This work examines plants overexpressing the putative membrane protein NDR1/HIN1-like 26 (NHL26), finding that overexpressor plants grew more slowly and accumulated more shoot biomass and leaf carbohydrates than wild-type plants. NHL26 localizes in the phloem plasmodesmata and endoplasmic reticulum, indicating a function in plasmodesmata permeability or sugar signaling in companion cells.
Abstract
The complex process of phloem sugar transport involves symplasmic and apoplasmic events. We characterized Arabidopsis thaliana lines ectopically expressing a phloem-specific gene encoding NDR1/HIN1-like26 (NHL26), a putative membrane protein. NHL26 overexpressor plants grew more slowly than wild-type plants, accumulated high levels of carbohydrates in mature leaves, and had a higher shoot biomass, contrasting with slower root growth and a lower seed yield. Similar effects were observed when NHL26 was overexpressed in companion cells, under the control of a companion cell–specific promoter. The soluble sugar content of the phloem sap and sink organs was lower than that in the wild type, providing evidence of a sugar export defect. This was confirmed in a phloem-export assay with the symplastic tracer carboxyfluorescein diacetate. Leaf sugar accumulation was accompanied by higher organic acid, amino acid, and protein contents, whereas analysis of the metabolite profile of phloem sap exudate revealed no change in amino acid or organic acid content, indicating a specific effect on sugar export. NHL26 was found to be located in the phloem plasmodesmata and the endoplasmic reticulum. These findings reveal that NHL26 accumulation affects either the permeability of plasmodesmata or sugar signaling in companion cells, with a specific effect on sugar export.
INTRODUCTION
Plants are photoautotrophic organisms, producing carbohydrates in photosynthetic organs and distributing them to heterotrophic organs. The phloem manages the partitioning of sugars in the plant, controlling the entry of sugars into the translocation stream, sugar transport from source to sink organs, and delivery to the various competing sink organs (van Bel, 2003). Sugar transport involves the delivery of these molecules from photosynthetic cells to the phloem sieve elements (SEs) (Sjölund, 1997; Lalonde et al., 2003). Three transport strategies have been described in different species: active loading from the apoplasm, passive diffusion via the symplasm, and passive symplasmic transfer followed by polymer trapping (Rennie and Turgeon, 2009). Plasmodesmata (PD) are directly involved in the symplasmic steps, whereas Suc symporters at the plasma membrane of the phloem cells are involved in apoplasmic loading (Dinant and Lemoine, 2010; Ayre, 2011). The sugar loading process comprises several steps (Lalonde et al., 2003; Ayre, 2011), including efflux into the apoplasm by transporters from the SWEET family (Chen et al., 2012) and influx into the companion cell (CC)–SE complex by transporters from the SUC/SUT family (Sauer, 2007). However, other aspects, such as the sugar signaling pathways involved in coordinating the symplasmic or apoplasmic steps and the mechanisms preventing the flow of Suc back through PD in various cell types (Turgeon, 2006) remain unclear. It is also possible that both the apoplasmic and symplasmic pathways contribute to photoassimilate flux (Turgeon and Ayre, 2005). A recent study showed that, in cantaloupe (Cucumis melo), a species defined as symplasmic, there may be a switch to the use of sugar transporters in response to viral infection (Gil et al., 2011).
Forward genetic screens have led to the identification of mutants affected in sugar export (high levels of carbohydrate accumulation in source organs). For instance, the sucrose export defective1 (sxd1/vte1) mutants of Arabidopsis thaliana display impaired sugar transfer from the bundle sheath to the phloem parenchyma, whereas those of maize (Zea mays) display impaired export into the SEs. These mutants lack the mitochondrial enzyme tocopherol cyclase (Russin et al., 1996; Maeda et al., 2006). They display an overaccumulation of callose either in the PD between CCs and SEs (Russin et al., 1996) or in the transfer cells between the bundle sheath and phloem parenchyma (Maeda et al., 2006). In maize, the tie dyed1 (Tdy1) mutant is impaired in a regulatory process that affects loading and consequently has a general defect of sugar export (Braun et al., 2006; Ma et al., 2008, 2009). The maize Tdy2 mutant, in which a callose synthase is defective, also displays impaired phloem transport (Slewinski et al., 2012). The Arabidopsis pho3 mutant, which has a weak allele of suc2, has a deficiency of the Suc transporter SUC2 (Gottwald et al., 2000; Zakhleniuk et al., 2001), which controls the influx of Suc into the CC-SE complex. Knockout suc2 mutants have a more severe phenotype, characterized by a much smaller rosette, an accumulation of starch and anthocyanin, and a lower seed yield (Srivastava et al., 2008, 2009). These studies have contributed to the description of some of the regulatory steps, but other approaches might identify additional components of the sugar export pathways.
Various surveys of genes preferentially expressed in the phloem have been performed, leading to the identification of subsets of genes encoding a range of putative membrane proteins of unknown function (Vilaine et al., 2003; Zhao et al., 2005). The classical approach to investigation of gene function involves the up- or downregulation of the gene and assessment of the effects on growth and development. We mined transcriptome profiling data for the vascular tissues of several plant species (Le Hir et al., 2008) to select candidate genes preferentially expressed in the phloem in celery (Apium graveolens) and Arabidopsis for further analyses of the function of Arabidopsis orthologs. We identified a member of the NDR1/HIN1-like (NHL) family in celery, HIN1, which was preferentially expressed in phloem cells (Vilaine et al., 2003; Divol et al., 2005) and encoded a predicted membrane protein of unknown function. Some distantly related members of the NDR1/HIN1 family have been shown to be involved in the hypersensitive response to pathogens, indicating a potential, but unclear, role in plant defense signaling pathways (Varet et al., 2002; Zheng et al., 2004). In total, 45 members of this family have been identified in Arabidopsis (Zheng et al., 2004), and NHL26 is the ortholog most closely related to celery HIN1. It encodes a protein that is 69% similar to the corresponding protein from celery.
Here, we report an analysis of the expression pattern of NHL26 and analyses of transgenic lines with deregulated NHL26 expression. The phenotype of NHL26-overexpressing lines suggests an impairment of sugar partitioning, leading to a substantial accumulation of Suc and other primary metabolites in mature source leaves. We also demonstrate that NHL26 is associated with PD and the endoplasmic reticulum (ER).
RESULTS
NHL26 Encodes a Putative Membrane Protein Present in All Organs
In Arabidopsis, NHL26 is encoded by At5g53730, which is composed of a single exon, as confirmed by ESTs and full-length cDNAs in The Arabidopsis Information Resource database. NHL26 is a 213–amino acid protein (23.9 kD) with a predicted transmembrane domain (amino acids 27 to 46) next to its N-terminal region, which is predicted to be cytosolic. NHL26 belongs to a plant-specific family for which no structural information is available. We searched for structural similarities to proteins of known structure, with the I-Tasser server (Zhang, 2008). NHL26 has a predicted structure similar to that of two late embryogenesis abundant (LEA) proteins of subgroup 2 (LEA_2), the LEA14 late embryogenesis protein (Protein Data Bank [PDB] code: 1x08) and a LEA desiccation-related protein (PDB code: lyccA), the structures of which have been determined by NMR spectroscopy, with normalized Z scores of 2.02 and 1.76, respectively (C-score = −2.24; T-M score = 0.45). A similar result was obtained with the PHYRE server, with a high degree of confidence (98%) (Kelley and Sternberg, 2009). Alignments with these proteins indicated that NHL26 has four α-helices and seven β-strands that could form two antiparallel β-sheets (see Supplemental Figure 1 online). In this model, the transmembrane domain toward the N terminus of the protein is predicted to fold as a large α-helix.
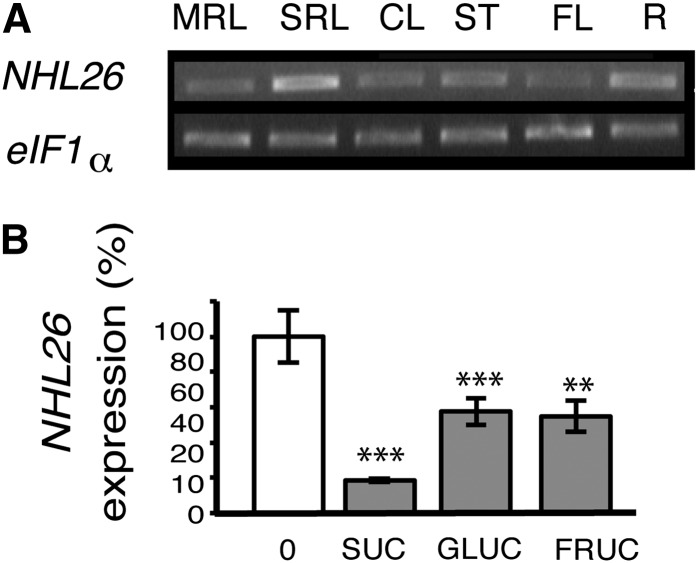
NHL26 is not present on the Affymetrix oligonucleotide array (ATH1 22k) but is found on CATMA arrays. A survey of the gene expression data provided by the CATdb database indicated that NHL26 was expressed in all organs. We investigated the pattern of expression of NHL26 in various organs by RT-PCR: NHL26 mRNA was detected in mature rosette leaves, senescent rosette leaves, cauline leaves, floral stems, flowers, and roots (Figure 1A). CATMA array analysis showed an upregulation in rosette leaves after nitrogen deprivation and a downregulation of the expression of this gene in starch branching enzyme II double-knockout mutants (sbe1 sbe2) (Dumez et al., 2006), indicating possible regulation by nitrogen or carbon availability. The transcriptional regulation of NHL26 by sugars was assessed by quantitative RT-PCR (qRT-PCR), which showed strong downregulation in response to Suc, Glc, and Fru (Figure 1B). Searches for cis-acting regulatory DNA elements in the NHL26 promoter, with AtcisDB, identified motifs associated with stress responses (MYB4 binding sites and a W-box motif) and light-responsive elements (SORLEP3, GATA motifs, and T-box motifs), suggesting additional transcriptional regulation of this gene by biotic stress and light.
Figure 1.
Pattern of Expression of NHL26.
(A) Expression of NHL26 in various organs. The accumulation of NHL26 RNA was analyzed by RT-PCR. EiF1α was used as an internal quantitative control. RT-PCR products were detected by ethidium bromide staining of the agarose gels used for electrophoresis of the PCR products. CL, cauline leaves; FL, flowers; MRL, mature rosette leaves; R, roots; SRL, senescent rosette leaves; ST, stem leaves.
(B) Transcriptional regulation of NHL26 by sugars. The expression of NHL26 was analyzed by qRT-PCR on 7-d-old Arabidopsis seedlings treated for 6 h with 10 mM Suc, Glc, or Fru. Expression levels are indicated as percentages of the values obtained for the untreated samples (0 mM). The levels of NHL26 transcript accumulation were normalized relative to levels of APT RNA. Asterisks indicate values that are significantly different, as assessed in a t test (**P < 0.02; ***P < 0.01). Error bars indicate the se (n = 6).
Phloem-Specific Expression of NHL26
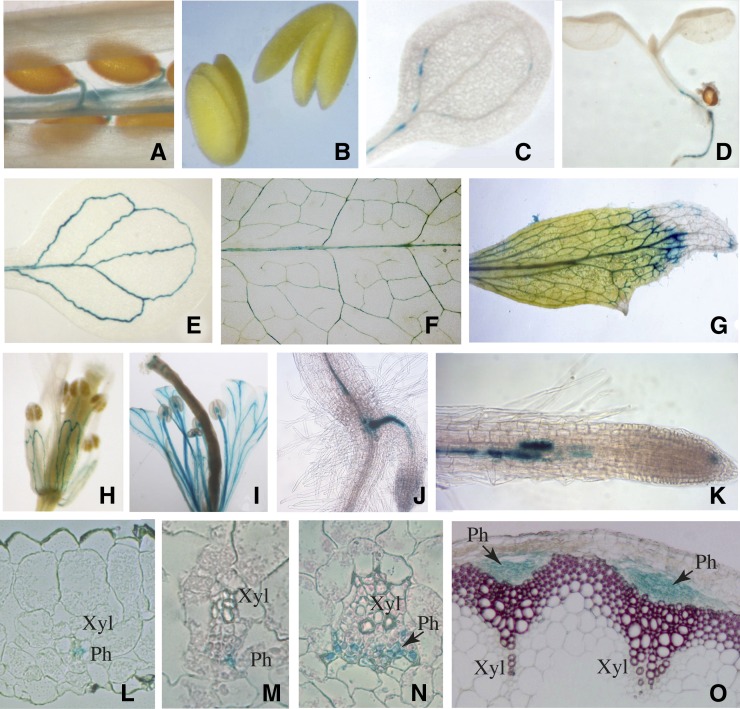
The tissue specificity of NHL26 expression was investigated with a transcriptional fusion of a 1.5-kb NHL26 promoter sequence and the UidA reporter gene, encoding the β-glucuronidase (GUS) reporter protein. Two representative Arabidopsis transgenic lines expressing the pNHL26:GUS construct were analyzed (Figure 2). No GUS activity was observed in nongerminating seeds or embryos (Figures 2A and 2B). Three days after germination, GUS activity was detected in the vasculature of the cotyledons (Figure 2C). It progressively extended after 8 d in the vasculature, to the base of the hypocotyl, the roots (Figure 2D), and the cotyledons (Figure 2E). In mature plants, GUS activity was present in the vascular system of all organs, including mature and senescent leaves (Figures 2F and 2G), flowers (Figures 2H and 2I), and roots (Figures 2J and 2K) The expression of pNHL26:GUS in flowers progressed from the sepals, to the petals, and the stamen filaments. The vasculature was also labeled in siliques (Figure 2A). In mature leaves and in the floral stem, GUS activity was strictly limited to the phloem tissue in major and secondary veins and to higher order veins, such as freely ending veinlets (Figures 2L to 2O). It was observed in all phloem cell types, including parenchyma cells and CCs.
Figure 2.
Organ and Tissue Localization of NHL26 Expression.
Representative GUS staining of pNHL:GUS-expressing plants. (A) Silique; (B) mature embryos; (C) cotyledon 3 d after germination; (D) 8-d-old seedling; (E) cotyledon 8 d after germination; (F) mature rosette leaf; (G) senescent cauline leaf; (H) and (I) flowers observed before and after pollination, respectively; (J) hypocotyl-root junction with emerging adventitious root; (K) primary root; (L) transverse thin section of minor vein from a mature leaf; (M) and (N) transverse thin section of a secondary vein from a mature leaf; (O) transverse thin section of floral stem (the lignin was stained with phloroglucinol). Arrows, phloem; Ph, phloem; Xyl, xylem.
Identification of Mutants Displaying Down- or Upregulation of NHL26 Expression
We first investigated the phenotype of nhl26 mutants. Three lines with an insertion in the NHL26 gene were available (nhl26-1, nhl26-2, and nhl26-3; see Supplemental Figure 2A online). In nhl26-1 and nhl26-2, the abundance of the NHL26 transcript was similar to that in the wild type, so these mutants were not studied further. In nhl26-3, we found a truncated transcript, ∼450 bases long, whereas the wild-type transcript is 642 bases long. There were no phenotypic differences between this mutant and the wild type. We also produced transgenic lines expressing an artificial microRNA (amiRNA) targeting an NHL26-specific sequence. Thirty independent lines were obtained, several of which had NHL26 transcript levels up to 90% lower than those of the wild type (see Supplemental Figure 2B online), but none showed any growth or developmental defect. The expression of the sugar transporter gene SUC2 and sugar accumulation were also normal in these plants (see Supplemental Figures 2C to 2F online). NHL26 belongs to a multigene family (Zheng et al., 2004), and the absence of an evident phenotype may reflect functional redundancy with other members of this family. A survey of the cell-specific expression of other members of the NHL family from available translatome data sets (Mustroph et al., 2009) revealed that several related genes, such as NHL12, NHL13, NHL14, and NHL18, were expressed in the CCs of the shoot (see Supplemental Figure 3 online).
As an alternative approach, we studied the effect of NHL26 overexpression under the control of the constitutive cauliflower mosaic virus (CaMV) 35S promoter or, more specifically, in the CCs, with the strong CC-specific promoter of the PP2-A1 gene (Dinant et al., 2003), hereafter named pPP2 (see Supplemental Figure 4 online). Several lines displaying higher levels of NHL26 mRNA accumulation were selected for each of these constructs. Further phenotyping and molecular characterization were performed on two representative lines for each construct, with similar growth and physiological effects on the plant phenotype associated with transgene expression in these plants (see Supplemental Figure 4 online). The relative amounts of NHL26 transcript in the lines expressing p35S:NHL and pPP2:NHL were determined by qRT-PCR. This transcript accumulated substantially in p35S:NHL lines (300 to 800 times normal levels) and, to a lesser extent, in pPP2:NHL lines (2 to 4 times higher) (see Supplemental Figure 4 online), reflecting the restriction of expression driven by the PP2-A1 promoter to phloem CCs. The two types of line presented consistent growth alterations, with the p35S:NHL-expressing lines presenting more severe effects than the pPP2:NHL lines. One representative T3 line homozygous for p35S:NHL and one homozygous for pPP2:NHL (p35S:NHL 5.6 and pPP2:NHL 4.1; see Supplemental Figure 4 online), displaying 500 times and 4 times higher than normal levels of transcripts, respectively, were chosen for subsequent analysis.
NHL26 Overexpression Leads to Growth Alterations
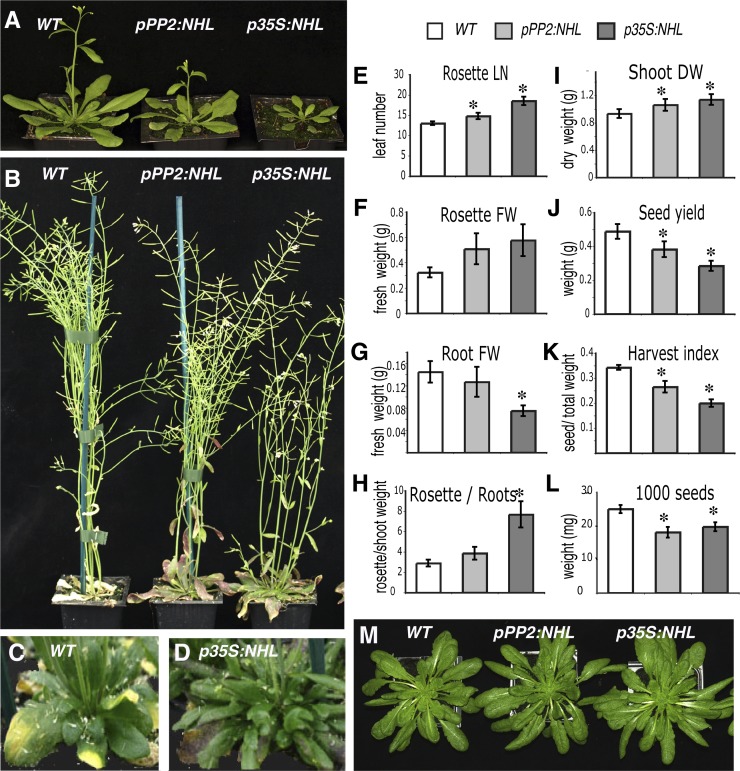
No differences between pPP2:NHL, p35S:NHL, and wild-type seedlings were observed in vitro. However, when grown in the greenhouse or in a growth chamber under long-day conditions (16-h photoperiod, 150 µE m−2 s–1), the pPP2:NHL lines grew more slowly than the wild-type lines and the p35S:NHL lines grew the most slowly of all (Figure 3A). Flowering time was delayed, by 1 week for the pPP2:NHL plants and 2 weeks for the p35S:NHL plants (Figure 3A). The growth of the flower stem of p35S:NHL plants was delayed (Figure 3B). Senescence was also delayed, with the leaves beginning to yellow 2 to 3 weeks later than for the wild type (Figures 3C and 3D). This slowing of flowering and senescence extended the growth period of the rosette, resulting in the final number of rosette leaves and rosette fresh weight being significantly higher in pPP2:NHL and p35S:NHL plants than in wild-type plants (Figures 3E and 3F). By contrast, root biomass was lower (Figure 3G) and the shoot-to-root fresh weight ratio in pPP2:NHL and p35S:NHL plants was 1.5 and 2.5 times higher, respectively, than that in the wild type (Figure 3H). At harvest time, the harvest index, defined as the ratio of the weights of seeds and total aerial parts, was significantly affected (Figures 3I to 3K). Seed yield and seed weight were lower than wild-type values (Figures 3J and 3L). The morphogenesis of the plant was not affected; leaf shape, stem architecture, flower, and silique formation were not altered. These effects on plant growth and biomass were not observed when plants were grown in short-day conditions (Figure 3M).
Figure 3.
Growth Defects in NHL26-Overexpressing Lines.
(A) to (D) Phenotype of NHL26-overexpressing plants, grown in long-day conditions. WT, the wild type.
(A) and (B) Plants grown in a growth chamber (16 h day, 150 µE s−1 m−1).
(A) Four-week-old plants.
(B) Eight-week-old plants.
(C) and (D) Ten-week-old plants grown in greenhouse.
(C) The wild type.
(D) p35S:NHL plants displayed no sign of senescence at the time at which the wild type started to senesce.
(E) to (L) Comparison of growth parameters between the transgenic pPP2:NHL and p35S:NHL plants and wild-type plants. Asterisks indicate values significantly different in a t test (P < 0.05).
(E) Number of rosette leaves at flowering time (LN). Error bars indicate the se (n = 12).
(F) to (H) Fresh weight (FW) of the rosette and root of plants grown in pots containing sand in the greenhouse.
(F) Rosette fresh weight.
(G) Root fresh weight.
(H) Rosette FW to root FW ratio. Error bars indicate the se (n = 12).
(I) to (L) Yield parameters for plants grown in long-day conditions in the greenhouse. Asterisks indicate values that are significantly different, as assessed in a t test (P < 0.05).
(I) Dry weight (DW) of shoots (rosette and stem) at harvest time.
(J) Seed weight.
(K) Harvest index, calculated as the ratio of seed weight to dry weight of the aerial part of the plant (rosette, stem, and seeds).
(L) Weight of 1000 seeds. Error bars indicate the se (n = 11).
(M) Ten-week-old plants grown in short-day conditions (8 h day, 150 µE s−1 m−1).
[See online article for color version of this figure.]
Leaf Reddening and Starch Accumulation
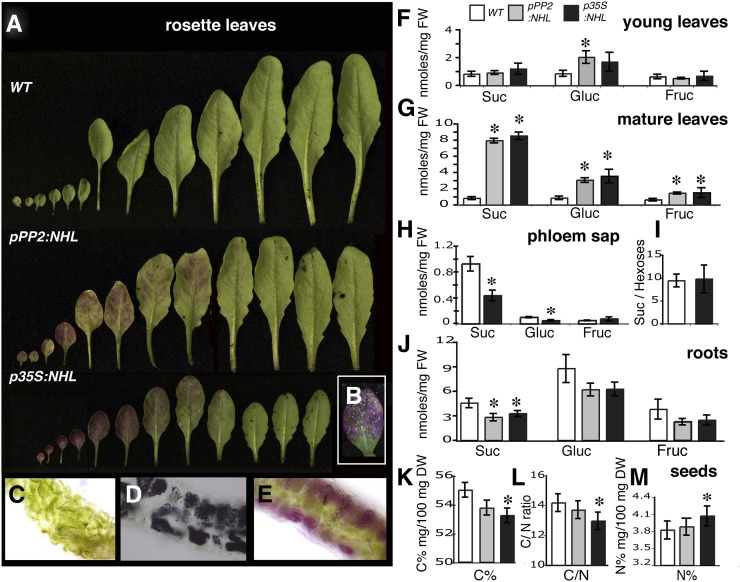
Another alteration observed in the rosette leaves in pPP2:NHL and p35S:NHL plants grown in a greenhouse or a growth chamber was a progressive reddening, which started at the six to eight expanded leaves stage. The cotyledons and the juvenile leaves began to redden first, probably due to anthocyanin accumulation, and the reddening then spread to adult leaves, progressing basipetally (tip to base) as the leaves matured (Figure 4A). It began at the leaf margin, subsequently spreading to the regions between the veins. In the most severe cases, the rosette leaves developed spontaneous necrotic lesions (Figure 4B). Reddening progressed acropetally as the plants aged, with the younger leaves that were still expanding remaining unaffected. Determinations of anthocyanin content in leaf extracts confirmed that pPP2:NHL and p35S:NHL plants contained significantly more anthocyanin (up to 10 times more) than the wild type. Thin sections of the leaves from pPP2:NHL and p35S:NHL plants also confirmed anthocyanin accumulation and showed starch storage (Figures 4C to 4E). Redness and anthocyanin and starch accumulation were more pronounced and appeared more rapidly in the p35S:NHL plants than in the pPP2:NHL plants.
Figure 4.
Differential Accumulation of Carbohydrates in the Source and Sink Organs of NHL26-Overexpressing Lines.
(A) to (E) Rosette leaves from adult plants grown in long-day conditions (stage 6.00).
(A) In the leaves of overexpressing lines, the gradient of reddening follows the sink-to-source transition in mature leaves of the rosette. WT, the wild type.
(B) The inset the appearance of necrotic spots on some old leaves from NHL26-overexpressing plants.
(C) to (E) Transverse sections of mature rosette leaves from the wild type (C) and from plants overexpressing NHL26 under the control of the PP2 promoter. NHL26-overexpressing leaves display starch (D) and anthocyanin (E) accumulation.
(F) and (G) Suc, Glc, and Fru in young rosette leaves that are still expanding (F) and fully expanded mature rosette leaves (G) of pPP2:NHL- and p35S:NHL-expressing and wild-type plants. Plants were harvested at stage 6.10. The data points and error bars represent the mean and se (n = 12). FW, fresh weight.
(H) and (I) Suc, Glc, and Fru content of the phloem sap exudate from mature fully expanded rosette leaves of p35S:NHL-expressing and wild-type plants. Plants were harvested at stage 6.0. The data points and error bars represent the mean and se (n = 9).
(J) Suc, Glc, and Fru content of the roots of pPP2:NHL- and p35S:NHL- expressing and wild-type plants. Plants were harvested at stage 6.0. The data points and error bars represent the mean and se (n = 6).
(K) to (M) Carbon and nitrogen contents and carbon-to-nitrogen ratio in the seeds of pPP2:NHL- and p35S:NHL-expressing and wild-type plants. The data points and error bars represent the mean and se (n = 9). DW, dry weight.
Asterisks indicate values significantly different in a t test (P < 0.05).
Sugar Accumulation in Source and Sink Organs
Sugar content was quantified in the rosette leaves of wild-type, p35S:NHL, and pPP2:NHL plants at bolting. At this stage, the first eight rosette leaves, which were typically fully expanded (i.e., source leaves), had reddened, whereas the last leaves to emerge, which were still expanding, remained green. The effect of NHL26 levels on sugar accumulation was determined either on leaves 3 to 6 (hereafter referred to as mature leaves) or on the leaves that were still expanding, leaves 8 and 9 (hereafter referred to as young leaves). The soluble sugar content of the young rosette leaves did not differ significantly between the transgenic lines and the wild type (Figure 4F). However, the sugar content of the mature, fully expanded leaves was significantly higher in the mutants than the wild type: There was 8 times as much Suc, three times as much Glc, and twice as much Fru (Figure 4G). These increases in sugar content were associated with a 50% decrease in the Suc content of phloem sap exudates (Figure 4H), with no significant effect on the Suc/hexose ratio (Figure 4I). The Suc content of the roots of NHL26-overexpressing plants was also lower than that of the wild type, but no significant difference between plant lines was observed for the Glc and Fru contents of the roots (Figure 4J). The proportion of carbon in the seeds of the p35S:NHL plants was low (Figure 4K), resulting in a low carbon/nitrogen ratio (Figure 4L) and an accumulation of nitrogen (Figure 4M). This suggests that the lower seed weight (Figure 3L) may result from a decrease in the export of carbohydrates rather than nitrogen compounds.
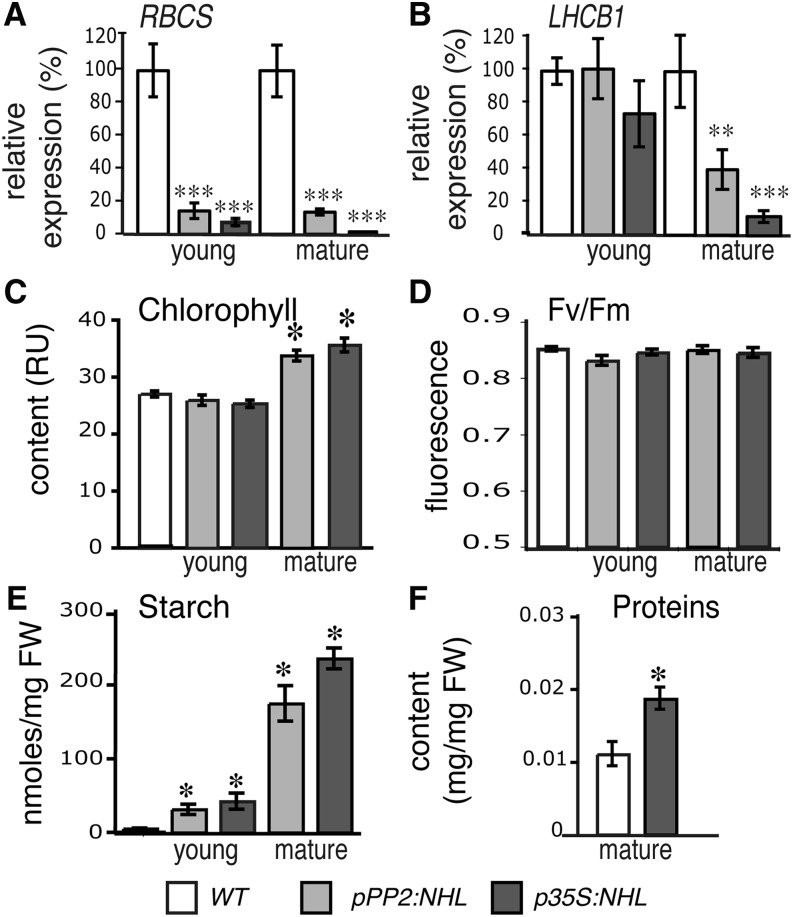
Photosynthesis and Chlorophyll Content
An excess of carbon metabolites can trigger the negative feedback regulation of leaf photosynthesis (Paul and Foyer, 2001; Ainsworth and Bush, 2011). We analyzed the expression, in mature and young leaves, of the photosynthetic genes RBCS (encoding the ribulose-1,5-bis-phosphate carboxylase/oxygenase small subunit) and LHCB1 (encoding a light-harvesting complex II protein from photosystem II [PSII]). RBCS transcript levels were significantly lower than wild-type levels (Figure 5A) in the mature and young leaves of NHL26-overexpressing plants. An effect in the mature leaves was also observed for LHCB1 transcripts, although no significant decrease was recorded in the young leaves (Figure 5B). We investigated the chlorophyll content and activity of PSII in NHL26-overexpressing plants. The chlorophyll content of the mature leaves of p35S:NHL and pPP2:NHL plants was slightly higher than that in wild-type plants (Figure 5C), with no change in the chlorophyll fluorescence of the PSII in the leaves of overexpressing plants (Figure 5D), but with the expected massive accumulation of starch (Figure 5E) and a high protein content (Figure 5F).
Figure 5.
Starch Accumulation and Photosynthetic Activity in NHL26-Overexpressing Lines.
(A) and (B) Changes in the transcript abundance of two photosynthesis genes in NHL26-overexpressing lines.
(A) Relative expression of RBCS in young and mature leaves.
(B) Relative expression of LHCB1 in young and mature leaves. Mature leaves presenting strong reddening were analyzed. The expression of these genes was assessed by qRT-PCR and normalized relative to that of APT. The data points and error bars represent the mean and se (n = 6).
(C) and (D) Photosynthetic parameters in p35S:NHL- and pPP2:NHL-expressing plants. The chlorophyll content, measured in Relative Units, RU (C) and chlorophyll fluorescence (Fv/Fm) of PSII (D) were measured in the rosette leaves of 8-week-old plants (stage 6.50) grown in the greenhouse. Chlorophyll content is expressed in relative SPAD units. The data points and error bars represent the mean and se of two measurements (n = 12).
(E) Starch content of the rosette leaves of pPP2:NHL- and p35S:NHL-expressing and wild-type plants. Plants were harvested at stage 6.0. The data points and error bars represent the mean and se (n = 12). FW, fresh weight.
(F) Protein content of the mature rosette leaves in wild-type and pPP2:NHL- and p35S:NHL-expressing plants. Plants were harvested at stage 6.0. The data points and error bars represent the mean and se (n = 6).
Asterisks indicate values significantly different in a t test (*P < 0.05; **P < 0.02; ***P < 0.01). WT, the wild type.
Metabolomic Changes in NHL26-Overexpressing Lines
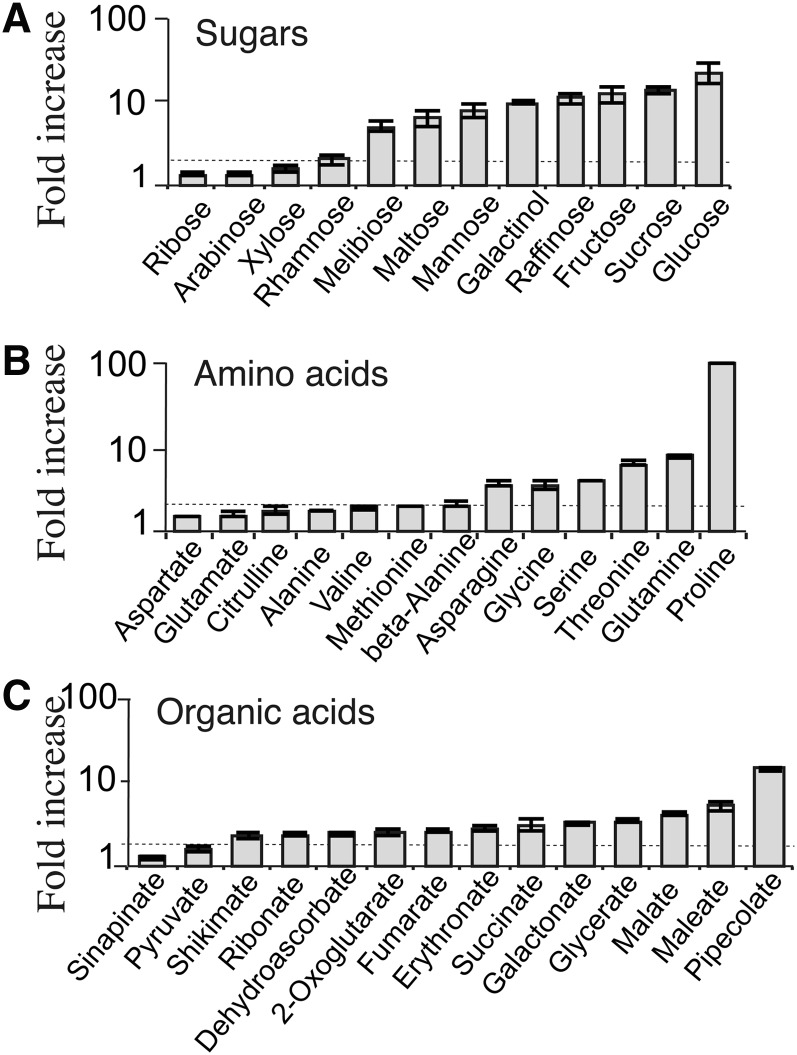
The metabolic compounds in the mature rosette leaves of pPP2:NHL, p35S:NHL, and wild-type plants were analyzed by gas chromatography–mass spectrometry (GC-MS). We unambiguously identified 84 compounds, the abundance of 58 of which differed significantly between the lines (P < 0.01). The metabolite profiles of the overexpressing and wild-type plants were significantly different (Figure 6A), and this effect was more pronounced in p35S:NHL than in pPP2:NHL plants. The compounds accumulating in p35S:NHL plants included sugars, amino acids, and organic acids (Figure 6B; see Supplemental Figure 5 online). Consistent with the findings presented above, Suc, Glc, and Fru were substantially more abundant in p35S:NHL plants, and other sugars, including maltose, galactinol, raffinose, and melibiose also accumulated in large amounts. Mannitol, a polyol that is barely detectable in wild-type plants, accumulated in the p35S:NHL plants. The concentrations of several amino acids, including Gln, Asn, Gly, Ser, and His, were also significantly higher (10 times higher for Gln). The concentration of Pro, a stress indicator, was very high (up to 115 times wild-type levels). Gamma-aminobutyric acid, a nonprotein amino acid that serves as a stress signal, was also slightly more abundant. Most organic acids, except for citrate, and various nitrogen derivatives, including allantoin, urea, and putrescine, accumulated in the leaves of the p35S:NHL plants. Pipecolate, an osmolyte, was abundant in both p35S:NHL and pPP2:NHL plants, as was α-tocopherol, an antioxidant, which was slightly more abundant. Five metabolites were less abundant in the mutants than the wild type (uracil, citrate, linolenic acid, glycerol monostearate, and glycerol monopalmitate) (see Supplemental Figure 5 online).
Figure 6.
Modification of the Metabolite Profile in NHL26-Overexpressing Lines.
Metabolites for which the abundance in the rosette leaves of p35S:NHL-overexpressing plants was significantly different from that in wild-type controls (P > 0.01). Sugars (A), amino acids (B), and organic acids (C). Data are presented as fold increases with respect to the wild type on a log scale, for each compound, of the mean value obtained for p35S:NHL-expressing plants (±se; n = 6). The dotted line indicates the threshold for a twofold increase.
Changes in Phloem Sap Exudate Composition
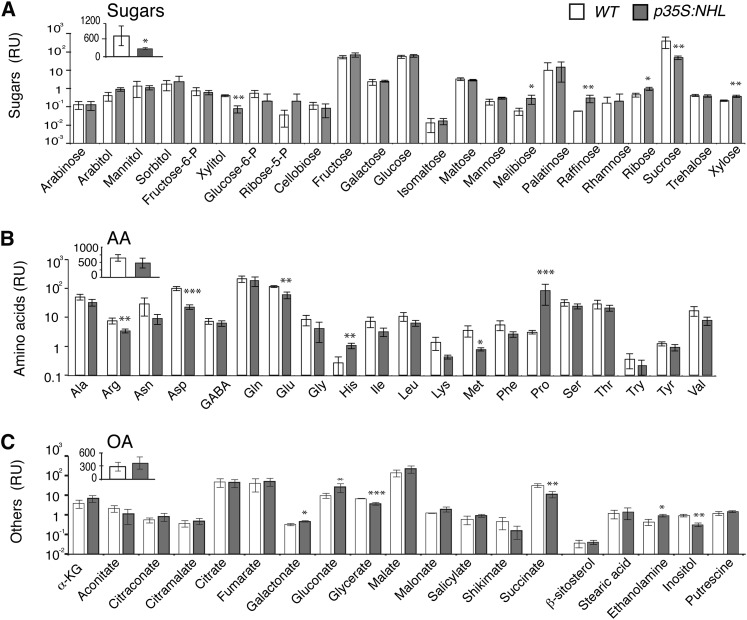
Given the much lower Suc content of the phloem sap exudates of the mutants (Figure 4H), we performed metabolite profiling on the exudates of mature rosette leaves from p35S:NHL and wild-type plants (Figure 7). The metabolites in the phloem sap exudate were analyzed by GC-MS and quantified, to estimate their content per milligram of fresh weight of leaves. We quantified 64 metabolites in the exudate. The data set was normalized by least-rectangle regression, making it possible to carry out pairwise comparisons of the metabolite profiles and to eliminate the effects of differences in exudation efficiency between samples. We confirmed the much lower Suc content of the mutant (Figure 7A), in which raffinose levels were higher than in the wild type. By contrast, the mutant displayed no significant decrease in total amino acid content (Figure 7B). Gln content was significantly higher in the mature leaves but unaffected in phloem exudate. A few amino acids, such as Glu, Asp, and Arg, were less abundant in the mutant, whereas Pro and His were more abundant. The lower Suc content of the mutant was therefore not correlated with a decrease in overall amino acid content. Total organic acid content was not significantly affected either, but the levels of succinate and glycerate were lower in the mutant than in the wild type.
Figure 7.
Metabolite Content in the Phloem Sap Exudate of Rosette Leaves from NHL26-Overexpressing Lines.
Mean metabolite content, in relative units in the phloem sap exudate of p35:NHL-overexpressing plants and wild-type (WT) plants, collected by EDTA-facilitated exudation. The data points and error bars represent the mean and se (n = 3) of the metabolite content, per unit fresh weight measured in pmol/mg fresh weight, normalized to the overall content of the phloem exudate. AA, amino acids; OA, organic acids; RU, relative units. Asterisks indicate values significantly different in a t test (*P < 0.05; **P < 0.01; ***P < 0.001).
(A) Sugar content. The inset indicates the total sugar content.
(B) Amino acid content. The inset indicates the total amino acid content.
(C) Other metabolites, including organic acid content. The inset indicates the total organic acid content.
NHL26 Overexpression Affects the Expression of Sugar-Responsive and Sugar Transporter Genes
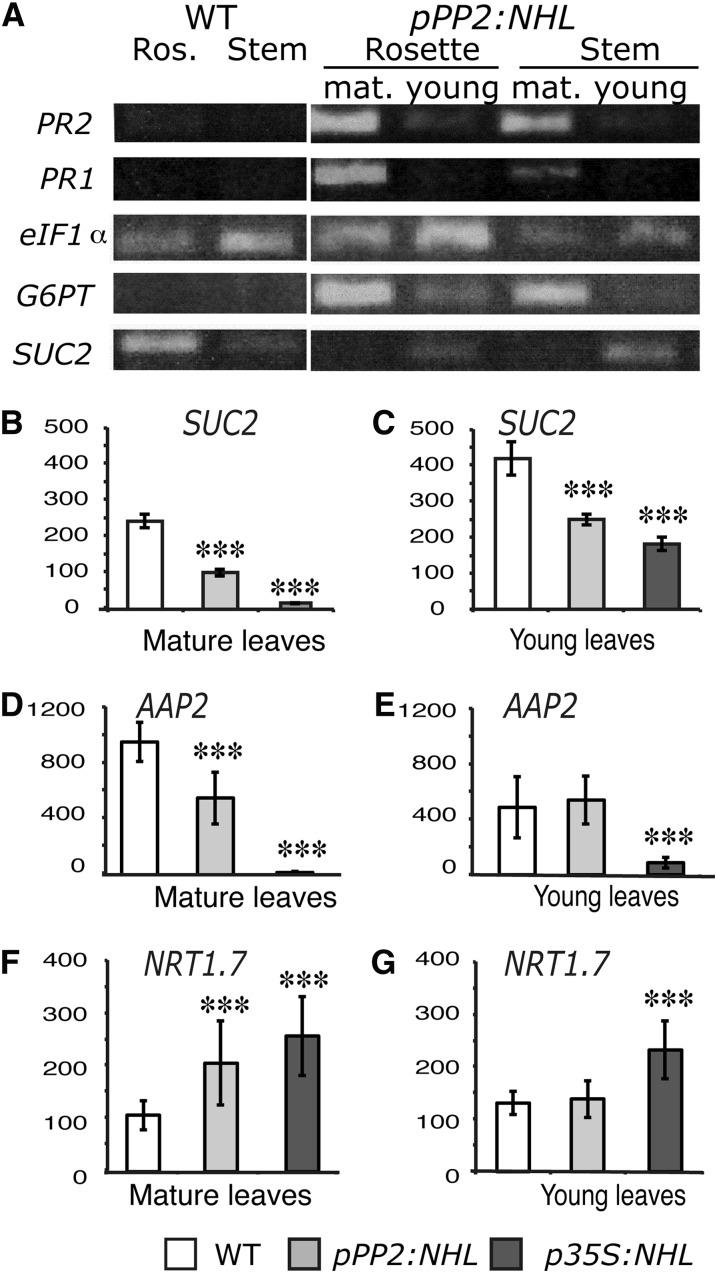
We assessed G6PT expression in the young and mature rosette leaves of NHL26-overexpressing plants. G6PT (At1g61800) encodes a Glc-6-phosphate/phosphate translocator that is reported to be Suc responsive (Lloyd and Zakhleniuk, 2004; Gonzali et al., 2006). This experiment was performed on the line that was less severely affected, pPP2:NHL, to limit the downstream effects that may result from an excessive accumulation of sugars. G6PT transcript content was higher in the mature reddened leaves from the rosette and in the stem leaves from the transgenic plants than in those from the wild type; no such difference was observed for the young leaves (Figure 8A). This is consistent with previous reports that G6PT expression is responsive only to high Suc concentrations (Gonzali et al., 2006). We also investigated the expression of PR1 (At3g57260) and PR2 (At3g57260), both defense-associated markers (Reuber et al., 1998) induced by high sugar content (Thibaud et al., 2004). As for G6PT, an accumulation of PR1 and PR2 transcripts was observed in the mature leaves of pPP2:NHL plants but not in the young leaves (Figure 8A).
Figure 8.
Changes in Transcript Abundance for Selected Genes in NHL26-Overexpressing Lines.
(A) Relative expression levels for stress markers and sugar metabolism or transport markers, determined by RT-PCR, in the rosette leaves or stem leaves from wild-type (WT) plants and fully expanded mature (1) or young (2) leaves from pPP2:NHL-expressing plants. In these plants, mature leaves were characterized by strong reddening. Transcript abundance was compared with that of eiF1α. Stress markers: PR1 and PR2, encoding pathogenesis-related proteins 1 and 2; sugar metabolism and transport markers: G6PT, encoding the chloroplast Glc-6-phosphate/phosphate translocator 2, and SUC2, encoding the Suc transporter SUC2.
(B) and (C) Relative expression of the gene encoding the SUC2 sugar transporter in the leaves of pPP2:NHL- and p35S:NHL-expressing plants was assessed by qRT-PCR. Expression values were normalized with respect to those of TIP41. SUC2 relative transcript abundance was studied in mature rosette leaves that had reddened (C) or young leaves (B) in the transgenic lines.
(D) to (G) The relative expression of nitrogen-responsive genes in the leaves of pPP2:NHL- and p35S:NHL-expressing plants was assessed by qRT-PCR. Expression values were normalized with respect to those for TIP41.
(D) and (E) AAP2 relative transcript abundance in mature and young rosette leaves, respectively.
(F) and (G) NRT1.7 relative transcript abundance in mature and young rosette leaves, respectively.
Data points and error bars represents the mean and se of the mean (n = 8). Asterisks indicate values significantly different in a t test (***P < 0.001).
We also investigated the transcription of SUC2 (At1g22710), which encodes the main influx Suc transporter involved in phloem loading in CCs (Truernit and Sauer, 1995; Stadler and Sauer, 1996; Srivastava et al., 2008). The level of SUC2 expression in the young leaves of transgenic overexpressing plants was moderately reduced (Figures 8A and 8B), whereas SUC2 transcript abundance was significantly lower in the mature leaves of NHL26 overexpressor plants than in the corresponding leaves of control plants (Figures 8A and 8B), by up to 80% reduction. The expression of two genes involved in nitrogen transport was also monitored: AAP2, which is involved in the transfer of amino acids from xylem to phloem and phloem loading (Zhang et al., 2010), and NRT1.7, involved in the source-to-sink remobilization of nitrate (Fan et al., 2009). AAP2 expression levels were lower, whereas NRT1.7 expression levels were higher in NHL26-overexpressing lines (Figures 8D to 8G). This effect was more pronounced in mature leaves than in young leaves and was stronger in p35S:NHL plants than in pPP2:NHL plants.
NHL26 Localization to the PD and ER
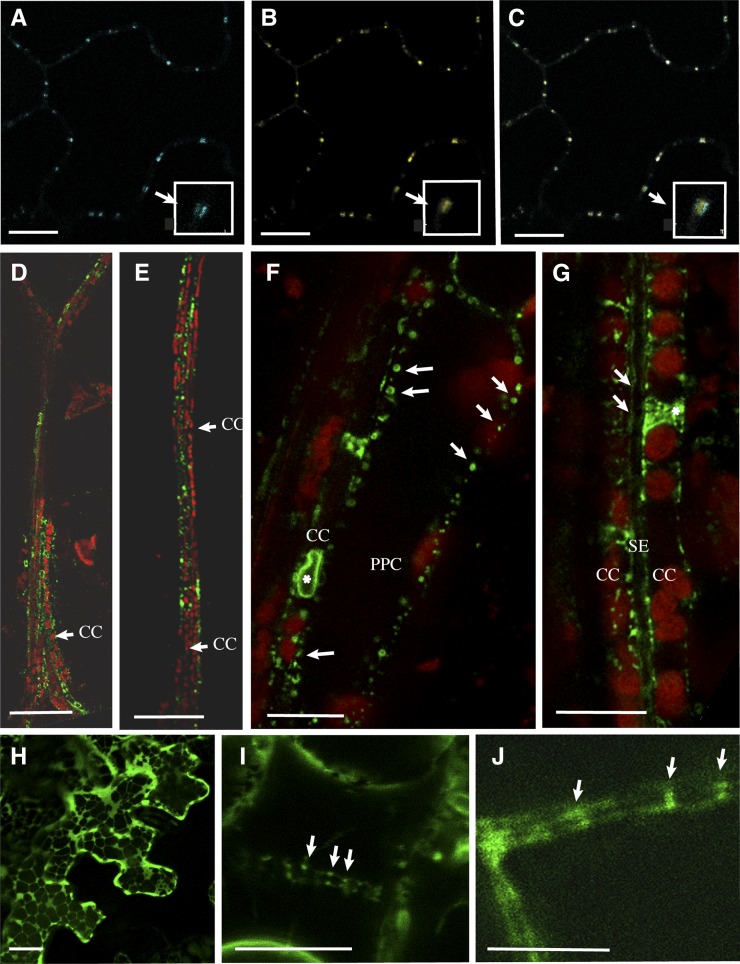
The subcellular distribution of NHL26 was first examined with a pNHL:NHL-CFP (for cyan fluorescent protein) construct in a transient expression assay on epidermal cells from the cotyledons of Arabidopsis plants. When observed by confocal laser scanning microscopy (CLSM), CFP fluorescence showed a punctate pattern at the periphery of the cells (Figure 9A). When performed on the p35S:PDLP1-GFP–expressing Arabidopsis line (Thomas et al., 2008), which produces a plasmodesmal protein fused to green fluorescent protein (GFP), we observed that the NHL-CFP fusion and PDLP1-GFP colabeled the same structures in epidermal cells (Figures 9A to 9C), demonstrating that NHL26 was located in PD. PDLP1 was observed within the PD pore, whereas NHL26 was located outside the pore (details; Figures 9A to 9C). Similar results were obtained with N- and C-terminal GFP translational fusions under the control of the 35S promoter in transient expression assays on epidermal cells from the leaves of Nicotiana benthamiana (data not shown). We confirmed this localization in transgenic Arabidopsis lines expressing the pNHL:NHL-GFP construct. Fluorescence was observed in the phloem cells of the veins, including phloem parenchyma cells, CCs, and SEs (Figures 9D and 9E). The signal in phloem parenchyma cells and CCs was punctate and peripheral (Figures 9F and 9G), consistent with a plasmodesmal location. In the SEs, a stacked fluorescence signal was observed at the cell periphery, with regular thickening reminiscent of the SE reticulum (Figure 9G). Labeling was also observed in the CCs, in a network surrounding the nucleus, consistent with an ER location. In epidermal cells from Arabidopsis lines expressing the NHL-GFP fusion under the control of the 35S constitutive promoter, based on use of the p35S:NHL-GFP construct, intense fluorescence was observed. This signal was localized to the ER. After plasmolysis, it remained attached to the cell wall (Figure 9I), consistent with its previous localization to PD.
Figure 9.
Localization of NHL26 in Phloem Cells.
Confocal microscopy of NHL26-CFP and NHL26-GFP. PPC, phloem parenchyma cells. Bars = 10 µm in (A) to (C), (F), and (G), 50 µm in (D) and (F).
(A) to (C) Colocalization of NHL26-CFP with PD-associated PDLP1-GFP, observed by confocal microscopy, after the agroinfiltration of pNHL:NHL-CFP into epidermal cells of the cotyledons of p35S:PDLP1-GFP-expressing Arabidopsis plants. PDLP1-GFP signal (A), NHL-CFP signal (B), and overlay of (A) and (B) in (C).
(D) and (E) Localization of the NHL26-GFP fusion protein, in the secondary (D) and minor (E) leaf veins of pNHL:NHL-GFP Arabidopsis plants.
(F) to (G) Subcellular localization of the NHL26-GFP fusion protein in phloem cells in pNHL:NHL-GFP Arabidopsis plants.
(F) Details of subcellular localization in phloem parenchyma and CCs.
(G) Details of subcellular localization in CCs and SEs. In these images, the CCs can be identified by the alignments of chloroplasts, typical of the organization of plastids in these cells. The SEs can be identified by the absence of plastids and the double membrane. Arrows indicate GFP-labeled punctate structures stacked at the periphery of CCs and SEs. Stars indicate a thin network surrounding the nucleus, indicative of a location within the ER.(H) Transient expression in Nicotiana benthamiana epidermal cells using the p35S:NHL-GFP construct. (I) Stable expression in Arabidopsis epidermal cells of p35S:NHL-GFP after plasmolysis treatment. (J) Stable expression in vascular cells in pNHL:NHL-GFP Arabidopsis plants. (I) shows, after plasmolysis, punctate structures on both sides of the cell walls in p35S:NHL-GFP, as observed in pNHL:NHL-GFP plants (J), without any plasmolysis treatment. CC, companion cells, SE, sieve elements, PPC, phloem parenchyma cells. Bars = 10 μm in A, B, C, F, H, I, J and G, 50 μm in D and F.
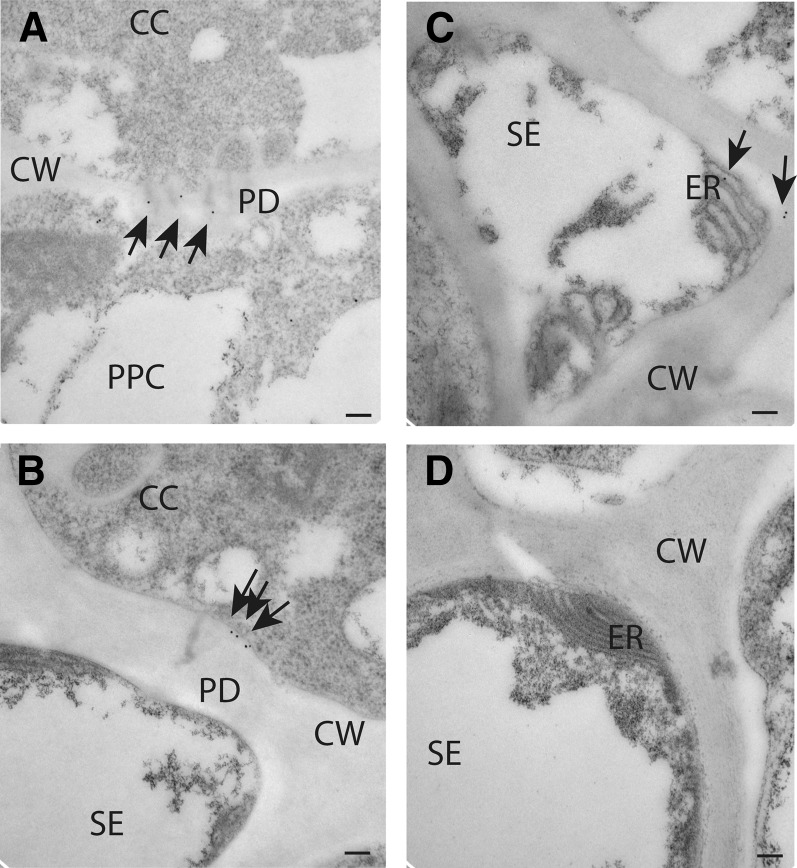
The association of NHL26 with the PD and the ER in phloem cells was confirmed by immunogold labeling with a polyclonal antibody against GFP on ultrathin sections of leaves from transgenic pNHL:NHL-GFP plants. High-resolution observations of the phloem by transmission electron microscopy showed that the antigen was present in the PD between phloem parenchyma cells and CCs (Figure 10A; see Supplemental Figure 6 online) and in branched PD between CCs and SEs (Figure 10B; see Supplemental Figure 6 online). Gold labeling was also found in the SE reticulum (Figure 10C). No background signal was obtained in the absence of the primary anti-GFP antibody (Figure 10D), confirming the specificity of the antibody.
Figure 10.
Localization of NHL26 in PD and in the SE Reticulum.
NHL26 was localized by immunogold labeling and transmission electron microscopy. Arrows indicate gold particles conjugated to an anti-GFP polyclonal antibody. Bars = 100 nm.
(A) to (C) Detection of the NHL26-GFP fusion protein with an anti-GFP antibody in phloem cells in pNHL:NHL-GFP Arabidopsis plants.
(A) Details of localization between phloem parenchyma and CCs, showing localization to PD.
(B) Details of localization between CCs and SEs, showing localization to PD.
(C) Details of localization in SEs, showing localization to the SE reticulum.
(D) Negative control for immunogold labeling.
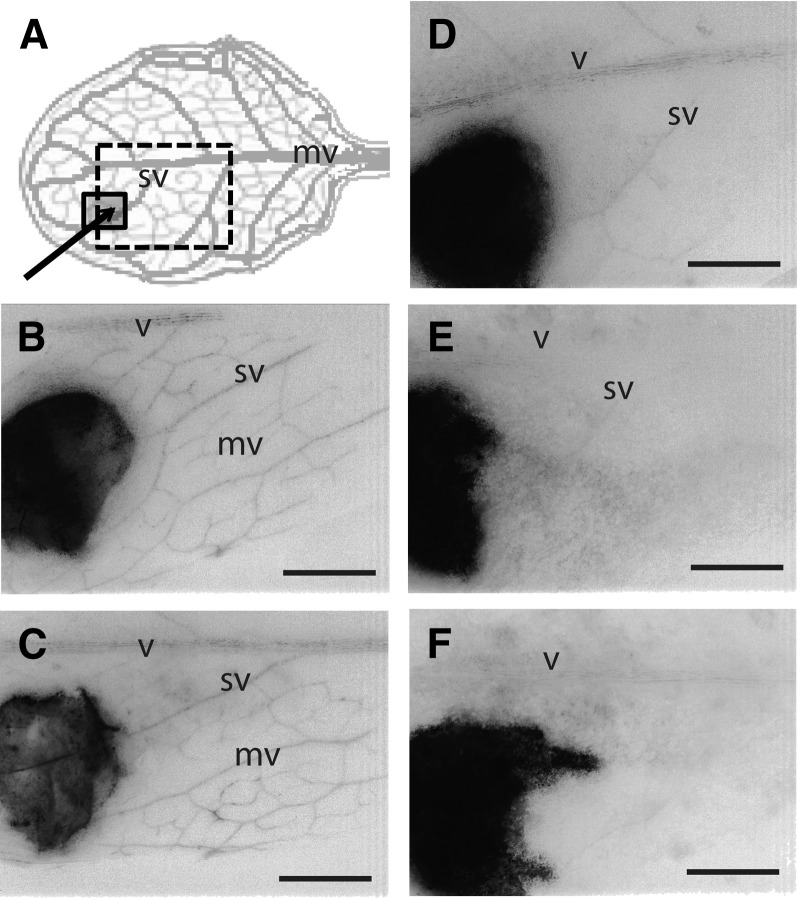
Phloem Transport Assay in Mature Leaves
We investigated the transport activity of the phloem in p35S:NHL-expressing plants, with 5,6-carboxyfluorescein diacetate (CFDA), a phloem-mobile, symplasmic tracer (Oparka et al., 1994). This tracer was spotted onto a small area that had been peeled immediately before application to remove the epidermal and palisade layers (Figure 11A). From the peeled area, the CFDA progressively diffused from cell to cell until it reached the minor veins, where it rapidly entered for transport to the adjacent main veins. The translocation of fluorescence in the leaf vein system was monitored with a binocular microscope coupled to a video camera. On wild-type leaves, fluorescence was rapidly transported in the minor veins of the surrounding unpeeled region and the dye was present in the main veins after 2 min (Figures 11B and 11C). In the young leaves of the NHL26-overexpressing plants, which had not yet reddened, loading of the veins was still observed, although to a much lesser extent than for the wild type, as demonstrated by the faintness of vein fluorescence (Figure 11D). By contrast, no translocation of the tracer was observed in the mature leaves of p35S:NHL26-overexpressing plants (Figures 11E and 11F; see Supplemental Movie 1 online), indicating an arrest of phloem transport.
Figure 11.
NHL26 Overexpression Blocks Phloem Export.
Defect in phloem export in p35S:NHL-expressing plants. The phloem transport assay was performed on mature leaves. The images represent the fluorescence observed 2 min after loading of the CFDA dye.
(A) Schematic representation of the vein pattern on a fully expanded leaf and the treated area.
(B) to (F) Representative binocular microscopy observations of the fluorescence 2 min after the application of CFDA on leaves. The images were obtained by negative conversion and the use of a grayscale in ImageJ. Wild-type leaves ([B] and [C]); green, mildly and strongly reddened leaves sampled from p35S:NHL-overexpressing plants ([D] to [F]). mv, minor vein; sv, secondary vein; v, midvein. Bars = 1 mm.
DISCUSSION
Deregulation of NHL26 Expression Affects Carbohydrate Partitioning and Biomass Production
NHL26 was initially identified as the ortholog of a phloem-specific gene in celery (Vilaine et al., 2003). In Arabidopsis, it encodes a membrane protein that also accumulates in phloem cells. It localizes to the PD and ER at the interfaces between the CCs, the SEs, and the phloem parenchyma cells. Knockout and knockdown lines with altered NHL26 expression presented no particular change in phenotype, suggesting possible redundancy between NHL26 and other members of the NHL family. By contrast, transgenic lines overexpressing NHL26 under the control of the CaMV 35S promoter displayed leaf reddening and slower growth, associated with the accumulation of substantial amounts of Suc, starch, and anthocyanin in mature leaves. At flowering time, the biomass of the rosette leaves was higher than that in the wild type. By contrast, the weight of root and seeds, two important sink organs, were low, resulting in a lower harvest index and a higher shoot-to-root ratio.
The abnormal sugar content of source and sink organs and the lower Suc content of the phloem sap indicated that the observed changes in growth resulted from defective carbohydrate partitioning due to a sugar export defect. Similar phenotypic alterations, with slower growth and sugar accumulation in leaves, have been reported in the Arabidopsis Suc2/pho3 sugar export mutants (Gottwald et al., 2000; Zakhleniuk et al., 2001; Srivastava et al., 2008, 2009), the antisense transgenic tobacco (Nicotiana tabacum) plants with downregulated expression of the Suc transporter SUT1 (Bürkle et al., 1998), and the maize Tdy1 (Braun et al., 2006; Ma et al., 2009), Tdy2 (Baker and Braun, 2008), and sxd1 mutants (Russin et al., 1996; Provencher et al., 2001). The slower shoot growth in the NHL26-overexpressing plants and their greater biomass production are consistent with recent studies in Arabidopsis showing that the growth rate is inversely related to the levels of sugars and starch (Cross et al., 2006; Smith and Stitt, 2007; Sulpice et al., 2009).
Limited Effects on the Phloem Export of Amino Acids and Organic Acids
The discovery of a Suc export defect raised questions about possible effects on the export of other metabolites transported by the phloem. We investigated the metabolite composition of phloem sap in the mature leaves of NHL26-overexpressing and wild-type plants by GC-MS, which confirmed the lower levels of Suc in the phloem sap exudate. Strikingly, the overall amino acid and organic acid contents of the sap were largely unaffected, although differences were observed for some metabolites, such as Pro. Levels of the most abundant amino acid in phloem sap, Gln, which was 10 times more abundant in the leaves of NHL26-overexpressing plants than in those of the wild type, remained stable in phloem sap exudate. Similar observations for the most abundant amino acids and organic acids demonstrated no impairment of their export. Thus, the main effect of NHL26 overexpression was a specific defect in Suc export by the phloem.
Additional Metabolic and Stress Responses Associated with Sugar Accumulation
In addition to Suc and starch, the mature leaves of NHL26-overexpressing plants had high contents of soluble sugars (hexoses, raffinose, galactinol, and maltose) together with high amino acid, organic acid, and protein contents. For example, levels of Gln and Asn, the two main amino acids involved in nitrogen assimilation, were high. Gly, Ser, Thr, and Pro contents were also high, these amino acids often being associated with high levels of raffinose and galactinol (Sulpice et al., 2009). This metabolite profile suggests that excess carbon triggers the synthesis of organic acids, amino acids, and proteins. The high levels of Pro and raffinose, metabolites often associated with stress responses, suggest that the overflow of sugars and metabolic products induced a stress response; indeed, necrotic spots appeared on the more severely affected leaves. We observed an upregulation of the stress response genes PR1 and PR2, consistent with the role of sugars as signal molecules in defense mechanisms (Badur et al., 1994; Herbers et al., 1996; Thibaud et al., 2004; Gómez-Ariza et al., 2007). High levels of α-tocopherol, a potent antioxidant involved in photooxidative stress in Arabidopsis (Havaux et al., 2005), also suggested that high levels of sugar accumulation triggered oxidative stress.
Sugar Accumulation and the Regulation of Transporter Genes
NHL26 overexpression led to an increase in the sugar content of mature leaves associated with downregulation of the expression of SUC2, the uptake transporter, consistent with previous observations that excess sugar in the source leaves can induce the transcriptional repression of sugar uptake transporters (Chiou and Bush, 1998; Vaughn et al., 2002; Wingenter et al., 2010). We also monitored the expression of other transporters involved in the remobilization of nitrogen in source leaves. The expression of AAP2, a phloem amino acid transporter involved in the transfer of amino acids from xylem to phloem (Zhang et al., 2010), was much weaker in NHL26-overexpressing than in wild-type plants. As the amino acid content of the source leaves of NHL26-overexpressing plants was high, our findings suggest that high amino acid content in the phloem may have a negative feedback effect on AAP2 expression. By contrast, the expression of NRT1.7, encoding a phloem nitrate transporter (Fan et al., 2009), was strong in mature leaves, suggesting that nitrate remobilization is triggered to respond to high nitrogen content in the source leaves. Thus, the defects in sugar partitioning observed in NHL26-overexpressing plants directly affected nitrate remobilization and amino acid exchanges between the xylem and phloem.
Feedback Effects on Photosynthesis and the Expression of Photosynthetic Genes
The observation of carbohydrate accumulation in the leaves raised questions about the possible feedback regulation of photosynthesis. We analyzed regulation of the transcription of RBCS in NHL26-overexpressing plants and observed a downregulation consistent with reports of the repression of photosynthetic genes by sugar (Pego et al., 2000). LHCB1 expression was also repressed in NHL26-overexpressing lines. However, chlorophyll content and Fv/Fm value were not affected, suggesting an absence of photosynthetic machinery impairment. The higher protein content of source leaves in the NHL26-overexpressing plants may be sufficient to ensure that ribulose-1,5-bis-phosphate carboxylase/oxygenase content is nonlimiting. As the accumulation of LHCII proteins is little affected by their mRNA levels (Flachmann and Kühlbrandt, 1995), these proteins may also be sufficiently abundant to support efficient photosynthesis. Similarly in Phaseolus vulgaris, sugar accumulation in source leaves, leading to a decrease in the RBCS and LHCII contents of source leaves, has no effect on chlorophyll content and Fv/Fm (Araya et al., 2006). Other mechanisms may thus be triggered, leading to the storage or metabolism of sugars rather than a shutdown of carbon fixation by photosynthesis, as already suggested for the pho3 mutants (Lloyd and Zakhleniuk, 2004). The large excess of amino acids and other nitrogen compounds observed in NHL26-overexpressing plants may prevent photosynthesis feedback regulation by sugars.
Evidence for Compromised Sugar Export between CCs and SEs
Arabidopsis belongs to the Gamalei Type 1-2a group of apoplasmic loaders (Haritatos et al., 2000), with carrier-mediated Suc uptake taking place in CCs. In transgenic lines, in which the overexpression of NHL26 was restricted in the CCs, the phenotypic changes were similar to, but less severe, than those in p35S-NHL–expressing lines. Thus, the misexpression of NHL26 in CCs is sufficient to impair sugar export and the initial events blocking sugar export take place in the CCs. The expression of SUC2, the CC influx transporter, was downregulated in these lines. As excess sugar induces the transcriptional repression of sugar uptake transporters (Chiou and Bush, 1998; Vaughn et al., 2002; Wingenter et al., 2010), this suggests that there is an accumulation of sugars in the CCs and that the step between the CC and SE is blocked. We tried to identify more precisely the steps altered in these plants by performing a phloem transport assay with CFDA. CFDA is a membrane-permeable, nonfluorescent dye that is cleaved by cellular esterases to release carboxyfluorescein (CF), a non-membrane-permeable fluorescent form of the dye. This tracer has been used as a symplasmic tracer (Oparka et al., 1994) and we used it to monitor phloem export in vivo by fluorescence microscopy. In overexpressor plants, the fluorescence signal was barely visible in young leaves and absent from the veins of mature leaves, demonstrating an absence of CF transport in the phloem. However, phloem transport in the sieve tubes was not blocked because we were able to collect phloem sap exudate and to analyze the metabolite content of exudates from NHL26-overexpressing plants. These observations indicate that the steps in sugar export occurring between the CC and the SE were impaired and suggest that the permeability of PD at the interface between CC and SE is compromised, blocking the symplasmic entry of Suc and CF in the SE. Since other classes of metabolites were found in phloem sap exudate, this further suggests that the apoplasmic loading of other solutes in the SE is mostly unchanged.
NHL26, a Phloem Protein Targeted to PD and the ER
We used GFP or CFP fusions to investigate the subcellular distribution of NHL26. In transient expression assays, we obtained a strong, punctate signal in the cell walls, typical of localization to PD. Similar results were obtained in transgenic lines expressing the NHL26:GFP fusion. This was confirmed by the colocalization of NHL26-CFP with PDLP1-GFP, a marker of PD (Thomas et al., 2008). Immunogold labeling confirmed that NHL26 was located in PD. A signal was also detected in the ER and confirmed by immunogold labeling in the sieve element reticulum, suggesting that NHL26 is associated with the ER in addition to PD or that the trafficking of NHL26 to the PD involves the ER. Alternatively as the desmotubule is an appressed ER section in the PD channel, this may indicate a desmotubule location for NHL26, although few desmotubule-associated proteins have been characterized to date, with the exception of a few viral movement proteins (Epel, 2009). The localization of NHL26 in the PD and the blocking of sugar export by its overaccumulation suggest a role for NHL26 in the opening of PD. This would be consistent with the phenotypic changes in overexpressing plants, resembling those observed in transgenic plants constitutively producing movement proteins, also leading to a disruption of sugar allocation (Balachandran et al., 1995; Olesinski et al., 1996; Almon et al., 1997; Herbers et al., 1997; Hofius et al., 2001; Shalitin et al., 2002; Rinne et al., 2005; Kronberg et al., 2007). The simplest explanation is that the overaccumulation of NHL26 modifies PD permeability. The flux of solutes within the PD is thought to occur in the cytoplasmic annulus, a space between the desmotubule and the plasma membrane, crossed by as yet unidentified proteins and predicted to be involved in the control of PD aperture. It has been suggested that the PDs in phloem cells are regulated by pressure, in some cases maintaining carbon balance by closure (Turgeon, 2006). Thus, an overaccumulation of NHL26 in the cytoplasmic annulus of the PD might lead to PD closure, preventing solute movement.
An alternative explanation accounting for the location of NHL26 in the ER is that an overaccumulation of NHL26 in an ER-dependent sorting route disrupts correct sugar compartmentalization, triggering a sugar signaling pathway that blocks sugar export. Sugar signaling plays an important role in the control of plant growth and development (Smeekens et al., 2010; Eveland and Jackson, 2012). For example, it has been shown in tmt1-overexpressing Arabidopsis lines that changes in sugar compartmentalization in the vacuole modify cellular sugar sensing, leading to defects in sugar allocation (Wingenter et al., 2010).
NHL26 Structural Domains
Alignment of the sequences of NHL26 and LEA14, a protein for which structural determinations have been performed (Singh et al., 2005), made it possible to establish a three-dimensional model, with a core α-β fold and two α-helices at its N terminus, one of which was predicted to be a transmembrane domain. NDR1, another member of the NHL family, was also recently shown to display structural similarity to LEA proteins (Knepper et al., 2011). LEA proteins constitute a large family, classified into several loosely defined groups, such as dehydrins, with various functions, protecting enzymatic activities, interacting with sugars, and protecting proteins or membranes (Shih et al., 2008; Caramelo and Iusem, 2009). A dehydrin has already been detected in the PD of Cornus sericea (Karlson et al., 2003). LEAs have been described as intrinsically disordered chaperones involved in the stress responses of plants and animals, with changes in their cellular environment triggering changes in the conformation of these proteins (Tompa and Kovacs, 2010). The core α-β fold of NHL26 is predicted to be located in the ER lumen, the N terminus domain in the cytosol annulus, and the first α-helix is thought to be the transmembrane domain anchoring the protein in the ER membrane. Based on structural similarities to LEA, we can speculate that changes in the conformation of the protein, particularly the core α-β fold present in the ER lumen or within the desmotubule, may be critical for the function of this protein in the PD aperture.
PD and Phloem Transport
The contribution of phloem PD to sugar export in apoplasmic species has been little investigated. It has been suggested that PDs are regulated by pressure, preventing Suc backflow into mesophyll cells or phloem parenchyma cells from the CC and sieve tubes (Turgeon, 2006). Our findings are consistent with NHL26 overexpression modifying PD permeability and with a role of PD in the control of sugar export at the interface between CC and SEs. However, we cannot exclude the possibility that other steps are affected because the effects in p35S:NHL plants were more substantial than those in pPP2:NHL plants. Sugar signaling in the CCs may be impaired in NHL26-overexpressing lines, possibly due to NHL26 being located in the ER of phloem cells. The ER is known to be involved in sorting toward other compartments, such as the vacuole, an important sugar storage compartment (Poustka et al., 2007). NHL26 expression was repressed by Suc and Glc, indicating a direct link with sugar signaling pathways. Strikingly, the export of amino acids and organic acids via the phloem was not blocked, contrasting with the observation that NHL26 overexpression triggered large metabolic changes, with the massive accumulation of amino acids, organic acids, and proteins in source leaves. These findings indicate that there is little diffusion of amino acids through the PD at the interface between CCs and SEs, further supporting the assumption that their loading is mainly performed by plasma membrane transporters (Tegeder and Rentsch, 2010).
Our results provide a new perspective with regard to the role of PD in the export of Suc and amino acids. Our results do not rule out the possibility of a role for as yet uncharacterized sugar signaling pathways triggered in the CCs and interfering with phloem loading, but they highlight the potential role of PD at the interface between CCs and SEs. They also highlight the close link between sugar export and nitrogen metabolism and the role of the phloem in controlling carbon and nitrogen balance.
METHODS
Sequences and Plant Material
Arabidopsis thaliana NHL26 (Arabidopsis Genome Initiative [AGI] accession number At5g53730) is the most closely related ortholog of the celery (Apium graveolens) gene Ag HIN1 (GenBank No. JF776633). The NHL26 gene model, EST, and cDNA were analyzed with The Arabidopsis Information Resource (http://www.Arabidopsis.org/). The promoter sequence was analyzed with the Arabidopsis cis-regulatory element database (AtcisDB, http://Arabidopsis.med.ohio-state.edu/AtcisDB/). The plant membrane protein database Aramemnon (http://aramemnon.botanik.uni-koeln.de/) was used to identify the protein domains, transmembrane regions of the deduced NHL26 protein, and the orientation of its N terminus. Genevestigator https://www.genevestigator.com/gv/plant.jsp and CATdb (http://urgv.evry.inra.fr/CATdb) were used for expression data mining. Arabidopsis mutants affected in the NHL26 gene were identified with T-DNA express (http://signal.salk.edu/cgi-bin/tdnaexpress). One Columbia-0 (Col-0) insertion line and one Wassilewskija-4 T-DNA insertion line were identified, nhl26-1 (Sail633_F02) and nhl26-2 (Flag238_C12), with insertions in the promoter and downstream from the 3′ untranslated region of the gene, respectively. One Landsberg erecta transposon-tagged line, nhl26-3 (CSHL_ET8225), harbored an insertion in the coding sequence (Supplemental Figure 2A online). Plants homozygous for nhl26-1, nhl26-2, and nhl26-3 were genotyped by PCR with specifically designed primers. Transgenic plants expressing the plasmodesmal p35S:PDLP1-GFP reporter gene (Thomas et al., 2008) were kindly provided by Andy Maule (Norwich, UK).
Expression Vectors and amiRNA
The binary vectors for plant transformation, for example, GUS fusions (pNHL:GUS), GFP/CFP fusions (p35S-NHL-GFP, p35S:GFP-NHL, pNHL-NHL-GFP, and pNHL-NHL-CFP), and overexpressors (pPP2:NHL and p35S:NHL), were obtained with Gateway cloning technology (Invitrogen). The At5g53730 gene contains no introns, so both the promoter and coding region were amplified from genomic DNA extracted from Arabidopsis (accession Col-0). The primers used for the first PCR are listed in Supplemental Table 1 online. These primers contain NHL26-specific sequences and recombination site-specific sequences. The second PCR step was performed with the primers attB1 and attB2 (AttB1, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′, AttB2, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′), which reconstituted intact attB recombination sites at the 5′ and 3′ ends of the PCR products. These PCR fragments were introduced into pDONR207 (Invitrogen) by BP recombination and then transferred by LR recombination into destination vectors (listed in Supplemental Table 1 online). GFP and CFP fusions were transferred into pMDC and pGHGWC binary vectors (Curtis and Grossniklaus, 2003; Zhong et al., 2008). The pMDC32 vector (Curtis and Grossniklaus, 2003) was used for overexpression driven by the 35S promoter. For overexpression driven by the CC-specific PP2-A1 promoter, the destination vector pBI101-pPP2A1-R1R2-tNOS was obtained by inserting a 1500-bp fragment carrying the promoter region of At PP2-A1 (AGI No. At4g19840) into the HindIII site of a modified pBI101R1R2-GUS destination vector (Divol et al., 2007), from which the GUS sequence had already been removed. An amiRNA (Schwab et al., 2006) was designed with WMD3-Web MicroRNA Designer (http://wmd3.weigelworld.org; Stephan Ossowski, Joffrey Fitz, Rebecca Schwab, Markus Riester, and Detlef Weigel, personal communication) and inserted into the pRS300 vector, according to the protocol described on WMD3. The targeted sequence was 5′-CTACTCGTATATGCAGCGTAT-3′, corresponding to nucleotide positions 395 to 415 of the coding region, and the amiRNA was 5′-GTACCCGTATATGCAGCGTAA-3′ (reverse complement). The amiRNA precursor was then transferred into the binary vector pMDC32 (Curtis and Grossniklaus, 2003), under the control of the strong constitutive CaMV 35S promoter. All binary vectors were introduced into Agrobacterium tumefaciens C58pMP90 (Koncz and Schell, 1986) by electroporation.
Arabidopsis wild-type (accession Col-0) plants were transformed by the floral dip method (Clough and Bent, 1998), in agrobacteria inoculation medium (5% Suc and 0.005% [v/v] Silvet L-77). Transformants were selected on kanamycin (50 mg/L) or hygromycin (15 mg/L), depending on the binary vector used. For the GUS lines, 14 transgenic lines were examined, eight of which displayed detectable GUS activity. Two T3 homozygous lines presenting a representative GUS pattern were chosen and used for the detailed analysis of expression patterns. For the overexpressor lines, 20 independent transgenic lines were produced for each of the two constructs and screened by RT-PCR to select 10 overexpressing lines. For both constructs, one representative line was selected for subsequent studies. Homozygous T3 seeds resulting from self-fertilization were used for further phenotypic analyses. For the GFP lines, T2 seeds resulting from self-fertilization were used for CLSM.
Growth Conditions
All developmental and growth stages were determined as described by Boyes et al. (2001). For growth in vitro, seeds were surface sterilized and grown on Murashige and Skoog medium supplemented with 1% Suc in growth chambers (18°C, 60% humidity, and 16 h light/8 h dark). Plants were grown in the greenhouse and growth chambers, in soil (Tref Substrates) or sand, and watered with Plant-Prod nutrient solution (Fertil). The number of rosette leaves was determined on plants grown in the greenhouse in long-day conditions (16-h-light/8-h-dark cycle) and harvested at stage 6.10. Seed yield was determined on plants maintained in long-day conditions in growth chambers (150 µE m–2 s–1, 16 h light 23°C, 8 h darkness 18°C, 70% humidity). The harvest index was the ratio of seed mass to total aerial plant mass measured at harvest time. Experiments in short-day conditions (8 h light 23°C, 16 h darkness 18°C, 70% humidity, 150 µE m–2 s–1) were performed in a growth chamber. Roots were sampled from plants grown in sand in the greenhouse, in long-day conditions (16-h-light/8-h-dark cycle). The pots were watered by immersion of their bases in a solution containing 10 mM nitrate, 2.75 mM potassium, 0.5 mM calcium, 0.7 mM chloride, 0.25 mM phosphate, 0.25 mM sulfate, 0.25 mM magnesium, and 0.20 mM sodium, pH 5.5. Shoots and roots were harvested at stage 6.0. For statistical testing, Student's t test was used, with P values <5 × 10−2 considered significant.
RNA Isolation and RT-PCR
Total RNA was isolated from frozen tissue, as previously described (Verwoerd et al., 1989). Reverse transcription was performed with 1 µg total RNA with the Superscript II enzyme (Invitrogen), after DNase treatment (Invitrogen). The primers used for PCR amplification are listed in Supplemental Table 2 online. Ef-1α was used as a reference gene in RT-PCR experiments. Programs were designed such that the PCR products were recovered during the exponential phase (30 cycles for NHL26 and 20 for Ef-1α PCR products, at 50°C). Control reactions were performed, omitting the reverse transcriptase from the initial step, to check for a lack of contamination with genomic DNA. qRT-PCR was performed with the MESA GREEN MasterMix Plus for SYBR assay, according to the manufacturer’s instructions (Eurogentec). Amplification was performed with 1 μL of a 1:10 or 1:20 dilution of cDNA in a total volume of 20 µL: 5 min at 95°C, and 40 cycles of 95°C for 5 s, 55°C for 15 s, and 72°C for 40 s, in an Eppendorf Realplex2 MasterCycler (Eppendorf SARL). A melting curve was obtained to confirm the specificity of the amplification. Relative expression was calculated as a percentage of the expression of three reference genes (TIP41, APT, and UBI10). The robustness of the results was verified with these reference genes, and the data are reported as percentages of TIP41 expression. The results were then recalculated as percentages of the expression in wild-type plants.
Phloem Sap Exudates
The petioles of the mature leaves sampled at flowering time, at stage 6.10, were cut off and recut in buffer and immediately immersed in the collection buffer (50 mM phosphate buffer, pH 7.5, and 10 mM EDTA), as previously described (Beneteau et al., 2010). Exudates of three replicates (three leaves per plant and three plants per replicate) were collected for 2 h and pooled, and the sugar content of the pooled samples was determined.
Anthocyanin Determination and Lugol’s and GUS Staining
The anthocyanin content of fully expanded mature leaves was measured by spectrophotometry, as previously described (Diaz et al., 2006). Starch was detected in mature leaves by direct dipping in Lugol's solution (2% KI and 1% I2 in 0.2 n HCl) for 2 min. Transverse sections (60 µm thick) were cut on a Leica-VT 1000S (Leica Rueil-Malmaison) vibratome after embedding in 8% agarose and were observed by differential interference contrast microscopy (Leica DMRB microscope). GUS activity was assayed histochemically, as previously described (Jefferson et al., 1987), by overnight incubation at 37°C in 1 mg/mL 5-bromo 4-chloro 3-indolyl glucuronide, 100 mM Na2HPO4-NaH2PO4 buffer, pH 7, 0.1% (v/v) Triton X-100, 0.5 mM K4Fe(CN)6, and 0.5 mM K3Fe(CN)6. Samples were washed in water and cleared in 96% ethanol. Thin sections (70 µm thick) were cut on a Leica-VT 1000S vibratome, as described above, and stained with phloroglucinol-HCl (1% [w/v] phloroglucinol in 6 n HCl; VWR Prolabo). Stained sections were observed under a binocular microscope or a Nikon Microphot FXA microscope and photographed with a Jenoptik ProgRes C10 plus digital camera (Clara Vision).
Carbohydrate, Nitrogen, and Starch Content
Soluble sugar (Glc, Fru, and Suc) concentrations were determined enzymatically (saccharose d-glucose d-fructose kit; Boehringer Mannheim). Leaf material was collected 4 h after the beginning of the light period and immediately frozen in liquid nitrogen. Tissues were ground in liquid nitrogen, and soluble sugars were extracted by incubation in 80% ethanol at 80°C for 20 min (500 μL 80% ethanol/100 mg fresh weight). The pellet, kept for starch determination, was resuspended in water and incubated at 100°C for 2 h, and starch was determined after the release of Glc by incubation with α-amylase and amyloglucosidase (Boehringer Mannheim) at 50°C for 3 h in 20 mM acetate buffer, pH 4.6. The protein content of fresh tissue was determined with Bradford reagent (Sigma-Aldrich). Total N and C contents of seeds were determined with an elemental analyzer (Thermoflash 2000; Thermo Scientific).
PSII Activity and Chlorophyll Content
PSII activity was measured with a Handy PEA (Hansatech Instruments), with a 3000 µm m−2 s−1 light flash. Chlorophyll content was measured with a SPAD-502 chlorophyll meter (Konica Minolta). Duplicate measurements were made for each genotype, on 12 individual leaves sampled from the rosette of 8-week-old plants grown in the greenhouse, with and without the redness characteristic of sugar accumulation.
Transient Expression in Nicotiana benthamiana and Arabidopsis
N. benthamiana leaves from 4-week-old plants were used for transient expression experiments (English et al., 1997) in a simplified infiltration buffer (10 mM MES, pH 5.6, 10 mm MgCl2, and 200 µM acetosyringone) with Agrobacterium strains harboring the GFP constructs (p35S:NHL-GFP and p35S:GFP-NHL), which were coexpressed with the P19 viral suppressor of gene silencing to enhance expression (Voinnet et al., 2003). GFP fluorescence was assessed 2 to 3 d after infiltration. For transient expression in Arabidopsis, we used the cotyledons of plantlets grown in vitro (Marion et al., 2008).
CLSM
The fluorescence of GFP and CFP fusions was visualized with a Leica TCS-SP2-AOBS spectral confocal laser scanning microscope. CFP was excited with a 405-nm diode laser, and GFP/chloroplast autofluorescence was excited with the 488-nm line of an argon laser. Emitted fluorescence was detected in the 450- to 520-nm range for CFP constructs, the 500- to 560-nm range for GFP, and the 650- to 700-nm range for chloroplast autofluorescence. Images were recorded and processed with LCS version 2.5 (Leica Microsytems). The colocalization experiments were performed in sequential mode. For plasmolysis treatment, to study fluorescence in the PD of epidermal cells, the leaves of seedlings grown in vitro were incubated for 30 min in 2.5% KCl, 0.2% CaCl2, and 0.7 g/L MES (657 mOsm/kg) and then observed by CLSM. For clear imaging of the leaf minor vein system, the abaxial epidermis was peeled off over a small area with a razor blade (Martens et al., 2006), rinsed, and mounted in water immediately before observation.
Phloem Transport Assay
For phloem transport imaging, a small area (∼25 mm2) from the abaxial surface of fully expanded source leaves, taken from the leaf margin and at some distance from the main vein, was gently peeled away with a razor blade, and 5 μL of CFDA mixed isomers (60 µg mL−1) (Invitrogen) was applied to the surface. The treated leaves were immediately observed under a Nikon SMZ1500 binocular microscope, under UV light with a GFP filter, and the loading of the fluorescent dye was monitored by video recordings every 1.5 s for 2 min.
Transmission Immunoelectron Microscopy
Petioles from 21-d-old pNHL:NHL-GFP Arabidopsis plants were cut in 0.1 M Sörensen phosphate buffer, pH 7.2, and transferred to small plates filled with 20% BSA in phosphate buffer for rapid, high-pressure freezing in a Leica EM-Pact high-pressure freezer. Freeze substitution was performed in a Leica AFS, in anhydrous acetone and 0.1% uranyl acetate, at −90°C for 73 h. After a 20-h linear warm-up to −50°C, samples were rinsed in acetone and then in ethanol. They were infiltrated with resin over a period of 20 h, in a stepwise procedure: 25 to 50 to 75% Lowicryl HM20 in ethanol to 100% HM20. Polymerization was obtained by incubation for 48 h at −50°C and then 48 h at +20°C, under UV light. For immunolabeling, 75-nm-thick sections, mounted on Parlodion-coated nickel grids, were blocked by incubation with 2% BSA and 0.05% Tween 20 in TBS (0.02 M Tris and 0.15 M NaCl) for 30 min. Thin sections were probed with anti-GFP antibodies (Torrey Pines; TP401) diluted 1:250 or 1:500 in TBS with 0.1% BSA and 0.05% Tween 20 (0.1 BT) for 1 h at room temperature. After washing in 0.1 BT, the sections were incubated with anti-rabbit IgG conjugated to 10-nm gold particles (EM.GAR 10; Biocell) diluted 1:30 in 0.1 BT for 1 h at room temperature. After washing, sections were viewed at 80 kV in a FEI CM 10 transmission electron microscope. Digital images were obtained with a lateral XR60 AMT camera.
GC-MS Metabolite Profiling of Leaves and Phloem Exudate
Chemical derivatization and GC-MS metabolite profiling analysis were performed essentially as previously described (Fiehn, 2006). For leaf samples, six independent biological replicates from each line were subjected to metabolic analysis and the experiments were repeated twice, with 50 mg of plant extract for each sample. Rosette leaves were sampled at stage 6.10 and immediately frozen in liquid nitrogen. After grinding in liquid nitrogen, 50 mg of powder was collected. The ground frozen samples were resuspended in 1 mL of frozen (−20°C) water:chloroform:methanol (1:1:2.5) and extracted for 10 min at 4°C, with shaking at 1400 rpm in an Eppendorf Thermomixer. Insoluble material was removed by centrifugation at 20,000g for 5 min and 4 µg of ribitol in 20 μL methanol was added to 900 μL of the supernatant. A 50-μL aliquot of the reaction mixture was dried for 3 h, under vacuum centrifugation, and stored at −80°C.
For phloem sap exudate, three independent biological replicates from each line were subjected to metabolic analysis and the experiments were repeated twice, with 250 μL of phloem exudate for each sample. We then added 1 mL of acetonitrile:isopropanol (1:1) supplemented with 4 µg of ribitol to the exudate. The mixture was incubated for 10 min shaking at 4°C with shaking and was then centrifuged. We dried 100 μL of the supernatant in a vacuum centrifuge.
Vacuum-dried samples were dissolved in 10 μL of methoxylamine (20 mg/mL pyridine) and incubated at 28°C for 90 min, with continuous shaking, in an Eppendorf thermomixer. We then added 90 μL of N-methyl-N-(trimethylsilyl)-trifluoroacetamide (Sigma-Aldrich) and incubated the mixture at 37°C for 30 min. The derivatized samples were kept at room temperature for 150 min before injection. Samples (1 µL) were injected in split-less mode into an Agilent 7890A GC coupled to an Agilent 5975C mass spectrometer. GC-MS analysis was performed on an Rxi-5SilMS column (Restek). The liner (Restek 20994) was changed before each series of 24 samples. The oven temperature ramp was 70°C for 7 min then 10°C/min to 325°C for 4 min (run length 36.5 min). The constant flow rate for helium was 0.7 mL/min. The temperatures were, for the injector, 250°C; the transfer line, 290°C; the source, 250°C; the quadripole, 150°C. Amino acid standards were injected at the beginning and end of the analysis, for the monitoring of derivatization stability. An alkane mixture (C10, C12, C15, C19, C22, C28, C32, and C36) was injected in the middle of the run, for external retention index calibration.
Raw Agilent data files were converted into NetCDF format and analyzed with AMDIS (http://chemdata.nist.gov/mass-spc/amdis/). A custom-built standard library with retention indices and mass spectral collections, generated from the NIST (http://www.nist.gov/srd/mslist.htm), Golm metabolome database (Kopka et al., 2005), Fiehn database (Kind et al., 2009), and standard compounds, was used for metabolite identification. Compounds were quantified using Quanlynx software (Waters). Peak areas were then determined with Quanlynx software (Waters), after conversion of the NetCDF file into masslynx format. TMEV software (http://www.tm4.org/mev.html) was used for statistical analyses of leaf extracts, including univariate analysis by permutation (one-way analysis of variance) to select the significant metabolites (P value < 0.01) and multivariate analysis (hierarchical clustering analysis and principal component analysis). For hierarchical clustering analysis graphical representation of the metabolic changes, we used Genesis (http://genome.tugraz.at/) after log2 transformation. Leaf metabolite data were normalized by dividing each raw data value by the median of all measurements of the experiment for a given metabolite.
For the analysis of phloem sap exudates, amino acid standards were injected at the beginning and end of the analysis, for the quantification of amino acid content, in picomoles per milligram leaf fresh weight. After log2 transformation, the entire data set was normalized by linear regression with the least-rectangles method available from the RVAideMemoire Package, in the R software environment.
Prediction of the Three-Dimensional Structure of NHL26
The three-dimensional structure of NHL26 was predicted by searching for similarity to proteins with solved structures, with I-TASSER (threading/assembly/refinement) (Zhang, 2008; Roy et al., 2010; http://zhanglab.ccmb.med.umich.edu/I-TASSER/). The model was confirmed with the Phyre2 server (Kelley and Sternberg, 2009; http://www.sbg.bio.ic.ac.uk/phyre2/). A three-dimensional model was prepared with MacPyMOL software. The accession numbers for late embryogenesis proteins showing structural similarities to NHL26 are as follows: AGI, At1g01470; PDB id, 1xo8A (LEA14), and AGI, At2g46140.1; PDB id, 1yycA. Both proteins belong to the LEA_2 subgroup (Hundertmark and Hincha, 2008).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers for Arabidopsis sequences: At5g53730 for NHL26, At5g60390 for EF-1αA4, At1g61800 for G6PT, At2g14610 for PR1, At3g57260 for PR2, At4g34270 for TIP41, At1g27450 for APT, At1g22710 for SUC2, At5g09220 for AAP2, At1g69870 for NRT1.7, At1g29920 for LHCB1, At5g38410 for RBCS, At1g01470 for LEA14, At2g46140 for LEA-desiccation-related protein, and JF776633 for celery HIN1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Predicted 3D Model of NHL26.
Supplemental Figure 2. Characterization of nhl26 Mutants.
Supplemental Figure 3. Scatterplot of the Expression of NHL Genes in Phloem Cells and Other Cell Types.
Supplemental Figure 4. Characterization of NHL26-Overexpressing Lines.
Supplemental Figure 5. Metabolic Profiling in NHL26-Overexpressing Lines.
Supplemental Figure 6. Localization of NHL26 in Plasmodesmata.
Supplemental Table 1. Description of the Binary Vectors Derived from the NHL26 Gene Sequence.
Supplemental Table 2. Primers Used for RT-PCR Experiments.
Supplemental Movie 1. CFDA Phloem-Export Assay on Mature Leaves of Wild-Type and p35S:NHL-Overexpressing Plants.
Supplementary Material
Acknowledgments
We thank Rémi Lemoine and Catherine Bellini for critical reading of the article. We also thank Andy Maule for the gift of Arabidopsis p35S:PDLP1-GFP–expressing plants. We thank Olivier Grandjean for his assistance with CLSM, Céline Masclaux-Daubresse and Marie Laure Martin-Magniette for stimulating discussions, Anne Marmagne for her assistance with C and N measurements in seeds, Nelly Wolff for her assistance with cytochemistry, and Maelle Jaouannet for her assistance with the initial characterization of the plant materials. We also thank Hervé Ferry and Bruno Letarnec for their work in the greenhouses. P.K. was supported by a Marie Curie fellowship. This work was sponsored by a grant from Institut National de la Recherche Agronomique–FORMAS.
AUTHOR CONTRIBUTIONS
F.V. was the principal investigator, although each author contributed to the experimental work. P.K. was involved in the initial physiological characterizations, G.C. conducted the metabolome studies, T.C. and L.G. did the CLSM studies, and L.B. was responsible for preparing the plant material and collecting phloem exudate. T.C. developed a protocol for the observation of the phloem tissues in vivo by CLSM and the phloem transport assay. B.B. carried out the transmission electron microscopy experiments. S.D. wrote the article and designed the experiments, together with F.V.
Glossary
- SE
sieve element
- CC
companion cell
- ER
endoplasmic reticulum
- PDB
Protein Data Bank
- qRT-PCR
quantitative RT-PCR
- GUS
β-glucuronidase
- amiRNA
artificial microRNA
- CaMV
cauliflower mosaic virus
- PSII
photosystem II
- GC-MS
gas chromatography–mass spectrometry
- CFP
cyan fluorescent protein
- CLSM
confocal laser scanning microscopy
- GFP
green fluorescent protein
- PD
plasmodesmata
- CFDA
5,6-carboxyfluorescein diacetate
- CF
carboxyfluorescein
- AGI
Arabidopsis Genome Initiative
- Col-0
Columbia-0
References
- Ainsworth E.A., Bush D.R. (2011). Carbohydrate export from the leaf: A highly regulated process and target to enhance photosynthesis and productivity. Plant Physiol. 155: 64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almon E., Horowitz M., Wang H.L., Lucas W.J., Zamski E., Wolf S. (1997). Phloem-specific expression of the tobacco mosaic virus movement protein alters carbon metabolism and partitioning in transgenic potato plants. Plant Physiol. 115: 1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya T., Noguchi K., Terashima I. (2006). Effects of carbohydrate accumulation on photosynthesis differ between sink and source leaves of Phaseolus vulgaris L. Plant Cell Physiol. 47: 644–652 [DOI] [PubMed] [Google Scholar]
- Ayre B.G. (2011). Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol. Plant 4: 377–394 [DOI] [PubMed] [Google Scholar]
- Badur R., Herbers K., Monke G., Ludewig F., Sonnewald U. (1994). Induction of pathogenesis-related proteins in sugar accumulating tobacco-leaves. Photosynthetica 30: 575–582 [Google Scholar]
- Baker R.F., Braun D.M. (2008). Tie-dyed2 functions with tie-dyed1 to promote carbohydrate export from maize leaves. Plant Physiol. 146: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S., Hull R.J., Vaadia Y., Wolf S., Lucas W.J. (1995). Alteration in carbon partitioning induced by the movement protein of tobacco mosaic virus originates in the mesophyll and is independent of change in the plasmodesmal size exclusion limit. Plant Cell Environ. 18: 1301–1310 [Google Scholar]
- Beneteau J., Renard D., Marché L., Douville E., Lavenant L., Rahbé Y., Dupont D., Vilaine F., Dinant S. (2010). Binding properties of the N-acetylglucosamine and high-mannose N-glycan PP2-A1 phloem lectin in Arabidopsis. Plant Physiol. 153: 1345–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun D.M., Ma Y., Inada N., Muszynski M.G., Baker R.F. (2006). tie-dyed1 regulates carbohydrate accumulation in maize leaves. Plant Physiol. 142: 1511–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürkle L., Hibberd J.M., Quick W.P., Kühn C., Hirner B., Frommer W.B. (1998). The H+-sucrose cotransporter NtSUT1 is essential for sugar export from tobacco leaves. Plant Physiol. 118: 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo J.J., Iusem N.D. (2009). When cells lose water: Lessons from biophysics and molecular biology. Prog. Biophys. Mol. Biol. 99: 1–6 [DOI] [PubMed] [Google Scholar]
- Chen L.-Q., Qu X.-Q., Hou B.-H., Sosso D., Osorio S., Fernie A.R., Frommer W.B. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211 [DOI] [PubMed] [Google Scholar]
- Chiou T.-J., Bush D.R. (1998). Sucrose is a signal molecule in assimilate partitioning. Proc. Natl. Acad. Sci. USA 95: 4784–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cross J.M., von Korff M., Altmann T., Bartzetko L., Sulpice R., Gibon Y., Palacios N., Stitt M. (2006). Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiol. 142: 1574–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C., Saliba-Colombani V., Loudet O., Belluomo P., Moreau L., Daniel-Vedele F., Morot-Gaudry J.-F., Masclaux-Daubresse C. (2006). Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol. 47: 74–83 [DOI] [PubMed] [Google Scholar]
- Dinant S., Clark A.M., Zhu Y., Vilaine F., Palauqui J.C., Kusiak C., Thompson G.A. (2003). Diversity of the superfamily of phloem lectins (phloem protein 2) in angiosperms. Plant Physiol. 131: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant S., Lemoine R. (2010). The phloem pathway: New issues and old debates. C. R. Biol. 333: 307–319 [DOI] [PubMed] [Google Scholar]
- Divol F., Vilaine F., Thibivilliers S., Amselem J., Palauqui J.-C., Kusiak C., Dinant S. (2005). Systemic response to aphid infestation by Myzus persicae in the phloem of Apium graveolens. Plant Mol. Biol. 57: 517–540 [DOI] [PubMed] [Google Scholar]
- Divol F., Vilaine F., Thibivilliers S., Kusiak C., Sauge M.H., Dinant S. (2007). Involvement of the xyloglucan endotransglycosylase/hydrolases encoded by celery XTH1 and Arabidopsis XTH33 in the phloem response to aphids. Plant Cell Environ. 30: 187–201 [DOI] [PubMed] [Google Scholar]
- Dumez S., Wattebled F., Dauvillee D., Delvalle D., Planchot V., Ball S.G., D’Hulst C. (2006). Mutants of Arabidopsis lacking starch branching enzyme II substitute plastidial starch synthesis by cytoplasmic maltose accumulation. Plant Cell 18: 2694–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English J., Davenport G., Elmayan T., Vaucheret H., Baulcombe D. (1997). Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J. 12: 597–603 [Google Scholar]
- Epel B.L. (2009). Plant viruses spread by diffusion on ER-associated movement-protein-rafts through plasmodesmata gated by viral induced host beta-1,3-glucanases. Semin. Cell Dev. Biol. 20: 1074–1081 [DOI] [PubMed] [Google Scholar]
- Eveland A.L., Jackson D.P. (2012). Sugars, signalling, and plant development. J. Exp. Bot. 63: 3367–3377 [DOI] [PubMed] [Google Scholar]
- Fan S.-C., Lin C.-S., Hsu P.-K., Lin S.-H., Tsay Y.-F. (2009). The Arabidopsis nitrate transporter NRT1.7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21: 2750–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. (2006). Metabolite profiling in Arabidopsis. Methods Mol. Biol. 323: 439–447 [DOI] [PubMed] [Google Scholar]
- Flachmann R., Kühlbrandt W. (1995). Accumulation of plant antenna complexes is regulated by post-transcriptional mechanisms in tobacco. Plant Cell 7: 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil L., Yaron I., Shalitin D., Sauer N., Turgeon R., Wolf S. (2011). Sucrose transporter plays a role in phloem loading in CMV-infected melon plants that are defined as symplastic loaders. Plant J. 66: 366–374 [DOI] [PubMed] [Google Scholar]
- Gómez-Ariza J., Campo S., Rufat M., Estopà M., Messeguer J., San Segundo B., Coca M. (2007). Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related PRms protein in rice plants. Mol. Plant Microbe Interact. 20: 832–842 [DOI] [PubMed] [Google Scholar]
- Gonzali S., Loreti E., Solfanelli C., Novi G., Alpi A., Perata P. (2006). Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. J. Plant Res. 119: 115–123 [DOI] [PubMed] [Google Scholar]
- Gottwald J.R., Krysan P.J., Young J.C., Evert R.F., Sussman M.R. (2000). Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. USA 97: 13979–13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haritatos E., Medville R., Turgeon R. (2000). Minor vein structure and sugar transport in Arabidopsis thaliana. Planta 211: 105–111 [DOI] [PubMed] [Google Scholar]
- Havaux M., Eymery F., Porfirova S., Rey P., Dörmann P. (2005). Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17: 3451–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbers K., Meuwly P., Métraux J.-P., Sonnewald U. (1996). Salicylic acid-independent induction of pathogenesis-related protein transcripts by sugars is dependent on leaf developmental stage. FEBS Lett. 397: 239–244 [DOI] [PubMed] [Google Scholar]
- Herbers K., Tacke E., Hazirezaei M., Krause K.-P., Melzer M., Rohde W., Sonnewald U. (1997). Expression of a luteoviral movement protein in transgenic plants leads to carbohydrate accumulation and reduced photosynthetic capacity in source leaves. Plant J. 12: 1045–1056 [DOI] [PubMed] [Google Scholar]
- Hofius D., Herbers K., Melzer M., Omid A., Tacke E., Wolf S., Sonnewald U. (2001). Evidence for expression level-dependent modulation of carbohydrate status and viral resistance by the potato leafroll virus movement protein in transgenic tobacco plants. Plant J. 28: 529–543 [DOI] [PubMed] [Google Scholar]
- Hundertmark M., Hincha D.K. (2008). LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlson D.T., Fujino T., Kimura S., Baba K., Itoh T., Ashworth E.N. (2003). Novel plasmodesmata association of dehydrin-like proteins in cold- acclimated Red-osier dogwood (Cornus sericea). Tree Physiol. 23: 759–767 [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J. (2009). Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kind T., Wohlgemuth G., Lee Y., Lu Y., Palazoglu M., Shahbaz S., Fiehn O. (2009). FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 81: 10038–10048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knepper C., Savory E.A., Day B. (2011). Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol. 156: 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396 [Google Scholar]
- Kopka J., et al. (2005). GMD@CSB.DB: The Golm metabolome database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Kronberg K., Vogel F., Rutten T., Hajirezaei M.-R., Sonnewald U., Hofius D. (2007). The silver lining of a viral agent: Increasing seed yield and harvest index in Arabidopsis by ectopic expression of the potato leaf roll virus movement protein. Plant Physiol. 145: 905–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S., Tegeder M., Throne-Holst M., Frommer W.B., Patrick J.W. (2003). Phloem loading and unloading of sugars and amino acids. Plant Cell Environ. 26: 37–56 [Google Scholar]
- Le Hir R., Beneteau J., Bellini C., Vilaine F., Dinant S. (2008). Gene expression profiling: Keys for investigating phloem functions. Trends Plant Sci. 13: 273–280 [DOI] [PubMed] [Google Scholar]
- Lloyd J.C., Zakhleniuk O.V. (2004). Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J. Exp. Bot. 55: 1221–1230 [DOI] [PubMed] [Google Scholar]
- Ma Y., Baker R.F., Magallanes-Lundback M., DellaPenna D., Braun D.M. (2008). Tie-dyed1 and sucrose export defective1 act independently to promote carbohydrate export from maize leaves. Planta 227: 527–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Slewinski T.L., Baker R.F., Braun D.M. (2009). Tie-dyed1 encodes a novel, phloem-expressed transmembrane protein that functions in carbohydrate partitioning. Plant Physiol. 149: 181–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Song W., Sage T.L., DellaPenna D. (2006). Tocopherols play a crucial role in low-temperature adaptation and Phloem loading in Arabidopsis. Plant Cell 18: 2710–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion J., Bach L., Bellec Y., Meyer C., Gissot L., Faure J.-D. (2008). Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 56: 169–179 [DOI] [PubMed] [Google Scholar]
- Martens H.J., Roberts A.G., Oparka K.J., Schulz A. (2006). Quantification of plasmodesmatal endoplasmic reticulum coupling between sieve elements and companion cells using fluorescence redistribution after photobleaching. Plant Physiol. 142: 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M.E., Jang C.J.H., Holtan H.E., Repetti P.P., Galbraith D.W., Girke T., Bailey-Serres J. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesinski A.A., Almon E., Navot N., Perl A., Galun E., Lucas W.J., Wolf S. (1996). Tissue-specific expression of the tobacco mosaic virus movement protein in transgenic potato plants alters plasmodesmal function and carbohydrate partitioning. Plant Physiol. 111: 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka K.J., Duckett C.M., Prior D.A.M., Fisher D.B. (1994). Real-time imaging of phloem unloading in the root tip of Arabidopsis. Plant J. 6: 759–766 [Google Scholar]
- Paul M.J., Foyer C.H. (2001). Sink regulation of photosynthesis. J. Exp. Bot. 52: 1383–1400 [DOI] [PubMed] [Google Scholar]
- Pego J.V., Kortstee A.J., Huijser C., Smeekens S.C. (2000). Photosynthesis, sugars and the regulation of gene expression. J. Exp. Bot. 51 (Spec. No.): 407–416 [DOI] [PubMed] [Google Scholar]
- Poustka F., Irani N.G., Feller A., Lu Y., Pourcel L., Frame K., Grotewold E. (2007). A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol. 145: 1323–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher L.M., Miao L., Sinha N., Lucas W.J. (2001). Sucrose export defective1 encodes a novel protein implicated in chloroplast-to-nucleus signaling. Plant Cell 13: 1127–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie E.A., Turgeon R. (2009). A comprehensive picture of phloem loading strategies. Proc. Natl. Acad. Sci. USA 106: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber T.L., Plotnikova J.M., Dewdney J., Rogers E.E., Wood W., Ausubel F.M. (1998). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16: 473–485 [DOI] [PubMed] [Google Scholar]
- Rinne P.L.H., van den Boogaard R., Mensink M.G.J., Kopperud C., Kormelink R., Goldbach R., van der Schoot C. (2005). Tobacco plants respond to the constitutive expression of the tospovirus movement protein NS(M) with a heat-reversible sealing of plasmodesmata that impairs development. Plant J. 43: 688–707 [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. (2010). I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russin W.A., Evert R.F., Vanderveer P.J., Sharkey T.D., Briggs S.P. (1996). Modification of a specific class of plasmodesmata and loss of sucrose export ability in the sucrose export defective1 maize mutant. Plant Cell 8: 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N. (2007). Molecular physiology of higher plant sucrose transporters. FEBS Lett. 581: 2309–2317 [DOI] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin D., Wang Y., Omid A., Gal-On A., Wolf S. (2002). Cucumber mosaic virus movement protein affects sugar metabolism and transport in tobacco and melon plants. Plant Cell Environ. 25: 989–997 [Google Scholar]
- Shih M.-d., Hoekstra F.A., Hsing Y.-I.C. (2008). Late embryogenesis abundant proteins. In Advances in Botanical Research, Vol. 48, J.-C. Kader and M. Delseny, eds (Amsterdam: Elsevier Academic Press), pp. 211–255 [Google Scholar]
- Singh S., Cornilescu C.C., Tyler R.C., Cornilescu G., Tonelli M., Lee M.S., Markley J.L. (2005). Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stress-related protein. Protein Sci. 14: 2601–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölund R.D. (1997). The phloem sieve element: A river runs through it. Plant Cell 9: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski T.L., Baker R.F., Stubert A., Braun D.M. (2012). Tie-dyed2 encodes a callose synthase that functions in vein development and affects symplastic trafficking within the phloem of maize leaves. Plant Physiol. 160: 1540–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S., Ma J., Hanson J., Rolland F. (2010). Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13: 274–279 [DOI] [PubMed] [Google Scholar]
- Smith A.M., Stitt M. (2007). Coordination of carbon supply and plant growth. Plant Cell Environ. 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Srivastava A.C., Dasgupta K., Ajieren E., Costilla G., McGarry R.C., Ayre B.G. (2009). Arabidopsis plants harbouring a mutation in AtSUC2, encoding the predominant sucrose/proton symporter necessary for efficient phloem transport, are able to complete their life cycle and produce viable seed. Ann. Bot. (Lond.) 104: 1121–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A.C., Ganesan S., Ismail I.O., Ayre B.G. (2008). Functional characterization of the Arabidopsis AtSUC2 sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol. 148: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R., Sauer N. (1996). The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot. Acta 109: 299–306 [Google Scholar]
- Sulpice R., et al. (2009). Starch as a major integrator in the regulation of plant growth. Proc. Natl. Acad. Sci. USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M., Rentsch D. (2010). Uptake and partitioning of amino acids and peptides. Mol. Plant 3: 997–1011 [DOI] [PubMed] [Google Scholar]
- Thibaud M.-C., Gineste S., Nussaume L., Robaglia C. (2004). Sucrose increases pathogenesis-related PR-2 gene expression in Arabidopsis thaliana through an SA-dependent but NPR1-independent signaling pathway. Plant Physiol. Biochem. 42: 81–88 [DOI] [PubMed] [Google Scholar]
- Thomas C.L., Bayer E.M., Ritzenthaler C., Fernandez-Calvino L., Maule A.J. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P., Kovacs D. (2010). Intrinsically disordered chaperones in plants and animals. Biochem. Cell Biol. 88: 167–174 [DOI] [PubMed] [Google Scholar]
- Truernit E., Sauer N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of beta-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 196: 564–570 [DOI] [PubMed] [Google Scholar]
- Turgeon R. (2006). Phloem loading: How leaves gain their independence. Bioscience 56: 15–24 [Google Scholar]
- Turgeon, R., and Ayre, B.G. (2005). Pathways and mechanisms of phloem loading. In Vascular Transport in Plants, N.M. Holbrook and M.A. Zwieniecki, eds (Amsterdam: Elsevier Academic Press), pp. 45–67. [Google Scholar]
- van Bel A.J.E. (2003). The phloem, a miracle of ingenuity. Plant Cell Environ. 26: 125–149 [Google Scholar]
- Varet A., Parker J., Tornero P., Nass N., Nürnberger T., Dangl J.L., Scheel D., Lee J. (2002). NHL25 and NHL3, two NDR1/HIN1-1ike genes in Arabidopsis thaliana with potential role(s) in plant defense. Mol. Plant Microbe Interact. 15: 608–616 [DOI] [PubMed] [Google Scholar]
- Vaughn M.W., Harrington G.N., Bush D.R. (2002). Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc. Natl. Acad. Sci. USA 99: 10876–10880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd T.C., Dekker B.M., Hoekema A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilaine F., Palauqui J.C., Amselem J., Kusiak C., Lemoine R., Dinant S. (2003). Towards deciphering phloem: A transcriptome analysis of the phloem of Apium graveolens. Plant J. 36: 67–81 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wingenter K., Schulz A., Wormit A., Wic S., Trentmann O., Hoermiller I.I., Heyer A.G., Marten I., Hedrich R., Neuhaus H.E. (2010). Increased activity of the vacuolar monosaccharide transporter TMT1 alters cellular sugar partitioning, sugar signaling, and seed yield in Arabidopsis. Plant Physiol. 154: 665–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhleniuk O.V., Raines C.A., Lloyd J.C. (2001). pho3: A phosphorus-deficient mutant of Arabidopsis thaliana (L.) Heynh. Planta 212: 529–534 [DOI] [PubMed] [Google Scholar]
- Zhang L., Tan Q., Lee R., Trethewy A., Lee Y.-H., Tegeder M. (2010). Altered xylem-phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. Plant Cell 22: 3603–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Craig J.C., Petzold H.E., Dickerman A.W., Beers E.P. (2005). The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 138: 803–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M.S., Takahashi H., Miyazaki A., Hamamoto H., Shah J., Yamaguchi I., Kusano T. (2004). Up-regulation of Arabidopsis thaliana NHL10 in the hypersensitive response to Cucumber mosaic virus infection and in senescing leaves is controlled by signalling pathways that differ in salicylate involvement. Planta 218: 740–750 [DOI] [PubMed] [Google Scholar]
- Zhong S., Lin Z., Fray R.G., Grierson D. (2008). Improved plant transformation vectors for fluorescent protein tagging. Transgenic Res. 17: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.