This work reports on the reconstruction of the three diploid Brassica genomes with seven chromosomes involved in the origin of the hexaploid Brassica ancestor with 42 chromosomes. The hexaploid genome evolved through extensive genome diploidization toward extant diploid-like Brassica genomes.
Abstract
The genus Brassica includes several important agricultural and horticultural crops. Their current genome structures were shaped by whole-genome triplication followed by extensive diploidization. The availability of several crucifer genome sequences, especially that of Chinese cabbage (Brassica rapa), enables study of the evolution of the mesohexaploid Brassica genomes from their diploid progenitors. We reconstructed three ancestral subgenomes of B. rapa (n = 10) by comparing its whole-genome sequence to ancestral and extant Brassicaceae genomes. All three B. rapa paleogenomes apparently consisted of seven chromosomes, similar to the ancestral translocation Proto-Calepineae Karyotype (tPCK; n = 7), which is the evolutionarily younger variant of the Proto-Calepineae Karyotype (n = 7). Based on comparative analysis of genome sequences or linkage maps of Brassica oleracea, Brassica nigra, radish (Raphanus sativus), and other closely related species, we propose a two-step merging of three tPCK-like genomes to form the hexaploid ancestor of the tribe Brassiceae with 42 chromosomes. Subsequent diversification of the Brassiceae was marked by extensive genome reshuffling and chromosome number reduction mediated by translocation events and followed by loss and/or inactivation of centromeres. Furthermore, via interspecies genome comparison, we refined intervals for seven of the genomic blocks of the Ancestral Crucifer Karyotype (n = 8), thus revising the key reference genome for evolutionary genomics of crucifers.
INTRODUCTION
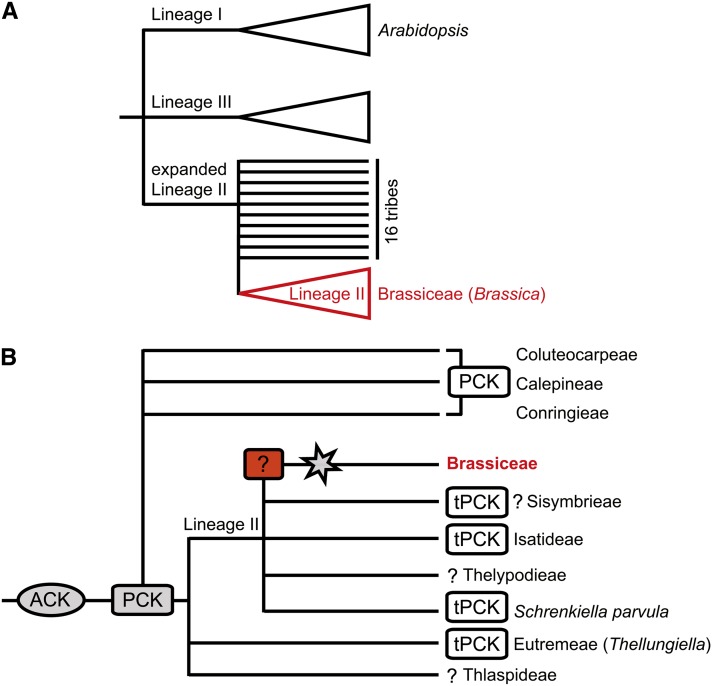
The genus Brassica currently comprises 38 species and numerous varieties, many of which are important crops or weeds. The six species that make up the so-called “Triangle of U” are of particular economic importance because of their cultivation as vegetables, condiments, and sources of oilseed. These species are the diploids Brassica rapa (AA genome), Brassica nigra (BB), and Brassica oleracea (CC) as well as the allotetraploid species Brassica juncea (AABB), Brassica napus (AACC), and Brassica carinata (BBCC). Brassica is one of 47 genera in the monophyletic tribe Brassiceae. This tribe, which contains ∼238 species primarily distributed in the Mediterranean region, southwestern Asia, and northern Africa (Prakash and Hinata, 1980; Al-Shehbaz, 2012), belongs to lineage II, one of three well-supported lineages recognized within the Brassicaceae family (Figure 1A). Within lineage II, the Brassiceae have been grouped together with tribes Sisymbrieae, Isatideae, and Thelypodieae (Schizopetaleae) (Al-Shehbaz et al., 2006; Beilstein et al., 2006, 2008; Franzke et al., 2011). However, the phylogenetic relationships within lineage II, in particular the position of the closely related tribes Eutremeae and Thlaspideae, remain unclear, and more than 20 loosely interrelated tribes have been described within an expanded lineage II (Figure 1B) (Franzke et al., 2011).
Figure 1.
Currently Hypothesized Phylogenetic Relationships among the Discussed Brassicaceae Taxa and Purported Evolutionary Relationships among Ancestral and Extant Genomes.
(A) Schematic diagram of phylogenetic relationships among the three major Brassicaceae lineages (I, II, and III).
(B) Tentative genome evolution in Lineage II and five closely related tribes. The star refers to the Brassiceae-specific whole-genome triplication. Question marks indicate unknown or assumed ancestral genomes. ACK (n = 8) and tPCK/PCK (n = 7). Adopted and modified from Franzke et al. (2011) and Mandáková and Lysak (2008).
[See online article for color version of this figure.]
Early chromosomal studies indicated that the diploid Brassica species might actually represent balanced secondary polyploids (Catcheside, 1934; Roebbelen, 1960). Later comparative restriction fragment length polymorphism mapping among these species and between Brassica species and Arabidopsis thaliana (Lagercrantz and Lydiate, 1996; Lagercrantz, 1998) suggested that diploid Brassica genomes descended from a hexaploid ancestor. All Brassiceae species analyzed to date contain either three or six copies of orthologous genomic regions of A. thaliana (Lysak et al., 2005, 2007; Parkin et al., 2005). These findings provide compelling evidence for an ancestral whole-genome triplication in the ancestry of the monophyletic tribe Brassiceae. More recently, analysis of the draft genome sequence of B. rapa (n = 10) revealed the three subgenomes of the hexaploid ancestor (Wang et al., 2011). A comparison of collinearity between the 10 B. rapa chromosomes and the five A. thaliana chromosomes using 24 ancestral genomic blocks (GBs) of the Ancestral Crucifer Karyotype (ACK; n = 8; Schranz et al., 2006) revealed three syntenic copies of each A. thaliana GB, with only one exception, in the B. rapa genome (Wang et al., 2011). Because the three GB copies differ regarding their rate of gene loss (fractionation), blocks were classified according to their gene density as belonging to the least fractionated (LF), the medium fractionated (MF1), and the most fractionated (MF2) subgenome. Wang et al. (2011) suggested a two-step origin of B. rapa involving a tetraploidization followed by substantial genome fractionation (subgenomes MF1 and MF2) and subsequent hybridization with a third, less-fractionated genome. This two-step model gained support from a comparison of the frequency of deletions in exon sequences between the three B. rapa subgenomes (Tang et al., 2012).
Karyotype and genome evolution in the expanded lineage II was analyzed by Mandáková and Lysak (2008). Comparative chromosome painting using A. thaliana BAC contigs demonstrated that the six analyzed tribes (i.e., Calepineae, Coluteocarpeae, Conringieae, Eutremeae, Isatideae, and Sisymbrieae) descended from a common ancestor with the Proto-Calepineae Karyotype (PCK; n = 7). While Calepineae, Coluteocarpeae (synonym of Noccaeeae), and Conringieae taxa retain the PCK genome structure, the tribes Eutremeae, Isatideae, and Sisymbrieae display an additional whole-arm translocation (Figures 1B and 2A). This younger ancestral genome is henceforth referred to as the translocation PCK (tPCK). The tPCK genome structure was recently confirmed by whole-genome sequencing of Schrenkiella parvula (synonym of Thellungiella parvula, formerly placed in the Eutremeae) (Dassanayake et al., 2011) and Thellungiella salsuginea (Eutremeae) (Wu et al., 2012). The PCK shares five chromosomes with the ACK and structurally closely resembles the extant karyotypes of Arabidopsis lyrata and Capsella rubella (Lysak et al., 2006; Schranz et al., 2006; Hu et al., 2011). It has therefore been suggested that either the PCK or ACK is descended from a common ancestor or, more likely, that the seven PCK chromosomes are derived from the eight chromosomes of the ACK (Mandáková and Lysak, 2008) (Figure 1B).
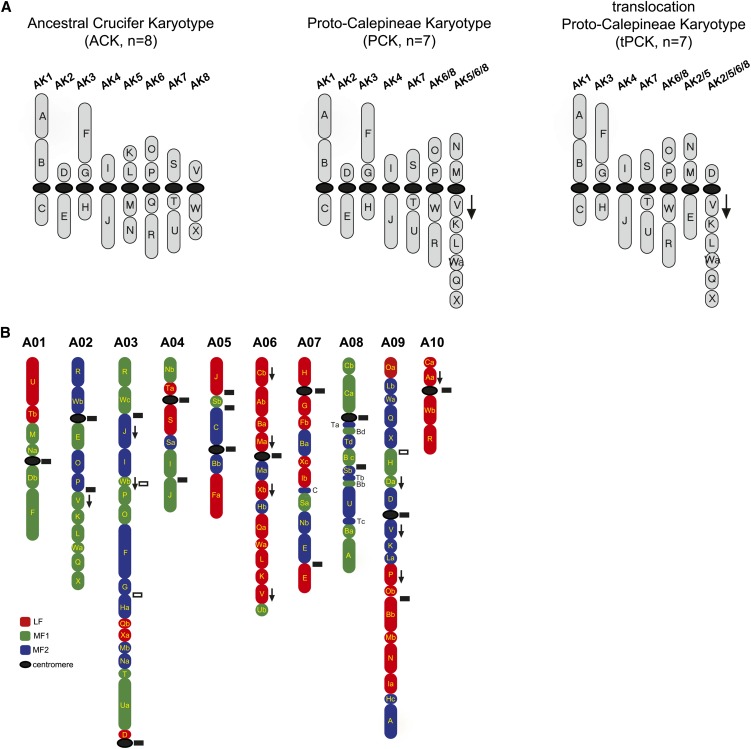
Figure 2.
Ancestral Brassicaceae Genomes and the Distribution of Ancestral GBs along 10 Chromosomes of B. rapa.
(A) The ancestral genomes ACK, PCK, and tPCK, each comprising 24 ancestral GBs. In the PCK and tPCK, chromosome labels reflect the presumed origin of the two karyotypes from the more ancestral ACK genome.
(B) Ten B. rapa chromosomes (A01 to A10) consisting of 24 triplicated GBs (71 blocks in total, as one copy of block G was lost) assigned to subgenomes LF (red), MF1 (green), and MF2 (blue). Two or more segments of a single block are labeled by lowercase letters (a, b, etc.). Downward-pointing arrows are adjacent to GBs that were inverted relative to other block(s), which originated from a single ACK chromosome. Detected and undetected ancestral centromeres in the B. rapa genome are shown as black and white small rectangles, respectively.
For more than half a century, the chromosome number and genomic structure of the hexaploid Brassica ancestor and its three parental subgenomes have been a matter of debate. Several chromosome numbers, ranging from x = 3 to 7, have been proposed for ancestral Brassica subgenomes (Roebbelen, 1960; Truco et al., 1996; reviewed in Prakash and Hinata, 1980). Based on a comparison of B. rapa, B. oleracea, and B. nigra linkage maps, Truco et al. (1996) suggested that all three subgenomes were derived from an ancestor with five, six, or seven chromosomes. Although those authors favored six ancestral chromosomes (W1 to W6), a seventh chromosome (W7) could not be ruled out. Comparison of the PCK with the structure of the B. rapa subgenome in B. napus (Parkin et al., 2005) revealed two shared rearrangements, thus suggesting that two or all three subgenomes of the hexaploid ancestor of the Brassiceae corresponded to either PCK or tPCK (Mandáková and Lysak, 2008).
The aim of this study was to elucidate the structure of the three ancestral subgenomes of the hexaploid ancestor of the Brassiceae. Toward that aim, structural genome analyses of B. rapa in comparison with ancestral and extant crucifer genomes were performed. Specifically, we tested whether the PCK and/or tPCK represent the karyotype of the diploid ancestors of the mesohexaploid B. rapa, the genus Brassica, and the tribe Brassiceae.
RESULTS
Deciphering the Ancestral Diploid Karyotypes of B. rapa
Based on the previously determined syntenic relationship of the three B. rapa subgenomes (LF, MF1, and MF2) to the A. thaliana genome (Wang et al., 2011; Cheng et al., 2012b), 71 of 72 expected GBs (3 × 24) were detected in the B. rapa genome (Figure 2B; see Supplemental Table 1 online). Of the 24 blocks, 23 were present in three copies and one (block G) in only two copies. Among the 71 GBs, 48 (67.6%) exhibited continuous sequence collinearity between B. rapa and A. thaliana, 21 (29.6%) were disrupted into at least two segments, and two (2.8%; one copy each of blocks E and T) contained sequence deletions (see Supplemental Tables 1 and 2 online). Analysis of GB integrity did not reveal significant differences in the degree of rearrangements among the three B. rapa subgenomes (see Supplemental Table 3 and Supplemental Data Set 1 online).
We compared the structure of the ACK, PCK, and tPCK (Figure 2A) with that of the three B. rapa subgenomes (Figure 2B). Using gene densities characteristic for each of the three B. rapa subgenomes and methods previously described (Wang et al., 2011; Cheng et al., 2012b), we then reconstructed three tentative B. rapa ancestral subgenomes by projecting the 71 GBs onto the ACK genome and the seven chromosomes of the PCK and tPCK. When a modern genome is compared with ancestral ones, the more recent the ancestor, the greater the expected degree of genomic continuity with the present-day genome. To examine genome continuity, we defined two adjacent GBs as a GB association and compared all GB associations between B. rapa and the ancestral genomes ACK, PCK, and tPCK. There are 16 GB associations in the ACK genome and 17 associations in the PCK and tPCK genomes (Table 1, Figure 2A). In B. rapa, 10, 15, and 16 GB associations were identified between ACK, PCK, and tPCK, respectively. Of the 10 ACK block associations, four were present in three copies in B. rapa, while six were present in two copies. Of the 15 GB associations in PCK, eight were present in three copies, six in two, and one as a single copy. Of the 16 tPCK block associations, eight were present in three copies, six in two copies, and two as single copies (Table 1; see Supplemental Table 4 online).
Table 1. Comparison of GB Associations Shared between B. rapa and Three Ancestral Crucifer Genomes (ACK, PCK, and tPCK).
|
ACK (n = 8) |
PCK (n = 7) |
tPCK (n = 7) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr.a | GB Assoc.b | No.c | B. rapa Chr. | Chr. | GB Assoc. | No. | B. rapa Chr. | Chr. | GB Assoc. | No. | B. rapa Chr. |
| AK1 | A/B | 2 | A06, A08 | AK1 | A/B | 2 | A06, A08 | AK1 | A/B | 2 | A06, A08 |
| B/C | 2 | A05, A08 | B/C | 2 | A05, A08 | B/C | 2 | A05, A08 | |||
| AK2 | D/E | 0 | – | AK2 | D/E | 0 | – | AK2/5 | N/M | 3 | A01, A03, A09 |
| AK3 | F/G | 2 | A03, A07 | AK3 | F/G | 2 | A03, A07 | M/E | 0 | – | |
| G/H | 2 | A03, A07 | G/H | 2 | A03, A07 | AK3 | F/G | 2 | A03, A07 | ||
| AK4 | I/J | 2 | A03, A04 | AK4 | I/J | 2 | A03, A04 | G/H | 2 | A03, A07 | |
| AK5 | K/L | 3 | A02, A06, A09 | AK7 | S/T | 2 | A04, A08 | AK4 | I/J | 2 | A03, A04 |
| L/M | 0 | – | T/U | 3 | A01, A03, A08 | AK7 | S/T | 2 | A04, A08 | ||
| M/N | 3 | A01, A03, A09 | AK6/8 | O/P | 3 | A02, A03, A09 | T/U | 3 | A01, A03, A08 | ||
| AK6 | O/P | 3 | A02, A03, A09 | P/W | 1 | A03 | AK6/8 | O/P | 3 | A02, A03, A09 | |
| P/Q | 0 | – | W/R | 3 | A02, A03, A10 | P/W | 1 | A03 | |||
| Q/R | 0 | – | AK5/6/8 | N/M | 3 | A01, A03, A09 | W/R | 3 | A02, A03, A10 | ||
| AK7 | S/T | 2 | A04, A08 | M/V | 0 | - | AK2/5/6/8 | D/V | 1 | A09 | |
| T/U | 3 | A01, A03, A08 | V/K | 3 | A02, A06, A09 | V/K | 3 | A02, A06, A09 | |||
| AK8 | V/W | 0 | – | K/L | 3 | A02, A06, A09 | K/L | 3 | A02, A06, A09 | ||
| W/X | 0 | – | L/Q | 3 | A02, A06, A09 | L/Q | 3 | A02, A06, A09 | |||
| Q/X | 3 | A02, A03, A09 | Q/X | 3 | A02, A03, A09 | ||||||
Ancestral chromosome (Chr.).
GB association.
Copy number of GB association in the B. rapa genome.
–, nonexistent GB association.
Block associations V/K/L/Wa/Q/X and O/P/W/R, specific for both PCK and tPCK, were identified in all three B. rapa subgenomes (Figure 2B, Table 1). Three genomic copies of the V/K/L/Wa/Q/X association were found on chromosomes A02 (MF1), A06 (LF), and A09 (MF2), although further rearrangements occurred on A06 and A09. Copies of the O/P/W/R association were found but rearranged on chromosomes A02 (MF2) and A03 (MF1). A third O/P/W/R copy was split between chromosomes A09 (blocks O/P) and A10 (W/R).
tPCK contained two specific GB associations, D/V and M/E, resulting from a whole-arm reciprocal translocation between chromosomes AK2 and AK5/6/8 within the PCK genome (Figure 2A; Mandáková and Lysak, 2008). In the MF2 subgenome, the D/V association was identified on chromosome A09. In the LF subgenome, blocks D and V were split between chromosomes A03 and A06 due to an intersubgenomic translocation between block D (LF) and a part of block U (MF1) (see Supplemental Figure 1A online). The M/E association is not present in B. rapa. However, we propose that the present-day block associations M(LF)/M(MF2) (A06) and E(LF)/E(MF2) (A07) are the products of intersubgenomic translocations between the duplicate M/E regions of the ancestral tPCK-like genomes (see Supplemental Figure 1B online). Furthermore, evidence that the MF1 subgenome also originated from the tPCK arises from a comparison of B. rapa and B. nigra genome structures with respect to the M/E association (Panjabi et al., 2008). Based on chromosome synteny, the block association R/W/M/O/P/E on B2 in B. nigra is syntenic to R/W/E(MF1)/O/P on chromosome A02 in B. rapa (see Supplemental Figure 2 online). Although the M/E association was interrupted by blocks O and P on chromosome B2, the colocalization of M/E with blocks O, P, R, and W on the same chromosome supports the idea that it originated from the ancestral associations O/P/W/R and M/E.
The larger number of GB associations shared between B. rapa and PCK/tPCK compared with those between B. rapa and ACK, as well as the tPCK-specific GB associations and chromosome breakpoints found in B. rapa, indicate that the three B. rapa subgenomes bear a close structural resemblance to the seven chromosomes of the tPCK.
To facilitate the retrieval of gene sets of the three tPCK-like subgenomes in B. rapa (see Supplemental Figure 3 online) with mutual homoeology and synteny to A. thaliana orthologs, we developed a Web tool, Search Syntenic Genes Br-tPCK, embedded in the BRAD Brassica Database (BRAD; http://brassicadb.org/brad/searchSyntenytPCK.php). There were a total of 12,914, 8905, and 7719 genes in the LF, MF1, and MF2 subgenomes, respectively, syntenic to A. thaliana (see Supplemental Table 5 online).
Paleocentromeres in the B. rapa Genome
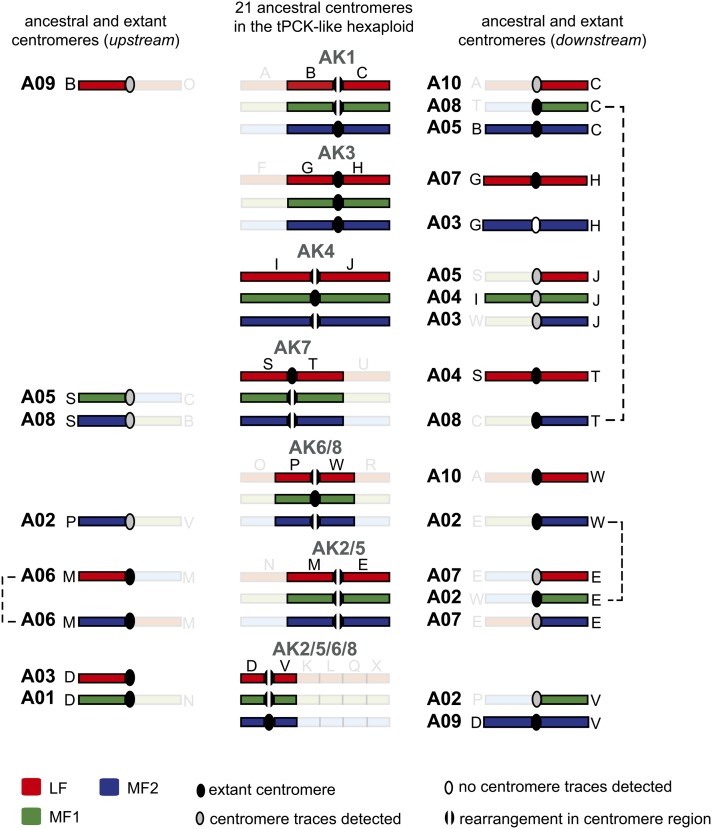
Paleocentromere sequence analysis indicated that the 10 extant B. rapa centromeres were all inherited from the triplicated tPCK-like genomes. Using the B. rapa genome sequence, we analyzed the fate of paleocentromeres corresponding to 21 ancestral chromosomes of the three tPCK-like genomes. After aligning (peri)centromere-specific repeats, including centromeric satellite repeats CentBr (Lim et al., 2005; Koo et al., 2011), PCRBr, and TR238 (Lim et al., 2007), to the B. rapa genome sequence, signals were detected for 18 paleocentromere regions. The paleocentromeres of three ancestral chromosomes [AK3(MF1), AK3(MF2), and AK6/8(MF1)] could not be located (Figures 2B and 3; see Supplemental Data Set 2 online).
Figure 3.
The Fate of 21 Paleocentromeres in the Triplicated Ancestral Genome of B. rapa.
The central column shows triplicated tPCK-like chromosomes with paleocentromeres and two adjacent GBs highlighted. The left and right columns display the extant or purported location of the ancestral centromeres on B. rapa chromosomes. All blocks that are not ancestrally adjacent to paleocentromeres are shown in semitransparent colors. Three instances of extant centromeres having an equally possible origin from two different paleocentromeres are marked by connecting dashed lines.
Among the 18 detected paleocentromeres, 10 were involved in the origin of the 10 B. rapa centromeres (Table 2, Figures 2B and 3). The centromeres on chromosomes A04, A05, A07, and A09 correspond to ancestral centromeres of AK7, AK1, AK3, and AK2/5/6/8, respectively, with two ancestrally associated GBs flanking the centromeres. The centromeres of A01, A03, and A10 were derived via a translocation or pericentric inversion event from paleocentromeres of AK2/5/6/8, AK2/5/6/8, and AK6/8, respectively, leaving only one of the two expected ancestral GBs associated with the centromere. The origin of centromeres on A02, A06, and A08 is directly linked to a reduction in chromosome number. For these centromeres, we inferred translocation events with breakpoints within the (peri)centromeric regions of two different ancestral chromosomes resulting in the loss of one of the two paleocentromeres. The available data do not allow us to determine which three of the six participating paleocentromeres remained active in B. rapa (Table 2, Figure 3).
Table 2. The Inheritance of 10 B. rapa Centromeres from the Triplicated Ancestral tPCK-Like Genome (n = 21).
| B. rapa Chromosome | Ancestral Chromosome | B. rapa Subgenome |
|---|---|---|
| A01 | AK2/5/6/8 | MF1 |
| A02 | AK6/8 or AK2/5 | MF2 or MF1 |
| A03 | AK2/5/6/8 | LF |
| A04 | AK7 | LF |
| A05 | AK1 | MF2 |
| A06 | AK2/5 or AK2/5 | LF or MF2 |
| A07 | AK3 | LF |
| A08 | AK1 or AK7 | MF1 or MF2 |
| A09 | AK2/5/6/8 | MF2 |
| A10 | AK6/8 | LF |
Of the eight remaining ancestral centromeres, six were disrupted by translocation events, with sequence traces detectable at four paleocentromere regions (Figure 3). Chromosomes A03 and A04 contained two entirely conserved ancestral chromosomes with inactivated/deleted centromeres. The AK3 centromere left no sequence remnants between blocks GMF2 and HMF2 within A03, whereas centromere sequence traces (PCRBr and TR238 in the boundary of J/IMF1) corresponding to AK4 (see Supplemental Data Set 2 online) were detected on A04 (Figures 2B and 3).
Comparison of B. rapa Ancestral Genomes with Other Brassicaceae Genomes
To obtain further insights into the origin and phylogenetic context of the B. rapa ancestral genomes, we investigated whether the tPCK-specific GB associations D/V/K/L/Wa/Q/X, O/P/W/R, M/E, and D/V are present in genomes of other Brassica and Brassiceae species, as well as other lineage II species (i.e., B. oleracea, B. nigra, radish (Raphanus sativus), Sinapis alba, Caulanthus amplexicaulis, S. parvula, and T. salsuginea; Figure 1B). Three copies of the first two tPCK-specific block associations were identified in B. oleracea assembled scaffolds (see Supplemental Table 6 online). In the B. nigra genome analyzed within the allotetraploid B. juncea, two copies of V/K/L/(Wa)/Q/X were located on chromosomes B1 and B6, whereas the third copy was split between B4 and B8. Similarly, two copies of O/P/W/R were located on B2 and B3, while the third copy was rearranged on chromosome B8 (Panjabi et al., 2008) (see Supplemental Figure 2 online). Furthermore, the D/V association was found on one scaffold of B. oleracea (see Supplemental Table 6 online) as well as on chromosomes B1 and B6 in B. nigra (Panjabi et al., 2008) (see Supplemental Figure 2 online). Similar to B. rapa, the M/E association was not found in B. oleracea; however, one copy of M/E was rearranged with O/P/W/R on B2 in B. nigra (Panjabi et al., 2008) (see Supplemental Figure 2 online). These analyses suggest that the three Brassica genomes (A, B, and C) descended from a common hexaploid ancestor that originated from the merger of three tPCK-like ancestral genomes.
For radish, we used an EST linkage map (Shirasawa et al., 2011) to align EST sequences to A. thaliana genes. These results were then used to determine the position of 24 GBs on the nine radish chromosomes (see Supplemental Table 7 online). Most of the 24 GBs (75%) were present in three copies in the radish genome. The two tPCK-specific block associations [V/K/L/(Wa)/Q/X and O/P/W/R] were detected in triplicate on chromosomes LG2, LG4, and LG6, and LG5, LG6, and LG8, respectively. In addition, two copies of V/K/L/(Wa)/Q/X blocks were associated with D on chromosomes LG2 and LG6, and one copy of O/P/W/R was rearranged with M/E on LG5 (see Supplemental Table 7 online). This suggests that at least two tPCK-like genomes were involved in the origin of the radish genome.
In the diploid genome of S. parvula, tPCK-specific GB associations V/K/L/Wa/Q/X, O/P/W/R, and M/E were located on chromosomes SpChr2, SpChr6, and SpChr5, respectively (Dassanayake et al., 2011). tPCK-specific block associations were found to be conserved also in the T. salsuginea genome (Wu et al., 2012).
Comparative genome mapping in white mustard (S. alba; Nelson et al., 2011) and C. amplexicaulis (Burrell et al., 2011) also revealed tPCK-specific arrangements of GBs. However, because of extensive genome rearrangements and the limited resolution of the linkage maps, no clear conclusions could be drawn for the two species. In S. alba, V/K/L/V/Q on chromosomes S10 and S11 and O/P/W/R on S09 were rearranged and not readily identifiable. The M/E association was identified on S02 (Nelson et al., 2011), but the D/V association was not found. In Caulanthus, the block association D/V/K/L/(Wa)/Q/X on LG3 was fragmented to such extent that it could not be clearly identified (i.e., blocks V, L, and Q were disrupted, while K remained undetected). The O/P/W/R association was observed on LG2 (with blocks P and R fragmented), whereas the M/E association was not present (Burrell et al., 2011).
Analysis of Synonymous Substitution Rates
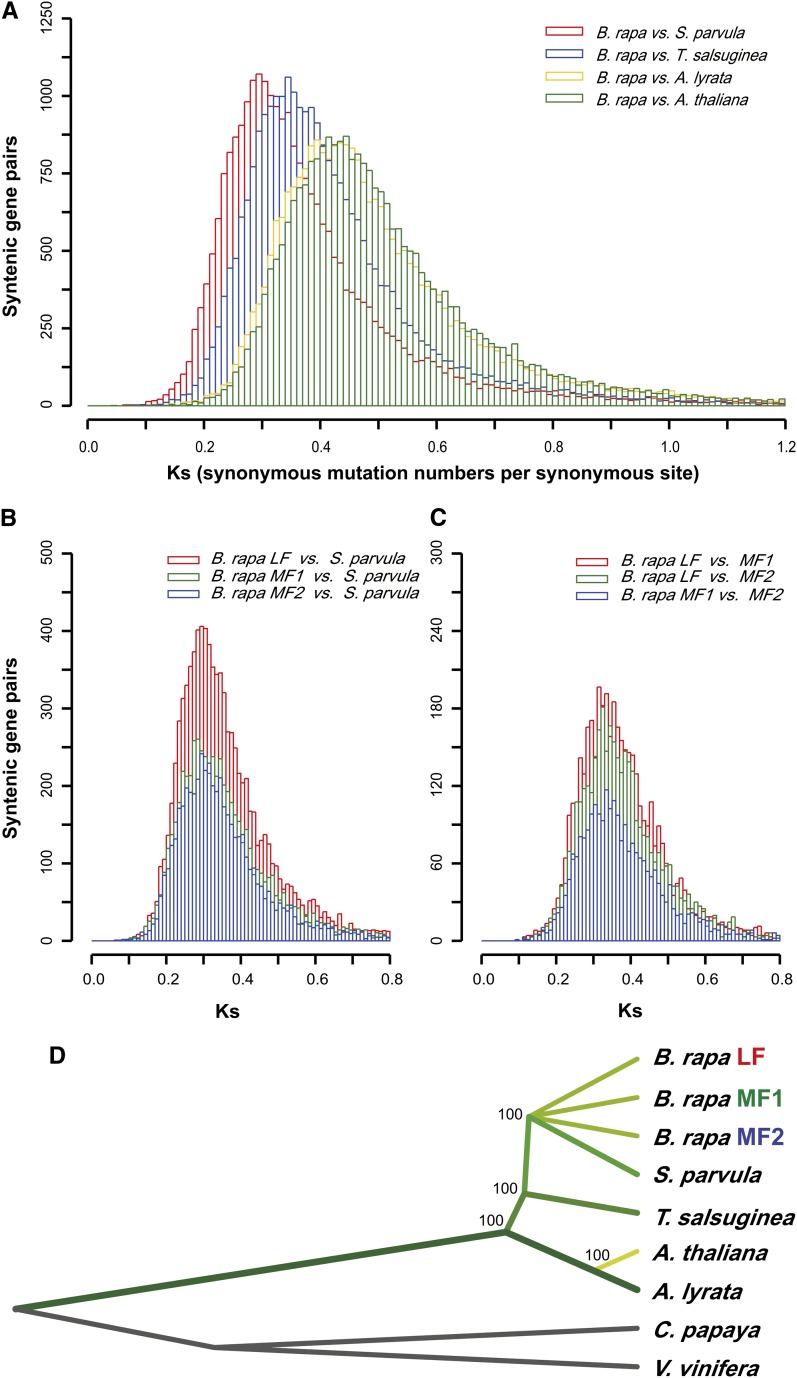
Using the syntenic orthologs identified for B. rapa, A. thaliana, A. lyrata, S. parvula, and T. salsuginea, we calculated Ks values (the number of substitutions per synonymous site) for B. rapa relative to the other crucifer species. Ks for B. rapa relative to S. parvula was smaller (∼0.29 to 0.31, ∼10 million years of divergence; Koch et al., 2000) than that relative to T. salsuginea (∼0.34 to 0.36, ∼11.7 million years), A. lyrata (∼0.41 to 0.45, ∼14.3 million years), and A. thaliana (∼0.42 to 0.45, ∼14.5 million years) (Figure 4A). Based on these calculations, B. rapa is thus phylogenetically closer to S. parvula than to T. salsuginea or to the genus Arabidopsis. Ks values for each B. rapa subgenome relative to S. parvula ranged from 0.29 to 0.31 (Figure 4B), while those for the subgenomes relative to one another ranged from 0.29 to 0.33 (i.e., ∼10.3 million years) (Figure 4C).
Figure 4.
Pairwise Comparison of Ks Values and a Phylogenetic Tree Based on Ks Loci.
(A) The distribution of Ks values between B. rapa and crucifer species T. salsuginea, S. parvula, A. thaliana, and A. lyrata.
(B) The distribution of Ks values between each of the three B. rapa subgenomes and S. parvula.
(C) The distribution of Ks values between pairs of the three B. rapa subgenomes.
(D) Phylogenetic tree for the four crucifer species and the three B. rapa subgenomes, with non-Brassicaceae species papaya and grape used as outgroups. The tree was constructed based on Ks loci for all syntenic orthologs shared among the seven species. Numbers refer to bootstrap values.
To investigate the evolutionary relationship of A. thaliana, A. lyrata, S. parvula, T. salsuginea, and the three B. rapa subgenomes, we performed a phylogenetic analysis using shared synonymous single nucleotide polymorphisms for these five genomes and the two outgroup species papaya (Carica papaya) and grape (Vitis vinifera). From the 591 syntenic orthologs shared by the three B. rapa subgenomes and the six other species, 63,239 Ks loci (i.e., synonymous single nucleotide polymorphisms; see Methods) were identified and concatenated into a data set that was subjected to phylogenetic analysis. Based on the resulting tree, Brassiceae and Arabidopsis ancestors diverged prior to the Brassiceae-Thellungiella split. The Brassiceae-specific whole-genome triplication occurred near the time of the split between the progenitors of the Brassiceae and Schrenkiella (Figure 4D).
Reevaluation of Block Association V/K/L/Wa/Q/X and Interval Refinement of Seven GBs
The results of the multispecies comparison prompted a reevaluation of the V/K/L/Q/X block association considered specific for the x = 7 tribes of expanded lineage II (Mandáková and Lysak, 2008). This association was identified on chromosome Sp2 in S. parvula, albeit with a fragment of block V between L and Q (V/K/L/V'/Q/X) (Dassanayake et al., 2011). The same rearrangement was found on B. rapa chromosomes A02, A06, and A09 (Figure 5A), B. oleracea (see Supplemental Table 6 online), and radish (Li et al., 2011; Shirasawa et al., 2011). We analyzed this chromosome region by performing an interspecies comparison between B. rapa, B. oleracea, S. parvula, A. thaliana, and A. lyrata, with the A. lyrata genome (Hu et al., 2011) serving as a proxy for the ACK genome (n = 8). The presumed V' region turned out to be part of block W, located on the bottom arm of the AK8 (Al8) chromosome and adjacent to the centromere (Figure 5A). The origin of the V/K/L/Wa/Q/X rearrangement, which is specific to the ancestral PCK genome, is consistent with the previously proposed scenario (Mandáková and Lysak, 2008), except that one of the breakpoints is actually positioned within block W (Figure 5B; see Supplemental Figure 4A online). An alternative scenario involves a paracentric inversion of blocks V/K/L followed by inactivation/removal of the AK8 paleocentromere (see Supplemental Figure 4B online). These results, together with multispecies comparisons of syntenic chromosomes, allowed us to redefine the boundaries of blocks V and W (Table 3, Figure 5A).
Figure 5.
Redefinition of Intervals for GBs H, I, Q, R, V, W, and X, and the Interspecies Collinearity of the Diagnostic Block Association V/K/L/Wa/Q/X.
Block intervals were redefined based on interspecies comparison among genomes of A. lyrata (Scaffold_8 on chromosome AK8), A. thaliana (At5), S. parvula (Sp2), and B. rapa (A02, A06, and A09).
(A) The diagnostic block association V/K/L/Wa/Q/X. The split of block W into Wa and Wb preceded the formation of the block association V/K/L/Wa/Q/X in B. rapa, S. parvula, and probably all species of the expanded lineage II. Block V is inverted in A. thaliana, S. parvula, and B. rapa compared with the ancestral ACK-like orientation in A. lyrata.
(B) Refined intervals of seven ancestral GBs shown as corresponding A. thaliana genes located on chromosomes At2 and At5. GB intervals as defined by Schranz et al. (2006) appear in black on the left, whereas newly refined intervals are listed in red on the right. The boundary between Wa and Wb is located between AT5G49630 and AT5G49640.
Table 3. Interval Redefinition of Seven GBs in the ACK.
| GB | AK Chromosome | Interval | |
|---|---|---|---|
| A | 1 | AT1G01560 | AT1G19330 |
| B | 1 | AT1G19850 | AT1G36240 |
| C | 1 | AT1G43600 | AT1G56120 |
| D | 2 | AT1G63770 | AT1G56530 |
| E | 2 | AT1G65040 | AT1G80420 |
| F | 3 | AT3G01040 | AT3G25520 |
| G | 3 | AT2G05170 | AT2G07690 |
| H* | 3 | AT2G15670 | AT2G20900* |
| I* | 4 | AT2G20920* | AT2G28910 |
| J | 4 | AT2G31040 | AT2G47730 |
| K | 5 | AT2G01250 | AT2G03750 |
| L | 5 | AT3G25855 | AT3G29770 |
| M | 5 | AT3G43740 | AT3G49970 |
| N | 5 | AT3G50950 | AT3G62790 |
| O | 6 | AT4G00030 | AT4G04955 |
| P | 6 | AT4G12070 | AT4G08690 |
| Q* | 6 | AT5G28885* | AT5G23010* |
| R* | 6 | AT5G23000* | AT5G01010* |
| S | 7 | AT5G41900 | AT5G33210 |
| T | 7 | AT4G12750 | AT4G16143 |
| U | 7 | AT4G16250 | AT4G38770 |
| V* | 8 | AT5G42130* | AT5G47810* |
| W* | 8 | AT5G47820* | AT5G60800* |
| X* | 8 | AT5G60805* | AT5G67385 |
Newly defined GB intervals based on multigenome comparison are indicated by an asterisk. Intervals of the 17 remaining blocks follow Schranz et al. (2006). Each GB interval is defined by two A. thaliana genes.
By comparing the boundaries of blocks H, I, Q, R, V, W, and X reported by Schranz et al. (2006) with corresponding regions and/or breakpoints within genomes of B. rapa, A. lyrata, S. parvula, and A. thaliana (see Supplemental Data Set 3 online), we revised intervals of the seven blocks of ACK (Table 3, Figure 5B).
DISCUSSION
We compared the whole-genome sequence of B. rapa (Wang et al., 2011) with genome sequences or genetic maps of other crucifer species, including A. thaliana, A. lyrata, B. oleracea, B. nigra, C. amplexicaulis, radish, S. alba, S. parvula, and T. salsuginea, as well as with previously proposed Brassicaceae ancestral genomes (Schranz et al., 2006; Mandáková and Lysak, 2008; Panjabi et al., 2008; Burrell et al., 2011; Dassanayake et al., 2011; Hu et al., 2011; Li et al., 2011; Nelson et al., 2011; Shirasawa et al., 2011; Wu et al., 2012). We provided conclusive evidence that the B. rapa genome arose via a whole-genome triplication event involving three n = 7 genomes structurally resembling the ancestral tPCK.
tPCK-Like Ancestral Genomes and the Origin of the Mesohexaploid Brassica Genome
Since diploid Brassica species were proposed to represent balanced secondary polyploids (Catcheside, 1934; Roebbelen, 1960), numerous lines of evidence have supported the occurrence of an ancestral whole-genome triplication prior to species radiation of the tribe Brassiceae (Lagercrantz, 1998; Lysak et al., 2005; Parkin et al., 2005; Wang et al., 2011). In this study, we inferred the tPCK-like genome with seven chromosomes to be the basic diploid genome of the hexaploid ancestor of Brassiceae. The hexaploid genome likely evolved through hybridization between a tetraploid and diploid progenitor genome. Evidence for this is based on genetic mapping (Lagercrantz, 1998; Babula et al., 2003), comparative cytogenetic analyses (Lysak et al., 2005; Ziolkowski et al., 2006), and, most convincingly, whole-genome sequence analysis. Wang et al. (2011) found that the three subgenomes in B. rapa have experienced different levels of gene loss. The more extensive fractionation of two subgenomes (MF1 and MF2) compared with the third subgenome (LF) suggest a two-step origin for the hexaploid genome. Several recent investigations (Cheng et al., 2012b; Tang and Lyons, 2012; Tang et al., 2012) convincingly support the origin of the Brassiceae ancestral genome according to the equation 4x × 2x → 3x → 6x. Here, we provide evidence that all three participating ancestral genomes had seven chromosomes and that the hexaploid ancestor probably possessed 42 chromosomes (2n = 6x = 42).
tPCK in a Phylogenetic Context
Because species of the genus Brassica and closely related genera, such as Raphanus and Sinapis, can be intercrossed, and generic circumscriptions based on morphological characters are unreliable, congeneric species have frequently been assigned to two different sister phylogenetic clades within Brassiceae: Rapa/Oleracea and Nigra (Warwick and Black, 1991). To better understand the structure of Brassica progenitor genomes, we compared tPCK-specific block associations with chromosome structures of Rapa/Oleracea (B. oleracea, B. rapa, and radish) and Nigra (B. nigra) species. We found that all three Rapa/Oleracea species (B. oleracea, B. rapa, and radish) retained three copies of the V/K/L/Wa/Q/X and O/P/W/R associations. D/V or M/E associations were also found in the three species. In addition, chromosome synteny among B. nigra, B. oleracea, and B. rapa and the existence of the tPCK-specific associations D/V and M/E in B. nigra and D/V in B. oleracea and radish indicate that B. nigra, B. oleracea, and radish had the same origin as B. rapa. Furthermore, some tPCK-specific associations observed within the comparative linkage map of S. alba (Nelson et al., 2011), a species from the Nigra clade, suggest that the genome of this species is also derived from a triplicated tPCK-like ancestral genome.
The tribes Brassiceae, Isatideae, Thelypodieae, and Sisymbrieae were designated as lineage II of the Brassicaceae (Al-Shehbaz et al., 2006; Beilstein et al., 2006, 2008). However, the relationship of these tribes to the two other Brassicaceae lineages and to the remaining 45 tribes remains uncertain. Franzke et al. (2011) grouped lineage II with 18 other tribes to form an expanded lineage II. Using comparative chromosome painting, Mandáková and Lysak (2008) provided valuable insight into the phylogenetics of the expanded lineage II. They demonstrated that tribes Calepineae, Coluteocarpeae, Conringieae, Eutremeae, Isatideae, the genus Ochthodium (formerly Sisymbrieae), and probably the Brassiceae descended from an ancestral PCK genome. In Eutremeae, Isatideae, and Ochthodium, PCK was altered by a whole-arm translocation giving rise to tPCK. The tPCK genome structure was recently confirmed by draft genome assemblies in T. salsuginea (from Eutremeae) (Wu et al., 2012) and in S. parvula (Dassanayake et al., 2011). Although the tribal placement of Schrenkiella is still uncertain (Al-Shehbaz, 2012), our results with respect to Ks values place Brassiceae closer to Schrenkiella than to Eutremeae (Thellungiella) and indicate that the Brassiceae triplication event occurred near the time of the divergence between Brassiceae and Schrenkiella (∼10 million years ago). Collectively, our data suggest that the ancestor of Brassiceae, Isatideae, Schrenkiella, Ochthodium, and Eutremeae, and probably also Thelypodieae (Caulanthus), descended from the same diploid tPCK-like ancestral genome. While the triplicated tPCK genome has undergone extensive repatterning in Brassiceae, the ancestral structure of the tPCK genome has been conserved in diploid x = 7 taxa, such as Schrenkiella or Thellungiella, where no diploidization of a polyploid genome could occur.
Descending Dysploidy in the Hexaploid Ancestor of Brassiceae
The chromosome number of 2n = 42 of the hexaploid ancestor has been reduced by a factor of 2 during the evolution of Brassiceae (Warwick and Al-Shehbaz, 2006). Such reductions are associated with the origin of composite chromosomes and loss of chromosome segments, such as centromeres, telomeres, and nucleolar organizing regions. In B. rapa, descending dysploidy led to the origin of 10 fusion chromosomes and the elimination of 11 paleocentromeres. Some paleocentromeres were eliminated through symmetric translocations resulting in products of unequal size: a large fusion chromosome and a minichromosome, which contains mainly a centromere and telomeres and usually gets lost during meiosis (Lysak et al., 2006; Schranz et al., 2006; Schubert and Lysak, 2011). Other ancestral chromosomes were combined by asymmetric end-to-end translocations (e.g., between AK6/8 and AK3 and in A03; Figure 2B). End-to-end translocation events must be accompanied by centromere loss or inactivation on one of the participating chromosomes to prevent unstable dicentric chromosomes. Centromeres were apparently lost from the collinear GBs F/G/H (AK3) of A03 and I/J (AK4) of A03 and A04 (Figure 2B). Nested chromosome fusions (i.e., insertion of one chromosome into the centromere of a recipient chromosome), which occurred frequently in the evolution of grass species (Luo et al., 2009; Salse et al., 2009; Murat et al., 2010), have not been detected in the karyotype of B. rapa. In Brassicaceae, single cases of such events apparently occurred in Pachycladon species (Mandáková et al., 2010) and in Hornungia alpina (Lysak et al., 2006).
We also analyzed whether recombination between homoeologous chromosome segments of the three B. rapa subgenomes exceeded recombination between nonhomoeologous segments. If recombination between homoeologous segments was prevalent, we would expect to find the same GBs located adjacent to one another [e.g., association A(MF1)/A(MF2)]. In B. rapa, only four such associations were detected [S(LF)/S(MF2) on A04, M(LF)/M(MF2) on A06, E(MF2)/E(LF) on A07, and D(MF1)/D(MF2) on A09]. These data suggest that interchromosomal rearrangements were not mediated preferentially by recombination between homoeologous GBs. Nevertheless, the true picture may have been blurred by numerous rearrangements (typically inversions) that may have occurred later and reshuffled originally adjacent homoeologous GBs.
Whole-Genome Sequence Assembly Improved the Resolution of Comparative Paleogenomics in Brassicaceae
The B. rapa genome sequence (Wang et al., 2011) provided much more detailed information on genome structure than previously constructed linkage maps. Although most GBs and parts thereof (68%) were detected by genetic mapping (Parkin et al., 2005; Panjabi et al., 2008; Kim et al., 2009; Trick et al., 2009), some blocks and block associations were missed or reported as being present in less than three copies. A comparison of the B. rapa genome sequence with previously reported linkage maps shows that block D on chromosome A01; blocks W, Q, X, M, and D on A03; S on A05; M, H, and U on A6; F, X, and S on A07; S on A08; K, L, and M on A09; and C on A10 were not detected by genetic mapping (Parkin et al., 2005; Panjabi et al., 2008; Kim et al., 2009; Trick et al., 2009). Although the assembly of B. rapa chromosome A03 by Mun et al. (2010) identified 14 GBs or block segments, three additional blocks on the same chromosome were detected within the B. rapa reference genome (Wang et al., 2011). Similarly, the 2.4-Mb fragment of block W within the V/K/L/Wa/Q/X association was not detected by genetic mapping. Moreover, two adjacent copies of the same GB belonging to different subgenomes have often been regarded as a single block in previous genetic maps, including blocks S(LF)/S(MF2), E(LF)/E(MF2), and D(MF1)/D(MF2) on chromosomes A04, A07, and A09, respectively (Panjabi et al., 2008; Figure 2B). Furthermore, we found that earlier reports on GBs represented by more than three copies were mostly due to misinterpretation of block segments as entire blocks. For instance, Panjabi et al. (2008) observed four copies of block A located on chromosomes A06, A08, A09, and A10. However, analysis of B. rapa genome sequences revealed that the two copies of block A on chromosomes A06 and A10 were in fact two segments of this block (Figure 2B). These examples demonstrate that whole-genome sequence assemblies largely eliminate the risk of misinterpreting low-resolution genetic data.
Refining Intervals of Ancestral GBs
The original concept of 24 GBs (Schranz et al., 2006) resulted from the synthesis of comparative genetic and cytogenetic data for A. thaliana, A. lyrata, C. rubella, the A and C genomes of B. napus, and other lineage I species (Boivin et al., 2004; Kuittinen et al., 2004; Parkin et al., 2005; Lysak et al., 2006). Because A. thaliana was the only crucifer species with a sequenced genome at that time, chromosome collinearity between all Brassicaceae species was inferred by mapping homoeologous genetic markers from A. thaliana and by chromosomal location of A. thaliana BAC clones and contigs. This approach was highly influenced by the density of genetic markers and the selection of chromosome-specific BAC contigs. Without sufficiently accurate information on cross-species collinearity, only approximate or minimal intervals could be defined for some GBs (Schranz et al., 2006) and GB associations. This is exemplified by the PCK/tPCK-specific chromosome rearrangement initially described as V/K/L/Q/X (Mandáková and Lysak, 2008) and V/K/L/V'/Q/X (Dassanayake et al., 2011) and eventually refined as V/K/L/Wa/Q/X in this study. The extensive repatterning of ancestral GBs within the B. rapa genome, along with newly available sequence information for Brassicaceae genomes, allowed us to refine the intervals of seven GBs. This was particularly feasible when individual GBs were found to be relocated and formed a nonancestral GB association with breakpoints confirmed by syntenic positions in two or three B. rapa subgenomes. The interval refinement of the ancestral GB structure will improve the resolution of interspecies comparisons of genome collinearity across Brassicaceae.
METHODS
Syntenic Ortholog Determination
Syntenic orthologs between Brassica rapa, Arabidopsis thaliana, Arabidopsis lyrata, Schrenkiella parvula, Thellungiella salsuginea, papaya (Carica papaya), and grape (Vitis vinifera) were identified using the tool SynOrths (Cheng et al., 2012a), with B. rapa as the query genome and each of the other species as the subject genome. SynOrths was run with default parameters (m = 20, n = 100, and r = 0.2). Under these settings, SynOrths first identifies homoeologous gene pairs between the two genomes using Basic Local Alignment Search Tool for Protein, defining gene pairs as those with an E-value more significant than 1 × 10−20 or representing the best hits through the entire genome. The 20 closest genes (m = 20) flanking either side of the gene in the query B. rapa genome are then compared with the 100 closest genes (n = 100) flanking either side of the gene in the subject genome. If at least 20% (r = 0.2) of the best hits for the 40 genes (20 × 2 for both sides) in the query B. rapa genome are found within the 200 genes (100 × 2 for both sides) in the subject genome, the original pair of homoeologous genes are designated as a syntenic ortholog candidate. The increase in the number of flanking genes searched in the subject genomes was necessary to compensate for the fractionation that has reduced the gene content of each B. rapa subgenome.
Comparison of Arrangements of GB Associations
We searched the distribution of predefined GBs in the three genomes of the ACK, PCK, and tPCK for adjacent pairs of GBs (GB associations). These GB associations were compared with the extant arrangement of GBs within B. rapa. GB associations in ACK, PCK, and tPCK that were also immediately adjacent to each other in the B. rapa genome were counted and recorded.
Reconstruction of Subgenomes Based on the tPCK
As previously reported (Wang et al., 2011; Cheng et al., 2012b), the B. rapa genome is composed of three subgenomes each of the 24 GBs. We annotated these blocks along the seven chromosomes of the tPCK and reconstructed three tPCK-like subgenomes. At first we identified all continuous chromosome segments corresponding to GBs and belonging to the same chromosome of the tPCK. Then, chromosome segments were concatenated according to the following rules: (1) the reconstructed chromosome could not contain redundant copies of the same syntenic segment, and (2) the concatenated chromosome segments had to have a comparable gene density corresponding to the three B. rapa subgenomes (LF, MF1, and MF2).
Ancestral Centromere Detection
Centromere-specific sequences, including CentBr, PCRBr, and TR238, were obtained from previous studies (Koo et al., 2004, 2011; Lim et al., 2005, 2007). These sequences were aligned to the B. rapa genome using Nucmer (Kurtz et al., 2004) with parameters “-maxmatch -g 500 -c 16 -l 16 ” (i.e., find all matches with minimal exact match size of 16 bp regardless of uniqueness, merge exact matches distributed <500 bp apart into clusters, and report all clusters >16 bp). The nearest genes to the best Nucmer hits were determined and used as labels of genomic coordinates for the detected centromere traces (see Supplemental Data Set 2 online).
Identification of tPCK-Specific Block Associations in B. oleracea
Because a pseudomolecule-level assembly for the B. oleracea genome is not yet available, we used a different method to determine local syntenic relationships between B. rapa and B. oleracea. Thirty B. rapa genes located on either side of the boundaries between GBs within the GB associations O/P/W/R and D/V/K/L/Q/X were selected. These genes were compared using BLAST against B. oleracea genes via the Web interface provided by BRAD (http://brassicadb.org/brad/blastPage.php) (Cheng et al., 2011). Two blocks were classified as being syntenic in B. oleracea when >60% of the B. rapa block boundary genes from each GB had BLAST hits (identity and coverage) on the same B. oleracea scaffold. For example, if (1) 60% of 30 B. rapa genes at the boundary of block O (near P) were homoeologous to genes on Scaffold000203 of B. oleracea, (2) 30 B. rapa genes at the block P boundary (near O) were also homoeologous to B. oleracea genes in Scaffold000203, and (3) these genes were adjacent to each other on Scaffold000203, then the O/P association was considered to be present also in the B. oleracea genome.
Ks Analysis
Protein sequences from B. rapa were aligned with those of A. thaliana, A. lyrata, S. parvula, and T. salsuginea using MUSCLE (Edgar, 2004). Protein alignments were translated into coding sequence alignments using an in-house Perl script. Ks values were calculated based on the coding sequence alignments using the method of Nei and Gojobori as implemented in KaKs_calculator (Zhang et al., 2006). Ks values of all syntenic orthologs between B. rapa and each of the four species were then plotted as histograms.
Phylogenetic Analysis
For phylogenetic analysis, the same MUSCLE alignments as for Ks analysis were used. The genotypes of synonymous loci in 591 syntenic orthologs among B. rapa, S. parvula, T. salsuginea, A. thaliana, A. lyrata, papaya (C. papaya), and grape (V. vinifera) were extracted from multiple alignments and concatenated into a single sequence for each species. Phylogenies were constructed, based on the character states of these 63,239 synonymous loci using the neighbor-joining method, and plotted under a linearized tree model as implemented in MEGA (Tamura et al., 2011).
Accession Numbers
Gene and protein sequences for B. rapa were obtained from BRAD (V1.2; http://brassicadb.org) (Cheng et al., 2011). Gene and genome data sets for A. thaliana were downloaded from The Arabidopsis Information Resource (TAIR9; http://www.Arabidopsis.org/index.jsp). Data sets for A. lyrata were downloaded from the Joint Genome Initiative database (Gene model 6; http://genome.jgi-psf.org/Araly1/Araly1.home.html) (Hu et al., 2011). S. parvula data sets (TpV7) were obtained from Dassanayake et al. (2011). T. salsuginea data were downloaded from the National Center for Biotechnology Information (accession number AHIU00000000) (Wu et al., 2012).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Purported Translocation Events Reshuffling tPCK-Specific Associations D/V and M/E in the B. rapa Genome.
Supplemental Figure 2. Chromosome Collinearity Comparison among A, B, and C Genomes, Revised from Panjabi et al. (2008).
Supplemental Figure 3. Reconstruction of Three tPCK-Like Ancestral Subgenomes in B. rapa.
Supplemental Figure 4. Two Alternative Origins of Translocation Chromosomes AK5/6/8 (PCK) and AK2/5/6/8 (tPCK).
Supplemental Table 1. Summary of Genomic Blocks Identified in B. rapa.
Supplemental Table 2. Genomic Blocks E, G, and T Showing Sequence Deletion in B. rapa (Br) as Compared with Arabidopsis thaliana (At).
Supplemental Table 3. Number of Genomic Blocks with Different Fragmental Status in B. rapa Subgenomes.
Supplemental Table 4. Frequency of tPCK-Specific Block Associations in Each of the Three B. rapa Subgenomes.
Supplemental Table 5. Number of Syntenic Genes Shared between Each B. rapa Subgenome and the Arabidopsis thaliana Genome (At).
Supplemental Table 6. The Frequency of tPCK-Specific Block Associations in B. rapa and B. oleracea.
Supplemental Table 7. List of Genomic Blocks Comprising the Nine Linkage Groups of Raphanus sativus (Shirasawa et al., 2011).
Supplemental Data Set 1. Syntenic Fragment Comparison between B. rapa and the Ancestral Crucifer Karyotype.
Supplemental Data Set 2. Centromere-Specific Sequences Detected in the B. rapa Genome.
Supplemental Data Set 3. Redefined Intervals of Seven Ancestral Genomic Blocks.
Supplementary Material
Acknowledgments
We thank Michael Freeling, Isobel A. Parkin, James Schnable, and Ingo Schubert for their helpful suggestions concerning this work. This study was funded by the National Program on Key Basic Research Projects (The 973 Program: 2012CB113900 and 2013CB127000) and the National High Technology R&D Program of China (2012AA100101). Research was carried out in the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, P.R. China. T.M. and M.A.L. were supported by a research grant from the Czech Science Foundation (Excellence Cluster P501/12/G090) and by the European Regional Development Fund (CZ.1.05/1.1.00/02.0068).
AUTHOR CONTRIBUTIONS
X.W., F.C., and M.A.L. designed the research. F.C., T.M., and J.W. performed the research and analyzed the data. F.C., M.A.L., and Q.X. contributed new computational tools and data. F.C., M.A.L., and X.W. wrote the article.
Glossary
- GB
genomic block
- ACK
Ancestral Crucifer Karyotype
- PCK
Proto-Calepineae Karyotype
- tPCK
translocation Proto-Calepineae Karyotype
- BRAD
BRAD Brassica Database
References
- Al-Shehbaz I.A. (2012). A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon 61: 931–954 [Google Scholar]
- Al-Shehbaz I.A., Beilstein M.A., Kellogg E.A. (2006). Systematics and phylogeny of the Brassicaceae (Cruciferae): An overview. Plant Syst. Evol. 259: 89–120 [Google Scholar]
- Babula D., Kaczmarek M., Barakat A., Delseny M., Quiros C.F., Sadowski J. (2003). Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: Complexity of the comparative map. Mol. Genet. Genomics 268: 656–665 [DOI] [PubMed] [Google Scholar]
- Beilstein M.A., Al-Shehbaz I.A., Kellogg E.A. (2006). Brassicaceae phylogeny and trichome evolution. Am. J. Bot. 93: 607–619 [DOI] [PubMed] [Google Scholar]
- Beilstein M.A., Al-Shehbaz I.A., Mathews S., Kellogg E.A. (2008). Brassicaceae phylogeny inferred from phytochrome A and ndhF sequence data: Tribes and trichomes revisited. Am. J. Bot. 95: 1307–1327 [DOI] [PubMed] [Google Scholar]
- Boivin K., Acarkan A., Mbulu R.S., Clarenz O., Schmidt R. (2004). The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae. A comparison of the Arabidopsis and Capsella rubella genomes. Plant Physiol. 135: 735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell A.M., Taylor K.G., Williams R.J., Cantrell R.T., Menz M.A., Pepper A.E. (2011). A comparative genomic map for Caulanthus amplexicaulis and related species (Brassicaceae). Mol. Ecol. 20: 784–798 [DOI] [PubMed] [Google Scholar]
- Catcheside D.G. (1934). The chromosomal relationships in the Swede and turnip groups of Brassica. Ann. Bot. (Lond.) 48: 601–633 [Google Scholar]
- Cheng F., Liu S., Wu J., Fang L., Sun S., Liu B., Li P., Hua W., Wang X. (2011). BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 11: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Wu J., Fang L., Sun S., Liu B., Lin K., Bonnema G., Wang X. (2012b). Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS ONE 7: e36442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Wu J., Fang L., Wang X. (2012a). Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front. Plant Sci. 3: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake M., Oh D.H., Haas J.S., Hernandez A., Hong H., Ali S., Yun D.J., Bressan R.A., Zhu J.K., Bohnert H.J., Cheeseman J.M. (2011). The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzke A., Lysak M.A., Al-Shehbaz I.A., Koch M.A., Mummenhoff K. (2011). Cabbage family affairs: The evolutionary history of Brassicaceae. Trends Plant Sci. 16: 108–116 [DOI] [PubMed] [Google Scholar]
- Hu T.T., et al. (2011). The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Choi S.R., Bae J., Hong C.P., Lee S.Y., Hossain M.J., Van Nguyen D., Jin M., Park B.S., Bang J.W., Bancroft I., Lim Y.P. (2009). Sequenced BAC anchored reference genetic map that reconciles the ten individual chromosomes of Brassica rapa. BMC Genomics 10: 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.A., Haubold B., Mitchell-Olds T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17: 1483–1498 [DOI] [PubMed] [Google Scholar]
- Koo D.H., Hong C.P., Batley J., Chung Y.S., Edwards D., Bang J.W., Hur Y., Lim Y.P. (2011). Rapid divergence of repetitive DNAs in Brassica relatives. Genomics 97: 173–185 [DOI] [PubMed] [Google Scholar]
- Koo D.H., Plaha P., Lim Y.P., Hur Y., Bang J.W. (2004). A high-resolution karyotype of Brassica rapa ssp. pekinensis revealed by pachytene analysis and multicolor fluorescence in situ hybridization. Theor. Appl. Genet. 109: 1346–1352 [DOI] [PubMed] [Google Scholar]
- Kuittinen H., de Haan A.A., Vogl C., Oikarinen S., Leppälä J., Koch M., Mitchell-Olds T., Langley C.H., Savolainen O. (2004). Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168: 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U. (1998). Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150: 1217–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U., Lydiate D.J. (1996). Comparative genome mapping in Brassica. Genetics 144: 1903–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Hasegawa Y., Saito M., Shirasawa S., Fukushima A., Ito T., Fujii H., Kishitani S., Kitashiba H., Nishio T. (2011). Extensive chromosome homoeology among Brassiceae species were revealed by comparative genetic mapping with high-density EST-based SNP markers in radish (Raphanus sativus L.). DNA Res. 18: 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.B., et al. (2005). Characterization of rDNAs and tandem repeats in the heterochromatin of Brassica rapa. Mol. Cells 19: 436–444 [PubMed] [Google Scholar]
- Lim K.B., et al. (2007). Characterization of the centromere and peri-centromere retrotransposons in Brassica rapa and their distribution in related Brassica species. Plant J. 49: 173–183 [DOI] [PubMed] [Google Scholar]
- Luo M.C., et al. (2009). Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc. Natl. Acad. Sci. USA 106: 15780–15785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M.A., Berr A., Pecinka A., Schmidt R., McBreen K., Schubert I. (2006). Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 103: 5224–5229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M.A., Cheung K., Kitschke M., Bures P. (2007). Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol. 145: 402–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M.A., Koch M.A., Pecinka A., Schubert I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15: 516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T., Heenan P.B., Lysak M.A. (2010). Island species radiation and karyotypic stasis in Pachycladon allopolyploids. BMC Evol. Biol. 10: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T., Lysak M.A. (2008). Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae). Plant Cell 20: 2559–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun J.H., et al. (2010). Sequence and structure of Brassica rapa chromosome A3. Genome Biol. 11: R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat F., Xu J.H., Tannier E., Abrouk M., Guilhot N., Pont C., Messing J., Salse J. (2010). Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res. 20: 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M.N., Parkin I.A., Lydiate D.J. (2011). The mosaic of ancestral karyotype blocks in the Sinapis alba L. genome. Genome 54: 33–41 [DOI] [PubMed] [Google Scholar]
- Panjabi P., Jagannath A., Bisht N.C., Padmaja K.L., Sharma S., Gupta V., Pradhan A.K., Pental D. (2008). Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: Homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics 9: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin I.A., Gulden S.M., Sharpe A.G., Lukens L., Trick M., Osborn T.C., Lydiate D.J. (2005). Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Hinata K. (1980). Taxonomy, cytogenetics and origin of crop Brassicas, a review. Opera Botanica 55: 1–57 [Google Scholar]
- Roebbelen G. (1960). [Contributions to the analysis of the Brassica-genome]. Chromosoma 11: 205–228 [DOI] [PubMed] [Google Scholar]
- Salse J., Abrouk M., Bolot S., Guilhot N., Courcelle E., Faraut T., Waugh R., Close T.J., Messing J., Feuillet C. (2009). Reconstruction of monocotelydoneous proto-chromosomes reveals faster evolution in plants than in animals. Proc. Natl. Acad. Sci. USA 106: 14908–14913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz M.E., Lysak M.A., Mitchell-Olds T. (2006). The ABC’s of comparative genomics in the Brassicaceae: Building blocks of crucifer genomes. Trends Plant Sci. 11: 535–542 [DOI] [PubMed] [Google Scholar]
- Schubert I., Lysak M.A. (2011). Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 27: 207–216 [DOI] [PubMed] [Google Scholar]
- Shirasawa K., et al. (2011). An EST-SSR linkage map of Raphanus sativus and comparative genomics of the Brassicaceae. DNA Res. 18: 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Lyons E. (2012). Unleashing the genome of Brassica rapa. Front. Plant Sci. 3: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Woodhouse M.R., Cheng F., Schnable J.C., Pedersen B.S., Conant G., Wang X., Freeling M., Pires J.C. (2012). Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics 190: 1563–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trick M., et al. (2009). Complexity of genome evolution by segmental rearrangement in Brassica rapa revealed by sequence-level analysis. BMC Genomics 10: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truco M.J., Hu J., Sadowski J., Quiros C.F. (1996). Inter- and infra-genomic homology of the Brassica genomes: Implications for their origin and evolution. Theor. Appl. Genet. 93: 1225–1233 [DOI] [PubMed] [Google Scholar]
- Wang X., et al. Brassica rapa Genome Sequencing Project Consortium (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Warwick S.I., Black L.D. (1991). Molecular systematics of Brassica and allied genera (subtribe Brassicinae, Brassiceae) - Chloroplast genome and cytodeme congruence. Theor. Appl. Genet. 82: 81–92 [DOI] [PubMed] [Google Scholar]
- Warwick S.I., Al-Shehbaz I.A. (2006). Brassicaceae: Chromosome number index and database on CD-Rom. Plant Syst. Evol. 259: 237–248 [Google Scholar]
- Wu H.J., et al. (2012). Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. USA 109: 12219–12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li J., Zhao X.Q., Wang J., Wong G.K., Yu J. (2006). KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4: 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski P.A., Kaczmarek M., Babula D., Sadowski J. (2006). Genome evolution in Arabidopsis/Brassica: Conservation and divergence of ancient rearranged segments and their breakpoints. Plant J. 47: 63–74 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.