This study shows that PIF1/PIF3 and HY5/HYH physically interact and coordinately regulate the expression of ROS-responsive genes. It reveals that the PIF1/PIF3-HY5/HYH transcriptional modules mediate crosstalk between light and ROS signaling pathways and suggests a mechanism by which plants optimize their growth in response to excess light.
Abstract
The critical developmental switch from heterotrophic to autotrophic growth of plants involves light signaling transduction and the production of reactive oxygen species (ROS). ROS function as signaling molecules that regulate multiple developmental processes, including cell death. However, the relationship between light and ROS signaling remains unclear. Here, we identify transcriptional modules composed of the basic helix-loop-helix and bZIP transcription factors PHYTOCHROME-INTERACTING FACTOR1 (PIF1), PIF3, ELONGATED HYPOCOTYL5 (HY5), and HY5 HOMOLOGY (HYH) that bridge light and ROS signaling to regulate cell death and photooxidative response. We show that pif mutants release more singlet oxygen and exhibit more extensive cell death than the wild type during Arabidopsis thaliana deetiolation. Genome-wide expression profiling indicates that PIF1 represses numerous ROS and stress-related genes. Molecular and biochemical analyses reveal that PIF1/PIF3 and HY5/HYH physically interact and coordinately regulate the expression of five ROS-responsive genes by directly binding to their promoters. Furthermore, PIF1/PIF3 and HY5/HYH function antagonistically during the seedling greening process. In addition, phytochromes, cryptochromes, and CONSTITUTIVE PHOTOMORPHOGENIC1 act upstream to regulate ROS signaling. Together, this study reveals that the PIF1/PIF3-HY5/HYH transcriptional modules mediate crosstalk between light and ROS signaling and sheds light on a new mechanism by which plants adapt to the light environments.
INTRODUCTION
Besides being the primary energy source for photosynthesis, light provides signals that regulate diverse aspects of plant growth and development (Chen et al., 2004). In the dark, seedlings undergo skotomorphogenesis and exhibit long hypocotyls and closed cotyledons with undifferentiated chloroplasts. Light-triggered seedling deetiolation (photomorphogenesis) is a particularly important process, as it allows plants to establish autotrophic growth, which is essential for survival. When light is perceived by photoreceptors, including the red/far-red light-absorbing phytochromes (phyA to phyE) and blue/UV-A light-absorbing cryptochromes (cry1 and cry2), the light signals are sequentially transduced to a series of downstream intermediates, leading to changes in transcriptional programs and, eventually, to physiological changes, including reduced hypocotyl growth, opening of cotyledons, and chloroplast development (Quail, 2002; Chen et al., 2004).
Genetic and molecular studies have uncovered dozens of intermediates that relay the light signal. A group of CONSTITUTIVE PHOTOMORPHOGENIC (COP)/DEETIOLATED/FUSCA proteins acts as central repressors of photomorphogenesis downstream of both phytochromes and cryptochromes (Wei and Deng, 1996). COP1, a RING (for Really Interesting New Gene) finger protein, possesses E3 ubiquitin ligase activity toward a number of photomorphogenesis-promoting factors, facilitating their targeted degradation through the 26S proteasome pathway (Yi and Deng, 2005). The transcription factors ELONGATED HYPOCOTYL5 (HY5) and its close homolog, HY5 HOMOLOG (HYH), and PHYTOCHROME-INTERACTING FACTORs (PIFs) are known to mediate two distinct signaling branches of the photomorphogenic response in Arabidopsis thaliana (Castillon et al., 2007; Lau and Deng, 2010). HY5 and HYH are a pair of bZIP transcription factors that play pivotal roles in positively regulating seedling deetiolation (Oyama et al., 1997; Holm et al., 2002). Genome-wide gene expression and chromatin immunoprecipitation (ChIP)–based sequencing analyses revealed that HY5 directly binds to thousands of genes and regulates the expression of a broad range of genes (Lee et al., 2007; Zhang et al., 2011). Consistent with their biological function, HY5 and HYH are degraded in the dark by the COP1-mediated degradation pathway but are stabilized in the light (Osterlund et al., 2000; Holm et al., 2002). HY5 has also been reported to mediate plant responses to hormones, cold, and UV-B (Ulm et al., 2004; Lau and Deng, 2010; Catalá et al., 2011), indicating that HY5 serves as a master modulator during plant growth and development. PIFs are a small subfamily of basic helix-loop-helix (bHLH) transcription factors that play multiple functions in processes such as seed germination, seedling deetiolation, and shade avoidance responses (Castillon et al., 2007; Leivar et al., 2008; Shin et al., 2009; Stephenson et al., 2009; Leivar and Quail, 2011). Contrary to HY5/HYH, PIFs accumulate in the dark to promote skotomorphogenesis, whereas light induces the rapid phosphorylation and degradation of PIFs, through the activity of phyB and phyA (Shen et al., 2005; Al-Sady et al., 2006; de Lucas et al., 2008; Henriques et al., 2009). Molecular studies showed that PIFs directly regulate the expression of downstream genes by binding to a G-box motif (CACGTG) present in their promoters (Martínez-García et al., 2000; Huq et al., 2004; Shin et al., 2007). PIF1 and PIF3 are also involved in facilitating the seedling greening process, in part by negatively regulating the chlorophyll biosynthetic pathway in the dark (Huq et al., 2004; Moon et al., 2008; Shin et al., 2009; Stephenson et al., 2009).
Reactive oxygen species (ROS) are common byproducts of normal metabolism in cells from bacteria to mammals and act as important signaling molecules that regulate multiple developmental processes, such as root growth, stress tolerance, senescence, pathogen defense, and hormonal responses (Apel and Hirt, 2004; Mittler et al., 2004; Gechev et al., 2006; Miller et al., 2008). ROS are produced in cellular compartments, particularly when plants are subjected to environmental stress conditions. For instance, excess light irradiation results in the generation of ROS, including hydrogen oxygen, superoxide, and singlet oxygen, in chloroplasts, and this leads to photooxidative damage in plant cells or even to cell death (Li et al., 2009). Consequently, the ability of plants to use light energy for photosynthesis is limited (op den Camp et al., 2003; Gechev et al., 2006). Singlet oxygen has a short lifetime and is a strong electrophilic reagent that reacts with many biological molecules, including DNA, proteins, and lipids (Triantaphylidès and Havaux, 2009). The intensity, duration, and localization of different ROS signals are determined by the interplay between the ROS-producing and ROS-scavenging pathways of cells (Mittler et al., 2004). Thus, plants have evolved mechanisms that tightly regulate the ROS gene network to modulate the steady state level of ROS (Miller et al., 2008). Some important signaling components of the ROS gene network, such as transcription factors and protein kinases, have been identified by genetic and molecular approaches (Mittler et al., 2004; Miller et al., 2008). Studies also suggested that ROS signaling is integrated with many other signaling networks in plants; however, the mechanisms that mediate these interactions are poorly understood (Mittler et al., 2011). Although light is known to affect ROS production, the manner by which light signals regulate ROS production and ROS signaling remains unknown.
Here, we demonstrate that PIF1 and PIF3 prevent singlet oxygen production and cell death during seedling deetiolation in Arabidopsis. PIF1 regulates the genome-wide expression of ROS-responsive genes. We show that PIF1/PIF3 and HY5/HYH physically interact to form transcriptional modules that directly bind to the G-box motif in the promoters of five representative ROS signaling genes and regulate their expression coordinately. In addition, the functions of PIF1 and PIF3 largely require the presence of HY5 and HYH. Furthermore, phyA, phyB, cry1, and cry2 photoreceptors and COP1 are also involved in regulating ROS signaling. Our study demonstrates that key components of the light signaling pathway modulate the ROS signaling network and thus affect plant fitness in excess light.
RESULTS
PIF1 and PIF3 Redundantly Promote Seedling Greening and Prevent Singlet Oxygen Production
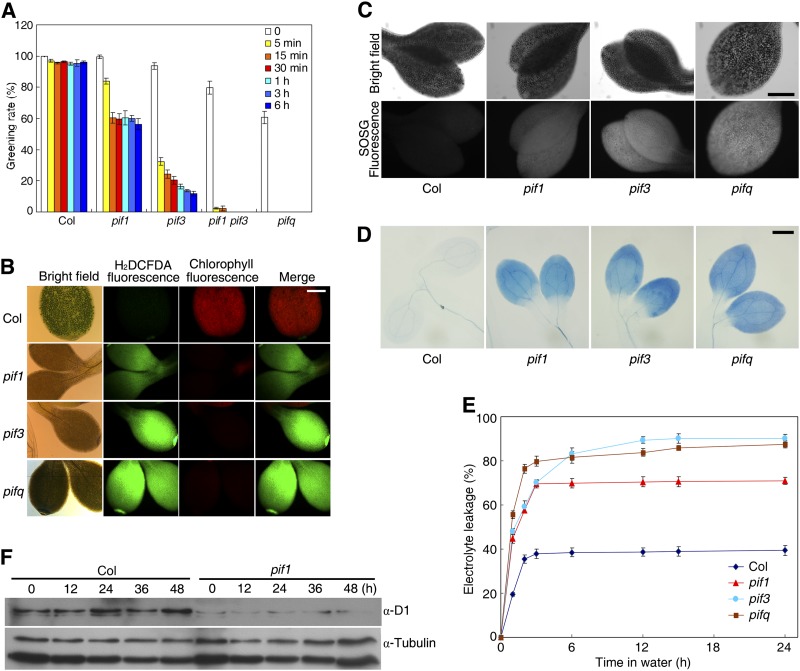
Previous studies documented that loss of either PIF1 or PIF3 caused overaccumulation of protochlorophyllide (Pchlide), a precursor of chlorophyll, in dark-grown seedlings and resulted in photobleaching upon transfer to light (Huq et al., 2004; Moon et al., 2008; Shin et al., 2009; Stephenson et al., 2009). The etiolated seedlings of the pif5 mutant also showed the photobleaching phenotype after light exposure (Shin et al., 2009). To examine whether these PIF proteins have redundant functions in regulating seedling greening, we examined the phenotypes of the pif1 pif3 double and pifq quadruple mutant (loss of PIF1, 3, 4, and 5) (Leivar et al., 2008). The levels of Pchlide of dark-grown seedlings were determined by monitoring the fluorescence emission of the samples with a fluorescence spectrophotometer (Tang et al., 2012). We found that pifq possessed the highest levels of Pchlide and that pif1 pif3 also accumulated more Pchlide than either of the single mutant parents and the Columbia (Col) wild type (see Supplemental Figure 1A online). We then tested the greening ability by subjecting 4-d-old dark-grown seedlings to increasing periods of growth in moderate light (60 µmol m−2 s−1) followed by 2 d of growth in weak light (10 µmol m−2 s−1). In the absence of moderate light treatment, ∼80 and 60% seedlings of pif1 pif3 and pifq, respectively, turned green normally, whereas pif1 and pif3 seedlings were indistinguishable from those of the wild type (Figure 1A). When the etiolated seedlings were exposed to moderate light for 5 min, ∼68 and 98% of seedlings of pif3 and pif1 pif3, respectively, were photobleached, while all of the pifq seedlings died. Moderate light treatments for 15 min to 6 h caused photobleaching in ∼40 and 85% of pif1 and pif3 seedlings, respectively (Figure 1A). These data indicate that PIF3 and PIF1 play major and redundant roles in promoting seedling greening and that the etiolated seedlings are extremely sensitive to the onset of relatively high light illumination. In the following experiments, all light treatments were performed in 60 µmol m−2 s−1 light.
Figure 1.
PIF1 and PIF3 Redundantly Promote Seedling Greening and Prevent Singlet Oxygen Production and Cell Death.
(A) Percentage of greening cotyledons in the wild type (Col) and various pif mutants under different light treatments. Four-day-old dark-grown seedlings were first exposed to growth light (60 µmol m−2 s−1) for the indicated periods of time and then transferred to weak light (10 µmol m−2 s−1) for 2 d. Data are mean ± sd, n = 3.
(B) to (D) Four-day-old etiolated seedlings were exposed to light (60 µmol m−2 s−1) for 24 h (B) or 6 h ([C] and [D]). Bars = 200 μm.
(B) Cellular ROS levels in the cotyledons of the wild type (Col) and various pif mutants. H2DCFDA fluorescence (green) indicates ROS, and chlorophyll autofluorescence is shown in red.
(C) Singlet oxygen production in the cotyledons as determined by SOSG fluorescence.
(D) Trypan blue staining of cotyledons of the wild type (Col) and pif1, pif3, and pifq.
(E) Electrolyte leakage of the pif mutants and wild-type seedlings. Four-day-old etiolated seedlings were exposed to light (60 µmol m−2 s−1) for 12 h and immersed in water, and electrolyte leakage was measured periodically. Data are mean ± sd, n = 3.
(F) Turnover of D1 protein in the pif1 mutant compared with the wild type. Immunoblot of a tubulin protein serves as a control. Seedlings were grown in darkness for 4 d before exposure to light (60 µmol m−2 s−1) for the indicated periods of time.
It was reported that the seedling photobleaching phenotype during the dark-to-light transition is largely caused by ROS (Reinbothe et al., 1996). We then examined the cellular ROS levels of pif1, pif3, pifq, and the wild type by detecting the fluorescence of 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFDA), a ROS-sensitive dye, in the cotyledons (Tang et al., 2012). When 4-d-old dark-grown seedlings were illuminated with light for 24 h, H2DCFDA fluorescence (Figure 1B, shown in green) was strong in pif1 and further increased in pif3 and pifq compared with the wild type, whereas chlorophyll autofluorescence was only observed in the wild-type cotyledons (Zhong et al., 2009; Figure 1B). Free Pchlide is a potent photosensitizer that generates singlet oxygen upon illumination (op den Camp et al., 2003). We thus investigated singlet oxygen production by these mutants using the fluorescent probe Singlet Oxygen Sensor Green (SOSG) (Flors et al., 2006). As shown in Figure 1C, the cotyledons of pif1, pif3, and pifq displayed SOSG fluorescence, whereas the wild type did not, indicating that the mutants release singlet oxygen. Superoxide and hydrogen peroxide are often simultaneously generated in cellular compartments. Surprisingly, the levels of both superoxide and hydrogen peroxide, as determined by nitroblue tetrazolium (NBT) and diaminobenzidine (DAB) staining, respectively, were reduced in pif1 and completely absent in pif3 and pifq, whereas they were abundant in the wild type (see Supplemental Figures 1B and 1C online). These results imply that PIF1 and PIF3 specifically prevent singlet oxygen production during seedling deetiolation.
PIF1 and PIF3 Prevent Cell Death during Deetiolation
To determine whether photooxidative damage resulted in cell death in the pif mutants, seedlings were stained with trypan blue, which marks dead or dying cells. We found that the cotyledons of pif1, pif3, and pifq mutants showed a prominent increase in trypan blue staining; however, such stress symptoms were weakly detected in the wild-type control (Figure 1D). To quantify the extent of cell death in these mutants, we analyzed the cell death–induced electrolyte leakage of etiolated seedlings subjected to various periods of light treatment. In agreement with the trypan blue staining results, electrolyte leakage during the first 3 h of light irradiation was significantly greater in pif1 and pif3 than in the wild type and was greatest in the pifq mutant (Figure 1E). After 6 h, the electrolyte leakage of the pif3 mutant was similar to that of pifq (Figure 1E). A previous study suggested that quenching of singlet oxygen is primarily linked to the rapid turnover of D1 protein of the photosystem II reaction center (Telfer et al., 1994). As expected, the steady state level of D1 protein in the pif1 seedlings was significantly reduced relative to that of wild-type seedlings (Figure 1F). Taken together, our results indicate that PIF1 and PIF3, and possibly also PIF5, repress cell death during seedling deetiolation.
PIF1 Regulates Genome-Wide ROS-Responsive Gene Expression
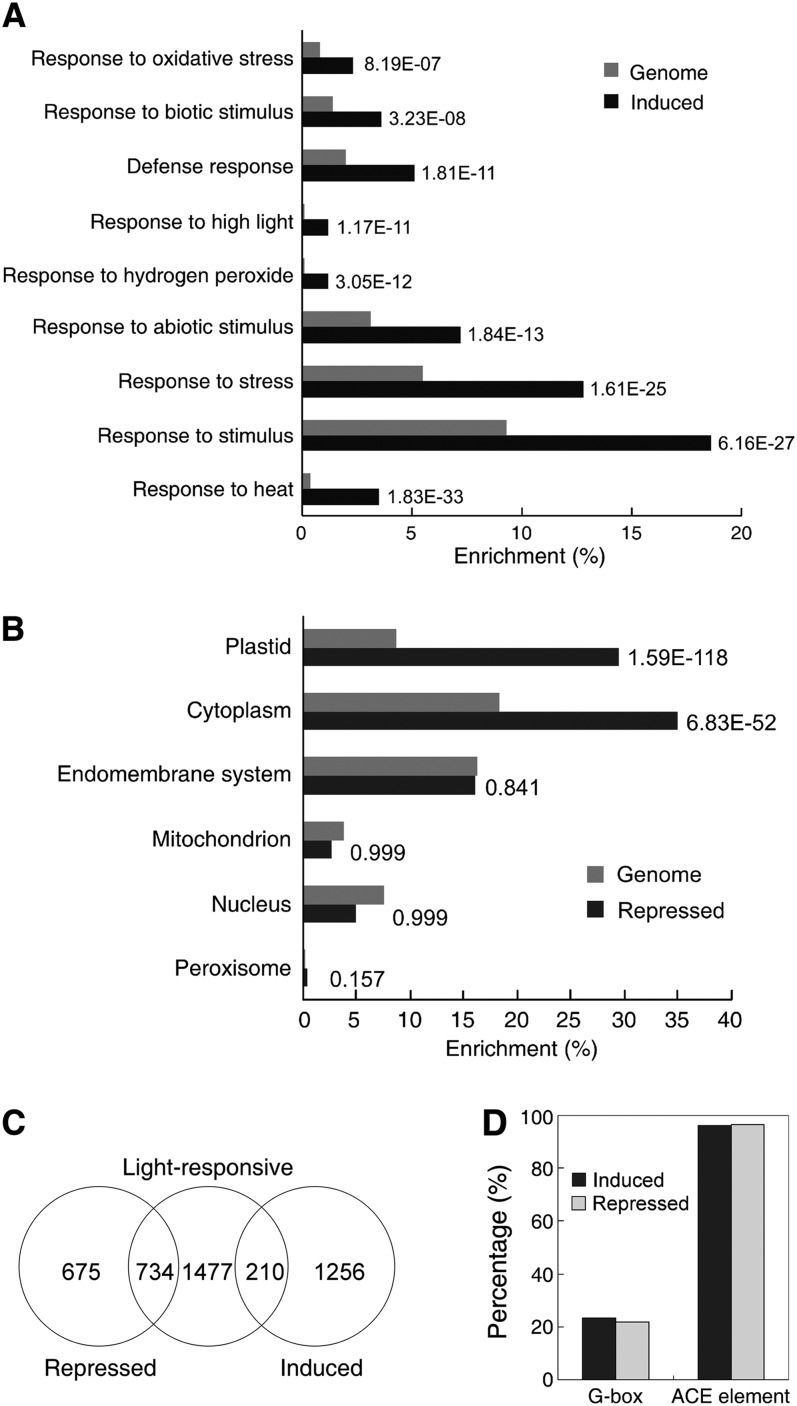
To obtain insight into the regulation of PIFs at the whole-genome level, we performed a microarray study to compare global gene expression changes between pif1 and Col of 4-d-old etiolated seedlings treated with light for 3 h. Compared with the wild type, 1466 genes were induced, whereas 1409 were downregulated twofold or more in pif1 (see Supplemental Data Set 1 online). A functional classification of differentially expressed genes was performed using GO::TermFinder (Boyle et al., 2004). Among the upregulated genes, categories of response to abiotic, biotic stimulus/stress, hydrogen peroxide, high light, oxidative stress, defense, and hormones were significantly overrepresented (Figure 2A; see Supplemental Data Set 2 online for the complete list). Notably, the frequency of genes involved in the response to high light and hydrogen peroxide was 11 times greater in pif1 than that found in the whole genome. These genes encode transcription factors, such as ethylene-responsive transcription factors (ERF13, ERF4, and ERF1) and WRKY transcription factors (WRKY51, WRKY67, and WRKY46), various regulatory proteins (zat zinc finger proteins, ZAT10 and ZAT12, sigma factor binding protein, SIB1), heat shock protein (HSP17, HSP90, and HSP21), and mitogen-activated protein kinases (MAPKKK19, MKK9, and MPK3), which are important components of ROS signaling pathways (Apel and Hirt, 2004; Mittler et al., 2004; Miller et al., 2008). We also observed differentially expressed genes involved in jasmonate signaling (JAZ7, JAZ8, and JAZ5), defense (BAP1, PDF1.2, and RPS6), and antioxidation (APX2, AOX1, and MDAR2).
Figure 2.
Microarray Analysis of PIF1-Regulated Genes.
(A) Enrichment of selected categories of GO biological process in genes induced in pif1. The numbers on the right are P values calculated based on their relative abundance in the wild-type genome. For a complete list of significant GO terms, see Supplemental Data Set 2 online.
(B) Enrichment of selected categories of GO cellular component in genes repressed in pif1. The numbers on the right are P values calculated based on their relative abundance in the wild-type genome. For a complete list of significant GO terms, see Supplemental Data Set 3 online.
(C) Venn diagram showing the overlap of differentially regulated genes in pif1 identified in this study with previously reported light-responsive genes (Charron et al., 2009).
(D) Distribution of the putative G-box motif (CACGTG) and ACE element (ACGT) in the 2-kb promoter regions of PIF1-regulated genes.
The repressed genes are mostly localized to the plastid and cytoplasm. Remarkably, the enrichment of plastid-localized proteins was more than 3 times that found in the whole genome (Figure 2B). These genes are involved in various aspects of photosynthesis and chloroplast development, such as the light reaction, light harvesting, chlorophyll biosynthesis, electron transport, and response to light, consistent with the photooxidative phenotype of the pif1 mutant (see Supplemental Data Sets 1 and 3 online).
A comparative analysis with previous light-regulated transcriptomes (Charron et al., 2009) revealed that 210 of the genes induced in pif1, and 734 of the genes repressed in pif1, were regulated by light (Figure 2C). PIF1 is capable of directly binding to the promoters of downstream targets through a G-box motif (Oh et al., 2009). Interestingly, 341 (23.2%) and 307 (21.8%) of the induced or repressed genes, respectively, possess at least one putative G-box motif within the 2-kb promoter regions (Figure 2D). These data suggest that PIF1 may directly regulate a wide range of ROS-responsive downstream genes by binding to their promoters.
PIF1 and PIF3 Bind to and Regulate ROS-Responsive Genes
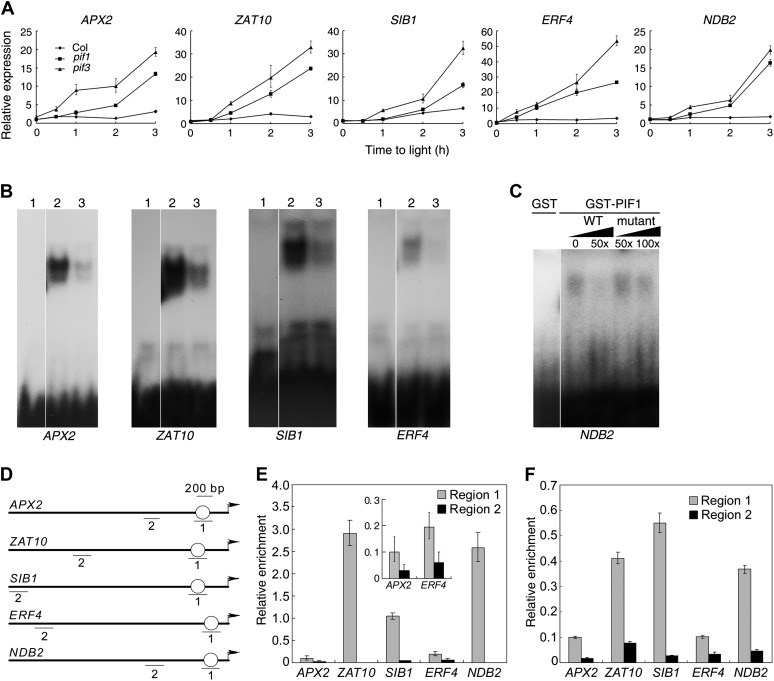
To test the above hypothesis, two genes with prominent roles in ROS signaling, ZAT10 and ASCORBATE PEROXIDASE2 (APX2) (Miller et al., 2008), two singlet oxygen-responsive genes, SIGMA FACTOR BINDING PROTEIN1 (SIB1) and ETHYLENE-RESPONSIVE TRANSCRIPTION FACTOR4 (ERF4) (Laloi et al., 2007), and an oxidative stress–induced gene, NDB2 [encoding a NAD(P)H dehydrogenase] (Ho et al., 2008), were selected for further analysis. Quantitative RT-PCR (qRT-PCR) analysis showed that these genes were upregulated in pif1 and pif3 compared with the wild type after more than 1 h of light exposure (60 µmol m−2 s−1 for 1 to 3 h; Figure 3A), suggesting that PIF1 and PIF3 repress ROS-responsive gene expression in the light. The expression of APX2 and ERF4 was even increased in the pif3 mutant relative to the wild type when seedlings were exposed to light for half an hour. By contrast, no obvious expression difference between the mutants and wild type was observed in the dark-grown seedlings (Figure 3A). Furthermore, we found that after low light (10 µmol m−2 s−1) irradiation, the expression of APX2 and ERF4 was also upregulated in the pif3 and pif1 pif3 mutants compared with the wild-type seedlings (see Supplemental Figure 2 online).
Figure 3.
PIF1 and PIF3 Directly Inhibit ROS-Responsive Gene Expression in the Light.
(A) Relative expression of various genes by qRT-PCR. Four-day-old etiolated seedlings were kept in darkness or transferred to light (60 µmol m−2 s−1) for up to 3 h. Data are mean ± sd, n = 3.
(B) EMSA of the binding of promoter fragments of the indicated genes to GST-PIF1 or GST-PIF3. Lane 1, GST protein only; lanes 2 and 3, GST-PIF1 fusion protein without (lane 2) or with (lane 3) cold competitor DNA. Signals at the bottom indicate free probes.
(C) EMSA of the binding of the NDB2 promoter fragment to GST-PIF1. WT, wild-type cold competitor; mutant, cold competitor with a mutation in the G-box. The numbers indicate the amount of excess cold competitor added to the reaction mix.
(D) Promoter diagrams of genes that function downstream of PIF1 and PIF3. Arrows indicate the translation start sites of the genes. Circles denote the position of the G-box motif. “1” and “2” indicate the approximate positions of primers used for ChIP amplification.
(E) and (F) ChIP assays showing the enrichment of regions 1 and 2 of DNA isolated from Pro35S:TAP-PIF1 (E) and Pro35S:Myc-PIF3 (F) plants following precipitation with an anti-Myc antibody. Seedlings were grown in darkness for 4 d and then were irradiated (60 µmol m−2 s−1) for 30 min. Data are mean ± sd, n = 3. Inset in (E) is the enlargement for APX2 and ERF4 genes.
We conducted electrophoretic mobility shift assays (EMSAs) to test if PIF1 could bind to the DNA of the downstream genes in vitro. GST-PIF1 (PIF1 fused with glutathione S-transferase) recombinant proteins were expressed in Escherichia coli and incubated with 32P-lableled oligonucleotide fragments that contain putative G-box sequences of the target promoters. As shown in Figures 3B and 3C, GST-PIF1, but not GST alone, caused a mobility shift of the promoter fragments of APX2, ZAT10, SIB1, and ERF4 (lane 2). Moreover, the amount of shifted band was significantly decreased by the addition of excess unlabeled wild-type DNA (lane 3) but not by that of the mutant competitors in NDB2 (CACGTG changed to CTTGTG; Figure 3C).
To analyze the protein-DNA binding in vivo, we performed ChIP experiments using transgenic seedlings expressing Pro35S:TAP-PIF1 (Moon et al., 2008) or Pro35S:Myc-PIF3 (see Supplemental Figure 3 online). After precipitation with anti-Myc antibody, the DNA fragments were quantified by real-time PCR using primers spanning a region of the promoter that contains the G-box motif (region 1) and primers spanning a region that is upstream of the G-box motif (region 2) (Figure 3D). We found that the occupancy of PIF1 at region 1 of APX2, ZAT10, SIB1, ERF4, and NDB2 was remarkably higher than at region 2 (Figure 3E). Similarly, PIF3 was strongly recruited to region 1 of those genes (Figure 3F). Hence, PIF1 and PIF3 directly associate with these key ROS-responsive genes.
HY5 and HYH Directly Bind to ROS-Responsive Genes
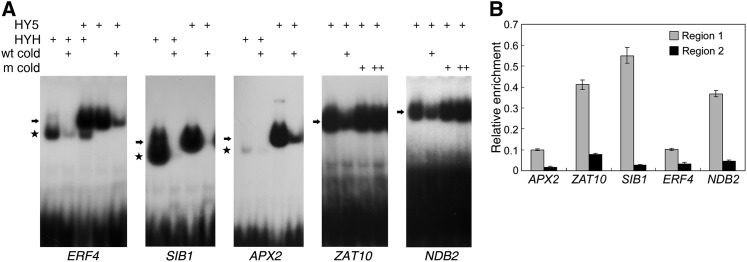
Previous studies documented that the ACE element (ACGT) and its special form G-box motif are putative binding sites of HY5 (Lee et al., 2007; Zhang et al., 2011). We found that the ACE element is overrepresented in the promoters of PIF1-regulated genes (Figure 2D). We then asked whether HY5 and its close homolog HYH could also bind to DNA of the ROS-responsive genes. GST-HY5 and GST-HYH recombinant fusion proteins were incubated with probes of ERF4, SIB1, APX2, ZAT10, and NDB2 genes in the absence or presence of cold competitors. As shown in Figure 4A, both GST-HY5 and GST-HYH bound strongly to these genes and caused bands to shift upwards on the gel, although the affinity of HYH for the promoter fragment of APX2 was weak. These bands were much fainter or completely absent in the presence of excess amounts of unlabeled wild-type oligonucleotides but not in the presence of unlabeled oligonucleotides containing the G-box mutation (ZAT10 and NDB2 are shown), demonstrating that HY5 and HYH bind to the promoters of these genes directly via the G-box in vitro. To substantiate the binding in vivo, we performed ChIP assays of DNA isolated from Col wild-type seedlings using an anti-HY5 antibody. We found that the region 1 fragments of APX2, ZAT10, SIB1, ERF4, and NDB2 were significantly enriched compared with region 2 fragments after precipitation with the HY5 antibody (Figure 4B), indicating that HY5, like PIF1 and PIF3, associates with the promoters of ROS-regulated genes in plant cells.
Figure 4.
HY5 Binds to the Promoter Regions of ROS-Responsive Genes in Vitro and in Vivo.
(A) EMSA of the binding of various promoter fragments to GST-HY5 or GST-HYH recombinant proteins. Arrows indicate HY5-DNA complexes; stars denote HYH-DNA complexes. wt cold, unlabeled wild-type competitor DNA; m cold, unlabeled competitor DNA with mutations (CttGTG) in the G-box motif. For cold DNA, “+” and “++” indicate a 50- and 100-fold excess, respectively.
(B) ChIP assays showing enrichment of regions 1 and 2 in DNA isolated from Col wild-type plants following precipitation with an anti-HY5 antibody. Regions 1 and 2 are defined in Figure 3D. Seedlings were grown in darkness for 4 d and then exposed to light (60 µmol m−2 s−1) for 30 min. Data are mean ± sd, n = 3.
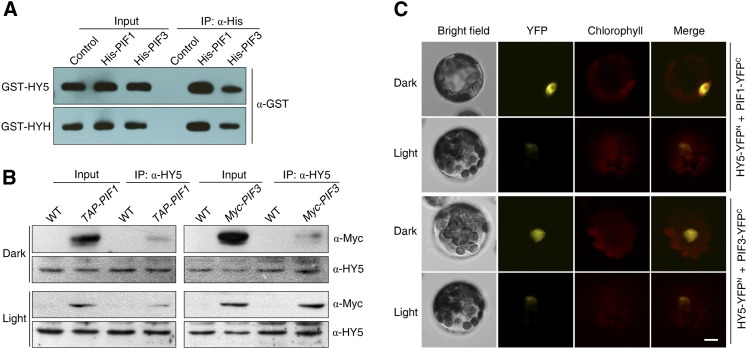
PIF1 and PIF3 Physically Interact with HY5 and HYH
The ability of PIF1, PIF3, HY5, and HYH to bind the same cis-element of the target genes prompted us to test whether PIF1/PIF3 and HY5/HYH could interact with each other. We thus performed an in vitro pull-down assay using His-fused PIF1 (His-PIF1) or His-PIF3, and GST-tagged HY5 (GST-HY5) or GST-HYH recombinant fusion proteins. Our results showed that His-PIF1 and His-PIF3, but not proteins extracted from E. coli expressing His empty vector, were able to coprecipitate GST-HY5 and GST-HYH (Figure 5A). Using a yeast two-hybrid assay, we found that a LexA DNA binding domain fusion of the C-terminal fragment of PIF1 (LexA-PIF1C) interacted with the C terminus of PIF1 or HY5 tagged with B42 activation domain (AD-PIF1C or AD-HY5C) (see Supplemental Figure 4 online). The full-length and N terminus of PIF1 showed strong transcriptional activation activity, as previously reported (Shen et al., 2008). This result indicates that the C-terminal portions of PIF1 and HY5, which contain the bHLH and bZIP domain, respectively, are responsible for mediating their interaction.
Figure 5.
PIF1 and PIF3 Physically Interact with HY5 and HYH.
(A) In vitro pull-down assay of His-PIF1 or His-PIF3 and GST-HY5 or GST-HYH. His-PIF1 or His-PIF3 fusion proteins were incubated with GST-HY5 or GST-HYH and pulled down by nickel-nitrilotriacetic acid agarose. The precipitated fractions were probed with an anti-GST antibody. Control, proteins extracted from E. coli expressing His empty vector. IP, immunoprecipitation.
(B) In vivo coimmunoprecipitation assay between TAP-PIF1 or Myc-PIF3 with HY5. Pro35S:TAP-PIF1, Pro35S:Myc-PIF3, or Col wild-type (WT) seedlings were grown in darkness for 4 d and then either kept in the dark or transferred to light (60 µmol m−2 s−1) for an additional 30 min. After precipitation with the anti-HY5 antibody, proteins were immunoblotted with anti-HY5 or anti-Myc antibodies.
(C) BiFC analysis of interactions between HY5, PIF1, and PIF3 in the nuclei of Arabidopsis protoplasts. After cotransformation, the protoplasts were incubated in darkness for 16 h and then kept in darkness or exposed to light (10 µmol m−2 s−1) for 1 h before observation. Chlorophyll autofluorescence is shown in red. YFPn and YFPc, the N-terminal or C-terminal fragment of YFP, respectively. Bar = 5 μm.
We next performed coimmunoprecipitation assays using proteins isolated from Pro35S:TAP-PIF1 or Pro35S:Myc-PIF3 transgenic plants or from the wild type to substantiate the interaction in vivo. As shown in Figure 5B, the HY5 antibody was able to immunoprecipitate TAP-PIF1 or Myc-PIF3 fusion proteins, as detected by the Myc antibody, both in dark-grown seedlings and in plants transferred to light for 30 min. Next, a bimolecular fluorescence complementation (BiFC) assay was conducted by transiently coexpressing the N terminus of yellow fluorescent protein (YFPn)– and the C terminus of YFP (YFPc)–fused constructs in Arabidopsis protoplasts (Walter et al., 2004). Coexpression of HY5-YFPn and PIF3-YFPc, or HY5-YFPn and PIF1-YFPc, reconstituted a functional YFP in the nucleus with strong fluorescence in plants grown in darkness, but weak fluorescence after exposure to light (Figure 5C). Similarly, a firefly luciferase (LUC) complementation imaging (LCI) assay further showed that coexpression of HY5-LUCn and LUCc-PIF1, or PIF3-LUCn and LUCc-HY5, caused high levels of LUC activity (see Supplemental Figure 5 online). PIF3-LUCn and LUCc-PIF1 showed strong interaction, as previously documented (Bu et al., 2011). Furthermore, a fluorescence resonance energy transfer (FRET) analysis was conducted by coexpressing PIF1-YFP and HY5-CFP in the protoplasts. The sharp drop in fluorescence intensity of the acceptor PIF1-YFP due to photobleaching was compensated for by the increase in the intensity of the donor HY5-CFP, and the FRET efficiency was ∼0.2 to ∼0.3, whereas the FRET efficiency of the control where HY5-CFP was cotransformed with YFP alone was below 0.03 (see Supplemental Figure 6 online). Collectively, these data demonstrate that PIF1 and PIF3 physically interact with HY5 in the nucleus to form heterodimers both in dark and light conditions.
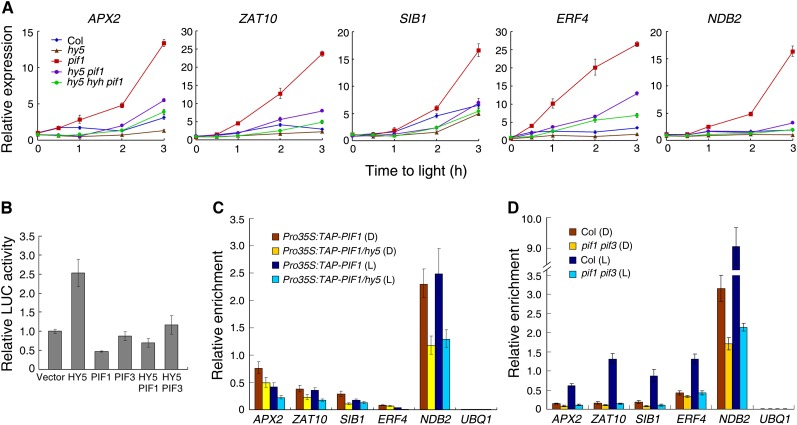
PIF1/PIF3 and HY5/HYH Coregulate ROS-Responsive Genes
The observation that PIF1/PIF3 and HY5/HYH physically interact and bind to common targets suggests that they might coregulate downstream genes. To address this possibility, we introduced hy5 and/or hyh mutations into the pif1 mutant background through genetic crossing, and double or triple homozygous mutants were used for analyses. When 4-d-old etiolated seedlings were transferred to light for a series of time (up to 3 h), the expression of APX2, ZAT10, SIB1, and ERF4 was lower in hy5 than in the wild type after 1 h of light exposure. However, the transcript levels of APX2, ZAT10, SIB1, ERF4, and NDB2 were drastically reduced in the hy5 pif1 double mutant and reduced even further in the hy5 hyh pif1 triple mutant compared with those in the pif1 single mutant background (Figure 6A), indicating that the repression activity of PIF1 is partly dependent on both HY5 and HYH in vivo. We found that after light exposure, HY5 protein level was gradually increased, while the level of PIF1 was drastically reduced (Shen et al., 2005; see Supplemental Figures 7A and 7B online). To determine whether the two types of proteins are sensitive to light quantity during dark-to-light transition, we conducted immunoblotting with 4-d-old etiolated seedlings exposed to various intensities of white light. The data indicate that the stability of HY5 and PIF1 is modulated in a light quantity–dependent manner (see Supplemental Figures 7C and 7D online). Consistently, PIF1 was found to be sensitive to the intensity of monomeric red and far-red light (Shen et al., 2005, 2008). As the direct targets of PIF1/PIF3 and HY5/HYH, the transcript levels of APX2, ZAT10, SIB1, ERF4, and NDB2 were gradually increased by increasing light intensities (see Supplemental Figure 7E online).
Figure 6.
PIF1/PIF3 and HY5/HYH Coregulate ROS-Responsive Genes.
(A) qRT-PCR showing the relative expression of various ROS-responsive genes. Four-day-old etiolated seedlings were kept in darkness or transferred to light (60 µmol m−2 s−1) for up to 3 h. Data are mean ± sd, n = 3.
(B) The relative activity of the ProERF4:LUC reporter in Arabidopsis protoplasts cotransformed with the indicated effector constructs. The relative LUC activities were normalized to the Pro35S:GUS internal control. Protoplast transformation, incubation, and protein extraction were performed in darkness. Mean ± sd, n = 3.
(C) and (D) Seedlings were grown in darkness (D) for 4 d or irradiated with light (L; 60 µmol m−2 s−1) for 30 min. Data are mean ± sd, n = 3. The enrichment of UBQ1 serves as a negative control.
(C) Relative enrichment of region 1 fragments (shown in Figure 3D) in DNA isolated from hy5 and Col wild-type plants harboring Pro35S:TAP-PIF1 and coimmunoprecipitated with the anti-Myc antibody.
(D) Relative enrichment of region 1 fragments (shown in Figure 3D) in DNA isolated from pif1 pif3 and Col wild-type plants coimmunoprecipitated with HY5 antibody.
Next, we transiently expressed HY5 and/or PIF1/PIF3, together with a LUC reporter gene driven by the ERF4 promoter (the 2.0-kb region upstream of the ATG start site), in Arabidopsis protoplasts. As shown in Figure 6B, HY5 greatly promoted LUC reporter gene expression, whereas PIF1 inhibited the expression of LUC. Remarkably, coexpression of PIF1 or PIF3 drastically suppressed the activation activity of HY5 on the ProERF4:LUC reporter, suggesting that HY5 and PIF1/PIF3 associate with the promoter and act together to fine-tune ERF4 expression. Consistently, HY5 and PIF1 were able to form a heterodimer and bind to the promoter of APX2 and ERF4 in vitro (see Supplemental Figure 8 online).
To determine whether the binding of PIF1 to the promoters of the target genes requires HY5, we performed ChIP assays using Pro35S:TAP-PIF1 transgenic plants in the wild-type and hy5 mutant backgrounds. Our data showed that relatively less target DNA was pulled down by the Myc antibody in the hy5 mutant than in the wild type both in dark-grown and light-treated seedlings (Figure 6C), suggesting that the DNA binding activity of PIF1, in part, requires the presence of HY5. Similarly, the binding activity of HY5 to the targets was also partly dependent on PIF1 and PIF3, as mutations in both PIF1 and PIF3 diminished the enrichment of downstream genes when pulled-down samples were analyzed with the HY5 antibody (Figure 6D). These data further indicate that HY5 and PIF1 are able to bind to these ROS-related genes both in darkness and after light irradiation.
PIF1/PIF3 and HY5/HYH Function Antagonistically
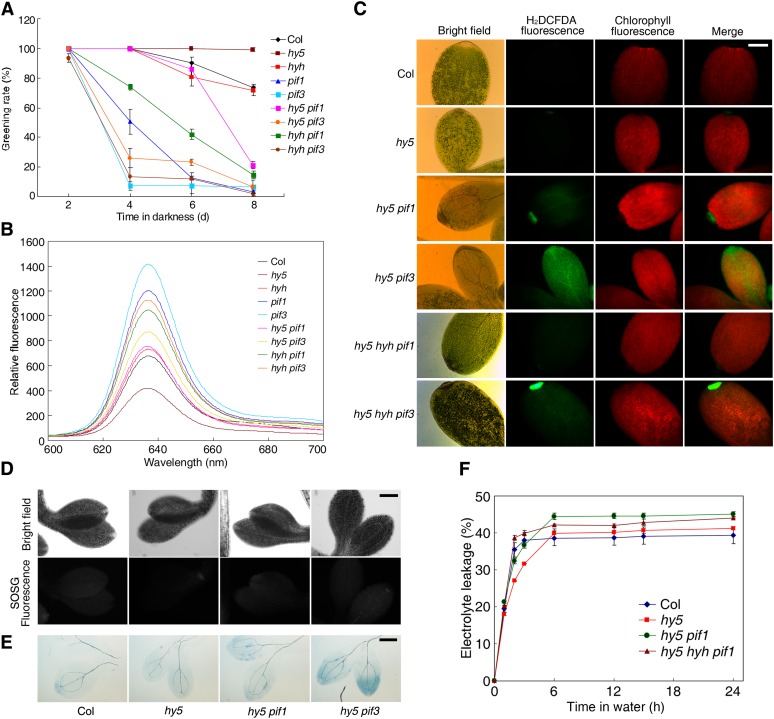
We next investigated how HY5 and HYH affect the function of PIF1 and PIF3. Mutation in HY5 partly inhibited the phenotypes of high Pchlide levels and low greening rates of pif1, and to a lesser extent of pif3. Relative to hy5, the hyh single mutant had minor effects on these phenotypes (Figures 7A and 7B). As shown in Figure 7C, we observed that H2DCFDA fluorescence was barely detected in the hy5 mutant seedlings after light treatment. The high level of H2DCFDA fluorescence in the pif1 mutant (Figure 1) was largely suppressed by the hy5 mutation in the hy5 pif1 double mutant, while the fluorescence in the hy5 hyh pif1 triple mutants was almost identical to that in the wild type. Compared with the pif3 single mutant (Figure 1), H2DCFDA fluorescence was also greatly reduced in the hy5 pif3 and hy5 hyh pif3 mutants. Consistent with this, singlet oxygen generation was largely blocked in the hy5 pif1 and hy5 pif3 double mutants relative to pif1 and pif3 (Figures 1 and 7D). Accordingly, trypan blue staining showed that the extent of cell death of the hy5 pif1 and hy5 pif3 double mutants was greatly reduced and close to the levels in the wild type (Figure 7E). Moreover, the electrolyte leakage of hy5 pif1 and hy5 hyh pif1 plants dropped to levels similar to those of the wild type (Figure 7F).
Figure 7.
Genetic Interaction between HY5/HYH and PIF1/PIF3.
(A) Seedling greening rate. Two- to eight-day-old etiolated seedlings were transferred to light (60 µmol m−2 s−1) for an additional 2 d. Mean ± sd, n = 3.
(B) Pchlide accumulation of 4-d-old dark-grown seedlings in the indicated mutants and the wild type.
(C) H2DCFDA fluorescence showing cellular ROS production of 4-d-old dark-grown seedlings after 24 h of light exposure (60 µmol m−2 s−1).
(D) and (E) SOSG fluorescence imaging (D) and trypan blue staining (E) of 4-d-old etiolated seedlings after light illumination (60 µmol m−2 s−1) for 6 h. Bars = 200 μm.
(F) Relative electrolyte leakage showing the extent of cell death. Four-day-old etiolated seedlings were exposed to light (60 µmol m−2 s−1) for 12 h and immersed in water, and electrolyte leakage was measured periodically. Data are mean ± sd, n = 3.
We further examined how the overexpression of PIF1, PIF3, or HY5 affects seedling greening and downstream gene expression. We found that the expression of ROS-responsive genes, including ZAT10, SIB1, ERF4, and NDB2, was decreased in the Pro35S:TAP-PIF1 and Pro35S:Myc-PIF3 transgenic lines, while the levels of APX2, ZAT10, SIB1, and ERF4 were increased in the Pro35S:HA-HY5 overexpression plants compared with the wild-type seedlings (see Supplemental Figure 9A online). Consistently, PIF1 and PIF3 overexpression lines have higher greening rate than the wild type when dark-grown seedlings were exposed to high light (250 µmol m−2 s−1). By contrast, HY5 overexpression plants showed reduced greening ability compared with the control (see Supplemental Figure 9B online). We further examined the responses of PIF1 and PIF3 overexpression in the hy5 mutant background and found that their phenotypes were even stronger than the single mutant/transgenic line, consistent with their antagonistic role (see Supplemental Figures 9C and 9D online). Together, these observations demonstrate that HY5/HYH and PIF1/PIF3 antagonistically regulate singlet oxygen production and cell death during the seedling greening process. Interestingly, we further observed that HY5 and PIF1/PIF3 had a slight regulatory effect on each other at the protein level (see Supplemental Figure 10 online).
Phytochromes and Cryptochromes Positively Regulate and COP1 Negatively Regulates ROS Signaling
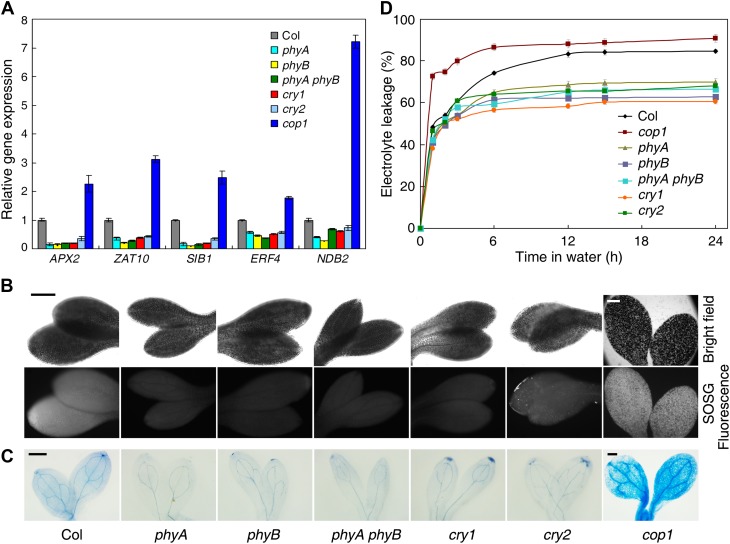
We speculated that the phytochrome and cryptochrome photoreceptors and COP1 might also contribute to the regulation of ROS signaling, as they are the upstream components in the light signaling pathway. To test this hypothesis, we grew seedlings of the phyA, phyB, phyA phyB, cry1, cry2, and cop1 mutants and also of the Col wild type in darkness for 4 d followed by 3 h of light exposure. qRT-PCR results showed that the transcript levels of APX2, ZAT10, SIB1, ERF4, and NDB2 were remarkably reduced in all of the photoreceptor mutants but were drastically increased in cop1 (Figure 8A), indicating that phyA, phyB, cry1, and cry2 promote ROS-responsive gene expression, whereas COP1 represses it. Surprisingly, the etiolated seedlings of phyA, phyB, phyA phyB, cry1, and cry2 exhibited low Pchlide levels relative to the wild type (see Supplemental Figure 11A online). Accordingly, the photoreceptor mutants had higher greening rates than the wild type when seedlings were grown in the dark for 4 d or longer and then exposed to light (see Supplemental Figure 11B online). A previous study showed that the cop1 mutant accumulated extremely high levels of Pchlide and was unable to turn green after light exposure (Zhong et al., 2009). Consistently, the photoreceptor mutants accumulated lower levels of singlet oxygen and underwent less cell death than the wild type. By contrast, the cop1 mutant displayed higher levels of singlet oxygen and cell death than the wild type (Figures 8B to 8D). These results indicate that the phyA, phyB, cry1, and cry2 photoreceptors and COP1 have opposite effects on ROS signaling and cell death.
Figure 8.
The Opposite Role of Photoreceptors and COP1 in Regulating ROS Production and Signaling.
(A) qRT-PCR showing the expression of the indicated ROS-responsive genes. Seedlings were grown in darkness for 4 d and transferred to light (60 µmol m−2 s−1) for 3 h. Data are from three biological replicates; bars indicate sd.
(B) and (C) SOSG fluorescence imaging (B) and trypan blue staining (C) of 6-d-old wild-type and mutant etiolated seedlings after light illumination (60 µmol m−2 s−1) for 6 h. Bars = 200 μm.
(D) Electrolyte leakage levels in the indicated photoreceptor and cop1 mutants. Data are mean ± sd, n = 4.
DISCUSSION
Key Light Signaling Components Regulate Singlet Oxygen Production and Cell Death
During the past decades, numerous studies have established the role of light signaling networks in regulating diverse plant growth and developmental processes (Quail, 2002; Chen et al., 2004; Lau and Deng, 2010). Recently, the light signaling pathways have been shown to interact with phytohormone signaling pathways to mediate distinct responses (Alabadí and Blázquez, 2009). In this study, we provide insight into the functional diversity of the key components in light signaling pathways that contribute either positively or negatively to singlet oxygen production and cell death. We show that the first 5 min of exposure to relatively high levels of light (60 µmol m−2 s−1) are critical for the survival of etiolated seedlings and that PIF1, PIF3, and possibly also PIF5 are required for survival during the onset of illumination (Figure 1). In addition, the survival rate of plants exposed to light correlates inversely with the levels of Pchlide, in agreement with our previous report (Tang et al., 2012). Pchlide is a potent photosensitizer in the chloroplast that leads to photooxidative damage in plants (op den Camp et al., 2003). Accordingly, the pif mutants displayed severe cell death, as determined by trypan blue staining as well as electrolyte leakage, during the dark-to-light transition (Figure 1). We demonstrate that PIF proteins, including PIF1, PIF3, and PIF5, specifically and redundantly prevent singlet oxygen production during seedling deetiolation, with PIF1 and PIF3 playing the major roles. However, the cotyledons of these mutants accumulated reduced amounts of superoxide and hydrogen peroxide, possibly due to the antagonistic effect on other ROS by singlet oxygen (Laloi et al., 2007). This study also shows that the pif1 and pif3 phenotypes largely depend on the presence of functional HY5 and HYH, supporting a negative role of HY5/HYH in regulating greening and plant survival (Figure 7). Thus, PIF1/PIF3 and HY5/HYH have opposite effects on the regulation of ROS and cell death.
The phyA, phyB, cry1, and cry2 photoreceptors promote Pchlide synthesis in the dark and induce ROS signaling in the light (Figure 8). We speculate that the phytochromes and cryptochromes might also transduce signals in the cytosol to regulate tetrapyrrole biosynthesis, even in darkness, although the underlying mechanism needs to be determined. cry1 has been shown to trigger blue light–dependent, singlet oxygen–mediated programmed cell death, as the cry1 mutant suppressed the cell death phenotype of fluorescent (flu) after the dark-to-blue light transition (Danon et al., 2006). The cop1 mutants accumulate a high level of singlet oxygen and almost die after transition from dark to light (Figure 8), consistent with a previous report showing high levels of Pchlide accumulation in cop1 (Zhong et al., 2009). These phenotypes of cop1 are similar to those observed for the pifq mutant, in agreement with their constitutive photomorphogenic response in darkness (Leivar et al., 2008; Shin et al., 2009). Therefore, light signaling networks not only remodel the morphological structures of plants (e.g., inhibit hypocotyl elongation and promote the unfolding of cotyledons), but also trigger cellular and metabolic changes that ensure plant survival and development.
ROS regulate many processes in plants; however, little is known about the specific ROS responses that underlie particular stimuli (Mittler et al., 2004, 2011; Miller et al., 2008). The flu mutant was previously shown to specifically generate singlet oxygen in etiolated seedlings irradiated with light or in plants grown under a dark–light cycle (op den Camp et al., 2003; Wagner et al., 2004). Our data demonstrate that PIF1 and PIF3 represent two components that predominantly regulate the production of singlet oxygen. However, the molecular mechanisms by which PIF1/PIF3 and FLU mediate singlet oxygen generation and signaling might be diverse, since the former proteins are transcription factors in the nucleus, whereas the latter one is localized in the plastid and its biochemical function is not well understood. It will be interesting to test whether (and how) PIF1/PIF3 and FLU could coact to regulate singlet oxygen signaling.
Interaction between bZIP and bHLH Transcription Factors
Direct interactions between PIF1 and PIF3 and between HY5 and HYH have been documented previously (Holm et al., 2002; Bu et al., 2011). Although HY5 and HYH are unstable in the dark, a small amount of HY5 is detectable in darkness (Hardtke et al., 2000; Osterlund et al., 2000; this study). Similarly, PIF1 and PIF3 proteins were detectable after 2 h of light exposure, despite their rapid degradation in the presence of light (Shen et al., 2005; Al-Sady et al., 2006; this study). Evidence from yeast two-hybrid, pull-down, coimmunoprecipitation, and LCI approaches consistently demonstrated that PIF1/PIF3 (bHLH transcription factors) and HY5/HYH (bZIP transcription factors) directly interact with each other in vitro and in vivo (Figure 5; see Supplemental Figures 4 and 5 online). Our BiFC and FRET assays further indicate that these transcription factors coexist and interact in the same nucleus at the same time in darkness or after light transition (Figure 5; see Supplemental Figure 6 online). From a structural point of view, the bHLH and bZIP proteins share some similar features: The short basic domain at the N-terminal region makes contact with the major groove of the DNA, and the HLH and ZIP domains mediate dimerization (Sibéril et al., 2001; Jones, 2004). Furthermore, both types of transcription factors are able to bind to the G-box cis-element (The bZIP Research Group, 2002; Toledo-Ortiz et al., 2003). These structural similarities between bHLH and bZIP might allow PIF and HY5 heterodimerization and the binding of their basic regions to the same site of the G-box. Consistent with this, the C-terminal regions containing the bHLH domain of PIF1 and the bZIP domain of HY5 are responsible for mediating their interaction in yeast cells (see Supplemental Figure 4 online), and PIF1 and HY5 formed heterodimers and bound DNA fragments of APX2 and ERF4 (see Supplemental Figure 8 online). The possibility that they form heterotetramers cannot be excluded. The formation of analogous complexes of bZIP and bHLH transcription factors have been observed in yeast, and these complexes were found to bind to a cis-element (TCACGTG) similar to the G-box motif (Kuras et al., 1997; Blaiseau and Thomas, 1998), indicating that the interaction and DNA binding activities of bZIP/bHLH proteins are likely conserved among organisms.
In addition, homo- or heterodimerization of the components of HY5/HYH and PIF1/PIF3 might also offer regulatory flexibility and diversity in the interactions with target genes in response to diverse signals. A previous study reported that PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner, although the authors failed to detect a direct interaction between PIF3 and HY5 (Shin et al., 2007). Moreover, ChIP-based sequencing analyses showed that some of the direct targets of HY5 overlap with those of PIF1, whereas others are distinct (Lee et al., 2007; Oh et al., 2009). Other transcription repressors or activators might also be involved in this process by interacting with PIFs and HY5 proteins. We recently found that FAR-RED ELONGATED HYPOCOTYL3 (FHY3) and FAR-RED-IMPAIRED RESPONSE1 (FAR1) participate in regulating Pchlide synthesis and seedling greening by interacting and cooperating with PIF1 (Tang et al., 2012). HY5 feedback regulates phyA signaling homeostasis by interacting with FHY3 and FAR1 (Li et al., 2010). FHY3 and FAR1 likely possess a similar function as HY5 in mediating ROS production and cell death. The discovery of more interacting factors will enable us to better understand the regulatory complexity underlying the actions of these bHLH and bZIP transcription factors in response to developmental and environmental cues.
The PIF1/PIF3-HY5/HYH Transcriptional Modules Define a Key Link between Light and ROS Signaling
PIF1/PIF3 and HY5/HYH have been studied individually as regulators of photomorphogenic responses. Here, we reveal that transcriptional modules composed of PIF1/PIF3 and HY5/HYH act as a key molecular node that directly connects light to ROS signaling during plant postgermination development. First, by analyzing genome-wide gene expression changes in the pif1 mutants, we found that a large number of ROS-related genes was induced during the first 3 h of light illumination of etiolated seedlings. Strikingly, many of the singlet oxygen–responsive genes (Laloi et al., 2007) were present in the group of genes induced in pif1. Similarly, when 4-d-old etiolated seedlings were exposed to weak red light for 1 h, 42 out of 139 upregulated genes (more than twofold change) in pif3 were found also to be induced in the flu mutant (Monte et al., 2004; Laloi et al., 2007). Second, the EMSA and/or ChIP results demonstrate that PIF1, PIF3, HY5, and HYH directly bind to the promoters of five ROS-responsive genes, including APX2, ZAT10, SIB1, ERF4, and NDB2, through the G-box motif (Figures 3 and 4). Third, nearly 23% and almost all of the upregulated genes contain the putative G-box motif and ACE elements in their promoters, respectively, suggesting that they are the potential direct targets of PIF1/PIF3 and HY5/HYH (Figure 2D). Moreover, qRT-PCR data show that PIF1 and PIF3 repress the expression of these ROS-responsive genes in light and that the repression also partly requires both HY5 and HYH (Figure 6A).
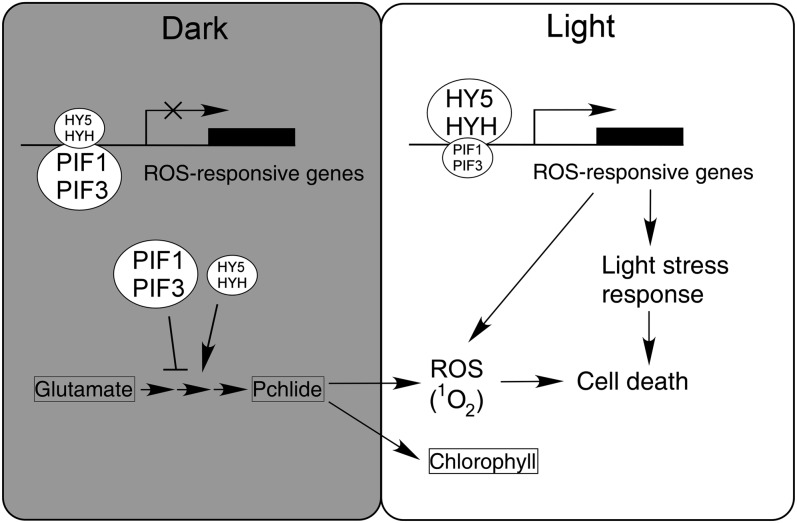
The observations that PIF1 and HY5 bound to the target DNAs and the binding ability was partly dependent on each other in vivo (Figures 6C and 6D) and that they formed both DNA-bound homodimers and heterodimers (see Supplemental Figure 8 online) suggest that homodimerization and heterodimerization between PIF1/PIF3 and HY5/HYH may exist together in plant cells in darkness and after light transition. In the dark, PIF1/PIF3 are abundant, and their homodimers repress ROS-responsive gene expression in the nucleus and meanwhile inhibit Pchlide synthesis and singlet oxygen production in the plastid by regulating the biosynthesis of chlorophyll intermediates. Because of the opposite effect of PIF1/PIF3 and HY5/HYH on downstream gene expression, the heterodimers of the bZIP and bHLH factors likely function as inactive forms, which maintain the ROS-related transcripts at a basal level (mild repression) (Figure 6B). Most strikingly, the protein levels of PIF1/PIF3 and HY5/HYH are sensitively degraded or stabilized, respectively, by light (Henriques et al., 2009). The heterodimeric states are hence dynamically regulated by the environmental light conditions. After short periods or low fluence of light exposure, where plants encounter less light stress, PIF1/PIF3 are partly degraded and relatively high amounts of the PIF-HY5/HYH heterodimers are formed, such that the downstream ROS-related genes are inhibited. However, extended or high-light exposure leads to almost complete degradation of PIF1/PIF3 and abolishes the PIF-HY5/HYH heterodimeric states, which consequently relieve their repressive effect. Under this circumstance, accumulated HY5/HYH form homodimers that predominantly elevate the transcripts of ROS-responsive genes and activate their network (Figure 9). Consistent with this notion, the protein stability of HY5/HYH and PIF1/PIF3 and the expression of downstream ROS-responsive genes are coordinately regulated by the exposure length and intensity of light (Figures 3A and 6A; see Supplemental Figure 7 online).
Figure 9.
A Model for the Function of PIF1/PIF3 and HY5/HYH in Integrating Light and ROS Signaling.
In the dark, PIF1 and PIF3 accumulate while HY5 and HYH are largely degraded, leading to less Pchlide accumulation. Meanwhile, HY5/HYH and PIF1/PIF3 interact and bind to the promoter regions of ROS-responsive genes, resulting in the inhibition of their gene expression. Upon light irradiation, photosensitized Pchlide generates singlet oxygen (1O2), which causes photooxidative damage and cell death in plants; on the other hand, light promotes the stabilization of HY5/HYH and the rapid turnover of PIF1/PIF3, which in turn activates the expression of ROS-responsive genes and the ROS signaling, thus allowing plants to adjust ROS level and to cope with cell death under unfavorable light stress conditions. Light also promotes the formation of chlorophyll from Pchlide. Arrows, positive effect; bar, negative regulation.
Therefore, we propose that the PIF1/PIF3-HY5/HYH transcription modules serve as rheostats to fine-tune the flow of the ROS signaling pathway. Since etiolated seedlings are sensitive to excess light and the heterotrophic-to-autotrophic switch is critical for plant survival, the integration and involvement of the transcriptional modules might be central for the regulation of ROS-mediated photoprotective machinery against high light stress during seedling deetiolation. Repression of ROS pathway under low-light conditions by the bHLH/bZIP transcription modules could have adaptive value, as it would save energy and nutrient resources. The efficiency of the plants to activate this pathway in response to light is further supported by studies showing that PIF3 acts transiently and mainly mediates phytochrome-induced signaling during the dark-to-light transition (Bauer et al., 2004; Monte et al., 2004). It will be interesting to investigate whether this regulatory mechanism is conserved for plants during day/night cycles. The ROS-related genes are likely simultaneously activated by the loss of PIF1/PIF3 together with the generation of singlet oxygen. Accordingly, when etiolated seedlings were exposed to light, singlet oxygen generation gradually increased in conjunction with increasing lengths of light illumination, with higher level found in pif1 pif3 compared with the wild type. The transcript levels of APX2, ZAT10, and SIB1 increased in a similar pattern to that of singlet oxygen (see Supplemental Figure 12 online). However, upon activation, the ROS network may adjust ROS levels through detoxifying and quenching systems, such as antioxidant molecules and enzymes (Triantaphylidès and Havaux, 2009).
Although studies suggested that ROS signaling is highly integrated with hormonal signaling networks, such as gibberellic acid, abscisic acid, salicylic acid, and auxin, to regulate various developmental processes and adaptive responses in plants, unraveling the components that connect ROS with other signaling pathways has been a challenge in the field (Mittler et al., 2011). A previous study reported that gibberellic acid signaling contributes to the fine-tuning of ROS levels by stimulating the degradation of DELLA proteins, which regulate the transcript levels of antioxidant enzymes (Achard et al., 2008). Light signaling can also interact with the ROS pathway, and the PIF1/PIF3-HY5/HYH transcriptional modules represent a key convergence point between these signaling pathways. Moreover, PIF1, PIF3, PIF4, and HY5 have independently been shown to be involved in integrating light with hormone signaling (Chen et al., 2008a; de Lucas et al., 2008; Feng et al., 2008; Oh et al., 2009; Catalá et al., 2011). It is speculated that the transcriptional module described in this study might constitute a common molecular point of crosstalk between light and other cellular pathways.
METHODS
Plant Materials and Growth Conditions
The pif1-2 (Huq et al., 2004), pif3 (Salk_030753; Kim et al., 2003), hy5-215 (Oyama et al., 1997), hyh (cs849765; Kleine et al., 2007), pifq (Leivar et al., 2008), phyA-211 (Reed et al., 1994), phyB-9 (Reed et al., 1993), cry1-304 (Mockler et al., 1999), cry2-1 (Guo et al., 1998), and cop1-4 (McNellis et al., 1994) mutants and the Pro35S:Myc-PIF3, Pro35S:TAP-PIF1 (Moon et al., 2008), and Pro35S:HA-HY5 (Lee et al., 2007) transgenic lines were derived from the Arabidopsis thaliana Col ecotype. Double and triple mutants were generated by genetic crossing and homozygous lines were confirmed by PCR genotyping and/or antibiotic selection. The original mutations in the transgenic lines were removed by crossing. Seedlings were grown on Murashige and Skoog medium containing 1% Suc and 0.8% agar. For light treatment, etiolated seedlings were transferred to moderate growth light (60 µmol m−2 s−1) for various periods of time or subjected to certain conditions, as described in the text.
Greening Rate and Pchlide Determination
Greening rate was determined by counting the number of dark-green cotyledons from 50 to 80 seedlings of each genotype. For Pchlide measurement, dark-grown seedlings were homogenized in 500 μL of ice-cold 80% acetone and incubated in darkness overnight. Samples were excited at 440 nm and scanned from 600 to 700 nm using a fluorescence spectrophotometer (Hitachi).
Fluorescence Imaging of ROS and Singlet Oxygen
ROS fluorescence determination was performed as previously described (Tang et al., 2012). Imaging of singlet oxygen production was performed as described (Flors et al., 2006). Briefly, dark-grown seedlings were immersed in a solution of 10 μM SOSG (Invitrogen) in 50 mM phosphate buffer, pH 7.5, for 2 h in darkness and then transferred to light for 3 h. Following excitation of SOSG by UV light, fluorescence images were acquired with a charge-coupled device camera (Olympus) with a GFPA interference filter in the objective. Fluorescence intensity was determined by Image J software (http://rsb.info.nih.gov/ij/), and the background was subtracted.
Trypan Blue, DAB, and NBT Staining
For all histochemistry studies, etiolated seedlings were exposed to white light for the indicated period of time before staining. Trypan blue and DAB staining were performed as described (Yang et al., 2007). For NBT staining, seedlings were submerged in solution (1 mg/mL NBT, 10 mM NaN3, and 10 mM potassium phosphate, pH 7.8) and stained for 30 min at room temperature. Samples were then boiled in 95% ethanol for 10 min and stored in 60% glycerol. After staining, all samples were mounted on slides and photographed through a dissecting microscope.
Electrolyte Leakage Measurement
Cell death was quantified by electrolyte leakage, using a method adapted from Laloi et al. (2007). Dark-grown seedlings were transferred to light for 12 h and then immersed in 5 mL of distilled water in a glass tube for up to 24 h. The conductivity of the solutions was determined at different time points with a conductivity meter (HANNA Instruments). The maximum electrolyte content was obtained by boiling the samples for 25 min at 100°C. The electrolyte leakage rate was expressed as percentage of the maximum content.
Plasmid Construction
To obtain the open reading frames and/or the N- or C-terminal fragments of PIF1, PIF3, HY5, and HYH, PCR was performed using the primers listed in Supplemental Data Set 4 online and pfu DNA polymerase, and the fragments were cloned into the pEASY-Blunt vector (TransGen), resulting in pEASY-PIF1, pEASY-PIF3, pEASY-HY5, pEASY-HYH, pEASY-PIF1N (1 to 271 amino acids), pEASY-PIF1C (272 to 478 amino acids), pEASY-HY5N (1 to 77 amino acids), and pEASY-HY5C (78 to 168 amino acids). All clones were validated by sequencing.
To generate constructs for the yeast two-hybrid assay, pEASY-PIF1C was digested with EcoRI and SalI, and the PIF1 C-terminal fragment was inserted into the pLexA vector (Clontech) cut by EcoRI and XhoI, to give rise to pLexA-PIF1C. The pEASY-PIF1, pEASY-PIF1N, and pEASY-PIF1C plasmids were digested with EcoRI and SalI, and the corresponding fragments were cloned into the pB42AD vector (Clontech) digested with EcoRI and XhoI, to generate pAD-PIF1, pAD-PIF1N, and pAD-PIF1C, respectively. To construct pAD-HY5N and pAD-HY5C, pEASY-HY5N and pEASY-HY5C, respectively, were cut with EcoRI and XhoI, and inserted into pB42AD.
To construct vectors for expressing recombinant protein, the corresponding fragments from pEASY-PIF1 (cut with EcoRI and SalI), pEASY-PIF3 (cut with EcoRI and XhoI), pEASY-HY5 (cut with EcoRI and XhoI), or pEASY-HYH (cut with EcoRI and SalI) were cloned into the pGEX-5X-1 vector (GE Healthcare) and digested with EcoRI and XhoI, resulting in pGEX-PIF1, pGEX-PIF3, pGEX-HY5, and pGEX-HYH, respectively. The same fragments from pEASY-PIF1 or pEASY-PIF3 were also inserted into the pET-28a vector (Novagen) digested with EcoRI and XhoI, to generate pHis-PIF1 and pHis-PIF3, respectively.
To make vectors for the BiFC experiment, the fragments from pEASY-PIF3 or pEASY-HY5 cut with EcoRI and XhoI were cloned into the pUC-SPYNE vector (Walter et al., 2004) digested with EcoRI and XhoI, to generate pSPYNE-PIF3 and pSPYNE-HY5, respectively. The corresponding fragments from pEASY-PIF1 (cut with EcoRI and SalI) or pEASY-HY5 (cut with EcoRI and XhoI) were cloned into the pUC-SPYCE vector digested with EcoRI and XhoI, to give rise to pSPYCE-PIF1 and pSPYCE-HY5, respectively.
To generate plasmids for LCI analysis, the fragments from pEASY-PIF3 or pEASY-HY5 cut with KpnI and XhoI were cloned into the pUC19-nLUC vector (Chen et al., 2008b) and digested with KpnI and SalI, to generate pLUCn-PIF3 and pLUCn-HY5, respectively. The corresponding fragments from pEASY-PIF1 (cut with KpnI and SalI) or pEASY-HY5 (cut with KpnI and XhoI) were cloned into the pUC19-cLUC vector digested with KpnI and SalI, to give rise to pLUCc-PIF1 and pLUCc-HY5, respectively.
To construct plasmids for the FRET assay, the HY5 fragment was PCR amplified from pEASY-HY5 digested with BglII and KpnI and inserted into pSAT6-CFP cut with BglII and KpnI, to give rise to pCFP-HY5. The PIF1 open reading frame was amplified using pEASY-PIF1 as template, digested with SalI and KpnI, and cloned into the SalI-KpnI site of pSAT6-YFP to generate pYFP-PIF1.
For the transient expression assay, the coding fragment from pEASY-HY5 (digested with EcoRI and XhoI), pEASY-PIF1 (EcoRI and SalI), or pEASY-PIF3 (EcoRI and XhoI) was inserted into the MfeI-XhoI site of pUC-3HA, resulting in the effectors Pro35S:HY5, Pro35S:PIF1, and Pro35S:PIF3, respectively. The promoter fragment (∼1.0 kb upstream of the ATG start site) of ERF4 was PCR amplified from Col genomic DNA and cloned into pEASY to generate pEASY-ERF4p. The LUC open reading frame was released from pGEM-LUC (Lin et al., 2007) and inserted into the BamHI-SacI site of pUC-CPYNE to generate pUC-35sLUC. pEASY-ERF4p was then cut with HindIII and BamHI, and the promoter fragment was inserted into the pUC-35sLUC vector digested with HindIII and BamHI, to generate ProERF4p:LUC.
qRT-PCR
Four-day-old dark-grown seedlings were transferred to white light or kept in darkness for up to 3 h. Plant total RNA was extracted using an RNAprep Pure plant kit (Tiangen), and the first-strand cDNA was synthesized by reverse transcriptase (Invitrogen). Real-time PCR was performed using the SYBR Premix ExTaq kit (Takara) following the manufacturer’s instructions. The expression levels were normalized to the expression of a UBIQUITIN (UBQ) gene. Primers are listed in Supplemental Data Set 4 online.
ChIP
The Col wild-type, Pro35S:TAP-PIF1, and Pro35S:Myc-PIF3 transgenic plants were used in a ChIP assay, following a previously described procedure (Lin et al., 2007). Briefly, the seedlings were cross-linked with 1% formaldehyde and ground to powder under liquid nitrogen. After isolation and sonication, the chromatin complexes were incubated with anti-Myc (Abcam) or anti-HY5 polyclonal antibodies. The precipitated DNA fragments were recovered and quantified by quantitative PCR with the primers shown in Supplemental Data Set 4 online.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed as previously described (Lin et al., 2007). Briefly, the respective combinations of AD and LexA fusions were cotransformed with the LexAop:LacZ (Clontech) reporter construct into yeast strain EGY48. Transformants were grown on SD/-Trp-Ura-His dropout plates containing X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for blue color development.
Preparation of Recombinant Proteins
GST and GST-PIF1, GST-PIF3, GST-HY5, GST-HYH, His-PIF1, and His-PIF3 recombinant fusion proteins were induced by isopropyl β-d-1-thiogalactopyranoside and expressed in the Escherichia coli BL21 (DE3) strain. The proteins were then purified using Glutathione Sepharose 4B beads (GE Healthcare; for GST fusion proteins) or nickel-nitrilotriacetic acid agarose (Qiagen; for His fusion proteins), following the manufacturer’s instructions.
EMSA
The EMSA analysis was performed as previously described (Tang et al., 2012). The oligonucleotide sequences of the probes are listed in Supplemental Data Set 4 online.
In Vitro Pull-Down, Coimmunoprecipitation, and Immunoblot Assays
The procedures for these assays were as described previously (Tang et al., 2012). Anti-GST (Abcam), anti-His (Abcam), anti-Myc (Abcam), and anti-D1 (Agrisera) were commercially available. Anti-HY5 and antitubulin antibodies were raised in rabbits (Jing et al., 2013).
BiFC Assay
Plasmids containing N- and C-terminal YFP fusions were cotransformed into Arabidopsis protoplasts as previously described (Walter et al., 2004). The protoplasts were incubated under darkness for 12 to 16 h before observation. YFP fluorescence was monitored using a Leica TCS SP5 confocal microscope.
LUC Activity Assay
For the transient reporter expression assay, the ProERF4:LUC reporter plasmid, effector constructs (Pro35S:HY5, Pro35S:PIF1, or Pro35S:PIF3), and the Pro35S:GUS internal control were cotransformed into Arabidopsis protoplasts. For the LCI assay, plastid combinations of various N- and C-terminal LUC fusions were cotransformed into protoplasts together with an internal control. The protoplasts were incubated under weak light for 12 to 16 h before harvesting. The LUC and GUS activities were determined with a luminometer/fluorometer (Promega) as described previously (Tang et al., 2012). The relative LUC activity was expressed as the ratio of LUC to GUS.
Microarray Analysis
The pif1 mutant and Col wild-type seedlings were grown in darkness for 4 d and exposed to light for 3 h, and total RNA was isolated using the RNAprep Pure plant kit. Hybridization to the Agilent Arabidopsis Oligo Microarray (44k; Agilent Technologies) was performed according to the manufacturer’s instructions. Three biological replicates were analyzed. Gene Ontology (GO) terms enriched in upregulated and downregulated genes were identified with GO::TermFinder (Boyle et al., 2004). Raw P values of GO term enrichment were corrected for multiple tests using false discovery rate (Benjamini and Hochberg, 1995).
Accession Numbers
Sequence data from this article can be found in the Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: PIF1 (At2g20180), PIF3 (AT1G09530), HY5 (At5g11260), HYH (At3g17609), APX2 (At3g09640), ERF4 (At3g15210), SIB1 (At3g56710), ZAT10 (At1g27730), NDB2 (At4g05020), and UBQ1 (At3g52590).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The pif Mutants Accumulate Increased Levels of Pchlide in Darkness but Have Reduced Superoxide and Hydrogen Peroxide Levels after Light Exposure.
Supplemental Figure 2. APX2 and ERF4 Expression under Weak Light Condition.
Supplemental Figure 3. Characterization of the Pro35S:Myc-PIF3 Transgenic Plants.
Supplemental Figure 4. Interaction between PIF1 and HY5 in Yeast Cells.
Supplemental Figure 5. HY5 Interacts with PIF1 and PIF3 in a Luciferase Complementation Imaging Assay.
Supplemental Figure 6. Fluorescence Resonance Energy Transfer Assay between HY5 and PIF1 in the Nucleus.
Supplemental Figure 7. Light-Dependent Regulation.
Supplemental Figure 8. HY5 and PIF1 Form Homo- and Heterodimers That Bind to DNA in the EMSA Assay.
Supplemental Figure 9. Analysis of PIF1, PIF3, and HY5 Overexpression Plants.
Supplemental Figure 10. The Mutual Regulation between PIF1 and HY5 at the mRNA and Protein Levels during Seedling Deetiolation.
Supplemental Figure 11. Greening Rate and Pchlide Levels of the Photoreceptor Mutants.
Supplemental Figure 12. Time-Course Analysis of Singlet Oxygen Production and ROS-Responsive Gene Expression.
Supplemental Data Set 1. List of PIF1-Regulated Genes Identified in the Microarray Analysis.
Supplemental Data Set 2. Overview of the Category of GO Biological Process Enriched of Genes Differentially Expressed in pif1.
Supplemental Data Set 3. Overview of the Category of GO Cellular Component Enriched in Genes Repressed in pif1.
Supplemental Data Set 4. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Haiyang Wang and the anonymous reviewers for insightful comments on the article. We thank Peter Quail, Xing Wang Deng, Enamul Huq, and the ABRC for providing seeds. We thank Jian-min Zhou for LCI vectors and Shanjin Huang for sharing the fluorescence microscope. This work was supported by grants from the National Natural Science Foundation of China (31170221), the State Basic Research Development Program (2009CB118500), the Solar Energy Initiative of the Chinese Academy of Sciences, and the Ministry of Agriculture of China (2011ZX08009-003) to R.L.
AUTHOR CONTRIBUTIONS
D.C. and R.L. designed the research. D.C., G.X., W.T., Q.J., and Y.J. performed the research. D.C., Z.F., and R.L. analyzed the data. D.C. and R.L. wrote the article.
Glossary
- bHLH
basic helix-loop-helix
- ROS
reactive oxygen species
- Pchlide
protochlorophyllide
- Col
Columbia
- H2DCFDA
2’,7’-dichlorodihydrofluorescein diacetate
- SOSG
Singlet Oxygen Sensor Green
- NBT
nitroblue tetrazolium
- DAB
diaminobenzidine
- qRT-PCR
quantitative RT-PCR
- EMSA
electrophoretic mobility shift assay
- GST
glutathione S-transferase
- ChIP
chromatin immunoprecipitation
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- LCI
luciferase complementation imaging
- FRET
fluorescence resonance energy transfer
- GO
Gene Ontology
References
- Achard P., Renou J.-P., Berthomé R., Harberd N.P., Genschik P. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Alabadí D., Blázquez M.A. (2009). Molecular interactions between light and hormone signaling to control plant growth. Plant Mol. Biol. 69: 409–417 [DOI] [PubMed] [Google Scholar]
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H.R. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C., Adám E., Fejes E., Schäfer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300 [Google Scholar]
- Blaiseau P.L., Thomas D. (1998). Multiple transcriptional activation complexes tether the yeast activator Met4 to DNA. EMBO J. 17: 6327–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G. (2004). GO::TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q.Y., Castillon A., Chen F.L., Zhu L., Huq E. (2011). Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol. Biol. 77: 501–511 [DOI] [PubMed] [Google Scholar]
- Castillon A., Shen H., Huq E. (2007). Phytochrome Interacting Factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Catalá R., Medina J., Salinas J. (2011). Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 16475–16480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron J.B., He H., Elling A.A., Deng X.W. (2009). Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhang J.Y., Neff M.M., Hong S.W., Zhang H.Y., Deng X.W., Xiong L.M. (2008a). Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc. Natl. Acad. Sci. USA 105: 4495–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Zou Y., Shang Y.L., Lin H.Q., Wang Y.J., Cai R., Tang X.Y., Zhou J.M. (2008b). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Danon A., Coll N.S., Apel K. (2006). Cryptochrome-1-dependent execution of programmed cell death induced by singlet oxygen in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 103: 17036–17041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Feng S.H., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flors C., Fryer M.J., Waring J., Reeder B., Bechtold U., Mullineaux P.M., Nonell S., Wilson M.T., Baker N.R. (2006). Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J. Exp. Bot. 57: 1725–1734 [DOI] [PubMed] [Google Scholar]
- Gechev T.S., Van Breusegem F., Stone J.M., Denev I., Laloi C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28: 1091–1101 [DOI] [PubMed] [Google Scholar]
- Guo H., Yang H., Mockler T.C., Lin C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Gohda K., Osterlund M.T., Oyama T., Okada K., Deng X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R., Jang I.C., Chua N.H. (2009). Regulated proteolysis in light-related signaling pathways. Curr. Opin. Plant Biol. 12: 49–56 [DOI] [PubMed] [Google Scholar]
- Ho L.H.M., Giraud E., Uggalla V., Lister R., Clifton R., Glen A., Thirkettle-Watts D., Van Aken O., Whelan J. (2008). Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol. 147: 1858–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C.H., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Jing Y., Zhang D., Wang X., Tang W., Wang W., Huai J., Xu G., Chen D., Li Y., Lin R. (2013). Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. (2004). An overview of the basic helix-loop-helix proteins. Genome Biol. 5: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.-S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T., Kindgren P., Benedict C., Hendrickson L., Strand Å. (2007). Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L., Barbey R., Thomas D. (1997). Assembly of a bZIP-bHLH transcription activation complex: formation of the yeast Cbf1-Met4-Met28 complex is regulated through Met28 stimulation of Cbf1 DNA binding. EMBO J. 16: 2441–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C., Stachowiak M., Pers-Kamczyc E., Warzych E., Murgia I., Apel K. (2007). Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2010). Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H.Y., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Gao S., Martinez C., He G., Zhou Z., Huang X., Lee J.-H., Zhang H., Shen Y., Wang H., Deng X.W. (2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K. (2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H.Y. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García J.F., Huq E., Quail P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Shulaev V., Mittler R. (2008). Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133: 481–489 [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V.B., Vandepoele K., Gollery M., Shulaev V., Van Breusegem F. (2011). ROS signaling: The new wave? Trends Plant Sci. 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Mockler T.C., Guo H., Yang H., Duong H., Lin C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Monte E., Tepperman J.M., Al-Sady B., Kaczorowski K.A., Alonso J.M., Ecker J.R., Li X., Zhang Y.L., Quail P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101: 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Zhu L., Shen H., Huq E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 9433–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- op den Camp, R.G.L., Przybyla, D., Ochsenbein, C., Laloi, C., Kim, C.H., Danon, A., Wagner, D., Hideg, E., Göbel, C., Feussner, I., Nater, M., and Apel, K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis Plant Cell 15: 2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagatani A., Elich T.D., Fagan M., Chory J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S., Reinbothe C., Apel K., Lebedev N. (1996). Evolution of chlorophyll biosynthesis—The challenge to survive photooxidation. Cell 86: 703–705 [DOI] [PubMed] [Google Scholar]
- Shen H., Moon J., Huq E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Park E., Choi G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49: 981–994 [DOI] [PubMed] [Google Scholar]
- Sibéril Y., Doireau P., Gantet P. (2001). Plant bZIP G-box binding factors. Modular structure and activation mechanisms. Eur. J. Biochem. 268: 5655–5666 [DOI] [PubMed] [Google Scholar]
- Stephenson P.G., Fankhauser C., Terry M.J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Wang W., Chen D., Ji Q., Jing Y., Wang H., Lin R. (2012). Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A., Bishop S.M., Phillips D., Barber J. (1994). Isolated photosynthetic reaction center of photosystem II as a sensitizer for the formation of singlet oxygen. Detection and quantum yield determination using a chemical trapping technique. J. Biol. Chem. 269: 13244–13253 [PubMed] [Google Scholar]
- The bZIP Research Group (2002). bZIP transcription factors in Arabidopsis Trends Plant Sci. 7: 106–111. [DOI] [PubMed]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantaphylidès C., Havaux M. (2009). Singlet oxygen in plants: Production, detoxification and signaling. Trends Plant Sci. 14: 219–228 [DOI] [PubMed] [Google Scholar]
- Ulm R., Baumann A., Oravecz A., Máté Z., Adám E., Oakeley E.J., Schäfer E., Nagy F. (2004). Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Przybyla D., Op den Camp R., Kim C., Landgraf F., Lee K.P., Würsch M., Laloi C., Nater M., Hideg E., Apel K. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wei N., Deng X.W. (1996). The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 112: 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yang S., Li Y., Hua J. (2007). The Arabidopsis BAP1 and BAP2 genes are general inhibitors of programmed cell death. Plant Physiol. 145: 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C., Deng X.W. (2005). COP1 - From plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 15: 618–625 [DOI] [PubMed] [Google Scholar]
- Zhang H.Y., He H., Wang X.C., Wang X.F., Yang X.Z., Li L., Deng X.W. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358 [DOI] [PubMed] [Google Scholar]
- Zhong S.W., Zhao M.T., Shi T.Y., Shi H., An F.Y., Zhao Q., Guo H.W. (2009). EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 106: 21431–21436 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.