A microscopy screen for endomembrane defects identified an Arabidopsis N-myristoyltransferase (NMT1) mutant exhibiting reduced activity of the expressed protein. Characterization of this mutant highlights a postembryonic role for NMT1 in the functional and morphological integrity of the Golgi predominantly challenging the Golgi membrane/cytosol partitioning of ADP-ribosylation factors.
Abstract
N-myristoylation is a crucial irreversible eukaryotic lipid modification allowing a key subset of proteins to be targeted at the periphery of specific membrane compartments. Eukaryotes have conserved N-myristoylation enzymes, involving one or two N-myristoyltransferases (NMT1 and NMT2), among which NMT1 is the major enzyme. In the postembryonic developmental stages, defects in NMT1 lead to aberrant cell polarity, flower differentiation, fruit maturation, and innate immunity; however, no specific NMT1 target responsible for such deficiencies has hitherto been identified. Using a confocal microscopy forward genetics screen for the identification of Arabidopsis thaliana secretory mutants, we isolated STINGY, a recessive mutant with defective Golgi traffic and integrity. We mapped STINGY to a substitution at position 160 of Arabidopsis NMT1 (NMT1A160T). In vitro kinetic studies with purified NMT1A160T enzyme revealed a significant reduction in its activity due to a remarkable decrease in affinity for both myristoyl-CoA and peptide substrates. We show here that this recessive mutation is responsible for the alteration of Golgi traffic and integrity by predominantly affecting the Golgi membrane/cytosol partitioning of ADP-ribosylation factor proteins. Our results provide important functional insight into N-myristoylation in plants by ascribing postembryonic functions of Arabidopsis NMT1 that involve regulation of the functional and morphological integrity of the plant endomembranes.
INTRODUCTION
The acquisition of cotranslational and posttranslational modifications on proteins is fundamental for the function of a large portion of the eukaryotic proteome. Protein N-myristoylation (MYR) is one of the most critical lipid modifications, and it is catalyzed by myristoyl CoA:protein N-myristoyltransferases (NMTs) (Giglione et al., 2004; Meinnel et al., 2006). Protein MYR is necessary to target crucial proteins to membranes; most of the time, such proteins belong to signal transduction cascades, including G proteins (Resh, 1999). MYR implies the irreversible addition of a saturated C:14 to an N-terminal Gly of some proteins (2 to 4%) after their N-terminal Met removal (Johnson et al., 1994; Martinez et al., 2008; Meinnel and Giglione, 2008). MYR occurs mostly cotranslationally, although a few cases of posttranslational MYR have also been reported (Wilcox et al., 1987; Glover et al., 1997; Martin et al., 2011). Lower and higher eukaryotes share very similar MYR enzyme systems, involving one or two NMTs (NMT1 and NMT2), among which NMT1, the major enzyme, has been studied in many organisms (Johnson et al., 1994; Bhatnagar et al., 2001; Boisson et al., 2003; Pierre et al., 2007). Interspecies studies of various NMTs indicate that they share common characteristics, although they are not completely redundant because of distinctive peptide substrate specificities (Heuckeroth et al., 1988; Towler et al., 1988; Rajala et al., 2000; Farazi et al., 2001a; Boisson et al., 2003; Pierre et al., 2007). For instance, compared with animals and fungi, plants have a higher number of N-myristoylated (MYRed) proteins (Martinez et al., 2008), suggesting that plants have widely adopted this modification most likely through evolution-dependent gene duplications. This makes the plant MYRed targets more frequent than other eukaryotes.
Complete inactivation of NMT1 genes has been shown to impair cell viability in protists and worms (Duronio et al., 1989; Kamath et al., 2003; Price et al., 2003, 2005a) and to affect early embryonic development in higher eukaryotes, including animals and plants (Ntwasa et al., 2001; Yang et al., 2005; Pierre et al., 2007). In Arabidopsis thaliana, using trans-kingdom complementation experiments in the nmt1-1 null background and in vitro substrate specificity of orthologous NMTs, it was shown that NMT1 defects of the shoot apical meristem development were directly linked to a unique target in the myristoylome: the protein kinase SnRK1 (Pierre et al., 2007). Depending on the organism, alteration of NMT expression or activity has also been shown to be tightly associated with several types of tumor progression or with aberrant postembryonic defects. In plants, a low level of Arabidopsis NMT1 impairs flower differentiation, fruit maturation, fertility, and innate immunity (Pierre et al., 2007) and induces dwarf phenotypes (Qi et al., 2000). The molecular bases of these phenotypes (i.e., the NMT primary targets that are responsible for the absolute requirement of NMT for cell growth at postembryonic stages) are unknown. The diversity of the observed phenotypes suggests that specific NMT target proteins are potentially involved in different processes involving lipid modification.
Here, we report the characterization of a partial loss-of-function of NMT Arabidopsis NMT1 identified through a confocal microscopy–based forward genetics screen aimed at identifying alleles related to the secretory pathway in 10-d-old cotyledons of Arabidopsis seedlings. The mutant bears a single base pair substitution in the NMT1 coding sequence, which is responsible for the transition of an Ala to Thr residue in position 160 corresponding to the catalytic region of NMT1 (NMT1A160T). In vitro kinetic studies with the purified mutated form NMT1A160T enzyme revealed significant reduction of its activity due to a strong decrease in the affinity for both myristoyl-CoA and substrate peptides. These data are supported by modeling the substitution on the Arabidopsis NMT1 three-dimensional structure. Despite a reduced in vitro activity, no massive differences were observed when the whole soluble and membrane proteome of the STINGY mutant was compared with that of the wild type, suggesting that the partial loss of function of NMT1A160T enzyme does not affect the whole set of MYRed proteins, the so-called myristoylome. We show here that this recessive mutation is responsible for the alteration in Golgi traffic and integrity by predominantly affecting the Golgi membrane/cytosol partitioning of ADP-ribosylation factor (Arf) proteins. Our results provide important functional insight into MYR in plants by ascribing postembryonic functions of Arabidopsis NMT1 that involve regulation of the functional and morphological integrity of the plant endomembranes.
RESULTS
Isolation of STINGY through a Confocal Microscopy Screen of Ethyl Methanesulfonate–Mutagenized Arabidopsis Seedlings Expressing SEC-RFP
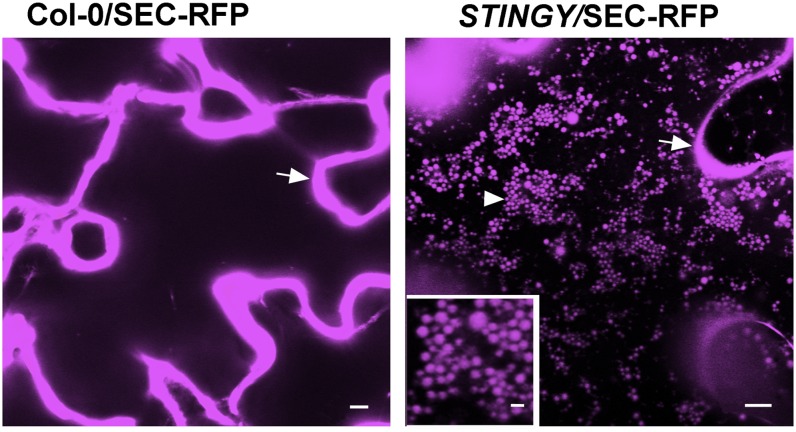
To identify regulatory factors in secretion, we performed a confocal microscopy–based forward genetics screen on Arabidopsis Columbia-0 (Col-0) ethyl methanesulfonate (EMS) mutants expressing the secreted (SEC)-red fluorescent protein fusion (Col-0/SEC-RFP). This is a bulk-flow reporter based on a fusion of monomeric red fluorescent protein 1 (mRFP1; Campbell et al., 2002) fused to the cleavable sporamin signal peptide, which accumulates in the apoplast in nonmutagenized Col-0 background (Figure 1) (Faso et al., 2009b). The aim of the screen was to identify key regulators of constitutive secretion mutants by scoring individuals with the marker partially retained within the epidermal cells of cotyledons. We screened 60 M2 seedlings collected from each of 303 M1 SEC-RFP lines. One of the mutants that we identified and named STINGY showed partial accumulation of SEC-RFP inside globular intracellular structures compared with nonmutagenized Col-0/SEC-RFP (Figure 1). With the exception of a late flowering phenotype, STINGY did not show obvious defects in plant growth and development under normal growth conditions (see Supplemental Figure 1 online).
Figure 1.
Identification of STINGY.
SEC-RFP localizes to the apoplast in Col-0/SEC-RFP (arrow); however, in the STINGY mutant (STINGY/SEC-RFP), SEC-RFP localizes also in intracellular globular structures (arrowhead). Inset: magnified area of the main image. Bars = 5 μm; bar in the inset = 1 μm.
STINGY Has Defects in Endoplasmic Reticulum/Golgi Traffic
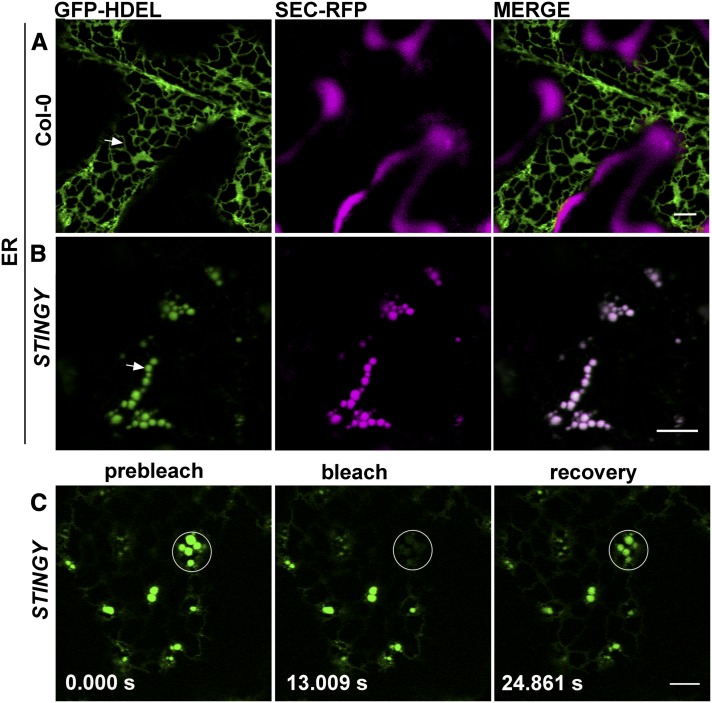
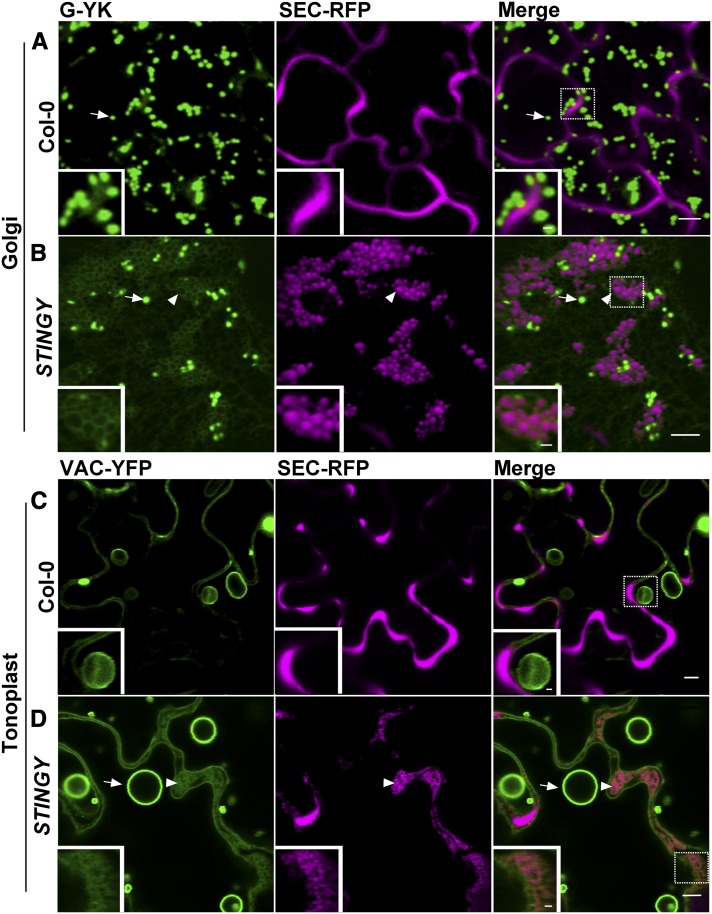
To establish the nature of the intracellular structures highlighted by SEC-RFP in STINGY, we crossed the mutant with Col-0 stably expressing the soluble endoplasmic reticulum (ER) lumen marker GFP-HDEL (i.e., sporamin signal peptide fused to green fluorescent protein [GFP] bearing the ER retention signal for soluble proteins; Brandizzi et al., 2003; Figure 2A). Analyses of the F2 segregants showed that the GFP-HDEL fluorescence overlapped with the intracellular SEC-RFP structures (Figure 2B), suggesting that the aberrant structures observed in the mutant are a modified ER network in which SEC-RFP is partially accumulated. To test whether the structures are interconnected or represent fragmented ER, we performed fluorescence recovery after photobleaching (FRAP) analyses of GFP-HDEL in STINGY (Brandizzi et al., 2002) with the expectation that recovery of fluorescence in the bleached areas would occur only if the structures were not isolated. Indeed, we found recovery of fluorescence within the bleached regions (Figure 2C), indicating that the aberrant structures labeled by SEC-RFP and GFP-HDEL belong to morphologically altered tubules within a modified ER network. The accumulation of soluble secretory proteins in the aberrant ER structures suggested the occurrence of defects in ER/Golgi traffic that could lead to partial retention of SEC-RFP in the ER. To test for traffic defects, we analyzed F2 segregants showing the STINGY phenotype derived from crosses of STINGY with Col-0 stably expressing either the Golgi marker G-YK (i.e., a yellow fluorescent protein [YFP] fusion to the Glycine max mannosidase I; Nelson et al., 2007) or the tonoplast marker VAC-YFP (a YFP fusion to a γ-tonoplast intrinsic protein; Nelson et al., 2007). Compared with the respective nonmutagenized Col-0 controls (Figures 3A and 3C), we found that in STINGY, G-YK and VAC-YFP signals surrounded the intracellular SEC-RFP structures (Figures 3B and 3D), which are part of an aberrant ER network (Figure 2). Together, these results confirm that the STINGY globular structures belong to a modified ER network and indicate that ER/Golgi traffic is partially disrupted in STINGY.
Figure 2.
SEC-RFP Distribution in STINGY Coincides with That of a Soluble ER Marker into Globular Structures That Are Interconnected.
(A) Confocal optical section of a SEC-RFP cotyledon epidermal cell expressing the ER marker GFP-HDEL, which localizes to the lumen of the ER and defines the ER network.
(B) Confocal optical section of STINGY cotyledon epidermal cells expressing the GFP-HDEL marker showing an overlap of the RFP and GFP signals in the globular structures of the mutant.
(C) Photobleaching of globular structures and recovery of fluorescence indicate that the globular structures are not isolated. Time of frame acquisition is indicated on individual panels in seconds.
Bars = 5 μm.
Figure 3.
The STINGY Globular Structures Are Contained in a Modified ER Membrane in Which Golgi and Tonoplast Proteins Are Partially Retained.
(A) and (B) Confocal optical section of Col0/SEC-RFP control coexpressing the Golgi marker G-YK showing that the marker is localized to the Golgi apparatus ([A], arrow), as expected. However, STINGY/SEC-RFP coexpressing G-YK shows partial accumulation of the Golgi marker into a modified ER network enwrapping the globular STINGY structures ([B], arrowhead and inset).
(C) and (D) Confocal images of Col-0/SEC-RFP coexpressing the tonoplast marker VAC-YFP showing tonoplast (arrow) and bulb structures (arrowhead). In the STINGY mutant, the marker partially accumulated in circular structures (arrowhead and inset).
Bars = 5 μm; bars in the insets = 1 μm. Insets show enlargements of the boxed regions.
The Number and Velocity of Golgi Stacks Are Reduced in STINGY Compared with the Wild Type
Next, we aimed to test whether STINGY had defects in other aspects of the biology of the Golgi apparatus besides traffic. The plant Golgi is composed of a large number of dispersed and polarized stacks that are motile (Boevink et al., 1998; Faso et al., 2009a). To test whether these features were compromised in STINGY, we compared the number and velocity of Golgi stacks between STINGY and the wild type. To do so, we first estimated the number of Golgi stacks in cotyledon epidermal cells of STINGY and nonmutagenized seedlings expressing the Golgi marker ST-GFP (Boevink et al., 1998). We found that in the mutant, the number of Golgi stacks was reduced compared with the wild type (Figure 4A). Then, to compare the velocity of the Golgi stacks between STINGY and the wild type, we performed time-lapse confocal microscopy on cortical regions of cotyledon epidermal cells and measured the average and maximal velocity of the stacks, as described earlier (Stefano et al., 2012b). We found that compared with the wild type, the velocity of the Golgi stacks in STINGY was largely reduced (Figures 4B and 4C). These data indicate that STINGY has defects both in secretory traffic as well as in Golgi number and movement.
Figure 4.
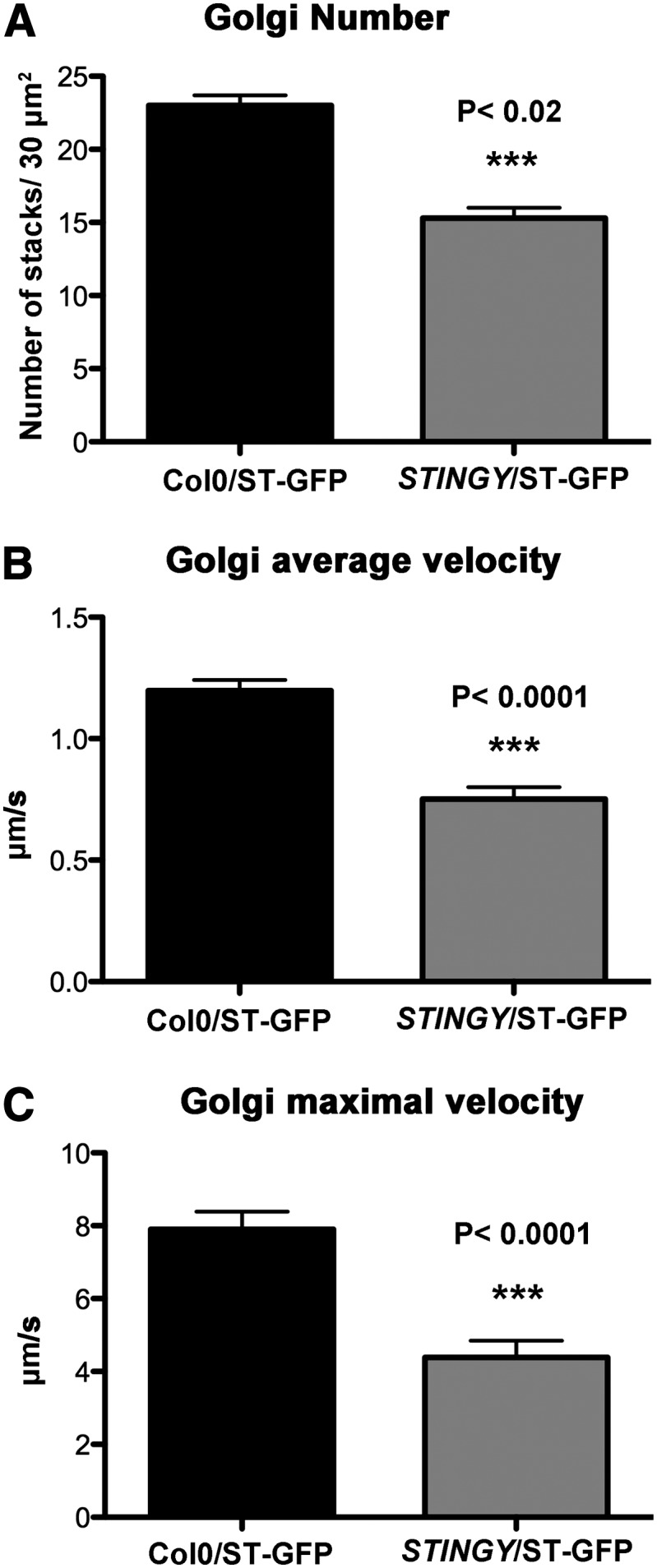
In STINGY, the Number and Velocity of Golgi Stacks Are Reduced Compared with the Wild Type.
(A) Measurements of the number of Golgi bodies show a reduced number of Golgi in the mutant compared with the control. Sample size = 75 regions analyzed.
(B) and (C) Mean and maximal velocity (μm/s) of Golgi stacks in nonmutagenized ST-GFP plants (Col-0, control) and the STINGY mutant measured by time-lapse confocal microscopy of cotyledon pavement cells. Sample size ≥3000 Golgi stacks.
Error bars are se.
STINGY Is Allelic to Arabidopsis NMT1
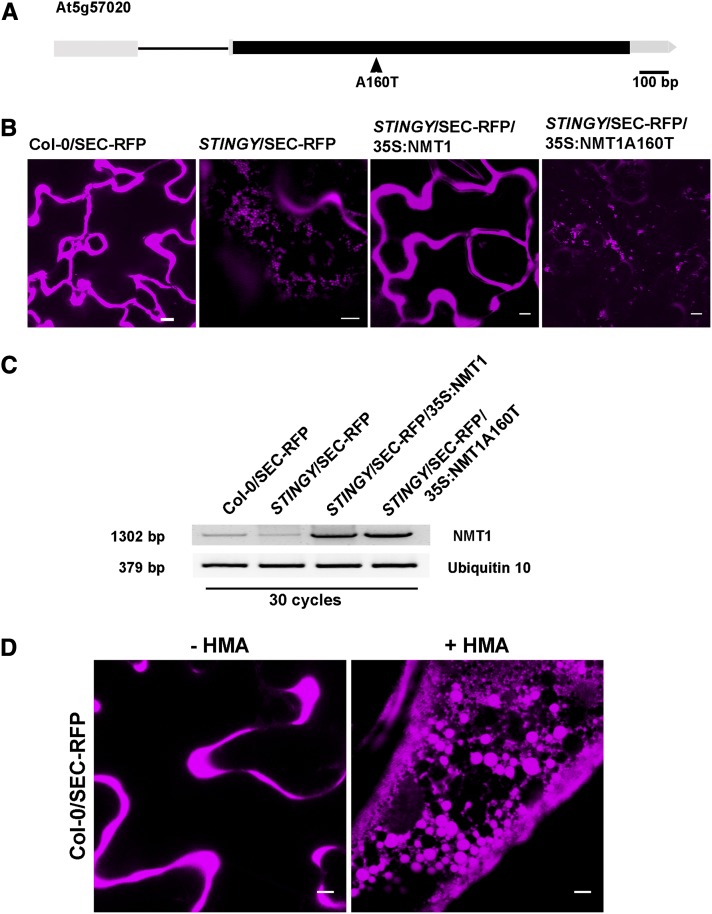
To identify the mutation responsible for the abnormal phenotype, we crossed STINGY with Landsberg erecta (Ler) to generate a mapping population that was used for rough mapping with a GeneChip Arabidopsis ATH1 genome array strategy (Borevitz, 2006; Stefano et al., 2012a, 2012b) (see Methods). The mutation was mapped on chromosome 5 between 22.9 and 24.6 Mb. Subsequently, we sequenced the genomic DNA of STINGY by next-generation sequencing, as described earlier (Marti et al., 2010; Stefano et al., 2012a, 2012b). Within the rough-mapped region, we identified only one typical G/C to A/T EMS transition (Maple and Møller, 2007). The mutation mapped to a GCT-to-ACT transition in the At5g57020 locus, which results in a missense mutation and an amino acid switch from an Ala residue to a Thr residue (A160T) at a transcriptional level (Figure 5A). The locus encodes NMT1. The Ala residue in position 160 corresponds to a Gly in a conserved region of the catalytic domain of the human NMT1 (see Supplemental Figure 2 online). To confirm that the identified mutation was indeed responsible for the STINGY phenotype, we stably transformed either NMT1 or NMT1A160T into the STINGY background and found that only wild-type NMT1 complemented the STINGY phenotype (Figures 5B and 5C). We also established that wild-type NMT1 complemented the late flowering phenotype of STINGY (see Supplemental Figure 1 online) in line with previous reports showing that low levels of NMT1 impair flower differentiation (Pierre et al., 2007) and create a whole dwarf phenotype (Qi et al., 2000). Furthermore, we established that wild-type NMT1 rescued a verified sensitivity of STINGY to salt stress (see Supplemental Figure 3 online), which is consistent with our proteomic analysis showing a downregulation of a pool of Myred and Paled calcium-dependent protein kinases (CDPKs) in the STINGY mutant (see below). Furthermore, we also analyzed the distribution of nonmutagenized Col-0/SEC-RFP seedlings treated with 2-hydroxymyristic acid (HMA), a potent inhibitor of N-myristoyltransferases (Paige et al., 1990). We found defects in the subcellular distribution of SEC-RFP that strongly resembled the phenotype of STINGY (Figure 5D). Together with the complementation of the flowering delay and salt sensitivity of STINGY by NMT1 (see Supplemental Figures 1 and 2 online), these data indicate that STINGY is linked to a partial loss-of-function allele of NMT1.
Figure 5.
Identification of STINGY and Allele Mapping of NMT1.
(A) Schematic diagram of the open reading frame of the At5g57020 locus (filled black box, exon; line, intron).
(B) Confocal optical sections of Col-0, STINGY, STINGY/NMT1, and STINGY/NMT1A160T showing that the subcellular phenotype of STINGY is rescued by wild-type NMT1 but not by the NMT1A160T mutant, as demonstrated by the differences in the subcellular distribution of SEC-RFP in control and complemented lines compared with STINGY and STINGY/NMT1A160T.
(C) RT-PCR analyses in SEC-RFP, STINGY, STINGY/35S:NMT1, and STINGY/35S:NMT1A160T, showing higher level of transcripts in STINGY/35S:NMT1 and STINGY/35S:NMT1A160T compared with the controls, as expected for transformed lines. Ubiquitin10 was used as a control.
(D) HMA treatment phenocopies the STINGY phenotype: SEC-RFP Arabidopsis seedlings were treated with the NMT1 inhibitor HMA. Confocal analyses of pavement cells of cotyledons showed that treated seedlings retain SEC-RFP in globular structures similar to those observed in the STINGY mutant.
Bars = 5 μm.
In Vitro and in Silico Comparison of Wild-Type NMT1 and NMT1A160T Highlights Altered Kinetic Properties Associated with the A160T Substitution
Based on the observed traffic phenotype, we propose that NMT1 is involved in membrane traffic in seedlings. To further support this hypothesis, we aimed to identify NMT1 targets in the secretory pathway by analyzing gene coresponse analyses. Genes with similar expression profiles are more likely to encode functionally interacting proteins than are random pairs (Alabadí et al., 2001). Therefore, we hypothesized that genes with products having biological activity in highly coordinated processes would significantly correlate with NMT1. To proceed with this approach, we used ARANET (http://www.functionalnet.org/aranet/) as a database for gene correlations. Within the top six of the most highly correlated genes with NMT1, we found listed four loci encoding Arf-GTPases (ArfA1C, ArfA1E, ArfA1F, and ArfA1D) (see Supplemental Table 1 online). These Arf-GTPases share high homology, particularly at their N terminus (Table 1; see Supplemental Figure 4 online). The submission of their N-terminal sequence to the latest version of the online prediction tool TermiNator (http://www.isv.cnrs-gif.fr/terminator3), software that predicts several N-terminal modifications including MYR (Frottin et al., 2006; Martinez et al., 2008), reveals that all these small Arf-GTPases can be MYRed as other small GTPases (Table 1). Since the reliability of TermiNator3 remains limited for MYR, we used a biochemical strategy (Traverso et al., 2013a) to confirm the above predictions. Specifically, we employed peptide derivatives of the N-terminal sequence of the Arf-GTPases predicted to undergo MYR, and we assessed their MYR in vitro using purified wild-type NMT1 or NMT1A160T. For each peptide, we could determine each associated kinetic parameter (Km, kcat, and kcat/Km), allowing us to compare the affinity and the efficiency of NMT1 and NMT1A160T. The data in Table 2 show that an N-terminal peptide derivate common to ArfA1A,D,F was efficiently MYRed in vitro by NMT1. By contrast, the mutant enzyme featured a twofold increase in the Km for the peptide compared with the wild type, which contributed to an observed 16-fold catalytic efficiency (kcat/Km) reduction. In our analyses, we also included peptides corresponding to additional tested substrates of NMT1, for which the catalytic efficiency for MYR was extremely variable (Table 2). We found that compared with the wild type, NMT1A160T had reduced affinity (increase of the Km value) for all tested peptides (Table 2). As observed with peptide derivative from Arf-GTPases, we also established that, compared with wild-type NMT1, NMT1A160T had generally a strongly reduced activity (kcat/Km); this was most noticeable for those peptides for which the kcat/Km was high with the wild-type enzyme compared with peptides with already low kcat/Km values (compared with RPS5-like peptide with a 47-fold reduction to TRXh7 peptide with a threefold reduction; Table 2). The differences observed between wild-type NMT1 and NMT1A160T were explored further by determining the kinetics parameters for the second substrate, myristoyl-CoA, for both mutant and wild-type enzymes (Table 2). Indeed, the kinetic analysis of NMT1A160T revealed a 22-fold diminished Km value and 54-fold decrease of the kcat/Km value for myristoyl-CoA, which strongly contributed to the reduced productive catalysis of the mutant.
Table 1. Prediction of N-Terminal Amino Acid Myr of Small GTPases of the Arf Family from the Arabidopsis Proteome.
| Protein | Predicted N Terminus of the Mature Protein | Likelihood (%) | Translation Efficiency | Predicted Half-Life (h) |
|---|---|---|---|---|
| ArfA1E (AT3G62290) MGLSFGKLFS | Myr-G(2) | 45 | 5 | 220 |
| ArfA1F (AT2G47170) MGLSFAKLFS | Myr-G(2) | 45 | 5 | 220 |
| ArfA1B (AT1G10630) MGLNFTKLFS | Myr-G(2) | 45 | 5 | 220 |
| Arf2 (AT1G70490) MGLSFAKLFS | Myr-G(2) | 45 | 5 | 220 |
Prediction was achieved using the TermiNator predictor (v3; http://www.isv.cnrs-gif.fr/terminator3/index.html) (Meinnel et al., 2005; Martinez et al., 2008).
Table 2. In Vitro MYR Assay of Various Peptides Using Recombinant Wild-Type NMT1 and NMT1A160T.
| NMT1a | |||||
|---|---|---|---|---|---|
| Protein of Originb | Substrate Assayed | Km (µM) | kcat (s−1) | kcat/Km (103 M−1⋅s−1) | |
| ARFA1adf | GLSFAKLFRR | 29 ± 5 | 0.65 ± 0.03 | 21.9 | |
| TRXh2 | GGALSTVF | 60 ± 17 | 0.28 ± 0.03 | 4.6 | |
| TRXh7 | GSNVSSVH | 301 ± 41 | 0.26 ± 0.02 | 0.88 | |
| TRXh8 | GANVSTPD | 545 ± 159 | 0.55 ± 0.11 | 1.0 | |
| TRXh9 | GGSCVSKGK | 18 ± 3 | 0.027 ± 0.011 | 1.5 | |
| PLL5 | GNGVTKLS | 12 ± 4 | 0.15 ± 0.07 | 11.9 | |
| RPS5-like | GGCVSVQV | 2.4 ± 0.4 | 0.075 ± 0.001 | 31.7 | |
| SOS3 | GCSVSKKK | 7.4 ± 2.2 | 0.091 ± 0.004 | 13.0 | |
| SOS3 | Myr-CoA | 1.7 ± 0.3 | 0.153 ± 0.007 | 89 | |
| NMT1A160Ta | |||||
| Protein of Originb | Substrate Assayed | Km (µM) | kcat (s−1) | kcat/Km (103 M−1⋅s−1) | Reduction of kcat/Km (Fold versus Wild Type) |
| ARFA1adf | GLSFAKLFRR | 68 ± 14 | 0.091 ± 0.006 | 1.3 | 16.5 |
| TRXh2 | GGALSTVF | 197 ± 53 | 0.044 ± 0.001 | 0.22 | 21 |
| TRXh7 | GSNVSSVH | 176 ± 54 | 0.05 ± 0.01 | 0.27 | 3.3 |
| TRXh8 | GANVSTPD | 926 ± 301 | 0.18 ± 0.03 | 0.20 | 5 |
| TRXh9 | GGSCVSKGK | 106 ± 28 | 0.029 ± 0.004 | 0.28 | 5.2 |
| PLL5 | GNGVTKLS | 36 ± 2 | 0.028 ± 0.002 | 0.76 | 16 |
| RPS5-like | GGCVSVQV | 50 ± 8 | 0.034 ± 0.002 | 0.67 | 47 |
| SOS3 | GCSVSKKK | 57 ± 11 | 0.069 ± 0.004 | 1.18 | 11 |
| SOS3 | Myr-CoA | 38 ± 11 | 0.062 ± 0.007 | 1.63 | 54 |
Kinetic parameters obtained from an in vitro continuous fluorigenic NMT assay and several N-terminal peptides (Traverso et al., 2013a).
N-terminal octapeptides (from amino acids 2 to 9) were derived from Arabidopsis proteins for which MYR was previously predicted and/or reported (Traverso et al., 2013a, 2013b). The kcat/Km value reflects the catalytic efficiency of each peptide to be MYRed by the NMT enzyme.
Using three-dimensional structure data available from NMT from yeast (Saccharomyces cerevisiae NMT) and humans (Homo sapiens NMT1) obtained in complex with various ligands (Farazi et al., 2001b), we built a model of Arabidopsis NMT1 in complex with myristoyl-CoA (see Supplemental Figure 5 online). The final model indicates that residue Ala-160 plays a key role at the interface between three antiparallel β-strands through a network of hydrophobic interactions between Val-150, Leu-195, and Val-186. Val-186 (the homolog of Val-174 in Sc NMT) is very close to the myristoyl-CoA moiety (shown in orange in Supplemental Figure 5 online). When Ala-160 is substituted into Thr, the hydrophilic nature of the hydroxyl group most likely induces a major modification of the aforementioned hydrophobic network, explaining the weaker affinity of the mutant enzyme for myristoyl-CoA and consequently of its lower catalytic efficiency compared with the wild type.
Immunoblot analyses of proteins extracted from the wild type and STINGY revealed a reduced amount of NMT1 in the mutant (24% versus 100% in the wild type; see Supplemental Figure 6 online). This suggests that the A160T substitution in NMT1 causes decreased stability of the protein and/or translation efficiency of the corresponding mRNA (Figure 5C). Together with the observed reduced in vitro activity of NMT1A160T, these data indicate partial loss of function, essentially due to reduced affinity of the enzyme for its substrates.
In Vivo Comparative Analyses Reveal No Massive Changes in the Proteome and Myristoylome of STINGY
Since no obvious changes in the distribution of fluorescent protein fusions of NMT1 or NMT1A160T were observed (see Supplemental Figure 7 online), we expected that the whole myristoylome in the STINGY background would be affected rather than only MYRed proteins residing at the Golgi. Thus, since the primary function of MYR is to allow proteins to anchor to a membrane compartment, we expected to recover an increased amount of proteins belonging to the Arabidopsis myristoylome in the soluble fraction of NMT1A160T. However, proteomic analyses of soluble protein extracts from STINGY leaves versus the SEC-RFP control did not reveal any massive differences in relative protein accumulation, even when focused only on the potential MYRed pool (see Supplemental Figure 8 and Supplemental Data Set 1 online).
Since Myred proteins are enriched in the plasma membrane (PM) (Marmagne et al., 2007) and we have shown an impairment in traffic with partial ER retention of Golgi and tonoplast marker proteins in the STINGY mutant (Figure 3), we then hypothesized that reduced activity of NMT1A160T could affect the composition of the STINGY mutant PM proteome. Thus, we performed a deep proteomic analysis of PM-enriched fractions of STINGY mutant versus the SEC-RFP control. This analysis yielded the identification of more than 4000 proteins in both SEC-RFP and the STINGY mutant (see Methods), with PM-localized proteins being most abundant and known plastid- or mitochondria-localized proteins being two orders of magnitude less abundant than the PM set (annotations of cellular subproteomes were obtained from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/) and Plant Proteomics databases (http://ppdb.tc.cornell.edu/dbsearch/subproteome.aspx) (Sun et al., 2009). In addition, a lower number of proteins annotated as ER/Golgi (∼100) were also retrieved. The relative quantification of the identified proteins showed no major changes between SEC-RFP and STINGY PM-enriched fractions (see Supplemental Figure 9A and Supplemental Data Set 2 online), as in the case of the proteomic analysis of the total protein fractions (see above), suggesting the absence of any massive PM proteome mistargeting induced by the mutation of NMT1. Nonetheless, a few interesting alterations affecting a small number of proteins could be detected, and most of them were previously shown to be associated with the inhibition of ER-to-Golgi transport. In this context, a subset of ER proteins, supporting the crucial roles of the ER in the biosynthesis and transport of proteins and lipids and in calcium regulation, showed a downregulation in the STINGY mutant (see Supplemental Data Set 2 online). Particularly, a reduction in the STINGY mutant was observed in a small set of proteins (n = 13) implicated in long-chain fatty acid synthesis (see Supplemental Data Set 2 online). Interestingly, the majority of this set of proteins present in our PM-enriched fraction were previously identified at the ER/Golgi membrane, and three of them (LACS8, LACS1, and LACS9) are predicted by plant ARANET functional interaction network (http://www.functionalnet.org/aranet/) to be functionally associate with NMT1. Finally, a strong downregulation in the STINGY mutant was observed for the Sec23/Sec24 protein transport family (AT4G14160.1 and AT1G05520.1) with a SEC-RFP/STINGY ratio >7. Taken together, this reduced accumulation of ER/Golgi proteins supports the notion that the effect of the mutation on NMT1 in the STINGY background might mostly center at the level of the ER/Golgi compartment in agreement with the observed subcellular phenotype.
Within the analyzed PM-enriched fractions of SEC-RFP and STINGY, we identified 457 proteins starting with an N-terminal Gly and within those we were able to distinguish 135 proteins belonging to the Arabidopsis myristoylome (see Supplemental Data Set 2 online). This is the largest number of Myred proteins identified so far. The relative protein accumulation ratio between SEC-RFP and STINGY remained below a threefold change for the majority of proteins with an N-terminal Gly, even when considering only the Myred pool (90%; see Supplemental Figure 9B online). Indeed, a reduction in protein accumulation was observed only for few abundant proteins in the membrane fraction of the STINGY mutant [∼15 proteins, Σ(precursor area) > 1.1008]. Among the identified MYRed proteins, PHOSPHATIDYLSERINE DECARBOXYLASE3 (PSD3), located in the endomembrane system (Nerlich et al., 2007), was the most affected. PSDs have been previously shown in yeast and animals to be involved in the formation of phosphatidylethanolamine from phosphatidylserine and to be part of a recognition and transport macromolecular complex committed in the mechanisms that facilitate the transport of this anionic lipid between membranes (Wu and Voelker, 2004; Schuiki et al., 2010). Differently to animals and yeast, it has been shown that Arabidopsis PSD3 does not contribute to the synthesis of phosphatidylethanolamine and that the knockout mutant for psd3 is indistinguishable from the wild type (Nerlich et al., 2007). Thus, the reduced accumulation of PSD3 cannot be considered the primary responsibility of the STINGY phenotype, but barely a consequence. Finally, downregulation was also observed for a few PM MYRed CDPK-related kinases and plant defense response proteins (see Supplemental Data Set 2 online), suggesting that the STINGY mutant might differently react to abiotic and/or biotic stress compared with the wild type. This hypothesis is in agreement with the observed high sensitivity of the STINGY mutant under salt stress conditions (see Supplemental Figure 3 online) and previous reports showing the mutants of these CDPK salt-responsive phenotypes (Wan et al., 2007; Zhu et al., 2007; Mehlmer et al., 2010; Wurzinger et al., 2011). Although the reduced activity of NMT1 in the STINGY mutant can influence the accumulation at the PM of the above proteins, once more none of them can be considered the major cause of the alteration of Golgi traffic and integrity. Indeed, alteration of these plant defense response proteins or CDPK-related proteins has never been associated with any defects in the ER-Golgi system. Taken together, our proteomic analyses suggest that the reduced activity of NMT1A160T is unlikely to induce pleiotropic effects on the majority of proteins, including those belong to the Arabidopsis myristoylome. Rather, our data strongly suggest that MYR of one or only a few ER/Golgi NMT1 substrates is critical for efficient ER/Golgi traffic.
The NMT1A160T Mutation Reduces the Association of Arfs onto the Golgi
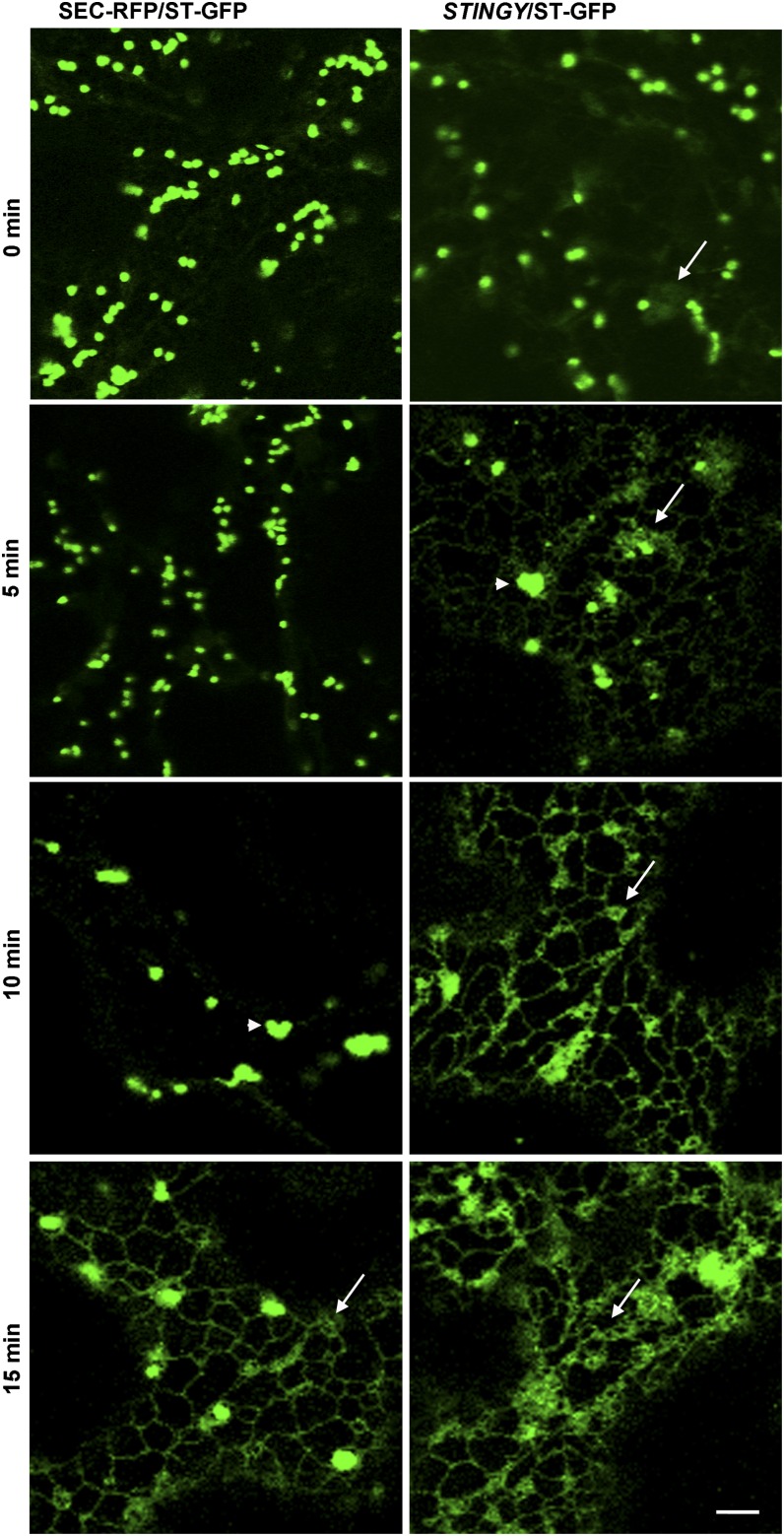
Interestingly, myristoylatable Arf proteins, which play important roles in secretion and endocytosis, have been suggested, indirectly, to be linked to the primary NMT targets responsible for NMT-altered phenotypes in unicellular organisms, including endocytic defects, which are observed in NMT knockdown mutants in fungi and trypanosomes (Shaw et al., 2002; Lee et al., 2008; Lee and Shaw, 2008a, 2008b; Price et al., 2010). Based on these data, we tested the sensitivity of STINGY, compared with the wild type, to brefeldin A (BFA), a fungal metabolite that specifically affects Arf-GEFs (for guanine exchange factors) responsible for Arf activation and causes redistribution of Golgi proteins in the ER (Robineau et al., 2000; Saint-Jore et al., 2002; Robinson et al., 2008). We analyzed the distribution of ST-GFP, a specific Golgi marker (Boevink et al., 1998), in nonmutagenized and STINGY backgrounds. We found that in nonmutagenized Col-0/ST-GFP cells, treatment with BFA led to clumping of Golgi stacks within 10 min of treatment, followed by the appearance of some ST-GFP fluorescence in the ER by 15 min (Figure 6). In contrast with the control, BFA treatment of STINGY caused complete distribution of ST-GFP into the ER within 15 min (Figure 6). Our results indicate that the defects in ER-Golgi traffic in STINGY strongly depend on Arf-GTPases.
Figure 6.
STINGY Shows Enhanced Sensitivity to BFA Treatment Compared with the Wild Type.
Confocal optical sections of Arabidopsis cotyledons from SEC-RFP (control) and STINGY plants expressing the Golgi marker ST-GFP subjected to a time-course BFA treatment. Left column shows a BFA time course for Col-0/SEC-RFP/ST-GFP. In this background, Golgi clumping occurs within 10 min (arrowheads) and protein redistribution within 15 min of treatment (arrows). Right column shows a BFA time course for STINGY/ST-GFP. In the mutant, Golgi clumping occurs within 5 min (arrowheads). By 10 min, the fluorescence associated with the Golgi bodies has largely diminished, and by 15 min, the ER appears enlarged, most likely due to the accumulation of ER and GA proteins consequent to BFA-induced ER export disruption (arrows). Note that for clarity, only the GFP fluorescence is shown. Bar = 5 μm.
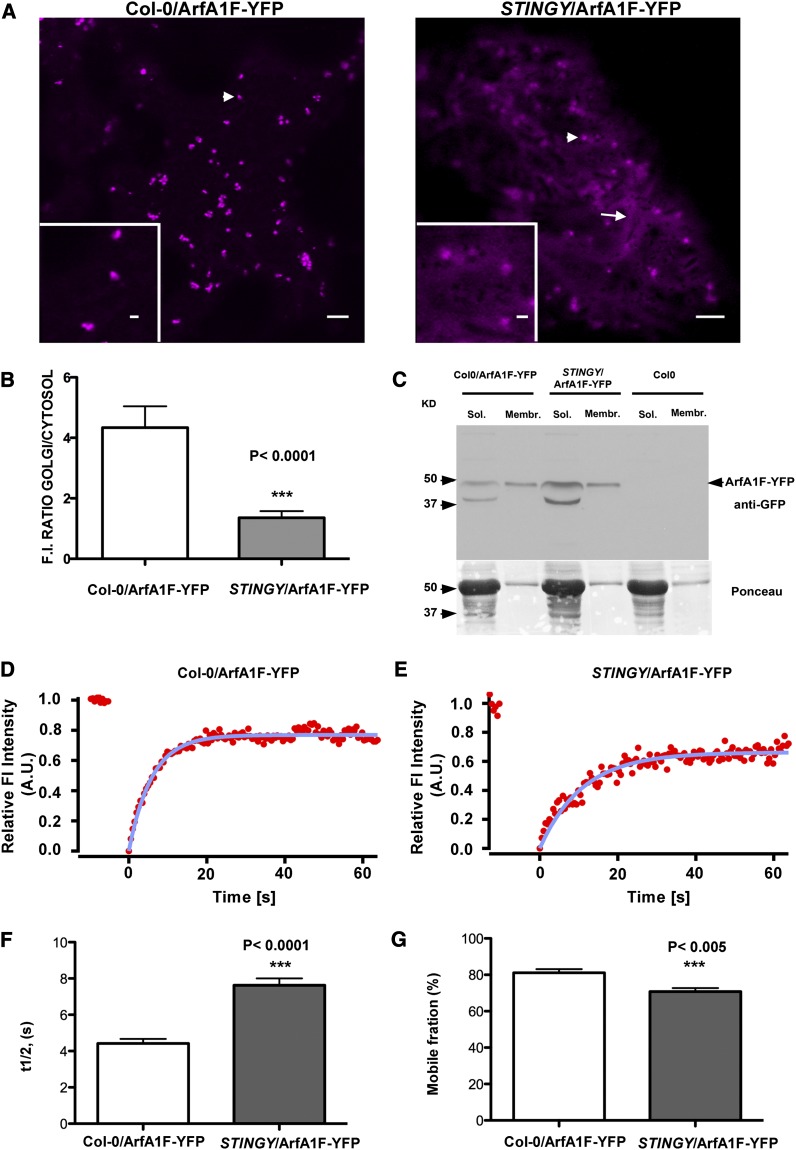
Partitioning between the endomembrane compartment and the cytosol correlates with the catalytic efficiency of the N-myristoyltransferase (i.e., low NMT catalytic efficiency induces an increase in the soluble fraction of the MYRed protein substrate) (Traverso et al., 2013b). Therefore, we wondered whether, owing to the reduced in vitro myristoylase activity of NMT1A160T (Table 2), we would observe an effect of the STINGY mutation on the distribution and membrane binding of Arf-GTPases. We stably transformed Col-0 and STINGY with one of the highly correlated Arf proteins (see Supplemental Table 1 online). Due to the strong degree of similarity of the protein sequences at the amino acid level (see Supplemental Figure 4 online; Table 1), we used ArfA1F fused to YFP. Confocal microscopy analyses of the distribution of ArfA1F-YFP in tobacco (Nicotiana tabacum) epidermal cells showed that ArfA1F was localized mainly to punctate structures of heterogeneous size (see Supplemental Figure 10 online). The largest ones represent Golgi stacks, as demonstrated by colocalization analyses with the Golgi marker ERD2-GFP (see Supplemental Figure 10 online) (daSilva et al., 2004; Renna et al., 2005; Hanton et al., 2007); the smaller ones are most likely trans-Golgi network, as shown for other Arf-GTPases in Arabidopsis (Robinson et al., 2011). In the Col-0 and STINGY background, ArfA1F-YFP was associated with punctate structures (Figure 7A). However, ArfA1F-YFP distribution appeared largely cytosolic in STINGY compared with the nonmutagenized background (Figure 7A), as also shown by the measurement of the ratio of fluorescence intensity at the Golgi and into the cytosol (Hanton et al., 2009) (Figure 7B). The larger amount of ArfA1F-YFP in the cytosol of STINGY compared with that of the wild type was further confirmed by immunoblot analyses on Col-0 and STINGY expressing ArfA1F-YFP (Figure 7C). In the soluble fraction, we verified the occurrence of a smaller band most likely due to partial protein degradation (Figure 7C). The presence of such a band in wild-type and mutant backgrounds argues that the larger distribution of ArfA1F-YFP in the STINGY cytosol (Figures 7A and 7B) is mainly linked to mistargeting of ArfA1F-YFP rather than exclusive visualization of degradation products. Together, these data indicated that the STINGY mutation affected the subcellular distribution of ArfA1F and correlated to low catalytic efficiency of the mutated NMT1. Because the Golgi markers were partially distributed in the ER in STINGY (Figures 3A and 3B), we reasoned that it could not be excluded that the distribution of ArfA1F-YFP was linked to an incapacity of NMT1A160T to myristoylate the entire set of ArfGTPases. Therefore, we sought additional evidence in support of a role of NMT1 in myristoylation of Arf proteins, and we performed FRAP on ArfA1F-YFP at Golgi stacks. This would allow measuring the time necessary for ArfA1F-YFP to recover on target membranes upon bleaching; it would also allow investigating protein mobility as well as exchange between cytosolic and membrane-bound Arf proteins in the mutant background compared with the wild type. Although ArfA1F-YFP fluorescence recovered in both backgrounds, we found that the half-time recovery of ArfA1F-YFP was longer in STINGY/ArfA1F-YFP compared with Col-0/ArfA1F-YFP (Figures 7D to 7F). Moreover, we established that the percentage of available Arf protein to bind membranes (mobile fraction) was slightly but significantly reduced in the mutant background compared with the wild type (Figures 7D, 7E, and 7G). These data indicate that, compared with the wild type, a portion of ArfA1F is not efficiently recruited onto membranes in the STINGY background. This is in agreement with a small proportion of ArfA1F to be not myristoylated as a result of too low MYR catalytic efficiency. An additional confirmation of these results was provided by HMA treatment, which induced cytosolic redistribution of ArfA1F-YFP compared with untreated plants (see Supplemental Figure 11A online). This is consistent with the cytosolic distribution of a YFP fusion of ArfA1F bearing an Ala substitution on the Gly that is normally myristoylated [ArfA1F(G2A)-YFP] (see Supplemental Figure 11A online). Furthermore, the ArfA1F(G2A)-YFP mutant does not act as a dominant negative, since transient expression of this construct in tobacco had no effect on Golgi distribution and number (see Supplemental Figure 12 online). Overall, our results indicate that the defects in myristoylation of Arf-GTPases largely contribute to the defects in ER-Golgi traffic in STINGY.
Figure 7.
The NMT1 Mutation Affects the Proper Localization and Dynamics of ArfA1F at the Golgi.
(A) Confocal live-cell images of a cotyledon epidermal cell of Col-0 and STINGY backgrounds stably expressing ArfA1F-YFP. Col0/ArfA1F-YFP shows ArfA1F localization mainly at punctate structures (left panel), which include Golgi stacks (see Supplemental Figure 10 online), while STINGY/ArfA1F-YFP also shows abundant ArfA1F localization in the cytosol. Bars = 5 μm; bar in the inset = 1 μm.
(B) The relative membrane/cytosol ArfA1F-YFP fluorescence distribution in both Col-0/ArfA1F-YFP and STINGY/ArfA1F-YFP is visualized as histograms, which report the ratio between the fluorescence intensity measured in regions of identical size for the punctate structures and the cytosol.
(C) Immunoblot analyses of ArfA1F-YFP (∼47 kD) using a GFP antibody on the soluble and membrane fractions of Col-0/ArfA1F, STINGY/ArfA1F, and Col-0. As supported by a comparison of the relative abundance of the bands in the soluble and membrane fractions in each sample, in STINGY, ArfA1F-YFP is more abundant in the soluble fraction than in the membrane fraction compared with the nonmutagenized Col-0-SEC-RFP control. Ponceau red staining represents the protein loading control.
(D) and (E) FRAP curves representing the time course of ArfA1F-YFP (starting at 0 s) for Col-0/ArfA1F-YFP and STINGY/ArfA1F-YFP.
(F) Representation of the half-time recovery (s) of ArfA1F-YFP in Col-0 and STINGY backgrounds. A.U., arbitrary units.
(G) Representation of the mobile fraction (%) for ArfA1F-YFP in Col-0 and STINGY backgrounds. The difference in fluorescence recovery half-times and the mobile fraction between the samples is significant, as indicated by the P value on the charts ([F] and [G]).
DISCUSSION
NMTs have been extensively characterized both biochemically and genetically in many unicellular organisms and a few multicellular organisms, including humans and higher plants. So far, much less is known about the relative roles and contributions of each of their many targets in development. It has been proposed that the Arabidopsis NMTs have nonoverlapping roles during growth and development, as supported by the evidence that loss of NMT1 causes death in late embryogenesis (Pierre et al., 2007). To date, only the SNF1-related kinase complex (SnRK1) has been identified as the primary target of MYR NMT1 in embryogenesis (Pierre et al., 2007), but the targets of the deleterious effect observed at later stages of development have not been identified. It is unknown whether SnRK1 still corresponds to the NMT1 target responsible for later developmental defects associated with the reduced expression of NMT1. NMT2 function is not essential, and a knockout of its gene causes only a modest reduction in flowering time (Pierre et al., 2007). This is unlike NMT1 knockdown, which induces clear and strong alterations of various organs and functions, including global growth, flowering, fruit maturation, as well as increased sensitivity to pathogens (Pierre et al., 2007). Here, we report on STINGY, a partial loss-of-function of NMT1 identified through a confocal microscopy–based screen for endomembrane traffic defects, which is viable and allowed pursuing further research on the roles of NMT1 in vivo in post embryonic stages of development. We demonstrated that STINGY is linked to a nucleotide change leading to an Ala–Thr transition in the putative catalytic site of NMT1. This, in turn, causes a reduction of affinity and catalytic activity of NMT1 on substrate peptides including myristoyl-CoA. We showed that STINGY has defects in Golgi traffic and Golgi movement and has a reduced abundance of Golgi stacks as well as that a lack of functional NMT1 causes defects in the association of Arf proteins to Golgi stacks. These data provide important insight into the biology of NMT1 by supporting experimentally the nonredundant involvement of NMT1 in the plant secretory pathway at least in postembryonic developmental stages, most likely through an alteration of the N terminus of Arf proteins, which are critical membrane traffic regulators.
Subcellular Characterization of a Viable Partial Loss-of-Function Allele of NMT1 Demonstrates the Involvement of NMT1 in the Plant Secretory Pathway
The high sensitivity to the NMT inhibitor HMA and the biochemical data showing that NMT1A160T has reduced affinity and activity on substrate peptides imply that STINGY is a partial loss-of-function allele of NMT1. Our live-cell imaging studies were conducted in epidermal cells of 10-d-old cotyledons. At this stage, cotyledon epidermal cells have ceased to grow (Zhang et al., 2011). Given that NMT1 is an essential gene both in late embryogenesis and later throughout organ development and plant growth (Pierre et al., 2007), the absence of obvious phenotypes in the vegetative tissues of STINGY indicates that STINGY is a weak allele of NMT1 that does not compromise growth. This is in agreement with our whole proteomics analysis in which no massive global differences were observed when comparing the general pattern of abundant proteins or when restricting the analysis to identify predicted MYRed proteins of NMT1A160T seedlings compared with the wild type.
Our plant phenotypic analyses revealed that partial loss of NMT1 causes flowering delay as well as salt sensitivity. Although we have not explored whether these phenotypes are linked to loss of activity of Arf proteins on membranes, flowering delay is in accordance with previous results that a low level of NMT1 impairs flower differentiation (Pierre et al., 2007). Furthermore, considering our proteomics analysis revealing as a consequence of the reduced activity of NMT1 a down-accumulation at the PM of several salt-sensitive CDPK proteins (Wan et al., 2007; Zhu et al., 2007; Mehlmer et al., 2010; Wurzinger et al., 2011), we speculated and confirmed that STINGY is hypersensitive to salt.
Our characterization of STINGY has established that NMT1 does not have redundant functions in the secretory pathway of seedlings in the postembryonic stages, as demonstrated by the evidence that secretory defects in STINGY are not compensated for by other proteins. Although in this study we did not examine the effects of the STINGY mutations during embryo development, we cannot exclude that NMT1 also has a role in endomembrane traffic during the early stages of plant development.
We showed that at a subcellular level STINGY partially accumulated apoplastic (SEC-RFP), Golgi (G-YK) and tonoplast (VAC-YFP) markers in the ER. Because post-Golgi markers (tonoplast and apoplast) were not detected in intermediate post-Golgi compartments to the vacuole and cell wall, it is possible that NMT1 has a primary role in ER-Golgi and Golgi protein traffic rather than post-Golgi traffic. However, in our work, we have not analyzed post-Golgi protein traffic organelles, such as the trans-Golgi network. Because our work supports that Arf proteins, which are localized in post-Golgi compartments (Robinson et al., 2011), are targets of NMT1, we cannot exclude the possibility that defects in NMT1 also affect later membrane traffic steps.
The Molecular Phenotype of STINGY, Which Causes Reduced Affinity for Myristoyl-CoA, Is Reminiscent of Most NMT Mutants Characterized in Unicellular Organisms
We report here that the NMT substitution in the STINGY background induces (1) a major increase (55-fold) of the Km value for myristoyl-CoA and (2) varying but substantial decreases of the Km values for model protein substrates, including Arfs (20-fold). This in vitro analysis led us to suggest that partial MYR of a number of proteins occurs in vivo in the STINGY background and that the function of the most essential MYRed targets is only partially assured as a result. Nevertheless, our proteomic investigation of the STINGY mutant supports that pleiotropic effects are unlikely to occur subsequent to the in vivo decrease of NMT1A160T activity on the whole myristoylome, suggesting that a general decrease of MYR is not the limiting factor in the mutant. Rather, our data reveal that Arf MYR mediated by NMT1 is critical for efficient ER/Golgi traffic.
Although the NMT gene was similarly described to be essential in the yeast S. cerevisiae (Duronio et al., 1989), single substitutions of Gly-451 and -412 (both residues lying next to the catalytic site) were already reported to induce a severe decrease of its affinity for myristoyl-CoA (Duronio et al., 1991). This causes a critical reduction of NMT catalysis and induces temperature-sensitive myristic acid auxotrophy in yeast (Duronio et al., 1990). Expression of human NMT1 in S. cerevisiae exhibiting a substitution of the structurally homologous residues of Gly-451 (Gly-412) revealed the same phenotype (Duronio et al., 1992). Finally, an identical substitution (Gly447Asp) in the pathogenic yeast Cryptococcus neoformans caused the same phenotype (Lodge et al., 1997). Interestingly, it was revealed that this effect induced a significant decrease in the MYR state of Arf proteins. In the filamentous fungus Aspergillus nidulans, a single mutation of the NMT gene (Asp369Tyr, swoF mutant) causes aberrant cell polarity (Shaw et al., 2002). Partial restoration of temperature sensitivity was also observed upon the addition of myristate, indicating that this mutant is likely related to the above yeast and human NMT mutants. In this latter case, it was hypothesized that an unknown target required for polar growth needed to be MYRed to be properly localized and fully active. A link between the swoF mutant and increased activity of the 26S proteasome was identified, suggesting that MYR attenuates proteasome activity in the wild type (Lee and Shaw, 2007). Several hypotheses were proposed to explain this link, but no possible molecular mechanisms could be identified. More recently, it was demonstrated that overexpression of A. nidulans ArfA, a myristoylatable Arf family protein localized to Golgi-equivalent bodies, partially suppresses the swoF phenotype (Lee and Shaw, 2008b). Together with the findings that another A. nidulans miristoylatable Arf family protein, ArfB, plays important roles in polarity establishment and maintenance, it was hypothesized that hyphal polarity in A. nidulans is linked to the MYR of Arf proteins and their proper localization (Lee et al., 2008; Lee and Shaw, 2008a, 2008b). Finally, recent work has shown that RNA interference knockdown of NMT in bloodstream forms of Trypanosoma brucei results in a defect in endocytic uptake (Price et al., 2010). Nonetheless, it is yet to be demonstrated whether the two MYRed proteins, Tb ARL1 and Tb ARF1, which have been proposed to be the NMT targets, are indeed responsible for the above phenotype (Price et al., 2005b). Therefore, there is clear evidence that MYR defects cause a deficiency of MYR of Arf-GTPases in various unicellular organisms. So far, however, no direct NMT1 target(s) has been identified as responsible per se for the aforementioned phenotypes in higher eukaryotes. Here, we show (1) the direct link between NMT1 and Arfs, (2) that the situation is similar in multicellular organisms, including plants at late developmental stages, and (3) that reduced MYR linked to abnormal NMT1 activity causes a number of defects in endomembrane traffic.
On the Role of NMT1 on Arf-GTPases
Efficient membrane traffic within the secretory pathway depends on the activity of Arfs, a subfamily of the Ras superfamily of GTP binding proteins that serve as molecular switches for the recruitment and release of coat proteins and tethering factors on target membranes of secretory organelles (Donaldson and Jackson, 2011). Arfs cycle between an inactive state (GDP bound) and active state (GTP bound); Arf-GEFs and GTPase activating proteins (Arf-GAPs) are responsible for the activation and inactivation of the Arf-GTPases, respectively (Cox et al., 2004; Randazzo and Hirsch, 2004; Anders and Jürgens, 2008). The Arabidopsis genome encodes a large number of Arf-GEFs and Arf-GAPs, and the identity and function of several of these proteins is being unraveled (Geldner et al., 2003; Koizumi et al., 2005; Richter et al., 2007; Tanaka et al., 2009; Stefano et al., 2010; Richter et al., 2011). Our current knowledge of Arf-GEFs and Arf-GAPs in Arabidopsis indicates not only a wide functional diversity but also substrate specificity of these proteins. Nonetheless, not much is known about the mechanisms that precede the recruitment of Arf-GTPases on membranes. In the inactive state, Arf-GTPases are cytosolic and reversibly associate with the membrane surface. The tight association of the active form of the Arf-GTPases depends on an MYRed α-helical extension from the basic G-protein fold, which is a unique structural feature among Ras family proteins (Antonny et al., 1997). In our study, we show that diminished NMT1 activity in vivo causes a reduced affinity of the protein for peptide substrates, which in turn induces an alteration of distribution between membrane and cytosol of Arf-GTPases. Although we analyzed the partition of one member of the Arf-GTPase family, we cannot exclude that secretory defects observed in STINGY are linked also to a lack of efficient myristoylation of Arfs proteins, including Arf-like proteins that are known to be localized on the plant Golgi (Latijnhouwers et al., 2005b; Stefano et al., 2006) and that are known to be myristoylated in nonplant species (Price et al., 2005b; Sahin et al., 2008). Furthermore, the combined evidence for an HMA-induced ArfA1F-YFP partial distribution in the cytosol as well as complete cytosolic distribution of an ArfA1F deficient in myristoylation due to a G2A mutation further supports our data that myristoylation, rather than other protein modifications and/or targeting domains, has an important role for the membrane binding of Arf.
Defects in NMT1 Lead to a Reduction in the Number of Golgi Stacks and Golgi Velocity
The observation that the number of Golgi stacks is reduced in STINGY compared with nonmutagenized controls supports that NMT1 has a role in Golgi biogenesis. It has been demonstrated that the plant Golgi is a highly dynamic organelle whose membranes and peripheral constituents are continuously remodeled through a cyclic exchange of membrane and peripheral components to and from the ER or the cytosol, respectively (Brandizzi et al., 2002; Matheson et al., 2007; Schoberer et al., 2010). It has also been shown that in conditions of increased cellular availability of Golgi membrane cargo, the ER domains involved in ER export to the Golgi (ER exit sites) increase in number compared with nonstimulated cells in plant cells (Hanton et al., 2007). Because ER exit sites and Golgi are associated in plant cells, it has been speculated that in conditions of increased Golgi membrane cargo, plant cells may also increase the number of Golgi stacks (Hanton et al., 2007). The reduction in Golgi number in STINGY could be linked to reduced ER protein export consequent to collapse of the COPI route that is regulated by Arf-GTPases, which function depends on proper membrane localization. Indeed, if these proteins cannot be efficiently MYRed in the absence of a fully functional NMT1, our data would imply that defects in the early secretory pathway of mutants with altered functions of traffic regulators not only cause partial accumulation of post-ER cargo in the ER but also a reduction in the number of Golgi stacks.
A unique characteristic of the plant Golgi is that it is highly motile on the underlying ER (Boevink et al., 1998). We found that in STINGY, the velocity of the Golgi stack was significantly reduced in comparison to nonmutagenized controls. It has been proposed that the ER and Golgi are physically attached, as demonstrated by laser trapping experiments showing that displacement of individual Golgi stacks was followed by rapid growth of the attached ER tubule and that the Golgi that were manipulated to be detached from the ER could be redocked to the ER (Sparkes et al., 2009). It has been proposed that a matrix surrounding the Golgi may be responsible for the ER-Golgi attachment (Sparkes et al., 2009). Although this hypothesis is yet to be experimentally validated, elements of the Golgi matrix have been identified (Latijnhouwers et al., 2005a, 2005b; Stefano et al., 2006; Matheson et al., 2007), and it has been shown that Arf proteins have a role in recruiting such elements onto the Golgi (Matheson et al., 2007). Therefore, it is not unreasonable to hypothesize that defects in Arf recruitment onto the Golgi due to partial loss of function of NMT1 may cause a reduction of MYRed Arf proteins in the Golgi pool of matrix elements responsible for an ER/Golgi attachment. This, in turn, would cause an ineffective anchorage of the Golgi stacks to the ER and a reduction in Golgi velocity.
METHODS
Molecular Cloning
Untagged wild-type and mutant NMT1 constructs were amplified from cDNA of wild-type Col-0 and STINGY, respectively, followed by subcloning for 35S-driven expression between the AscI and XbaI restriction sites in the multiple cloning site of a pFGC5941 binary vector (McGinnis et al., 2005), which was modified to remove the CHSA intron present in the multiple cloning site. The cDNAs were also subcloned into the binary vector pEARLY-Gate 104 to generate YFP fusions at the N terminus of cDNAs. ArfA1F and ArfA1F(G2A) were amplified from cDNA and subcloned into the binary vector pEARLY-GATE 101 to generate a YFP fusion at the C terminus of the cDNAs (oligonucleotides used for the above-described constructs are listed in Supplemental Table 2 online). Constructs were confirmed by sequencing. The available pET16b-NMT1 was employed for protein overexpression purposes (Boisson et al., 2003). Generation of the A160T mutant was achieved by site-directed mutagenesis of pET16b-NMT1 using the QuikChange II site-directed mutagenesis kit (Stratagene). Two oligonucleotides (5′-CTTCGAAGAAACTCGTTACTTTCATCAGCGGCGTG-3′; 5′-CACGCCGCTGATGAAAGTAACGAGTTTCTTCGAAG-3′) were designed using the QuikChange PrimerDesign software (www.stratagene.com/sdmdesigner) and obtained by custom oligonucleotide synthesis (Invitrogen). PCR amplification was performed following the manufacturer’s standard procedure (one cycle of denaturation at 95°C for 30 s; 16 cycles of denaturation at 95°C for 30 s followed by annealing at 55°C for 30 s and final elongation at 68°C for 6 min 30 s). At completion, 10 units of DpnI enzyme were added and the reaction was incubated at 37°C for 1 h. XL-1Blue Supercompetent cells (Stratagene) were transformed using 5 μL of the reaction solution. Plasmids carrying the desired mutation were selected by plating cells on Luria-Bertani-agar plates containing 100 µg/mL ampicillin. Plasmids from different clones were extracted (QIAprep Spin Miniprep kit; Qiagen), and their DNA sequence was verified by sequencing the gene on both strands (GATC Biotech). The confirmed pET16b-NMT1A160T plasmid was used to transform Rosetta2 (DE3)–competent cells (Novagen) to perform protein expression.
RNA Extraction and PCR Analysis
RNA was extracted using the RNeasy plant mini kit (Qiagen). Reverse transcription was performed using the Superscript III first-strand synthesis kit (Invitrogen). PCR experiments were performed using 0.2 mM deoxynucleotide triphosphates, 0.2 μM primers, and 1 unit of Taq polymerase (Promega).
Plant Material and Growth Conditions
Wild-type tobacco (Nicotiana tabacum cv Petit Havana) plants and wild-type plants of the Arabidopsis thaliana Col-0 and Ler ecotypes as well as EMS-mutagenized Col-0 plants expressing SEC-RFP were used in this study. Seedlings were grown at 21°C (Arabidopsis) or 25°C (tobacco) under a 16-h-light/8-h-dark regime. Salt sensitivity assays were performed on Col-0/SEC-RFP, STINGY, and STINGY/NMT1 stable lines in media containing 50, 100, and 150 mM NaCl through measurements of fresh weight of whole seedlings as well root growth measurements. The latter were performed on seedlings germinated for 4 d on half-strength Murashige and Skoog (MS) vertical plates, followed by transfer of the seedlings to half-strength MS vertical plates with various NaCl concentrations and subsequent 180° rotation. Root measurements were acquired from the edge of the root turning point upon an additional 7 d of growth (Wu et al., 1996).
Plant Crosses and Transformation
For analyses of the distribution of the various fluorescence markers of the endomembranes in Arabidopsis, homozygous transgenic Arabidopsis Col-0 lines stably expressing the various markers were used for crossing with STINGY (Col-0). For the confocal microscopy analyses, segregating F2 mutants with the STINGY phenotype were used. Complementation of STINGY with wild-type and mutated NMT1 was achieved by stable transformation through the floral dip method (Clough and Bent, 1998) followed by antibiotic selection on half-strength MS agar plates. For transient expression in tobacco, we used 4-week-old N. tabacum (cv Petit Havana) plants and Agrobacterium tumefaciens (strain GV3101; OD600 = 0.05), according to an established protocol (Batoko et al., 2000).
Isolation of STINGY
A homozygous Arabidopsis Col-0/SEC-RFP transgenic line was used for EMS mutagenesis. M1 SEC-RFP seeds were soaked for 16 h in 0.25% (v/v) methanesulfonic acid ethyl ester (Sigma-Aldrich) and then washed for 11 h in running water. The M1 seeds were grown after self-fertilization, and the M2 seeds were collected from individual M1 plants to generate M2 lines (303 independent lines). Sixty seedlings from each SEC-RFP M2 line were screened using a confocal microscope to identify mutants with an abnormal distribution of the SEC-RFP marker compared with the wild-type control. STINGY was backcrossed three times. To generate a mapping population, STINGY was crossed with Ler. For mapping procedures, a 0.60-mm-diameter leaf disc was collected from each of 80 F2 individuals bearing the abnormal phenotype and 80 F2 individuals showing a wild-type phenotype using a hole punch. The leaf discs were grouped in clusters of five and processed for genomic DNA extraction. The genomic DNA was then quantified (Borevitz, 2006) and prepared for GeneChip Arabidopsis ATH1 hybridization for rough mapping, as described earlier (Stefano et al., 2012a). Identification of the single nucleotide polymorphisms in the rough-mapped region was achieved by Next Generation genome sequencing, as described previously (Marti et al., 2010), using an Illumina Genome Analyzer II (Bentley et al., 2008).
Confocal Laser Scanning Microscopy
Confocal analyses were performed using an inverted laser scanner confocal microscope (LSM510 META; Zeiss) on 10-d-old cotyledon epidermal cells (Arabidopsis) or the lower epidermis of leaves (tobacco). The fluorescent proteins used in this study were GFP5 (Haseloff et al., 1997), enhanced YFP (Clontech), and mRFP (Campbell et al., 2002). Imaging of markers was achieved as previously described (Brandizzi et al., 2002; Hanton et al., 2007; Faso et al., 2009b). The results presented in this work are representative of at least five independent experiments.
For FRAP experiments, fluorochrome photobleaching was performed on Golgi stacks (n = 30) in cells treated with 25 µM latrunculin B (Calbiochem; stock solution 10 mM in DMSO to disrupt Golgi movement (Brandizzi et al. 2002). Mobile fraction and half-time recovery analyses were performed as described by Brandizzi et al. (2002). Student`s two-tailed t test was used for statistical analysis, assuming equal variance and data with P value < 0.05 were considered significant.
To estimate the number of Golgi, we counted the number of Golgi stacks highlighted by ST-GFP within regions (n of regions = 3 in each cell; n of cells = 25; n of independent seedlings = 5; total number of regions = 75) of identical area (30 µm × 30 µm) at the cortex of cotyledon epidermal cells.
To analyze the velocity of the Golgi stacks, a protocol described earlier was followed (Stefano et al., 2012b). Briefly, 50 time-lapse sequences with 50 frames for each sequence in the cortical region (3.0 to 5.0 μm depth) of cotyledon pavement cells were recorded. Time-lapse frames at a 512 × 512-pixel resolution were captured using a 2-μm pinhole at low laser power (i.e., 10% of an argon 488-nm laser line) to avoid photobleaching, and 3 digital zoom using an EC Plan-Neofluar ×40/1.30 objective. Velocity was calculated in postacquisition using plug-ins available through Image J version 1.45k (http://rsbweb.nih.gov/) using the manual tracking plug-in, and data were confirmed using Image Pro-Plus 6 (Media Cybernetics). Velocity values were calculated by averaging the velocity of all the Golgi stacks in the 90 512 × 512 frames in each time-lapse sequence; then, mean values were calculated as the mean of the velocities estimated in the 50 time-lapse sequences. Maximal velocity values were estimated by averaging maximal data values of each time-lapse sequence for each sample. Over 3000 Golgi stacks in the STINGY mutant and the control were used.
HMA Treatment
HMA (Santa Cruz Biotechnology) was dissolved in DMSO. Five-day-old SEC-RFP seedlings were grown for 2 d on sterile filter paper soaked in MS growth medium containing either 1 mM HMA or DMSO.
BFA Time-Course Treatment
BFA treatment was performed by mounting cotyledons on microscope glass slides with 100 µg/mL BFA followed by immediate observation with the confocal microscope.
Cell Fractionation and Immunoblot
One-week-old seedlings grown on half-strength MS of Col-0/ArfA1F-YFP and STINGY/ArfA1F-YFP were harvested and homogenated at 4°C with a mortar and pestle in buffer (300 mM Suc, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1× P9599 Protease inhibitors). The homogenate was filtered using Miracloth paper and then centrifuged at 5000g for 5 min at 4°C. To pellet the microsomal fraction, the supernatant was centrifuged for 1 h at 100,000g. The supernatant (soluble fraction) was concentrated using Amicon ultra-4 filter units up to 1 mL. The membrane fraction was solubilized using 1 mL of PBS buffer, 0.05% Triton, and 1× P9599 Protease inhibitors. To test whether Arf1AF-YFP distribution in the cytosol is larger in STINGY compared with the nonmutagenized control, the soluble fractions were normalized to the respective membrane fractions. To ensure that an equal membrane fraction was extracted, quantification was carried out using a Bio-Rad protein assay. Ponceau staining of the blotting membrane further confirmed equal loading of the soluble and membrane fractions. Samples were separated by SDS page, and the blotted nitrocellulose was incubated overnight with blocking solution (5% powdered milk, 1× PBS, 0.02% sodium azide, and 0.5% Tween20). The blotted membrane was incubated with a GFP antibody (1:3000 dilution) for 4 h. Immunoblot analyses and quantification of NMT and NMTA160T were performed using NMT1 antibodies as described (Pierre et al., 2007; Adam et al., 2011).
Expression and Purification of NMT1 and NMT1A160T
Recombinant NMT1 and NMT1A160T carrying a 6-His tag in their N terminus were expressed and purified as previously described (Boisson and Meinnel, 2003; Pierre et al., 2007). Purified NMTs (>90% pure as estimated by SDS-PAGE) were dialyzed against 20 mM sodium phosphate buffer, pH 7.3, 0.5 M NaCl, 10 mM 2-mercaptoethanol, and 55% glycerol and were stored at −20°C prior to use.
MYR Kinetic Assays
The kinetics of MYR catalyzed by NMT or NMT1A160T were monitored by a modification of the coupled assay described by Boisson and Meinnel (2003), in which absorbance detection of NADH production was replaced by fluorescence detection (Traverso et al., 2013a). The range of peptide concentrations for the determination of the kinetic parameters was 2.5 to 200 µM. Kinetic parameters were derived as described earlier (Boisson and Meinnel, 2003).
Liquid Chromatography–Mass Spectrometry and Protein Identification and Quantification
Total soluble protein extracts from leaves of Arabidopsis SEC-RFP and STINGY were prepared as previously described (Espagne et al., 2007). Forty-five micrograms of protein was run in a one-dimensional SDS-PAGE gel, and each lane was cut in four bands and analyzed by nano-liquid chromatography–tandem mass spectrometry using LTQ-Orbitrap-Velos after in-gel digestion (Shevchenko et al., 1996). Quantification of each identified protein was based using a label-free Proteome Discoverer analysis (ThermoFinnegan). The PM-enriched fraction was purified as described previously (Marmagne et al., 2007) based on Dextran/polyethylene glycol membrane vesicles enrichment using microsomal fractions prepared from SEC-RFP and STINGY seedlings. One-hundred-and-fifty micrograms of purified protein fractions were separated on a one-dimensional SDS-PAGE gel, and each lane was cut into eight bands, which were analyzed as the soluble proteins. In both cases, Easy-nLC (Thermo Scientific) peptides were loaded on a guard column (Proxeon Easy column, i.d. 100 µm, C18-A1) followed by a separation on a Nikkyo Technos analytical column (NTCC-360/100-5-153) using 80-min gradients with 0.1% FA in water (solvent A) and 0.1% FA in ACAN (solvent B) at a flow rate of 300 nL/min. Each sample injection and analysis was followed by a blank injection to prevent carryover. The acquisition cycle consisted of a survey Fourier transform mass spectrometry scan at the highest resolving power (100,000) followed by top 20 data-dependent tandem mass spectrometry scans acquired in the LTQ with a dynamic exclusion set at 30 s.
All the spectral data were searched against TAIR (version 10) of the Arabidopsis annotated genome (http://www.arabidopsis.org/) using Mascot. The relative quantification of identified proteins was performed based on the integrated precursor area in mass spectrometry using Proteome Discoverer (Thermo Scientific v. 1.3) and combined with peptide identification results from Mascot (Matrix Science v2.3). For the soluble extract, the identification yielded 532 proteins. For PM-enriched fractions, the analysis yielded the identification of 5210 proteins in both SEC-RFP and the STINGY mutant. After removing ambiguous gene models (1000 accessions), the identified protein was represented by 4210 accessions. The proteins with a low number of spectral counts (<10 spectral counts) and/or low precursor area (<1.107) counts were considered to be of low confidence for quantifications and are indicated in gray (see Supplemental Table 2 online).
The PM set of annotations of cellular subproteomes was obtained from TAIR (http://www.arabidopsis.org/) and the Plant Proteomics database (http://ppdb.tc.cornell.edu/dbsearch/subproteome.aspx). In addition, a lower number of proteins annotated as ER/Golgi (∼100) were also retrieved. To functionally assign proteins within cellular functions we used the MapManBin functional classification (Thimm et al., 2004).
Homology Modeling and Structural Analyses
A three-dimensional model (residues 45 to 432) of Arabidopsis NMT1 was computed using available coordinates of Homo sapiens NMT1 (Hs NMT1; Protein Data Bank code 3IU1, chain B; 1.42-Å resolution) using SWISS-MODEL (Schwede et al., 2003; Arnold et al., 2006). Both NMT1 and Hs NMT1 display 56% homology. The A160T substitution was next introduced in the Arabidopsis NMT1 sequence, and the model was refined using the wild-type protein structure. The model was next aligned with the various complexes available of the Saccharomyces cerevisiae NMT three-dimensional structure (Farazi et al., 2001b) using magic and iterative fit functions (1IIC, chain A). The Sc NMT:myristoyl-CoA complex was selected as it revealed close vicinity between the myristoyl-CoA binding site lying in the N-terminal domain and the location of the substitution.
Accession Numbers
Sequence data for the genes used in this article can be found in TAIR under the following accession numbers: NMT1 (At5g57020), ArfA1F (At1g10630), and Ubiquitin 10 (At4g05320); other genes mentioned are LACS8 (AT2G04350.1), LACS1 (AT2G47240.1), LACS9 (AT1G77590.1), and PSD3 (AT4G25970).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Plant Phenotype.
Supplemental Figure 2. Alignment of Arabidopsis, H. sapiens, and S. cerevisiae Protein Sequences.
Supplemental Figure 3. Complementation of STINGY Salt Sensitivity by NMT1.
Supplemental Figure 4. Multiple Sequence Alignment of Arabidopsis Arf1A1C, ArfA1F, and ArfA1E.
Supplemental Figure 5. Model of NMT1A160T in Complex with Myristoyl-CoA.
Supplemental Figure 6. STINGY Shows Reduced Accumulation of NMT1A160T Protein.
Supplemental Figure 7. YFP-NMT1 and YFP-NMT1A160T Are Distributed in the Cytosol.
Supplemental Figure 8. Proteome Analysis of Total Soluble Proteins from Leaves of 9-Week-Old Plantlets.
Supplemental Figure 9. Comparison of SEC-RFP and STINGY Plasma Membrane Proteomes.
Supplemental Figure 10. ArfA1F-YFP Is Distributed to the Golgi.
Supplemental Figure 11. HMA Treatment and the ArfA1F(G2A) Mutation Compromise ArfA1F Distribution to Endomembranes.
Supplemental Figure 12. Overexpression of ArfA1F(G2A) Does Not Affect the Distribution of the Golgi Marker ST-GFP and the Golgi Number in Cortical Region Areas.
Supplemental Table 1. Probabilistic Functional Gene Network of NMT1.
Supplemental Table 2. Oligonucleotides Used for the Construct Preparation.
Supplemental Data Set 1. List and Quantification of the Whole Proteins Identified in STINGY and Control (Classification According to Protein Function).
Supplemental Data Set 2. Summarizing Table of All Identifications and Quantifications.
Supplementary Material
Acknowledgments
We thank Eileen Morey (Michigan State University–Department of Energy Plant Research Lab, Michigan State University) for editing the article. We acknowledge support by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (Award DE-FG02-91ER20021), National Science Foundation (MCB 0948584 and 1243792), and the Agence Nationale de la Recherche (PalMyPRot). We thank Christelle Espagne and Willy Bienvenut (C.G. laboratory) for assistance with mass spectrometry analysis.
AUTHOR CONTRIBUTIONS
F.B. conceived the project. L.R. and F.B. designed and analyzed the cell biology experiments; G.S. and L.R. isolated, mapped and complemented STINGY, and performed the cell biology experiments (Figs. 1-7; Supplemental Figures 1-4, 7, 10-12, Supplemental Tables 1 and 2). T.M. and C.G. designed and analyzed the proteomics experiments; W.M. and C.M. performed the proteomics experiments (Tables 1 and 2; Supplemental Figures 5 and 6; 8-9; Supplemental Data Set 1 and 2). L.R., G.S., T.M., G.C. and F.B. wrote the article.
Glossary
- MYR
N-myristoylation
- NMT
N-myristoyltransferase
- MYRed
N-myristoylated
- Col-0
Columbia-0
- RFP
red fluorescent protein
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- FRAP
fluorescence recovery after photobleaching
- YFP
yellow fluorescent protein
- Ler
Landsberg erecta
- CDPK
calcium-dependent protein kinase
- HMA
2-hydroxymyristic acid
- PM
plasma membrane
- TAIR
The Arabidopsis Information Resource
- BFA
brefeldin A
- MS
Murashige and Skoog
- EMS
ethyl methanesulfonate
References
- Adam Z., Frottin F., Espagne C., Meinnel T., Giglione C. (2011). Interplay between N-terminal methionine excision and FtsH protease is essential for normal chloroplast development and function in Arabidopsis. Plant Cell 23: 3745–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Anders N., Jürgens G. (2008). Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell. Mol. Life Sci. 65: 3433–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B., Beraud-Dufour S., Chardin P., Chabre M. (1997). N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry 36: 4675–4684 [DOI] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Batoko H., Zheng H.Q., Hawes C., Moore I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D.R., et al. (2008). Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar, R.S., Ashrafi, K., Futterer, K., Waksman, G., and Gordon, J.I. (2001). Biology and enzymology of protein N-myristoylation. In The enzymes, F. Tamanoi and D.S. Sigman, eds (San Diego, CA: Academic Press), pp. 241–286. [Google Scholar]
- Boevink P., Oparka K., Santa Cruz S., Martin B., Betteridge A., Hawes C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15: 441–447 [DOI] [PubMed] [Google Scholar]
- Boisson B., Giglione C., Meinnel T. (2003). Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J. Biol. Chem. 278: 43418–43429 [DOI] [PubMed] [Google Scholar]
- Boisson B., Meinnel T. (2003). A continuous assay of myristoyl-CoA:protein N-myristoyltransferase for proteomic analysis. Anal. Biochem. 322: 116–123 [DOI] [PubMed] [Google Scholar]
- Borevitz J. (2006). Genotyping and mapping with high-density oligonucleotide arrays. Methods Mol. Biol. 323: 137–145 [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Hanton S., DaSilva L.L., Boevink P., Evans D., Oparka K., Denecke J., Hawes C. (2003). ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J. 34: 269–281 [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Snapp E.L., Roberts A.G., Lippincott-Schwartz J., Hawes C. (2002). Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell 14: 1293–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cox R., Mason-Gamer R.J., Jackson C.L., Segev N. (2004). Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol. Biol. Cell 15: 1487–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- daSilva L.L., Snapp E.L., Denecke J., Lippincott-Schwartz J., Hawes C., Brandizzi F. (2004). Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16: 1753–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J.G., Jackson C.L. (2011). ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12: 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio R.J., Jackson-Machelski E., Heuckeroth R.O., Olins P.O., Devine C.S., Yonemoto W., Slice L.W., Taylor S.S., Gordon J.I. (1990). Protein N-myristoylation in Escherichia coli: Reconstitution of a eukaryotic protein modification in bacteria. Proc. Natl. Acad. Sci. USA 87: 1506–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio R.J., Reed S.I., Gordon J.I. (1992). Mutations of human myristoyl-CoA:protein N-myristoyltransferase cause temperature-sensitive myristic acid auxotrophy in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89: 4129–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio R.J., Rudnick D.A., Johnson R.L., Johnson D.R., Gordon J.I. (1991). Myristic acid auxotrophy caused by mutation of S. cerevisiae myristoyl-CoA:protein N-myristoyltransferase. J. Cell Biol. 113: 1313–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio R.J., Towler D.A., Heuckeroth R.O., Gordon J.I. (1989). Disruption of the yeast N-myristoyl transferase gene causes recessive lethality. Science 243: 796–800 [DOI] [PubMed] [Google Scholar]
- Espagne C., Martinez A., Valot B., Meinnel T., Giglione C. (2007). Alternative and effective proteomic analysis in Arabidopsis. Proteomics 7: 3788–3799 [DOI] [PubMed] [Google Scholar]
- Farazi T.A., Waksman G., Gordon J.I. (2001a). The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 276: 39501–39504 [DOI] [PubMed] [Google Scholar]
- Farazi T.A., Waksman G., Gordon J.I. (2001b). Structures of Saccharomyces cerevisiae N-myristoyltransferase with bound myristoylCoA and peptide provide insights about substrate recognition and catalysis. Biochemistry 40: 6335–6343 [DOI] [PubMed] [Google Scholar]
- Faso C., Boulaflous A., Brandizzi F. (2009a). The plant Golgi apparatus: Last 10 years of answered and open questions. FEBS Lett. 583: 3752–3757 [DOI] [PubMed] [Google Scholar]
- Faso C., Chen Y.N., Tamura K., Held M., Zemelis S., Marti L., Saravanan R., Hummel E., Kung L., Miller E., Hawes C., Brandizzi F. (2009b). A missense mutation in the Arabidopsis COPII coat protein Sec24A induces the formation of clusters of the endoplasmic reticulum and Golgi apparatus. Plant Cell 21: 3655–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frottin F., Martinez A., Peynot P., Mitra S., Holz R.C., Giglione C., Meinnel T. (2006). The proteomics of N-terminal methionine cleavage. Mol. Cell. Proteomics 5: 2336–2349 [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Giglione C., Boularot A., Meinnel T. (2004). Protein N-terminal methionine excision. Cell. Mol. Life Sci. 61: 1455–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover C.J., Hartman K.D., Felsted R.L. (1997). Human N-myristoyltransferase amino-terminal domain involved in targeting the enzyme to the ribosomal subcellular fraction. J. Biol. Chem. 272: 28680–28689 Erratum. J. Biol. Chem. 273: 5988. [DOI] [PubMed] [Google Scholar]
- Hanton S.L., Chatre L., Renna L., Matheson L.A., Brandizzi F. (2007). De novo formation of plant endoplasmic reticulum export sites is membrane cargo induced and signal mediated. Plant Physiol. 143: 1640–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton S.L., Matheson L.A., Chatre L., Brandizzi F. (2009). Dynamic organization of COPII coat proteins at endoplasmic reticulum export sites in plant cells. Plant J. 57: 963–974 [DOI] [PubMed] [Google Scholar]
- Haseloff J., Siemering K.R., Prasher D.C., Hodge S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuckeroth R.O., Towler D.A., Adams S.P., Glaser L., Gordon J.I. (1988). 11-(Ethylthio)undecanoic acid. A myristic acid analogue of altered hydrophobicity which is functional for peptide N-myristoylation with wheat germ and yeast acyltransferase. J. Biol. Chem. 263: 2127–2133 [PubMed] [Google Scholar]
- Johnson D.R., Bhatnagar R.S., Knoll L.J., Gordon J.I. (1994). Genetic and biochemical studies of protein N-myristoylation. Annu. Rev. Biochem. 63: 869–914 [DOI] [PubMed] [Google Scholar]
- Kamath R.S., et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Koizumi K., Naramoto S., Sawa S., Yahara N., Ueda T., Nakano A., Sugiyama M., Fukuda H. (2005). VAN3 ARF-GAP-mediated vesicle transport is involved in leaf vascular network formation. Development 132: 1699–1711 [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M., Hawes C., Carvalho C. (2005a). Holding it all together? Candidate proteins for the plant Golgi matrix. Curr. Opin. Plant Biol. 8: 632–639 [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M., Hawes C., Carvalho C., Oparka K., Gillingham A.K., Boevink P. (2005b). An Arabidopsis GRIP domain protein locates to the trans-Golgi and binds the small GTPase ARL1. Plant J. 44: 459–470 [DOI] [PubMed] [Google Scholar]
- Lee S.C., Schmidtke S.N., Dangott L.J., Shaw B.D. (2008). Aspergillus nidulans ArfB plays a role in endocytosis and polarized growth. Eukaryot. Cell 7: 1278–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Shaw B.D. (2007). A novel interaction between N-myristoylation and the 26S proteasome during cell morphogenesis. Mol. Microbiol. 63: 1039–1053 [DOI] [PubMed] [Google Scholar]
- Lee S.C., Shaw B.D. (2008a). ArfB links protein lipidation and endocytosis to polarized growth of Aspergillus nidulans. Commun. Integr. Biol. 1: 51–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Shaw B.D. (2008b). Localization and function of ADP ribosylation factor A in Aspergillus nidulans. FEMS Microbiol. Lett. 283: 216–222 [DOI] [PubMed] [Google Scholar]
- Lodge J.K., Jackson-Machelski E., Devadas B., Zupec M.E., Getman D.P., Kishore N., Freeman S.K., McWherter C.A., Sikorski J.A., Gordon J.I. (1997). N-myristoylation of Arf proteins in Candida albicans: An in vivo assay for evaluating antifungal inhibitors of myristoyl-CoA:protein N-myristoyltransferase. Microbiology 143: 357–366 [DOI] [PubMed] [Google Scholar]
- Maple J., Møller S.G. (2007). Mutagenesis in Arabidopsis. Methods Mol. Biol. 362: 197–206 [DOI] [PubMed] [Google Scholar]
- Marmagne A., Ferro M., Meinnel T., Bruley C., Kuhn L., Garin J., Barbier-Brygoo H., Ephritikhine G. (2007). A high content in lipid-modified peripheral proteins and integral receptor kinases features in the Arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 6: 1980–1996 [DOI] [PubMed] [Google Scholar]
- Marti L., Stefano G., Tamura K., Hawes C., Renna L., Held M.A., Brandizzi F. (2010). A missense mutation in the vacuolar protein GOLD36 causes organizational defects in the ER and aberrant protein trafficking in the plant secretory pathway. Plant J. 63: 901–913 [DOI] [PubMed] [Google Scholar]
- Martin D.D., Beauchamp E., Berthiaume L.G. (2011). Post-translational myristoylation: Fat matters in cellular life and death. Biochimie 93: 18–31 [DOI] [PubMed] [Google Scholar]
- Martinez A., Traverso J.A., Valot B., Ferro M., Espagne C., Ephritikhine G., Zivy M., Giglione C., Meinnel T. (2008). Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics 8: 2809–2831 [DOI] [PubMed] [Google Scholar]
- Matheson L.A., Hanton S.L., Rossi M., Latijnhouwers M., Stefano G., Renna L., Brandizzi F. (2007). Multiple roles of ADP-ribosylation factor 1 in plant cells include spatially regulated recruitment of coatomer and elements of the Golgi matrix. Plant Physiol. 143: 1615–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K., Chandler V., Cone K., Kaeppler H., Kaeppler S., Kerschen A., Pikaard C., Richards E., Sidorenko L., Smith T., Springer N., Wulan T. (2005). Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol 392: 1–24 [DOI] [PubMed] [Google Scholar]
- Mehlmer N., Wurzinger B., Stael S., Hofmann-Rodrigues D., Csaszar E., Pfister B., Bayer R., Teige M. (2010). The Ca(2+)-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 63: 484–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Giglione C. (2008). Protein lipidation meets proteomics. Front. Biosci. 13: 6326–6340 [DOI] [PubMed] [Google Scholar]
- Meinnel T., Peynot P., Giglione C. (2005). Processed N-termini of mature proteins in higher eukaryotes and their major contribution to dynamic proteomics. Biochimie 87: 701–712 [DOI] [PubMed] [Google Scholar]
- Meinnel T., Serero A., Giglione C. (2006). Impact of the N-terminal amino acid on targeted protein degradation. Biol. Chem. 387: 839–851 [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nerlich A., von Orlow M., Rontein D., Hanson A.D., Dörmann P. (2007). Deficiency in phosphatidylserine decarboxylase activity in the psd1 psd2 psd3 triple mutant of Arabidopsis affects phosphatidylethanolamine accumulation in mitochondria. Plant Physiol. 144: 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntwasa M., Aapies S., Schiffmann D.A., Gay N.J. (2001). Drosophila embryos lacking N-myristoyltransferase have multiple developmental defects. Exp. Cell Res. 262: 134–144 [DOI] [PubMed] [Google Scholar]
- Paige L.A., Zheng G.Q., DeFrees S.A., Cassady J.M., Geahlen R.L. (1990). Metabolic activation of 2-substituted derivatives of myristic acid to form potent inhibitors of myristoyl CoA:protein N-myristoyltransferase. Biochemistry 29: 10566–10573 [DOI] [PubMed] [Google Scholar]
- Pierre M., Traverso J.A., Boisson B., Domenichini S., Bouchez D., Giglione C., Meinnel T. (2007). N-myristoylation regulates the SnRK1 pathway in Arabidopsis. Plant Cell 19: 2804–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price H.P., Goulding D., Smith D.F. (2005a). ARL1 has an essential role in Trypanosoma brucei. Biochem. Soc. Trans. 33: 643–645 [DOI] [PubMed] [Google Scholar]
- Price H.P., Güther M.L., Ferguson M.A., Smith D.F. (2010). Myristoyl-CoA:protein N-myristoyltransferase depletion in trypanosomes causes avirulence and endocytic defects. Mol. Biochem. Parasitol. 169: 55–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price H.P., Menon M.R., Panethymitaki C., Goulding D., McKean P.G., Smith D.F. (2003). Myristoyl-CoA:protein N-myristoyltransferase, an essential enzyme and potential drug target in kinetoplastid parasites. J. Biol. Chem. 278: 7206–7214 [DOI] [PubMed] [Google Scholar]
- Price H.P., Panethymitaki C., Goulding D., Smith D.F. (2005b). Functional analysis of TbARL1, an N-myristoylated Golgi protein essential for viability in bloodstream trypanosomes. J. Cell Sci. 118: 831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q., Rajala R.V., Anderson W., Jiang C., Rozwadowski K., Selvaraj G., Sharma R., Datla R. (2000). Molecular cloning, genomic organization, and biochemical characterization of myristoyl-CoA:protein N-myristoyltransferase from Arabidopsis thaliana. J. Biol. Chem. 275: 9673–9683 [DOI] [PubMed] [Google Scholar]
- Rajala R.V., Datla R.S., Moyana T.N., Kakkar R., Carlsen S.A., Sharma R.K. (2000). N-myristoyltransferase. Mol. Cell. Biochem. 204: 135–155 [DOI] [PubMed] [Google Scholar]
- Randazzo P.A., Hirsch D.S. (2004). Arf GAPs: Multifunctional proteins that regulate membrane traffic and actin remodelling. Cell. Signal. 16: 401–413 [DOI] [PubMed] [Google Scholar]
- Renna L., Hanton S.L., Stefano G., Bortolotti L., Misra V., Brandizzi F. (2005). Identification and characterization of AtCASP, a plant transmembrane Golgi matrix protein. Plant Mol. Biol. 58: 109–122 [DOI] [PubMed] [Google Scholar]
- Resh M.D. (1999). Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451: 1–16 [DOI] [PubMed] [Google Scholar]
- Richter S., Geldner N., Schrader J., Wolters H., Stierhof Y.D., Rios G., Koncz C., Robinson D.G., Jürgens G. (2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448: 488–492 [DOI] [PubMed] [Google Scholar]
- Richter S., Müller L.M., Stierhof Y.D., Mayer U., Takada N., Kost B., Vieten A., Geldner N., Koncz C., Jürgens G. (2011). Polarized cell growth in Arabidopsis requires endosomal recycling mediated by GBF1-related ARF exchange factors. Nat. Cell Biol. 14: 80–86 [DOI] [PubMed] [Google Scholar]
- Robineau S., Chabre M., Antonny B. (2000). Binding site of brefeldin A at the interface between the small G protein ADP-ribosylation factor 1 (ARF1) and the nucleotide-exchange factor Sec7 domain. Proc. Natl. Acad. Sci. USA 97: 9913–9918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Langhans M., Saint-Jore-Dupas C., Hawes C. (2008). BFA effects are tissue and not just plant specific. Trends Plant Sci. 13: 405–408 [DOI] [PubMed] [Google Scholar]
- Robinson D.G., Scheuring D., Naramoto S., Friml J. (2011). ARF1 localizes to the golgi and the trans-Golgi network. Plant Cell 23: 846–849, author reply 849–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin, A., Espiau, B., Tetaud, E., Cuvillier, A., Lartigue, L., Ambit, A., Robinson, D.R., and Merlin, G. (2008). The leishmania ARL-1 and Golgi traffic. PLoS ONE 3: e1620. [DOI] [PMC free article] [PubMed]
- Saint-Jore C.M., Evins J., Batoko H., Brandizzi F., Moore I., Hawes C. (2002). Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29: 661–678 [DOI] [PubMed] [Google Scholar]
- Schoberer J., Runions J., Steinkellner H., Strasser R., Hawes C., Osterrieder A. (2010). Sequential depletion and acquisition of proteins during Golgi stack disassembly and reformation. Traffic 11: 1429–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuiki I., Schnabl M., Czabany T., Hrastnik C., Daum G. (2010). Phosphatidylethanolamine synthesized by four different pathways is supplied to the plasma membrane of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1801: 480–486 [DOI] [PubMed] [Google Scholar]
- Schwede T., Kopp J., Guex N., Peitsch M.C. (2003). SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw B.D., Momany C., Momany M. (2002). Aspergillus nidulans swoF encodes an N-myristoyl transferase. Eukaryot. Cell 1: 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Ketelaar T., de Ruijter N.C., Hawes C. (2009). Grab a Golgi: Laser trapping of Golgi bodies reveals in vivo interactions with the endoplasmic reticulum. Traffic 10: 567–571 [DOI] [PubMed] [Google Scholar]
- Stefano, G., Renna, L., and Brandizzi, F. (2012a). Fluorescence-microscopy screening and next-generation sequencing: useful tools for the identification of genes involved in organelle integrity. J. Vis. Exp. 62: 3809. [DOI] [PMC free article] [PubMed]
- Stefano G., Renna L., Hanton S.L., Chatre L., Haas T.A., Brandizzi F. (2006). ARL1 plays a role in the binding of the GRIP domain of a peripheral matrix protein to the Golgi apparatus in plant cells. Plant Mol. Biol. 61: 431–449 [DOI] [PubMed] [Google Scholar]
- Stefano G., Renna L., Moss T., McNew J.A., Brandizzi F. (2012b). In Arabidopsis, the spatial and dynamic organization of the endoplasmic reticulum and Golgi apparatus is influenced by the integrity of the C-terminal domain of RHD3, a non-essential GTPase. Plant J. 69: 957–966 [DOI] [PubMed] [Google Scholar]
- Stefano G., Renna L., Rossi M., Azzarello E., Pollastri S., Brandizzi F., Baluska F., Mancuso S. (2010). AGD5 is a GTPase-activating protein at the trans-Golgi network. Plant J. 64: 790–799 [DOI] [PubMed] [Google Scholar]
- Sun Q., Zybailov B., Majeran W., Friso G., Olinares P.D., van Wijk K.J. (2009). PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37 (Database issue): D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19: 391–397 [DOI] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Towler D.A., Gordon J.I., Adams S.P., Glaser L. (1988). The biology and enzymology of eukaryotic protein acylation. Annu. Rev. Biochem. 57: 69–99 [DOI] [PubMed] [Google Scholar]
- Traverso J.A., Giglione C., Meinnel T. (2013a). High-throughput profiling of N-myristoylation substrate specificity across species including pathogens. Proteomics 13: 25–36 [DOI] [PubMed] [Google Scholar]
- Traverso J.A., Micalella C., Martinez A., Brown S.C., Satiat-Jeunemaître B., Meinnel T., Giglione C. (2013b). Roles of N-terminal fatty acid acylations in membrane compartment partitioning: Arabidopsis h-type thioredoxins as a case study. Plant Cell 25: 1056–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan B., Lin Y., Mou T. (2007). Expression of rice Ca(2+)-dependent protein kinases (CDPKs) genes under different environmental stresses. FEBS Lett. 581: 1179–1189 [DOI] [PubMed] [Google Scholar]
- Wilcox C., Hu J.S., Olson E.N. (1987). Acylation of proteins with myristic acid occurs cotranslationally. Science 238: 1275–1278 [DOI] [PubMed] [Google Scholar]
- Wu S.J., Ding L., Zhu J.K. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.I., Voelker D.R. (2004). Reconstitution of phosphatidylserine transport from chemically defined donor membranes to phosphatidylserine decarboxylase 2 implicates specific lipid domains in the process. J. Biol. Chem. 279: 6635–6642 [DOI] [PubMed] [Google Scholar]
- Wurzinger B., Mair A., Pfister B., Teige M. (2011). Cross-talk of calcium-dependent protein kinase and MAP kinase signaling. Plant Signal. Behav. 6: 8–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Shrivastav A., Kosinski C., Sharma R.K., Chen M.H., Berthiaume L.G., Peters L.L., Chuang P.T., Young S.G., Bergo M.O. (2005). N-myristoyltransferase 1 is essential in early mouse development. J. Biol. Chem. 280: 18990–18995 [DOI] [PubMed] [Google Scholar]
- Zhang C., Halsey L.E., Szymanski D.B. (2011). The development and geometry of shape change in Arabidopsis thaliana cotyledon pavement cells. BMC Plant Biol. 11: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S.Y., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.