This work identifies the bHLH transcription factor, JA-ASSOCIATED MYC2-LIKE1 (JAM1), which negatively regulates jasmonate responses independently from known regulators, namely, JASMONATE-ZIM DOMAIN proteins. JAM1 is involved in JA-mediated male fertility and the defense response to insect attack and represses the transcription of the target genes of MYC2 by competing for the binding sequences.
Abstract
Jasmonates (JAs) are plant hormones that regulate the balance between plant growth and responses to biotic and abiotic stresses. Although recent studies have uncovered the mechanisms for JA-induced responses in Arabidopsis thaliana, the mechanisms by which plants attenuate the JA-induced responses remain elusive. Here, we report that a basic helix-loop-helix–type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1 (JAM1), acts as a transcriptional repressor and negatively regulates JA signaling. Gain-of-function transgenic plants expressing the chimeric repressor for JAM1 exhibited substantial reduction of JA responses, including JA-induced inhibition of root growth, accumulation of anthocyanin, and male fertility. These plants were also compromised in resistance to attack by the insect herbivore Spodoptera exigua. Conversely, jam1 loss-of-function mutants showed enhanced JA responsiveness, including increased resistance to insect attack. JAM1 and MYC2 competitively bind to the target sequence of MYC2, which likely provides the mechanism for negative regulation of JA signaling and suppression of MYC2 functions by JAM1. These results indicate that JAM1 negatively regulates JA signaling, thereby playing a pivotal role in fine-tuning of JA-mediated stress responses and plant growth.
INTRODUCTION
As sessile organisms, plants have to adapt their physiological and morphological properties to the fluctuating environment. Plants perceive exogenous and endogenous stimuli, decode them, and alter their gene expression patterns via various plant hormonal signal transduction pathways. Jasmonates (JAs) are lipid-derived hormones that regulate plant responses to stresses such as wounding and herbivore attack and also act in various developmental processes, including male fertility (Devoto and Turner, 2003; Wasternack, 2007; Balbi and Devoto, 2008). JA negatively regulates plant growth and is considered to modulate the distribution of energy to defense responses (Yan et al., 2007; Zhang and Turner, 2008; Yang et al., 2012). Therefore, sophisticated mechanisms to control JA biosynthesis and the JA response at an appropriate level are necessary for optimal plant growth and development. These mechanisms must include both rapid induction of JA responses to provide effective defenses against stresses, such as herbivore attack, and effective negative regulation to temper these responses and limit negative effects on plant growth.
Biosynthesis of JAs starts with release of linolenic acid from chloroplast membranes and is mediated by JA biosynthetic enzymes, including phospholipase A1-type lipases, lipoxygenase, allene oxide synthase, allene oxide cyclase, and 12-oxophytodienoic acid reductase (Wasternack, 2007; Schaller and Stintzi, 2009). Mutants for ALLENE OXIDE SYNTHASE (AOS) or 12-OXOPHYTODIENOIC ACID REDUCTASE3 (OPR3)/DELAYED DEHISCENCE1 have defects in endogenous JA production and reduced growth inhibition in response to wounding (Sanders et al., 2000; Stintzi and Browse, 2000; Park et al., 2002; Yan et al., 2007; Zhang and Turner, 2008). Wounding or JA application induces the expression of genes encoding JA biosynthetic enzymes and thus is expected to further increase JA production. However, the induction of expression of JA biosynthetic genes is suppressed within few hours after treatment (Koo et al., 2011). This suggests that JA signaling regulates JA biosynthesis both positively and negatively.
Indeed, substantial research has identified both positive and negative regulators of the JA response and has shown the key role of ubiquitin-mediated protein degradation in this regulation. For example, CORONATINE INSENSITIVE1 (COI1), which encodes an F-box protein, was identified through analysis of a mutant that lacks responses to coronatine and methyl jasmonate (MeJA) (Feys et al., 1994; Xie et al., 1998). Identification of JASMONATE ZIM-DOMAIN (JAZ) proteins as targets of the E3 ligase SCFCOI1 also facilitated elucidation of the JA signaling pathway (Chini et al., 2007; Thines et al., 2007). In the absence of JA, the activity of transcription factors that regulate JA responses is repressed by binding of JAZ and a corepressor complex consisting of NOVEL INTERACTOR OF JAZ (NINJA) and TOPLESS (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Pauwels et al., 2010). When the bioactive JA-Ile conjugate is produced in response to endogenous and exogenous stimuli, formation of a COI1-JAZ coreceptor complex is promoted by JA-Ile, and JAZ is polyubiquitinated by the E3 ligase SCFCOI1 and is then degraded by the 26S proteasome pathway (Chini et al., 2007; Thines et al., 2007; Katsir et al., 2008; Fonseca et al., 2009; Sheard et al., 2010). Degradation of JAZ leads to release of the transcription factor from the JAZ complex and induction of the expression of JA-responsive genes and JA responses.
MYC2 is a target of JAZ proteins and a major regulator of JA signaling. myc2 mutants are defective in JA-mediated root growth inhibition, the expression of JA-responsive genes, and responses to pathogens (Boter et al., 2004; Lorenzo et al., 2004; Chini et al., 2007; Thines et al., 2007). MYC2 directly activates the expression of JAZs and several JA biosynthetic genes (Chini et al., 2007; Thines et al., 2007; Hou et al., 2010). The MYC3 and MYC4 transcription factors are closely related to MYC2 and act additively with MYC2 to regulate different subsets of JA response genes (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011). However, myc2 myc3 myc4 triple mutants are partially responsive to JA and, unlike the male sterile coi1 and JAZ gain-of-function mutants, are male fertile (Feys et al., 1994; Thines et al., 2007; Chung and Howe, 2009; Fernández-Calvo et al., 2011). This indicates that transcription factors other than MYC2/3/4 may also regulate a subset of JA responses (Kazan and Manners, 2008; Fernández-Calvo et al., 2011).
Mechanisms to attenuate JA signaling are presumably important for plants to limit fitness and metabolic costs associated with sustained defense responses. One of the mechanisms for limiting the JA response is inactivation of bioactive JA-Ile by the cytochrome P450 CYP94B3 (Kitaoka et al., 2011; Koo et al., 2011; Heitz et al., 2012; Koo and Howe, 2012). cyp94B3 loss-of-function mutants exhibit increased production of JA-Ile and enhanced expression of JA-responsive genes after wounding, indicating a crucial role for JA-Ile catabolism in attenuation of JA responses. However, because downregulation of the expression of early JA-responsive genes initiates while the JA-Ile level remains high (Koo et al., 2009, 2011), additional mechanisms may act to attenuate transcription. For example, stable JAZ proteins produced by alternative splicing desensitize JA responsiveness in cells containing high JA-Ile levels (Yan et al., 2007; Chung and Howe, 2009; Chung et al., 2010). Overexpression of an alternatively spliced form of JAZ10, which is resistant to COI1-mediated degradation, considerably suppressed JA responses. Conversely, jaz10 loss-of-function mutants exhibit enhanced JA responses (Yan et al., 2007; Chung and Howe, 2009; Demianski et al., 2012). JAZ8, which lacks a canonical degron and is stabilized against JA-mediated degradation, performs a crucial role in negative regulation of JA signaling (Shyu et al., 2012). The contribution of stable JAZ proteins in restraining JA responses remains unclear, and other mechanisms for negative regulation of JA signaling remain to be discovered.
To find transcription factors that act as regulators of JA signal transduction, we applied the chimeric repressor silencing technology (CRES-T) system (Hiratsu et al., 2003; Matsui et al., 2004; Mitsuda et al., 2005, 2007; Koyama et al., 2007) and identified a basic helix-loop-helix (bHLH) transcription factor previously called ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR (AIB; Li et al., 2007) and herein renamed JA-ASSOCIATED MYC2-LIKE1 (JAM1), which functions in JA responses, including JA-mediated male fertility and defense response to insect attack. We show that JAM1 modulates the balance between defense responses and plant growth.
RESULTS
Identification of CRES-T Plants Affected in JA Responses
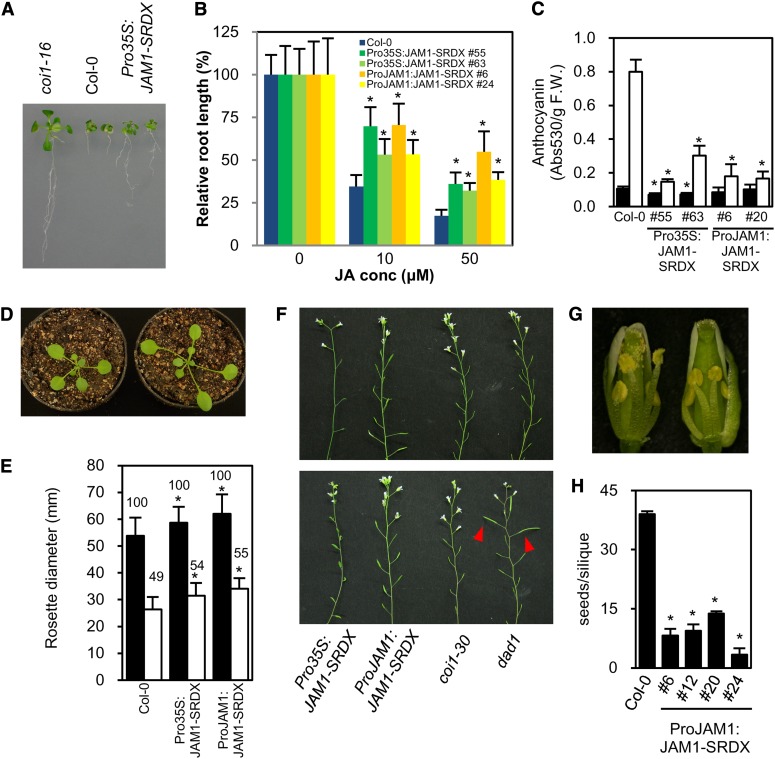
To identify transcription factors that are involved in the regulation of JA signaling, we applied the CRES-T system, in which a transcription factor is converted to a chimeric repressor by fusion to the SRDX repression domain (SUPERMAN Repression Domain X; Hiratsu et al., 2003, 2004). Because exogenous JA is known to inhibit root elongation, we screened for CRES-T lines that had shorter or longer roots than wild-type seedlings grown in the presence of 50 μM MeJA. MYC2 CRES-T plants (Pro35S:MYC2-SRDX) exhibited a JA-insensitive phenotype similar to myc2 loss-of-function mutants (see Supplemental Figure 1 online) (Boter et al., 2004; Lorenzo et al., 2004), indicating that the CRES-T system and our screening conditions are effective in identifying transcription factors related to JA signaling. The JA-insensitive mutant coi1-16 was included in the screen as a positive control (Ellis and Turner, 2002). The screen identified a JA-insensitive CRES-T line that expresses the chimeric repressor for a MYC2-related bHLH-type transcription factor (At2g46510), which we named JAM1 (Figure 1A). We observed JA insensitivity in two independent Pro35S:JAM1-SRDX T3 lines and confirmed that the JA insensitivity of the transgenic plants is a heritable phenotype (Figure 1B). JAM1 has been previously reported as a transcription factor, AIB, which is involved in abscisic acid (ABA) signaling (Li et al., 2007). To exclude potential artifacts that may arise from ectopic expression of the JAM1 chimeric repressor by the constitutive cauliflower mosaic virus (CaMV) 35S promoter, we prepared transgenic plants expressing JAM1-SRDX under the control of the JAM1 promoter (ProJAM1:JAM1-SRDX). Similar to the Pro35S:JAM1-SRDX plants, ProJAM1:JAM1-SRDX plants exhibited a JA-insensitive root growth phenotype (Figure 1B), confirming that expression of JAM1-SRDX from a native promoter is sufficient to inhibit JA responses.
Figure 1.
JA-Insensitive Phenotypes of JAM1 CRES-T Plants.
(A) The coi1-16 mutant (left), Col-0 (middle), and Pro35S:JAM1-SRDX plants (right) grown for 17 d on MS medium supplemented with 50 μM MeJA.
(B) Root lengths of two independent T3 lines for each JAM1 CRES-T construct grown for 14 d on MS medium supplemented with MeJA at the indicated concentration. Error bars indicate sd (n > 10). Asterisks indicate significant difference from Col-0 (Welch’s t test, P < 0.001).
(C) Accumulation of anthocyanin in JAM1 CRES-T plants. Plants were grown in liquid MS medium supplemented with (white bars) or without (black bars) 50 μM MeJA for 7 d. Error bars indicate sd of results from three biological replicates. Asterisks indicate significant difference from Col-0 (Welch’s t test, P < 0.05). F.W., fresh weight.
(D) Three-week-old Col-0 (left) and ProJAM1:JAM1-SRDX plants (right) grown in long-day conditions.
(E) Diameter of rosettes of 4-week-old plants with (white bars) and without (black bars) wounding. The value above each bar indicates the growth ratio in each line when values for untreated plants were set to 100. Error bars indicate sd (n > 14). Asterisks indicate significant difference from Col-0 (Welch’s t test, P < 0.05).
(F) Inflorescences of 8-week-old plants with (bottom panel) or without (top panel) MeJA treatment for 10 d. Arrowheads indicate siliques that were rescued to fertility by MeJA treatment.
(G) Opening flowers of 6-week-old wild-type (Col-0, left) and ProJAM1:JAM1-SRDX (right) plants. Front petals and sepals were removed.
(H) Number of seeds in a silique in ProJAM1:JAM1-SRDX plants. More than 10 plants in each line and at least 25 siliques per plant were examined. Error bars indicate se. Asterisks indicate significant difference from Col-0 (Welch’s t test, P < 0.001).
Quantitative RT-PCR analyses showed that the expression of JAM1 was induced by JA and wounding within 1 h of treatment, similar to other JA-responsive genes, such as MYC2 and JAZs (see Supplemental Figure 2A online) (Lorenzo et al., 2004; Chung et al., 2008). A promoter-reporter gene expression assay using transgenic plants expressing the β-glucuronidase (GUS) reporter gene under the control of JAM1 promoter (ProJAM1:GUS) revealed that the promoter activity of JAM1 was induced within 1 h of MeJA treatment or wounding (see Supplemental Figures 2B and 2C online). JAM1 expression was also observed in floral organs, including styles, anthers, and pollen, with weaker expression observed in filaments (see Supplemental Figure 2D online).
JAM1 Affects JA Responses and Growth
The JA-induced anthocyanin accumulation observed in Columbia-0 (Col-0) plants was reduced in both ProJAM1:JAM1-SRDX and Pro35S:JAM1-SRDX plants (collectively designated as JAM1 CRES-T plants) (Figure 1C). The expression of anthocyanin-related genes DIHYDROFLAVONOL 4-REDUCTASE (DFR), LEUCOANTHOCYANIDIN DIOXYGENASE, and PRODUCTION OF ANTHOCYANIN PIGMENT1 was also suppressed in JAM1 CRES-T plants (see Supplemental Figures 3A to 3C online). Trichome formation, which is not affected in myc2 mutants (Yoshida et al., 2009), was reduced in ProJAM1:JAM1-SRDX plants (see Supplemental Figure 4 online). These phenotypes are probably due to the impaired JA responsiveness of JAM1 CRES-T plants because JA mediates anthocyanin biosynthesis and trichome formation (Shan et al., 2009; Yoshida et al., 2009; Qi et al., 2011).
Repeated wounding stunts plant growth by a process that depends on JA synthesis and signaling (Yan et al., 2007; Zhang and Turner, 2008). We performed wound-induced growth inhibition assays to evaluate whether the JA insensitivity of JAM1 CRES-T plants affects rosette growth. Unwounded JAM1 CRES-T plants were significantly larger than unwounded Col-0 plants (Figures 1D and 1E). Wounding reduced the growth rate of both Col-0 and JAM1 CRES-T plants, but JAM1 CRES-T plants were less affected than Col-0 plants (Figure 1E). These results indicate that JA insensitivity of JAM1 CRES-T plants affects their growth rate, consistent with previous reports that JA negatively regulates plant growth (Yan et al., 2007; Zhang and Turner, 2008).
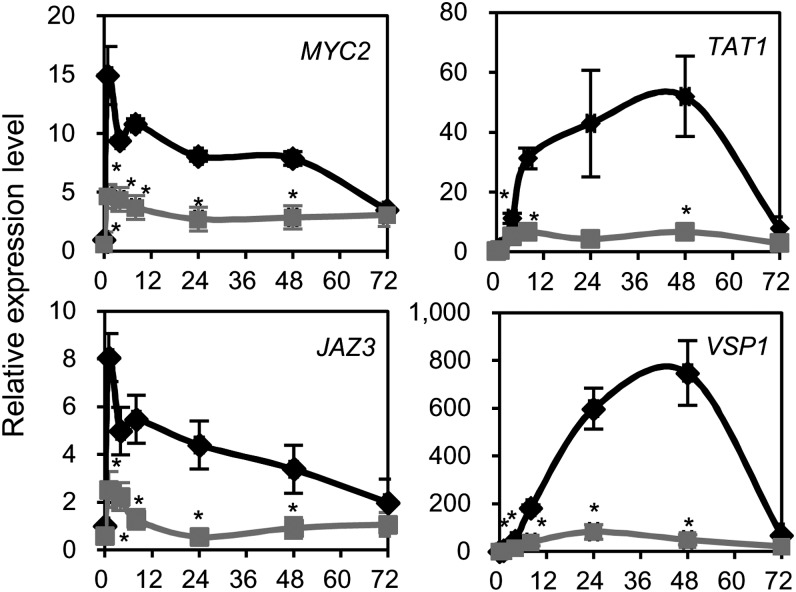
To investigate the effect of JAM1 on the expression of JA-responsive genes, we examined the expression level of several JA-responsive genes in ProJAM1:JAM1-SRDX plants. As shown in Figure 2, MYC2 and JAZ3 expression was induced within 1 h of JA treatment and then gradually decreased. The expression of TYROSINE AMINOTRANSFERASE1 (TAT1) and VEGETATIVE STORAGE PROTEIN1 (VSP1) gradually increased and reached maximal levels at 48 h after treatment in Col-0 plants, whereas the expression of these genes was significantly lower in ProJAM1:JAM1-SRDX plants.
Figure 2.
JA-Responsive Gene Expression in JAM1 CRES-T Plants.
Expression of JA-inducible genes. RNAs were extracted from 7-d-old seedlings grown in liquid MS medium treated with 50 μM MeJA. Black and gray lines indicate relative expression in Col-0 and ProJAM1:JAM1-SRDX plants, respectively. The value for Col-0 at time 0 was set to 1. Horizontal axis indicates hours after MeJA treatment. Error bars represent sd of results from three biological replicates. Asterisks indicate significant difference from Col-0 (Welch’s t test, P < 0.05).
JAM1 CRES-T Plants Show JA-Insensitive Male Sterility
We found that JAM1 CRES-T plants exhibit delayed anther dehiscence and reduced fertility and also have larger flowers than those of Col-0 (Figures 1F and 1G) (see Supplemental Figures 5A to 5D online). Such phenotypes are similar to those reported for aos, opr3, defective in anther dehiscence1 (dad1), and coi1 mutants (Feys et al., 1994; Stintzi and Browse, 2000; Ishiguro et al., 2001; Park et al., 2002). The severity of male reproductive defects in Pro35:JAM1-SRDX lines correlated with transgene expression levels (see Supplemental Figures 5E and 5F online). These effects on anther dehiscence are consistent with high levels of JAM1 transcripts in anthers (see Supplemental Figure 2D online).
The number of seeds produced by ProJAM1:JAM1-SRDX plants was approximately one-fourth that of Col-0 (Figure 1H). In addition to the delayed anther dehiscence phenotype, ProJAM1:JAM1-SRDX plants exhibited a short filament phenotype that is not observed in Pro35S:JAM1-SRDX plants (Figure 1G) (see Supplemental Figures 5B and 6 online). These observations suggest that JAM1 is involved in anther dehiscence and filament elongation and regulates JA-mediated male fertility. By contrast, neither ProMYC2:MYC2-SRDX plants nor the myc2 myc3 myc4 triple mutant were affected in fertility (see Supplemental Figure 5G online) (Fernández-Calvo et al., 2011), suggesting that MYC2/3/4 do not function in JA-mediated male fertility.
To analyze whether the reduced fertility of JAM1 CRES-T plants is due to defects in JA biosynthesis or impaired JA signaling, we applied exogenous MeJA to inflorescences of these plants. Application of MeJA restored fertility of the JA-deficient mutant dad1 (arrowheads in Figure 1F), but fertility of JAM1 CRES-T plants was not restored by exogenous MeJA, similar to the T-DNA–tagged coi1-30 mutant (SALK_035548). In addition, the amount of bioactive JA-Ile in control (unwounded) and wounded leaves of JAM1 CRES-T plants was not significantly different from that in wild-type plants (see Supplemental Figure 7A online). These collective results suggested that the reduced fertility of JAM1 CRES-T plants results from impaired JA signaling rather than a defect in JA synthesis.
JAM1 Acts as a Transcriptional Repressor
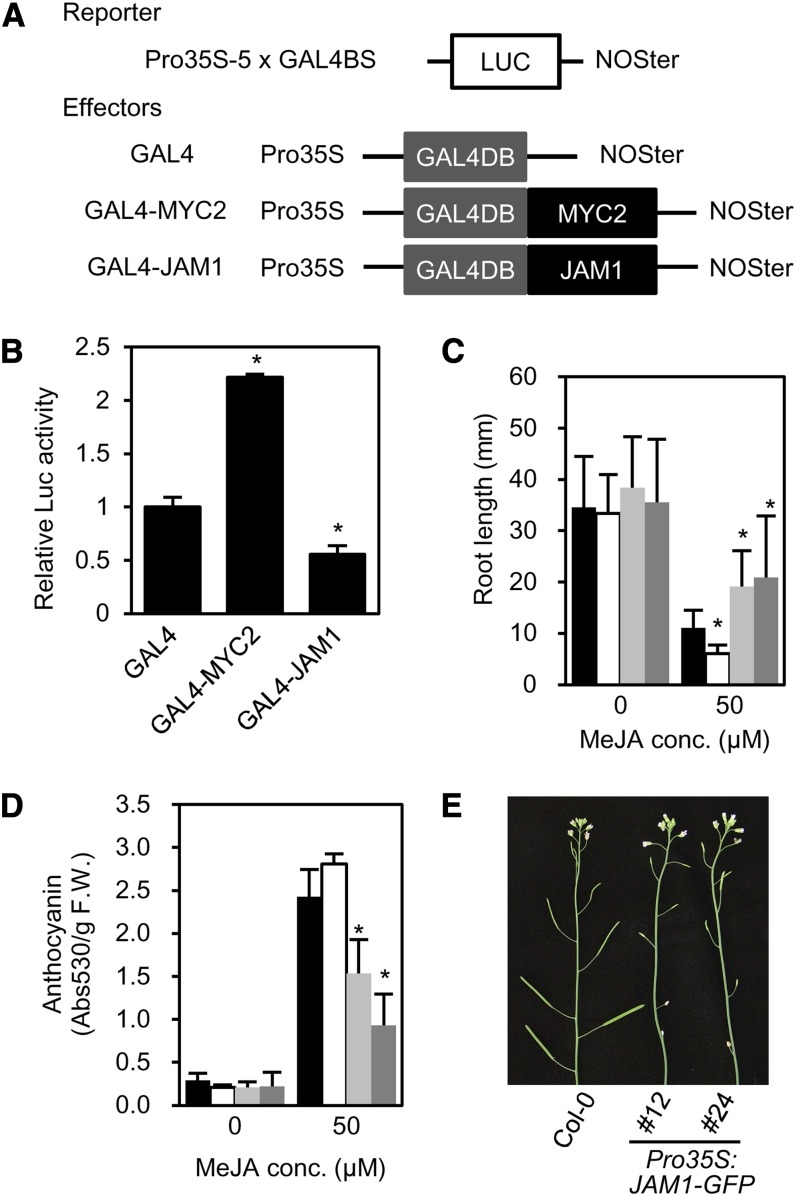
JAM1 belongs to the IIId bHLH group, which is closely related to IIIe group that includes the transcriptional activator MYC2 (Heim et al., 2003; Niu et al., 2011). Alignment of the amino acid sequence of JAM1, MYC2, and closely related bHLHs revealed a high degree of sequence conservation throughout the entire length of the proteins. Interestingly, however, the acidic region of MYC2/3/4, which may act as an activation domain (Fernández-Calvo et al., 2011), is not conserved in JAM1 (see Supplemental Figure 8 online). This prompted us to test the idea that JAM1 may not act as a transcriptional activator. To examine the transcriptional activity of JAM1, we performed reporter-effector transient expression assays using the luciferase (LUC) reporter driven by the CaMV 35S promoter with five repeats of the GAL4 binding sequence (Figure 3A). The MYC2 effector, in which the protein coding region of MYC2 was fused to yeast GAL4 DNA binding domain (GAL4-MYC2), increased LUC activity, consistent with the previous report (Niu et al., 2011). By contrast, the JAM1 effector (GAL4-JAM1) repressed LUC activity (Figure 3B), indicating that JAM1 has a transcriptional repression activity.
Figure 3.
Transcriptional Repression Activity of JAM1 and JA-Insensitive Phenotypes in JAM1-GFP–Overexpressing Plants.
(A) Schematic representation of the constructs used for transient expression assays. The reporter construct consists of the CaMV 35S promoter, five repeats of the GAL4 binding sequence (5xGAL4BS), NOS terminator (NOSter), and firefly luciferase (LUC) coding sequence. Effector constructs express GAL4 DNA binding domain (GAL4DB)-fused protein under the control of CaMV 35S promoter.
(B) Transient expression assays of GAL4-JAM1 and GAL4-MYC2. Constructs for the assay are shown in (A). The value for GAL4 was set to 1, and relative values are shown. Error bars indicate sd of results from three replicates. Asterisks indicate significant difference from GAL4 (Welch’s t test, P < 0.005).
(C) Root lengths of 14-d-old Col-0 (black), Pro35S:GFP (white), Pro35S:JAM1-GFP #12 (light gray), and Pro35S:JAM1-GFP #24 (dark gray) plants grown on MS medium containing 0 or 50 μM MeJA. Error bars indicate sd (n > 10). Asterisks indicate significant difference from Col-0 (Welch’s t test, P < 0.05).
(D) JA-induced anthocyanin accumulation. Plants were grown on MS medium containing 0 or 50 μM MeJA for 7 d. Black, white, light-gray, and dark-gray bars indicate Col-0, Pro35S:GFP, Pro35S:JAM1-GFP #12, and Pro35S:JAM1-GFP #24, respectively. Error bars indicate sd of results from more than three biological replicates. Asterisks indicate significant difference from Col-0 (Welch’s t test, P < 0.05). F.W., fresh weight.
(E) Inflorescences of 6-week-old Col-0 (left), Pro35S:JAM1-GFP #12 (middle), and Pro35S:JAM1-GFP #24 (right).
[See online article for color version of this figure.]
Because fusion of a native repressor to SRDX often phenocopies ectopic expression of the transcription factor (Matsui et al., 2008; Ikeda and Ohme-Takagi, 2009), we compared the phenotype of plants that constitutively express JAM1 with that of JAM1 CRES-T plants. In the transgenic plants that ectopically express JAM1 fused to green fluorescent protein (GFP) under the control of CaMV 35S promoter (Pro35S:JAM1-GFP), the GFP signal was observed in nuclei, confirming that JAM1 is a nuclear protein (see Supplemental Figure 9 online). Pro35S:JAM1-GFP plants exhibited a JA-insensitive root growth phenotype that was slightly weaker than that of 35S:JAM1-SRDX plants (Figures 1B and 3C). This is consistent with the finding that repression activity of JAM1 was enhanced by fusion with SRDX (see Supplemental Figure 10 online). Accumulation of anthocyanin in response to MeJA was also downregulated in Pro35S:JAM1-GFP plants compared with Col-0 plants (Figure 3D). In addition, Pro35S:JAM1-GFP plants showed reduced fertility (Figure 3E). Plants expressing GFP alone did not show any JA-insensitive phenotypes, indicating that JA-insensitive phenotypes in Pro35S:JAM1-GFP plants are due to the activity of JAM1. These results are consistent with those of transient assays, which suggest transcriptional repression activity of JAM1 (Figure 3B).
Loss-of-Function Mutants of JAM1 Are Hypersensitive to JA
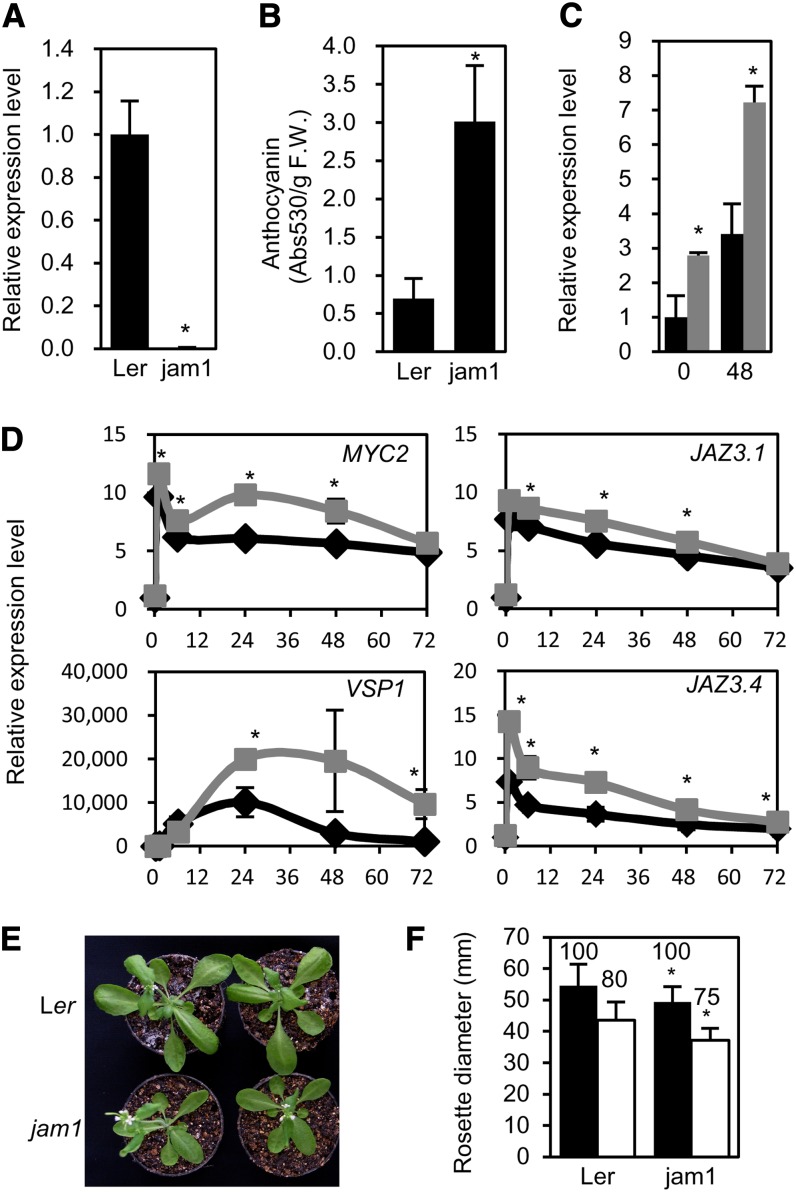
Next, we analyzed the functional role of JAM1 in JA signaling using a loss-of-function allele (jam1; CS170389) in the Landsberg erecta (Ler) genetic background (see Supplemental Figure 11A online). jam1 appears to be a null allele because no JAM1 transcript was detected in the jam1 mutants (Figure 4A). Although two T-DNA insertion lines of JAM1 in the Col-0 background (SAIL_536_F09 and GABI285E09) were available, both lines expressed a part of JAM1 cDNA, albeit not full length (Y. Sasaki-Sekimoto and K. Shirasu, personal communication). We thus used jam1 for further study. First, we confirmed that ProJAM1:JAM1-SRDX confers root growth insensitivity to JA and male sterility in the Ler background, as it does in the Col-0 background (see Supplemental Figures 11B and 11C online). Because JAM1 is hypothesized to be a negative regulator of JA signaling, loss-of-function mutants are expected to exhibit JA-hypersensitive phenotypes. Indeed, JA-induced accumulation of anthocyanin and the expression of DFR, MYC2, VSP1, JAZ3, and JAZ10 were increased in jam1 compared with that in Ler (Figures 4B to 4D) (see Supplemental Figure 12A online). Interestingly, the expression of a truncated JAZ3 and JAZ10 splice variant (JAZ3.4 and JAZ10.4) was also increased in jam1 (Figure 4D) (see Supplemental Figure 12B online). In addition, a weak dwarf phenotype and hypersensitivity to wounding were observed in jam1 (Figures 4E and 4F). The jam1 mutant did not show obvious alterations of fertility or root growth inhibition by JA (see Supplemental Figures 11D and 11E online). JA-Ile content in jam1 plants was comparable to that in Ler (see Supplemental Figure 7B online), indicating that sensitivity to JA-Ile, rather than JA-Ile production, is enhanced in jam1 mutants. Together, these data confirm that JAM1 acts as a negative regulator of JA signaling and that loss of function of JAM1 results in JA-hypersensitive phenotypes and growth inhibition in the absence of exogenous JA.
Figure 4.
JA-Hypersensitive Phenotype of jam1.
(A) The expression of JAM1 in 7-d-old seedlings of jam1 mutant grown in liquid MS medium. The value for Col-0 was set to 1, and the relative value is shown. Error bars represent sd of results from three biological replicates. Asterisk indicates significant difference from Ler (Welch’s t test, P < 0.001).
(B) Amount of anthocyanin in 8-d-old seedlings of Ler and jam1 plants treated with 50 μM MeJA for 3 d. Error bars indicate sd of results from nine biological replicates. Asterisks indicate significant difference from Ler (Welch’s t test, P < 0.005). F.W., fresh weight.
(C) and (D) Relative expression of DFR (C) and JA-responsive genes (D) in 7-d-old seedlings of Ler (black) and jam1 (gray) grown in liquid MS medium treated with 50 μM MeJA. The value for Ler at 0 h was set to 1. The horizontal axis indicates hours after JA treatment. Error bars represent sd of results from three biological replicates. Asterisks indicate significant difference from Ler (Welch’s t test, P < 0.05).
(E) Four-week-old wild-type (Ler; top) and jam1 plants (bottom).
(F) Diameter of rosettes of 4-week-old plants with (white bar) and without (black bar) wounding. The value above each bar indicates the growth ratio in each line when values for untreated plants were set to 100. Error bars indicate sd (n > 30). Asterisks indicate significant difference from Ler (Welch’s t test, P < 0.01).
[See online article for color version of this figure.]
JAM1 Has Differential Activity from MYC2 and Functions Downstream of COI1
To analyze functional differences between JAM1 and MYC2 in vivo, we expressed JAM1 under the control of the MYC2 promoter in the myc2 mutant (jin1-8). The jin1-8 ProMYC2:MYC2 transgenic plants recovered JA-responsive inhibition of root elongation, confirming that the promoter region of the MYC2 gene used for this analysis is sufficient to complement MYC2 functions (see Supplemental Figure 13 online). By contrast, the root length of jin1-8 ProMYC2:JAM1 transgenic plants was comparable to that of jin1-8 when grown on MeJA-containing medium, indicating that ProMYC2:JAM1 does not rescue the activity of MYC2 and that JAM1 and MYC2 have differential activities.
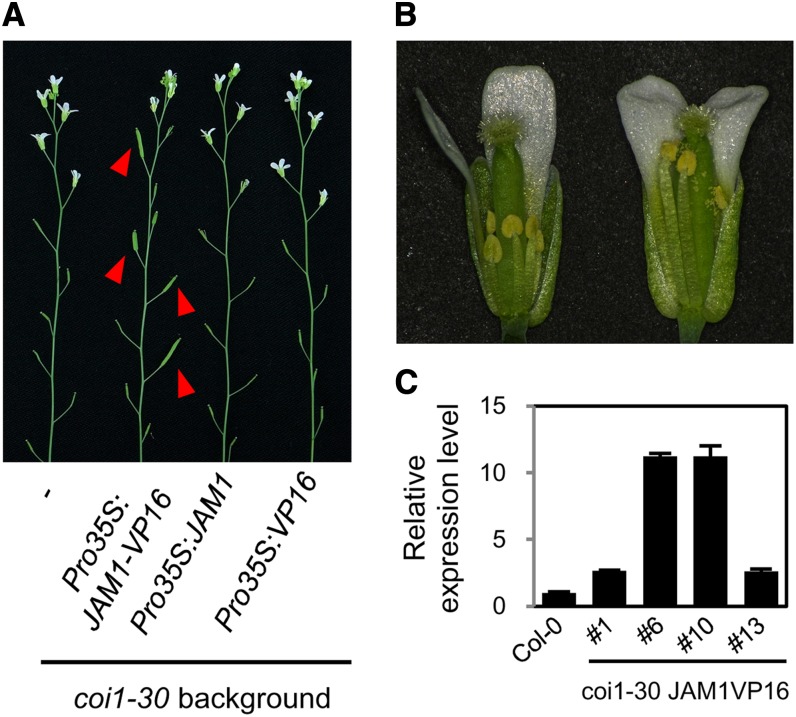
To analyze whether JAM1 acts downstream of COI1, we performed epistasis experiments with the coi1-30 mutant. The JAM1 activation form, Pro35S:JAM1-VP16, in which VP16 activation domain (Triezenberg et al., 1988) was fused to JAM1, Pro35S:JAM1, and Pro35S:VP16 were transformed into the coi1-30 mutant. Transcriptional activation activity of JAM1-VP16 fusion protein in vivo was confirmed by transient assays (see Supplemental Figure 14 online). Of the Pro35S:JAM1-VP16 plants homozygous for coi1-30, as confirmed by genomic PCR analysis (see Supplemental Figure 15 online), four of 10 exhibited fertility at later reproductive phases (Figure 5A). Anthers of opening flowers in fertile coi1-30 Pro35S:JAM1-VP16 plants were dehisced and their pollen was able to pollinate the pistils (Figure 5B). Also, higher expression of JAM1-VP16 was detected in the fertile plants (Figure 5C), suggesting that acquisition of fertility in coi1-30 mutants depended on the expression level of JAM1-VP16. The expression of VP16 alone did not restore the fertility of coi1-30. As expected, ectopic expression of JAM1 (Pro35S:JAM1) also did not restore fertility of coi1-30 (Figure 5A), although the expression levels of JAM1 in these transgenic plants were almost comparable to fertile coi1-30 plants expressing JAM1-VP16 (see Supplemental Figure 16 online). JAM1 is a transcriptional repressor and Pro35S:JAM1 appeared to suppress genes required for male fertility, similar to Pro35S:JAM1-SRDX. By contrast, JAM1-VP16 acted as a transcriptional activator to recover fertility. Our results suggest that JAM1 acts downstream of COI1 and regulates the signaling cascade involved in JA-mediated male fertility.
Figure 5.
JAM1 Functions Downstream of COI1.
(A) Inflorescences of 7-week-old coi1-30, coi1-30 Pro35S:JAM1-VP16, coi1-30 Pro35S:JAM1, and coi1-30 Pro35S:VP16 plants (left to right). Arrowheads indicate siliques with viable seeds.
(B) Opening flowers of 7-week-old coi1-30 (left) and coi1-30 Pro35S:JAM1-VP16 (right) plants. Front petals and sepals were removed.
(C) Relative expression levels of JAM1 in coi1-30 Pro35S:JAM1-VP16 plants. #6 and #10 were partially fertile, and #1 and #13 were sterile lines. The value for Col-0 was set to 1 and relative values are shown. Error bars indicate sd of results from three replicates.
JAM1 Affects the Defense Response to the Insect Herbivore Spodoptera exigua
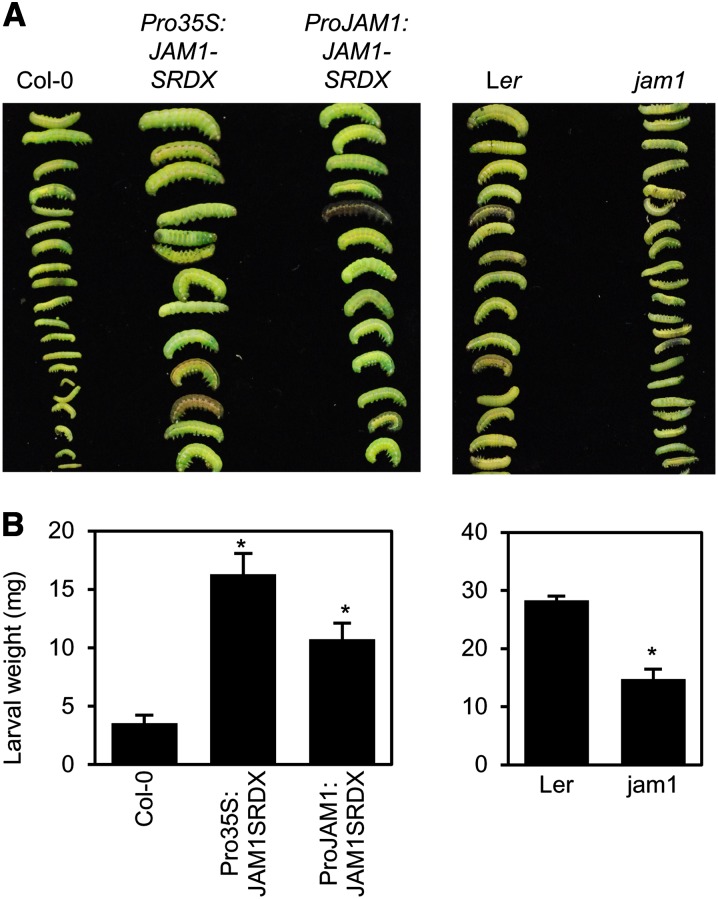
JA plays a critical role in defense responses to herbivore attack. To evaluate the effects of gain and loss of function of JAM1 on the resistance to herbivore attack, we compared the weight gain of the generalist herbivore S. exigua on JAM1 CRES-T plants, jam1 mutants, and their respective wild-type parents. Larvae reared on Pro35S:JAM1-SRDX and ProJAM1:JAM1-SRDX plants for 16 d were 4.5 and 3.0 times heavier than those grown on Col-0 plants, respectively (Figures 6A and 6B). Conversely, the weight of larvae grown on jam1 was approximately half of that of insects grown on Ler. These results are consistent with JA-insensitive and -hypersensitive responses in JAM1 CRES-T plants and jam1, respectively, and indicate that JAM1 has crucial functions in defense responses to herbivore attack via modulation of JA signaling.
Figure 6.
JAM1 Regulates Defense Responses to S. exigua Feeding.
(A) Photograph of representative S. exigua larvae after 16 d of feeding.
(B) Weight of S. exigua larvae reared on plants for 16 d. Error bars represent se (n > 25 larvae per plant genotype). Asterisks indicate significant difference from wild-type plants (Welch’s t test, P < 0.001).
[See online article for color version of this figure.]
JAM1 Regulates Early JA-Responsive Genes
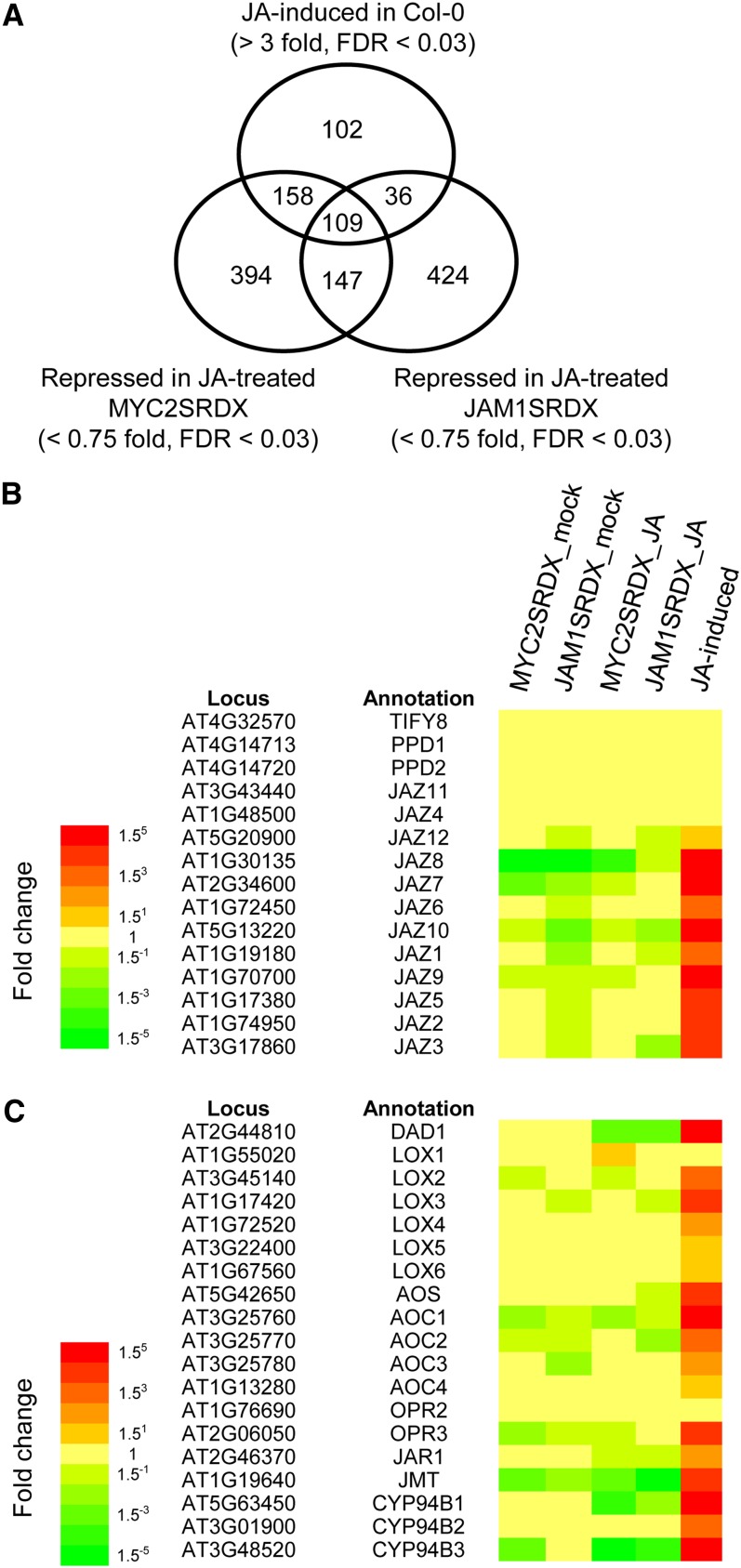
To obtain information regarding the mechanisms by which JAM1 suppresses the JA response, we performed genome-wide expression analyses using microarrays. RNAs isolated from 11-d-old seedlings of Col-0 and ProJAM1:JAM1-SRDX plants treated with or without 50 μM MeJA for 1 h were used for microarray analysis (Figure 7A). In addition, we included ProMYC2:MYC2-SRDX plants in the analysis and compared JAM1- and MYC2-regulated downstream genes because MYC2 is a well-characterized transcription factor in JA signaling and JAM1 and MYC2 are closely related proteins (see Supplemental Figure 8 online) (Heim et al., 2003). Results of microarray analysis revealed that 403 genes were upregulated (threefold more than mock-treated Col-0 with false discovery rate [FDR] < 0.03) by JA treatment within 1 h in Col-0; these upregulated genes were referred to as “early JA-responsive genes.” A total of 716 and 806 genes were downregulated (<0.75-fold of JA-treated Col-0 with FDR < 0.03) in JA-treated ProJAM1:JAM1-SRDX and ProMYC2:MYC2-SRDX plants, respectively. A total of 36.0% (145/403) and 65.8% (265/403) of early JA-responsive genes were downregulated in ProJAM1:JAM1-SRDX and ProMYC2:MYC2-SRDX plants, respectively. The set of early JA-responsive genes downregulated by both of the transcription factors contained several genes for JA metabolic enzymes and for JA signaling components (see Supplemental Table 1 online); this category represented 27.0% of the early JA-responsive genes (109/403). JA-responsive expression of most JAZ genes and JA metabolic enzyme genes was downregulated in ProJAM1:JAM1-SRDX and ProMYC2:MYC2-SRDX plants, indicating their expression is under the control of MYC2 and JAM1 (Figures 7B and 7C). The expression of eight JA-responsive genes downregulated in JA-treated ProJAM1:JAM1-SRDX and ProMYC2:MYC2-SRDX plants was confirmed by quantitative RT-PCR (see Supplemental Figure 17 online). These results suggest that JAM1 functions as a component of the core JA signal transduction pathway, as MYC2 does, and that JAM1 and MYC2 share a large portion of their target genes.
Figure 7.
Analyses of JA-Inducible Genes Regulated by MYC2 and/or JAM1.
(A) Venn diagram showing JA-induced genes in Col-0 and genes downregulated in JA-treated ProJAM1:JAM1-SRDX or ProMYC2:MYC2-SRDX plants.
(B) and (C) Fold changes of JAZ (B) and JA metabolic enzyme (C) genes in ProMYC2:MYC2SRDX and ProJAM1:JAM1SRDX plants treated with MeJA or mock treated for 1 h in comparison with those in Col-0 plants with the same treatment. Fold change of those genes in Col-0 treated with MeJA for 1 h is also shown. Induced and reduced genes are shown in red and green, respectively.
JAM1 and MYC2 Have Similar DNA Binding Preferences
One of the possible mechanisms of JAM1 repression of JA responses is that JAM1 forms a heterodimer with MYC2 and suppresses MYC2 function. However, yeast two-hybrid analysis indicated that JAM1 does not interact with MYC2 (see Supplemental Figure 18A online). In addition, we did not observe a JAM1-MYC2 interaction in a GAL4DB-based transient expression assay in Arabidopsis thaliana leaves (see Supplemental Figures 18B to 18D online). In this expression system, JA-deficient aos mutant leaves were used to eliminate potential effects of endogenous JA-Ile production on the JAM1-MYC2 interaction. Coexpression of GAL4-JAM1 and MYC-VP16 did not activate LUC activity compared with GAL4-JAM1 alone, but coexpression of GAL4-JAZ3 and MYC-VP16 significantly increased LUC activity (see Supplemental Figure 18C online). These results indicate that MYC2 interacts with JAZ3 but not with JAM1 in the absence of endogenous JA. Moreover, the interaction between JAM1 and MYC2 was not detected in the presence of exogenous JA (see Supplemental Figure 18D online). These results are consistent with a previous report that MYC2 forms a heterodimer with MYC3 and MYC4 but not with AIB (JAM1) (Fernández-Calvo et al., 2011).
Another possibility is that JAM1 may repress transcription by competitively binding to MYC2 target sequences. Because the amino acid sequence of the JAM1 bHLH domain is very similar (83.9% identity) to the bHLH domain of MYC2 (see Supplemental Figure 8 online), JAM1 and MYC2 are predicted to have similar DNA binding affinities, as reported for MYC2/3/4 (Fernández-Calvo et al., 2011). We used electrophoretic mobility shift assays to analyze the binding affinity of JAM1 to the MYC2 target sequences in vitro. IRDye-labeled oligo DNA probes containing G-box (CACGTG), TG-box (CACGTT), or MYC2 binding sequence (MBS) in rd22 (CACATG) (Abe et al., 1997) and His-tagged MYC2 and JAM1 expressed in Escherichia coli were used for the experiments (see Supplemental Figure 19A online). The electrophoretic mobility shift assay results showed that both MYC2 and JAM1 bind to G-box, TG-box, and MBS sequences. The sequence-specific binding of MYC2 and JAM1 to these cis-elements was confirmed by the addition of unlabeled oligo DNA probes and of unlabeled mutated oligo DNA probe (sees Supplemental Figures 19B and 19C online).
Transient expression analysis using the LUC reporter construct in which upstream regulatory region contained five repeats of the G-box or MBS fused to TATA-box revealed that coexpression of JAM1-VP16 or MYC2-VP16 effectors activated the reporter gene (see Supplemental Figures 20A to 20C online). The results of the transient assay showed that both JAM1 and MYC2 bind to G-box and MBS in plant cells.
JAM1 Competes with MYC2 for Binding to Its Target Sequence
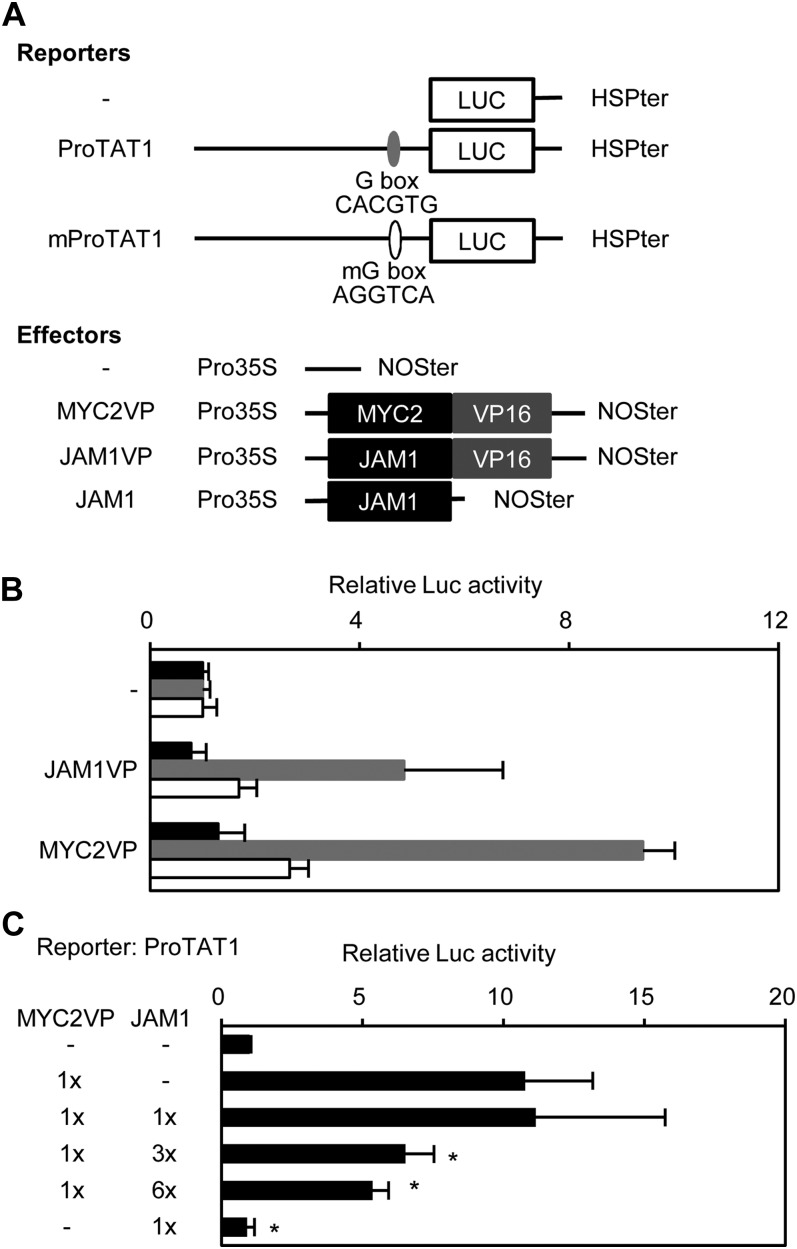
TAT1 and JAZ3 are known to be direct targets of MYC2 and contain putative MYC2 target site(s) within 1000 and 418 bp upstream of the translational start site, respectively (Figure 8A) (see Supplemental Figures 20D and 20E online) (Chini et al., 2007; Hou et al., 2010). To examine binding of JAM1 to putative MYC2 target sites in the TAT1 and JAZ3 upstream regions, we prepared reporter plasmids in which the LUC reporter was fused with 1000 and 418 bp upstream regions of TAT1 and JAZ3, respectively (Figure 8A) (see Supplemental Figure 20E online). Coexpression of JAM1-VP16 increased LUC expression, as did expression of MYC2-VP16 (Figure 8B) (see Supplemental Figure 20F online). Mutation of the G-box in the TAT1 promoter mostly eliminated the activation of LUC reporter by JAM1-VP16 and MYC2-VP16 (Figure 8B). Neither effector activated reporter expression when the putative MYC2 target sites in the JAZ3 promoter were mutated (see Supplemental Figure 20F online). These results indicate that both JAM1 and MYC2 regulate JAZ3 and TAT1 expression by binding to the G-box and its derivative cis-elements in the promoters. Next, we performed competition assays by transient expression of JAM1 and MYC2-VP16. Transcriptional activation of the ProTAT1-regulated LUC reporter by MYC2-VP16 was reduced in a dose-dependent manner by coexpression of JAM1, providing direct evidence for competitive binding of MYC2 and JAM1 to the same cis-element (Figure 8C).
Figure 8.
MYC2 and JAM1 Bind the G Box in the TAT1 Promoter.
(A) Schematic representation of the constructs used for transient expression assays. The reporter construct consists of TAT1 promoter, HSP terminator (HSPter), and firefly LUC coding sequence. Effector constructs express MYC2-VP16, JAM1-VP16, or JAM1 under the control of the CaMV 35S promoter.
(B) Binding of JAM1 and MYC2 on the G-box in the TAT1 promoter by transient expression assays. Constructs for the assay are shown in (A). Bars indicate values for no promoter (black), TAT1 promoter (gray), and mutated TAT1 promoter (white). The value without effector in each reporter was set to 1 and relative values are shown. Error bars indicate sd of results from four replicates.
(C) Competition of JAM1 and MYC2 for binding to the G-box by transient expression assays. Constructs for the assay are shown in (A). Relative amounts of effector construct are shown on the left. The value without effector was set to 1 and relative values are shown. Error bars indicate sd of results from four replicates. Asterisks indicate significant difference from the value of MYC2-VP16 (Welch’s t test, P < 0.05).
DISCUSSION
Recent advances have established a simple transcriptional regulatory model for JA signaling consisting of MYC2, JAZs, NINJA, TOPLESS, and SCFCOI1 complexes (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Pauwels et al., 2010). However, new insights have recently emerged on the regulatory mechanisms of JA signaling, including identification of JAZ binding partners, and the influence of circadian rhythms and hormonal crosstalk (Hou et al., 2010; Qi et al., 2011; Song et al., 2011; Goodspeed et al., 2012; Hong et al., 2012; Shin et al., 2012; Yang et al., 2012). These suggest that JA signaling encompasses multiple complicated regulatory mechanisms. In this study, we identified JAM1, a bHLH transcription factor, as a negative regulator of JA signaling. JAM1 possesses transcriptional repression activity, and the jam1 mutant exhibits a JA-hypersensitive phenotype and elevated JA responsiveness. Compared with myc2 mutants and MYC2 CRES-T plants, JAM1 CRES-T plants exhibited more diverse JA-related phenotypes, including effects on male fertility, suggesting broad functions of JAM1 in the JA signaling pathway. In addition, we demonstrated that the two transcription factors MYC2 and JAM1 cooperatively regulate JA responses by competitive binding to their target sequence. Our findings provide insights into the transcriptional regulatory mechanisms of JA signaling.
JAM1 Is a Passive Repressor
Transcriptional repressors can be categorized as active or passive repressors (Krogan and Long, 2009). Passive repressors do not directly influence transcription but physically interfering with activators, such as by preventing their binding to target DNA sequence. We found that JAM1 is likely to be a passive repressor because it does not possess a known repression domain such as ERF-associated amphiphilic repression motif or B3 repression domain (Ikeda and Ohme-Takagi, 2009). The repression activity of JAM1 was relatively weak (Figure 3B) and addition of SRDX to the C terminal of JAM1 enhanced repression activity in the GAL4DB-based transient expression assay (see Supplemental Figure 10 online). This is consistent with severity of phenotypes in Pro35S:JAM1-SRDX and Pro35S:JAM1-GFP plants. We also demonstrated that JAM1 bound to MYC2 target sequences and inhibited binding of MYC2 to those sequences (Figure 8C).
JAM1 Functions as a Negative Regulator of JA Signaling
Continuous activation of JA responses results in plant growth arrest; therefore, negative regulation of JA production and JA signaling is necessary to limit the JA response. Molecular mechanisms for attenuating JA signaling were uncovered by identification of JAZ proteins, which negatively regulate JA responses by binding to transcriptional activators, such as MYC2 (Chini et al., 2007; Thines et al., 2007). The existence of degradation-resistant JAZs, including JAZ splice variants, further supports the importance of negative regulation of JA signaling by JAZ proteins (Chung and Howe, 2009; Chung et al., 2010; Shyu et al., 2012). However, JAM1 CRES-T plants exhibit JA-insensitive phenotypes, even when the expression of most of JAZ genes, including JAZ8, was reduced (Figures 2 and 7B) (see Supplemental Figure 17 online). Moreover, the jam1 mutant exhibits elevated and prolonged JA responses even though the expression of alternatively spliced JAZ3.4 and JAZ10.4 was increased in the plants (Figure 4D) (see Supplemental Figure 12B online). Inhibition of MYC2 activity by binding of the JAZs and other mechanisms, such as transcriptional repression by JAM1, would be required to regulate JA responses (Figure 9).
Figure 9.
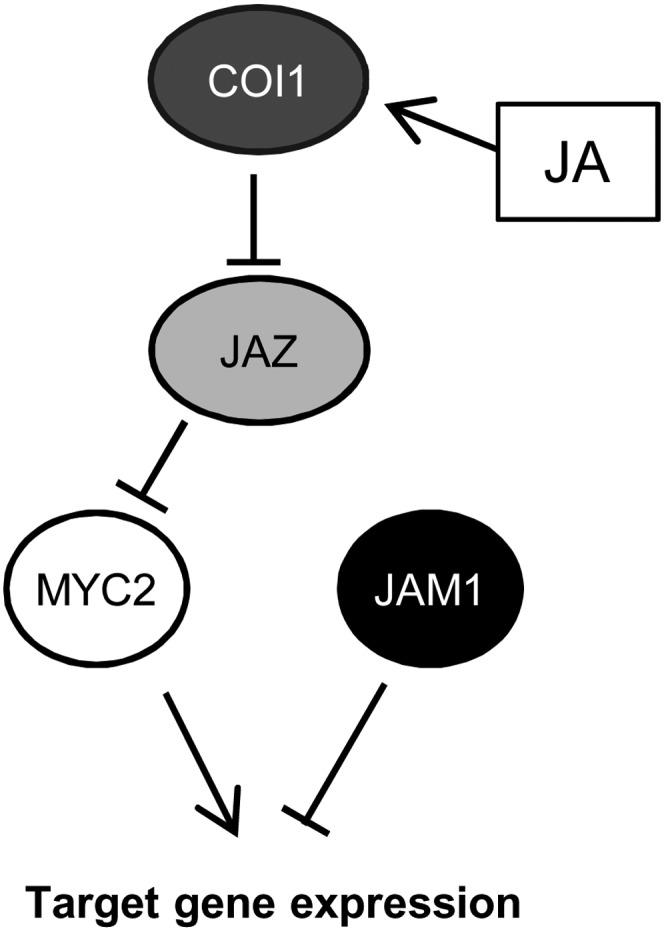
Model of JAM1-Mediated Suppression of JA Signaling.
JAM1 and MYC2 are expressed at low levels in the absence of JA due to suppression of MYC2 function by JAZ. JA-Ile promotes formation of the SCFCOI1-JAZ receptor complex and leads to degradation of JAZ protein. Released MYC2 activates expression of its target genes, including JAZs and JA biosynthetic genes and possibly MYC2 and JAM1. JAM1 binds MYC2’s target sequences and represses JA-responsive gene expression and JA responses.
There may be redundant factors with similar function to JAM1 because the jam1 single gene mutation shows relatively weak hypersensitivity to JA, although JAM1 appears to be major factor for the negative regulation of JA signaling. Two bHLHs (bHLH13/At1g01260 and bHLH003/At4g16430), which are closely related to JAM1, are supposed to be redundant factors of JAM1 (Y. Sasaki-Sekimoto and K. Shirasu, personal communication).
JAM1 induction by JA treatment occurs within 1 h (see Supplemental Figure 2A online), and JA-induced JAM1 expression follows MYC2 induction in the anthers of opr3 mutants (Mandaokar et al., 2006). These data indicate the possibility that JA-activated MYC2 induces expression of early JA-responsive target genes, including JAZs, MYC2, and JAM1, and then de novo–synthesized JAM1 negatively regulates the upregulated genes by competition for the same cis-regulatory elements as MYC2. Several JA-biosynthetic enzyme genes are directly regulated by MYC2 and likely to be bound by JAM1 (Figure 7C) (Hou et al., 2010). Once the high expression of these genes activated by MYC2 begins to be downregulated by JAM1 and JAZ, production of JA and JA-Ile is also downregulated. Reduction of JA-Ile production and induction of JA-Ile catabolism reduce JA-Ile levels and result in accumulation of JAZ protein (Koo et al., 2011; Koo and Howe, 2012), allowing further suppression of the expression of early JA-responsive genes. Suppression of gene expression by JAM1, production of stable JAZs, and increased JA-Ile catabolism provide distinct but coordinated mechanisms to terminate the JA response.
Although we demonstrated that JAM1 suppresses MYC2 activity through competitive binding to the target sequences of MYC2 (Figure 8C), effects of JAM1 and MYC2 on development differed. For example, male fertility was severely impaired in JAM1 CRES-T plants, but not in jin1-8 mutants, myc2 myc3 myc4 triple mutants, or MYC2-SRDX plants (Figures 1F to 1H) (see Supplemental Figure 5G online) (Fernández-Calvo et al., 2011). On the other hand, there is considerable overlap between genes repressed in ProJAM1:JAM1-SRDX and ProMYC2:MYC2-SRDX plants (Figure 7A). The strong induction of JAM1 and MYC2 transcripts in JA-treated or wounded tissues (see Supplemental Figure 2A online) would imply that competitive regulation of JA response by the activator MYC2 and repressor JAM1 may take place mainly in response to exogenous stimuli.
Recently, posttranslational regulation of MYC2 by mechanisms other than JAZ binding was reported. For example, binding of the circadian clock component TIME FOR COFFEE to MYC2 promotes degradation of MYC2 through the 26S proteasome pathway and negatively regulates JA signaling (Shin et al., 2012). Binding of MYC2 to DELLA proteins, which act as negative regulators of gibberellin (GA) signaling, inhibits MYC2-mediated expression of terpene synthase genes (Hong et al., 2012). In addition, the MYC2 protein level is gradually increased by JA treatment and reaches a peak at 12 to 24 h after treatment (Hou et al., 2010). In parallel, JA-induced transcripts of MYC2 target genes, including MYC2 itself, reduce within few hours after JA treatment (Figure 2). Together, these results indicate that the accumulation of MYC2 protein and its function are not correlated with its transcriptional level. This may be applicable to JAM1 and to JAZs and regulation of the activity of these factors may also be modulated by posttranscriptional mechanisms.
JAM1 Is Involved in Male Fertility
Recent work has revealed a large part of the regulatory network for JA-mediated male fertility. During stamen development, auxin and GA stimulate the expression of the JA biosynthetic gene DAD1 (Yu et al., 2004; Nagpal et al., 2005; Ito et al., 2007; Cheng et al., 2009; Tabata et al., 2010; Reeves et al., 2012). Elevated biosynthesis of JA in anthers leads to expression of MYB21, MYB24, and MYB57, which are involved in filament elongation, pollen maturation, and anther dehiscence (Mandaokar et al., 2006; Mandaokar and Browse, 2009; Reeves et al., 2012). The expression of MYB21 was significantly suppressed in coi1-1 mutants, indicating that JA-Ile perception by COI1 and subsequent JA signal transduction is necessary for induction of MYB21 in male organs (Song et al., 2011; Reeves et al., 2012). The expression of MYB21 and MYB24 in opr3 stamens is induced within 2 h and reaches a peak at 8 h after JA treatment, but JAM1 and MYC2 induction occurs earlier than that (Mandaokar et al., 2006). In addition, the expression of MYB21 was significantly suppressed in ProJAM1p:JAM1-SRDX plants (see Supplemental Figure 17 online). We examined whether JAM1 interacts with these MYBs and coordinately regulates downstream genes because some bHLH transcription factors form bHLH/MYB/WD-repeat complexes during anthocyanin biosynthesis and trichome formation (Payne et al., 2000). However, our yeast two-hybrid analysis did not show any interaction of JAM1 either with MYB21 or MYB24 (data not shown). These results suggest that JAM1 acts upstream of MYBs to regulate JA signaling for their induction.
The existence of transcriptional activators that partner with JAM1 to participate in JA-mediated male fertility has been suggested because JAM1 acts as transcriptional repressor and MYC2/3/4 are not required for JA-mediated male fertility (see Supplemental Figure 5G online) (Fernández-Calvo et al., 2011). Although overexpression of undegradable JAZ10.4 caused JA-insensitive root growth and male sterile phenotypes (Chung and Howe, 2009), overexpression of a JAZ10 genomic fragment caused only a JA-insensitive root growth phenotype (Chung et al., 2010). These results suggest that the mechanisms that promote JA responses in flower organs are different from those in other organs. In addition, recent reports showed that MYB21 and MYB24 directly interact with JAZs, which attenuate their transcriptional functions (Song et al., 2011), and that MYB21 negatively regulates expression of JA biosynthetic enzyme genes to decrease JA levels in the flower (Reeves et al., 2012), indicating that JA-mediated male fertility is regulated by a complicated transcriptional network.
JAM1 Function and Hormonal Crosstalk
Several AP2/ERF-type transcription factors, including ETHYLENE RESPONSE FACTOR1 (ERF1), have been reported to integrate JA and ethylene signaling pathways (Lorenzo et al., 2003; Pré et al., 2008). The expression of ERF1 is upregulated in MeJA-treated myc2 mutant, indicating that MYC2 negatively regulates ERF1 (Dombrecht et al., 2007). Because ERF1 was induced in MYC2-SRDX plants in our microarray analysis (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42552), these findings imply the existence of unidentified negative regulators that function downstream of MYC2 and suppress the expression of ERF1. Our microarray analysis showed that ERF1 was also induced in JAM1-SRDX plants, as in MYC2-SRDX. These results indicate that JAM1 and MYC2 functions upstream of ERF1 in JA signaling.
AIB/JAM1 was previously reported to be a positive regulator of ABA signaling (Li et al., 2007). By contrast, our results clearly demonstrated that JAM1 acts as a negative regulator in JA signaling. Not only synergistic, but also antagonistic, interactions between JA and ABA signaling were observed in response to various biotic and abiotic stresses in Arabidopsis (Anderson et al., 2004; Kazan and Manners, 2008). The Arabidopsis ABA receptor mutants pyl4 and pyl5 showed JA-hypersensitive shoot growth as well as reduced anthocyanin accumulation in response to JA (Lackman et al., 2011). JAM1 may act oppositely in JA and ABA signaling and fine-tune the responses to these two plant hormones.
The effects of JA on plant growth and defense responses antagonize the effects of GA (Kazan and Manners, 2012). Recently, it was reported that direct binding of DELLA proteins to JAZ proteins promotes JA signaling via competitive inhibition of the JAZ–MYC2 interaction (Hou et al., 2010). By contrast, JA treatment simultaneously elevates JAZ degradation and DELLA accumulation and suppresses GA responses by facilitating DELLA–PHYTOCHROME INTERACTING FACTOR (PIF) interactions to suppress PIF-mediated growth (Yang et al., 2012). These findings demonstrate a molecular mechanism underlying growth inhibition by JA. Our microarray experiments demonstrated that the expression of three DELLA genes, REPRESSOR OF ga1-3-LIKE1 (RGL1), RGL2, and RGL3, but not GA INSENSITIVE and REPRESSOR OF ga1-3, is suppressed in JA-treated JAM1 CRES-T plants (see Supplemental Table 1 online; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42552). This may be the reason that JA-mediated growth retardation is reduced in JAM1 CRES-T plants.
Our findings provide a model of the JA signaling pathway in which the negative regulator JAM1 acts in the MYC2-regulated pathway to attenuate JA responses. The complex mechanism of JA signal transduction involving JAM1, MYC2, JAZs, and possibly other unidentified transcription factors, JA-Ile production/catabolism, and crosstalk with other hormone networks enables strict regulation of the JA response. Plants have evolved such complex mechanisms so that they can survive environmental stresses while also optimizing growth in a fluctuating environment.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (Col-0 or Ler) was used for generating transgenic plants. Unless otherwise indicated, plants were grown at 22°C under long-day conditions (16 h light and 8 h dark). To measure root length, seeds were sown on Murashige and Skoog (MS) medium containing 0.8% agar supplemented with the indicated concentrations of MeJA, and the seedlings were grown in vertically held Petri dishes. For time-course experiments, ∼10 surface-sterilized seeds were placed in each well of a 24-well plate with 2 mL of 1× MS liquid medium. The plates were incubated with gentle agitation (100 rpm) under continuous light at 22°C. Seven-day-old seedlings were treated with 50 μM MeJA and were collected at the indicated times after treatment. coi1-30 (SALK_035548), jin1-8 (SALK_061267), aos (CS6149), SAIL_536_F09, and jam1 (CS170389) were provided by the ABRC. GABI285E09 was obtained from The European Arabidopsis Stock Centre. coi1-16 (Ellis and Turner, 2002) and dad1 (Ishiguro et al., 2001) seeds were provided by Alessandra Devoto (University of London, UK) and Sumie Ishiguro (Nagoya University, Japan), respectively.
Along with our group, Y. Sasaki-Sekimoto and K. Shirasu (RIKEN, Japan), who also study AIB/JAM1 function, support renaming this MYC2-related bHLH-type transcription factor (At2g46510) as JAM1 because both groups have found that JAM1 is essential in JA signaling rather than ABA signaling.
Preparation of Constructs
Primers used for this study are shown in Supplemental Table 2 online. The 5′ upstream regions of 2978 and 3034 bp, extending from the translational start sites of the JAM1 and MYC2 genes, were used for preparation of ProJAM1:GUS and ProJAM1:JAM1-SRDX, and ProMYC2:MYC2-SRDX, respectively. The same promoter region of MYC2 was fused to JAM1 and MYC2 coding sequences to construct ProMYC2:JAM1 and ProMYC2:MYC2, respectively. To prepare a plasmid expressing JAM1 fused with GFP at the C terminus, a DNA fragment encoding JAM1-GFP was inserted into the SmaI site of p35SG. Pro35S:JAM1-VP16 was prepared by insertion of the JAM1 coding region without the stop codon into the SmaI site of p35SVP16 (Mitsuda et al., 2006). Pro35S:JAM1 and Pro35S:VP16 were prepared by insertion of the JAM1 coding region with the stop codon and VP16 with a stop codon into SmaI site of p35SG. The region corresponding to the transgene was transferred to the pBCKH plant expression vector by LR clonase reaction (Invitrogen) (Mitsuda et al., 2006). For His-tagged protein expression in Escherichia coli, coding sequences of JAM1 and MYC2 were inserted into the NcoI site of pETHis vector (Chen and Hai, 1994).
The coding sequences of JAZ3, JAM1, and MYC2 and JAM1-VP16 without the stop codon were amplified from an Arabidopsis cDNA library and Pro35S:JAM1-VP16, respectively, with appropriate primers containing attB1 or attB2 sequences, and the resultant DNA fragments were cloned into pDONR207 (Invitrogen) by BP clonase reaction (Invitrogen). Effector plasmids for the expression of the GAL4-fused protein in plant cells (Ohta et al., 2001) were modified for Gateway destination vector by insertion of the ccdB cassette and stop codons in all three reading frames into the SmaI site. Effector plasmids for the expression of VP16-fused protein in plant cells were constructed by insertion of the ccdB cassette for Gateway cloning (Invitrogen) into the SmaI site of p35SVP16 from which the attL1 and attL2 sequences were removed. For yeast two-hybrid analysis, the pBTM116 and pVP16S1 vectors were modified by insertion of Gateway ccdB cassette and stop codons in all three reading frames into the SmaI and BamHI site, respectively. Genes cloned into pDONR207 were transferred to these destination vectors by LR clonase reaction (Invitrogen).
The pGL4.1HSP vector was constructed as follows. Approximately 250 bp of the 3′ region of Arabidopsis HSP18.2 (At5g59720) (Nagaya et al., 2010) was amplified using Xba_HSP_F and Bam_HSP_R primers and inserted between the XbaI and BamHI sites of pGL4.10 vector (Promega). Annealed primers for the five repeats of the G-box and MBS, 418-bp region of JAZ3 promoter, or 1000-bp region of TAT1 promoter were inserted between the SacI and XhoI sites, SalI site, or SacI and XhoI sites of pGL4.1HSP, respectively. The 35S-5xGAL4:LUC reporter plasmid was described previously (Hiratsu et al., 2002).
Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was performed as described previously (Matsui et al., 2008). In brief, Saccharomyces cerevisiae strain L40 was transformed by the LiAc/DNA/PEG method. Cotransformed yeast cells were plated on dropout medium without Trp, His, and Leu to test for His prototrophy and protein interactions.
Insect Feeding Assay
Spodoptera exigua eggs (Benzon Research) were hatched at 30°C. One day after hatching, larvae were transferred to fully expanded rosette leaves of 6-week-old plants grown at 20°C under short-day conditions as described (Herde et al., 2013). Five larvae were reared on each plant and 14 or more plants were used per genotype. Larval weights were measured 16 d after challenge. The experiment was independently repeated twice.
Electrophoresis Mobility Shift Assay
His-tagged JAM1 and MYC2 proteins expressed in E. coli were purified using Ni-NTA agarose (Qiagen) and stored in storage buffer (10 mM Tris-HCl, pH 8.0, 250 mM NaCl, 0.05% Triton X-100, and 50% glycerol). Oligonucleotides end-labeled with or without IRDye 800 infrared dye were annealed with complementary oligonucleotides for probes or competitors, respectively. Labeled probe (50 fmol) was incubated with 10 ng of purified protein in 20 μL of binding buffer (10 mM Tris-HCl, pH 7.5, 50 mM KCl, 3.5 mM DTT, 0.25% Tween 20, 5% glycerol, 5 mM MgCl2, and 50 μM EDTA) and 1 μg poly (dI-dC), in the presence or absence of nonlabeled competitor (50 pmol) for 20 min at room temperature and then run on to 4% polyacrylamide gels in 0.5× TBE buffer at 4°C. The infrared dye signal was detected with an Odyssey CLx imaging system (LI-COR).
GUS Staining
Plant tissues were immersed in 100 mM sodium phosphate, pH 7.0, 0.1% Triton X-100, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, and 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide and vacuum infiltrated for 30 min, followed by incubation at 37°C for up to 2 h. To examine the response to wounding, rosette leaves were wounded with forceps and stained 1 h later.
Quantification of Anthocyanin Content
Surface-sterilized seeds were sown in liquid 1× MS medium containing 0 or 50 μM MeJA and grown under continuous light at 22°C for 1 week with gentle agitation. MeJA (50 μM) was added to 8-d-old seedlings, and the seedlings were harvested 3 d after treatment. Whole plants frozen with liquid nitrogen were ground in a mortar and pestle, then resuspended into five volumes (based on fresh weight) of 45% methanol and 5% acetic acid. After removing cell debris by centrifugation, the relative level of anthocyanin was calculated from the absorbance at 530 and 637 nm, as described previously (Matsui et al., 2004).
RNA Extraction and Quantitative RT-PCR
Total RNA was extracted with the RNeasy Plant Mini kit (Qiagen). Genomic DNA was removed by DNase digestion during RNA extraction according to the manufacturer’s instructions. One microgram of total RNA was subjected to first-strand cDNA synthesis using the PrimeScript RT reagent kit (Takara). Quantitative RT-PCR was performed by the SYBR green method using ABI7300 real-time PCR system (Applied Biosystems) as described previously (Mitsuda et al., 2005). The gene-specific primers used for RT-PCR are shown in Supplemental Table 2 online. Relative amounts of transcripts were calculated by an absolute quantification method, with the PP2AA3 gene as an internal control. At least three biological replicates were included in each experiment.
Wounding Treatment and JA-Ile Analysis
Mechanical wounding and quantification of JA-Ile was performed as described previously (Koo et al., 2009, 2011; Herde et al., 2013). Briefly, four rosette leaves of 24-d-old plants were wounded by crushing twice across the mid-rib with a hemostat. Damaged leaves were harvested 2 h after wounding, together with undamaged leaves from untreated plants as a control. Approximately 200 mg leaf tissue pooled from four plants was used to generate each data point for one replicate. Tissue was weighed, immediately frozen in liquid nitrogen, and then stored at −80°C until use. [13C6]JA-Ile (Chung et al., 2008) was included during JA extraction as an internal standard for the quantification of endogenous JA-Ile. Compounds in plant extracts were separated on an Ascentis Express C18 column (50 × 2.1 mm, 2.7 μm; Supelco) attached to an Acquity ultraperformance liquid chromatography system (Waters) and analyzed by Quattro Premier XE tandem quadrupole mass spectrometer (Waters) using electrospray ionization (negative mode) as described previously (Koo et al., 2009).
Transient Expression Assay
Transient expression assays were performed as described previously (Ohta et al., 2001; Hiratsu et al., 2002). Reporter plasmid (1.2 μg) and 0.8 μg of each effector plasmid were used for each bombardment. For normalization of reporter gene activity, 0.4 μg of reference plasmid, pRL, was cobombarded. Bombarded leaves were incubated for 12 h in darkness, and then luciferase activity was quantified. At least three biological replicates were included in each experiment.
Measurement of Plant Diameter
Two leaves of 2-week-old plants were wounded by forceps three times at 2-d intervals. Rosette diameter was measured 6 d after the final treatment.
Microarray Analysis
The microarray experiments were performed using the Agilent Arabidopsis 4 (44k) microarray (Agilent Technologies) according to the manufacturer’s instructions. Four biological replicates were tested with a one-color method. Spot signal values were calculated with Feature Extraction version 9.1 software (Agilent). The quality control (QC) value was defined as 1 when a spot passed the “FeatNonUnifOL” filter and as 2 when the spot further passed the “FeatPopnOL” filter. The detection value was defined as 1 when a spot passed the “IsPosAndSignif” filter and as 2 when the spot further passed the “IsWellAboveBG” filter. All signal values were divided by the median value among spots with QC = 2 to enable comparison with other microarray data. Spot-to-gene conversion was accomplished based on a table provided by The Arabidopsis Information Resource (ftp://ftp.Arabidopsis.org/home/tair/Microarrays/Agilent/agilent_array_elements-2010-12-20.txt). The average values were used for the genes corresponding to two or more probes. The P value for each gene was calculated using Welch’s t test. To control type I family-wise error, we calculated the Q-value (FDR) from the P value using QVALUE software using the default settings (Storey and Tibshirani, 2003) in genes that have average QC and detection value among all hybridizations >1.5. The up- or downregulated genes were selected using Q-value and fold-change filters (Q < 0.03 and greater than threefold or <0.75-fold). All data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE42552.
Microscopy
Light microscopy and fluorescence microscopy for detection of GFP were performed using the Axioskop2 Plus system (Carl Zeiss). Stereomicroscopy was performed using MZ FL III (Leica). For scanning electron microscopy, fresh samples were observed using a scanning electron microscope (real 3D system model VE8800; Keyence) at an accelerating voltage of 1.3 kV.
Accession Numbers
Arabidopsis Genome Initiative numbers described in this article are as follows: AOS (At5g42650), bHLH003 (At4g16430), bHLH013 (At1g01260), COI1 (At2g39940), DAD1 (At2g44810), DFR (At5g42800), JAM1 (At2g46510), JAZ3 (At3g17860), JAZ10 (At5g13220), LEUCOANTHOCYANIDIN DIOXYGENASE (At4g22880), LOX3 (At1g17420), MYB21 (At3g27810), MYC2 (At1g32640), MYC3 (At5g46760), MTC4 (At4g17880), OPR3 (At2g06050), PRODUCTION OF ANTHOCYANIN PIGMENT1 (At1g56650), PP2AA3 (At1g13320), TAT1 (At4g23600), and VSP1 (At5g24780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. JA-Insensitive Root Growth of Pro35S:MYC2-SRDX Plants.
Supplemental Figure 2. Expression Pattern of JAM1.
Supplemental Figure 3. Reduced Expression of Anthocyanin-Related Genes in JAM1 CRES-T Plants.
Supplemental Figure 4. Reduced Trichome Phenotype of ProJAM1:JAM1-SRDX Plants.
Supplemental Figure 5. Effect of JAM1-SRDX on Floral Development.
Supplemental Figure 6. Filament Length in ProJAM1:JAM1-SRDX Flowers.
Supplemental Figure 7. The Level of Wound-Induced JA-Ile Production Is Similar in JAM1 CRES-T and jam1 Plants.
Supplemental Figure 8. Alignment of Amino Acid Sequences of JAM1, MYC2, and Closely Related bHLHs.
Supplemental Figure 9. Subcellular Localization of JAM1-GFP.
Supplemental Figure 10. Enhanced Repression Activity of JAM1-SRDX.
Supplemental Figure 11. jam1 Transposon Insertion Line and Effects of ProJAM1:JAM1-SRDX on Ler.
Supplemental Figure 12. RT-PCR Analysis of JAZ10 in jam1.
Supplemental Figure 13. Complementation Analysis of jin1-8 by MYC2 and JAM1.
Supplemental Figure 14. Activation Activity of JAM1-VP16.
Supplemental Figure 15. Determination of Genotype of coi1-30 JAM1-VP16 Plants.
Supplemental Figure 16. Expression of JAM1 in coi1-30 Pro35S:JAM1-VP16 and coi1-30 Pro35S:JAM1 Plants.
Supplemental Figure 17. RT-PCR Analysis of JA-Inducible Genes after 1 h of JA Treatment in Col-0, ProMYC2:MYC2-SRDX, and ProJAM1:JAM1-SRDX Plants.
Supplemental Figure 18. JAM1 and MYC2 Did Not Interact in Yeast and Plants.
Supplemental Figure 19. Electrophoretic Mobility Shift Assay of MYC2 and JAM1.
Supplemental Figure 20. The Binding Affinity of JAM1 and MYC2 to the G-Box and MYC2 Target Site.
Supplemental Table 1. List of Early JA-Responsive Genes Downregulated by Both JAM1-SRDX and MYC2-SRDX.
Supplemental Table 2. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Alessandra Devoto (University of London, UK) and Sumie Ishiguro (Nagoya University, Japan) for providing coi1-16 and dad1 seeds, respectively. We thank members of the Ohme-Takagi laboratory for helpful discussions and technical assistance. This research was supported in part by the National Institutes of Health Grant R01GM57795 (to G.A.H.). M.H. was supported by a fellowship (HE 5949/1-1) from the German Research Foundation (Deutsche Forschungsgemeinschaft). The College of Natural Science at Michigan State University is acknowledged for their support of a student exchange program between the Ohme-Takagi and Howe labs.
AUTHOR CONTRIBUTIONS
M.N., K.S., G.A.H., and M.O.-T. designed the research. M.N., N.M., M.H., A.J.K.K., and J.E.M. performed experiments. M.N., G.A.H., and M.O.-T. wrote the article.
Glossary
- MeJA
methyl jasmonate
- CRES-T
chimeric repressor silencing technology
- bHLH
basic helix-loop-helix
- JA
jasmonates
- ABA
abscisic acid
- CaMV
cauliflower mosaic virus
- Col-0
Columbia-0
- GFP
green fluorescent protein
- Ler
Landsberg erecta
- FDR
false discovery rate
- MBS
MYC2 binding sequence
- GA
gibberellin
- MS
Murashige and Skoog
- QC
quality control
References
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.P., Badruzsaufari E., Schenk P.M., Manners J.M., Desmond O.J., Ehlert C., Maclean D.J., Ebert P.R., Kazan K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbi V., Devoto A. (2008). Jasmonate signalling network in Arabidopsis thaliana: Crucial regulatory nodes and new physiological scenarios. New Phytol. 177: 301–318 [DOI] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.P., Hai T. (1994). Expression vectors for affinity purification and radiolabeling of proteins using Escherichia coli as host. Gene 139: 73–75 [DOI] [PubMed] [Google Scholar]
- Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Sun L., Qi T., Zhang B., Peng W., Liu Y., Xie D. (2011). The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4: 279–288 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chung H.S., Cooke T.F., Depew C.L., Patel L.C., Ogawa N., Kobayashi Y., Howe G.A. (2010). Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Koo A.J., Gao X., Jayanty S., Thines B., Jones A.D., Howe G.A. (2008). Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146: 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demianski A.J., Chung K.M., Kunkel B.N. (2012). Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol. Plant Pathol. 13: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A., Turner J.G. (2003). Regulation of jasmonate-mediated plant responses in Arabidopsis. Ann. Bot. (Lond.) 92: 329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis C., Turner J.G. (2002). A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215: 549–556 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Goodspeed D., Chehab E.W., Min-Venditti A., Braam J., Covington M.F. (2012). Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. USA 109: 4674–4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Heitz T., Widemann E., Lugan R., Miesch L., Ullmann P., Désaubry L., Holder E., Grausem B., Kandel S., Miesch M., Werck-Reichhart D., Pinot F. (2012). Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 287: 6296–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herde M., Koo A.J.K., Howe G.A. (2013). Elicitation of jasmonate-mediated defense responses by mechanical wounding and insect herbivory. Methods Mol. Biol. 1011: 51–61. [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Mitsuda N., Matsui K., Ohme-Takagi M. (2004). Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem. Biophys. Res. Commun. 321: 172–178 [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Ohta M., Matsui K., Ohme-Takagi M. (2002). The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514: 351–354 [DOI] [PubMed] [Google Scholar]
- Hong G.J., Xue X.Y., Mao Y.B., Wang L.J., Chen X.Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24: 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Lee L.Y., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Ikeda M., Ohme-Takagi M. (2009). A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50: 970–975 [DOI] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Ng K.H., Lim T.S., Yu H., Meyerowitz E.M. (2007). The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell 19: 3516–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2008). Jasmonate signaling: Toward an integrated view. Plant Physiol. 146: 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2012). JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17: 22–31 [DOI] [PubMed] [Google Scholar]
- Kitaoka N., Matsubara T., Sato M., Takahashi K., Wakuta S., Kawaide H., Matsui H., Nabeta K., Matsuura H. (2011). Arabidopsis CYP94B3 encodes jasmonyl-L-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 52: 1757–1765 [DOI] [PubMed] [Google Scholar]
- Koo A.J., Cooke T.F., Howe G.A. (2011). Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc. Natl. Acad. Sci. USA 108: 9298–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo A.J., Gao X., Jones A.D., Howe G.A. (2009). A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 59: 974–986 [DOI] [PubMed] [Google Scholar]
- Koo A.J., Howe G.A. (2012). Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front Plant Sci 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.T., Long J.A. (2009). Why so repressed? Turning off transcription during plant growth and development. Curr. Opin. Plant Biol. 12: 628–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackman P., et al. (2011). Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc. Natl. Acad. Sci. USA 108: 5891–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Sun J., Xu Y., Jiang H., Wu X., Li C. (2007). The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol. Biol. 65: 655–665 [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sánchez-Serrano J.J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A., Browse J. (2009). MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 149: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A., Thines B., Shin B., Lange B.M., Choi G., Koo Y.J., Yoo Y.J., Choi Y.D., Choi G., Browse J. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46: 984–1008 [DOI] [PubMed] [Google Scholar]
- Matsui K., Tanaka H., Ohme-Takagi M. (2004). Suppression of the biosynthesis of proanthocyanidin in Arabidopsis by a chimeric PAP1 repressor. Plant Biotechnol. J. 2: 487–493 [DOI] [PubMed] [Google Scholar]
- Matsui K., Umemura Y., Ohme-Takagi M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55: 954–967 [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Hiratsu K., Todaka D., Nakashima K., Yamaguchi-Shinozaki K., Ohme-Takagi M. (2006). Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol. J. 4: 325–332 [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Iwase A., Yamamoto H., Yoshida M., Seki M., Shinozaki K., Ohme-Takagi M. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaya S., Kawamura K., Shinmyo A., Kato K. (2010). The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 51: 328–332 [DOI] [PubMed] [Google Scholar]
- Nagpal P., Ellis C.M., Weber H., Ploense S.E., Barkawi L.S., Guilfoyle T.J., Hagen G., Alonso J.M., Cohen J.D., Farmer E.E., Ecker J.R., Reed J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Niu Y., Figueroa P., Browse J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Halitschke R., Kim H.B., Baldwin I.T., Feldmann K.A., Feyereisen R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C.T., Zhang F., Lloyd A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pré M., Atallah M., Champion A., De Vos M., Pieterse C.M.J., Memelink J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P.H., et al. (2012). A regulatory network for coordinated flower maturation. PLoS Genet. 8: e1002506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P.M., Lee P.Y., Biesgen C., Boone J.D., Beals T.P., Weiler E.W., Goldberg R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A., Stintzi A. (2009). Enzymes in jasmonate biosynthesis - Structure, function, regulation. Phytochemistry 70: 1532–1538 [DOI] [PubMed] [Google Scholar]
- Shan X., Zhang Y., Peng W., Wang Z., Xie D. (2009). Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 60: 3849–3860 [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Heidrich K., Sanchez-Villarreal A., Parker J.E., Davis S.J. (2012). TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24: 2470–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu C., Figueroa P., Depew C.L., Cooke T.F., Sheard L.B., Moreno J.E., Katsir L., Zheng N., Browse J., Howe G.A. (2012). JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24: 536–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Browse J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. (2003). Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol. Biol. 224: 149–157 [DOI] [PubMed] [Google Scholar]
- Tabata R., Ikezaki M., Fujibe T., Aida M., Tian C.E., Ueno Y., Yamamoto K.T., Machida Y., Nakamura K., Ishiguro S. (2010). Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 51: 164–175 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Triezenberg S.J., Kingsbury R.C., McKnight S.L. (1988). Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 2: 718–729 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.-X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.L., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Sano R., Wada T., Takabayashi J., Okada K. (2009). Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 136: 1039–1048 [DOI] [PubMed] [Google Scholar]
- Yu H., Ito T., Zhao Y., Peng J., Kumar P., Meyerowitz E.M. (2004). Floral homeotic genes are targets of gibberellin signaling in flower development. Proc. Natl. Acad. Sci. USA 101: 7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Turner J.G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.