This work investigates how metabolism and oil storage capacity of the growing embryo of oilseed rape is adjusted to developmental changes in its architecture. It shows that locally distinct growth conditions due to the folding of cotyledons cause metabolic heterogeneity that reflects at the level of pathway activity, metabolites, and storage products (oil/protein).
Abstract
Constrained to develop within the seed, the plant embryo must adapt its shape and size to fit the space available. Here, we demonstrate how this adjustment shapes metabolism of photosynthetic embryo. Noninvasive NMR-based imaging of the developing oilseed rape (Brassica napus) seed illustrates that, following embryo bending, gradients in lipid concentration became established. These were correlated with the local photosynthetic electron transport rate and the accumulation of storage products. Experimentally induced changes in embryo morphology and/or light supply altered these gradients and were accompanied by alterations in both proteome and metabolome. Tissue-specific metabolic models predicted that the outer cotyledon and hypocotyl/radicle generate the bulk of plastidic reductant/ATP via photosynthesis, while the inner cotyledon, being enclosed by the outer cotyledon, is forced to grow essentially heterotrophically. Under field-relevant high-light conditions, major contribution of the ribulose-1,5-bisphosphate carboxylase/oxygenase–bypass to seed storage metabolism is predicted for the outer cotyledon and the hypocotyl/radicle only. Differences between in vitro– versus in planta–grown embryos suggest that metabolic heterogeneity of embryo is not observable by in vitro approaches. We conclude that in vivo metabolic fluxes are locally regulated and connected to seed architecture, driving the embryo toward an efficient use of available light and space.
INTRODUCTION
The shape, size, and architecture of the seed affect many aspects of a plant's ecology and evolution (Moles et al., 2005; Gegas et al., 2010; Muller-Landau, 2010). The size of the seed is of high relevance for crop production, and seed yield has been and remains the key trait in many breeding programs. Small-seeded species have evolved to produce large numbers of progeny, among which only few will complete their life cycle; by contrast, large-seeded species produce few seeds, but the surviving seedlings are generally robust enough to ensure that a high proportion completes its life cycle. Seed development in the small-seeded species Arabidopsis thaliana has been thoroughly investigated (Spencer et al., 2007; Niu et al., 2009; DeSmet et al., 2010; North et al., 2010; Sun et al., 2010; Xiang et al., 2011), allowing for some of the molecular bases for determining seed size and pattern formation to be elucidated (Garcia et al., 2003; Ohto et al., 2005; Ingouff et al., 2006; Adamski et al., 2009). After fertilization, the growing embryo establishes a bilateral symmetry, followed by bending and folding of cotyledons due to the physical restrictions imposed by the testa and the endosperm. It is relevant to ask whether these morphological changes in themselves have an effect on the metabolism of the embryo and whether embryo bending in turn affects local growth conditions. Similarly, positional/architectural cues may underlie some of the well-established gradients of gene expression (Belmonte et al., 2013) as well as those for storage products and metabolite concentrations (Borisjuk et al., 2005; Rolletschek et al., 2005; Li et al., 2006; Andriotis et al., 2010). It has recently been possible to demonstrate that the architecture of the cereal caryopsis induces variation in the severity of these constraints, forcing the endosperm to adjust to localized concentrations of Suc and oxygen (Melkus et al., 2011; Rolletschek et al., 2011). This adjustment causes metabolic heterogeneity within the starchy endosperm and provides the means to substantially improve the nitrogen and carbon use efficiency of the caryopsis.
Unlike Arabidopsis, its close relative oilseed rape (Brassica napus) produces seeds of high economic value. The lipid content of the seed of elite varieties is ∼50%, and, consequently, oilseed rape has become the third most valuable oil crop on a global basis. The storage metabolism in its seed is therefore of great interest, particularly in the context of making further improvements in lipid content (Abbadi and Leckband, 2011). The regulation of lipid storage has been reviewed repeatedly (Baud and Lepiniec, 2009; Weselake et al., 2009; Wallis and Browse 2010), and there is evidence that the metabolic machinery used by Arabidopsis is readily transferable to oilseed rape (Niu et al., 2009). Although most of the genes involved in lipid storage have been well characterized, the control of metabolic flux in vivo remains obscure. Recent data suggest that lipid assembly exerts significant control over oil accumulation in oilseed rape (Taylor et al., 2009; Tang et al., 2012), which, however, contrasts with other species (Ramli et al., 2009), and the final levels of triacylglycerol seem to be only poorly correlated to the transcriptional activity of the corresponding genes (Troncoso-Ponce et al., 2011). Neither the spatial regulation of lipid storage nor the existence of any positional cues imposed by seed architecture have been explored to date, even though it has been shown that distinct embryo components (radicle/hypocotyl versus cotyledons) in both Arabidopsis and oilseed rape accumulate different levels of lipid at the mature stage (Li et al., 2006).
The absence of the required positional information is in part attributable to a lack of appropriate analytical methods. However, recent developments in magnetic resonance imaging (MRI; Borisjuk et al., 2012) and mass spectrometry have begun to remove this limitation, thereby allowing for the spatial mapping of storage lipids (Neuberger et al., 2009; Fuchs et al., 2013) and their individual components (Horn et al., 2012). In this article, we designed an array of spatial high-resolution techniques, which allowed us not only to visualize steep gradients in oil deposition within the embryo of oilseed rape but also to define their spatial/temporal relations to those established for the accumulation of starch and storage proteins, cellular growth, photosynthetic activity, and metabolite pattern. A high metabolic heterogeneity became apparent and was further explored by applying tissue-specific metabolic modeling. Our findings demonstrate how the embryo is able to make local metabolic adjustments to its environmental and growing conditions and provide a mechanistic view on the role of seed architecture for embryo metabolism in vivo.
RESULTS
Embryo Topology Analyzed by MRI-Based Modeling
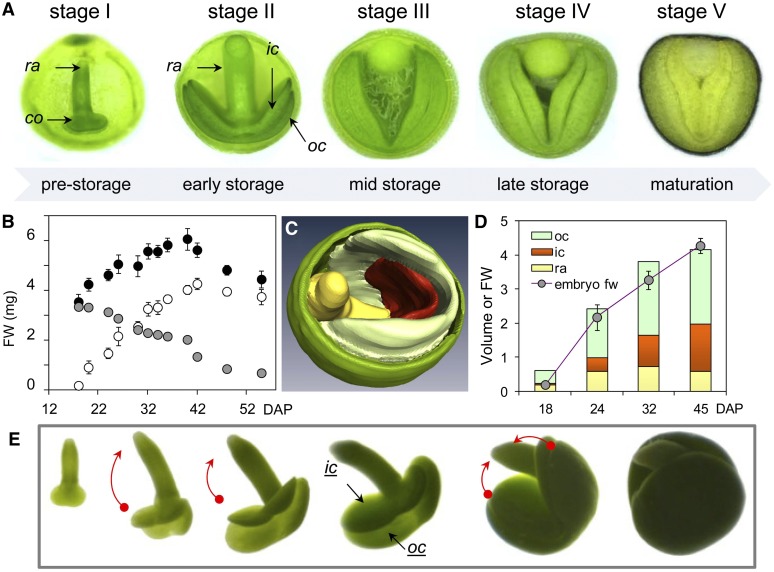
The final seed size (∼2 mm) of oilseed rape is established initially by the growth of the testa and endosperm; later, the embryo becomes the major site for assimilate storage (Figures 1A and 1B). As the amount of space available for expansion becomes limited, the embryo is forced to bend and fold (Figures 1A and 1E). To monitor the changes in the shape and volume of the embryo and its various components we applied noninvasive MRI (see Methods). At ∼20 d after pollination (DAP), the testa, both cotyledons, and the embryo axis are clearly distinguishable (Figure 1C). The MRI-based estimates of embryo volume were in good agreement with weight-based determinations (Figure 1D). The outer cotyledon expanded more rapidly than either the radicle or the inner cotyledon, growing to become the largest single component of the seed (Figure 1D). As development proceeded, the outer cotyledon expanded to cover both the inner cotyledon and (partly) the embryo axis, thereby limiting the latter’s access to light and restricting their space. Such subordination in growth causes distinct local conditions inside of the seed, possibly having feedback effects on embryo metabolism in a tissue-specific manner. We therefore tested the growing embryo for metabolic heterogeneity at the level of photosynthetic activity, storage product deposition, component-specific growth rates, and metabolites.
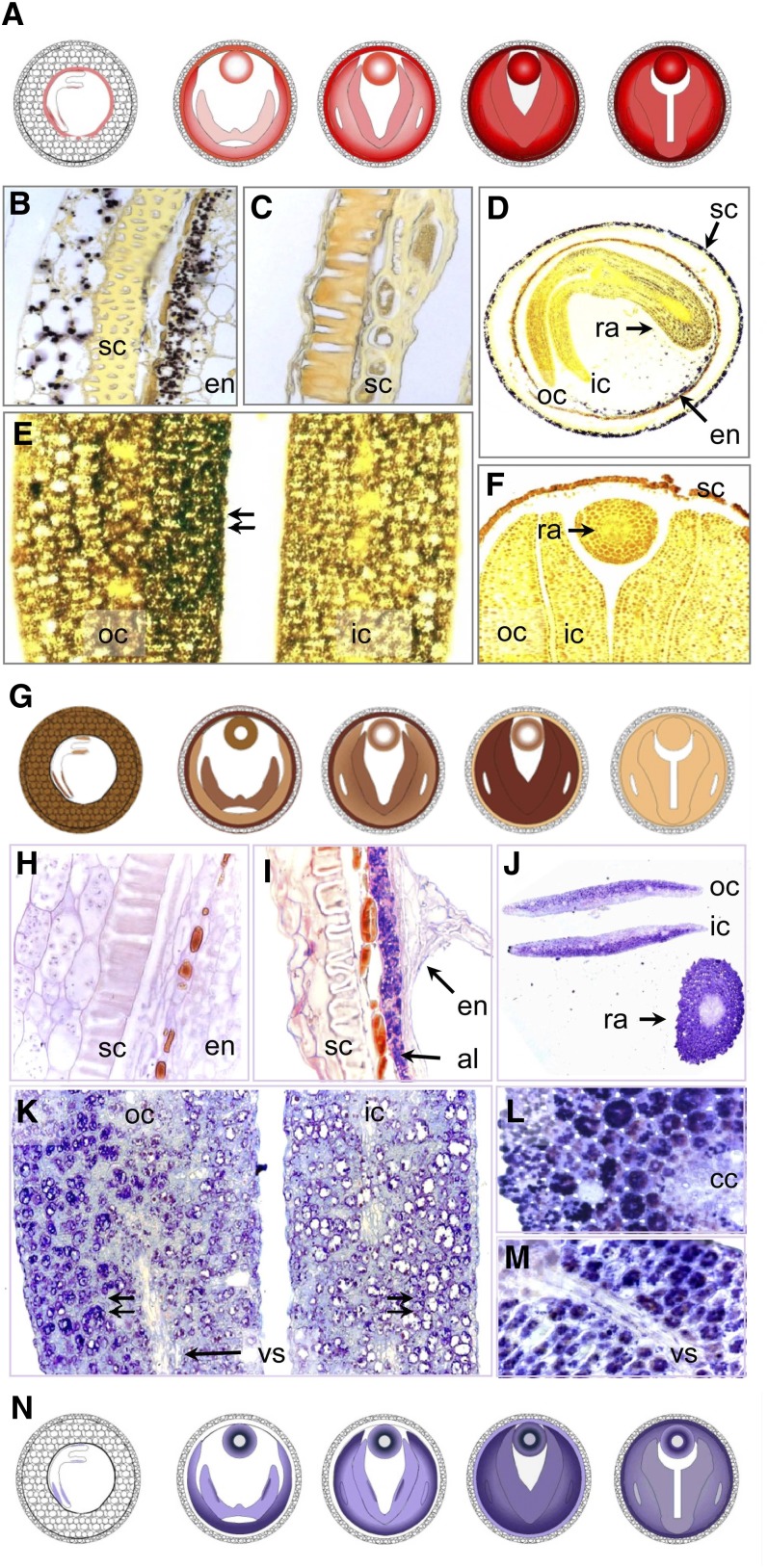
Figure 1.
Modeling Embryo Architecture in Oilseed Rape.
(A) The development of embryo size and shape. co, cotyledon; ic, inner cotyledon; oc, outer cotyledon; ra, radicle.
(B) Developmental changes in whole seed (black circle), embryo (white), and endosperm/seed coat (gray) fresh weight (FW); means ± sd (n = 10) are provided.
(C) A cut-away three-dimensional model of the seed (20 DAP) showing the seed coat (green), outer cotyledon (light green), inner cotyledon (red), and hypocotyl/radicle (yellow); the volumes of each embryo component were estimated from the model.
(D) Volumes of the various embryo components (in mm3) and whole embryo fresh weight (FW in mg) at various storage stages.
(E) Embryos isolated at distinct storage stages, showing the deformation of the embryo and the folding of the cotyledons (indicated by red arrows).
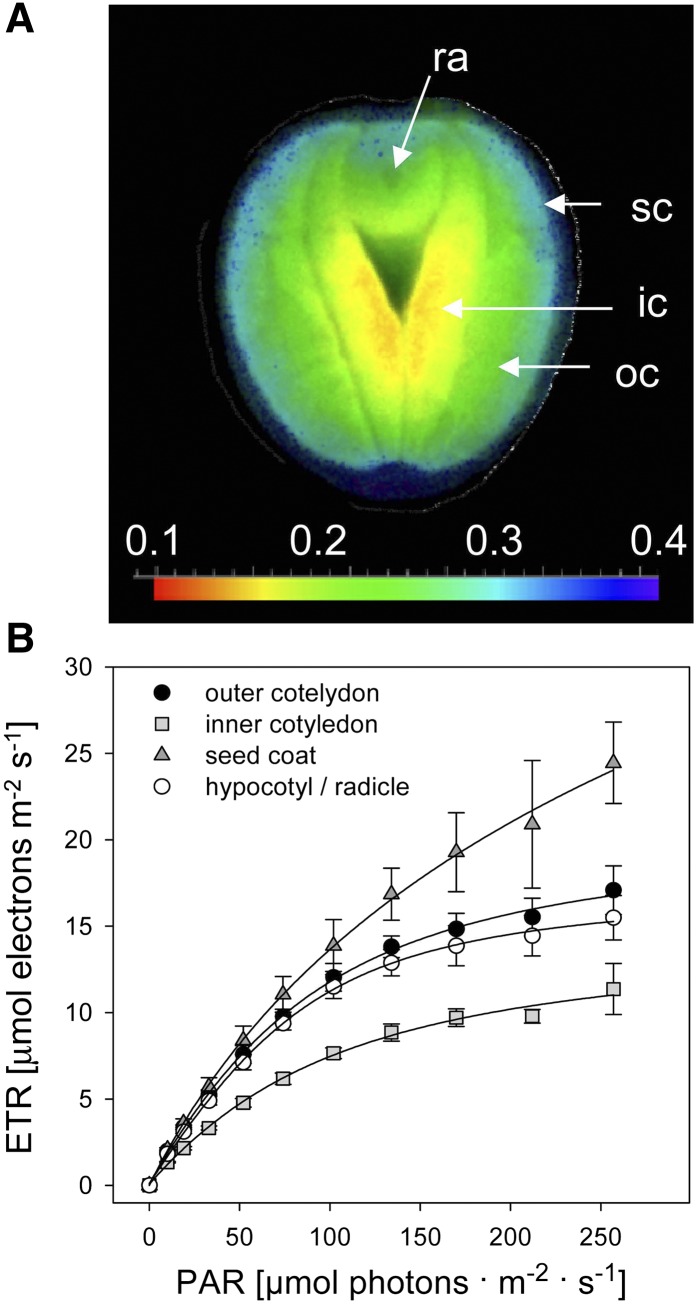
High-Resolution Quantification of the Photosynthetic Capability of Various Seed Tissues
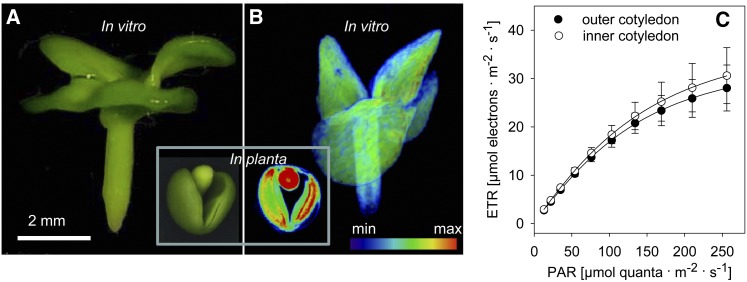
The photosynthetic energy transfer occurring within the seed was assessed by the measurement of the linear electron transport rate (ETR). Both the testa and the outer cotyledon displayed substantially higher levels of ETR during the main storage phase compared with the inner embryo region (Figure 2A). When exposed to close to saturating light levels, the ETR in the inner cotyledon was only 67% of that obtaining in the outer cotyledon (Figure 2B) and much higher in seed coat. This gradient in photosynthetic performance is even fortified in planta by the gradient in light supply from the outside to the inside of the seed.
Figure 2.
Gradients in Seed Photosynthesis as Measured by the Imaging-PAM Chlorophyll Fluorometer.
(A) Color-coded map of the effective quantum yield of photosystem II measured at 23 µmol quanta per m2 per second, showing the distribution of photosynthetic capability across the seed.
(B) Rapid light response profiles of the photosynthetic ETR in distinct regions of the seed. Each data point represents the mean ± sd (n = 5). ic, inner cotyledon; oc, outer cotyledon; ra, radicle; sc, seed coat.
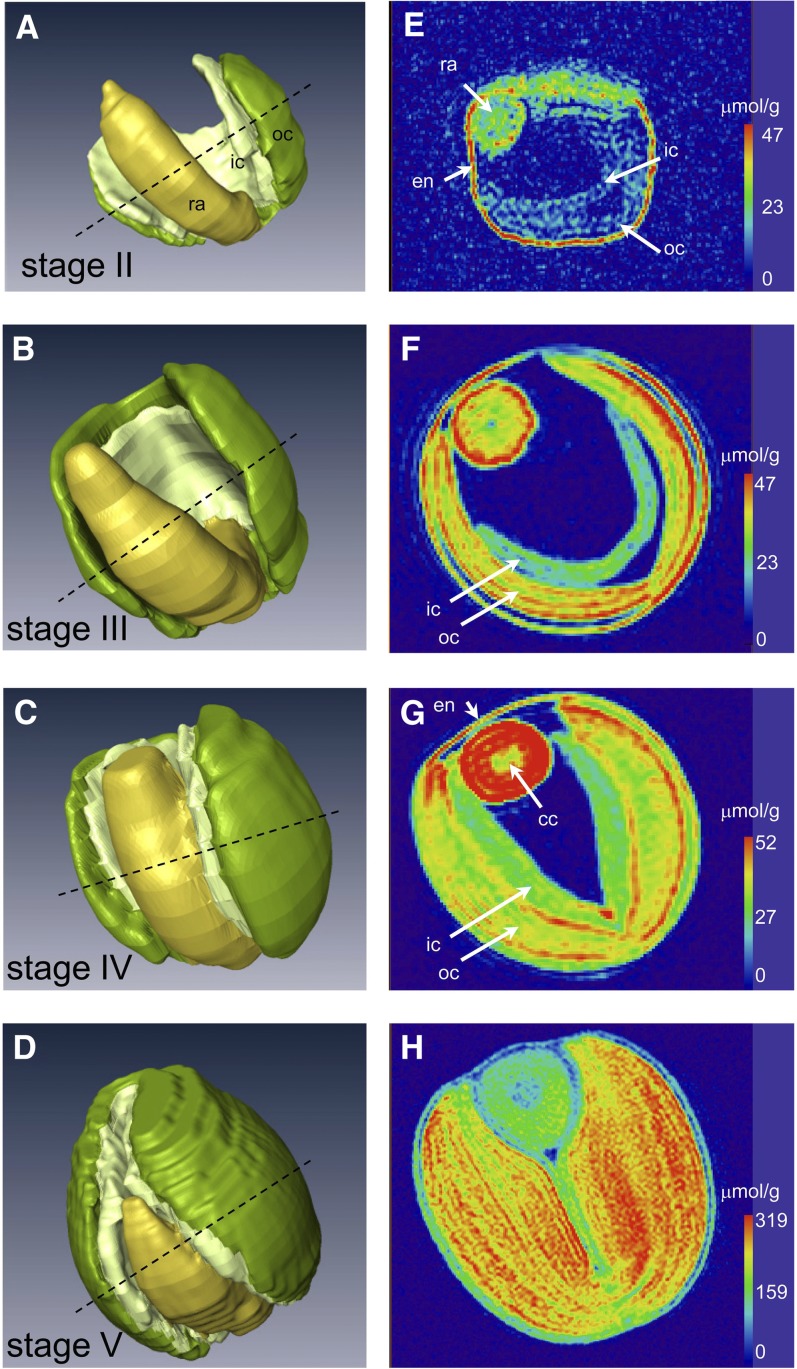
Spatial Profile of Lipid Deposition during Embryo Development
In vivo MRI-based lipid mapping (Neuberger et al., 2009) applied at distinct developmental stages allowed the distribution of the lipids within an individual seed to be documented in a three dimensional format (see Supplemental Movies 1 to 3 online). This also allowed us to relate structural changes of the embryo to storage pattern of lipids during development (Figure 3). In the early stage (Figures 3A and 3E), lipids were recognized first within the outer region of the endosperm (aleurone) and the endosperm layer surrounding the embryo, as well as in the embryo proper. Within the latter, the highest lipid concentration was in the radicle. Endosperm was the major location for lipid deposition at this stage. As development continued, a similar pattern of lipid deposition was retained, although the lipid level in the embryo increased substantially (Figures 3B and 3F). The outer cotyledon lipid content was much higher than that in the inner cotyledon. A pronounced lipid gradient was established within the outer cotyledon and also within the embryo axis, with twofold concentration differences within distances of only ∼300 µm. During the late storage stage, the lipid level of all of the embryo components rose (Figures 3C and 3G). Within the embryo axis, the level achieved in the root apex was higher than elsewhere, but declining markedly toward the hypocotyl. The parenchymal tissue within the axis was consistently more lipid rich than the cells within its central cylinder. The lipid gradient through the outer cotyledon became less marked, but retained its higher level than in the inner cotyledon. The outer tissue layers facing the endosperm and testa contained the highest content of lipid. The fatty acid composition of the lipid fraction within the specific embryo components sampled at this stage (obtained from dissected material analyzed by gas chromatography) was rather uniform, but differed from that found in the seed coat (with attached endosperm; see Supplemental Figure 1 online). By the maturation stage (Figures 3D and 3H), the lipid level throughout the embryo was substantially increased. The earlier differences between the inner and outer cotyledon with respect to lipid content had essentially disappeared. Ongoing lipid accumulation in the cotyledons caused much higher lipid level than in the hypocotyl/radicle.
Figure 3.
Embryo Modeling and MRI-Based Quantitative Imaging of Lipid Distribution in the Developing Oilseed Rape Seed.
Three-dimensional models of the embryo at the early stage (A), the mid storage stage (B), the late storage stage (C), and the maturation stage (D). The dotted line indicates the virtual section shown in (E) to (H), in which the lipid content (color-coded) is shown. Please note the different color scales. The corresponding three-dimensional models of lipid distribution are given in Supplemental Movies 1 to 3 online. cc, central cylinder; en, endosperm; ic, inner cotyledon; oc, outer cotyledon; ra, radicle; sc, seed coat.
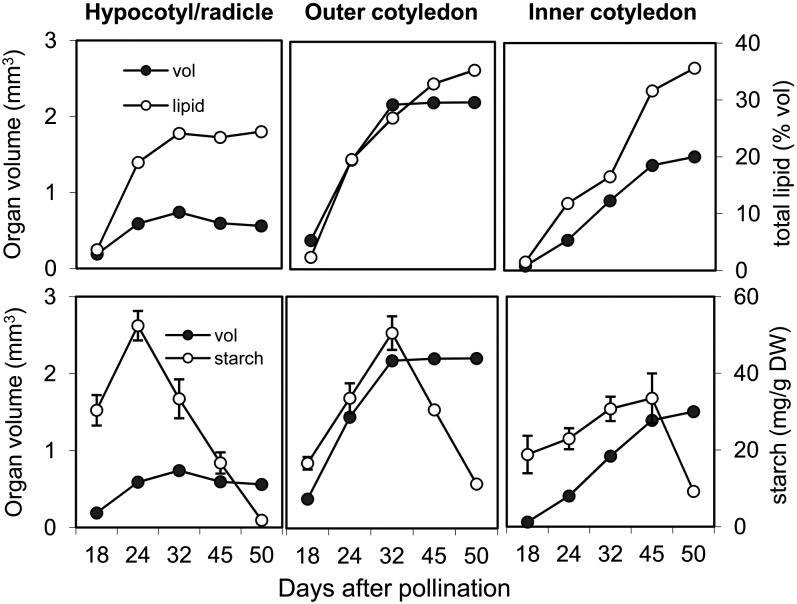
Based on above MRI analysis, we followed the dynamics in the relationship between lipid concentration and volume of the respective embryo organ during development (Figure 4). It appeared that the increases in lipid level could be correlated with the expansion in the volume of each organ. When the embryo organs reached maximal volume (at different time points in the organs), lipid accumulation in embryo was largely completed.
Figure 4.
Temporal Pattern of Lipid and Starch Accumulation in Various Embryo Components in Relation to Their Volume.
Top panels: lipid concentration versus component volume; bottom panels: starch concentration versus component volume. Error bars indicate sd (n = 6). The MRI model used for calculation of lipid/volume ratios at mid storage stage is given in Supplemental Movie 4 online. DW, dry weight.
Starch Deposition Is Spatially Regulated and Correlated with the Volume of the Embryo
During early development, the increase of starch levels was correlated with an increase in the volume of each component (Figure 4). Once the cotyledons had reached their maximum size, the starch level within them began to decline, indicating its degradation. This conclusion was reinforced by detailed histological analysis of seed development (Figures 5B to 5G). Starch first became visible in the testa, followed by the endosperm and finally the embryo. Within the embryo, starch deposition began in the radicle parenchyma, then spread first toward the outer and finally to the inner cotyledon. During the early to mid storage phase, starch was mostly present in completely differentiated and expanded cells, suggesting that starch storage at this time was associated with cell growth. This pattern corresponds to that of lipid (Figure 3). Starch degradation followed the same sequence, so that by the later storage stages, no starch remained in the seed (Figure 5F).
Figure 5.
Colocalization of Oleosin, Cruciferin, and Starch in the Developing Oilseed Rape Seed.
(A) Schematic representation of oleosin distribution (for details, see Supplemental Figure 2 online).
(B) to (F) Starch deposition (iodine staining).
(B) Starch grains are visible within the seed coat and endosperm during early storage stage.
(C) Starch is no longer detectable in the seed coat during the later stages.
(D) A cross section through the seed shows the presence of starch in the seed coat, endosperm, and embryo, visible as a gradient from the radicle toward the cotyledon. The outer cotyledon is more heavily stained than the inner one.
(E) A gradient in starch content within the outer cotyledon (cf. with the protein gradient shown in [K]).
(F) A cross section through the mature seed shows the complete absence of starch.
(G) Schematic representation of starch distribution.
(H) to (M) Cruciferin deposition (immunostaining).
(H) A cross-section through the seed coat and endosperm during the early storage stage shows no cruciferin to be present;
(I) The mid storage stage endosperm contains plenty of cruciferin.
(J) A cross section of the young seed demonstrates the presence of cruciferin in the embryo and cotyledons. A gradient in concentration is visible in both radicle and cotyledons.
(K) A cross section through the cotyledons during the mid storage stage shows a cruciferin gradient within the outer cotyledon. The storage vacuoles in the abaxial region are full of protein (double arrow left), those closer to the seed's interior are less filled, while those in inner cotyledon are empty (double arrow right).
(L) Cortical tissue of radicle showing cells completely full of cruciferin.
(M) The cotyledonary vascular tissue is relatively free of cruciferin.
(N) Schematic representation of cruciferin distribution.
al, aleurone layer; cc, central cylinder; en, endosperm; ic, inner cotyledon; oc, outer cotyledon; ra, radicle; sc, seed coat; vs, vascular tissue.
The Storage of Protein and Lipid Is Colocalized
The same seeds analyzed for their lipid distribution by MRI were also used to immunohistochemically locate the sites for deposition of oleosin (Figure 5A; for details, see Supplemental Figure 2 online), cruciferin (Figures 5H to 5N), and napin (see Supplemental Figure 3 online). The storage of cruciferin protein was confined to the embryo and endosperm (Figures 5H to 5N), with the radicle accumulating more heavily than the cotyledons during the early/mid storage stage (Figure 5J). A gradient was established from the outer to the inner cotyledon (Figure 5K). By the late storage stage, accumulation had ceased in the embryo, and the gradients had dissipated. Both the temporal and spatial patterns of cruciferin accumulation were very similar to those for napin and oleosin. Notably, these patterns also corresponded to those observed for lipid analyzed by MRI (Figure 3). Thus, the protein and lipid synthetic machinery needs to compete for substrate, energy, and cellular space. A more detailed analysis of the proteome was done using mass spectrometry (Supplemental Data Set 1 online) as discussed below.
Metabolite Composition Indicates Component-Specific Carbohydrate and Energy Metabolism
We next tested whether spatially distinct activities in photosynthesis and storage product deposition are reflected in the steady state level of central metabolites (see Supplemental Figure 4 online). The quantitatively major metabolites Suc, hexoses, and most free amino acids were evenly distributed between the outer and inner cotyledons and the hypocotyl/radicle. The latter, however, contained statistically significantly lower levels of glycolytic intermediates, the maturation-related sugars raffinose, stachyose, and verbascose, the organic acids isocitrate and oxoglutarate, pentose-5-phosphates, and the redox equivalents NAD, NADPH, and NADP and higher levels of ADP, ATP, NADH, γ-aminobutyric acid, Suc-6-phosphate, and trehalose-6-phosphate. Apart from a few compounds (e.g., citrate and citrulline), the metabolite contents of the two cotyledons resembled one another, and both were markedly different from that of the hypocotyl/radicle. The overall picture was one where similar steady state levels for key metabolites were maintained across the embryo, while some intermediates of carbohydrate and energy metabolism in hypocotyl/radicle differed substantially from that in the cotyledons.
Component-Specific Metabolic Modeling Based on Flux Balance Analysis
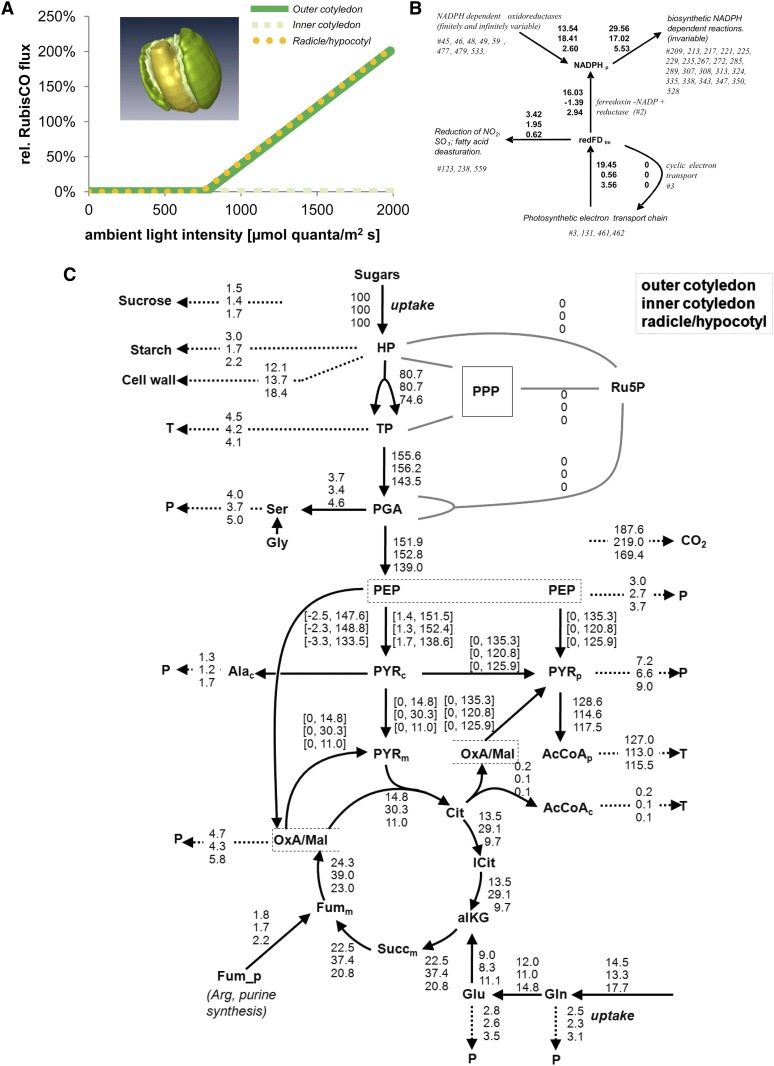
The above data provide evidence that a significant metabolic heterogeneity is established during development. In the next step, flux balance analysis (FBA) was applied to the data to predict in vivo metabolic fluxes at the main storage phase in the hypocotyl/radicle, inner cotyledon, and outer cotyledon (models A, B, and C, respectively). Modeling constraints and predicted flux values (variability intervals) for each tissue specific model are given in Table 1 (additional information is given in Supplemental Data Sets 2, 3 and 4 online). The models predicted a marginal improvement in carbon conversion efficiency (CCE) due to light usage, although the contribution of photosynthesis to seed metabolism was sizeable when a comparison was made between the rate of photolysis and the demand for NADPH. Only in the inner cotyledon was the effect of photosynthesis negligible (i.e., the component was growing heterotrophically). In all three components, ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) was predicted to have been essentially inactive, presumably because it is only when the light level exceeds a certain threshold that the light-dependent improvement of CCE depends on an increasing flux through Rubisco (Hay and Schwender, 2011b). Here, the predictions were based on a light intensity of 400 µmol quanta per m2 per second (as in the growth chamber); had the light level been increased to full sunlight, then the contribution of Rubisco to seed metabolism was predicted to have become substantial for both the outer cotyledon and the hypocotyl/radicle (Figure 6A). With increasing light, CCE can be expected to rise and the net CO2 emission to fall, reaching one in the outer cotyledon at ∼1930 µmol quanta per m2 per second. This extrapolation suggests that under high light conditions, the Rubisco bypass (Schwender et al., 2004) substantially contributes to seed storage metabolism in oilseed rape, yet only in the outer cotyledon and the hypocotyl/radicle.
Table 1. Model Constraints and Model Predicted Exchange Fluxes and Other Rates for Various Organs of Developing Oilseed Rape Embryos in Planta.
| A | B | C | ||
|---|---|---|---|---|
| Sub-Model (In Planta, 32 DAF) | ||||
| Flux value constraints | Radicle/Hypocotyl | Inner Cotyledon | Outer Cotyledon | Units |
| Growth rate (Biomass_exch) a | 0.0005 | 0.00136 | 0.00226 | mg DW h−1 |
| Photon uptake flux (Ph_tm_exch) | 0.014222 | 0.0022556 | 0.07781 | μmol h−1 |
| ATPdrain (ATP_c:H2O_c::ADP_c:Pi_c) | 0.014905 | 0.055864 | 0.098175 | μmol h−1 |
| Nitrate uptake (NO3_ap_exch) | 0 | 0 | 0 | μmol h−1 |
| Ammonia uptake (NH4_ap_exch) | 0 | 0 | 0 | μmol h−1 |
| Biomass composition b | ||||
| Total lipids | 34.8 | 40.8 | 42.8 | % DW |
| Total proteins | 24.2 | 21.4 | 21.8 | % DW |
| DNA | 0.10 | 0.10 | 0.10 | % DW |
| RNA | 0.10 | 0.10 | 0.10 | % DW |
| Starch | 3.35 | 3.07 | 5.06 | % DW |
| Free metabolites | 10.00 | 10.00 | 10.00 | % DW |
| Cell wall (glucose polymer) | 27.42 | 24.48 | 20.15 | % DW |
| Fatty acid composition, mid stage b | ||||
| C16:0 (C16H31O2) | 11.49 | 7.13 | 6.65 | g/ 100 g fatty acids |
| C18:0 (C18H35O2) | 2.33 | 2.95 | 2.47 | g/ 100 g fatty acids |
| C18:1 (C18H33O2) | 41.73 | 51.35 | 47.13 | g/ 100 g fatty acids |
| C18:2 (C18H31O2) | 31.36 | 24.71 | 27.88 | g/ 100 g fatty acids |
| C18:3 (C18H29O2) | 12.12 | 12.57 | 14.49 | g/ 100 g fatty acids |
| C20:0(C20H39O2) | 0.55 | 0.62 | 0.62 | g/ 100 g fatty acids |
| C20:1(C20H37O2) | 0.42 | 0.67 | 0.76 | g/ 100 g fatty acids |
| Model predicted exchange fluxes and flux ratios c | ||||
| Gln uptake | −0.00082 | −0.00200 | −0.00337 | μmol h-1 |
| Suc uptake | −0.00230 | −0.00749 | −0.01163 | μmol h-1 |
| CO2 exchange | 0.00779 | 0.03281 | 0.04365 | μmol h-1 |
| O2 exchange | −0.00297 | −0.01776 | −0.01759 | μmol h-1 |
| Relative Rubisco flux d | 0.000264 | 0.000203 | 0.000218 | NA |
| CCE e | 75.43% | 67.15% | 72.09% | NA |
| CCE (no light) | 69.70% | 66.87% | 66.07% | NA |
| Relative contribution of photosynthetic electron transport to demands of plastidic syntheses via NADPH, ferredoxin | 57.8% | 3.0% | 59.0% | NA |
| Relative contribution of light driven ATP production to plastidic ATP consumption | 78.6% | 4.1% | 80.6% | NA |
Reaction names, Supplemental Data Set 3.
Based on biomass composition, model specific equations for the reactions “TAGsynth_c”, “Lipase_c” and “ Biomasssynth_nc, ap, c, p” are specified in Suppl. Table 1.
Negative flux values denote influx.
as defined in Schwender et al. (2004)
carbon conversion efficiency
DAP, days after pollination; DW, dry weight; NA, not applicable.
Figure 6.
Simulation of Component-Specific Metabolism in the Oilseed Rape Embryo.
(A) Relative flux through Rubisco.
(B) Plastidial reducing equivalents (NADP and ferredoxin, redFD) redox balance in the outer cotyledon, inner cotyledon, and radicle (from top to bottom) at a simulated illumination level of 400 µmol quanta m−2 s−1. Each component is indicated by a specific color. All influxes and effluxes are shown, as derived from flux values of all reactions that share NADPH_p and redFD_tm. To assess the contribution to plastidial synthetic processes of photosynthetic electron transport, the principal electron sources and sinks were first defined and their combined electron flows derived. The identified producers were photosynthetic electron transport (photosystem I, #461) and various plastidial NADP-dependent oxidoreductases (Glu synthase, #45; Glu dehydrogenase, #46; malate dehydrogenase, #48; malic enzyme, #49; glyceraldehyde-3-phosphate dehydrogenase, #59; Glc-6-phosphate dehydrogenase, #477; phosphogluconate dehydrogenase, #479; isocitrate dehydrogenase, #533).
(C) Metabolic fluxes within each embryo component (from top to bottom: outer cotyledon, inner cotyledon, and radicle/hypocotyl) scaled on the basis of its sugar uptake rate (=100%). Arrowheads denote the direction of net flux. Dashed arrows represent the summed fluxes into protein (P) or triacylglycerol (T). For details, see Supplemental Data Set 3 online (table “B normalized fluxes”).
The effect on the plastids on the availability of NADPH/ferredoxin was assessed in all three components (Figure 6B). In the outer cotyledon and hypocotyl/radicle, ∼58% of the plastidic reductant was derived from photosynthetic electron transport compared with just 3% in the more heavily shaded inner cotyledon (Table 1). The analysis indicated that photosynthesis contributes essentially nothing to synthesis in the inner cotyledon. With respect to plastidic ATP, ∼80% was derived from photosynthesis (thylakoid H+ transporting ATPase, #92) in the outer cotyledon and hypocotyl/radicle, compared with just 4% in the inner cotyledon (Table 1).
Using a lumped network projection and normalizing metabolic fluxes relative to the sugar uptake rate (=100%), the flux distribution within the central carbon metabolism was compared between the various embryo components (Figure 6C). This showed that the relative flux associated with the majority of the reactions was comparable across the three components. However, clear differences were noted for the tricarboxylic acid (TCA) cycle, where the inner cotyledon had a rather higher flux than was present in either the outer cotyledon or in the radicle/hypocotyl (Figure 6C). In addition, mitochondrial ATP synthesis (H+-transporting two-sector ATPase, #91) was higher in the inner cotyledon than in either the outer cotyledon or the hypocotyl/radicle (see Supplemental Data Set 3 online).
Lipid Deposition and Photosynthetic Electron Transport in the Embryo Both Respond to Growing Conditions
To test how embryo architecture and light supply affect the tissue specificity of metabolism, we manipulated growth conditions in two ways. First, developing embryos were isolated and cultured in vitro. As the growing embryo experienced no space restrictions imposed by the endosperm/testa, the cotyledons did not fold/deform (Figure 7A), allowing each to receive a uniform level of light. Under these circumstances, no lipid gradient was established in the embryo nor was there any statistically significant variation in ETR across the cotyledons (Figures 7B and 7C; see Supplemental Movie 5 online). Second, the developing silique in planta was covered with a nontransparent material (aluminum foil) for 10 d. As light was excluded from reaching the embryo, there could be no photosynthetic contribution to its growth. Instead, there was a component-specific repression of lipid storage (31% in the outer cotyledon, 18% in inner cotyledon, and 0% in the radicle/hypocotyl; see Supplemental Figure 5 online). Analysis of the fatty acid composition of the lipid fraction showed that the contribution of unsaturated fatty acids (linoleic and α-linolenic acid) was depressed in the absence of light. The experiment demonstrated that photosynthesis in the embryo has the most pronounced effect on the outer cotyledon.
Figure 7.
Lipid Distribution and Photosynthetic Performance in the in Vitro–Grown Embryo.
(A) Embryo morphology at 32 DAP; inset shows in planta–grown embryo (left) and its lipid distribution (right) at the same developmental stage.
(B) Lipid distribution, as visualized by MRI; the corresponding three-dimensional model is given in Supplemental Movie 5 online.
(C) The light response of the photosynthetic ETR in the cotyledons. Data represent means ± sd (n = 5).
The Combined Response to Light of the Embryo Proteome and Metabolome
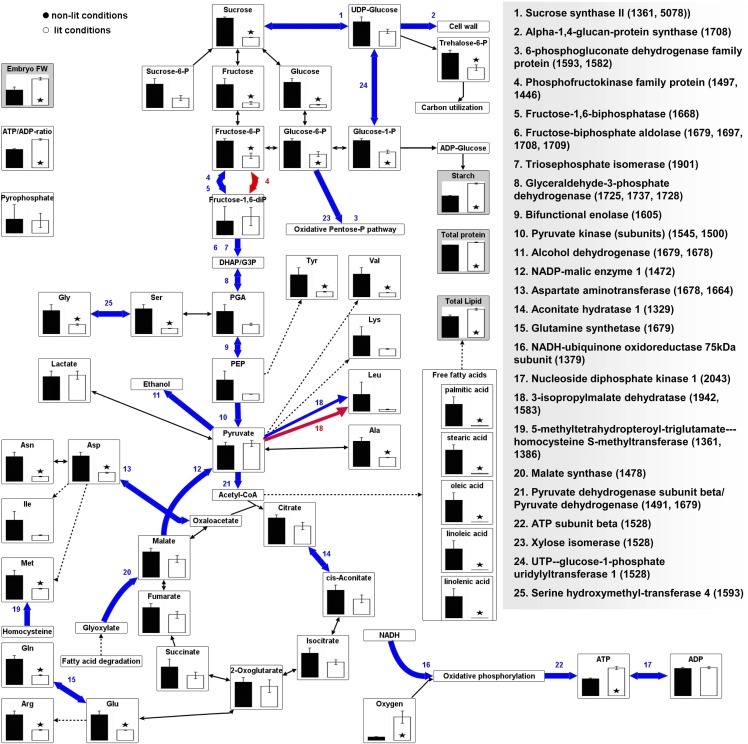
An analysis of the total embryo proteome identified 96 spots as differing between the illuminated and the nonilluminated developing seed (see Supplemental Data Set 5 online). Mass spectrometry was able to identify 13 distinct proteins belonging to eight functional categories that were of increased abundance under light; among these were several storage proteins. Under the nonlit conditions, 77 proteins were produced in increased abundance; most of these belong to the categories “carbohydrate and amino acid metabolism” and “genetic information processing.” Several enzymes associated with glycolysis, fatty acid oxidation, and amino acid metabolism featured among the 77 identified proteins (Figure 8), and the conversion between ATP and ADP via nucleoside diphosphate kinase was of greater importance in the absence of light. The overall pattern suggested that the absence of light induced the glycolytic pathway and generated a shift in amino acid metabolism, while its presence favored the transcriptional/translational activation of storage metabolism.
Figure 8.
Proteomic and Metabolomic Response of the Oilseed Rape Embryo to Illumination.
Changes in protein abundance are indicated in blue (upregulated under nonlit conditions) and red (upregulated under lit conditions). Steady state metabolite levels are given as means ± se (n = 5). Asterisks indicate statistically significant (P < 0.05) differences according to a t test. Raw proteomic and metabolomic data are given in Supplemental Data Sets 5 and 6 online. DHAP, dihydroxyacetone-phosphate; FW, fresh weight; PGA, phosphoglycerate; PEP, phosphoenolpyruvate.
The illuminated and the nonilluminated developing seed were also compared at the steady state metabolite level (Figure 8; see also Supplemental Data Set 6 online). Liquid chromatography–mass spectrometry–based analysis demonstrated that sugars and hexose phosphates persisted at much lower levels under lit conditions and that the content of most of the free amino acids and of all of the free fatty acids was drastically reduced by the presence of light. We further noticed differences in the levels of trehalose-6-phosphate (known to regulate carbon partitioning; Ponnu et al., 2011), ATP, and oxygen under lit versus nonlit conditions. Other energy metabolites, such as ADP and pyrophosphate, were maintained at comparable levels. The overall picture is one where metabolite levels (so presumably metabolic pathway activity) differed substantially between the light regimes. The reduced levels of sugars, free amino acids, and free fatty acids might indicate a higher turnover rate for enhanced accumulation of storage products under lit conditions.
The application of FBA to the outer cotyledon indicated a rising flux of glycolytic and other enzymes, including Suc synthase, phosphofructokinase, Fru-biphosphate aldolase, triosephosphate isomerase, enolase, and malic enzyme (reactions #30, #63, #25, #24, #536, and #49 in Supplemental Data Set 4 online) in response to a decrease in light availability. Correspondingly, our proteome data set confirmed an elevated abundance for each of these enzymes under nonlit conditions (Figure 8). Concentration changes were also noted for the relevant metabolic intermediates. Assuming relative enzyme abundance to be usable as a proxy for changes in flux (Sweetlove and Ratcliffe, 2011), the experimental data validated the predictions derived from the FBA approach.
Comparison of Embryo Growth in Planta versus in Vitro
We performed comparative analysis of in planta– versus in vitro–grown embryos (∼30 DAP) at the level of biomass composition, proteome, and metabolome. The in vitro–cultivated embryos showed fully opened, equally sized cotyledons and continued to expand, reaching higher biomass than the in planta–grown ones and also accumulated more starch (see Supplemental Figure 6 online). At the metabolite level, in vitro–grown embryos showed higher levels of most sugars and free amino acids, as well as various changes in glycolytic and TCA cycle intermediates in comparison to in planta–grown embryos (see Supplemental Figure 6 online). At the proteome level, we found significantly decreased abundance of proteins belonging to storage and photosynthesis under in vitro conditions but increased abundance of proteins involved in stress response and glycolysis (see Supplemental Data Set 7 online).
DISCUSSION
Owing to their sessile lifestyle, plants have developed various adaptations to cope with a changing environment (light, stress, and physical space). Our study demonstrates that the tiny rapeseed (Brassica napus) embryo exercises local metabolic adjustments already during its development inside of the seed. The spatially resolved analysis of gene expression in seeds can provide essential cues on cellular metabolic functions (Spencer et al., 2007; Belmonte et al., 2013) but only limited information on metabolic activities in vivo (Fernie and Stitt, 2012). Getting the whole picture therefore requires one to follow metabolism at the level of metabolic intermediates, storage products, and fluxes, all if possible at a high spatial resolution and in vivo. Here, we analyzed embryo metabolism at a high level of spatial resolution by applying noninvasive lipid mapping and various other techniques. The outcome served as input to a tissue-specific metabolic modeling approach, predicting how the embryo components adjust to local growth conditions inside the living seed. We argue that the folding of cotyledons, as observed in planta, causes a tremendous metabolic heterogeneity in the developing embryo of oilseed rape.
A Mechanistic View on the Timing and Causes of Metabolic Heterogeneity in the Oilseed Rape Embryo
The growing embryo has to adjust its shape, size, and metabolism to the environment afforded it by the mother plant, which involves restrictions in physical space and in the amount of incident light. At the early stage of its development, the embryo is surrounded by a liquid endosperm in which the necessary sugars and amino acids are homogeneously distributed (Melkus et al., 2009). As soon as space restrictions induce the bending of the embryo, the cotyledons grow unequally and gradients of various storage products are generated. The folding of the cotyledons results in the inner one becoming ever more enclosed by and thus controlled by the outer one. This necessarily includes that the outer embryo regions receive more light energy, which can be transmitted toward growth and storage metabolism. The outer cotyledon grew more rapidly, driving tissue differentiation and thereby forming gradients of starch, protein, and lipid (Figures 3B and 3C). Both absolute flux rates and relative flux distribution differed between the embryo components (Figure 6), which eventually caused distinct contributions of the embryo components to final seed biomass, oil storage, etc.
Space Use Efficiency as a Strategy for Embryogenesis
Starch synthesis and degradation are contemporaneous in the embryo (Da Silva et al., 1997), but here were shown to be spatially separated (Figure 5). Once growth has ceased, starch levels visibly declined at the same time as the accumulation of lipid and protein continued. The significance of transient starch storage in oilseeds is controversial. Starch has a low energy density (i.e., it requires more space per unit energy than does either lipid or protein). However, since the accumulation of assimilate in the form of starch is energetically highly efficient (Schwender, 2008), it is unsurprising to find its accumulation is associated with the rapidly growing young embryo (Figure 4). The antisense repression of starch synthesis in the oilseed rape embryo inhibits Suc import, glycolysis, and respiration and acts as a brake on growth (Vigeolas et al., 2004). Starch synthesis thus can be considered as representing a mechanism for creating a sink (Andriotis et al., 2010) during early embryogenesis, but when space becomes limiting, it is better for the embryo to accumulate lipid rather than starch. Once the oilseed embryo stopped its growth, its starch level declined (Figure 4). This switch was reached at differing times in the various components assayed. Under in vitro conditions, the spatially unrestricted growth of the embryo was associated with a continuous accumulation of starch, resulting in a lesser calorific value biomass than represented by the in planta–grown seed. The strategy of redirecting carbon from starch into lipid pushes the embryo toward a more efficient use of space and ultimately increases the energy density of the seed. The same strategy is currently applied in the engineering of plants as a feedstock for bioenergy production (Ohlrogge et al., 2009; Sanjaya et al., 2011).
Metabolic Modeling Highlights the Tissue Specificity of the Central Metabolism and Predicts Local Flux Adjustments in Vivo
Integrating experimental data sets with metabolic models is necessary to obtain a predictive understanding of metabolism (Sweetlove and Ratcliffe, 2011; Fernie and Stitt, 2012). Here, high-resolution measurements of in planta developing embryos were combined with FBA based on the bna572 metabolic model for cultured oilseed rape embryos (Hay and Schwender, 2011a, 2011b). The predicted contribution of photosynthesis to growth and synthesis markedly differed between the outer and the inner cotyledon (Table 1, Figure 6B). These changes reflected the embryo's adjustment to the local level of incident light and the dynamics of ATP/NADPH provision. The light-induced stimulation of lipid storage is well known (Ruuska et al., 2004; Goffman et al., 2005; Li et al., 2006). Here, we demonstrate how this translates into gradients in oil deposition inside the developing seed and highlight the necessary metabolic adjustments that the embryo makes at a high level of precision up to the cellular level.
The in planta models also predicted that Rubisco is essentially inactive in all three components. The lack of Rubisco flux relates to a metabolic phase formerly described to be specific for low-light conditions in embryo cultures (Hay and Schwender, 2011b). In simulations of bna572 that raise the photon flux from zero to higher levels, the seed carbon balance initially improves only due to a reduction in decarboxylation reactions like isocitrate dehydrogenase. Only with rising of the light flux above a threshold value, Rubisco flux is predicted to increase and to improve the carbon balance by refixing CO2 (Hay and Schwender, 2011b). In phase one, there is a negative relationship between photosynthetic activity and TCA cycle activity, which has been corroborated by comparing flux distributions derived by 13C-metabolic flux analysis for developing embryos of rapeseed and soy (Glycine max, light grown) and nonphotosynthetic embryos of sunflower (Helianthus annuus) and maize (Zea mays) (O’Grady et al., 2012). Similarly, the flux map in Figure 6C shows that the much more shaded inner cotyledon has a substantially higher TCA cycle activity than the photosynthetically active outer cotyledon. This corresponds to the general view on TCA cycle activity under the light/dark shift (Lee et al., 2010; Nunes-Nesi et al., 2013).
Given the experimental evidence that the bypassing of the upper reactions of glycolysis by Rubisco is an active process in the cultured oilseed rape embryo (Schwender et al., 2004), the predicted inactivity of Rubisco in planta was a surprising outcome. The difference between cultured and our in planta growing embryos is mainly due to the difference in light levels (see Supplemental Data Set 2 Flux Appendix online). Extrapolation of the in planta models to higher light levels suggested that in both outer cotyledon and hypocotyl/radicle the Rubisco becomes active at incident light beyond ∼750 µmol quanta per m2 per s (Figure 6A). This defines the tissues and conditions for the contribution of the Rubisco bypass in nature. We conclude that under the high light intensities found in the field, Rubisco is expected to substantially contribute to lipid synthesis and the carbon economy in the outer cotyledon and the hypocotyl/radicle, but likely not for the inner cotyledon.
Certain cofactors have recently been shown to be related to storage capacity for both lipids and proteins (Hayden et al., 2011), reflecting their higher biosynthetic energy requirements compared with starch (Schwender, 2008). As a result of being shaded, the inner cotyledon grows heterotrophically, while the growth of the outer one is photoheterotrophic. This is in contrast with the in vitro situation (Figure 7) where both cotyledons receive the same light and achieve similar photosynthetic ETR and similar oil content/distribution. Another prediction of the FBA model was that the outer cotyledon switches from net oxygen uptake to release at a light intensity above ∼920 µmol quanta m−2 s−1 (while the inner cotyledon remains as a net oxygen consumer irrespective of the incident light level). Thus, the embryo's photosynthetic activity has a considerable effect on the seed's oxygen balance, in particular, helping it to avoid the onset of hypoxia, which is inimical to seed metabolism (Verboven et al., 2013).
Advantages of in Planta– versus in Vitro–Based Metabolic Modeling Approaches
To get estimates (predictions) on metabolic activities of the embryo components in planta, we here applied FBA, while previous modeling attempts largely used 13C-metabolic flux analysis (13C-MFA). In general, 13C-MFA is superior in both quantitative and diagnostic cases (Schwender, 2008) and has provided valuable quantitative information (e.g., on the effect of nutrient source or light supply on embryo metabolism) (Junker et al., 2007; Allen et al., 2009; Allen and Young, 2013). However, 13C-MFA essentially relies on in vitro cultivation systems. Thus, its validity depends on how exact it can resemble the in planta situation. For nonphotosynthetic seeds/embryos, such as maize and sunflower, culture conditions might in general be closer to in planta since they are cultivated under continuous dark, and only minor compositional differences between in planta versus in vitro are reported (Alonso et al., 2007, 2010, 2011). For green seeds/embryos, the situation is more complicated as they have to grow under continuous light. The growth rate is often faster and the embryos accumulate more starch (Arabidopsis; Lonien and Schwender, 2009). This was also evident here for oilseed rape, and we additionally demonstrated how this reflects at the proteome and metabolite level (see Supplemental Data Set 7 and Supplemental Figure 6 online). Under in vitro conditions, where the developing cotyledons unfold and receive equal amounts of light, they also showed a uniform pattern of growth, photosynthesis, and lipid accumulation (Figure 7). From that we conclude that in vitro–based modeling is less suited to uncover metabolic heterogeneity of oilseeds and/or to quantify metabolic rearrangements due to developmentally changing seed architecture as occurring in planta.
In this study, we defined three models to differentiate between three embryo components, differing in their biomass composition, growth rate, etc. It should be noted that for in planta growing embryos, we also reported marked spatial gradients within the components. Thus, for interpretation of the flux distributions, one should keep in mind that the three submodels of embryo metabolism represent a coarse spatial resolution. Yet, we deem the approach to be a valid approximation that can be refined in the future, in particular based on the MRI-based technology presented here.
The FBA modeling framework accounts for metabolic processes related to the biosynthesis of cellular biomass synthesis but does not predict cellular maintenance functions. These metabolic expenses consume energy cofactors and therefore are represented as a generic ATP hydrolyzing reaction (ATPdrain). As it was impossible to accurately measure carbon balances for the various embryo components in planta, we had to estimate the ATPdrain flux based on the procedures detailed earlier (Hay and Schwender, 2011a). The ATPdrain estimate applied in our FBA is a rather conservative estimate of the size of this flux. Even twofold increases in ATPdrain in each model did not affect our major conclusions.
Topology of Lipid Storage and Implications for Oilseed Improvement
Increasing both seed lipid content and quality are important breeding goals in oilseed rape. The application of the MRI-based lipid imaging tool has allowed for the topological study of lipid deposition in a small seed to a resolution of ∼30 µm isotropic, thereby enabling the definition of both “where” and “when” lipids accumulate. The identification of regions of the embryo that differ in lipid content provides opportunities to uncover the transcription factors responsible for the initiation of lipid storage. Appropriate analytical methods based on laser microdissection applicable to oilseed rape have recently been established (Schiebold et al., 2011). Our data have also shown that the rate of lipid synthesis can vary severalfold between adjacent tissues/components and depends heavily on the local environment. To what extent this behavior requires differential gene expression remains to be established. In this context, however, it may be relevant that temporal variability in gene expression in the Arabidopsis embryo is much higher than its spatial variability (Spencer et al., 2007), so that the variability shown by the oilseed rape embryo may be in part at least governed by the posttranscriptional regulation of in vivo fluxes. A candidate mechanism is phosphorylation, and notably most of glycolytic and TCA cycle enzyme proteins in rapeseed are phosphorylated (Meyer et al., 2012). The broad scale significance of protein phosphorylation for central metabolism was recently demonstrated (Oliveira et al., 2012).
Lipid deposition starts in the endosperm and radicle early during embryogenesis. During the mid storage stage, lipid gradients are established both within and between the cotyledons (Figure 3). By maturity, the cotyledons have become uniformly lipid rich, while the lipid content of the hypocotyl/radicle remains low, as it does also in other species (Borisjuk et al., 2005; Neuberger et al., 2009; Horn et al., 2012). A possible approach to increasing the lipid yield of oilseed rape would therefore be to upregulate lipid synthesis in the hypocotyl/radicle so as to reach a level equivalent to that achieved in the cotyledons. Based simply on the volume occupied by the hypocotyl/radicle, one can estimate that this would increase the total lipid yield of the crop by ∼8%. Whether increasing the lipid content of this embryo component is even feasible, and if so whether there would be any negative side effects on maturation/germination remains to be seen (Shen et al., 2010).
In contrast with what has been concluded previously from whole-embryo analyses (Murphy and Cummins, 1989), we here demonstrated that lipid and protein storage occur simultaneously. This spatial and temporal overlap appears to be part of tissue differentiation and represents the basis for competition among the corresponding biosynthetic pathways for the same substrates, energy, and physical space. In mature seeds, lipid and protein content are generally negatively related (Grami et al., 1977), which most likely reflects this competition. While the redirection of substrates from lipid to protein synthesis appears to be feasible (Chen et al., 2009), the opposite has proven difficult to engineer to date (Abbadi and Leckband, 2011). Rather, selection for the summed content of protein and lipid has been shown to be more effective in raising lipid content than has selection based on just lipid content alone (Grami et al., 1977).
Since achieving any increase in lipid content appears to be challenging, other strategies may come into play. The metabolic models predict that the outer cotyledon of oilseed rape is particularly efficient in using light energy for lipid synthesis. Thus, increasing the surface area of the seed should increase the amount of light captured by the embryo and, therefore, the amount of cofactors/assimilates produced by the embryo photosynthesis. It might therefore be advantageous to manipulate the shape of the seed to increase its surface area; this implies selection against spherical seed. We believe that exploring the role of seed architecture in storage metabolism could open new avenues for crop improvement.
METHODS
Plant Growth
Plants of oilseed rape (Brassica napus var HS144B) were grown in a phytochamber at 18°C with 16 h of light (400 µmol quanta m−2 s−1) and a relative air humidity of 60%. At the time of flowering, plants were tagged for determination of developmental stages (DAP). Embryos were isolated at distinct stages, dissected, and instantly frozen.
MRI Experiments
All MRI data sets were acquired on a 17.6 tesla Avance III (Bruker) wide-bore system using an absolute quantification method described elsewhere (Neuberger et al., 2009). In brief, four experiments with seeds of different developmental stages were conducted. The seed was wrapped in Teflon tape to prevent moisture loss and inserted into a home built 3-mm ID solenoid coil. Before the imaging sequence, a global T1 measurement of the lipid signal was conducted to determine the minimum repetition time (TR) necessary to avoid for T1 correction during the quantification process. As a result of this measurement (T1 ∼700 ms; data not shown), the TR was chosen to be at least ∼3*T1 or longer. A standard multi-slice multi-echo spin echo (MSME) sequence with a CHESS suppression module on the water resonance was applied to acquire high-resolution lipid images of the seed. An in-plane resolution of up to 20 µm isotropic and a slice thickness of 105 µm in the seed of the late developmental stage could be achieved (experimental time 7.5 h). Seeds in the other developmental stages had slightly lower resolutions to reduce the experimental time as care had to be taken that no seed shrinkage due to degeneration occurred (shrinkage of the seed would have resulted in distorted images).
Either six or eight echoes with a maximum echo time of up to 47 ms were acquired in the MSME sequence. Apparent T2 fitting on a pixel-by-pixel basis was conducted using MATLAB (The Mathworks). As TR was chosen to be relatively long, a T1 correction was not applied. Furthermore, due to the uniform B1 field of the used solenoid coil (data not shown), a correction for B1 inhomogeneity was not necessary. To generate the volumes of the different organs in the seed, the first image from the echo train of each slice was imported into AMIRA (Mercury) and segmentation was conducted. The average signal in each organ could be calculated according to Neuberger et al. (2009). This result and the use of a calibration curve shown by Neuberger et al. (2009) enabled a pixel-by-pixel determination of the absolute lipid content within the seed.
For the measurement of the in vitro culture, the embryo was placed in a D2O-filled NMR tube with an inner diameter of 4 mm. The tube was inserted into an in-house-built saddle coil with an inner diameter of 5 mm and measured using a three-dimensional MSME method (echo time, 4.9 ms; TR, 2 s) with solvent suppression. The achieved isotropic resolution was 89 µm during the experimental time of 1.75 h. Reconstruction and visualization was performed using in-house Java-based software.
In Vitro Culture of Oilseed Rape Embryos
Intact embryos were isolated at 20 DAP and kept in liquid culture for 10 d under photoheterotrophy (50 µmol quanta m−2 s−1) at 20°C with organic nitrogen sources according to previous protocols (Schwender et al., 2006).
Chlorophyll Fluorescence Imaging and Light Transmission
Images of chlorophyll fluorescence parameters were obtained using an Imaging-PAM chlorophyll fluorometer as detailed earlier (Borisjuk et al., 2005). The obtained images of the effective quantum yield of photosystem II can be interpreted as a map of the relative rate of photosynthetic electron transport (ETR) at identical light supply across the seed tissue section. The obtained data were also used to create rapid light response curves. Measurements of light absorbance of pod wall, seed coat, and outer cotyledon were done using a photometer (LI-250; LICOR). The effective ETR of embryo organs was calculated based on the light transmitted to the surface of the individual organ, the organ-specific light response curve (Figure 2B), and the organ-specific surface area (derived from the NMR-based embryo models in Figure 3).
Experimental Treatment of Plants
To study the effect of lit versus nonlit conditions on embryo growth, developing pods were covered by black paper at 10 DAP. After 20 d, the embryos were isolated and weighed, followed by proteome and metabolite analysis. In some experiments, isolated embryos were dissected into various organs before analysis.
Biochemical Analysis
Freeze-dried embryo material was used for biochemical analysis. Metabolic intermediates were extracted and analyzed using liquid chromatography–mass spectrometry as detailed by Rolletschek et al. (2011). Starch content was assessed spectrophotometrically in the insoluble residue following extraction. Total lipid was quantified by NMR (MQ-60; Bruker) (Borisjuk et al., 2011). The fatty acid composition of total lipids and free fatty acids were extracted and measured by gas chromatography as described by Borisjuk et al. (2005). Oxygen concentration inside the seed was measured using microsensors (Rolletschek et al., 2011).
Proteome Studies
Proteome analysis of embryo material was analyzed as outlined in the Supplemental Data Set 1 Proteome Appendix online.
Histological Procedures
Histochemical techniques applied to seeds as well as immunostaining were performed as described (Radchuk et al., 2012). Immunolocalization procedure was performed with an affinity-purified anticruciferin (Tiedemann et al., 2008), antioleosin (20-kD class; Agrisera), and antinapin (Tiedemann et al., 2008) polyclonal antibody.
FBA
FBA was based on bna572, a model for cultured oilseed rape embryos (Hay and Schwender, 2011a, 2011b). The model was simulated in a photoheterotrophic mode with organic nitrogen sources available (Hay and Schwender, 2011a). Three submodels were derived for hypocotyl/radicle, inner cotyledon, and outer cotyledon, designated as models A, B, and C, respectively. These differed by constraints derived from organ-specific in planta physiological data specific for 30 DAP. An overview on submodel-specific constraints is given in workbook C of Supplemental Data Set 3 online. Biomass composition, growth rate, photosynthetic ETR, and light supply were determined at 30 DAP for each of the three organs (for details, see Supplemental Data Set 2 online). In particular, the biomass composition of outer and inner cotyledon and radicle/hypocotyl were estimated based on NMR measurements of organ volumes, as well as water, lipid, protein, and carbohydrate content (see Supplemental Data Set 3C online). Photon flux rates were based on the ambient light intensity in the growth chamber (400 µmol photons m−2 s−1), taking into consideration measured light absorbance of pod wall, seed coat, and outer cotyledons at 32 d after fertilization and measured ETRs. Then, for each submodel, a fixed value for non-growth-associated ATP drain (ATPdrain) was determined in same way as described before (Hay and Schwender, 2011a). ATPdrain is a generic reaction summarizing the effect of ATP-consuming processes that would take place without growth. In each case, the ATPdrain flux was determined by simulation of dark heterotrophy (photon uptake flux set to zero) while imposing a carbon balance to the model as measured before for oilseed rape embryos grown in liquid cultures in the dark (Goffman et al., 2005). Flux variability was calculated for all three submodels (for details, see Supplemental Data Set 2 Appendix Flux and Supplemental Data Set 3 online). Flux variability was also calculated as network projection on a lumped reaction network in order to visualize selected lumped fluxes of central carbon metabolism.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Fatty Acid Profile of Various Seed Organs in B. napus at Mid Storage Stage; Measured as Fatty Acid Methyl Esters Using Gas Chromatography.
Supplemental Figure 2. Oleosin Deposition in Developing Seeds of B. napus.
Supplemental Figure 3. Spatial and Temporal Pattern of Napin Accumulation in Developing Seeds of B. napus.
Supplemental Figure 4. Metabolite Distribution in Various Organs of the B. napus Embryo (30 DAP).
Supplemental Figure 5. Effect of Pod Shading on Lipid Content and Fatty Acid Composition in Various Components of the B. napus Embryo.
Supplemental Figure 6. Comparison of Biomass Composition and Steady State Metabolite Levels of B. napus Embryos Grown in Planta versus in Vitro.
Supplemental Data Set 1. Appendix Proteome.
Supplemental Data Set 2. Appendix Flux.
Supplemental Data Set 3. Flux Values for Three Submodels Representing Various Embryo Organs.
Supplemental Data Set 4. Flux Values for Outer Cotyledon under Varying Light Supply from 0 to 2000 µmol Quanta m−2 s−1 Ambient Light Intensity.
Supplemental Data Set 5. Comparative Analyses of Total Proteins Extracted from Embryos Grown under Lit and Nonlit Conditions: Image of 2D IEF/SDS-PAGE, List, and Functional Classification of Proteins.
Supplemental Data Set 6. List of Metabolite Data Measured for Developing B. napus Embryos.
Supplemental Data Set 7. Comparative Analyses of Total Proteins Extracted from Embryos Grown in Vitro and in Planta: Image of 2D IEF/SDS-PAGE, List, and Functional Classification of Proteins.
Supplemental Movie 1. Three-Dimensional Model of Lipid Distribution in the Embryo of B. napus at Stage III (Mid Storage Phase).
Supplemental Movie 2. Three-Dimensional Model of Lipid Distribution in the Embryo of B. napus at Stage IV (Late Storage Phase).
Supplemental Movie 3. Three-Dimensional Model of Lipid Distribution in the Embryo of B. napus at Stage V (Maturation Phase).
Supplemental Movie 4. Example Showing the Reassembling of Component-Specific Lipid Signal within the Corresponding Volume (B. napus Seed at Mid Developmental Stage).
Supplemental Movie 5. Three-Dimensional Model of Lipid Distribution in Cultured Embryos of B. napus.
Supplementary Material
Acknowledgments
We thank Christin Haase for support in proteomics, Steffen Wagner, Sabine Herrmann, and Katrin Blaschek for excellent technical assistance, and Karin Lipfert for artwork. This research was financially supported by Bayer CropScience. L.B., H.R., and P.M.J. thank the German Federal Ministry of Education and Research (Deutsche Forschungsgemeinschaft, BO-1917/4-1) for financial support. J.S. and J.O.H. thank the U.S. Department of Energy (Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, Field Work Proposal BO-133).
AUTHOR CONTRIBUTIONS
L.B. and H.R. designed the research, performed research, analyzed data, and wrote the article. T.N. and J.F. performed research (MRI). J.S. and J.O.H. performed research (FBA). S.S. performed research (proteomics). N.H. performed research (metabolite analysis). H.T. performed research (PAM analysis). H.-P.B, P.M.J., P.D., and B.L. analyzed data.
Glossary
- MRI
magnetic resonance imaging
- DAP
days after pollination
- ETR
electron transport rate
- FBA
flux balance analysis
- CCE
carbon conversion efficiency
- Rubisco
ribulose-1,5-bis-phosphate carboxylase/oxygenase
- TCA
tricarboxylic acid
- 13C-MFA
13C-metabolic flux analysis
- TR
repetition time
- MSME
multi-slice multi-echo spin echo
References
- Abbadi A., Leckband G. (2011). Rapeseed breeding for oil content, quality, and sustainability. Eur. J. Lipid Sci. Technol. 113: 1198–1206 [Google Scholar]
- Adamski N.M., Anastasiou E., Eriksson S., O’Neill C.M., Lenhard M. (2009). Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc. Natl. Acad. Sci. USA 106: 20115–20120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen D.K., Ohlrogge J.B., Shachar-Hill Y. (2009). The role of light in soybean seed filling metabolism. Plant J. 58: 220–234 [DOI] [PubMed] [Google Scholar]
- Allen D.K., Young J.D. (2013). Carbon and nitrogen provisions alter the metabolic flux in developing soybean embryos. Plant Physiol. 161: 1458–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A.P., Dale V.L., Shachar-Hill Y. (2010). Understanding fatty acid synthesis in developing maize embryos using metabolic flux analysis. Metab. Eng. 12: 488–497 [DOI] [PubMed] [Google Scholar]
- Alonso A.P., Goffman F.D., Ohlrogge J.B., Shachar-Hill Y. (2007). Carbon conversion efficiency and central metabolic fluxes in developing sunflower (Helianthus annuus L.) embryos. Plant J. 52: 296–308 [DOI] [PubMed] [Google Scholar]
- Alonso A.P., Val D.L., Shachar-Hill Y. (2011). Central metabolic fluxes in the endosperm of developing maize seeds and their implications for metabolic engineering. Metab. Eng. 13: 96–107 [DOI] [PubMed] [Google Scholar]
- Andriotis V.M.E., Pike M.J., Kular B., Rawsthorne S., Smith A.M. (2010). Starch turnover in developing oilseed embryos. New Phytol. 187: 791–804 [DOI] [PubMed] [Google Scholar]
- Belmonte, M.F., et al. (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA 110: E435–444. [DOI] [PMC free article] [PubMed]
- Baud S., Lepiniec L. (2009). Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol. Biochem. 47: 448–455 [DOI] [PubMed] [Google Scholar]
- Borisjuk L., Nguyen T.H., Neuberger T., Rutten T., Tschiersch H., Claus B., Feussner I., Webb A.G., Jakob P., Weber H., Wobus U., Rolletschek H. (2005). Gradients of lipid storage, photosynthesis and plastid differentiation in developing soybean seeds. New Phytol. 167: 761–776 [DOI] [PubMed] [Google Scholar]
- Borisjuk L., Rolletschek H., Fuchs J., Melkus G., Neuberger T. (2011). Recent applications of ‘low field’ and ‘high field’ magnetic resonance in seed research. Materials 4: 1426–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisjuk L., Rolletschek H., Neuberger T. (2012). Surveying the plant’s world by magnetic resonance imaging. Plant J. 70: 129–146 [DOI] [PubMed] [Google Scholar]
- Chen M., Mooney B.P., Hajduch M., Joshi T., Zhou M., Xu D., Thelen J.J. (2009). System analysis of an Arabidopsis mutant altered in de novo fatty acid synthesis reveals diverse changes in seed composition and metabolism. Plant Physiol. 150: 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva P.M.F.R., Eastmond P.J., Hill L.M., Smith A.M., Rawsthorne S. (1997). Starch metabolism in developing embryos of oilseed rape. Planta 203: 480–487 [Google Scholar]
- De Smet I., Lau S., Mayer U., Jürgens G. (2010). Embryogenesis - The humble beginnings of plant life. Plant J. 61: 959–970 [DOI] [PubMed] [Google Scholar]
- Fernie A.R., Stitt M. (2012). On the discordance of metabolomics with proteomics and transcriptomics: Coping with increasing complexity in logic, chemistry, and network interactions scientific correspondence. Plant Physiol. 158: 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs J., Neuberger T., Rolletschek H., Schiebold S., Nguyen T.H., Borisjuk N., Börner A., Melkus G., Jakob P., Borisjuk L. (2013). A noninvasive platform for imaging and quantifying oil storage in submillimeter tobacco seed. Plant Physiol. 161: 583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D., Saingery V., Chambrier P., Mayer U., Jürgens G., Berger F. (2003). Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 131: 1661–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegas V.C., Nazari A., Griffiths S., Simmonds J., Fish L., Orford S., Sayers L., Doonan J.H., Snape J.W. (2010). A genetic framework for grain size and shape variation in wheat. Plant Cell 22: 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffman F.D., Alonso A.P., Schwender J., Shachar-Hill Y., Ohlrogge J.B. (2005). Light enables a very high efficiency of carbon storage in developing embryos of rapeseed. Plant Physiol. 138: 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grami B., Baker R.J., Stefansson B.R. (1977). Genetics of protein and oil content in summer rape: heritability, number of effective factors, and correlations. Can. J. Plant Sci. 57: 937–943 [Google Scholar]
- Hay J.O., Schwender J. (2011a). Metabolic network reconstruction and flux variability analysis of storage synthesis in developing oilseed rape (Brassica napus L.) embryos. Plant J. 67: 526–541 [DOI] [PubMed] [Google Scholar]
- Hay J.O., Schwender J. (2011b). Computational analysis of storage synthesis in developing Brassica napus L. (oilseed rape) embryos: Flux variability analysis in relation to ¹³C metabolic flux analysis. Plant J. 67: 513–525 [DOI] [PubMed] [Google Scholar]
- Hayden D.M., Rolletschek H., Borisjuk L., Corwin J., Kliebenstein D.J., Grimberg A., Stymne S., Dehesh K. (2011). Cofactome analyses reveal enhanced flux of carbon into oil for potential biofuel production. Plant J. 67: 1018–1028 [DOI] [PubMed] [Google Scholar]
- Horn P.J., Korte A.R., Neogi P.B., Love E., Fuchs J., Strupat K., Borisjuk L., Shulaev V., Lee Y.-J., Chapman K.D. (2012). Spatial mapping of lipids at cellular resolution in embryos of cotton. Plant Cell 24: 622–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M., Jullien P.E., Berger F. (2006). The female gametophyte and the endosperm control cell proliferation and differentiation of the seed coat in Arabidopsis. Plant Cell 18: 3491–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker B.H., Lonien J., Heady L.E., Rogers A., Schwender J. (2007). Parallel determination of enzyme activities and in vivo fluxes in Brassica napus embryos grown on organic or inorganic nitrogen source. Phytochemistry 68: 2232–2242 [DOI] [PubMed] [Google Scholar]
- Lee C.P., Eubel H., Millar A.H. (2010). Diurnal changes in mitochondrial function reveal daily optimization of light and dark respiratory metabolism in Arabidopsis. Mol. Cell. Proteomics 9: 2125–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Beisson F., Pollard M., Ohlrogge J.B. (2006). Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Lonien J., Schwender J. (2009). Analysis of metabolic flux phenotypes for two Arabidopsis mutants with severe impairment in seed storage lipid synthesis. Plant Physiol. 151: 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkus G., et al. (2011). Dynamic ¹³C/¹ H NMR imaging uncovers sugar allocation in the living seed. Plant Biotechnol. J. 9: 1022–1037 [DOI] [PubMed] [Google Scholar]
- Melkus G., Rolletschek H., Radchuk R., Fuchs J., Rutten T., Wobus U., Altmann T., Jakob P.M., Borisjuk L. (2009). The metabolic role of the legume endosperm: A noninvasive imaging study. Plant Physiol. 151: 1139–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer L.J., Gao J., Xu D., Thelen J.J. (2012). Phosphoproteomic analysis of seed maturation in Arabidopsis, rapeseed, and soybean. Plant Physiol. 159: 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moles A.T., Ackerly D.D., Webb C.O., Tweddle J.C., Dickie J.B., Westoby M. (2005). A brief history of seed size. Science 307: 576–580 [DOI] [PubMed] [Google Scholar]
- Muller-Landau H.C. (2010). The tolerance-fecundity trade-off and the maintenance of diversity in seed size. Proc. Natl. Acad. Sci. USA 107: 4242–4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D.J., Cummins I. (1989). Biosynthesis of seed storage products during embryogenesis in rapeseed, Brassica napus. J. Plant Physiol. 135: 63–69 [Google Scholar]
- Neuberger, T., Rolletschek, H., Webb, A., and Borisjuk, L. (2009). Non-invasive mapping of lipids in plant tissue using magnetic resonance imaging. In Lipidomics, Volume 1: Methods and Protocols, D. Armstrong, ed (New York: Humana Press) pp. 485–496. [DOI] [PubMed] [Google Scholar]
- Niu Y., Wu G.Z., Ye R., Lin W.-H., Shi Q.-M., Xue L.-J., Xu X.-D., Li Y., Du Y.-G., Xue H.-W. (2009). Global analysis of gene expression profiles in Brassica napus developing seeds reveals a conserved lipid metabolism regulation with Arabidopsis thaliana. Mol. Plant 2: 1107–1122 [DOI] [PubMed] [Google Scholar]
- North H., et al. (2010). Arabidopsis seed secrets unraveled after a decade of genetic and omics-driven research. Plant J. 61: 971–981 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A., Araújo W.L., Obata T., Fernie A.R. (February 22, 2013). Regulation of the mitochondrial tricarboxylic acid cycle. Curr. Opin. Plant Biol. . [DOI] [PubMed] [Google Scholar]
- O’Grady J., Schwender J., Shachar-Hill Y., Morgan J.A. (2012). Metabolic cartography: Experimental quantification of metabolic fluxes from isotopic labelling studies. J. Exp. Bot. 63: 2293–2308 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J., Allen D., Berguson B., Dellapenna D., Shachar-Hill Y., Stymne S. (2009). Energy. Driving on biomass. Science 324: 1019–1020 [DOI] [PubMed] [Google Scholar]
- Ohto M.A., Fischer R.L., Goldberg R.B., Nakamura K., Harada J.J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 102: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira A.P., Ludwig C., Picotti P., Kogadeeva M., Aebersold R., Sauer U. (2012). Regulation of yeast central metabolism by enzyme phosphorylation. Mol. Syst. Biol. 8: 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnu, J., Wahl, V., and Schmid, M. (2011). Trehalose-6-phosphate: Connecting plant metabolism and development. Front. Plant Sci. 2: 70. [DOI] [PMC free article] [PubMed]
- Radchuk V., Kumlehn J., Rutten T., Sreenivasulu N., Radchuk R., Rolletschek H., Herrfurth C., Feussner I., Borisjuk L. (2012). Fertility in barley flowers depends on Jekyll functions in male and female sporophytes. New Phytol. 194: 142–157 [DOI] [PubMed] [Google Scholar]
- Ramli U.S., Salas J.J., Quant P.A., Harwood J.L. (2009). Use of metabolic control analysis to give quantitative information on control of lipid biosynthesis in the important oil crop, Elaeis guineensis (oilpalm). New Phytol. 184: 330–339 [DOI] [PubMed] [Google Scholar]
- Rolletschek H., Koch K., Wobus U., Borisjuk L. (2005). Positional cues for the starch/lipid balance in maize kernels and resource partitioning to the embryo. Plant J. 42: 69–83 [DOI] [PubMed] [Google Scholar]
- Rolletschek H., Melkus G., Grafahrend-Belau E., Fuchs J., Heinzel N., Schreiber F., Jakob P.M., Borisjuk L. (2011). Combined noninvasive imaging and modeling approaches reveal metabolic compartmentation in the barley endosperm. Plant Cell 23: 3041–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska S.A., Schwender J., Ohlrogge J.B. (2004). The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol. 136: 2700–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjaya D., Durrett T.P., Weise S.E., Benning C. (2011). Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol. J. 9: 874–883 [DOI] [PubMed] [Google Scholar]
- Schiebold S., Tschiersch H., Borisjuk L., Heinzel N., Radchuk R., Rolletschek H. (2011). A novel procedure for the quantitative analysis of metabolites, storage products and transcripts of laser microdissected seed tissues of Brassica napus. Plant Methods 7: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J. (2008). Metabolic flux analysis as a tool in metabolic engineering of plants. Curr. Opin. Biotechnol. 19: 131–137 [DOI] [PubMed] [Google Scholar]
- Schwender J., Goffman F., Ohlrogge J.B., Shachar-Hill Y. (2004). Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432: 779–782 [DOI] [PubMed] [Google Scholar]
- Schwender J., Shachar-Hill Y., Ohlrogge J.B. (2006). Mitochondrial metabolism in developing embryos of Brassica napus. J. Biol. Chem. 281: 34040–34047 [DOI] [PubMed] [Google Scholar]
- Shen B., Allen W.B., Zheng P., Li C., Glassman K., Ranch J., Nubel D., Tarczynski M.C. (2010). Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 153: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer M.W.B., Casson S.A., Lindsey K. (2007). Transcriptional profiling of the Arabidopsis embryo. Plant Physiol. 143: 924–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Shantharaj D., Kang X., Ni M. (2010). Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr. Opin. Plant Biol. 13: 611–620 [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Ratcliffe R.G. (2011). Flux-balance modeling of plant metabolism. Front. Plant Sci. 38: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M., Guschina I.A., O’Hara P., Slabas A.R., Quant P.A., Fawcett T., Harwood J.L. (2012). Metabolic control analysis of developing oilseed rape (Brassica napus cv Westar) embryos shows that lipid assembly exerts significant control over oil accumulation. New Phytol. 196: 414–426 [DOI] [PubMed] [Google Scholar]
- Taylor D.C., et al. (2009). Molecular modification of triacylglycerol accumulation by over-expression of DGAT1 to produce canola with increased seed oil content under field conditions. Botany 87: 533–543 [Google Scholar]
- Tiedemann J., Rutten T., Mönke G., Vorwieger A., Rolletschek H., Meissner D., Milkowski C., Petereck S., Mock H.P., Zank T., Bäumlein H. (2008). Dissection of a complex seed phenotype: Novel insights of FUSCA3 regulated developmental processes. Dev. Biol. 317: 1–12 [DOI] [PubMed] [Google Scholar]
- Tschiersch H., Borisjuk L., Rutten T., Rolletschek H. (2011). Gradients of seed photosynthesis and its role for oxygen balancing. Biosystems 103: 302–308 [DOI] [PubMed] [Google Scholar]
- Troncoso-Ponce M.A., Kilaru A., Cao X., Durrett T.P., Fan J., Jensen J.K., Thrower N.A., Pauly M., Wilkerson C., Ohlrogge J.B. (2011). Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 68: 1014–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboven P., Herremans E., Borisjuk L., Helfen L., Ho Q.T., Tschiersch H., Fuchs J., Nicolaï B.M., Rolletschek H. (May 21, 2013). Void space inside the developing seed of Brassica napus and the modelling of its function. New Phytologist, 10.1111/nph.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigeolas H., Möhlmann T., Martini N., Neuhaus H.E., Geigenberger P. (2004). Embryo-specific reduction of ADP-Glc pyrophosphorylase leads to an inhibition of starch synthesis and a delay in oil accumulation in developing seeds of oilseed rape. Plant Physiol. 136: 2676–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis J.G., Browse J. (2010). Lipid biochemists salute the genome. Plant J. 61: 1092–1106 [DOI] [PubMed] [Google Scholar]
- Weselake R.J., Taylor D.C., Rahman M.H., Shah S., Laroche A., McVetty P.B.E., Harwood J.L. (2009). Increasing the flow of carbon into seed oil. Biotechnol. Adv. 27: 866–878 [DOI] [PubMed] [Google Scholar]
- Xiang D., Venglat P., Tibiche C., Yang H., Risseeuw E., Cao Y., Babic V., Cloutier M., Keller W., Wang E., Selvaraj G., Datla R. (2011). Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiol. 156: 346–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.