Abstract
Human immunodeficiency virus type 1 (HIV-1)-infected individuals who develop drug-resistant virus during antiretroviral therapy may derive benefit from continued treatment for two reasons. First, drug-resistant viruses can retain partial susceptibility to the drug combination. Second, therapy selects for drug-resistant viruses that may have reduced replication capacities relative to archived, drug-sensitive viruses. We developed a novel single-cell-level phenotypic assay that allows these two effects to be distinguished and compared quantitatively. Patient-derived gag-pol sequences were cloned into an HIV-1 reporter virus that expresses an endoplasmic reticulum-retained Env-green fluorescent protein fusion. Flow cytometric analysis of single-round infections allowed a quantitative analysis of viral replication over a 4-log dynamic range. The assay faithfully reproduced known in vivo drug interactions occurring at the level of target cells. Simultaneous analysis of single-round infections by wild-type and resistant viruses in the presence and absence of the relevant drug combination divided the benefit of continued nonsuppressive treatment into two additive components, residual virus susceptibility to the drug combination and selection for drug-resistant variants with diminished replication capacities. In some patients with drug resistance, the dominant circulating viruses retained significant susceptibility to the combination. However, in other cases, the dominant drug-resistant viruses showed no residual susceptibility to the combination but had a reduced replication capacity relative to the wild-type virus. In this case, simplification of the regimen might still allow adequate suppression of the wild-type virus. In a third pattern, the resistant viruses had no residual susceptibility to the relevant drug regimen but nevertheless had a replication capacity equivalent to that of wild-type virus. In such cases, there is no benefit to continued treatment. Thus, the ability to simultaneously analyze residual susceptibility and reduced replication capacity of drug-resistant viruses may provide a basis for rational therapeutic decisions in the setting of treatment failure.
Treatment of human immunodeficiency virus type 1 (HIV-1)-infected patients with highly active antiretroviral therapy (HAART) can reduce plasma virus levels to below the detection limit (19, 20, 40) and can allow a significant degree of immune reconstitution when control of viremia is maintained (2, 33). However, eradication of HIV-1 infection has not been achieved despite suppression of viremia to below detection limits for as long as 7 years (53). A viral reservoir in latently infected resting memory CD4+ T cells has shown remarkable stability and can support life-long persistence of replication-competent HIV-1 (8-10, 17, 18, 41, 53, 57, 59; reviewed in reference 5). This reservoir in resting CD4+ T cells can serve as a permanent archive for all major forms of the virus present during the entire course of infection, including the original drug-sensitive forms as well as drug-resistant viruses that arise due to inadequate suppression of viral replication by antiretroviral drugs (41, 49).
Although HAART can effectively suppress viremia to below the limit of detection for prolonged periods in some infected individuals, virologic failure, as evidenced by consistently detectable viremia, is also common (32, 34). Failure is frequently associated with the development of resistance to one or more of the drugs in the regimen (15, 22), and drug resistance has emerged as a major problem in the management of HIV-1 infection.
Several assays can monitor the development of drug resistance. Population-level sequencing of viruses in plasma can reveal the existence of characteristic mutations associated with drug resistance (reviewed in reference 51). Genotypic data can be used to predict drug resistance phenotypes by using compiled databases and established algorithms (50). Direct phenotypic assays of drug resistance have also been developed (25, 42) and are of particular value when multiple mutations are present. These assays use pooled HIV-1 reverse transcriptase (RT) and protease sequences amplified from plasma to measure susceptibility to individual antiretroviral drugs. The interpretation of these assays is complicated by the fact that viruses replicating in vivo experience simultaneous selection by each of the drugs in the regimen. The possible synergy and antagonism that may occur with treatment with multiple agents are not reflected in current assays. A particular problem is that current assays do not provide a clear indication of whether or not multiple antiretroviral drugs acting synergistically might still have some residual activity against viruses with resistance mutations. Thus, phenotypic assays that can compare the susceptibility of viral isolates to drug combinations, rather than to individual drugs, would be a valuable tool for choosing alternative regimens in the setting of treatment failure.
The choice of treatment regimens in the setting of failure is further complicated by the issue of replication capacity. Elegant studies by Deeks et al. (13, 14) have demonstrated that some patients who are failing therapy maintain relatively high CD4 counts despite detectable viremia. Interruption of therapy leads to the loss of this immunologic benefit. Because some drug resistance mutations can reduce the fitness of the virus relative to wild-type virus in the absence of drugs (reviewed in references 39 and 47), Deeks et al. suggested that the immunologic benefit of continued treatment in the presence of virologic failure may reflect selection for drug-resistant mutants with diminished replication capacities (4, 12, 14). This benefit is entirely dependent upon the assumption that the wild-type virus with higher fitness is preserved and will reappear if therapy is stopped. Indeed, wild-type viruses do reappear several weeks after treatment interruption (14). The reappearance of wild-type virus is unlikely to be simply genetic reversion because different forms of resistance involving either single mutations or accumulations of multiple mutations disappear with similar kinetics (14). Phylogenetic evidence suggests that the reemerging wild-type viruses are archival (29). At the present time, the only site in which wild-type viruses have been shown to persist despite prolonged replication of and selection for drug-resistant viruses is the latent reservoir in resting memory CD4+ T cells (35, 41, 49).
The considerations presented above suggest that patients failing HAART may derive benefit from continued treatment for two reasons, namely the residual susceptibility of the resistant viruses to the drug regimen and the diminished replication capacities of the resistant viruses. Current assays do not provide a simple way to determine the relative importance of these two effects. We describe here a single-cell-level phenotypic assay that allows analytical comparison of the contributions of residual susceptibility and reduced replication capacity, thereby providing a rational basis for treatment decisions in the setting of virologic failure.
MATERIALS AND METHODS
Vectors.
The green fluorescent protein (GFP)-tagged HIV-1 vector pNL4-3-ΔE-GFP was modified from a previously described reporter virus construct (43). The KpnI-NheI fragment of the HIV-1 NL4-3 env gene (nucleotides 6351 to 7260 in HXB2 coordinates) was replaced with a 745-bp fragment containing the GFP gene. The deleted env region was downstream of the N-terminal Env signal peptide coding sequence and did not overlap with other HIV-1 open reading frames or the Rev-response element (RRE). The GFP-encoding fragment was amplified from the pEGFP-N1 plasmid (Clontech) with primers containing KpnI and NheI sites (GFP 5′primer, ATTGGGTACCTGTCGCCACCATGGTGAGC; GFP 3′primer, GTCCGTGCTAGCTTACAGCTCGTCCTTGTACAGCTCGTCCATGCC). The 3′ primer introduced an in-frame endoplasmic reticulum (ER) retention signal (KDEL) followed by a TAA stop codon at the end of the GFP gene. To insert the KpnI/NheI-flanked GFP fragment into the pNL4-3 backbone, a three-way ligation was set up, involving the KpnI/NheI-digested GFP PCR product, the 13.3-kb EcoRI/NheI fragment, and the 605-bp EcoRI/KpnI fragment of pNL4-3. Correct construction was verified by MfeI digestion and expression of GFP in transfected 293T cells, as detected by flow cytometry. The GFP-KDEL-stop sequence was inserted so as to preserve splice junctions as well as the RRE.
Patient samples.
gag-pol sequences with drug resistance mutations were obtained from the latent reservoir or plasma of compliant pediatric and adult patients who were failing HAART with consistently detectable viremia, as previously described (24, 41, 49). Isolates from the latent reservoir were obtained from replication-competent viruses grown out of the reservoir at a limiting dilution in cultures in which resting cells from patients were stimulated in vitro with mitogens and then cocultured in the presence of CD4+ lymphoblasts from healthy donors (18).
Insertion of patient-derived HIV-1 gag-pol sequences into pNL4-3-ΔE-GFP.
Recombinant HIV-1 vectors containing patient-derived gag-pol sequences were made by replacing the 1.5-kb ApaI/AgeI fragment of pNL4-3-ΔE-GFP with corresponding patient-derived sequences amplified by RT-PCR from plasma virus (24) or by PCR from proviral DNA in latently infected resting CD4+ T cells (49). This portion of the gag-pol gene includes a sequence encoding the Gag protein p7 C terminus, p1 and p6 (Gag codons 406 to 500), full-length protease, and the first 314 amino acids of RT. Viral RNA and proviral DNA were obtained as previously described (24, 49). The following nested sets of primers were used for PCR amplification: 5′outer, GCAAGAGTTTTGGCTGAAGCAATGAG (HXB2 positions 1867 to 1892); 3′outer, CCTTGCCCCTGCTTCTGTATTTCTGC (HXB2 positions 3528 to 3553); 5′inner, TGCAGGGCCCCTAGGAAAAAGGGCTG (HXB2 positions 2002 to 2027); 3′inner, CATGTACCGGTTCTTTTAGAATCTCTCTGTT (HXB2 positions 3465 to 3495).
ApaI and AgeI sites were incorporated in the 5′ and 3′ inner primers. PCR was performed with high-fidelity Platinum Pfx DNA polymerase (Invitrogen). The thermocycling protocol was denaturing at 94°C for 3 min, 30 rounds of denaturing-annealing-extension cycles (94°C for 20 s, 60°C for 30 s, and 68°C for 1.5 min), and a final extension at 68°C for 5 min. The outer PCR products were diluted 1:200 and used as templates in the second-round inner PCR. The 1.5-kb final PCR products were resolved in a 0.7% agarose gel and purified by use of a Qiaquick PCR purification kit (Qiagen). The patient-derived gag-pol PCR products were then cloned into pNL4-3-ΔE-GFP by ligation of ApaI/AgeI-digested PCR products with the 13.2-kb ApaI/AgeI fragment of pNL4-3-ΔE-GFP at an insert/vector molar ratio of 5:1. Ligation products were then transformed into STBL-2 competent cells (Invitrogen). Transformants were plated on Luria-Bertani agarose selection medium containing 50 μg of carbenicillin (Sigma)/ml. Positive clones were identified by MfeI digestion and were sequenced by using the following primers: PR, CAGAAAGGCAATTTTAGGAACC; RT5′, ACCTACACCTGTCAACATAATTGG; and RT3′, GATAAATTTGATATGTCCATTG.
Pseudotype virus production and infection.
The vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped HIV-1 virus was produced as described previously (43). Briefly, 293T cells were cotransfected with wild-type or recombinant pNL4-3-ΔE-GFP and pVSV-G by use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Four hours after transfection, the medium was replaced with RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum (Gemini) and 50% normal human serum (Gemini). Where appropriate, protease inhibitors (PIs) were added at this step. Supernatants containing VSV-G-pseudotyped HIV-1 virus were collected 48 h after transfection. Cell debris was removed from the supernatant by spinning at 450 × g for 5 min and filtering through Steriflip filters (Millipore). Viral supernatants were then used for infection or stored at −80°C.
The viral supernatants were standardized based on the number of GFP-positive transfected 293T cells per unit volume, which was calculated as the concentration of 293T cells times the fraction of GFP-positive 293T cells at 24 h posttransfection. Specific volumes of viral supernatants that were equivalent to a given number of virus-producing 293T cells were used to infect 0.5 × 106 Jurkat cells. The infection was induced by mixing each viral supernatant with 0.5 × 106 washed Jurkat cells in a constant final volume and spinning at 1,800 × g at 30°C for 2 h. Where appropriate, PIs, nucleoside analogue RT inhibitors (NRTIs), and nonnucleoside RT inhibitors (NNRTIs) were added during the infection and maintained throughout the culture. When NRTIs were used, the Jurkat cells were precultured in the presence of NRTIs for 16 h before infection in order to allow for intracellular phosphorylation to produce the active triphosphate forms of the drugs. After the 2-h spin infection, Jurkat cells were washed, resuspended in 2 ml of culture medium (RPMI 1640, 10% fetal bovine serum, 50% human serum), and incubated in 24-well plates at 37°C for 48 h before analysis by flow cytometry.
Analysis of Jurkat cells infected with recombinant NL4-3-ΔE-GFP.
For quantification of the replication of recombinant HIV-1 containing patient-derived gag-pol sequences, the percentage of infected Jurkat cells was measured. Jurkat cells were collected at 48 h postinfection, washed, and fixed with 1% paraformaldehyde in phosphate-buffered saline for 30 min on ice. Flow cytometry was performed in a FACScan instrument (Becton Dickinson) and analyzed with Cell Quest software (Becton Dickinson). Replication was quantified as the percentage of GFP-positive Jurkat cells after gating for live cells. The 50% inhibitory concentration (IC50) for each antiretroviral drug was determined by drug titration in this system. Minimum and maximum concentrations in plasma (Cmin and Cmax, respectively) for current antiretroviral drugs were obtained from the manufacturers and from the Micromedex database. Viral isolates were compared with respect to the replication capacity index, defined as the ratio of the fraction of target cells infected by the test isolate to the fraction of target cells infected by the reference wild-type NL4-3 clone in the absence of drugs, and the drug resistance index, defined as the ratio of the fraction of infected target cells in the presence of drugs to the fraction of infected target cells in the absence of drugs. The replication index was defined as the product of the replication capacity index and the drug resistance index. This value sums the two effects on a log scale and represents the total treatment benefit.
RESULTS
Rationale.
The goal of this study was to compare the relative contributions of two treatment effects that benefit patients with drug resistance, namely the residual susceptibility of resistant viruses to inhibition by drug combinations and the selection pressure to maintain drug-resistant mutants with reduced replication capacities relative to the wild-type virus. Both effects reduce viral replication relative to the replication of wild-type virus in the absence of drugs, and thus both effects can be measured on the same scale in replication assays. However, because of the potent inhibitory effects of multiple antiretroviral drugs used in combination, an assay with a wide dynamic range is essential. To this end, patient-derived gag-pol sequences, including sequences for protease and part of RT, were cloned into a novel HIV-1 vector carrying an ER-retained form of the fluorescent reporter GFP. This vector was used to generate pseudotyped virus particles that were used to infect CD4+ T cells in the presence or absence of relevant drug combinations. In this system, viral replication can be measured over a wide dynamic range (4 logs) by the detection of infection at the single-cell level by flow cytometry.
Production and characterization of a recombinant HIV-1 vector for single-cell phenotypic analysis.
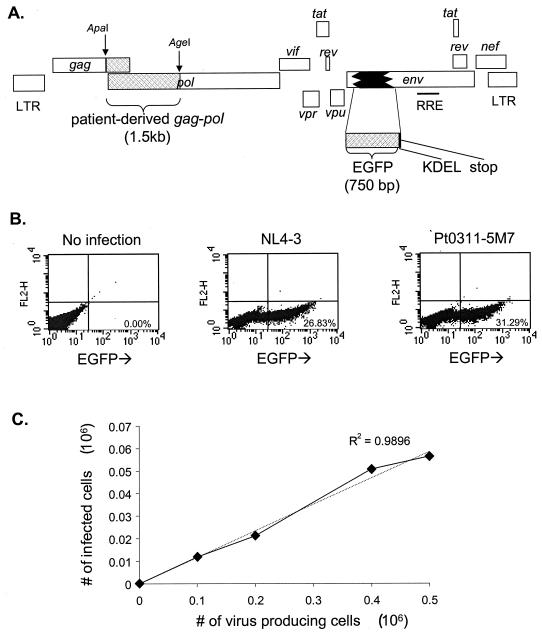
A portion of the env gene of the pNL4-3 proviral clone was replaced with an in-frame insert encoding an enhanced form of GFP followed by in-frame codons for a KDEL ER retention signal (38) and a stop codon (Fig. 1A). The resulting vector, pNL4-3-ΔE-GFP, expresses an Env-GFP fusion protein that is translocated into the ER and retained there, resulting in the accumulation of high levels of intracellular GFP. The expression of GFP fused to a virion structural protein under transcriptional control of the HIV-1 long terminal repeat allowed a high level of expression in infected cells.
FIG. 1.
Single-cell phenotypic assay for drug susceptibility and replication capacity. (A) Proviral construct used to generate pseudotyped virus for infections. Patient-derived gag-pol sequences were cloned in frame into a pNL4-3 proviral clone with the coding sequence for GFP replacing a portion of the env gene. The GFP sequence was followed by a KDEL ER retention signal and a stop codon. As a result, cells transfected with this vector or infected with pseudotyped viruses generated from this vector express an Env-GFP fusion protein that is directed into the ER by the Env signal peptide and retained in the ER by the KDEL sequence. (B) Expression of GFP by infected CD4+ T cells. The CD4+-T-cell Jurkat line was infected in vitro with GFP-encoding HIV-1 pseudotyped with VSV-G. Representative dot plots of GFP expression are shown for uninfected Jurkat cells (left), Jurkat cells infected with pseudovirions carrying the reference NL4-3 gag-pol sequence (center), and Jurkat cells infected with pseudovirions carrying a patient-derived wild-type gag-pol sequence, Pt0311-5M7 (right). (C) Linear relationship between the number of infected Jurkat cells and the amount of input viral inoculum. Jurkat cells were infected under standard conditions with increasing amounts of viral supernatant equivalent to the indicated numbers of virus-producing cells. GFP expression in Jurkat cells was measured by flow cytometry on day 2 after infection. The dotted line represents a fitted linear regression.
After cotransfection of 293T cells with the Env-negative pNL4-3-ΔE-GFP vector and a construct encoding VSV-G, VSV-G-pseudotyped HIV-1 virions were harvested and used to infect cells of the CD4+ human T-cell Jurkat line in single-round infections. Figure 1B shows the results of infection of Jurkat cells with pseudotyped viruses carrying gag-pol sequences from the reference HIV-1 clone NL4-3 or from a patient isolate. Infected cells were readily detected by flow cytometry. By confocal microscopy, infected cells showed bright perinuclear staining, consistent with ER localization of the Env-GFP fusion protein (not shown). Because of the high levels of fluorescence in infected cells and the low background fluorescence (<0.01%) (Fig. 1B), the dynamic range of the assay is limited principally by the number of cells analyzed. In this system, a dynamic range of up to 4 logs can be readily achieved.
Measurement of replication capacity.
In order to compare the replication capacities of viruses with different gag-pol sequences, it was necessary to normalize viral stocks to control for differences in transfection efficiency. Transfection supernatants could not be normalized based on p24 levels in the supernatants since virus release is influenced by protease (27) and since Gag cleavage to generate p24 is dependent on protease activity and can be influenced by protease mutations (60). Similarly, RT assays could not be used since drug resistance mutations in RT can reduce the function of the enzyme (3). We normalized stocks of pseudotyped virus based on the number of transfected cells, as determined by GFP expression. GFP expression was measured in an aliquot of transfected 293T cells to determine how many cells were successfully transfected with the vectors and were capable of expressing viral genes and thus producing pseudovirions. CD4+ T cells were infected with normalized viral supernatants representing fixed numbers of virus-producing cells, and the number of infected target cells, as measured by flow cytometry, was used as a readout for viral replication. This approach ensures that the efficiency of every step in the viral life cycle, including viral assembly, release, maturation, entry, reverse transcription, integration, and viral gene expression, is captured in the measurement of replication capacity. We observed a direct linear relationship between the number of target cells infected and the input amount of pseudotyped viral stock representing a known number of virus-producing cells (Fig. 1C). This linear relationship was observed for all isolates tested, including wild-type and drug-resistant isolates from patients in the presence and absence of drugs. This allowed us to use the number of infected cells as a direct readout for viral replication. Measurements of replication capacity by this method showed a very low variation coefficient (0.05 ± 0.02).
Effective concentration and estimated in vivo potency for individual drugs.
To demonstrate the usefulness of this phenotypic assay for measuring the inhibition of viral replication by antiretroviral drugs, we determined by drug titration the concentrations of protease and RT inhibitors needed to inhibit the replication of the reference wild-type NL4-3 clone in this system. In order to make the system mimic in vivo conditions as closely as possible, we performed assays with 50% human serum to account for the propensity of some antiretroviral drugs to bind to plasma proteins. PIs were added 4 h after transfection and were maintained in the culture during virus production and maturation and the infection of target cells. Target cells were preincubated with NRTIs for 16 h to allow these drugs to be converted to active triphosphate forms via intracellular phosphorylation. Longer preincubation times (24 h) did not further increase the inhibition by NRTIs (not shown). NRTIs, PIs, and NNRTIs were added to the viral supernatants during spin infections and were maintained in the culture medium during the subsequent incubation.
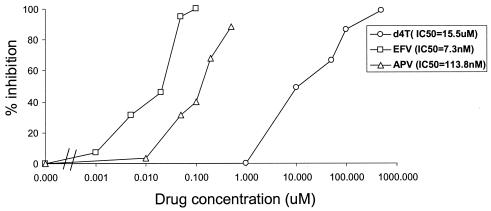
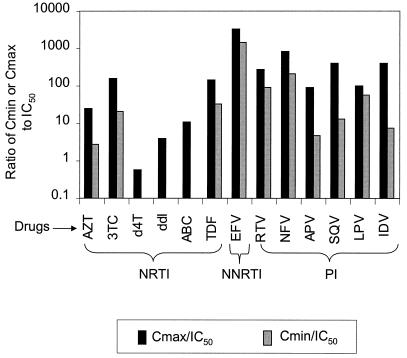
Figure 2 shows typical titration curves for representative drugs from the three major classes of antiretroviral drugs, the NRTI stavudine (d4T), the NNRTI efavirenz (EFV), and the PI amprenavir (APV). Each drug produced the expected sigmoidal dose-response curve for the inhibition of viral replication. The IC50 values were calculated from the titration curves by fitting of a median-effect pharmocokinetic model (6, 7). Differences in potency revealed by this analysis take on additional significance when viewed in the context of the different Cmin and Cmax values achieved by each drug under normal dosing. We estimated the in vivo potency for the antiretroviral drugs in current use by calculating the ratios of published Cmin and Cmax values to the IC50 values measured in this system (Fig. 3). Consistent with the clinical potency of the NNRTI EFV (1, 21, 55), this drug demonstrated extraordinary potency in that even the Cmin was >1,000-fold higher than the IC50 in this assay. The PIs were relatively potent, with Cmin values that were at least 10-fold higher than the IC50 values (except for APV and indinavir). For some of the NRTIs, particularly d4T and didanosine (ddI), Cmax values were close to or below the IC50 values.
FIG. 2.
Measurement of viral susceptibility to antiretroviral drugs using the pNL4-3-ΔE-GFP-derived pseudoviruses. The ability of pseudovirions carrying wild-type NL4-3 gag-pol sequences to infect Jurkat cells was measured in the presence of increasing concentrations of the NRTI d4T, the NNRTI EFV, and the PI APV. APV was added to cultures of virus-producing cells beginning 4 hours after transfection and was maintained throughout the course of viral assembly, release, maturation, and spin inoculation into the target cells. d4T was added to target cells beginning 16 h before infection and was maintained in all steps thereafter. EFV was added at the time of spin infection and was maintained thereafter. The IC50 for each drug was calculated by fitting data to the median-effect pharmacokinetic model (6, 7).
FIG. 3.
Ratios of Cmin and Cmax to IC50 for individual antiretroviral drugs. The in vitro IC50 was measured for each drug by using pseudoviruses carrying the reference NL4-3 sequence as described in Fig. 3. The ratios between published Cmax and Cmin values and the IC50 for each drug are plotted. ABC, abacavir; TDF, tenofovir disoproxil fumarate; RTV, ritonavir; NFV, nelfinavir; SQV, saquinavir; LPV, lopinavir; IDV, indinavir.
Inhibition by multiple drugs in combination.
In infected individuals undergoing HAART, HIV-1 evolves in the simultaneous presence of multiple antiretroviral drugs. Most phenotypic assays measure the capacity of viruses to replicate in the presence of individual drugs. Such assays, therefore, do not take into account the complex synergistic and antagonistic interactions between antiretroviral drugs in the setting of HAART. Antiretroviral drugs can affect each other's efficacies at the level of absorption, systemic metabolism and elimination, prodrug activation, and the targeted enzymatic reaction (31). Interactions affecting absorption and metabolism alter drug concentrations in the blood and are compensated for clinically by adjustments in dosage so that optimal blood levels are achieved. Interactions affecting prodrug activation and enzyme inhibition occur at the level of the target cells and can be directly assessed by using in vitro assays such as the one described here. Table 1 shows that zidovudine (AZT) and d4T strongly antagonize each other in this system, as previously reported for other in vitro (26) and in vivo (23) assays. The inhibition of viral replication observed in the presence of both drugs is much less than expected based on the fraction product principle (58) This antagonism reflects the fact that both are thymidine analogue prodrugs that share the same intracellular phosphorylation pathway (26). These results suggest that drug interactions that are operative in target cells can be accurately modeled in this in vitro system.
TABLE 1.
Measurement of intracellular drug interactions between AZT and d4T
| Drug treatmenta | % GFP-positive target cellsb | Mean % GFP-positive target cells | Drug resistance indexc | P value for antagonismd |
|---|---|---|---|---|
| No drug | 18.62 | 20.09 | ||
| 24.16 | ||||
| 20.08 | ||||
| 17.49 | ||||
| 10 μM AZT | 7.61 | 7.15 | 0.356 | |
| 6.70 | ||||
| 7.59 | ||||
| 6.71 | ||||
| 10 μM d4T | 6.83 | 6.54 | 0.325 | |
| 7.04 | ||||
| 5.74 | ||||
| 10 μM AZT plus 10 μM d4T | 7.14 | 7.25 | 0.361 | <0.001 |
| 8.76 | ||||
| 5.78 | ||||
| 7.33 |
Jurkat cells were infected in replicate with pseudotyped reporter viruses carrying the NL4-3 gag-pol sequence in the presence of the indicated drug(s). The drug(s) was added to target cells 16 h before infection and was maintained throughout the experiment.
Measured 48 h after infection.
The drug resistance index is the ratio of the measured replication in the presence of drug(s) to that in the absence of drugs.
Antagonism is scored according to the fraction product principle. f0 represents the fraction of infected cells in the absence of drug; fAZT and fd4T represent the fractions of infected cells in the presence of 10 μM AZT and 10 μM d4T, respectively; f(AZT+d4T) represents the fraction of infected cells in the presence of the combination of 10 μM AZT and 10 μM d4T. If AZT and d4T function independently, then f(AZT+d4T)/f0 should equal fAZT/f0 × fd4T/f0; if f(AZT+d4T)/f0 > fAZT/f0 × fd4T/f0, then there is antagonism, and if f(AZT+d4T)/f0 < fAZT/f0 × fd4T/f0, then there is synergism. f0, fAZT, fd4T, and f(AZT+d4T) were measured as the means of three or four replicates for each condition. The P value is of the coefficient for the interaction term ln (fAZT/f0) × ln (fd4T/f0) being equal to zero in a multilinear regression.
Analysis of drug susceptibility and replication capacity.
For the reasons described above, we were able to use the percentage of target cells infected by a standardized viral inoculum as a direct readout for the replication of viruses with different gag-pol inserts. Thus, we could compare the abilities of patient-derived resistant HIV-1 isolates and wild-type HIV-1 to replicate in the absence and presence of drugs.
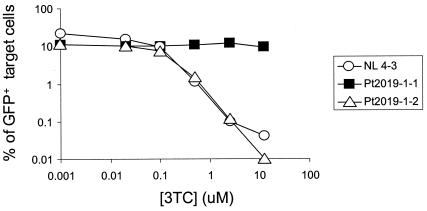
To facilitate these comparisons, we defined a replication capacity index, a drug resistance index, and a replication index (see Materials and Methods). A replication capacity index of <1 indicates a diminished replication capacity relative to that of the reference virus, NL4-3. A drug resistance index of <1 indicates susceptibility to the drug. The replication index, defined as the product of the replication capacity index and the drug resistance index, sums the two effects on a log scale and represents the total treatment benefit. For example, Fig. 4 compares the abilities of pseudoviruses carrying two patient-derived drug-resistant HIV-1 sequences to replicate in the presence and absence of the NRTI lamivudine (3TC). Both isolates are from the same patient and contain an almost identical spectrum of multiple drug resistance mutations in protease and RT, differing only by the presence of the characteristic 3TC resistance mutation M184V in one isolate. Both mutants exhibited a slightly diminished replication capacity relative to the wild-type NL4-3 virus. The replication capacity index of each was ∼0.5. Yet, as expected from the genotype, the isolate containing the M184V mutation was fully resistant to 3TC (drug resistance index of 1 up to 2.5 μM 3TC and of 0.8 at 12.5 μM 3TC). The isolate lacking this mutation showed the same high degree of susceptibility to 3TC as wild-type NL4-3 over about 3 logs of inhibition (drug resistance index of <0.002 at 12.5 μM 3TC). This result demonstrates that the phenotypic assay can simultaneously measure replication capacity and drug resistance for HIV-1 gag-pol isolates from patients. Interestingly, for the multidrug-resistant isolate pt2019-1-2 analyzed here, the magnitude of the decrease in replication capacity relative to that of NL4-3 in the absence of drugs was small compared to the profound degree of inhibition by 3TC of the viruses lacking the M184V mutation.
FIG. 4.
Simultaneous measurement of susceptibility to 3TC and replication capacity for different HIV-1 clones. Pseudovirions carrying patient-derived isolates and NL4-3 gag-pol sequences were used to infect Jurkat cells in the presence of the indicated concentrations of 3TC. Drug resistance mutations in the protease and RT of patient-derived HIV-1 clones are shown in Table 2.
Dissecting the benefits of nonsuppressive HAART into residual drug susceptibility and selection for resistant variants with diminished viral replication capacities.
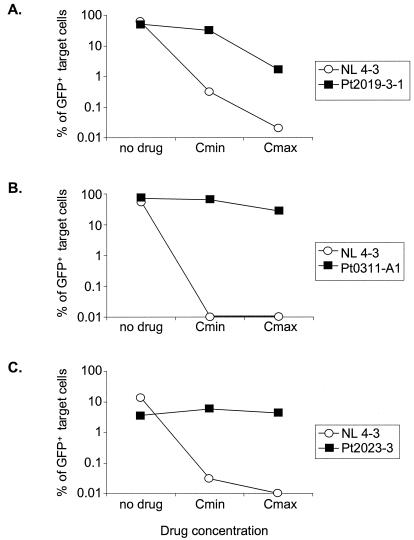
To demonstrate the utility of this assay for distinguishing residual suppression from diminished replication capacity, we analyzed drug-resistant viruses from patients who were failing HAART regimens. Three general patterns emerged (Fig. 5 and Table 2). In the first pattern, as shown in Fig. 5A, the resistant variant exhibited both a diminished replication capacity (replication capacity index of <1) and partial susceptibility to the drug combination being used (drug resistance index of <1), particularly when each drug was present at its Cmax. In this case, the defect in replication capacity relative to NL4-3 was slight (replication capacity index = 0.8), indicating that the multiple mutations in protease and RT did not substantially decrease the capacity of these enzymes to function, possibly due to the compensatory effects of some of the secondary mutations. The replication of the wild-type clone NL4-3 was strongly inhibited by the four-drug combination that constituted the patient's regimen at the time of virus isolation (drug resistance index = 0.0003 at the Cmax). The drug-resistant isolate was only partially inhibited (drug resistance index = 0.63 at the Cmin and 0.03 at the Cmax). This partial inhibition represents the residual suppression of the drug-resistant virus by the regimen. Therefore, this HAART regimen may exert some control on viremia via both partial suppression and selection for isolates with diminished replication capacities.
FIG. 5.
Three different patterns of replication of drug-resistant HIV-1 clones reflecting differential contributions of residual drug susceptibility and reduced replication capacity. gag-pol sequences amplified from the plasma of patients failing therapy were used to generate pseudovirions that were then used to infect Jurkat cells in the absence of drugs and in the presence of failing drug combinations at the Cmin and Cmax of each drug. Drug resistance mutations present in each isolate are indicated in Table 2. The replication of each isolate was compared to that of the wild-type NL4-3 sequence. (A) Resistant virus with significant residual susceptibility and marginally reduced replication capacity. (B) Resistant virus with minimal residual susceptibility and high replication capacity. (C) Resistant virus with no residual susceptibility and significantly reduced replication capacity.
TABLE 2.
Comparison of NL4-3 and patient-derived isolates with respect to replication capacity and drug resistance
| Isolatea | Mutationsb
|
Replication capacity indexc | Drug(s)d | Concne (μM) | Drug resistance indexf | Replication indexg | |
|---|---|---|---|---|---|---|---|
| Protease | RT | ||||||
| NL4-3 | Wild type | Wild type | 1 | 3TC | 12.5 | 0.002 | 0.002 |
| Pt2019-1-1 | L10I, K20R, M36I, M46L, I54V, L63A, A71V, I84V, L90M | M41L, M184V, H208Y, R211K, T215Y | 0.52 | 3TC | 12.5 | 0.8 | 0.42 |
| Pt2019-1-2 | L10I, K20R, M36I, I54V, L63A, A71V, I84V, L90M | M41L, H208Y, R211K, T215Y | 0.53 | 3TC | 12.5 | 0.001 | 0.00053 |
| NL4-3 | Wild type | Wild type | 1 | ddI, d4T, RTV, SOV | Cmin | 0.005 | 0.005 |
| Cmax | 0.0003 | 0.0003 | |||||
| Pt2019-3-1 | K20R, M36I, M46L, I54V, L63A, A71V, I84V, L90M | M41L, M184V, H208Y, R211K, T215Y | 0.8 | ddI, d4T, RTV, SOV | Cmin | 0.63 | 0.504 |
| Cmax | 0.03 | 0.02 | |||||
| NL4-3 | Wild type | Wild type | 1 | d4T, 3TC, NFV | Cmin | 0.0002 | 0.0002 |
| Cmax | 0.0002 | 0.0002 | |||||
| Pt0311-A1 | D30N, N37D, M46I, L63P, A71T, V77I, N88D, I93L | M41L, D67N, V118I, M184V, L210W, R211K, T215Y | 1.34h | d4T, 3TC, NFV | Cmin | 0.94 | 1.26 |
| Cmax | 0.39 | 0.52 | |||||
| NL4-3 | Wild type | Wild type | 1 | ddI, 3TC, ABC, EFV | Cmin | 0.005 | 0.005 |
| Cmax | 0.004 | 0.004 | |||||
| Pt2023-3 | Wild type | M41L, L74V, V75I, M184V, G190E, K219N | 0.21 | ddI, 3TC, ABC, EFV | Cmin | 1.74 | 0.37 |
| Cmax | 1.31 | 0.28 | |||||
The indicated patient-derived drug-resistant isolates are shown in Fig. 5 and 6 and are compared with the reference NL4-3 sequence with respect to replication capacity and susceptibility to the indicated combinations of antiretroviral drugs. Isolates were obtained from the plasma or latent reservoir of patients on the relevant regimens.
Characterized drug resistance mutations in protease and RT based on the International AIDS Society—USA compilation (11).
The ratio of the replication capacity of the test isolate to the replication capacity of the reference wild-type NL4-3 clone in the absence of drugs.
3TC, lamivudine; ddI, didanosine; d4T, stavudine; ABC, abacavir; EFV, efavirenz; RTV, ritonavir; SQV, saquinavir; NFV, nelfinavir.
For drug combinations, each drug was used at its Cmin or Cmax. The Cmin and Cmax values (μM) for the drugs used in this study are as follows: 3TC, 1.2 and 8.6; ddI, 0.04 and 6.78; d4T, 0.001 and 8.6; abacavir, 0.04 and 10.48; EFV, 8.57 and 13; ritonavir, 5.3 and 16; saquinavir, 0.3 and 9.3; nelfinavir, 1.74 and 6.97. Cmin and Cmax values were obtained from the Micromedex database and the manufacturer's package insert.
The ratio of the measured replication of an HIV-1 isolate in the presence of the indicated drug regimen to replication in the absence of drugs.
The product of the replication capacity index and the drug resistance index. The replication index is a measure of the capacity of the indicated isolate to replicate in the presence of the indicated drug regimen relative to the replication of the reference NL4-3 isolate in the absence of drug.
This drug-resistant isolate replicated more efficiently than NL4-3 in the absence of drugs. If the replication capacity of this drug-resistant isolate is compared to that of a wild-type isolate derived from the same patient, then the replication capacity index is 1.02.
In the second pattern, the patient-derived drug-resistant HIV-1 had a high replication capacity and minimal drug susceptibility. Figure 5B shows data for a drug-resistant isolate with a replication capacity slightly higher than that of NL4-3. The replication capacity index relative to NL4-3 was 1.34. We also compared the replication capacity of this resistant virus to that of a wild-type virus isolated from the latent reservoir of the same patient. In this case, the replication capacity index was 1.02. Thus, this resistant isolate was highly fit despite the presence of several major drug resistance mutations (Table 2). In addition, the resistant isolate was only minimally suppressed by the drug combination (drug resistance index = 0.94 and 0.39 at the Cmin and Cmax, respectively). Thus, for this isolate, treatment provides little benefit.
In the third pattern, shown in Fig. 5C, the patient-derived isolate was fully resistant to the HAART regimen (drug resistance index of ≥1 at both the Cmin and Cmax), yet it had a diminished replication capacity (replication capacity index = 0.21) relative to that of the wild-type NL4-3 isolate. Therefore, if the patient harbors a wild-type virus that is similar in replication capacity to NL4-3, the HAART regimen may benefit the patient mainly by selecting for a resistant variant with a reduced replication capacity.
In vitro analysis of partial treatment interruptions.
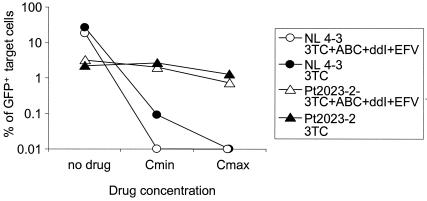
In cases for which the clinical benefit of continued treatment is solely due to the selection for resistant variants with diminished replication capacities, as shown in Fig. 5C, the drug regimen could potentially be simplified to keep only the minimum number of drugs needed to select for the resistant variants over the wild-type virus. In this situation, none of the drugs would still exert any direct suppressive effect on the relevant viral enzymes; they would function only to suppress replication of the wild-type virus. This scenario can be modeled in the in vitro system described here. For the specific example shown in Fig. 5C, the infections were repeated, using the original drug combination (ddI, abacavir, 3TC, and EFV) and 3TC alone (Fig. 6). As expected, the replication of the resistant virus was not affected, while the replication of the wild-type virus was strongly suppressed by 3TC alone, to almost the same extent as with the four-drug combination. Even in the presence of 3TC alone, the drug-resistant clone with a reduced replication capacity was still 30- to 100-fold more fit than the wild-type virus; thus, this analysis predicts that 3TC therapy alone is sufficient to maintain the resistant variant. Analysis of this kind could be used to find the simplest regimen that provides the best balance between reduced toxicity and prolonged suppression of wild-type virus.
FIG. 6.
In vitro demonstration of selection for a drug-resistant virus with reduced replication capacity by a simplified regimen. The drug-resistant virus analyzed for Fig. 5C was tested for replication in the absence of drugs, in the presence of the failing regimen, and in the presence of a simplified regimen consisting of only 3TC. Replication of this isolate was compared to that of the wild-type NL4-3 sequence.
DISCUSSION
We described a novel phenotypic assay that can simultaneously measure, on the same scale, HIV-1 susceptibility to drug combinations and changes in replication capacity relative to reference or patient-specific wild-type sequences. This provides a quantitative tool for analyzing the efficacy of antiretroviral therapy, especially the mechanism of the clinical benefit of HAART in the setting of virologic failure.
It is important to note that no in vitro assay can fully duplicate the in vivo conditions under which the antiretroviral drugs mediate suppression of viral replication. Nevertheless, in vitro phenotypic assays of drug resistance have potential clinical utility (28, 36, 46, 52). Results from single-cycle assays of replication capacity generally parallel results of virus culture assays for fitness (45, 48), although the correlation is not always perfect (48). In the assay described here, several steps have been taken to ensure that the cultures mimic in vivo conditions as closely as possible. First, we accounted for the protein-binding properties of some antiretroviral drugs by supplementing the culture medium with 50% normal human serum. Second, we have accounted for the prodrug activation required for the function of all NRTIs. Because all NRTIs require multiple steps of intracellular phosphorylation to be converted to active nucleoside triphosphate analogues (56), CD4+ T cells were pretreated with NRTIs 16 h prior to infection. This time is sufficient for intracellular levels of the active forms of these drugs to reach a steady state, as evidenced by the fact that pretreatment for longer times does not increase inhibition. Third, drugs were tested at their Cmin and Cmax values under the conditions described above. This effectively circumvents issues related to drug absorption and metabolism and exposes target cells to concentrations of drugs that bracket the concentrations that should be experienced by cells in vivo. Finally, and most significantly, the drugs were tested in the same combinations that are used in vivo. Because many combinations of antiretroviral drugs produce a profound synergistic inhibition of wild-type virus, quantitative analysis of drug inhibition is only possible with assays that have a wide dynamic range. The flow cytometric assay described here has a dynamic range of up to 4 logs, allowing quantification of the synergistic inhibitory effects of drug combinations as well as of individual components of the regimen. The assay faithfully reproduced reported drug interactions that occur at the level of target cells. For example, the reported antagonism between AZT and d4T caused by competition at the step of prodrug activation was readily observed with this assay (Table 1). Taken together, these results suggest that the phenotypic assay described here provides a reasonable first approximation of drug inhibitory effects in vivo.
Using this assay, we compared the potencies of available antiretroviral drugs by examining the ratio of the Cmin and Cmax values to the IC50 determined in this system. Our data highlight the extraordinary potency of the NNRTI EFV (1, 21, 55), which has a Cmin/IC50 ratio of >1,000 in our system. In contrast, the commonly used NRTIs d4T and ddI are relatively inefficient at inhibiting viral replication in this system. Cmax values for these drugs are actually below the IC50 and IC90 values, respectively. Because the actual IC50 depends on the viral strain, the target cell type, the culture medium, and the multiplicity of infection in specific phenotypic assay systems (37, 44), direct comparison of Cmin/IC50 and Cmax/IC50 ratios between different assay systems is not possible. It is worth noting that drug susceptibility measured in this system was also dependent on the properties of the virus-producing cells, 293T cells, and the target cells, the Jurkat CD4+-T-cell line. These cells may differ from primary CD4+ T cells in the absorption and metabolism of antiretroviral drugs. They may differ from primary cells in the expression of transporters, such as the P glycoprotein, that can export PIs from the cytoplasm (30). In addition, the in vivo efficacy of a drug is dependent upon more than its potency in inhibiting a single round of viral replication. Additional factors such as genetic barriers to resistance, tolerability, and pharmacokinetics are important. Therefore, these in vitro results should be interpreted with caution.
Another caveat is related to the heterogeneity of replication capacities of wild-type HIV-1 isolates relative to that of a reference sequence, NL4-3. The replication capacities of wild-type HIV-1 clones from patients can vary up to 2.5-fold from that of NL4-3 in our system. The mean replication capacity index relative to NL4-3 was 0.81 ± 0.34 (n = 7). Similar variations have been observed by other groups (54). These results suggest that in order to most accurately assess changes in viral fitness in vivo, it is necessary to compare the replication capacity of the patient's drug-resistant virus to that of the drug-sensitive virus obtained from the same patient. This can be readily done in the system described here provided that the wild-type sequence is available. In compliant patients who are failing therapy and have drug-resistant viruses, wild-type viruses are typically not found in the plasma but do persist in the latent reservoir in resting memory CD4+ T cells (49). It is also important to point out that viral clones with different mutations are likely to be present in each patient with drug resistance and that the results of this type of analysis may be different for each clone. Ideally, a large number of distinct clones representing the full range of variation in pol would be analyzed, although this may not be practical as a routine clinical test. Alternatively, it may be possible to analyze selected clones that represent extremes on the spectrum of wild-type to fully resistant viruses. A final caveat is that compensatory mutations outside of the gag-pol region studied here could potentially affect viral fitness. Our construct did include the Gag p7/p1 and p1/p6 cleavage sites that frequently accumulate compensatory mutations in response to PIs (16).
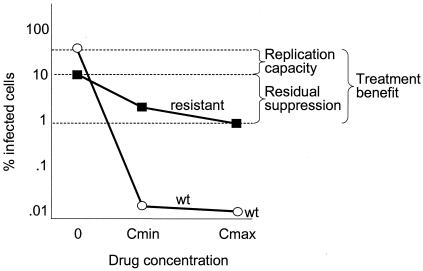
The ability of this assay to simultaneously measure HIV-1 drug susceptibility and replication capacity allowed us to study the mechanisms of the apparent clinical benefit of HAART in the setting of virologic failure. Our data show that the benefit of nonsuppressive HAART can be quantitatively deconstructed into two additive effects. This is illustrated graphically in Fig. 7 and numerically in Table 2. One effect is the residual suppression of replication of the resistant variants by antiretroviral drugs. This is a benefit that operates in real time, reflecting direct inhibition of viral enzymes by the drugs. The other beneficial effect is selection for drug-resistant variants with a diminished replication capacity. This is a potential benefit that only becomes apparent if the drugs are stopped and archived drug-sensitive variants with higher replication capacities emerge. A recent study by Ruff et al. (49) demonstrated the persistence of archival wild-type HIV-1 in the latent reservoir in resting memory CD4+ T cells even after years of selection for drug-resistant variants by failing drug regimens. Further evidence for the persistence of wild-type viruses in the setting of failure comes from the work of Deeks et al. (14) demonstrating simultaneous loss of all drug-resistant variants accompanied with the appearance of wild-type HIV-1 in patients with multidrug resistance who interrupt therapy. These data suggest that the selection pressure exerted by drugs in failing regimens can prevent drug-sensitive variants with potentially higher replication capacities from emerging.
FIG. 7.
Schematic plot showing decomposition of the clinical benefit of nonsuppressive HAART into two additive effects, the residual suppression of viral replication and the selection for resistant virus with diminished replication capacity. The difference in viral replication in the absence of drug represents the diminished replication capacity of the selected resistant virus versus the counterselected archival wild-type virus. The different replication capacities of the resistant virus in the absence and presence of the failing drug regimen represent residual suppression of the resistant virus. The addition of these two effects on a log scale represents the total inhibition of potential viral replication by nonsuppressive HAART or the treatment benefit.
This method of analysis of the benefits of HAART could provide the basis for the rational management of antiretroviral therapy in the problematic setting of virologic failure. For example, in circumstances in which the clinical benefit of the drug combination is solely due to selection for resistant variants with diminished replication capacities, as shown in Fig. 5C, the drug regimen could potentially be simplified, retaining the minimum number of drugs needed to provide selection pressure favoring the resistant variants over the wild-type virus. On the other hand, in cases in which the HAART regimen exerts little suppression on viral replication and the evolved resistant virus has achieved a replication capacity equivalent to that of the archived wild-type viruses present in the latent reservoir (Fig. 5B), continued treatment with the same regimen provides no obvious benefit.
In summary, we have described a novel single-cell-level phenotypic assay that can simultaneously analyze HIV-1 drug susceptibility and intrinsic replication capacity. This allows quantitative dissection of the functions of antiretroviral drugs into suppression of viral replication and selection of resistant viruses with diminished replication capacities. Although its application in clinical management remains to be tested, this experimental approach provides a tool for the rational evaluation of treatment decisions for patients failing antiretroviral therapy.
Acknowledgments
We thank Charles Flexner and current and previous members of the lab for helpful suggestions and Scott Barnet and Mike Paradise for help with scheduling patients.
This work was supported by NIH grants AI43222 and AI51178 and by a grant from the Doris Duke Charitable Foundation.
REFERENCES
- 1.Arribas, J. R., F. Pulido, J. M. Miro, M. A. Costa, J. Gonzalez, R. Rubio, J. M. Pena, M. Torralba, M. Lonca, A. Lorenzo, A. Del Palacio, J. J. Vazquez, and J. M. Gatell. 2002. High effectiveness of efavirenz-based highly active antiretroviral therapy in HIV-1-infected patients with fewer than 100 CD4 cells/μl and opportunistic diseases: the EfaVIP study (efavirenz in very immunosuppressed patients). AIDS 16:1554-1556. [DOI] [PubMed] [Google Scholar]
- 2.Autran, B., G. Carcelain, T. S. Li, C. Blanc, D. Mathez, R. Tubiana, C. Katlama, P. Debre, and J. Leibowitch. 1997. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science 277:112-116. [DOI] [PubMed] [Google Scholar]
- 3.Back, N. K., M. Nijhuis, W. Keulen, C. A. Boucher, E. B. Oude, A. B. van Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 4.Barbour, J. D., T. Wrin, R. M. Grant, J. N. Martin, M. R. Segal, C. J. Petropoulos, and S. G. Deeks. 2002. Evolution of phenotypic drug susceptibility and viral replication capacity during long-term virologic failure of protease inhibitor therapy in human immunodeficiency virus-infected adults. J. Virol. 76:11104-11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 6.Chou, T. C. 1976. Derivation and properties of Michaelis-Menten type and Hill type equations for reference ligands. J. Theor. Biol. 59:253-276. [DOI] [PubMed] [Google Scholar]
- 7.Chou, T. C., and P. Talalay. 1981. Generalized equations for the analysis of inhibitions of Michaelis-Menten and higher-order kinetic systems with two or more mutually exclusive and nonexclusive inhibitors. Eur. J. Biochem. 115:207-216. [DOI] [PubMed] [Google Scholar]
- 8.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 9.Chun, T. W., D. Finzi, J. Margolick, K. Chadwick, D. Schwartz, and R. F. Siliciano. 1995. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 1:1284-1290. [DOI] [PubMed] [Google Scholar]
- 10.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Aquila, R. T., J. M. Schapiro, F. Brun-Vezinet, B. Clotet, B. Conway, L. M. Demeter, R. M. Grant, V. A. Johnson, D. R. Kuritzkes, C. Loveday, R. W. Shafer, and D. D. Richman. 2002. Drug resistance mutations in HIV-1. Top. HIV Med. 10:21-25. [PubMed] [Google Scholar]
- 12.Deeks, S. G. 2001. Durable HIV treatment benefit despite low-level viremia: reassessing definitions of success or failure. JAMA 286:224-226. [DOI] [PubMed] [Google Scholar]
- 13.Deeks, S. G., J. D. Barbour, J. N. Martin, M. S. Swanson, and R. M. Grant. 2000. Sustained CD4+ T cell response after virologic failure of protease inhibitor-based regimens in patients with human immunodeficiency virus infection. J. Infect. Dis. 181:946-953. [DOI] [PubMed] [Google Scholar]
- 14.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 15.Descamps, D., P. Flandre, V. Calvez, G. Peytavin, V. Meiffredy, G. Collin, C. Delaugerre, S. Robert-Delmas, B. Bazin, J. P. Aboulker, G. Pialoux, F. Raffi, and F. Brun-Vezinet. 2000. Mechanisms of virologic failure in previously untreated HIV-infected patients from a trial of induction-maintenance therapy. Trilege (Agence Nationale de Recherches sur le SIDA 072) Study Team. JAMA 283:205-211. [DOI] [PubMed] [Google Scholar]
- 16.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 18.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 19.Gulick, R. M., J. W. Mellors, D. Havlir, J. J. Eron, C. Gonzalez, D. McMahon, D. D. Richman, F. T. Valentine, L. Jonas, A. Meibohm, E. A. Emini, and J. A. Chodakewitz. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337:734-739. [DOI] [PubMed] [Google Scholar]
- 20.Hammer, S. M., K. E. Squires, M. D. Hughes, J. M. Grimes, L. M. Demeter, J. S. Currier, J. J. J. Eron, J. E. Feinberg, H. H. J. Balfour, L. R. Deyton, J. A. Chodakewitz, and M. A. Fischl. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N. Engl. J. Med. 337:725-733. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann, M., A. Rump, J. Brust, D. Schuster, F. Mosthaf, M. Procaccianti, H. Klinker, and D. Petzoldt. 2001. Evaluation of an efavirenz-containing regimen: an open-label, multicenter study. HIV Clin. Trials 2:421-428. [DOI] [PubMed] [Google Scholar]
- 22.Havlir, D. V., N. S. Hellmann, C. J. Petropoulos, J. M. Whitcomb, A. C. Collier, M. S. Hirsch, P. Tebas, J. P. Sommadossi, and D. D. Richman. 2000. Drug susceptibility in HIV infection after viral rebound in patients receiving indinavir-containing regimens. JAMA 283:229-234. [DOI] [PubMed] [Google Scholar]
- 23.Havlir, D. V., C. Tierney, G. H. Friedland, R. B. Pollard, L. Smeaton, J. P. Sommadossi, L. Fox, H. Kessler, K. H. Fife, and D. D. Richman. 2000. In vivo antagonism with zidovudine plus stavudine combination therapy. J. Infect. Dis. 182:321-325. [DOI] [PubMed] [Google Scholar]
- 24.Hermankova, M., S. C. Ray, C. Ruff, M. Powell-Davis, R. Ingersoll, R. T. D'Aquila, T. C. Quinn, J. D. Siliciano, R. F. Siliciano, and D. Persaud. 2001. HIV-1 drug resistance profiles in children and adults with viral load of <50 copies/ml receiving combination therapy. JAMA 286:196-207. [DOI] [PubMed] [Google Scholar]
- 25.Hertogs, K., M.-P. de Béthune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoggard, P. G., S. Kewn, M. G. Barry, S. H. Khoo, and D. J. Back. 1997. Effects of drugs on 2′,3′-dideoxy-2′,3′-didehydrothymidine phosphorylation in vitro. Antimicrob. Agents Chemother. 41:1231-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan, A. H., M. Manchester, and R. Swanstrom. 1994. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J. Virol. 68:6782-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katzenstein, D. A., R. J. Bosch, N. Hellmann, N. Wang, L. Bacheler, and M. A. Albrecht. 2003. Phenotypic susceptibility and virological outcome in nucleoside-experienced patients receiving three or four antiretroviral drugs. AIDS 17:821-830. [DOI] [PubMed] [Google Scholar]
- 29.Kijak, G. H., V. Simon, P. Balfe, J. Vanderhoeven, S. E. Pampuro, C. Zala, C. Ochoa, P. Cahn, M. Markowitz, and H. Salomon. 2002. Origin of human immunodeficiency virus type 1 quasispecies emerging after antiretroviral treatment interruption in patients with therapeutic failure. J. Virol. 76:7000-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, R. B. 2003. Drug transporters in HIV therapy. Top. HIV Med. 11:136-139. [PubMed] [Google Scholar]
- 31.Kosel, B. W., and F. Aweeka. 2000. Drug interactions of antiretroviral agents. AIDS Clin. Rev. 2000:193-227. [PubMed] [Google Scholar]
- 32.Ledergerber, B., M. Egger, M. Opravil, A. Telenti, B. Hirschel, M. Battegay, P. Vernazza, P. Sudre, M. Flepp, H. Furrer, P. Francioli, and R. Weber. 1999. Clinical progression and virological failure on highly active antiretroviral therapy in HIV-1 patients: a prospective cohort study. Swiss HIV Cohort Study. Lancet 353:863-868. [DOI] [PubMed] [Google Scholar]
- 33.Lederman, M. M., and H. Valdez. 2000. Immune restoration with antiretroviral therapies: implications for clinical management. JAMA 284:223-228. [DOI] [PubMed] [Google Scholar]
- 34.Lucas, G. M., R. E. Chaisson, and R. D. Moore. 1999. Highly active antiretroviral therapy in a large urban clinic: risk factors for virologic failure and adverse drug reactions. Ann. Intern. Med. 131:81-87. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Picado, J., M. P. DePasquale, N. Kartsonis, G. J. Hanna, J. Wong, D. Finzi, E. Rosenberg, H. F. Gunthard, L. Sutton, A. Savara, C. J. Petropoulos, N. Hellmann, B. D. Walker, D. D. Richman, R. Siliciano, and R. T. D'Aquila. 2000. Antiretroviral resistance during successful therapy of HIV type 1 infection. Proc. Natl. Acad. Sci. USA 97:10948-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazzotta, F., C. S. Lo, C. Torti, C. Tinelli, P. Pierotti, F. Castelli, A. Lazzarin, G. Angarano, R. Maserati, N. Gianotti, N. Ladisa, E. Quiros-Roldan, A. R. Rinehart, and G. Carosi. 2003. Real versus virtual phenotype to guide treatment in heavily pretreated patients: 48-week follow-up of the Genotipo-Fenotipo di Resistenza (GenPheRex) trial. J. Acquir. Immune Defic. Syndr. 32:268-280. [DOI] [PubMed] [Google Scholar]
- 37.Molla, A., S. Vasavanonda, G. Kumar, H. L. Sham, M. Johnson, B. Grabowski, J. F. Denissen, W. Kohlbrenner, J. J. Plattner, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1998. Human serum attenuates the activity of protease inhibitors toward wild-type and mutant human immunodeficiency virus. Virology 250:255-262. [DOI] [PubMed] [Google Scholar]
- 38.Munro, S., and H. R. Pelham. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48:899-907. [DOI] [PubMed] [Google Scholar]
- 39.Nijhuis, M., S. Deeks, and C. Boucher. 2001. Implications of antiretroviral resistance on viral fitness. Curr. Opin. Infect. Dis. 14:23-28. [DOI] [PubMed] [Google Scholar]
- 40.Perelson, A. S., P. Essunger, Y. Cao, M. Vesanen, A. Hurley, K. Saksela, M. Markowitz, and D. D. Ho. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387:188-191. [DOI] [PubMed] [Google Scholar]
- 41.Persaud, D., T. Pierson, C. Ruff, D. Finzi, K. R. Chadwick, J. B. Margolick, A. Ruff, N. Hutton, S. Ray, and R. F. Siliciano. 2000. A stable latent reservoir for HIV-1 in resting CD4(+) T lymphocytes in infected children. J. Clin. Investig. 105:995-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierson, T. C., Y. Zhou, T. L. Kieffer, C. T. Ruff, C. Buck, and R. F. Siliciano. 2002. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J. Virol. 76:8518-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piliero, P. J. 2002. The utility of inhibitory quotients in determining the relative potency of protease inhibitors. AIDS 16:799-800. [DOI] [PubMed] [Google Scholar]
- 45.Prado, J. G., T. Wrin, J. Beauchaine, L. Ruiz, C. J. Petropoulos, S. D. Frost, B. Clotet, R. T. D'Aquila, and J. Martinez-Picado. 2002. Amprenavir-resistant HIV-1 exhibits lopinavir cross-resistance and reduced replication capacity. AIDS 16:1009-1017. [DOI] [PubMed] [Google Scholar]
- 46.Price, N., E. Smit, and D. Pillay. 2002. Resistance tests: what do clinical trials tell us? J. HIV Ther. 7:80-86. [PubMed] [Google Scholar]
- 47.Quiñones-Mateu, M. E., and E. J. Arts. 2001. HIV-1 fitness: implications for drug resistance, disease progression and global epidemic evolution, p. 134-170. In C. Kuiken, B. Foley, B. Hahn, P. Marx, F. McCuthan, J. Mellors, S. Wolinsky, and B. Korber (ed.), HIV sequence compendium. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 48.Resch, W., R. Ziermann, N. Parkin, A. Gamarnik, and R. Swanstrom. 2002. Nelfinavir-resistant, amprenavir-hypersusceptible strains of human immunodeficiency virus type 1 carrying an N88S mutation in protease have reduced infectivity, reduced replication capacity, and reduced fitness and process the Gag polyprotein precursor aberrantly. J. Virol. 76:8659-8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruff, C. T., S. C. Ray, P. Kwon, R. Zinn, A. Pendleton, N. Hutton, R. Ashworth, S. Gange, T. C. Quinn, R. F. Siliciano, and D. Persaud. 2002. Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J. Virol. 76:9481-9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidt, B., H. Walter, N. Zeitler, and K. Korn. 2002. Genotypic drug resistance interpretation systems—the cutting edge of antiretroviral therapy. AIDS Rev. 4:148-156. [PubMed] [Google Scholar]
- 51.Shafer, R. W. 2002. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin. Microbiol. Rev. 15:247-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shulman, N. S., M. D. Hughes, M. A. Winters, R. W. Shafer, A. R. Zolopa, N. S. Hellmann, M. Bates, J. M. Whitcomb, and D. A. Katzenstein. 2002. Subtle decreases in stavudine phenotypic susceptibility predict poor virologic response to stavudine monotherapy in zidovudine-experienced patients. J. Acquir. Immune Defic. Syndr. 31:121-127. [DOI] [PubMed] [Google Scholar]
- 53.Siliciano, J. D., J. Kajdas, D. Finzi, T. C. Quinn, K. Chadwick, J. B. Margolick, C. Kovacs, S. J. Gange, and R. F. Siliciano. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4(+) T cells. Nat. Med. 9:727-728. [DOI] [PubMed] [Google Scholar]
- 54.Simon, V., N. Padte, D. Murray, J. Vanderhoeven, T. Wrin, N. Parkin, M. Di Mascio, and M. Markowitz. 2003. Infectivity and replication capacity of drug-resistant human immunodeficiency virus type 1 variants isolated during primary infection. J. Virol. 77:7736-7745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staszewski, S., J. Morales-Ramirez, K. T. Tashima, A. Rachlis, D. Skiest, J. Stanford, R. Stryker, P. Johnson, D. F. Labriola, D. Farina, D. J. Manion, and N. M. Ruiz. 1999. Efavirenz plus zidovudine and lamivudine, efavirenz plus indinavir, and indinavir plus zidovudine and lamivudine in the treatment of HIV-1 infection in adults. Study 006 Team. N. Engl. J. Med. 341:1865-1873. [DOI] [PubMed] [Google Scholar]
- 56.St. Clair, M. H., C. A. Richards, T. Spector, K. J. Weinhold, W. H. Miller, A. J. Langlois, and P. A. Furman. 1987. 3′-Azido-3′-deoxythymidine triphosphate as an inhibitor and substrate of purified human immunodeficiency virus reverse transcriptase. Antimicrob. Agents Chemother. 31:1972-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strain, M. C., H. F. Gunthard, D. V. Havlir, C. C. Ignacio, D. M. Smith, A. J. Leigh-Brown, T. R. Macaranas, R. Y. Lam, O. A. Daly, M. Fischer, M. Opravil, H. Levine, L. Bacheler, C. A. Spina, D. D. Richman, and J. K. Wong. 2003. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc. Natl. Acad. Sci. USA 100:4819-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Webb, J. L. 1963. Enzymes and metabolic inhibitors, p. 66-79, 487-512. Academic Press, New York, N.Y.
- 59.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 60.Zennou, V., F. Mammano, S. Paulous, D. Mathez, and F. Clavel. 1998. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J. Virol. 72:3300-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]