This work shows the conservation in eudicots of a miRNA-TAS-PPR-siRNA pathway characterized by a set of 22-nucleotide miRNAs that trigger secondary siRNAs using distinct mechanistic strategies. This super-miR7122 family of miRNAs includes miR173 and originates from miR390, emerging via gene duplication and neofunctionalization, which involved yet another miRNA superfamily (super-miR4376).
Abstract
Trans-acting small interfering RNAs (tasiRNAs) are a major class of small RNAs performing essential biological functions in plants. The first reported tasiRNA pathway, that of miR173-TAS1/2, produces tasiRNAs regulating a set of pentatricopeptide repeat (PPR) genes and has been characterized only in Arabidopsis thaliana to date. Here, we demonstrate that the microRNA (miRNA)-trans-acting small interfering RNA gene (TAS)-pentatricopeptide repeat-containing gene (PPR)-small interfering RNA pathway is a highly dynamic and widespread feature of eudicots. Nine eudicot plants, representing six different plant families, have evolved similar tasiRNA pathways to initiate phased small interfering RNA (phasiRNA) production from PPR genes. The PPR phasiRNA production is triggered by different 22-nucleotide miRNAs, including miR7122, miR1509, and fve-PPRtri1/2, and through distinct mechanistic strategies exploiting miRNA direct targeting or indirect targeting through TAS-like genes (TASL), one-hit or two-hit, or even two layers of tasiRNA–TASL interactions. Intriguingly, although those miRNA triggers display high sequence divergence caused by the occurrence of frequent point mutations and splicing shifts, their corresponding MIRNA genes show pronounced identity to the Arabidopsis MIR173, implying a common origin of this group of miRNAs (super-miR7122). Further analyses reveal that super-miR7122 may have evolved from a newly defined miR4376 superfamily, which probably originated from the widely conserved miR390. The elucidation of this evolutionary path expands our understanding of the course of miRNA evolution, especially for relatively conserved miRNA families.
INTRODUCTION
Small RNAs (sRNAs), typically 20 to 24 nucleotides long, are important gene and chromatin regulators in plants. MicroRNAs (miRNAs) are 20- to 22-nucleotide RNAs that negatively regulate target genes through homology-directed mRNA cleavage or translational inhibition at the posttranscriptional level (Bartel, 2004; Voinnet, 2009). Another class of sRNAs, small interfering RNAs (siRNAs), account for the majority of the sRNA repertoire in cells and are implicated in a variety of processes, including combating viral and transposon replication, establishment and maintenance of heterochromatin, and posttranscriptional gene regulation (Baulcombe, 2004; Jones-Rhoades et al., 2006; Matzke et al., 2009; Law and Jacobsen, 2010). Trans-acting siRNAs (tasiRNAs) are a class of 21-nucleotide siRNAs that direct sequence-specific cleavage of their target genes, as do microRNAs (miRNAs). TasiRNA biogenesis from trans-acting siRNA transcripts (TAS) requires an initial cleavage of the mRNA transcripts by a specific miRNA, after which one of the cleaved products is made double stranded by RNA-DEPENDENT RNA POLYMERASE6 (RDR6) and subsequently diced by DICER-LIKE ENZYME4 (DCL4) into 21-nucleotide species; these 21-nucleotide secondary siRNAs have a characteristic phased pattern initiating at the miRNA cleavage site (Vazquez et al., 2004; Allen et al., 2005; Yoshikawa et al., 2005). Some of the phased siRNAs (phasiRNAs) may further target their parental genes in cis or other genes in trans, distinguishing them as cis-acting RNAs or tasiRNAs, respectively (Zhai et al., 2011).
To date, four TAS gene families have been well characterized in Arabidopsis thaliana, including the one-hit TAS1/2/4 initiated by a single target site of a 22-nucleotide miRNA (122, miR173, or miR828) and the two-hit TAS3 with two target sites of the 21-nucleotide miR390 (221) (Yoshikawa et al., 2005; Axtell et al., 2006; Rajagopalan et al., 2006). miR173-targeted TAS1/2 produce tasiRNAs, targeting a few genes coding for pentatricopeptide repeat–containing proteins (PPRs) (Yoshikawa et al., 2005; Howell et al., 2007). The miR828-TAS4–derived siRNA81(-) targets genes encoding MYB transcription factors (MYBs) (Rajagopalan et al., 2006; Luo et al., 2012; Xia et al., 2012; Zhu et al., 2012), whereas miR390-TAS3–spawned tasiARFs regulate genes coding for auxin responsive factors (Williams et al., 2005; Fahlgren et al., 2006; Xia et al., 2012). Although miR828 was originally identified as a trigger of phasiRNA production solely for the noncoding TAS4 in Arabidopsis, it was later found to trigger phasiRNA production in a few MYB-encoding genes as well (Xia et al., 2012; Zhu et al., 2012). In fact, phasiRNA production has been demonstrated to occur in numerous protein-coding genes, including those coding for nucleotide binding site Leucine-rich repeat proteins (NB-LRRs) (Klevebring et al., 2009; Zhai et al., 2011; Shivaprasad et al., 2012), MYBs (Xia et al., 2012; Zhu et al., 2012), PPRs (Chen et al., 2007; Howell et al., 2007), Ca2+-ATPase (Wang et al., 2011), and TIR/AFB2 (for TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX PROTEIN2) auxin receptors (Si-Ammour et al., 2011). In most cases, the miRNA triggers are 22 nucleotides and act through the 122 mode, but a few of them, such as miR156 and miR172, are 21 nucleotides and act together to function (like miR390) via the 221 mode (Zhai et al., 2011). Although originally described for 21-nucleotide miRNAs, the two-hit mechanism can also employ 22-nucleotide miRNAs, as in the case of miR1509 in Medicago truncatula that triggers phasiRNA biogenesis through binding and cleavage at two sites in its target gene (Zhai et al., 2011). Intriguingly, but apparently rarely, 22-nucleotide tasiRNAs can also serve as effective triggers of phasiRNAs, as in the case of a predominantly 22-nucleotide TAS2-derived tasiRNA 3′ D6(-) (also termed tasiR2140) that is capable of initiating phasiRNA biogenesis in PPRs, while one of the TAS1c-derived 21-nucleotide tasiRNAs works together with miR173 to delimit the boundary of phasiRNA production along the cleaved transcript (Yoshikawa et al., 2005; Howell et al., 2007; Chen et al., 2010; Rajeswaran et al., 2012). Evidently, both miRNA and tasiRNA can exploit different mechanisms to precisely trigger and define phasiRNA production in specific gene regions.
In contrast with the broad conservation of the miR390-TAS3 pathway in seed plants and the miR828-TAS4 pathway in eudicots, the miR173-TAS1/2 pathway has not been reported in species beyond Arabidopsis (Axtell et al., 2006; Cuperus et al., 2011; Luo et al., 2012). Several models have been proposed for the evolution of plant MIRNAs (Allen et al., 2004; Felippes et al., 2008; Piriyapongsa and Jordan, 2008); however, a complete view of how most individual MIRNAs originated and evolved is still lacking, especially for the relatively conserved MIRNAs. In this study, we characterized a widely adapted miRNA-TAS-PPR-siRNA regulatory circuit in eudicots. The initial miRNA triggers of PPR phasiRNA production share a common ancestor with miR173. We also demonstrate that these miRNA triggers are evolutionarily related to the highly conserved miR390 family, revealing a previously unknown miRNA evolutionary history.
RESULTS
PHAS Gene Identification with Reverse Computation
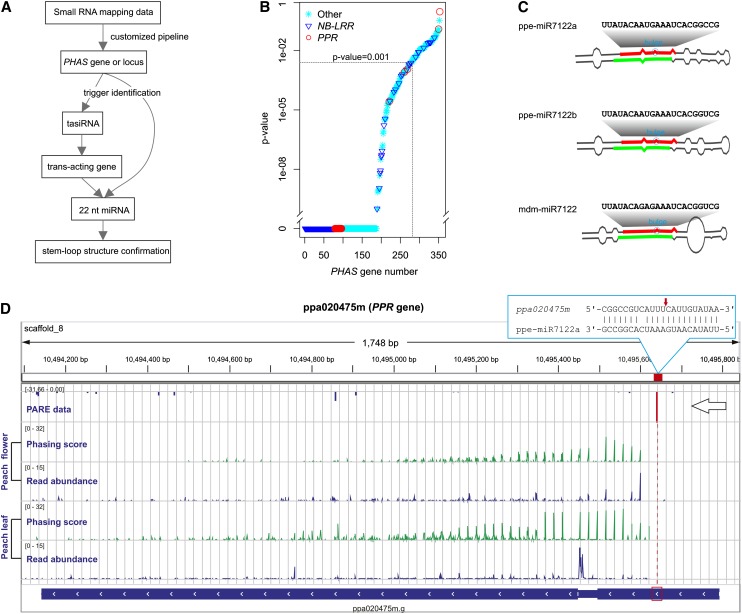
Plant genomes are rich in both coding and noncoding genomic loci capable of producing 21-nucleotide siRNAs (Howell et al., 2007; Johnson et al., 2009; Zhai et al., 2011; Zhang et al., 2012). Twenty-two-nucleotide miRNAs can trigger the production of secondary phasiRNAs, which are generally 21 nucleotides in length and in phase with the miRNA cleavage site, from the 5′ end of the 3′ cleavage product of their targeted transcripts (Chen et al., 2010; Cuperus et al., 2010). Both coding genes and noncoding transcripts capable of phasiRNA production (PHAS genes) have been identified in many species, but efficient identification of these PHAS genes and their miRNA trigger remains challenging. We developed customized computation pipelines for genome-wide identification of PHAS genes and their corresponding miRNA triggers. The process, referred to as reverse computation, is illustrated in Figure 1A, and includes a P value–based strategy similar to that of Chen et al. (2007) to first retrieve PHAS genes and then identify their corresponding triggers.
Figure 1.
Identification of PHAS PPR Genes and the Trigger miRNA in Peach.
(A) Workflow of identifying PHAS genes/loci and miRNA triggers (reverse computation). nt, nucleotides.
(B) P value distribution of all putative PHAS genes in peach. The P value threshold of 0.001 is indicated with a dotted line.
(C) Stem-loop structures of the miR7122 homologs in peach (ppe-miR7122a and b) and apple (mdm-miR7122). Positions of miRNA and miRNA* sequences are marked in red and light blue, respectively; position of bulge in each stem-loop structure is marked with a light-blue circle.
(D) Phasing score and read abundance distribution along a PHAS PPR gene in peach flower and leaf tissues viewed together with PARE data in the IGV. Phasing score was calculated based on the mapping results of 21-nucleotide sRNAs (see Methods). Paring of miR7122 and target site is denoted in the top-right corner with the cleavage site marked with a small red arrow; gray gridlines show the 21-nucleotide phasing pattern set up by the miR7122 cleavage site. The gene transcription direction and miR7122 cleavage site confirmed by PARA data are denoted by a large arrow and a red bar, respectively.
We first tested our pipelines in peach (Prunus persica) using its annotated transcriptome. From the four peach deep-sequencing sRNA libraries available, we were able to identify 350 PHAS gene candidates, of which 263 (∼75%) protein-coding genes were capable of secondary siRNA production with a P value ≤ 0.001 (Figure 1B), a threshold proposed by Chen et al. (2007), including 94 NB-LRR defense genes, 20 PPR genes, 10 kinase-like genes, and other genes coding for transcription factors (MYB, NAC, and TIR/AFB) or sRNA biogenesis-related genes (SGS, AGO, and DCL) (Table 1; see Supplemental Data Set 1A online). Members of these gene families have been reported to produce secondary siRNAs in other plants, like M. truncatula and soybean (Glycine max; Zhai et al., 2011). Additional PHAS genes encode other proteins like UDP-glucosyl transferase and the dehydration responsive protein RD22 (Table 1). Two TAS genes that produce tasiRNAs, miR390-targeted TAS3 and miR828-targeted TAS4 (Zhu et al., 2012), were retrieved with an extremely low P value in a genome-wide profiling of noncoding PHAS loci (Table 1). Our reverse computation analysis also helped identify triggers for these PHAS genes, including miR482/2118 as the trigger for NB-LRRs, miR828 for MYBs, miR393 for TIR/AFBs, and miR168 for AGOs (Table 1). These results verified our computation pipelines as a valid and efficient means for the identification of PHAS genes and their miRNA triggers.
Table 1. PHAS Loci and Their Triggers Identified in Peach.
| Trigger miRNA | PHAS Loci or Gene Families | Loci or Gene No. | P Valuea | References |
|---|---|---|---|---|
| ppe-miR390 | TAS3 | 1 | 0 | Allen et al. (2005, 2006); Zhu et al. (2012) |
| ppe-miR828 | TAS4 | 1 | 0 | Rajagopalan et al. (2006); Zhu et al. (2012) |
| ppe-miR828 | MYB | 5 (1)b | 0 | Xia et al. (2012); Zhu et al. (2012) |
| ppe-miR482/2118 | NB-LRR | 94 (14) | 0 | Zhai et al. (2011) |
| ppe-miR7122 | PPR | 20 (3) | 0 | Chen et al. (2007, 2010); Howell et al. (2007); Zhai et al. (2011) |
| ppe-miR393 | TIR/AFB | 2 | 0 | Chen et al. (2010); Si-Ammour et al. (2011); Zhai et al. (2011) |
| TasiARF | ARF | 3 | 9.09E-06 | Axtell et al. (2006) |
| ppe-miR408 | Laccase | 2 | 4E-10 | |
| ppe-miR4376 | Ca2+-ATPase | 3 | 0 | Wang et al. (2011) |
| NAc | NAC | 6 (3) | 3.67E-05 | Zhai et al. (2011) |
| ppe-miR168 | AGO | 2 (1) | 0.000743 | Chen et al. (2010); Zhai et al. (2011) |
| NA | SGS | 2 | 0 | Zhai et al. (2011) |
| NA | DCL | 1 | 0.000386 | Zhai et al. (2011) |
| NA | ATP TR | 5 | 0 | Zhai et al. (2011) |
| NA | UDP-glucosyl transferase | 5 (3) | 0.000533 | |
| NA | RD22 | 17 (1) | 4.5E-10 | |
| NA | Kinase-like | 10 (4) | 5.97E-06 | Zhai et al. (2011) |
Median P value was used for gene families with greater than or equal to two PHAS genes identified.
The value in parentheses indicates the number of additional PHAS genes with a P value > 0.001 in the corresponding gene family
NA, not available; ARF, AUXIN-RESPONSIVE FACTOR; RD22, RESPONSIVE TO DEHYDRATION22; ATP TR, ATP binding/protein binding/transmembrane receptor.
miR7122 as a Trigger for PhasiRNA Production in Both PPRs and TAS-Like Loci in Peach
Our initial analysis showed that 23 peach PPR genes produced phasiRNAs (including three with a P value > 0.001; see Supplemental Data Set 1B online). miR161, miR400, and miR173 directly or indirectly trigger the phasiRNA production of PPR genes in Arabidopsis (Axtell et al., 2006; Chen et al., 2007; Howell et al., 2007). In searching for homologs of these miRNAs, however, using the current criterion for miRNA families (greater than or equal to four base differences) (Meyers et al., 2008), these miRNAs appears to be absent from peach (Zhu et al., 2012), suggesting different miRNAs might trigger the PPR phasiRNAs. To address this, we profiled all the possible sRNA complementary sites for the 23 PPR genes and identified a high-confidence target site (alignment score [AS] ≤ 5) in 10 out of the 23 PPRs for ppe-miR7122, with cleavage in three putative PHAS PPRs supported by parallel analysis of RNA ends (PARE) data (see Supplemental Data Set 1B online) (Zhu et al., 2012). Detailed examination of the mapping profile revealed that the miR7122 cleavage site set the phase of the phasiRNA production (Figure 1D). In peach, two miR7122 variants (a/b) were identified, both of which possess all the features required to trigger phasiRNA biogenesis: 22 nucleotides in length, 5′ terminal U, and a bulge-bearing stem-loop precursor (Figure 1C) (Chen et al., 2010; Manavella et al., 2012; Zhu et al., 2012).
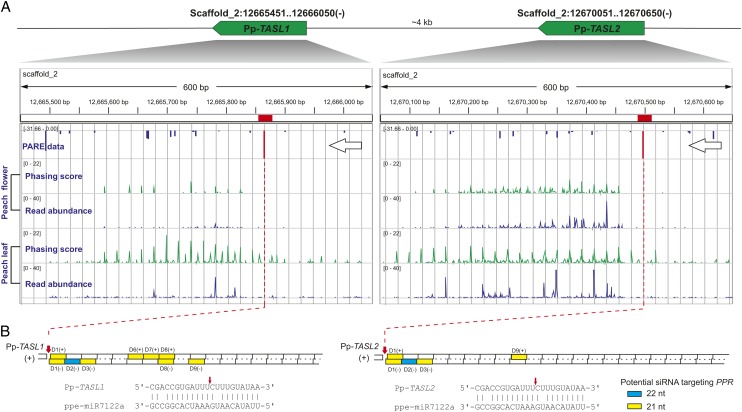
In Arabidopsis, PPRs are targeted not only by miR161, miR400, and miR173, but also by TAS1/2-derived tasiRNAs. To ascertain whether ppe-miR7122 also indirectly targets PPRs through tasiRNAs, we performed a genome-wide search for ppe-miR7122–targeted genomic loci and found that two loci, separated in the genome by ∼4500 bp, were cleaved by ppe-miR7122 and generated robust siRNAs (Figure 2). These loci, which share 78% sequence identity and appear to be non-protein coding, are likely to represent TAS-like genes, hereafter referred to as Pp-TASL1 and Pp-TASL2. The majority of tasiRNAs generated at the poly(A) proximal end fell into the second phase (Figure 2A; see Supplemental Figure 1A online), and several phased siRNAs, as expected, were predicted to target PPRs in trans (Figure 2B; see Supplemental Data Set 1C online). In particular, two tasiRNAs [Pp-TASL1-3′ D2(-) and Pp-TASL2-3′ D2(-)], predominantly 22 nucleotides in length, potentially target 14 PHAS PPRs (AS ≤ 5) and trigger subsequent phasiRNA production, based on the siRNA matching pattern (see Supplemental Figure 1B online). Six out of these 14 PHAS PPRs have either no or less cleavage-favorable (AS > 5) target sites for ppe-MIR7122 (see Supplemental Data Set 1B online), indicating synergistic and complementary roles of these TASL-derived tasiRNAs with the ppe-miR7122. Similar 22-nucleotide tasiRNA triggering of siRNA production in PPRs and reinforcement of the silencing effect have also been reported in the miR173-TAS2-PPR pathway in Arabidopsis (Yoshikawa et al., 2005; Howell et al., 2007; Chen et al., 2010). Thus, as illustrated in Figure 3 (category I), the ppe-miR7122 in peach is, like miR173 in Arabidopsis, able to trigger siRNA production from PPR genes by direct targeting or through an additional indirect TASL-tasiRNA path.
Figure 2.
miR7122-Triggered TasiRNA Production from TASL1 and TASL2 in Peach.
(A) Genomic configuration of TASL1/2 and their phasing score and read abundance distribution in peach flower and fruit tissues viewed together with PARE data in IGV. Phasing score was calculated based on the mapping results of 21-nucleotide sRNAs (see Methods). Gray gridlines show the 21-nucleotide phasing pattern set up by the miR7122 cleavage site. The gene transcription direction and miR7122 cleavage site confirmed by PARA data are denoted by large arrows and red bars, respectively.
(B) Diagrams illustrating the pattern and position of secondary siRNAs (green squares for 22 nucleotides [nt] and yellow for 21 nucleotides) potentially targeting PPR genes. The miR7122 cleavage site (red bar) confirmed by PARE data set the phase for the phasiRNA production in both TASL1 and TASL2. Generated siRNAs are numbered in order (D1, 2, 3, etc.) with strand information indicated in parentheses (“+” for plus strand and “−” for minus strand). miR7122-mRNA parings are denoted below with the cleavage site marked with a small red arrow.
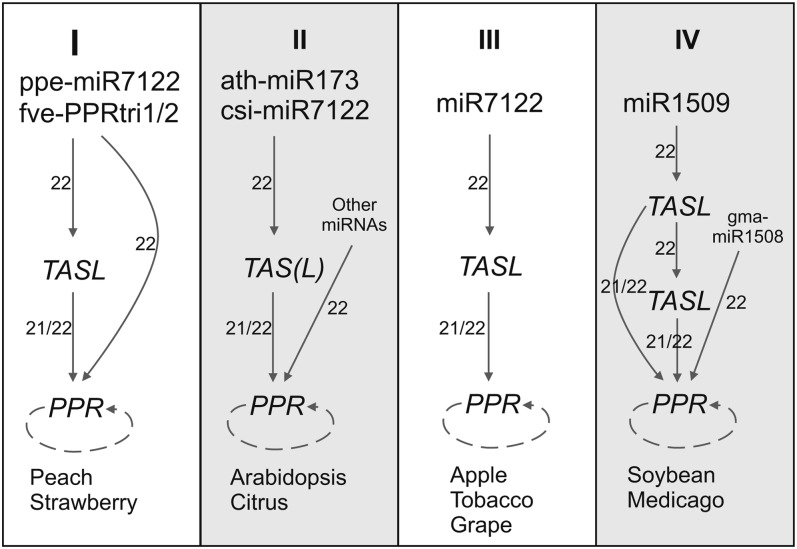
Figure 3.
Diverse miR7122-TAS(L)-PPR-siRNA Pathways Identified.
Four categories of miR7122-TAS(L)-PPR-siRNA pathways identified in nine plants are summarized. The dotted arc around the “PPR” represents the ability of processing secondary siRNAs. The length of trigger sRNA is indicated near the arrows. Category I: In peach and strawberry, PPR phasiRNA production was triggered by ppe-miR7122 and fve-PPRtri1/2, respectively, through direct targeting or the intermediation of TASL genes. Category II: In Arabidopsis and citrus, PPR phasiRNA generation was triggered by miR173 and csi-miR7122, respectively, through the intermediation of TAS(L) genes or by directly targeting of other miRNAs. Category III: In apple, tobacco, and grape, PPR siRNA production was triggered by miR7122 solely through the intermediation of TASL genes. Category IV: In soybean and M. truncatula, PPR phasiRNA generation was triggered by miR1509 through one or two layers of TASL–tasiRNA interactions or by direct targeting of gma-miR1508 in soybean as well.
Apple miR7122 Triggers the PhasiRNA Production of PPR Genes Only through TasiRNAs with a Two-Hit 222 Mode
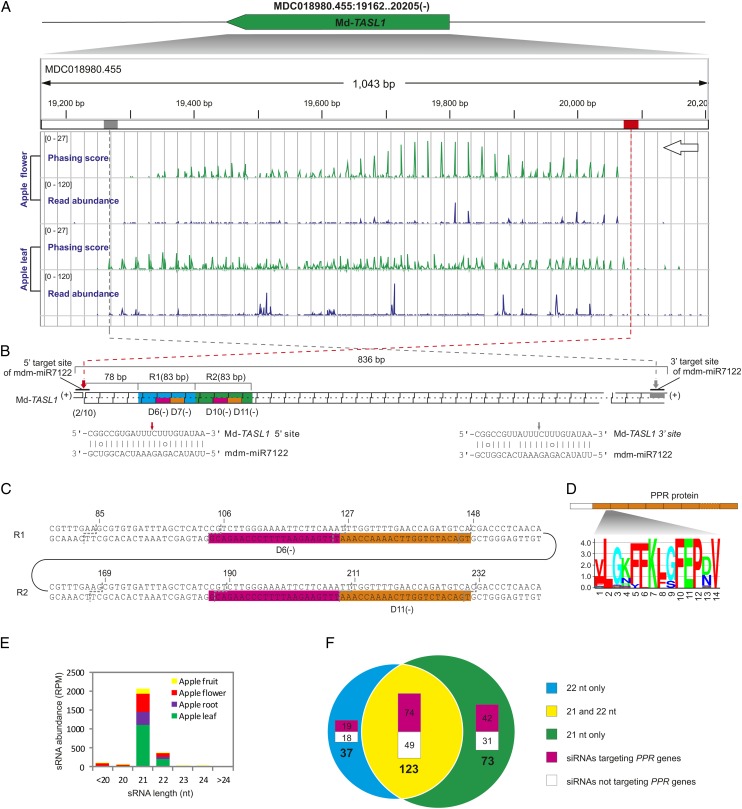
The identification of a homologous miR7122 (mdm-miR7122; Figure 1C) in our previous study in apple (Malus domestica; Xia et al., 2012) prompted us to evaluate whether an analogous miR7122-(TASL)-PPR-siRNA pathway was conserved in apple. After examining the apple small RNA data, 15 PPRs (three with a P value > 0.001) were retrieved from our phasing analysis (see Supplemental Data Set 1D online), indicating that apple PPRs are channeled into phasiRNA biogenesis as well. However, reverse computation failed to identify any complementary target sites for mdm-miR7122 within these PHAS PPR genes. Instead, two 22-nucleotide sRNAs located tandemly in the apple genome were found to be potential triggers. Analysis of sequence folding (e.g., secondary structure) failed to identify possible stem-loop structures for the precursor locus, suggesting these two sRNAs are unlikely to be miRNAs, for which biosynthesis requires a canonical stem-loop structure. The question of how these two sRNAs originated inspired us to examine the sRNA population matched to the precursor locus. We found that extremely abundant siRNAs were produced from this locus (Figure 4A). Trigger identification for this locus revealed that mdm-miR7122 was the potential trigger of siRNA production. Curiously, two target sites of mdm-miR7122, 792 bp apart, were found in this locus, and siRNAs predominantly 21 nucleotides in length with some 22-nucleotide siRNAs (Figure 4E) were profusely produced only from the region bordered by the two mdm-miR7122 target sites (Figure 4A). Both target sites are predicted to be cleavable based on the sequence pairing, but we confirmed only the cleavage of the 5′ site using RNA ligation mediated rapid amplification of 5′ cDNA ends (5′-RLM-RACE), which was consistent with the bulk of siRNAs in phase with the 5′ cleavage site (Figures 4A and 4B; see Supplemental Figure 1C online). However, the majority of 21 nucleotide tasiRNAs fell into the third register (a two nucleotide shift), instead of the first register that is in phase with the 5′ cleavage site (see Supplemental Figure 1C online). This out-of-phase phenomenon was also observed for other TAS or PHAS genes, perhaps due to slippage of the Dicer activity or reinforcement of cis-acting tasiRNAs (Axtell et al., 2006; Zhai et al., 2011; Rajeswaran et al., 2012; Xia et al., 2012). Since the nucleotide sequence of this PHAS locus appears not to code a polypeptide of any significant length or homology, we denoted it as Md-TASL1.
Figure 4.
The miR7122-TASL-PPR-siRNA Pathway Identified in Apple Has Distinct Features.
(A) Genomic configuration of Md-TASL1 and its phasing score and read abundance distribution in apple flower and fruit tissues viewed in IGV. Phasing score was calculated based on the mapping results of 21-nucleotide siRNAs (see Methods). Position of two mdm-miR7122 target sites are denoted in red (5′ site) and gray (3′ site) boxes; gray gridlines show the 21-nucleotide phasing pattern set up by the 5′ cleavage site of mdm-miR7122.
(B) A diagram illustrating the pattern of secondary siRNA biogenesis initiated by the 5′ target site. Parings of mdm-miR7122 with target sites are denoted below with the cleavage site marked by small red (5′ site) and gray (3′ site) arrows, respectively; two repetitive regions (R1 and R2) and tasiRNAs that trigger PPR phasiRNA production are individually labeled and indicated in boxes of different colors. Result of 5′-RLM-RACE is included in parentheses right below the 5′ target site; the numbers separated by the backslash indicate the number of sequenced DNAs with 5′-end corresponding to the mdm-miR7122 cleavage site (10th position) and the number of total sequenced DNAs.
(C) Double-stranded sequences of the two identical 83-bp regions (R1 and R2) with sites in phase with the 5′ cleavage site indicated with gray dotted lines and the two identical tasiRNAs that trigger PPR phasiRNA production marked in pink or orange.
(D) Position and conservation logo of the amino acid sequence encoded by the sequence targeted by the two trigger tasiRNAs. PPR domain is denoted by orange boxes.
(E) Length distribution of mdm-miR7122–triggered, Md-TASL1–derived phasiRNA population. RPM, reads per million.
(F) Categories of Md-TASL1–derived phasiRNAs and their function. siRNAs are classified according to their size and their ability to target PPRs. nt, nucleotides.
In total, 233 siRNA species were produced from Md-TASL1, 123 of which had both 21-nucleotide and 22-nucleotide variants (Figure 4F); over half of these siRNAs (135 out of 233) were predicted to be capable of targeting PPR genes in apple (AS ≤ 5; Figure 4F; see Supplemental Data Set 1E online). Deeper sequence analyses revealed that Md-TASL1 has a duplicated 83-nucleotide sequence (R1 and R2) 78 nucleotides away from the 5′ cleavage site (Figure 4B), and the two 22-nucleotide tasiRNAs triggering siRNA production from PPR genes are located in tandem within this repetitive region (Figures 4B and 4C). One is in phase with the 5′ cleavage site in the R1 repeat, while the other is in phase in the R2 repeat (Figure 4C). Accordingly, we designated these two siRNAs as 3′ D6(-) and 3′ D11(-) in accordance with their phase position (Figure 4C). They potentially cotarget a 44-nucleotide region in PPR genes that encodes a peptide of 14 amino acids spanning the adjacent parts of two PPR domains (Figure 4D); the 3′ D11(-)-mediated cleavage of three PPR genes was confirmed by the PARE data (see Supplemental Data Set 1E online). Taken together, we conclude that, distinct from the ppe-miR7122 that targets PPR directly or indirectly through TAS-like genes, mdm-miR7122 triggers PPR phasiRNA production solely by targeting Md-TASL1 through a potential two-hit 222 mode or an alternative 122 mode with the 3′ site not functioning biologically (Figure 3, category III).
The miRNA-TASL-PPR-siRNA Pathway Is More Extensive in Strawberry
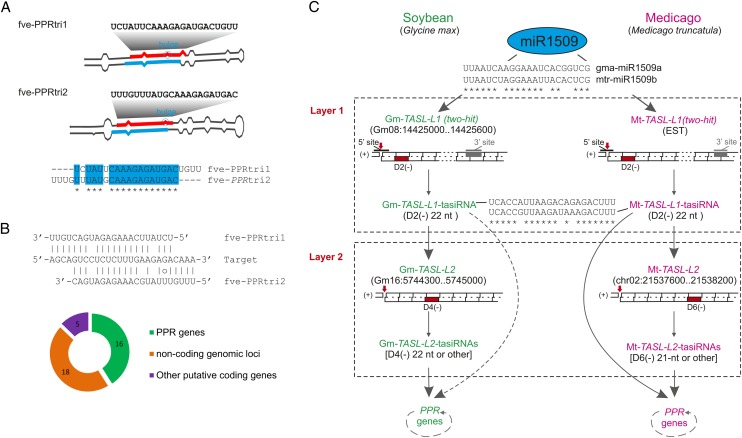
To ascertain whether analogous pathways are conserved in Rosaceae species, we conducted sRNA deep sequencing and similar reverse computation analyses in strawberry (Fragaria vesca). Nineteen PPR genes (see Supplemental Data Set 1F and Supplemental Figure 2A online; including eight PPRs not annotated accurately and one with a P value > 0.001) were shown to produce phasiRNAs; two 22-nucleotide miRNAs with critical structural features (22 nucleotides in length, 5′ terminal U, and a bulge-bearing stem-loop precursor) were identified as potential triggers. Pairwise comparison of their sequences revealed that they share 16 identical nucleotides with a four- nucleotide mutual shift (Figure 5A), but both show high sequence divergence (more than seven base differences) relative to the miR7122 conserved in peach and apple. We designated these as the strawberry PPR triggers 1 and 2 (fve-PPRtri1/2). The high similarity of these two trigger miRNAs showed an 18-base overlap of their target sites, with the target site of fve-PPRtri1 shifted four nucleotides toward the 5′ end of target genes relative to fve-PPRtri2 (Figure 5B). Apart from the PPR genes, fve-PPRtri1/2 appeared to target and trigger phasiRNA production from five other putative coding genes and 18 noncoding TAS-like loci in the strawberry genome, suggesting fve-PPRtri1/2 have gained targets outside the PPR family and thus are involved in a more extensive trans-acting network (Figure 5B; see Supplemental Data Set 1G online). It is noteworthy that five TAS-like loci are clustered in the same scaffold (scf05113146), with three residing within an ∼9-kb region (see Supplemental Figure 2B and Supplemental Data Set 1G online). They all show low similarity to PPR genes, and several phasiRNAs derived from these TAS-like loci have the potential to target PPR genes, indicative of a trans-acting role in line with that of TAS1/2 in Arabidopsis. Thus, as shown in Figure 3 (category I), fve-PPRtri1/2 directly or indirectly trigger the phasiRNA production of PPR genes, like ppe-miR7122, but in a much more extensive way.
Figure 5.
Distinct miRNA-TASL-PPR-siRNA Pathways Defined in Strawberry and Legume Plants.
(A) Stem-loop structures of strawberry miRNA triggers (fve-PPRtri1 and fve-PPRtri2) of PPR phasiRNA production. miRNA and miRNA* are marked in red and light blue, respectively, and bulges in stem-loop structures are indicated with a light-blue circle. Sequence comparison of fve-PPRtri1/2 is shown below.
(B) Illustration of sequence cotargeted by fve-PPRtri1 and fve-PPRtri2. Numbers of genes potentially targeted by fve-PPRtri1/2 is illustrated in a doughnut chart below.
(C) A diagram illustrating the miR1509-TASL-PPR-siRNA pathway with one or two layers of TASL-tasiRNA interaction in soybean and M. truncatula. TASL genes and pattern of their phasiRNA biogenesis are individually denoted. The 22-nucleotide (nt) tasiRNAs that are capable of targeting or triggering phasiRNA production in TASL transcripts or PPRs are marked in red. The potential direct cleavage of PPR transcripts by the 22-nucleotide tasiRNA, GmTASL-L1-D2(-), is denoted with a dotted line. The dotted arc around the “PPR genes” represents the ability to process secondary siRNAs.
Legume Plants Evolved a Similar miRNA-TASL-PPR-siRNA Pathway with Distinct Mechanistic Modes
Regulation of PPR transcripts via secondary siRNA production has been reported for several plant species, including Arabidopsis (Howell et al., 2007), soybean (Zhai et al., 2011), and poplar (Populus spp; Klevebring et al., 2009), indicating a broad existence for this pathway. Zhai et al. (2011) reported several PHAS PPR genes, but their miRNA triggers are currently unknown. To explore whether the pathway(s) by which PPR genes produce phasiRNAs in soybean shares features with those in the Rosaceae, we analyzed publicly available sRNA data from soybean. This identified a regulatory pathway consisting of two layers of TASL–tasiRNA interactions (Figure 5C; see Supplemental Figure 3 online). First, a 22-nucleotide miR1509 targets a transcript derived from an intergenic locus (Gm-TASL-L1) and initiates a first layer of siRNA biogenesis via a possible 222 mode. Subsequently, a 22-nucleotide tasiRNA [GmTASL-L1-D2(-)] from this locus triggers a second layer of siRNA biogenesis from another intergenic noncoding locus Gm-TASL-L2, previously reported by Zhai et al. (2011) (Figure 5C; see Supplemental Figure 3 online). Gm-TASL-L2 generates 21- or 22-nucleotide phased tasiRNAs, some of which are able to target PPR genes and subsequently initiate siRNA production (Figure 5C; see Supplemental Data Set 1H online). Eight of 13 PHAS PPRs have a target site complementary to gma-miR1508, for which cleavage in five PPRs was confirmed in the PARE data and also resulted in phasiRNA production (see Supplemental Figure 3 and Supplemental Data Set 1H online). A similar two-layer pathway was also identified in M. truncatula, another legume species (Figure 5C; see Supplemental Figure 3 online), although we failed to identify gma-miR1508 homologs for that species. An additional layer of complexity in soybean and M. truncatula is that the tasiRNA TASL-L1-D2(-) is capable of directly targeting PPR genes and initiating subsequent siRNA production, with the cleavage of a M. truncatula PHAS PPR gene (Medtr1g043080) by Mt-TASL-L1-D2(-) confirmed by PARE data (see Supplemental Data Set 1H online). In summary, PPR phasiRNA biogenesis can be initiated indirectly by miR1509 through one or two layers of TASL–tasiRNA interactions in both soybean and M. truncatula, with the miR1508-PPR-siRNA pathway present only in soybean (Figure 3, category IV).
miRNA Triggers Share a Common Origin and the miRNA-TASL-PPR-siRNA Pathway Is Conserved in Many Other Species
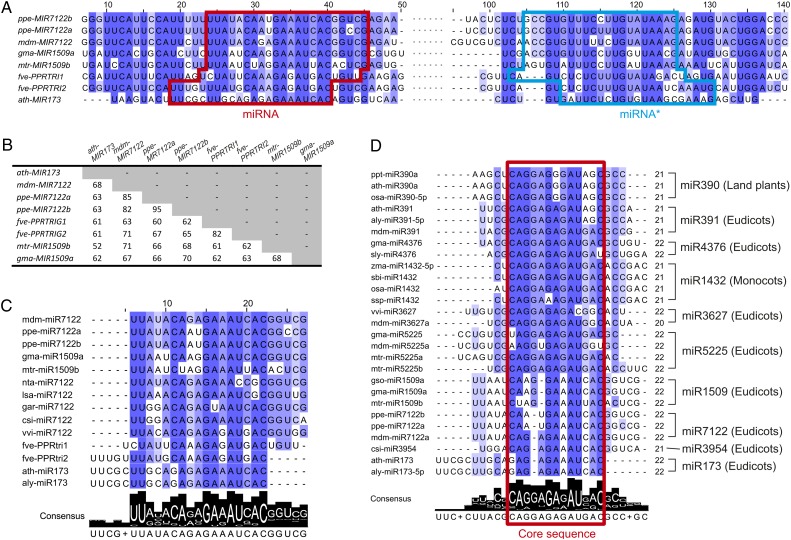
The functional similarity of miR173, miR7122, fve-PPRtri1/2, and miR1509 as triggers of the production of tasiRNAs targeting PPR genes raised the question of whether these miRNA triggers are evolutionarily related. To address this question, we performed a multiple alignment using the foldback sequences of eight relevant MIRNA genes (Figure 6A). Surprisingly, we found that with the exception of the ath-MIR173/mtr-MIR1509b pair for which the identity was only 52%, all interspecies pairwise comparisons revealed high levels of identity, ranging from 60 to 85% (Figure 6B). In particular, the miRNA and miRNA* as well as their adjacent sequences are highly conserved (Figure 6A; see Supplemental Figure 4A online). Three of the eight MIRNAs show different processing positions with a five-nucleotide shift toward the 5′ end occurring for ath-miR173 and fve-PPRtri2, and a one-nucleotide shift for fve-PPRtri1 (Figure 6A). Our results strongly suggest that these eight MIRNA genes, including MIR173, MIR7122s, fve-PPRTRI1/2, and MIR1509s, evolved from a common ancestor.
Figure 6.
miRNA Triggers Share a Common Origin and Show Sequence Similarity to Many Other miRNAs.
(A) Conservation profile of foldback sequences of eight MIRNA genes with miRNA and miRNA* marked in red and light-blue boxes, respectively. The sequence of the 61- to ∼94-bp region is denoted as dots, and full view of the alignment can be found in Supplemental Figure 4A online. The degree of conservation for each nucleotide along the MIRNA transcripts, as calculated by frequency, is represented by the color, with dark color denoting a high level of conservation and light color denoting a low level.
(B) Pairwise alignment scores (percentage) for each MIRNA gene.
(C) Conservation profile of all the miR7122 homologs identified. Consensus sequence and conservation logo are included below.
(D) Conservation profile of miR7122-related miRNAs. All the miRNA sequences were retrieved from miRbase (version 19). Position of the core sequences is marked with a red box.
To examine the conservation of these miR7122-related miRNAs and their functions in plants, we used publicly available deep-sequencing data to retrieve miR7122 homologs from five additional species, including tobacco (Nicotiana tabacum; nta-miR7122), grape (Vitis vinifera; vvi-miR7122), citrus (Citrus sinensis; csi-miR7122), tree cotton (Gossypium arboreum; gar-miR7122), and lettuce (Lactuca sativa; lsa-miR7122), all of which are of the same processing pattern as ppe-miR7122 without any miRNA shift (Figure 6C). Using reverse computation, we identified the miR7122-TAS-PPR-siRNA pathway in these species, except in tree cotton and lettuce due to the absence of their sequenced genomes. In citrus, in addition to the miR7122 homolog (a 22-nucleotide isoform of csi-miR3954; derived from a different MIRNA gene), we identified another miRNA trigger (csi-miRC1; see Supplemental Figure 5 online), which is predicted to directly target a PHAS PPR gene (see Supplemental Data Set 1I online). Similarly, we identified in poplar another 22-nucleotide miRNA as the trigger of the TAS-PPR-siRNA pathway, ptc-miR6427, instead of a miR7122 homolog (see Supplemental Data Set 1I online). Evidently, regulation of PPR genes mediated by miR7122 homologs is widespread in plants but with great mechanistic plasticity and diversity. Taken together, these pathways appear to fall into four categories as outlined in Figure 3.
Next, to examine the relationships between the PHAS PPRs from different species, 91 PHAS PPR gene sequences retrieved from eight species were subjected to phylogenetic analysis. These PPRs generally clustered together into species-specific subclasses (see Supplemental Figure 6A online), and the target site for miRNA or tasiRNAs appeared to be randomly distributed along or outside the PPR domain–coding region (see Supplemental Figure 6B online), implying that the strategy of targeting PPRs by miRNAs or tasiRNAs may have evolved relatively quickly after the divergence of these plant species, consistent with the extraordinary evolutionary plasticity of PPR genes in plants (O’Toole et al., 2008).
miR7122 Shares, to Various Degrees, Sequence Identity with Many Other miRNAs
The elucidation of the relationships among miR7122-homologous miRNAs indicates that miRNAs of a common ancestor and similar function are often divergent (more than four bases) or shifted along the precursor (more than two bases). Therefore, we applied more permissive search parameters to identify close relatives of miR7122. We found that miR7122 shares substantial identity with miR4376, especially in grape, in which vvi-miR4376 (see Supplemental Figure 7 online) and vvi-miR7122 share 17 nucleotides with 60% sequence identity detected over the length of the MIRNA foldback sequences (see Supplemental Figure 4B online). A deeper search in miRbase uncovered several other miRNAs that shared identity with miR7122 or miR4376, including the widely conserved miR390, and the lineage/species-specific miRNAs such as miR391, miR1432, miR5225, and miR3627. Multiple alignments revealed a 13-nucleotide core sequence highly conserved among these miRNAs (Figure 6D), suggestive of a potential common origin. The core sequence, which shows relatively more sequence divergence among the miR7122 homologs (including miR7122, miR1509, miR173, and miR3954; Figure 6D), is not consistently positioned within the miRNAs from different families. For instance, miR1432 is shifted three nucleotides in the 3′ direction, while miR173 is shifted five nucleotides in the 5′ direction, compared with miR390 (Figure 6D), indicating the common occurrence of sequence mutation and miRNA shifts during evolution within this group of plant miRNAs.
Potential Evolution of MIR7122s from MIR390s via MIR4376s
Next, we asked how these miRNA families evolved in the plant kingdom. The sequence similarity described above for these miRNAs provided a strong clue that they may have evolved from a common ancestral MIRNA gene. To test this possibility, we performed phylogenetic analysis for all the MIRNA genes from these families. Given that only the sequences of miRNA and miRNA* in the foldback sequences are relatively conserved during evolution (Jones-Rhoades et al., 2006; Fahlgren et al., 2010; Ma et al., 2010) and that frequent miRNA sequence shift was observed in this group of miRNAs (Figure 6D), to obtain a high-confidence multiple alignment, which is required for accurate phylogenetic analysis, we used the sequences consisting of the miRNA, miRNA*, and five nucleotides of their flanking sequences at each end to perform alignment and subsequent phylogenetic tree construction.
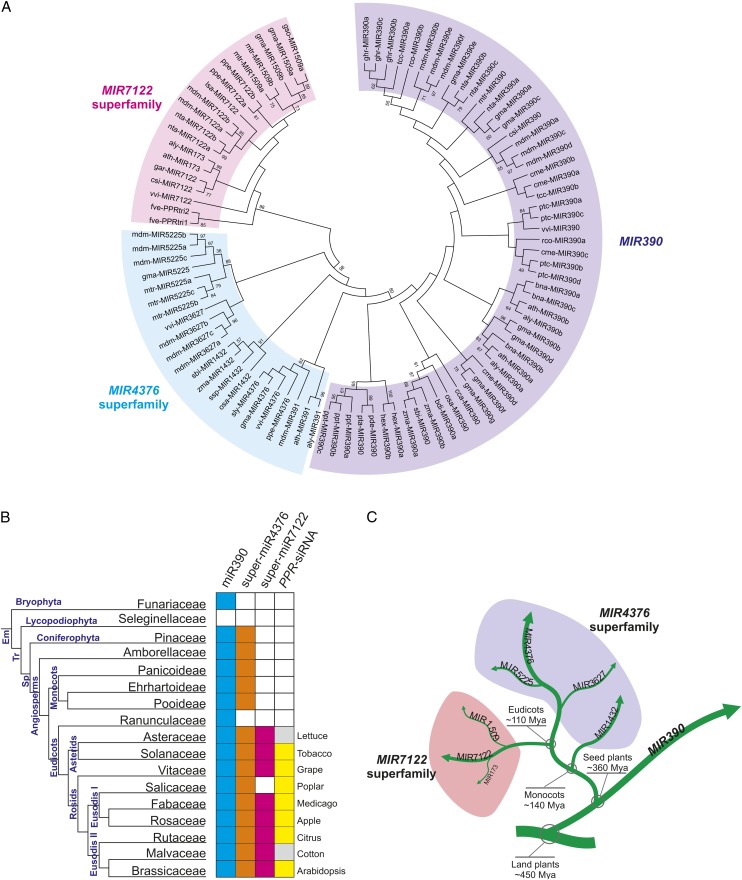
As shown in the unrooted tree in Figure 7A, the MIRNA genes of miR390, a highly conserved miRNA family of great sequence consistency (greater than or equal to two-nucleotide differences) and of extremely conserved biological function in plants, represented a basal clade and a separate group independent of other MIRNA genes. As expected, MIR7122 homologs constituted one of the two major clades other than MIR390 members, with MIR7122 genes dispersed among the MIR173 and MIR1509 taxa (Figure 7A). We designated this clade as the MIR7122 superfamily (super-MIR7122). Unexpectedly, MIRNA genes of miR391 members, previously classified into the miR390 family (Xie et al., 2005; Cuperus et al., 2011), were grouped into a separate clade with MIR4376, MIR1432, MIR3627, and MIR5225. Pairwise sequence comparisons revealed that miR391 showed greater identity to miR4376 than to miR390, especially for mdm-miR391 and gma-miR4376, which had only two nucleotide differences (Figure 7A; see Supplemental Figure 4C online). We identified a complementary site for ath-miR391 in the Arabidopsis gene coding for Ca2+-ATPase 10 (ACA10 [AT4G29900]) (see Supplemental Data Set 1J online), which is also suggestive of the closer evolutionary relationship of MIR391 to MIR4376 because miR4376 has been proven to target ACA10-homologous genes in members of the Solanaceae (Wang et al., 2011). The monocot-specific miR1432 also possesses high degree of sequence identity to the miR4376 (Figure 6D), which was reported to be present in eudicots in a lineage-specific manner (Wang et al., 2011), implying miR1432 is likely to be the variant of miR4376 in monocots. This is further supported by the observation that osa-miR1432 is also predicted to target a gene coding for Ca2+-ATPase (Os04g51610) (see Supplemental Data Set 1J online) (Lu et al., 2008; Sunkar et al., 2008). Similarly, miR5225 and miR3627 showed a high degree of sequence identity to miR4376 and were predicted to target ACA10 homologs (see Supplemental Figures 4D and 4E and Supplemental Data Set 1J online). These observations support the accuracy of our phylogenetic tree analysis (Figure 7A), thereby classifying MIR391, MIR1432, MIR4376, MIR3627, and MIR5225 as members of the MIR4376 superfamily (super-miR4376).
Figure 7.
Super-MIR7122s Are Potentially Evolved from MIR390s via super-MIR4376s.
(A) Phylogenetic analysis of MIR7122-related MIRNA genes. Excerpted nucleotide sequences encompassing the miRNA, miRNA*, and five nucleotide flanking sequences of them were aligned using CluastalW2 (see alignment in Supplemental Data Set 2 online). The bootstrap consensus tree was inferred using the maximum likelihood method and 1000 bootstraps with bootstrap values (above 50) indicated near the nodes (see detailed steps in Methods). Each major group is denoted with distinct colors.
(B) Conservation extent of miR7122-related miRNAs and PPR-siRNA pathway in plants. Boxes are highlighted if a miRNA or pathway was identified in at least one species for each of the plant families listed. Plant taxonomy was designated according to the Angiosperm Phylogeny Group classification III, as described by Cuperus et al. (2010). Em, embryophyta; Tr, tracheophyta; Sp, spermatophyta. Gray squares indicate that the existence of PPR-siRNA pathway cannot be determined due to the absence of sequenced genome.
(C) A diagram illustrating the possible evolutionary history of MIR7122-related MIRNA genes. Branch width represents the antiquity of the corresponding MIRNA gene family.
Overall, super-MIR7122 showed a close relationship with super-MIR4376, which was more closely related to MIR390 (Figure 7A). Therefore, we propose that super-MIR7122 evolved from super-MIR4376, which originated from the MIR390 family (Figure 7A).
miR7122 and the TAS-PPR-siRNA Pathway Emerged Only in Eudicot Plants
To further explore the proposed evolutionary path from MIR390, we examined the extent of conservation of the three (super)families using extensive publicly available data (small RNAs and genomes). As previously reported, miR390 is present in the common ancestor of all embryophytes (land plants; Cuperus et al., 2011) (Figure 7B). For the miR4376 superfamily, we were able to identify 24 homologs (representing miR4376, miR1432, miR3627, and miR5225) in 16 plant species besides those deposited in miRbase (see Supplemental Figure 7 online). The most ancient super-miR4376 homolog was identified from Picea abies, a Pinaceae plant belonging to the Coniferophyta. Two super-miR4376 homologs were also retrieved from Amborella trichopoda, a basal flowering plant (angiosperm). Thus, the super-MIR4376s were probably present in the common ancestor of all spermatophyta (seed plants) (Figure 7B). However, super-MIR7122 emerged much later and was detected only in the common ancestor of eudicots (Figure 7B), in agreement with the restricted appearance of the PPR-siRNA pathway in the eudicots. Thus, our results further revealed MIR390 as the common ancestor of both super-MIR4376 and super-MIR7122 and confirmed the delineated evolutionary course of super-MIR7122 from MIR390 through super-MIR4376.
Accordingly, we inferred the evolutionary history for these three (super)families to be that the MIR4376 superfamily probably originated from MIR390 after the divergence of seed plants ∼360 million years ago (Mya), after which the MIR1432 family evolved from MIR4376 following the origin of monocots ∼140 Mya (Figure 7C), and the MIR7122 superfamily arose after the split of the eudicots from its nearest relative ∼110 Mya (Figure 7C).
DISCUSSION
We identified in many eudicots a miRNA-TASL-PPR-siRNA pathway that is triggered by the miR7122 superfamily. We also elucidated the potential evolutionary path of this miRNA superfamily, thereby expanding our understanding of the process of miRNA evolution.
22-Nucleotide sRNAs Trigger Secondary siRNA Production in Plants
We identified a group of miRNAs, including miR7122, fve-PPRtri1/2, and miR1509, that are able to trigger secondary siRNA production in either TASL genes or PPRs. We also discovered their homologous relationship with miR173 in Arabidopsis and classified them all together into a miRNA superfamily, super-miR7122. All the members of super-miR7122, including the well-characterized miR173, are of 22 nucleotides in length and have an initial nucleotide “U” (Figure 6C); their miRNA/miRNA* duplexes are of asymmetrical structure with a bulge in the mature miRNA strand (see Supplemental Figure 8 online). The initial nucleotide “U” is vital for loading the miRNA to AGO1, the major effector protein directing RNA target cleavage (Mi et al., 2008; Chen et al., 2010). Even when shift in the processing sites of the mature miRNA has occurred, as for fve-PPRtri1/2 and miR173 (Figure 6A), the initial nucleotide “U” was preserved, consistent with the importance of AGO1 loading for secondary siRNA production (Montgomery et al., 2008b). Although sequence mutation frequently occurred for this group of miRNAs (only four nucleotides are completely conserved; Figure 6C), leading to nucleotide and position variation in the single bulge in the mature miRNA strand (see Supplemental Figure 8 online), the asymmetric miRNA/miRNA* duplex persisted, in agreement with the requirement of an asymmetric structure for processing miRNAs of 22 nucleotides in length, an important feature for a canonical miRNA trigger capable of initiating secondary siRNA production (Chen et al., 2010; Cuperus et al., 2010; Manavella et al., 2012).
The production of 22-nucleotide tasiRNAs was found to be prevalent in plants, and many of these 22-mers were capable of triggering secondary siRNA production in either TASL genes or PPRs in different species. This was described for TAS2 in Arabidopsis (Yoshikawa et al., 2005; Chen et al., 2007; Howell et al., 2007), TASL1/2 in peach, TASL1 in apple, TASL-L1/2 in soybean, and TASL-L1 in M. truncatula, as well as TASL1/2 in tobacco (see Supplemental Data Set 1I online). This observation is consistent with the importance of the 22-nucleotide length in triggering secondary siRNA production, but it is hard to reconcile with a recent proposal that an asymmetric bulge in the miRNA-duplex structure is necessary for secondary siRNA production, regardless of miRNA or miRNA* length (Manavella et al., 2012), because tasiRNAs are derived from double-stranded RNAs that are converted from miRNA-cleaved single-stranded RNAs and therefore are perfectly paired and presumably free of any asymmetric bulge (Peragine et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Yoshikawa et al., 2005). Furthermore, miR828, which is able to trigger siRNA production in TAS4 transcripts and MYB genes (Rajagopalan et al., 2006; Xia et al., 2012; Zhu et al., 2012), is produced from a symmetrical duplex without an asymmetric bulge. Thus, it appears that mechanisms underlying miRNA triggering of phasiRNA production in plants are far more complex.
miR7122-Mediated Regulation of PPR Genes Is Ubiquitous but Mechanistically Complex in Eudicots
We were able to identify the miR7122-TAS-PPR-siRNA pathway in nine different eudicots representing six plant families, indicating that this pathway is widely present in eudicots. For those pathways (as summarized in Figure 3), the initial miRNA triggers are homologous to each other and the processing of PPR genes into phasiRNA is conserved among different species. However, the intermediate components forming these regulatory circuits are highly dynamic from species to species. The miR7122 homologs can either target the PPR genes directly or act through TASL genes to initiate the phasiRNA production from PPR genes; the TASL-tasiRNA interaction can be multilayered, as in legume plants, and the miRNA triggers function through either 122 or 222 modes to activate TASL or PPR phasiRNA biogenesis. We found that two layers of TASL–tasiRNA interactions are integrated into a single trans-acting circuit in soybean and M. truncatula [i.e., miR1509 acts through the TASL-L1-tasiRNA-D2(-) (a first layer) and TASL-L2-tasiRNAs (a second layer) to initiate phasiRNA production from PPR genes] (Figure 5C). This second layer could have an advantage of adding more regulatory flexibility to the trans-acting circuit. Whereas most of the TASL or PHAS PPR genes are triggered via 122-acting miRNAs, we were able to identify three 222 PHAS loci, including Md-TASL1, Gm-TASL-L1, and Mt-TASL-L1. In contrast with the first 222 PHAS gene (Medtr7g012810) reported in M. truncatula in which two target sites are cleavable by miR1509 and phasiRNAs are produced only from the region flanked by the two sites (Zhai et al., 2011), we identified cleavage at only the 5′ target site of miR1509 in Gm-TASL-L1 and Mt-TASL-L1, while phasiRNA production was also detected in the region downstream from the 3′ site (see Supplemental Figure 3 online). A similar phenomenon was observed for a PHAS NB-LRR gene in tomato (Solanum lycopersicum), with two target sites of the trigger miR482 (5′ cleavable target site and 3′ noncleavable site) and siRNA produced all the way through the 3′ site (Shivaprasad et al., 2012). For Md-TASL1, although we failed to detect the cleavage of the 3′ site, phasiRNAs were produced only from the region flanked by the two target sites, and low-abundance siRNAs were found to be in phase with the 3′ cleavage site, implying that Md-TASL1 is more similar to Medtr7g012810, in which both sites are cleavable. Comparing these two types of 222 loci, it is conceivable that two target sites might be of unequal importance, as indicated in Md-TASL1 where the 5′ mdm-miR7122 target site may have more functional importance than the 3′ site since it is the 5′ site that sets the phase for the biogenesis of the two functional essential tasiRNA triggers of PPR siRNA production. Thus, the less important site may become nonfunctional or even lost over time, unlike the classic TAS3 221 triggers in which the noncleaving 5′ site is required (Axtell et al., 2006; Montgomery et al., 2008a). An alternative explanation is that the 3′ site of Md-TASL1 may serve as a protein binding site to set the boundary of siRNA production, like the 5′ noncleavable miR390 site in At-TAS3, which is essential for miR390:AGO7 complex binding to terminate the RDR6-mediated synthesis of complementary RNA (Rajeswaran and Pooggin, 2012).
In addition to miR7122 homologs, other miRNAs able to trigger PPR phasiRNA production have been identified in some species, including miR400 and miR161 in Arabidopsis (Howell et al., 2007), csi-miRC1 in citrus, and miR1508 in soybean. Intriguingly, miR1508 is missing from M. truncatula, a close relative of soybean, and miR6427, instead of a miR7122 homolog, was identified as the trigger of PPR phasiRNA biogenesis in poplar, illustrating both the dynamism and prevalence of the miRNA-TASL-PPR-siRNA pathway in eudicots.
Evolution of the MIR7122 Superfamily from the Widely Conserved MIR390 via the MIR4376 Superfamily Is Driven by Target Variation
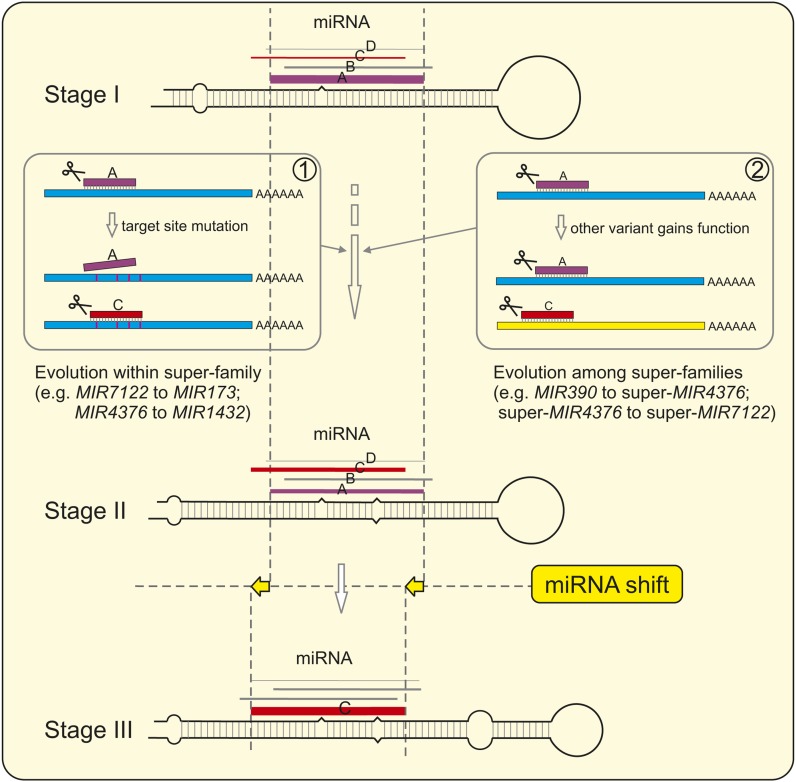
We demonstrated that the miRNA triggers of the PPR-siRNA pathway (miR7122, miR1509, fve-PPRtri1/2, and miR173) in different species are of a common origin and belong to the same miR7122 superfamily. Unlike other miRNA (super)families targeting a large family of PHAS genes, members of the miR7122 superfamily display great sequence diversity, which could be attributed to two causes: (1) processing shifts, particularly apparent in miR173 and fve-PPRtri2, in which the miRNA sequence shifts as much as five nucleotides toward the 5′ end (Figures 6A and 6C); and (2) point mutations, for example, only eight nucleotide positions were highly conserved (over 90% identity) across all of the species examined (Figure 6C). It is known that miR828 and the miR482/2118 superfamily directly cleave their target genes (MYBs and NB-LRRs, respectively), initiating phasiRNA production without an intermediate role of TAS genes. These two miRNA (super)families share the common feature of targeting highly conserved regions in their target genes (Zhai et al., 2011; Xia et al., 2012). However, the target sequences for miR7122 homologs and their ensuing tasiRNAs show high sequence diversity and are randomly distributed within PHAS PPR genes, which have low levels of sequence similarity in any given genome (see Supplemental Figure 6 online). In contrast with the MYB and NB-LRR families, which are defined by highly conserved functional domains, the large PPR family typically consists of hundreds of members and is characterized by two to 35 tandem repeats of a highly degenerate, 35–amino acid motif (Lurin et al., 2004; Shivaprasad et al., 2012). Thus, the PPR genes generally share less identity among family members at the nucleotide level. This dynamic nature of PPR genes could drive the rapid evolution of miRNA or other sRNA triggers for phasiRNA production to achieve proper regulation of gene function, which could account for the significant sequence divergence observed in the super-miR7122 and the requirement of a variety of tasiRNAs via one or two layer(s) of regulatory complexity. In fact, the sequence diversity of noncoding TASL genes would provide more flexibility for the derived phasiRNAs or tasiRNAs to meet rapid sequence change in the PPR genes in order to downregulate them effectively. Hence, we propose that the rapid variation of targeting sequences in PPR or TASL genes could eventually drive the evolution of miRNA triggers (Figure 8), leading to the high sequence diversity within the super-miR7122. The change of targeting sequences caused by point mutation (scenario 1 in Figure 8) would result in the switch of a functional, mature miRNA (from “A” to “C”) and give rise to the preferential processing of the functionally advantageous variant “C,” substituting the loss-of-function “A” variant (Stage I to Stage II). Continued selection would further reinforce the preferential processing of “C,” ultimately generating a new miRNA (Stage III).
Figure 8.
Model Proposed for miRNA Evolution via Duplication and Neofunctionalization.
For a given MIRNA gene, many miRNA variants are typically processed in addition to the most abundant one, which we referred to as mature miRNA “A” (Stage I). This mature miRNA “A” is the functional variant able to cleave the target gene, and the other variants are normally of low-level abundance and are nonfunctional (Stage I). Occasionally, for miRNAs from the same superfamily (scenario 1), like super-miR7122 and super-miR4376, mutations occurring in the target site would make the original mature miRNA “A” unable to cleave its target gene, but one of the variant “C” might happen to complementarily pair with the target gene and cleave it. Alternatively, for miRNAs from different superfamilies (scenario 2), the variant “C” might gain a function, targeting a gene different from the authentic target gene of the mature miRNA “A.” Conceivably, the functional variant switch (from “A” to “C” in scenario 1) or the newly gained function (scenario 2), conveying adaptive advantages, could give rise to the preferential processing of the variant “C,” and “A” would become less abundant (Stage II). After a long period, variant “C” could evolve as the predominant mature miRNA, while the previous “A” would be almost lost (Stage III). During this long evolutionary process, more frequent mutations would be present in regions of MIRNA genes outside of the miRNA/miRNA* duplex because of lower selection pressure imposed on those regions. Point mutations in the foldback sequences, occurring as well in the miRNA/miRNA* region but at a much lower frequency, would cause changes of the stem-loop structure and subsequent splicing shifts, leading to miRNA sequence shifts.
We also elucidated a miRNA evolutionary pathway in which the miR7122 superfamily is potentially evolved from another superfamily (super-miR4376), which itself originated from the conserved family miR390 (Figure 7). These three miRNA (super)families regulate distinct genes of diverse biological functions, with the miR390 regulating several auxin-responsive factors through TAS3-derived tasiARFs and the miR4376 superfamily targeting genes coding for Ca2+-ATPase (Axtell et al., 2006; Wang et al., 2011), while the miR7122 superfamily regulates PPR genes. Undoubtedly, diversification of target genes and selection pressure to diversify the proteins they encode can diversify the miRNA targeting sequences, serving to drive miRNA evolution. Therefore, the MIRNA evolution from MIR390 to super-MIR7122 via super-MIR4376 is as well driven by the target variation (Figure 8). Briefly, after the initial gene duplication, one of the duplicated MIRNA genes would be preserved to maintain the original essential function, while the other would be subjected to extensive mutation over time to create different miRNA variants, some of which could match or acquire new gene targets (scenario 2 in Figure 8). The new function for the miRNA variant “C,” initially gained by chance, may confer adaptive advantages under conditions of stress or reproductive isolation and stimulate the preferential processing of variant “C” (Stage II). This neofunctionalization process would eventually lead to the fixation of a new miRNA (Stage III). This model of target-driven miRNA evolution also fits to the super-miR4376 homologs, whose target sites are not well conserved, and instead are of relatively high diversity in their target genes (see Supplemental Data Set 1J online) (Wang et al., 2011).
In both inter- and intrasuperfamily evolution, sequence mutations would evenly occur in the foldback sequence of MIRNA genes, but fewer mutations would be retained in the miRNA/miRNA* duplex than other regions of MIRNA foldback sequences because of a higher purifying selection pressure (Fahlgren et al., 2010; Ma et al., 2010). This is apparent in the low sequence identity outside of the 13-bp core sequence among all the miRNAs from these three (super)families (Figure 6D). Mutations outside of the core sequence are not without effect, however, as they would impact the formation of secondary stem-loop structure (e.g., the loop position and stem length) (Song et al., 2010), possibly causing shifts in processing sites and giving rise to various miRNA variants. But only those mutations beneficial to the preferential processing of the advantageous new miRNA (“C” in Figure 8) would be selectively preserved. In the long term, the sequence similarity between the newly spawned MIRNA and its ancestral MIRNA would be gradually diluted due to mutation in the foldback sequences and miRNA processing changes.
The identification of two plant miRNA superfamilies, super-miR7122 and super-miR4376, and the discovery of the evolutionary path from MIR390 to super-MIR7122 via super-MIR4376 provide strong evidence for a sustained, deep evolutionary history of these plant miRNAs. We have linked one of the most conserved plant miRNAs (miR390) to narrowly conserved or lineage-specific miRNAs (miR173). Variation of target sequences has served as one of the major driving forces in this divergence. To date, the origin of most plant MIRNAs, especially highly conserved MIRNAs, remains largely unknown, although several evolutionary routes have been proposed. This includes the generation of new MIRNA genes from inverted gene duplication of their target genes (Allen et al., 2004), random foldback sequences (Felippes et al., 2008), or miniature inverted-repeat transposable elements (Piriyapongsa and Jordan, 2008). All of these routes require the accumulation of unpaired bases in the foldback sequences, acquisition of DCL1 as a processing enzyme, and subsequent production of discrete miRNA species, all in the context of coevolution with the miRNA targets (Voinnet, 2009). By contrast, generating a new MIRNA from a preexisted MIRNA would dramatically reduce the “cost” of adaption to the miRNA-biogenesis pathway because the preexisting MIRNA has already provided a “draft” of the structure for the new MIRNA: the canonical stem-loop structure and the DCL1 dependency required for miRNA biogenesis. Therefore, this route represents a new way of spawning new MIRNA genes, resembling the birth-and-death cycle for many protein-coding genes (Nei and Rooney, 2005).
Possible Function of the miR7122-TAS-PPR-siRNA Pathway in Plants
The wide conservation of the miR7122-TAS-PPR-siRNA pathway in eudicots suggests it assumes important biological functions. Most functionally characterized PPR proteins have nucleotide sequence–specific binding activity and have been proposed to be molecular adaptors directing RNA processing complexes to target RNAs in mitochondria or chloroplasts (Lurin et al., 2004; Schmitz-Linneweber and Small, 2008). PPR proteins can be separated into two major classes based on the nature of their PPR motifs, the P and PLS classes (Schmitz-Linneweber and Small, 2008). In plants, the P class of PPR proteins, to which phasiRNA production is restricted, may be involved in plant fertility, as discerned from roles of these proteins in cytoplasmic male sterility (CMS) (Howell et al., 2007; Schmitz-Linneweber and Small, 2008). To produce hybrid seed, nuclear restorer (Rf) genes, encoding P class PPR proteins in most cases, are capable of preventing the expression of mitochondrial CMS-inducing genes, which encode hydrophobic CMS-specific polypeptide (Schmitz-Linneweber and Small, 2008). Intriguingly, we failed to identify a similar PPR-siRNA pathway in monocots, although most of these Rf genes seem to share common characteristics in plants, even from species as divergent as rice and Arabidopsis (Schmitz-Linneweber and Small, 2008). The reason for the absence of PPR-siRNA pathway is probably the lack of miRNA triggers in monocots, in which the super-miR4376 ancestor has evolved into a 21-nucleotide miR1432 instead of a 22-nucleotide miR7122 homolog. Thus, the biological function of the PPR-siRNA pathway may be substituted by other sRNA-involved mechanisms in monocots, as suggested by a recent study in which a noncoding RNA locus producing a 21-bp sRNA, likely a phasiRNA, was characterized as an important regulator of male sterility in rice (Ding et al., 2012; Zhou et al., 2012). Additionally, miR4376, a miRNA evolutionarily related to miR7122, is also involved in reproductive growth through regulating an ACA10 gene and triggering phasiRNA production (Wang et al., 2011). Taken together, we speculate that the miR7122-TAS-PPR-siRNA pathway may play essential roles in the reproductive processes of plants, an important topic for future investigations.
METHODS
Plant Material and RNA Isolation for Strawberry
Plant tissue collection, RNA isolation, and sRNA sequencing for peach (Prunus persica cv Lovell) and apple (Malus × domestica cv Golden delicious) were reported and described previously by Zhu et al. (2012) and Xia et al. (2012), respectively. Sequencing of sRNAs in diploid strawberry (Fragaria vesca) was similarly performed. Total RNAs were isolated from 1-d-open flowers of the inbred line ‘Yellow Wonder 5AF7’ grown in growth chambers (Hollender et al., 2012) using the RNeasy plant mini kit (Qiagen). sRNAs were enriched by precipitating the supernatant passing through Qiashedder spin column. The recovered small RNA samples were used for library construction and deep sequencing at the Genomics Resources Core Facility at Weill Cornell Medical College (New York, NY).
Bioinformatics Analysis
Publicly available deep-sequencing data from peach, apple, soybean (Glycine max), Medicago truncatula, grape (Vitis vinifera), and other species were downloaded from the GenBank Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), and original SRA files were dumped into FASTQ files using the SRA toolkit (http://www.ncbi.nlm.nih.gov/sra). FASTA toolkits (http://hannonlab.cshl.edu/fastx_toolkit/) were used for data processing, including format conversion, adaptor trimming, and read collapsing. Read mapping was conducted using Bowtie (Langmead et al., 2009) with no mismatches allowed, and Vienna RNA package (Hofacker, 2003) or Mfold (Zuker, 2003) was used for the secondary structure prediction of miRNA precursors. Multiple alignments were performed using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with default settings and viewed with Jalview (http://www.jalview.org/). Sequence logo (Figure 4D) was produced by GENIO/logo (http://www.biogenio.com/logo/logo.cgi). miRNA target prediction and confirmation were conducted by combining the utilization of Targetfinder (http://carringtonlab.org/resources/targetfinder) and Cleveland 2.0 (Addo-Quaye et al., 2009). The P value plot (Figure 1B) was produced using the R package (http://www.r-project.org/). sRNA abundance was normalized to reads per million, and PARE reads were normalized to transcripts per 10 million.
Computational Calculation of P Value and Phasing Score
After mapping sRNA reads to the reference genome, unique sRNAs were denoted with their matching coordinates. Two-nucleotide positive offset was added for sRNA matching to the antisense strand because of the existence of two-nucleotide overhang at the 3′ end of sRNA duplex. A transcriptome- or genome-wide search was performed using a nine-cycle sliding window (189 bp) with each shift of three cycles (63 bp), and windows were reported when ≥10 unique reads fell into a nine-cycle window, ≥50% of matched unique reads were 21 nucleotides in length, and with greater than or equal to three unique reads fell into a certain register. Next reported windows with overlapping region were combined into a single longer window. Then, a P value was calculated for each window based on the mapping results using an algorithm similar that of Chen et al. (2007) with optimization.
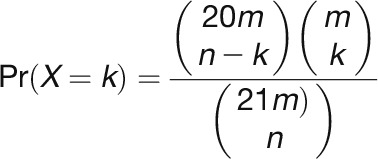 |
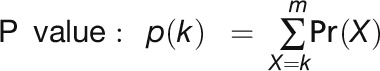 where n = number of total unique 21-nucleotide sRNAs matched within a window; k = number of unique 21-nucleotide sRNAs falling into the maximum register (k ≥ 3); and m = number of 21-nucleotide phases within a window.
where n = number of total unique 21-nucleotide sRNAs matched within a window; k = number of unique 21-nucleotide sRNAs falling into the maximum register (k ≥ 3); and m = number of 21-nucleotide phases within a window.
Phasing score was calculated using the algorithm developed by De Paoli et al. (2009) within a nine-cycle window (phase cycle length was set to 21 nucleotides).
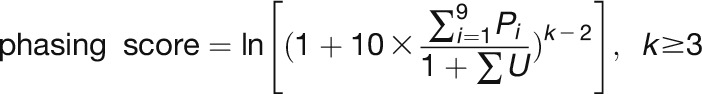 |
where n = number of phase cycle positions occupied by at least one 21-nucleotide sRNA within a nine-cycle window; P = total number of reads for all 21-nucleotide sRNAs falling into a given phase within a nine-cycle window; and U = total number of reads for all 21-nucleotide sRNA falling out of the given phase.
Phasing score data were viewed using the Integrative Genomics Viewer (IGV) (Robinson et al., 2011).
Deep-Sequencing Data and Reference Sequence Files
All sRNA deep-sequencing and PARE data are accessible in the GenBank Gene Expression Omnibus with the corresponding accession numbers listed in Supplemental Data Set 1K online. Transcriptome and genome files for the Rosaceae plants (apple, peach, and strawberry) and other plants were downloaded from the Genome Database for Rosaceae (http://www.rosaceae.org) and Phytozome (http://www.phytozome.net), respectively. miRNA and foldback sequences were retrieved from the miRbase (version 19; www.mirbase.org).
Phylogenetic Analysis
Nucleotide sequences, consisting of the miRNA, miRNA*, and five-nucleotide flanking sequences at each end, of 95 MIRNA genes were aligned using the IUB DNA weight matrix by ClustalW2, with an open gap penalty of 10 and an extension gap penalty of 0.1 in pairwise alignments, an extension gap penalty of 0.2 in multiple alignments, and a gap distances penalty of 5. Manual refinement was conducted for the nucleotide alignment by Jalview (shown in Supplemental Data Set 2 online). The phylogenetic tree was inferred using the maximum likelihood method based on the Tamura-Nei model by MEGA5 (Tamura et al., 2011). The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in <50% bootstrap replicates are collapsed. Uniform rate variation among site was enabled. Initial tree for the heuristic search was obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood approach and then selecting the topology with superior log likelihood value. All positions with <50% site coverage were eliminated. That is, fewer than 50% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 62 positions in the final data set.
Multiple alignment of PPR genes was performed with ClustalW2 with similar settings using deduced amino acid sequences (see Supplemental Data Set 3 online). Phylogenetic tree analysis for PPR genes was conducted using the neighbor-joining method by MEGA5 with 1000 bootstrap replicates.
5′-RLM-RACE
Following the manufacturer’s instructions for the First-Choice RLM-RACE kit (Ambion), 5 μg of total RNA isolated from apple flower was used for ligating 5′ RNA adaptors at 15°C overnight. Two specific primers (1st, 5′-CTGCCCTCCACCATTTCTTTG-3′, and 2nd, 5′-AGCTAAATCACACGCTTCAAAC-3′) were designed to conduct nested PCRs, and PCR products were cloned to the pGEM-easy vector (Promega) and sequenced by Beckman Coulter Genomics.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases or Phytozome (www.phytozome.net) with accession numbers listed in Supplemental Data Set 1K online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. siRNA Distribution along Pp-TASL1/2, a Peach PPR Gene and Md-TASL1.
Supplemental Figure 2. The miRNA-TASL-PPR-siRNA Pathway Is Conserved in Strawberry.
Supplemental Figure 3. Two Layers of Trans-acting Interaction Involved in the miR1509-TAS-PPR-siRNA Pathway in Soybean and Medicago.
Supplemental Figure 4. Alignment of MIRNA Genes.
Supplemental Figure 5. Stem-Loop Structure of miRC1 in Citrus.
Supplemental Figure 6. Phylogenetic Analysis of PHAS PPR Genes and Distribution of Target Sites of miRNAs or TasiRNAs along PPR Domains.
Supplemental Figure 7. Stem-Loop Structures of Newly Identified miRNA Homologs of the miR4376 Superfamily.
Supplemental Figure 8. Stem-Loop Structures of Identified MIR7122 Homologs.
Supplemental Data Set 1A. PHAS Genes Identified in Peach.
Supplemental Data Set 1B. PHAS PPR Genes Identified in Peach.
Supplemental Data Set 1C. Pp-TASL1/2-Derived TasiRNAs Predicted to Target PPR Genes.
Supplemental Data Set 1D. PHAS PPR Genes Identified in Apple.
Supplemental Data Set 1E. Md-TASL1–Derived TasiRNAs Predicted to Target PPR Genes.
Supplemental Data Set 1F. PHAS PPR Genes Identified in Strawberry.
Supplemental Data Set 1G. PHAS Genomic Loci or Other Genes Targeted by fve-PPRtri1/2 in Strawberry.
Supplemental Data Set 1H. PHAS PPR Genes Identified in Soybean and Medicago.
Supplemental Data Set 1I. PHAS PPR Genes or Genomic Loci Related to PPR-siRNA Pathways in Other Species.
Supplemental Data Set 1J. Predicted Target Genes for miR391, miR1432, miR5225, and miR3627.
Supplemental Data Set 1K. Deep Sequencing Data Used for Analysis.
Supplemental Data Set 2. Multiple Alignment of MIRNA Excerpted Sequences.
Supplemental Data Set 3. Multiple Alignment of Deduced Amino Acid Sequences of PHAS PPR Genes.
Supplementary Material
Acknowledgments
We thank Tony Wolf and the Department of Horticulture at Virginia Tech for financial support to R.X. We also thank members of the Liu and Meyers labs for critical discussions and valuable suggestions.
AUTHOR CONTRIBUTIONS
R.X. and Zo.L. designed the research, analyzed the data, and wrote the article. B.C.M. analyzed the data and wrote the article. Zh.L. and S.Y. performed the research. E.P.B. designed the research and wrote the article.
Glossary
- sRNA
small RNA
- miRNA
microRNA
- siRNA
small interfering RNA
- tasiRNA
trans-acting siRNA
- phasiRNA
phased siRNA
- AS
alignment score
- 5′-RLM-RACE
RNA ligation mediated rapid amplification of cDNA ends
- PARE
parallel analysis of RNA ends
- Mya
million years ago
- CMS
cytoplasmic male sterility
- IGV
Integrative Genomics Viewer
References
- Addo-Quaye C., Miller W., Axtell M. (2009). CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25: 130–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Sung G.H., Spatafora J.W., Carrington J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36: 1282–1290 [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Baulcombe D. (2004). RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Li Y.H., Wu S.H. (2007). Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3318–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Fahlgren N., Carrington J.C. (2011). Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paoli E., Dorantes-Acosta A., Zhai J., Accerbi M., Jeong D.H., Park S., Meyers B.C., Jorgensen R.A., Green P.J. (2009). Distinct extremely abundant siRNAs associated with cosuppression in petunia. RNA 15: 1965–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Shen J., Mao H., Xie W., Li X., Zhang Q. (2012). RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol. Plant 5: 1210–1216 [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Jogdeo S., Kasschau K.D., Sullivan C.M., Chapman E.J., Laubinger S., Smith L.M., Dasenko M., Givan S.A., Weigel D., Carrington J.C. (2010). MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22: 1074–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Felippes F.F., Schneeberger K., Dezulian T., Huson D.H., Weigel D. (2008). Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA 14: 2455–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L. (2003). Vienna RNA secondary structure server. Nucleic Acids Res. 31: 3429–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender C.A., Geretz A.C., Slovin J.P., Liu Z. (2012). Flower and early fruit development in a diploid strawberry, Fragaria vesca. Planta 235: 1123–1139 [DOI] [PubMed] [Google Scholar]
- Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C., Kasprzewska A., Tennessen K., Fernandes J., Nan G.-L., Walbot V., Sundaresan V., Vance V., Bowman L.H. (2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19: 1429–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. (2006). MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Klevebring D., Street N.R., Fahlgren N., Kasschau K.D., Carrington J.C., Lundeberg J., Jansson S. (2009). Genome-wide profiling of Populus small RNAs. BMC Genomics 10: 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., et al. (2008). Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc. Natl. Acad. Sci. USA 105: 4951–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q.J., Mittal A., Jia F., Rock C.D. (2012). An autoregulatory feedback loop involving PAP1 and TAS4 in response to sugars in Arabidopsis. Plant Mol. Biol. 80: 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Coruh C., Axtell M.J. (2010). Arabidopsis lyrata small RNAs: Transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22: 1090–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavella P.A., Koenig D., Weigel D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 109: 2461–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M., Kanno T., Daxinger L., Huettel B., Matzke A.J.M. (2009). RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21: 367–376 [DOI] [PubMed] [Google Scholar]
- Meyers B.C., et al. (2008). Criteria for annotation of plant MicroRNAs. Plant Cell 20: 3186–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. (2008a). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Montgomery T.A., Yoo S.J., Fahlgren N., Gilbert S.D., Howell M.D., Sullivan C.M., Alexander A., Nguyen G., Allen E., Ahn J.H., Carrington J.C. (2008b). AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl. Acad. Sci. USA 105: 20055–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Rooney A.P. (2005). Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39: 121–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole N., Hattori M., Andres C., Iida K., Lurin C., Schmitz-Linneweber C., Sugita M., Small I. (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25: 1120–1128 [DOI] [PubMed] [Google Scholar]
- Peragine A., Yoshikawa M., Wu G., Albrecht H.L., Poethig R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J., Jordan I.K. (2008). Dual coding of siRNAs and miRNAs by plant transposable elements. RNA 14: 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeswaran R., Aregger M., Zvereva A.S., Borah B.K., Gubaeva E.G., Pooggin M.M. (2012). Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res. 40: 6241–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeswaran R., Pooggin M.M. (2012). RDR6-mediated synthesis of complementary RNA is terminated by miRNA stably bound to template RNA. Nucleic Acids Res. 40: 594–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Small I. (2008). Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 13: 663–670 [DOI] [PubMed] [Google Scholar]
- Shivaprasad P.V., Chen H.M., Patel K., Bond D.M., Santos B.A., Baulcombe D.C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Ammour A., Windels D., Arn-Bouldoires E., Kutter C., Ailhas J., Meins F., Jr., andVazquez F. (2011). miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 157: 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Axtell M.J., Fedoroff N.V. (2010). RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 20: 37–41 [DOI] [PubMed] [Google Scholar]
- Sunkar R., Zhou X., Zheng Y., Zhang W., Zhu J.K. (2008). Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 8: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crété P. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Voinnet O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang Y., Itaya A., Zhong X., Wu Y., Zhang J., van der Knaap E., Olmstead R., Qi Y., Ding B. (2011). Function and evolution of a MicroRNA that regulates a Ca2+-ATPase and triggers the formation of phased small interfering RNAs in tomato reproductive growth. Plant Cell 23: 3185–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L., Carles C.C., Osmont K.S., Fletcher J.C. (2005). A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. USA 102: 9703–9708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R., Zhu H., An Y.Q., Beers E.P., Liu Z. (2012). Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13: R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25: 2540–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Li G., Wang J., Fang J. (2012). Identification of trans-acting siRNAs and their regulatory cascades in grapevine. Bioinformatics 28: 2561–2568 [DOI] [PubMed] [Google Scholar]
- Zhou H., et al. (2012). Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 22: 649–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Xia R., Zhao B., An Y.Q., Dardick C.D., Callahan A.M., Liu Z. (2012). Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol. 12: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.