaic virus. Plant J. 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Serino G., Deng X.-W., Dinesh-Kumar S.P. (2002b). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14: 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Vailleau F., Balagué C., Roby D. (2003). Lesion mimic mutants: Keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8: 263–271 [DOI] [PubMed] [Google Scholar]
- Love A.J., Laird J., Holt J., Hamilton A.J., Sadanandom A., Milner J.J. (2007). Cauliflower mosaic virus protein P6 is a suppressor of RNA silencing. J. Gen. Virol. 88: 3439–3444 [DOI] [PubMed] [Google Scholar]
- Love A.J., Geri C., Laird J., Carr C., Yun B.-W., Loake G.J., Tada Y., Sadanandom A., Milner J.J. (2012). Cauliflower mosaic virus protein P6 inhibits signaling responses to salicylic acid and regulates innate immunity. PLoS ONE 7: e47535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R., Rosas-Díaz T., Gusmaroli G., Luna A.P., Taconnat L., Deng X.W., Bejarano E.R. (2011). Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 23: 1014–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E., Bruce T.J.A., Roberts M.R., Flors V., Ton J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158: 844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D., Belkhadir Y., Alonso J.M., Ecker J.R., Dangl J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Mackey D., Holt B.F., III, Wiig A., Dangl J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Mandadi K.K., Scholthof K.-B.G. (2012). Characterization of a viral synergism in the monocot Brachypodium distachyon reveals distinctly altered host molecular processes associated with disease. Plant Physiol. 160: 1432–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mise, K., and Pocsai, E. (2004). Brome mosaic. In Viruses and Virus Diseases of Poaceae (Gramineae). H. Lapierre and P. Signoret, eds (Paris INRA), pp. 735–739. [Google Scholar]
- Mithöfer A., Fliegmann J., Neuhaus-Url G., Schwarz H., Ebel J. (2000). The hepta-β-glucoside elicitor-binding proteins from legumes represent a putative receptor family. Biol. Chem. 381: 705–713 [DOI] [PubMed] [Google Scholar]
- Moeder W., Yoshioka K. (2008). Lesion mimic mutants: A classical, yet still fundamental approach to study programmed cell death. Plant Signal. Behav. 3: 764–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P. (2009). Mechanisms of recognition in dominant R gene mediated resistance. Adv. Virus Res. 75: 1–33 [DOI] [PubMed] [Google Scholar]
- Mur L.A.J., Kenton P., Lloyd A.J., Ougham H., Prats E. (2008). The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59: 501–520 [DOI] [PubMed] [Google Scholar]
- Nezames C.D., Deng X.-W. (2012). The COP9 signalosome: Its regulation of cullin-based E3 ubiquitin ligases and role in photomorphogenesis. Plant Physiol. 160: 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V., German-Retana S., Sanjuán R., Dubrana M.-P., Mazier M., Maisonneuve B., Candresse T., Caranta C., LeGall O. (2003). The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol. 132: 1272–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima K., Taniyama T., Yamanaka T., Ishikawa M., Naito S. (1998). Isolation of a mutant of Arabidopsis thaliana carrying two simultaneous mutations affecting Tobacco mosaic virus multiplication within a single cell. Virology 243: 472–481 [DOI] [PubMed] [Google Scholar]
- Omarov R.T., Qi D., Scholthof K.-B.G. (2005). The capsid protein of satellite panicum mosaic virus contributes to systemic invasion and interacts with its helper virus. J. Virol. 79: 9756–9764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki J., Takahashi S., Komatsu K., Kagiwada S., Yamashita K., Mori T., Hirata H., Yamaji Y., Ugaki M., Namba S. (2006). A single amino acid in the RNA-dependent RNA polymerase of Plantago asiatica mosaic virus contributes to systemic necrosis. Arch. Virol. 151: 2067–2075 [DOI] [PubMed] [Google Scholar]
- Pacheco R., García-Marcos A., Manzano A., de Lacoba M.G., Camañes G., García-Agustín P., Díaz-Ruíz J.R., Tenllado F. (2012). Comparative analysis of transcriptomic and hormonal responses to compatible and incompatible plant-virus interactions that lead to cell death. Mol. Plant Microbe Interact. 25: 709–723 [DOI] [PubMed] [Google Scholar]
- Padmanabhan M.S., Dinesh-Kumar S.P. (2010). All hands on deck—The role of chloroplasts, endoplasmic reticulum, and the nucleus in driving plant innate immunity. Mol. Plant Microbe Interact. 23: 1368–1380 [DOI] [PubMed] [Google Scholar]
- Pallas V., García J.A. (2011). How do plant viruses induce disease? Interactions and interference with host components. J. Gen. Virol. 92: 2691–2705 [DOI] [PubMed] [Google Scholar]
- Park S.-W., Kaimoyo E., Kumar D., Mosher S., Klessig D.F. (2007). Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318: 113–116 [DOI] [PubMed] [Google Scholar]
- Parker D., Beckmann M., Enot D.P., Overy D.P., Rios Z.C., Gilbert M., Talbot N., Draper J. (2008). Rice blast infection of Brachypodium distachyon as a model system to study dynamic host/pathogen interactions. Nat. Protoc. 3: 435–445 [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh M., Dieterle M., Marrocco K., Lechner E., Berry B., Brault V., Hemmer O., Kretsch T., Richards K.E., Genschik P., Ziegler-Graff V. (2006). F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA 103: 1994–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peraldi A., Beccari G., Steed A., Nicholson P. (2011). Brachypodium distachyon: A new pathosystem to study Fusarium head blight and other Fusarium diseases of wheat. BMC Plant Biol. 11: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piron F., Nicolaï M., Minoïa S., Piednoir E., Moretti A., Salgues A., Zamir D., Caranta C., Bendahmane A. (2010). An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS ONE 5: e11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postnikova O.A., Nemchinov L.G. (2012). Comparative analysis of microarray data in Arabidopsis transcriptome during compatible interactions with plant viruses. Virol. J. 9: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmann S., De Vos M., Casteel C.L., Tian D., Halitschke R., Sun J.Y., Agrawal A.A., Felton G.W., Jander G. (2012). Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 158: 854–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redinbaugh, M., and Pratt, R. (2009). Virus resistance. In Handbook of Maize: Its Biology, J. Bennetzen and S. Hake, eds (New York: Springer), pp. 251–270. [Google Scholar]
- Reichel C., Beachy R.N. (2000). Degradation of Tobacco mosaic virus movement protein by the 26S proteasome. J. Virol. 74: 3330–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C., Caranta C. (2006). Translation initiation factors: A weak link in plant RNA virus infection. Trends Plant Sci. 11: 40–45 [DOI] [PubMed] [Google Scholar]
- Ross A.F. (1961a). Localized acquired resistance to plant virus infection in hypersensitive hosts. Virology 14: 329–339 [DOI] [PubMed] [Google Scholar]
- Ross A.F. (1961b). Systemic acquired resistance induced by localized virus infections in plants. Virology 14: 340–358 [DOI] [PubMed] [Google Scholar]
- Routledge A.P., Shelley G., Smith J.V., Talbot N.J., Draper J., Mur L.A. (2004). Magnaporthe grisea interactions with the model grass Brachypodium distachyon closely resemble those with rice (Oryza sativa). Mol. Plant Pathol. 5: 253–265 [DOI] [PubMed] [Google Scholar]
- Ruffel S., Dussault M.-H., Palloix A., Moury B., Bendahmane A., Robaglia C., Caranta C. (2002). A natural recessive resistance gene against Potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32: 1067–1075 [DOI] [PubMed] [Google Scholar]
- Ruffel S., Gallois J.L., Lesage M.L., Caranta C. (2005). The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Genet. Genomics 274: 346–353 [DOI] [PubMed] [Google Scholar]
- Russell, G.E. (1978). Plant Breeding for Pest and Disease Resistance. (London: Butterworths). [Google Scholar]
- Sacco M.A., Mansoor S., Moffett P. (2007). A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J. 52: 82–93 [DOI] [PubMed] [Google Scholar]
- Samuel G. (1934). The movement of tobacco mosaic virus within the plant. Ann. Appl. Biol. 21: 90–111 [Google Scholar]
- Satoh K., Kondoh H., Sasaya T., Shimizu T., Choi I.-R., Omura T., Kikuchi S. (2010). Selective modification of rice (Oryza sativa) gene expression by Rice stripe virus infection. J. Gen. Virol. 91: 294–305 [DOI] [PubMed] [Google Scholar]
- Schoelz, J.E. (2006). Viral determinants of resistance versus susceptibility. In Natural Resistance Mechanisms of Plants to Viruses. G. Loebenstein and J. Carr, eds (Dordrecht, The Netherlands: Springer), pp. 13–43. [Google Scholar]
- Scholthof H.B. (2006). The Tombusvirus-encoded P19: From irrelevance to elegance. Nat. Rev. Microbiol. 4: 405–411 [DOI] [PubMed] [Google Scholar]
- Scholthof H.B., Scholthof K.B., Jackson A.O. (1995). Identification of Tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a Potato virus X vector. Plant Cell 7: 1157–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof K.-B.G. (1999). A synergism induced by satellite panicum mosaic virus. Mol. Plant Microbe Interact. 12: 163–166 [Google Scholar]
- Scholthof K.-B.G. (2004). Tobacco mosaic virus: A model system for plant biology. Annu. Rev. Phytopathol. 42: 13–34 [DOI] [PubMed] [Google Scholar]
- Scholthof K.-B.G. (2011). TMV in 1930: Francis O. Holmes and the local lesion assay. Microbe 6: 221–225 [Google Scholar]
- Scholthof K.-B.G. (March 15, 2013). Making a virus visible: Francis O. Holmes and a biological assay for Tobacco mosaic virus. J. Hist. Biol. http://dx.doi.org/10.1007/s10739-013-9353-0 [DOI] [PubMed] [Google Scholar]
- Scholthof, K.-B.G., Shaw, J.G., and Zaitlin, M. (1999). Tobacco Mosaic Virus: One Hundred Years of Contributions to Virology. (St. Paul, MN: APS Press). [Google Scholar]
- Schwessinger B., Ronald P.C. (2012). Plant innate immunity: Perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63: 451–482 [DOI] [PubMed] [Google Scholar]
- Shirasu K. (2009). The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60: 139–164 [DOI] [PubMed] [Google Scholar]
- Spoel S.H., Dong X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12: 89–100 [DOI] [PubMed] [Google Scholar]
- Takahashi A., Casais C., Ichimura K., Shirasu K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 11777–11782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling W.I.L., Nooijen C., Ludwig N., Boter M., Slootweg E., Goverse A., Shirasu K., Joosten M.H.A.J. (2010). RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell 22: 4176–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottet, M., and Gouis, J.L. (2004). Manipulation of resistance genes. In Viruses and Virus Diseases of Poaceae (Gramineae). H. Lapierre and P. Signoret, eds (Paris INRA), pp. 180–186. [Google Scholar]
- Truman W.M., Bennett M.H., Turnbull C.G.N., Grant M.R. (2010). Arabidopsis auxin mutants are compromised in systemic acquired resistance and exhibit aberrant accumulation of various indolic compounds. Plant Physiol. 152: 1562–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truniger V., Aranda M.A. (2009). Recessive resistance to plant viruses. Adv. Virus Res. 75: 119–159 [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y., Numaga T., Ohshima K., Yano M.-A., Ohsawa R., Goto D.B., Naito S., Ishikawa M. (2003). Arabidopsis TOBAMOVIRUS MULTIPLICATION (TOM) 2 locus encodes a transmembrane protein that interacts with TOM1. EMBO J. 22: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turina M., Desvoyes B., Scholthof K.-B.G. (2000). A gene cluster encoded by Panicum mosaic virus is associated with virus movement. Virology 266: 120–128 [DOI] [PubMed] [Google Scholar]
- Turina M., Maruoka M., Monis J., Jackson A.O., Scholthof K.-B.G. (1998). Nucleotide sequence and infectivity of a full-length cDNA clone of Panicum mosaic virus. Virology 241: 141–155 [DOI] [PubMed] [Google Scholar]
- Ueki S., Citovsky V. (2005). Identification of an interactor of cadmium ion-induced glycine-rich protein involved in regulation of callose levels in plant vasculature. Proc. Natl. Acad. Sci. USA 102: 12089–12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme E.J.M., Barre A., Rougé P., Peumans W.J. (2004). Cytoplasmic/nuclear plant lectins: A new story. Trends Plant Sci. 9: 484–489 [DOI] [PubMed] [Google Scholar]
- Van der Does D., Leon-Reyes A., Koornneef A., Van Verk M.C., Rodenburg N., Pauwels L., Goossens A., Körbes A.P., Memelink J., Ritsema T., Van Wees S.C.M., Pieterse C.M.J. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn R.A.L., Kamoun S. (2008). From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot A.C., Klessig D.F., Park S.-W. (2008a). Systemic acquired resistance: The elusive signal(s). Curr. Opin. Plant Biol. 11: 436–442 [DOI] [PubMed] [Google Scholar]
- Vlot A.C., Liu P.-P., Cameron R.K., Park S.-W., Yang Y., Kumar D., Zhou F., Padukkavidana T., Gustafsson C., Pichersky E., Klessig D.F. (2008b). Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J. 56: 445–456 [DOI] [PubMed] [Google Scholar]
- Vogel, J., and Bragg, J. (2009). Brachypodium distachyon, a new model for the Triticeae In Genetics and Genomics of the Triticeae, C. Feuillet and G.J. Muehlbauer, eds (Berlin: Springer), pp. 427–449. [Google Scholar]
- Weststeijn E.A. (1978). Permeability changes in the hypersensitive reaction of Nicotiana tabacum cv. Xanthi nc. after infection with Tobacco mosaic virus. Physiol. Plant Pathol. 13: 253–258 [Google Scholar]
- Whitham S., Dinesh-Kumar S.P., Choi D., Hehl R., Corr C., Baker B. (1994). The product of the Tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78: 1101–1115 [DOI] [PubMed] [Google Scholar]
- Whitham S.A., Anderberg R.J., Chisholm S.T., Carrington J.C. (2000). Arabidopsis RTM2 gene is necessary for specific restriction of Tobacco etch virus and encodes an unusual small heat shock-like protein. Plant Cell 12: 569–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham S.A., Quan S., Chang H.-S., Cooper B., Estes B., Zhu T., Wang X., Hou Y.-M. (2003). Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33: 271–283 [DOI] [PubMed] [Google Scholar]
- Whitham S.A., Yang C., Goodin M.M. (2006). Global impact: Elucidating plant responses to viral infection. Mol. Plant Microbe Interact. 19: 1207–1215 [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Wildman, S. (1959). Discussion of Chapters XLIII and XLIV. In Plant Pathology: Problems and Progress 1908–1958. C.S. Holton, G.W. Fischer, R.W. Fulton, H. Hart, and S.E.A. McCallan, eds (Madison, WI: University of Wisconsin Press), pp. 501–502. [Google Scholar]
- Willment J.A., Brown G.D. (2008). C-type lectin receptors in antifungal immunity. Trends Microbiol. 16: 27–32 [DOI] [PubMed] [Google Scholar]
- Xu P., Roossinck M.J. (2000). Cucumber mosaic virus D satellite RNA-induced programmed cell death in tomato. Plant Cell 12: 1079–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Wang H., Coker F., Ma J.Y., Tang Y., Taylor M., Roossinck M.J. (2012). Genetic loci controlling lethal cell death in tomato caused by viral satellite RNA infection. Mol. Plant Microbe Interact. 25: 1034–1044 [DOI] [PubMed] [Google Scholar]
- Yamaji Y., Maejima K., Komatsu K., Shiraishi T., Okano Y., Himeno M., Sugawara K., Neriya Y., Minato N., Miura C., Hashimoto M., Namba S. (2012). Lectin-mediated resistance impairs plant virus infection at the cellular level. Plant Cell 24: 778–793 Erratum. Plant Cell 24: 3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T., Ohta T., Takahashi M., Meshi T., Schmidt R., Dean C., Naito S., Ishikawa M. (2000). TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. USA 97: 10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii M., Nishikiori M., Tomita K., Yoshioka N., Kozuka R., Naito S., Ishikawa M. (2004). The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 78: 6102–6111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaliev R., Ueki S., Epel B.L., Citovsky V. (2011). Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma 248: 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., et al. (2011). BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23: 273–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Jeong R.-D., Venugopal S.C., Lapchyk L., Navarre D., Kachroo A., Kachroo P. (2011). SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against Turnip crinkle virus. PLoS Pathog. 7: e1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]