Symmetric division of chloroplasts ensures consistent chloroplast morphology during leaf growth and development. This study reveals that Arabidopsis ACCUMULATION AND REPLICATION OF CHLOROPLASTS3 negatively regulates chloroplast division-site placement in concert with MinD1 and MinE1 by inhibiting FtsZ ring assembly at nondivision sites primarily through interaction with FtsZ2.
Abstract
Chloroplast division is initiated by assembly of a mid-chloroplast FtsZ (Z) ring comprising two cytoskeletal proteins, FtsZ1 and FtsZ2. The division-site regulators ACCUMULATION AND REPLICATION OF CHLOROPLASTS3 (ARC3), MinD1, and MinE1 restrict division to the mid-plastid, but their roles are poorly understood. Using genetic analyses in Arabidopsis thaliana, we show that ARC3 mediates division-site placement by inhibiting Z-ring assembly, and MinD1 and MinE1 function through ARC3. ftsZ1 null mutants exhibited some mid-plastid FtsZ2 rings and constrictions, whereas neither constrictions nor FtsZ1 rings were observed in mutants lacking FtsZ2, suggesting FtsZ2 is the primary determinant of Z-ring assembly in vivo. arc3 ftsZ1 double mutants exhibited multiple parallel but no mid-plastid FtsZ2 rings, resembling the Z-ring phenotype in arc3 single mutants and showing that ARC3 affects positioning of FtsZ2 rings as well as Z rings. ARC3 overexpression in the wild type and ftsZ1 inhibited Z-ring and FtsZ2-ring assembly, respectively. Consistent with its effects in vivo, ARC3 interacted with FtsZ2 in two-hybrid assays and inhibited FtsZ2 assembly in a heterologous system. Our studies are consistent with a model wherein ARC3 directly inhibits Z-ring assembly in vivo primarily through interaction with FtsZ2 in heteropolymers and suggest that ARC3 activity is spatially regulated by MinD1 and MinE1 to permit Z-ring assembly at the mid-plastid.
INTRODUCTION
Chloroplasts descended from an ancestral cyanobacterium through endosymbiosis and are responsible for photosynthesis and numerous essential metabolic processes (Gray, 1992; Pyke, 1999). Plastid continuity during plant development, growth, and reproduction is maintained by division of preexisting organelles. Chloroplasts divide by binary fission, similar to their bacterial ancestors, to produce daughter organelles of nearly equal size and maintain the appropriate population density during cell division and expansion (Plattaloia and Thomson, 1977; Leech et al., 1981; Pyke, 1999). Chloroplast division involves the assembly of a number of proteins into ring-shaped complexes at the chloroplast division site (Yang et al., 2008; Yoshida et al., 2012). Consistent with the origin of chloroplasts, several chloroplast division proteins are homologs of bacterial cell division proteins (Osteryoung and Vierling, 1995; Colletti et al., 2000; Itoh et al., 2001; Maple et al., 2002; Vitha et al., 2003), and models of bacterial cell division have guided studies and understanding of chloroplast division mechanisms in plants (Okazaki et al., 2010; Miyagishima, 2011). During evolution, plants have also acquired new chloroplast division components of eukaryotic origin (Gao et al., 2003; Miyagishima et al., 2006; Nakanishi et al., 2009).
In both bacteria and chloroplasts, assembly of the FtsZ ring (Z-ring) at the division site is the initial event in formation of the division complex (Bi and Lutkenhaus, 1991; Vitha et al., 2001). FtsZ is a self-assembling, tubulin-related cytoskeletal protein that polymerizes into protofilaments in vitro and forms the mid-cell or mid-plastid Z-ring in vivo (Bi and Lutkenhaus, 1991; Erickson et al., 1996; Vitha et al., 2001; Olson et al., 2010; Smith et al., 2010). Though FtsZ is a soluble protein, its interaction with membrane proteins tethers the Z ring to the membrane, where it generates contractile force for membrane constriction during division (Osawa et al., 2009). In bacteria, the protofilaments making up the Z ring are composed of a single FtsZ protein that assembles as a homopolymer, but the chloroplast Z ring, which is localized in the stroma, is composed of two FtsZ proteins, termed FtsZ1 and FtsZ2 (Osteryoung et al., 1998; Vitha et al., 2001). Like bacterial FtsZ, FtsZ1 and FtsZ2 are each capable of assembling as homopolymers in vitro and in a heterologous yeast (Schizosaccharomyces pombe) expression system, but they preferentially coassemble as heteropolymers (El-Kafafi et al., 2005; Olson et al., 2010; Smith et al., 2010; TerBush and Osteryoung, 2012). This is consistent with their colocalization to the mid-plastid Z ring in vivo and with genetic analysis conclusively demonstrating that both are required for normal chloroplast division in Arabidopsis thaliana (Vitha et al., 2001; Schmitz et al., 2009). However, in vivo, FtsZ2 can still assemble into long filaments and occasional FtsZ2 rings in the absence of FtsZ1, as shown in an Arabidopsis ftsZ1 null mutant (Yoder et al., 2007), whereas FtsZ1 does not assemble in mutants lacking FtsZ2 (Schmitz et al., 2009) even though it does so in vitro and in S. pombe (El-Kafafi et al., 2005; Olson et al., 2010; Smith et al., 2010; TerBush and Osteryoung, 2012). This suggests that FtsZ1 and FtsZ2 may contribute differentially to their coassembly in the Z ring in vivo.
Z-ring positioning at the cell division site in bacteria is achieved by a negative regulatory system called the Min system. In Escherichia coli, the Min system comprises Minicell C (MinC), MinD, and MinE, which cooperate in a complex set of interactions that inhibit Z-ring assembly everywhere but at the cell center (Raskin and de Boer, 1999; Margolin, 2005; Lutkenhaus, 2007; de Boer, 2010). MinC, a soluble protein, interacts directly with FtsZ and functions as the direct assembly inhibitor (Hu et al., 1999; Shiomi and Margolin, 2007; Shen and Lutkenhaus, 2010). MinD, an ATPase, oligomerizes on the membrane toward one end of the cell and recruits MinC, forming a MinCD complex that inhibits Z-ring assembly (de Boer et al., 1992; Bi and Lutkenhaus, 1993; Hu and Lutkenhaus, 2003). MinE is subsequently recruited by MinD, which displaces MinC and stimulates MinD ATPase activity, releasing MinC and MinD from the membrane (Hu and Lutkenhaus, 2001; Park et al., 2012). Following nucleotide exchange, MinD then reassembles on the membrane at the opposite end of the cell, recruiting MinC and inhibiting Z-ring assembly at that end (Lutkenhaus, 2008). MinE is again recruited by MinD to the opposite membrane, releasing MinC and MinD, and the cycle repeats (Rowland et al., 2000; Lutkenhaus, 2008). In this way, MinE causes MinC and MinD to oscillate from pole to pole, creating a time-averaged concentration gradient of membrane-tethered MinC that is high at the cell poles and low at the cell center, allowing Z-ring formation and cell division only at the mid-cell (Fu et al., 2001; Hu and Lutkenhaus, 2001; Meinhardt and de Boer, 2001; Lutkenhaus, 2008; Park et al., 2012). Thus, in E. coli, the inhibition of Z-ring assembly by MinCD is topologically suppressed at the cell center by the action of MinE.
MinC, MinD, and MinE are also present in cyanobacteria, and homologs of MinD and MinE acquired through endosymbiosis have conserved functions in regulating Z-ring and division-site placement in green-lineage chloroplasts (Colletti et al., 2000; Itoh et al., 2001; Fujiwara et al., 2004; Mazouni et al., 2004; Aldridge and Møller, 2005). However, plants have lost MinC as the direct inhibitor of Z-ring assembly. Instead, the plant-specific stromal protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS3 (ARC3) (Pyke and Leech, 1992) has been postulated as a functional replacement (Maple et al., 2007). In support of this hypothesis, overexpression of ARC3 in Arabidopsis inhibits chloroplast division, similar to the effect of MinC overexpression on bacterial cell division (de Boer et al., 1992; Maple et al., 2007), and chloroplasts in Arabidopsis arc3 mutants have misplaced and multiple constrictions (Maple et al., 2007; Zhang et al., 2009) and multiple Z rings (Glynn et al., 2007; Wilson et al., 2011). The latter phenotypes resemble those in arc11 mutants, which bear a mutation in Arabidopsis MinD1, and in transgenic plants overexpressing Arabidopsis MinE1 (Fujiwara et al., 2004; Glynn et al., 2007; Fujiwara et al., 2008), implicating ARC3 as a contributor to the regulation of Z-ring placement in plants. Furthermore, ARC3 was reported to interact directly with FtsZ1 (Maple et al., 2007) and to inhibit formation of FtsZ1 cytoskeletal filaments in S. pombe, consistent with a MinC-like function (TerBush and Osteryoung, 2012).
Here, we performed a series of studies in Arabidopsis to investigate the role of ARC3 in regulating chloroplast Z-ring positioning in vivo. Our findings provide strong evidence of a central role for ARC3 in spatial regulation of chloroplast division through negative regulation of Z-ring assembly and show that ARC3 functions primarily through interaction with FtsZ2. The results provide insights into the functioning of the chloroplast Min (cpMin) system in plants.
RESULTS
A C-Terminally Tagged ARC3 Fusion Protein Complements the Phenotypes of an ARC3 T-DNA Mutant
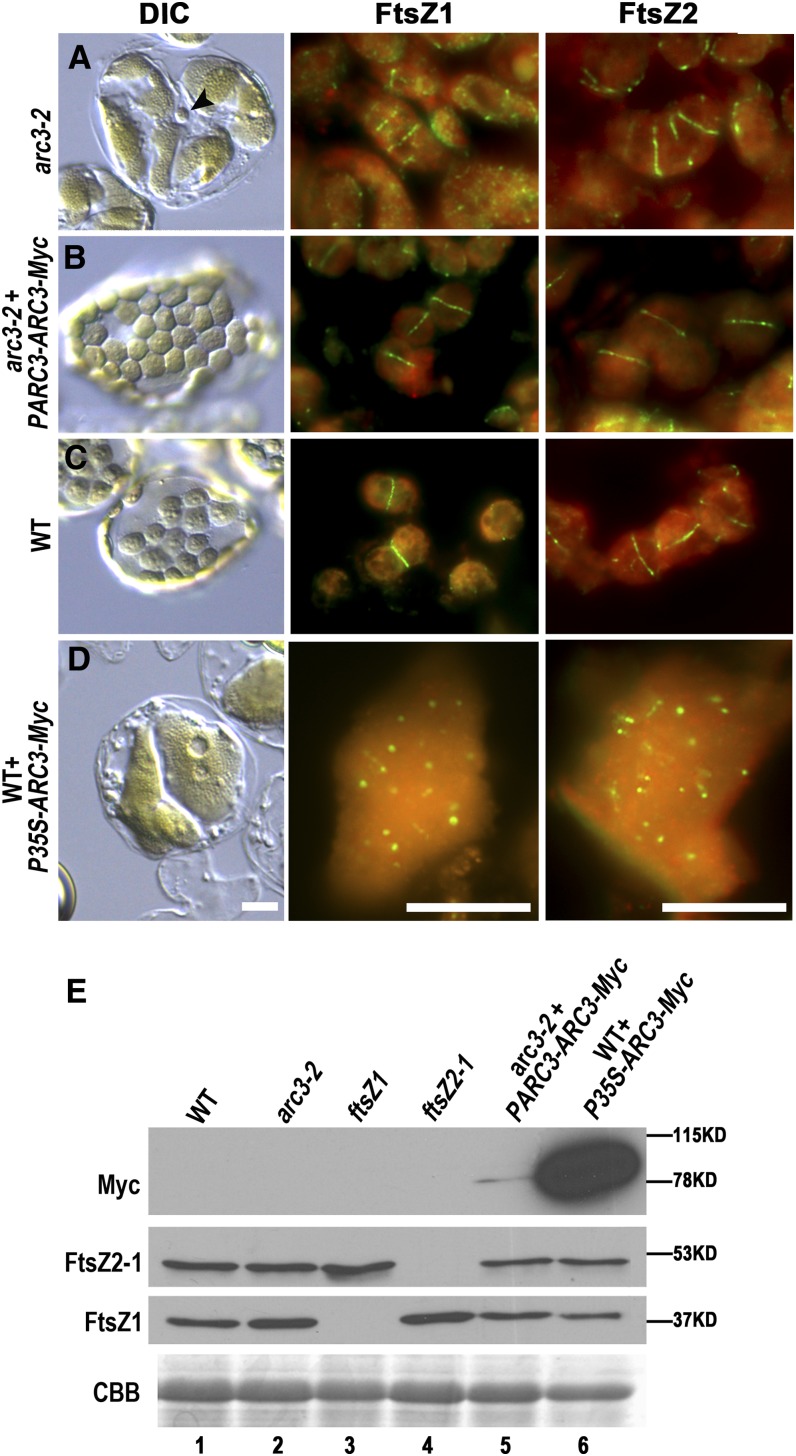
The ARC3 T-DNA insertion mutant arc3-2 (SALK_057144; Columbia-0 [Col-0]) (Shimada et al., 2004) contains a decreased number of elongated and expanded chloroplasts with irregular shapes and round mini-chloroplasts in mesophyll cells of young expanding leaves (Figure 1A, left panel), as observed previously (Pyke and Leech, 1992; Maple et al., 2007). Immunofluorescence labeling of FtsZ1 and FtsZ2 revealed multiple Z rings in this mutant (Figure 1A, middle and right panels), similar to the phenotype of the Landsberg erecta ethyl methanesulfonate mutant arc3-1 (Glynn et al., 2007). By contrast, single mid-plastid Z rings were observed in the wild type (Figure 1C, middle and right panels). FtsZ1 and FtsZ2 levels in arc3-2 were equivalent to those in the wild type (Figure 1E, lanes 1 and 2), indicating that ectopic Z-ring formation in the mutant was not the result of altered FtsZ accumulation. The presence of multiple Z rings in chloroplasts of arc3-2 supports a role for ARC3 as an inhibitor of Z-ring formation in the wild type.
Figure 1.
Overexpression of ARC3 Inhibits Z-Ring Assembly in Arabidopsis.
(A) to (D) Chloroplast phenotypes imaged with differential interference contrast (DIC) optics and immunofluorescence localization of FtsZ in mesophyll cells of the indicated genotypes. arc3-2 + PARC3-ARC3-Myc, arc3-2 mutants complemented with PARC3-ARC3-Myc; wild type + P35S-ARC3-Myc, wild-type (WT) plants expressing P35S-ARC3-Myc. FtsZ1 and FtsZ2 were immunolabeled with antibodies specific for FtsZ1 and FtsZ2-1, respectively. Green, Alexa Fluor 488–labeled FtsZ1 or FtsZ2-1; red, chlorophyll autofluorescence. Bars = 10 μm.
(E) Immunoblot analysis showing the relative ARC3, FtsZ1, and FtsZ2-1 levels in the indicated lines. Total protein from 2 mg fresh leaf tissue was loaded in each lane. The anti-Myc antibodies specifically recognized a single ARC3-Myc band (∼84 kD). Ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) in the Coomassie blue–stained gel (CBB) served as a loading control.
As a foundation for experiments described below, we fused a Myc tag to a genomic fragment of ARC3 bearing the native ARC3 promoter, generating PARC3-ARC3-Myc, and expressed the transgene in the arc3-2 mutant. In several independent T1 seedlings expressing ARC3-Myc (Figure 1E, lane 5), both the chloroplast morphology and Z-ring misplacement phenotypes were complemented (Figure 1B; see Supplemental Figure 1 online), indicating the ARC3-Myc fusion protein is functionally equivalent to ARC3.
FtsZ Assembly Is Sensitive to ARC3 Overexpression
To further characterize the role of ARC3 in chloroplast division, we generated an overexpression construct, P35S-ARC3-Myc, and expressed it in wild-type Col-0. Out of 43 T1 transgenic lines analyzed, 24 exhibited larger and fewer chloroplasts in mesophyll cells of young expanding leaves (two to three chloroplasts per cell) (Figure 1D, left panel), similar to the phenotype reported previously in plants expressing an untagged ARC3 cDNA (Maple et al., 2007). Immunoblot analysis showed that ARC3-Myc accumulation in these plants was much higher than in the complemented arc3-2 plants expressing PARC3-ARC3-Myc at the same growth stage (Figure 1E, lanes 5 and 6), demonstrating that elevated ARC3 accumulation inhibits chloroplast division.
To investigate the effect of ARC3 overexpression on FtsZ assembly, we performed immunofluorescence labeling of FtsZ1 and FtsZ2. In plants with extremely high ARC3-Myc levels (Figure 1E, lane 6), both FtsZ1 and FtsZ2 were localized in dots and occasionally very short filaments within chloroplasts (Figure 1D, middle and right panels). In plants with lower ARC3-Myc accumulation, some FtsZ filaments as well as dots were observed, though Z rings still could not assemble (see Supplemental Figure 2 online). FtsZ1 and FtsZ2 levels in all ARC3-Myc transgenics were nearly equal to those in the wild type (Figure 1E, lanes 1 and 6; see Supplemental Figure 2D online). These data provide evidence that the chloroplast division defects in the ARC3 cDNA (Maple et al., 2007) and ARC3-Myc overexpression lines are a consequence of inhibited FtsZ assembly and indicate the inhibition is dose-dependent.
ARC3 Is Required for MinD1 and MinE1 Function in Chloroplasts
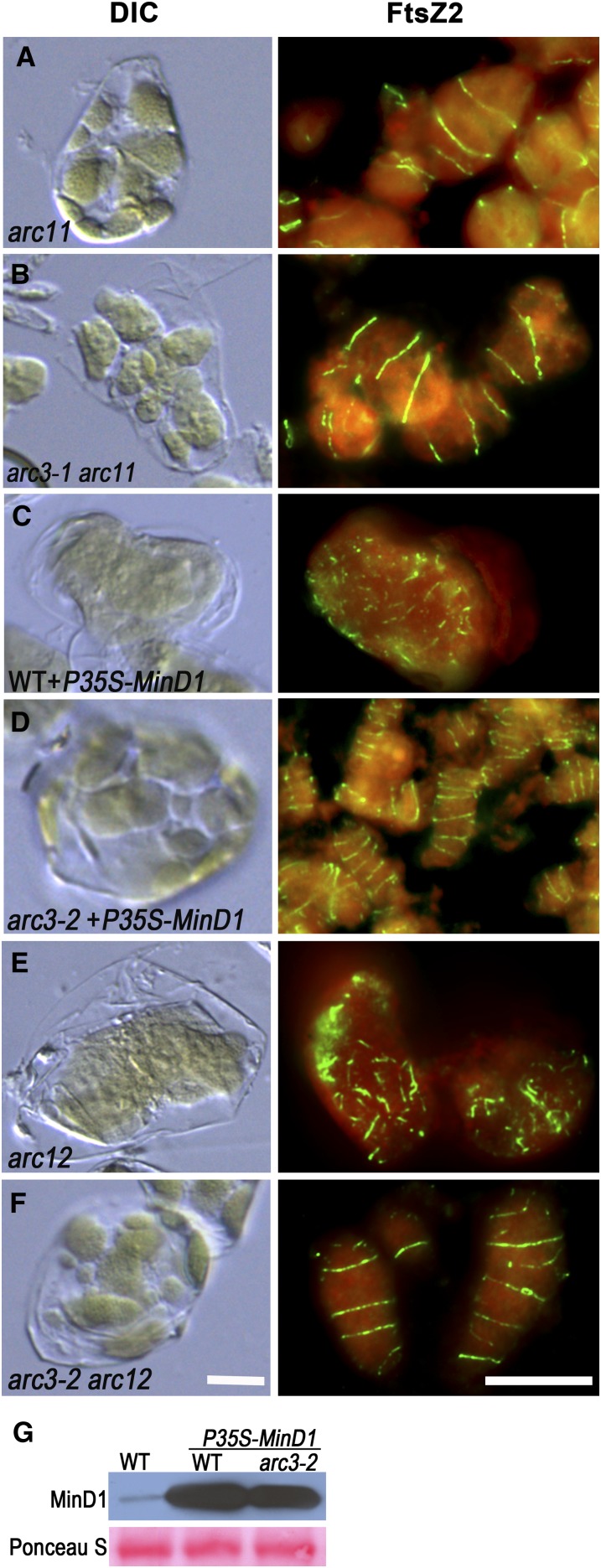
We used immunofluorescence labeling of FtsZ in a series of experiments to investigate the genetic relationships between ARC3, MinD1, and MinE1. We first examined double mutants homozygous for both arc3-1 and the MinD1 allele arc11 (Marrison et al., 1999; Fujiwara et al., 2004). Chloroplasts in both single mutants have very similar multiple Z-ring phenotypes (Wilson et al., 2011), and the phenotype of the arc3-1 arc11 double mutant (Figure 2B) was not notably distinct from that in the arc11 (Figure 2A), arc 3-1 (Wilson et al., 2011), or arc3-2 (Figure 1A) single mutants. Therefore, to further probe the relationship between ARC3 and MinD1, we overexpressed MinD1 in arc3-2 mutants and in wild-type plants as a control. As observed previously (Vitha et al., 2003), MinD1 overexpression in wild-type plants inhibited FtsZ assembly, producing plants with drastically enlarged chloroplasts and very short FtsZ filaments and puncta (Figures 2C and 2G). By contrast, chloroplasts in arc3-2 plants overexpressing MinD1 resembled those in arc3-2, both with respect to the heterogeneous chloroplast morphology phenotype and the presence of multiple Z rings (Figures 1A and 2D). Overexpression of MinD1 in both wild-type and arc3-2 plants was confirmed by immunoblotting (Figure 2G). These results reveal that MinD1 cannot inhibit FtsZ assembly in the absence of ARC3.
Figure 2.
ARC3 Is Required for MinD1- and MinE1-Mediated Z-Ring Positioning.
(A) to (F) Chloroplast phenotypes imaged by DIC and FtsZ immnunolocalization in the indicated genotypes. Anti-FtsZ2-1 antibody was used for FtsZ localization. WT, the wild type. Bars = 10 μm.
(G) Immunoblot analysis of relative MinD1 levels in the wild type, wild-type plants expressing P35S-MinD1, and arc3-2 plants expressing P35S-MinD1. Total protein from 0.5 mg of fresh leaf tissue was loaded in each lane. Ponceau S staining of Rubisco on the blot is shown as a loading control.
[See online article for color version of this figure.]
The Arabidopsis mutant arc12 bears a mutation in MinE1 and has one to two drastically enlarged chloroplasts and very short FtsZ filaments and dots (Pyke, 1999; Glynn et al., 2007) (Figure 2E). These phenotypes suggest that MinE1 functions in division-site placement by promoting FtsZ assembly. To test whether MinE1 functions in concert with ARC3, we generated an arc3-2 arc12 double mutant and compared the chloroplast morphology and FtsZ localization phenotypes with those in the single mutants. Chloroplasts were heterogeneous in size and number in arc3-2 arc12 plants (Figure 2F, left panel) and bore multiple Z rings (Figure 2F, right panel). These phenotypes clearly resembled those in arc3-2 (Figure 1A) but not arc12 (Figure 2E) mutants, indicating that MinE1, like MinD1, regulates Z-ring assembly through ARC3.
Additionally, we analyzed the genetic relationship between Arabidopsis MinD1 and MinE1. We first identified a MinD1 knockout mutant, minD1-1 (Wassilewskija background), harboring a T-DNA insertion near the beginning of the coding sequence (see Supplemental Figure 3A online). Immunoblot analysis showed that MinD1 was undetectable in the mutant (see Supplemental Figure 3B online), demonstrating that minD1-1 is a null allele of MinD1. The minD1-1 mutants exhibited heterogeneous chloroplasts and multiple Z rings in mesophyll cells (see Supplemental Figures 3D and 3G online), similar to the phenotype of arc11 (Marrison et al., 1999; Glynn et al., 2007). Subsequently, we generated a minD1-1 arc12 double mutant and found that the chloroplast and FtsZ morphology phenotypes were similar to those in minD1-1 and arc11 mutants, but not arc12 mutants (see Supplemental Figures 3E and 3H online). These results indicate that MinE1 function is dependent on MinD1 in vivo. Together with the analyses above (Figure 2), they also suggest that MinD1 and MinE1 may modulate ARC3 activity in a manner resembling the effects of MinD and MinE on MinC in bacteria.
FtsZ1 Is Not Required for Mid-Plastid Positioning of FtsZ2
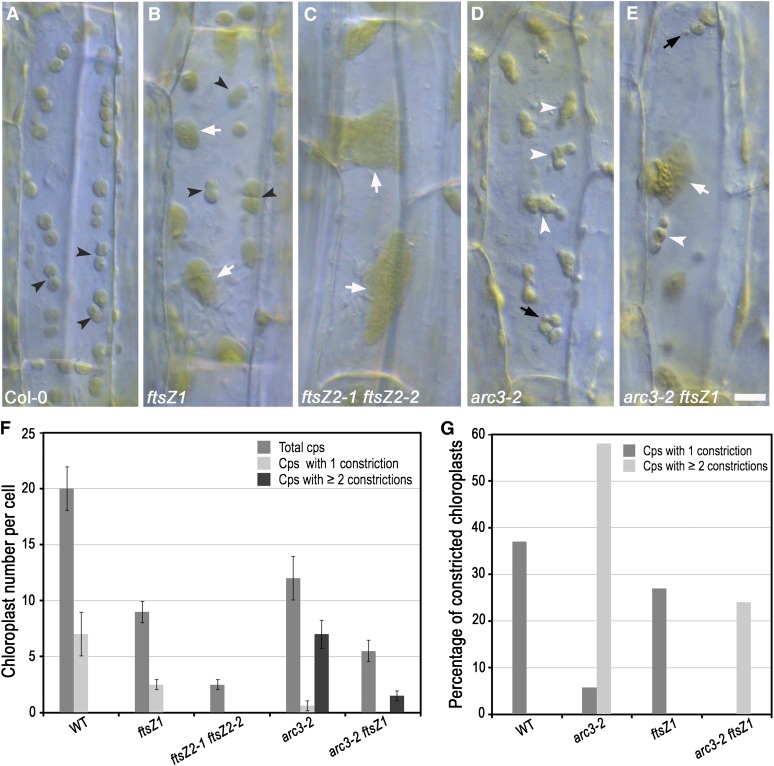
In Arabidopsis, FtsZ1 is encoded by a single gene, while FtsZ2 is encoded by two functionally redundant genes, FtsZ2-1 and FtsZ2-2 (Osteryoung et al., 1998; Schmitz et al., 2009). Although ftsZ1 and ftsZ2-1 ftsZ2-2 knockout mutants are both impaired in chloroplast division, we showed previously that leaf mesophyll cells in ftsZ2-1 ftsZ2-2 double mutants have only one or two drastically enlarged chloroplasts, whereas those in ftsZ1 mutants have multiple, somewhat smaller chloroplasts, indicating a more severe chloroplast division defect in mutants lacking FtsZ2 (Yoder et al., 2007; Schmitz et al., 2009). To better characterize the chloroplast division phenotypes in these mutants, we imaged live epidermal cells of hypocotyls, in which chloroplasts are fewer in number than in leaf mesophyll cells and division states can be more readily visualized (Fujiwara et al., 2004). Because chloroplast division is not synchronized, cells contained a combination of constricted and nonconstricted chloroplasts. In wild-type plants, cells contained an average of 20 chloroplasts, seven (37%) of which had single mid-plastid constrictions (Figures 3A, 3F, and 3G). By contrast, ftsZ1 mutants had fewer and mostly larger chloroplasts than wild-type plants, but all the chloroplasts in ftsZ1 mutants in which constrictions were visible (27%, n = 298) had only a single mid-plastid constriction, indicating proper division-site placement in these organelles (Figures 3B and 3F). These results indicate that FtsZ2 is able to sustain some degree of symmetric chloroplast division in the absence of FtsZ1 and reveal that FtsZ1 is not required for mid-plastid positioning of FtsZ2. By contrast, hypocotyl cells in ftsZ2-1 ftsZ2-2 mutants contained only two to three drastically enlarged chloroplasts (Figures 3C and 3F), indicating severely disrupted chloroplast division, and constrictions were not clearly detectable. Together, these findings are consistent with the presence of occasional FtsZ2 rings in mesophyll cells of ftsZ1 mutants (Yoder et al., 2007) and the lack of FtsZ1 rings in ftsZ2-1 ftsZ2-2 mutants (Schmitz et al., 2009).
Figure 3.
ARC3 Regulates Division-Site Positioning by FtsZ2 in the Absence of FtsZ1.
Live epidermal cells of hypocotyls in 5-d-old seedlings were imaged by DIC.
(A) to (E) Chloroplast phenotypes in the indicated genotypes. White and black arrowheads indicate chloroplasts with multiple and single constrictions, respectively; white and black arrows indicate expanded chloroplasts with no obvious constriction and mini-chloroplasts, respectively. Bar = 10 μm.
(F) Quantitative analysis of constricted chloroplasts in hypocotyls of the indicated genotypes. Total chloroplasts, chloroplasts with single constrictions, and chloroplasts with multiple constrictions in the plane of imaging were visually counted. Chloroplasts were counted as constricted whether the constriction was shallow or deep. Data show the mean ± sd (n ≥ 30 living cells per genotype). Cps, chloroplasts; WT, the wild type.
(G) Percentage of the total number of chloroplasts from all cells counted in (F) with single and multiple constrictions. A total of 620, 372, 298, and 165 chloroplasts were analyzed in wild-type, arc3-2, ftsZ1, and arc3-2 ftsZ1 plants, respectively. No constricted chloroplasts were observed in ftsZ2-1 ftsZ2-2 double mutants.
ARC3 Regulates Positioning of FtsZ2 Rings
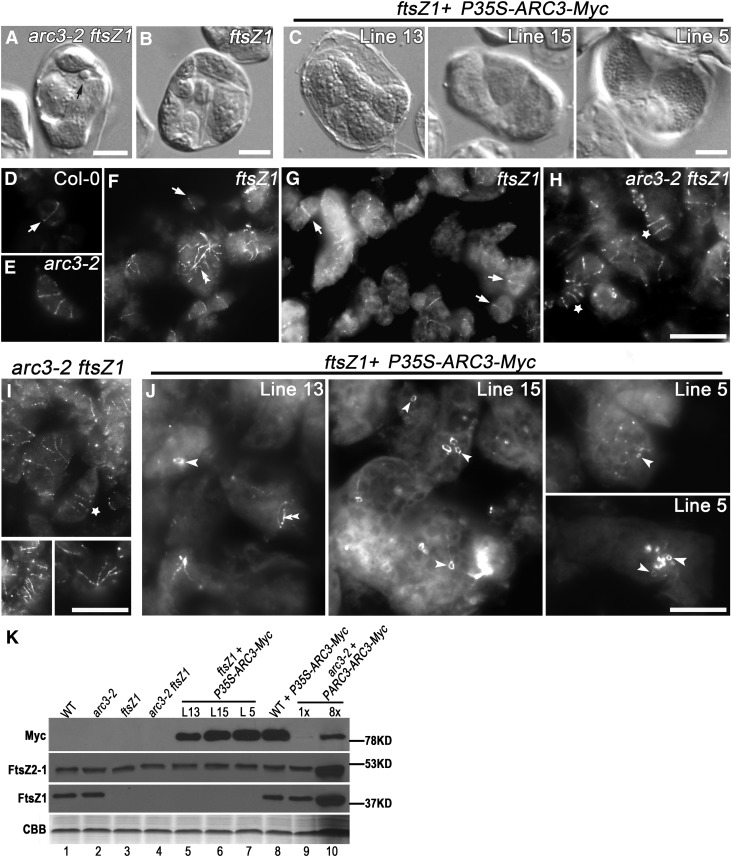
To test the effect of ARC3 on mid-plastid positioning of FtsZ2, we generated an arc3-2 ftsZ1 double mutant and observed chloroplast division profiles in epidermal cells of hypocotyls. Cells in the arc3-2 parent contained 12 chloroplasts per cell on average (Figures 3D and 3F), including some mini-chloroplasts and elongated chloroplasts with multiple constrictions (Figures 3D and 3G), similar to previous observations in petioles (Maple et al., 2007) and consistent with the presence of multiple Z rings in this mutant (Figure 1A, middle and right panels). Occasionally, chloroplasts with single constrictions were also observed in arc3-2 mutants, as seen in leaf mesophyll cells of arc3-1 (Wilson et al., 2011), but these constrictions were not positioned at the middle of the chloroplasts (Figures 3D, 3F, and 3G). arc3-2 ftsZ1 double mutants had an average of 5.5 chloroplasts per hypocotyl cell, fewer than in arc3-2 and ftsZ1 mutants, and enlarged chloroplasts, mini-chloroplasts, and elongated chloroplasts with multiple constrictions were observed simultaneously in the same cells (Figures 3E and 3F). However, among the 165 chloroplasts observed in 30 arc3-2 ftsZ1 cells, none had a single mid-plastid constriction, in contrast with chloroplasts in wild-type and ftsZ1 plants, while 24% had multiple constrictions (Figure 3G). Similarly, in young leaf tissue, mini-chloroplasts were observed in fixed mesophyll cells of arc3-2 ftsZ1 mutants (Figure 4A), similar to the phenotype of arc3-2 (Figure 1A), indicative of asymmetric chloroplast division (Colletti et al., 2000; Fujiwara et al., 2004).
Figure 4.
ARC3 Affects Assembly and Positioning of FtsZ2 Rings in the Absence of FtsZ1.
(A) to (J) Chloroplast phenotypes ([A] to [C]) and immunolocalization of FtsZ2 ([D] to [J]) in mesophyll cells of the indicated genotypes. Black arrow in (A) indicates a mini-chloroplast in arc3-2 ftsZ1 double mutants. In ([D] to [J]), the strong fluorescence signals (white) indicate FtsZ2 labeling by an anti-FtsZ2-1 antibody, and the dim background in gray shows the chloroplast shapes. White arrows indicate single Z rings in wild-type plants (D) and single FtsZ2 rings in ftsZ1 mutants ([F] and [G]). Asterisks indicate elongated chloroplasts in arc3-2 ftsZ1 plants bearing multiple FtsZ2 rings ([H] and [I]). Double arrowheads indicate long FtsZ2 filaments in ftsZ1 mutants (F) and ftsZ1 plants with lower levels of ARC3-Myc ([J], line 13). Single arrowheads indicate mini-rings of FtsZ2 in giant chloroplasts of ftsZ1 plants expressing P35S-ARC3-Myc (J). Bars = 10 μm.
(K) Immunoblot analysis showing the relative levels of ARC3-Myc, FtsZ1, and FtsZ2 in individual plants of the indicated lines. Total protein from 0.5 mg fresh leaf tissue was loaded in lanes 1 to 9 and eightfold more was loaded in lane 10. L, Transgenic line; WT, the wild type. Rubisco in the Coomassie blue–stained gel (CBB) served as a loading control.
To further investigate the effect of ARC3 on FtsZ2, we compared FtsZ2 filament morphologies in mesophyll cells of ftsZ1 mutants, arc3-2, and arc3-2 ftsZ1 mutants using immunofluorescence microscopy. In ftsZ1 mutants, long disorganized FtsZ2 filaments were seen within enlarged chloroplasts (Figure 4F), as observed previously (Yoder et al., 2007), but in addition FtsZ2 rings localized at the middle of smaller chloroplasts were evident (Figures 4F and 4G, arrows). These results show that FtsZ2 is able to assemble into a ring at the division site in the absence of FtsZ1. However, arc3-2 ftsZ1 double mutants exhibited arc3-2–like multiple FtsZ2 rings along elongated chloroplasts (Figures 4E, 4H, and 4I, asterisks, and Figure 1A) and displayed branched multiple FtsZ2 rings in some enlarged and expanded chloroplasts (Figures 4H and 4I), which were distinct from the more randomly distributed long FtsZ2 filaments in ftsZ1 mutants (Figure 4F). Together with the analysis of division-site placement described above, these results show that ARC3 is required for FtsZ2-ring positioning to the mid-plastid independent of FtsZ1.
Overexpression of ARC3 in ftsZ1 Mutants Inhibits FtsZ2 Filament Assembly
To investigate whether ARC3 influences FtsZ2 assembly as well as positioning, we transformed P35S-ARC3-Myc into ftsZ1 plants and analyzed 23 T1 transgenics. Lines 5 and 15, with high ARC3 accumulation, exhibited only two or three large chloroplasts in mesophyll cells (Figures 4C and 4K, lanes 6 and 7), similar to wild-type plants with high accumulation of ARC3-Myc (Figures 1D and 4K, lane 8) but quite different from the heterogonous chloroplast phenotype in ftsZ1 mutants (Figure 4B). These results indicate that chloroplast division in the transgenics is more impaired than in the ftsZ1 parent line. Line 13, with somewhat lower ARC3-Myc accumulation, had approximately five to seven moderately enlarged chloroplasts (Figures 4C and 4K, lane 5), suggesting that the severity of the division defect was correlated with the extent of ARC3-Myc accumulation. All these ftsZ1 transgenic lines accumulated much more ARC3-Myc than did arc3-2 plants complemented by PARC3-ARC3-Myc (Figure 4K, lanes 5 to 10).
To assess whether FtsZ2 assembly was affected by ARC3-Myc overexpression, we performed immunofluorescence labeling of FtsZ2 in ftsZ1 plants expressing P35S-ARC3-Myc. In line 13 (Figure 4J), a few long FtsZ2 filaments similar to those in the parent ftsZ1 mutant (Figure 4F) could be observed in enlarged chloroplasts, but their abundance was less than in ftsZ1 and the centrally localized FtsZ2 rings were not observed. Interestingly, small FtsZ2 rings (mini-rings) were occasionally seen in these chloroplasts as well (Figure 4J, arrowhead in right panel). Lines 5 and 15, both with higher ARC3-Myc levels, had many mini-rings within giant chloroplasts (Figures 4J and 4K, arrowheads). The diameter of the mini-rings ranged from 0.2 to 1.97 μm (n = 20), significantly smaller than the centrally localized rings formed at the early stages of chloroplast division in wild-type and ftsZ1 plants (Figures 4D, 4F, 4D, 4G, and 4J). Intriguingly, rings of similar size composed of either FtsZ2 or coassembled FtsZ1 and FtsZ2 were also seen in S. pombe (TerBush and Osteryoung, 2012). In line 5, many FtsZ2 puncta were also observed, similar to those seen in wild-type plants overexpressing ARC3-Myc at similar levels (Figures 1D, 4J, and 4K, lanes 7 and 8). FtsZ2 levels in all transgenic lines were equivalent to those in the wild type, ftsZ1, arc3-2, arc3-2 ftsZ1, wild type overexpressing ARC3-Myc, and complemented arc3-2 plants (Figure 4K), indicating that the FtsZ2 level was not altered by accumulation or depletion of ARC3. Based on these results, we conclude that ARC3 inhibits assembly of FtsZ2 filaments and mid-plastid FstZ2 rings.
The significance of the FtsZ2 mini-rings described above is unclear at present, though related structures have been reported for bacterial FtsZ in S. pombe and chloroplast FtsZ in Arabidopsis minE1 mutants (Srinivasan et al., 2008; Fujiwara et al., 2009). One possibility is that ARC3-Myc overexpression interferes with tethering of short FtsZ2 fragments to the membrane, increasing their concentration in the stroma and leading to annealing of filament ends to produce bundled mini-rings (Mingorance et al., 2005; Popp et al., 2009). A similar situation could explain mini-ring formation in minE1 (Fujiwara et al., 2009). In plants, mini-rings have only been observed when FtsZ2 is present, consistent with evidence that FtsZ2 is the main structural determinant of the chloroplast Z-ring (TerBush and Osteryoung, 2012; described below).
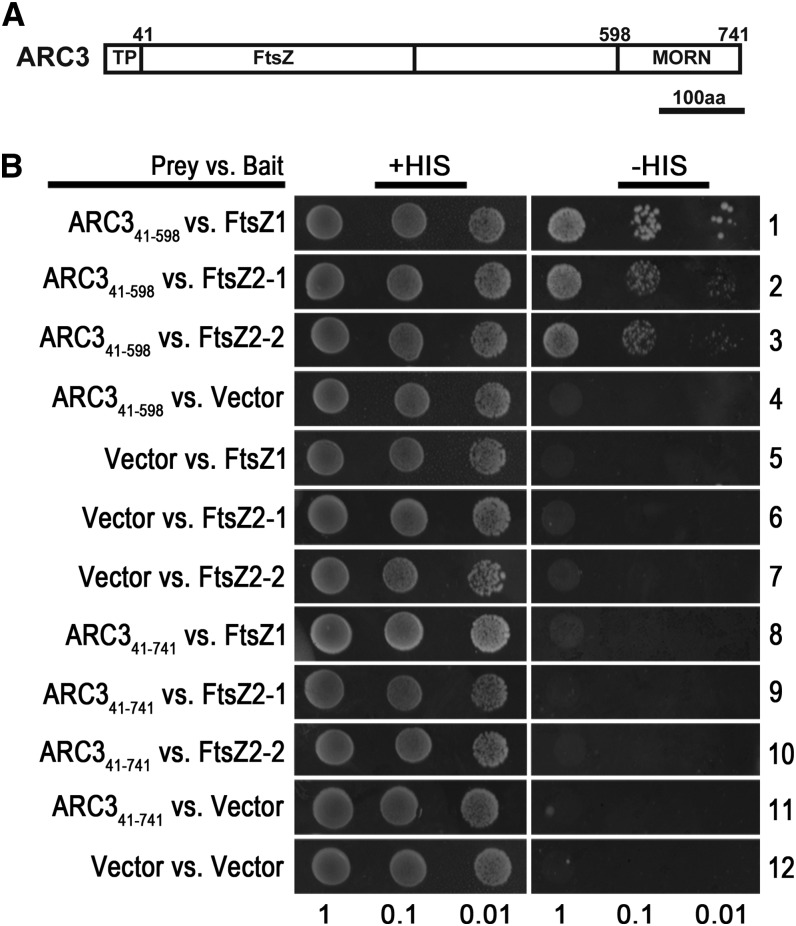
ARC3 Interacts with FtsZ2 in Yeast Two-Hybrid Assays
ARC3 is a chimeric protein consisting of an N-terminal FtsZ-like domain and a C-terminal Membrane Occupation and Recognition Nexus (MORN) domain and is targeted to the chloroplast stroma by a cleavable transit peptide (amino acids 1 to 41) (Shimada et al., 2004; Maple et al., 2007) (Figure 5A). Maple et al. (2007) reported that an ARC3 construct lacking the MORN domain (ARC31-598) interacted with FtsZ1 but not FtsZ2 in yeast two-hybrid assays. However, our finding that ARC3 affects FtsZ2 positioning and morphology in the absence of FtsZ1 in vivo suggested the possibility that ARC3 might also interact with FtsZ2. We revisited this issue in a new set of yeast two-hybrid experiments. First, we confirmed that ARC341-598 interacts with FtsZ1 (Figure 5B, row 1). However, in contrast with Maple et al. (2007), we detected an interaction between ARC341-598 and both FtsZ2-1 and FtsZ2-2 (Figure 5B, rows 2 and 3). The lack of ARC341-598-FtsZ2 interaction in the earlier report was probably due to toxicity when FtsZ2 was used in the prey vector, as done in that study, because yeast cells cotransfected with ARC341-589 and FtsZ2-1 or FtsZ2-2 only grew on synthetic dropout medium lacking His when FtsZ2 was used as bait (Figure 5). Consistent with published results for FtsZ1 (Maple et al., 2007), we also found that neither FtsZ1 nor FtsZ2 could interact with the MORN domain–containing construct ARC341-741 (Figure 5B, rows 9 and 10). These data implicate amino acids 41 to 598 of ARC3 as the region responsible for interaction with FtsZ2 as well as FtsZ1 (Figure 5B, row 8).
Figure 5.
Yeast Two-Hybrid Assays between FtsZ2-1 and ARC3.
(A) Schematic diagram showing the FtsZ-like (FtsZ) domain and MORN domain in ARC3. aa, amino acids; TP, transit peptide.
(B) Yeast-two hybrid assays between the indicated ARC3 constructs and FtsZ2-1 and FtsZ2-2 lacking their predicted transit peptides. Interaction with FtsZ1 shown previously was included as a positive control.
ARC3 Inhibits FtsZ2 Filament Formation in a Heterologous Expression System
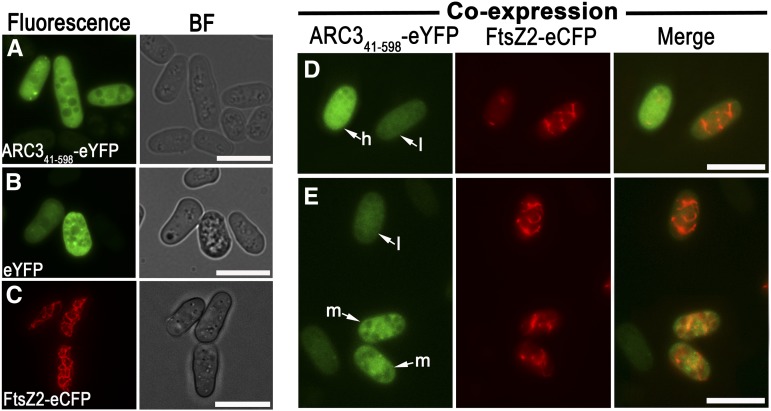
Recently, we used the fission yeast S. pombe as a heterologous system in which to investigate chloroplast FtsZ assembly and dynamics and showed that ARC341-598 disrupted FtsZ1 assembly in that system (TerBush and Osteryoung, 2012). To investigate whether ARC341-598 also inhibits FtsZ2 assembly, we generated a fluorescent fusion construct encoding ARC341-598-eYFP (for enhanced yellow fluorescent protein) and tested its effects on assembly of FtsZ2-eCFP (for enhanced cyan fluorescent protein) in S. pombe. Consistent with our earlier study (TerBush and Osteryoung, 2012), when expressed by themselves, FtsZ2-eCFP assembled into filament networks in the cytoplasm (Figure 6C, left panel), whereas ARC341-598-eYFP was distributed throughout the cytoplasm, similar to eYFP (Figures 6A and 6B, left panel), though the expression levels as indicated by fluorescence intensities were not uniform throughout the cultures. When ARC341-598-eYFP was coexpressed with FtsZ2-eCFP, the FtsZ2-eCFP signal was more diffuse and few FtsZ2-eCFP filaments were apparent in cells with relatively high accumulation of ARC341-598-eYFP (Figure 6D, h cell; see Supplemental Figure 3 online). By contrast, in cells with relatively low ARC341-598-eYFP levels, less FtsZ2-eCFP was dispersed in the cytoplasm and more FtsZ2-eCFP filaments were present (Figures 6D and 6E, l cells). Cells with moderate accumulation of ARC341-598-eYFP also appeared to have an intermediate degree of FtsZ2-eCFP filament assembly (Figure 6E, m cells). In the latter cells, ARC341-598-eYFP was largely colocalized with FtsZ2-eCFP filaments (Figure 6E, m cells, Merge). These results provide evidence that ARC341-598 inhibits FtsZ2 assembly in a dose-dependent manner, and, in combination with the yeast two-hybrid data, suggest that it does so through direct interaction with FtsZ2.
Figure 6.
ARC3 Inhibits FtsZ2 Filament Assembly in a Heterologous Expression System (S. pombe).
(A) to (C) Epifluorescence micrographs showing ARC341-598-eYFP (A), eYFP (B), and FtsZ2-eCFP (C) localization in S. pombe. BF, phase contrast images of cells of the indicated strain.
(D) and (E) Coexpression of ARC341-598-eYFP and FtsZ2-1-eCFP in S. pombe. Cells in (D) and (E) were taken from the same identically processed image; hence, differences in fluorescence intensity reflect differences in protein level. h, a cell with a relatively high signal from ARC341-598-eYFP; m and l, cells with more moderate (m) and low (l) signals from ARC341-598-eYFP. Fluorescence signals from eYFP and eCFP are false-colored green and red, respectively. The merged signals in (D) and (E) are shown on the right. Bars = 10 μm.
Overexpression of ARC31-598 Disrupts Z-Ring Assembly in Vivo
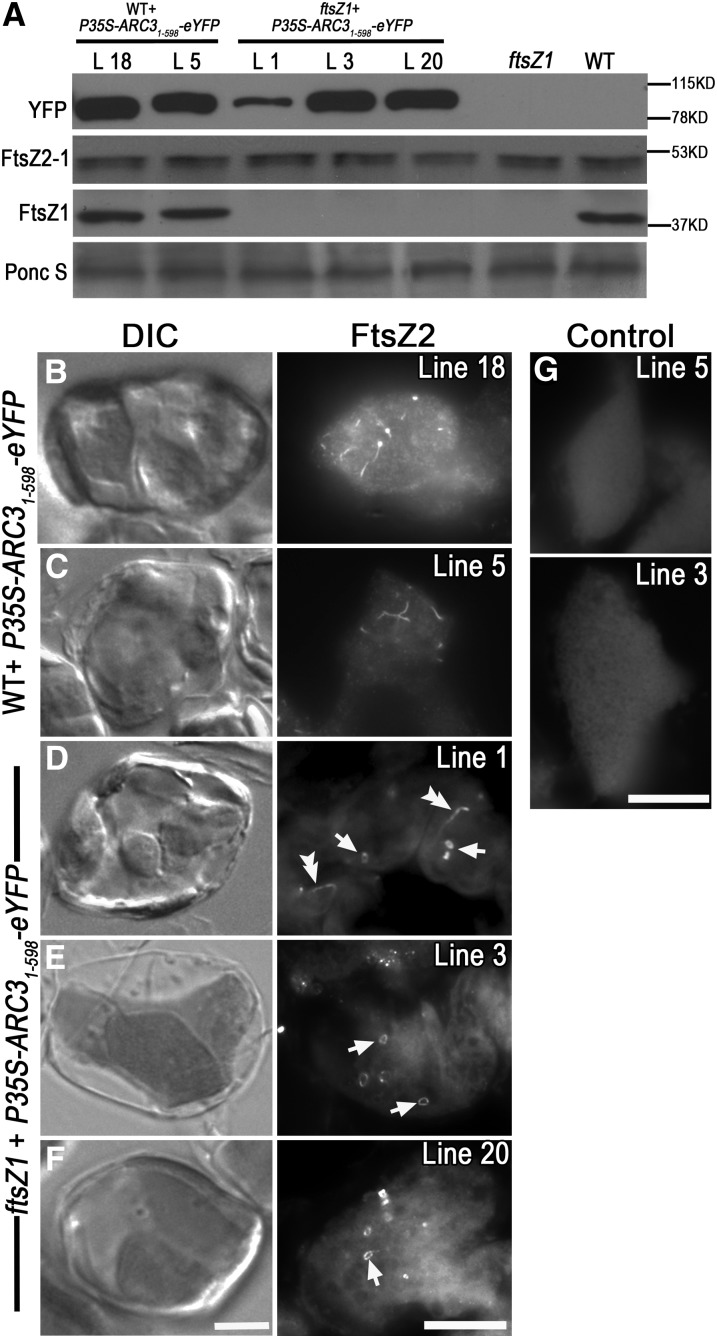
To test whether ARC341-598 has an inhibitory activity in plants, we generated a transgene encoding ARC31-598-eYFP, which includes the ARC3 N-terminal chloroplast transit peptide, and expressed it in wild-type Arabidopsis plants under control of the 35S promoter. Out of 36 T1 transgenic plants analyzed, 15 accumulated ARC31-598-eYFP to high levels, as indicated by immunoblotting (Figure 7A, lanes 1 and 2). Light microscopy on mesophyll cells in two of these lines (lines 5 and 18) showed they contained a reduced number of enlarged chloroplasts (Figures 7B and 7C, left panels), indicating chloroplast division was impaired. Immunofluorescence labeling of FtsZ2-1 to reveal FtsZ localization in the same lines showed the presence of short FtsZ fragments and puncta in the enlarged chloroplasts (Figures 7B and 7C, right panels). ARC31-598-eYFP fluorescence was detected in living cells but not in the fixed samples (Figure 7G; see Supplemental Figure 5A and 5D online), indicating the observed fluorescence in fixed tissue was from the immunolabeled FtsZ2-1. These results suggest that the ARC31-598 retains the ability to inhibit Z-ring assembly in wild-type Arabidopsis.
Figure 7.
Overexpression of ARC31-598-eYFP Inhibits Z-Ring Assembly in Wild-Type Plants and FtsZ2 Assembly in ftsZ1 Mutants.
(A) Immunoblot analysis of relative ARC31-598-eYFP and FtsZ levels in various lines of wild-type (WT) and ftsZ1 plants expressing P35S-ARC31-598-eYFP. Total protein from 1 mg fresh leaf tissue was loaded in each lane. Anti-GFP was used for detecting ARC31-598-eYFP, where it recognized a single band at ∼91 kD. Ponc S, Ponceau S staining of Rubisco on the blot as a loading control. L, transgenic lines.
(B) to (F) Chloroplast phenotypes (DIC) and immunofluorescence localization of FtsZ2 in single mesophyll cells of various lines of wild-type and ftsZ1 plants expressing P35S-ARC31-598-eYFP, as indicated at left. FtsZ2 was labeled with an FtsZ2-1–specific antibody. In right panels, the strong signals (white) and the dim background signals (gray) represent FtsZ2 localization and chloroplast shape, respectively. Double arrowheads and arrows indicate long FtsZ2 filaments and FtsZ2 mini-rings in giant chloroplasts, respectively.
(G) Controls. Tissue samples of wild-type plants expressing P35S-ARC31-598-eYFP (line 5) and ftsZ1 plants expressing P35S-ARC31-598-eYFP (line 3) were incubated with goat serum instead of anti-FtsZ2-1 as the primary antibody. Bars = 10 μm.
Overexpressing ARC31-598 Disrupts FtsZ2 Assembly in the ftsZ1 Knockout Mutant
We also examined the chloroplast phenotypes and FtsZ2 morphologies in mesophyll cells of ftsZ1 mutants expressing ARC31-598-eYFP at different levels (Figures 7D to 7F). In line 1, with a relatively low level of ARC31-598-eYFP (Figure 7A, lane 3), some FtsZ2 filaments were observed (Figure 7D, double arrowheads), but these were fewer than in the ftsZ1 parent mutant (Figure 4F). FtsZ2 mini-rings were also observed in this line (Figure 7D, arrows). In lines 3 and 20, which accumulated ARC31-598-eYFP to similar levels as wild-type lines 18 and 5 (Figure 7A, lanes 1, 2, 4, and 5), chloroplasts were fewer and larger than in line 1 and ftsZ1 mutants (Figures 4B, 7E, and 7F, left panels). These chloroplasts had many FtsZ2 mini-rings but very few FtsZ2 filaments (Figures 7D and 7E, right panels). The similarity of these phenotypes to those of ftsZ1 plants overexpressing full-length ARC3-Myc at high levels (Figure 4J) suggests that the region of ARC3 between the end of the predicted transit peptide and amino acid 598 is sufficient and solely responsible for the inhibition of FtsZ2 filament assembly in vivo.
Altered ARC3 Levels Do Not Change FtsZ1 Morphology in the ftsZ2-1 ftsZ2-2 Double Knockout Mutant
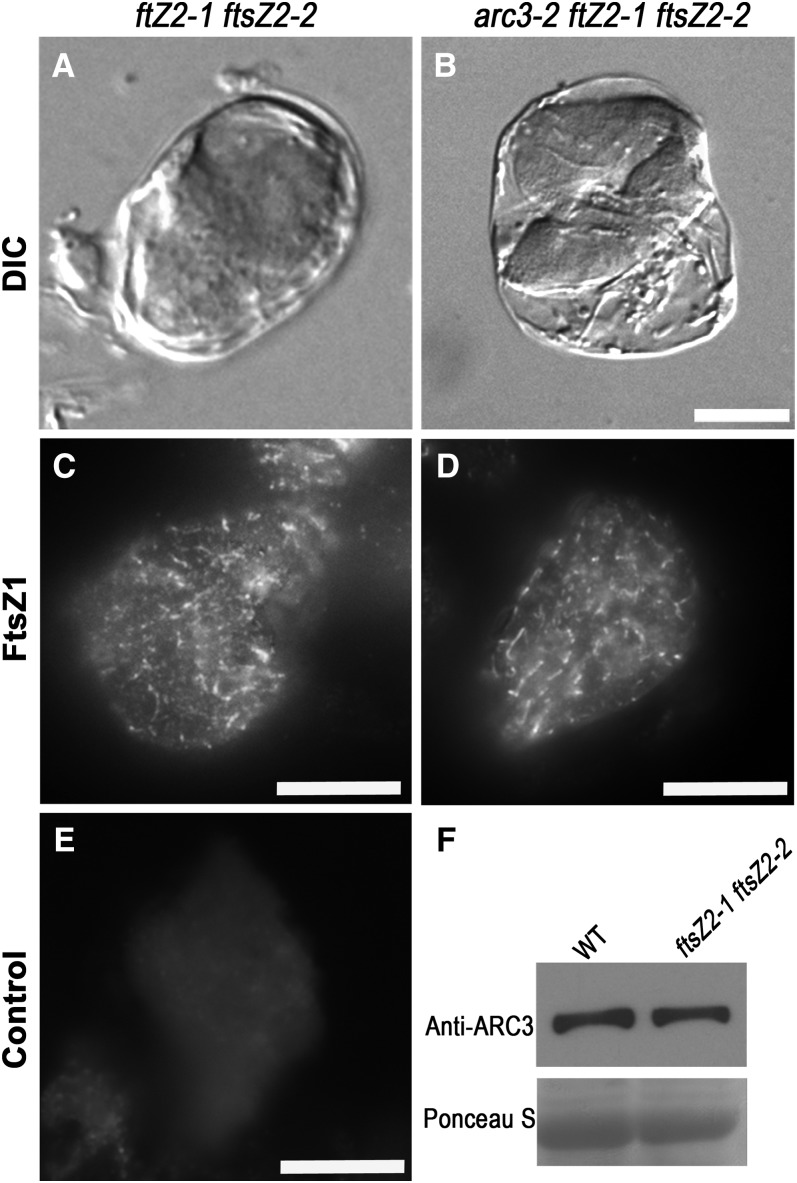
Previously, it was reported that mesophyll cells in ftsZ2-1 ftsZ2-2 double mutants exhibit one or two enlarged chloroplasts containing short FtsZ1 filaments (Schmitz et al., 2009). Due to the interaction between FtsZ1 and ARC3 and the ability of ARC341-598 to inhibit FtsZ1 assembly in S. pombe (Maple et al., 2007; TerBush and Osteryoung, 2012), we asked whether endogenous ARC3 might be responsible for the FtsZ1 fragmentation phenotype in the ftsZ2-1 ftsZ2-2 background. Toward this end, we created an arc3-2 ftsZ2-1 ftsZ2-2 triple mutant. One or two giant chloroplasts containing dots or short filaments of FtsZ1 were observed in mesophyll cells of this mutant (Figures 8B and 8D). These phenotypes were indistinguishable from those of the ftsZ2-1 ftsZ2-2 double mutant (Figures 8A and 8C). Immunoblot analysis showed wild-type and ftsZ2-1 ftsZ2-2 plants accumulated similar levels of ARC3 (Figure 8F). This finding indicates that the short FtsZ1 filaments in ftsZ2-1 ftsZ2-2 mutants were not due to inhibition of FtsZ1 assembly by ARC3, but rather were a result of the absence of FtsZ2.
Figure 8.
FtsZ1 Assembly Is Not Affected by ARC3 in the Absence of FtsZ2.
(A) to (D) Chloroplast phenotypes (DIC) and immunolocalization of FtsZ1 in single mesophyll cells of the ftsZ2-1 ftsZ2-2 double mutant and the arc3-2 ftsZ2-1 ftsZ2-2 triple mutant. FtsZ1 was immunolabeled with an anti-FtsZ1–specific antibody.
(E) Control. Tissue of ftsZ2-1 ftsZ2-2 mutants was incubated with goat serum instead of anti-FtsZ1 as the primary antibody. Bars = 10 μm.
(F) Immunoblot analysis of ARC3 levels in ftsZ2-1 ftsZ2-2 mutants with an ARC3-specific antibody. Total protein from 3 mg fresh leaf tissue was loaded in each lane. A single ARC3 band was detected at ∼83 kD. Ponceau S staining of Rubisco on the blot served as a loading control. WT, the wild type.
DISCUSSION
In bacteria, spatial regulation of division-site placement occurs through negative regulatory systems that inhibit Z-ring assembly everywhere but at the division site (Lutkenhaus, 2008; de Boer, 2010). In E. coli, MinC functions as the direct assembly inhibitor (Hu et al., 1999), and its localization and activity are regulated by MinD and MinE (Hu and Lutkenhaus, 1999; Fu et al., 2001; Pichoff and Lutkenhaus, 2001). Plants have retained MinD and MinE from their cyanobacterial ancestors and these proteins have been shown to regulate division-site placement in chloroplasts (Colletti et al., 2000; Itoh et al., 2001; Maple et al., 2002; Maple and Møller, 2007). However, although MinC-like sequences have been detected in Physcomitrella patens and Ostreococcus (Yang et al., 2008), vascular plants and at least some green algae lack MinC homologs, and the plastid-division protein ARC3 was proposed as a possible functional replacement based on the division-site misplacement phenotype in Arabidopsis arc3 mutants and on the ability of ARC3 to interact with MinD1, MinE1, and FtsZ1 (Maple et al., 2007). The work presented here provides compelling experimental support of a MinC-like role for ARC3 in Z-ring assembly and positioning in vascular plants and suggests that ARC3 functions in this capacity primarily through interaction with FtsZ2 in vivo.
ARC3 Functions as an Inhibitor of FtsZ Polymer and Z-Ring Assembly
In wild-type plants, chloroplast Z rings and division sites almost always localize to the mid-plastid (Vitha et al., 2001; Wilson et al., 2011) (Figure 3A). The presence of multiple Z rings in arc3-2 mutants (Figure 1A), as observed previously for arc3-1 mutants (Glynn et al., 2007), is consistent with the presence of multiple chloroplast constrictions in these plants (Maple et al., 2007) (Figure 3D) and shows that, in the wild type, ARC3 is required for restricting Z-ring formation to the mid-plastid. Our finding that FtsZ is detected in puncta or very short filaments in plants overexpressing ARC3-Myc at high levels, which were also severely inhibited in chloroplast division (Figure 1D), provides evidence that ARC3 functions in this capacity by inhibiting FtsZ filament and Z-ring assembly. The inhibitory effect of ARC3 on FtsZ filament formation and chloroplast division is probably dose-dependent because plants with lower ARC3-Myc levels had more mild chloroplast division defects and visibly detectable FtsZ filaments (see Supplemental Figure 2 online). The effect of ARC341-598-eYFP on assembly of FtsZ2-eCFP in the heterologous S. pombe expression system (Figures 7D and 7E) also suggests that ARC3 inhibits FtsZ assembly in a dose-dependent manner. These results parallel the effects of MinC overexpression on cell division in E. coli (de Boer et al., 1992) and suggest that overexpression allows ARC3 to interfere with Z-ring assembly at the mid-plastid where in wild-type plants it is prevented from doing so, probably by other cpMin system components.
ARC3 Regulates Z-Ring Positioning through FtsZ2
In contrast with a previous report that ARC341-598 interacts only with FtsZ1 in yeast two-hybrid assays (Maple et al., 2007), we have shown here that the same region of ARC3 also interacts with FtsZ2 (Figure 6). Our finding that ARC341-598 also affects formation of FtsZ1 and FtsZ2 homopolymers in S. pombe in the absence of all other chloroplast division proteins (TerBush and Osteryoung, 2012) (Figure 6; see Supplemental Figure 4 online) is consistent with this result and suggests the interaction is direct. But because our previous work indicates that chloroplast Z rings are probably composed of FtsZ1-FtsZ2 heteropolymers (Vitha et al., 2001; Olson et al., 2010; TerBush and Osteryoung, 2012), it seems likely that in wild-type plants, ARC3 functions at the level of heteropolymer assembly and/or disassembly in chloroplasts. However, despite the ability of ARC3 to inhibit both FtsZ1 and FtsZ2 homopolymer formation in S. pombe and to interact with both proteins, several genetic lines of evidence suggest its activity on presumed heteropolymer assembly in vivo may be primarily through interaction with FtsZ2. First, upon reexamination of FtsZ2 in ftsZ1 mutants, shown previously to assemble into long, disorganized FtsZ2 filaments and occasional FtsZ2 rings (Yoder et al., 2007), we found that, when FtsZ2 rings were detectable, they were localized at the mid-plastid (Figures 4F and 4G), consistent with the occasional mid-plastid constrictions observed in hypocotyls of these mutants (Figure 3B). Second, in arc3-2 ftsZ1 double mutants, we observed some chloroplasts with multiple parallel FtsZ2 rings (Figure 4H) resembling the multiple Z-rings composed of both FtsZ1 and FtsZ2 in arc3-2 mutants (Figure 1A). Together, these findings reveal that ARC3 is capable of regulating positioning of Z rings composed only of FtsZ2 homopolymers. Third, we found that overexpression of ARC3-Myc or ARC31-598-eYFP inhibits formation of FtsZ2 filaments and mid-plastid FtsZ2 rings in ftsZ1 mutants and enhances the severity of the ftsZ1 mutant chloroplast division defect (Figures 4C, 4J, and 7). These results indicate that ARC3 is capable of inhibiting FtsZ2 assembly independently of FtsZ1 in vivo. Finally, we observed that loss of ARC3 in plants lacking FtsZ2 did not alter the morphology of FtsZ1, which appeared in dots and very short filaments in both the ftsZ2-1 ftsZ2-2 and arc3-2 ftsZ2-1 ftsZ2-2 mutant backgrounds (Figure 8). The latter result could reflect the possibility that FtsZ1 filaments are too unstable in the absence of FtsZ2 for an effect of ARC3 to be seen. Nevertheless, while we do not rule out the possibility that ARC3 interacts with and influences heteropolymer formation partly through FtsZ1 in vivo, as suggested by the ability of ARC341-598 to interact with FtsZ1 in two-hybrid assays and inhibit its assembly in S. pombe (Maple et al., 2007; TerBush and Osteryoung, 2012), our genetic analyses strongly suggest that ARC3 regulates Z-ring positioning largely if not solely through FtsZ2 in chloroplasts. Two recent studies provide clues regarding the functional significance of these results for heteropolymer formation in wild-type plants. In one study, analysis of FtsZ1 and FtsZ2 polymer morphology and dynamics in S. pombe led to the hypothesis that FtsZ2 plays a dominant role in imparting stability and structural integrity to the Z-ring as a result of both its intrinsically slow dynamics and its role in Z-ring tethering through interaction with ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6 (ARC6). By contrast, FtsZ1 was proposed to enhance heteropolymer dynamics, which may be necessary for sustained Z-ring constriction (TerBush and Osteryoung, 2012). In the other study, in vitro assembly assays suggested that heteropolymers may assemble with variable stoichiometry and therefore may not be composed of strictly alternating FtsZ1 and FtsZ2 subunits (Olson et al., 2010). If this is also true in vivo, then heteropolymers might assemble with stretches of multiple FtsZ2 subunits. Because of the stabilizing influence of FtsZ2, interaction of ARC3 with FtsZ2 may be particularly important in vivo for inhibiting the formation of stable heteropolymers and Z rings at nondivision sites.
ARC3 Functions as a Central Negative Regulator of Z-Ring Assembly and Positioning in Arabidopsis
Previous studies have shown that MinD1 and MinE1 also function in chloroplast division-site placement in plants (Colletti et al., 2000; Itoh et al., 2001; Fujiwara et al., 2004). Similar to the phenotypes reported here in arc3 mutants and ARC3 overexpression lines, the Arabidopsis MinD1 mutants arc11 and minD1-1 exhibit multiple Z rings, and MinD1 overexpression inhibits FtsZ filament and Z-ring formation (McAndrew et al., 2001; Vitha et al., 2003; Glynn et al., 2007) (see Supplemental Figure 3G online). This indicates that MinD1, like ARC3, restricts Z-ring placement to the mid-plastid through negative regulation of FtsZ assembly. The Z-ring assembly and chloroplast division defects in the ARC3-Myc overexpression lines could reflect a stoichiometric imbalance between ARC3 and other cpMin-system components that allows ARC3 to interfere with assembly at the mid-plastid where it is normally prevented from doing so. However, unlike ARC3, MinD1 does not interact directly with FtsZ1 or FtsZ2 (Maple et al., 2005), and we have now shown that inhibition of FtsZ assembly by MinD1 overexpression requires ARC3 activity (Figures 2C and 2D). In combination with our finding that ARC3 inhibits FtsZ assembly in S. pombe (TerBush and Osteryoung, 2012) (Figure 6; see Supplemental Figure 4 online), these results indicate that ARC3 is the direct FtsZ assembly inhibitor in chloroplasts. Support for this hypothesis is further reinforced by analysis of the genetic relationship between ARC3 and MinE1. The MinE1 mutant arc12 and a MinE1 T-DNA insertion mutant both exhibit short FtsZ filaments, dots and mini-rings, and enlarged chloroplasts, whereas MinE1 overexpressors exhibit multiple Z-rings, indicating that, in the wild type, MinE1 positively regulates Z-ring assembly and positioning (Glynn et al., 2007; Fujiwara et al., 2008, 2009). Our finding that loss of Z-ring assembly in arc12 requires ARC3 (Figures 2E and 2F) also supports a role for ARC3 as the direct assembly inhibitor, similar to the role of MinC in bacteria.
Potential Mechanistic Similarities and Differences between ARC3 and MinC as FtsZ Assembly Inhibitors
Although our data are consistent with the proposed role for ARC3 as a functional replacement for bacterial MinC (Maple et al., 2007), ARC3 is a chimeric protein that probably arose during the evolution of green algae and bears no obvious sequence or structural similarity to MinC (Shimada et al., 2004; Yang et al., 2008). This suggests it may have a different mode of action. Recent models indicate that E. coli MinC does not inhibit FtsZ polymer and Z-ring assembly per se, but rather destabilizes preassembled polymers through multiple activities (de Boer, 2010). One is by competing with the membrane-associated cell division proteins ZipA and FtsA for binding to FtsZ (Shen and Lutkenhaus, 2009). FtsZ binding to all three proteins requires the C-terminal peptide that is conserved in FtsZ2 (Osteryoung and McAndrew, 2001). Though chloroplasts lack homologs of ZipA and FtsA, the conserved FtsZ2 C-terminal peptide mediates a specific interaction with the inner envelope chloroplast Z-ring stabilizing protein ARC6 (Osteryoung and McAndrew, 2001; Vitha et al., 2003; Maple et al., 2005). However, in contrast with the FtsZ-MinC interaction in E. coli, FtsZ2–ARC3 interaction does not require the FtsZ2 C-terminal peptide in yeast two-hybrid assays (see Supplemental Figure 6B online, rows 1 and 2), suggesting that ARC3 does not compete with ARC6 for interaction with FtsZ2 and potentially indicating a significant mechanistic difference between ARC3 and MinC. In E. coli, MinC also functions by attacking and weakening FtsZ subunit interfaces, causing polymer breakage (Shen and Lutkenhaus, 2010) and interferes with lateral interactions between polymers (Dajkovic et al., 2008). Though we do not yet know how ARC3 inhibits Z-ring assembly in chloroplasts, the presence of the FtsZ-like region toward the N terminus of ARC3 (Figure 5A), which mediates its interaction with FtsZ1 (Maple et al., 2007), speculatively suggests that, unlike MinC, ARC3 might assemble directly onto FtsZ subunits or polymer ends, perhaps inhibiting polymerization or destabilizing nascent polymers at nondivision sites. Surprisingly, we have found that ARC341-598, which contains the FtsZ-like region, not only inhibits FtsZ1 and FtsZ2 assembly in S. pombe (TerBush and Osteryoung, 2012) (Figure 6), but also inhibits E. coli FtsZ assembly in this system (see Supplemental Figure 7 online). This finding suggests that ARC3 interaction with FtsZ1 and FtsZ2 occurs through conserved domains, also present in bacterial FtsZs, which are required for FtsZ assembly. However, our current data suggest that this interaction affects Z-ring positioning in plants primarily through FtsZ2.
Complexity of the Chloroplast Min System
Notwithstanding the potential mechanistic differences between ARC3 and MinC, the conservation of MinD and MinE in chloroplast Z-ring and division-site placement indicates a significant degree of functional conservation with the bacterial Min system. Similar chloroplast phenotypes and FtsZ morphologies in arc3 and minD1 (arc11) mutants (Glynn et al., 2007; Wilson et al., 2011) and the ability of ARC3 to interact with MinD1 (Maple et al., 2007) suggest ARC3 may function in a complex with MinD1 to inhibit Z-ring assembly in vivo, reminiscent of the MinCD complex in E. coli (Bi and Lutkenhaus, 1993; Pichoff and Lutkenhaus, 2001). Similarly, the FtsZ morphology phenotypes described above for the MinE1 mutant and overexpression lines (Glynn et al., 2007; Fujiwara et al., 2008, 2009) (Figure 2D), along with work showing that chloroplast MinD1, like E. coli MinD, is an ATPase whose activity can be stimulated by MinE1 (Hu and Lutkenhaus, 2001; Hu et al., 2002; Aldridge and Møller, 2005) and that MinE1 functions through MinD1 in Z-ring positioning (see Supplemental Figure 3 online), are consistent with a role for MinE1 similar to that of E. coli MinE in suppressing the inhibitory activity of ARC3 at the mid-plastid to permit Z-ring formation at the division site. Our data showing that MinD1 and MinE1 lack functionality in the absence of ARC3 (Figure 2) are further consistent with roles for MinD1 and MinE1 as regulators of ARC3 in vivo activity. However, whether their activities involve regulation of ARC3 localization as is that case in bacteria remains to be established. While there is some evidence that localization of ARC3, MinD1, and MinE1 is dynamic (Maple and Møller, 2007; Maple et al., 2007), it is not yet known whether these proteins oscillate as occurs in E. coli (Lutkenhaus, 2007); one could imagine that the thylakoid membranes might pose a physical barrier to an oscillatory mechanism. Furthermore, unlike MinC, ARC3 has been shown to interact with MinE1 as well as MinD1 (Maple et al., 2007), and the Arabidopsis cpMin system includes at least two additional plant-specific proteins, MULTIPLE CHLOROPLAST DIVISION SITE1, which is required for MinD1 localization to the membrane (Nakanishi et al., 2009), and Paralog of ARC6, which may play an additional role in ARC3 localization and activity (Glynn et al., 2009; Zhang et al., 2009). Thus, plants appear to have evolved a unique and complex system for regulating Z-ring placement in chloroplasts. Future studies, including detailed biochemical analysis, will be important for understanding the mechanistic details of ARC3 action on chloroplast Z-ring assembly and fully elucidating how the cpMin system regulates chloroplast division-site placement in plants.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana T-DNA insertion mutants ftsZ1, ftsZ2-2, arc3-2, and ftsZ2-1 and the ethyl methanesulfonate mutant arc12, all in the Col-0 background, were described previously (Pyke, 1999; Shimada et al., 2004; Yoder et al., 2007; McAndrew et al., 2008; Schmitz et al., 2009), as were the ftsZ2-1 ftsZ2-2 (Col-0) and arc3-1 arc11 (Landsberg erecta background) double mutants (Pyke and Leech, 1994; Schmitz et al., 2009).
We generated the arc3-2 ftsZ2-1 ftsZ2-2 triple mutant and arc3-2 ftsZ1 and arc3-2 arc12 double mutants. Homozygous mutants among F2 progeny were identified by PCR using the following primers: LBb1.3 for the SALK T-DNA left border, ARC3LP and ARC3RP for arc3-2, Z1LP and Z1RP for ftsZ1, Z2-2LP and Z2-2RP for ftsZ2-2, G-DNA for the GABI-Kat T-DNA left border, and Z2-1LP and Z2-1RP for ftsZ2-1. Derived Cleaved Amplified Polymorphic Sequences (Neff et al., 1998) markers were designed to identify homozygous arc12: A 169-bp PCR product was amplified by primers A12F and A12R such that only fragments from the wild-type background could produce 140- and 29-bp products upon digestion by DdeI. All primer sequences are available in Supplemental Table 1 online. PCR reactions were performed in 25-μL volumes with GoTaq Flexi DNA Polymerase (Promega). Reaction conditions were as follows: one cycle at 96°C for 4 min; 30 cycles at 94°C for 30 s, 50 to 55°C for 30 s, and 72°C for 1 min; and one cycle at 72°C for 4 min.
The minD1-1 T-DNA insertional allele in the Wassilewskija background was identified by PCR-based screening of an activation-tagged population at the University of Wisconsin Arabidopsis Knockout Facility (Sussman et al., 2000) using the primers KOY09, KOY10, and KOY11 shown in Supplemental Table 1 online. The same primers were used for genotyping of individual plants and minD1-1 arc12 double mutants. PCR reaction conditions were the same as above. The insertion site was determined by sequencing the region flanking the T-DNA left border.
After vernalization for 3 d in the dark, seeds were sown in soil and grown at 21°C, 70% humidity, and in broad-spectrum light (110 μmol m−2 s−1) with a 16 h-light/8 h-dark photoperiod.
Plant Transformation Vectors and Screening
To obtain a full-length genomic ARC3 fragment with a Myc tag, genomic DNA spanning the coding region of ARC3 from the start codon to base pair 1204 was first amplified by primers MZ3 and MZ4 and cloned into the pGEM-T easy (Promega) to generate pT-Part1. Fragments of genomic ARC3 from 1205 to 2190 bp and 2191 bp to the end of the coding sequence without the stop codon were then amplified using primer sets MZ5 and MZ6 and MZ7 and MZ19, digested, and inserted into pT-part1 sequentially to produce the plasmid pT-ARC3-Myc. To obtain the P35S-ARC3-Myc overexpression vector, the ARC3-Myc fragment from pT-ARC3-Myc was digested via XbaI-SacI and used to replace eYFP in pCAMBIA1300-20, which is a version of the binary vector pCAMBIA1300 already containing a 35S-MCS-eYFP-Ter cassette. For complementation of the arc3-2 mutant, a 1469-bp fragment of the ARC3 native promoter region upstream of the start codon was amplified using primers MZ1 and MZ2 and cloned into the P35S-ARC3-Myc overexpression vector, replacing the 35S promoter. To make pP35S-ARC31-598-eYFP, an ARC3 genomic fragment from the start codon to base pair 3898 was obtained from pT-ARC3-Myc via XbaI-SpeI digestion and inserted into pCAMBIA1300-20. Then, the sequence from 3898 to 4089 bp of ARC3 was amplified using primers MZ9 and MZ22 and inserted before eYFP in pCAMBIA1300-20 after digestion with SpeI-SalI. To generate P35S-MinD1, MinD1 was amplified from Arabidopsis genomic DNA using primers MZ109 and MZ110, then digested with XbaI-SacI and cloned into pCAMBIA1300-20. Primer sequences are available in Supplemental Table 1 online. All clones were verified by sequencing. The vectors were introduced into plants by the floral dip method using Agrobacterium tumefaciens GV3101 (Clough and Bent, 1998). T1 seeds were sterilized and selected on half-strength Linsmaier Skoog medium (Caisson Laboratories) containing 30 μg/mL hygromycin.
Yeast Two-Hybrid Analysis
pGADT7 and pGBKT7 (Clontech) were used for making yeast two-hybrid constructs. The constructs pGADT7-ARC341-741, pGADT7-ARC341-598, pGBKT7-FtsZ2-1, pGBKT7-FtsZ2-2, and pGBKT7-FtsZ1 have been described previously (Glynn et al., 2009; Schmitz et al., 2009). Pairs of prey and bait constructs were cotransformed into yeast strain AH109 according to the manufacturer’s recommendations (Clontech). HIS3 reporter assays were performed as described previously (Glynn et al., 2008).
Arabidopsis Genes Expressed in Schizosaccharomyces pombe
Splicing by overlapping extension PCR (Warrens et al., 1997) was used to make the ARC341-598-eYFP fusion gene, using primers MZ38 and M169 and MZ170 and MZ171 (see Supplemental Table 1 online). The resultant PCR product was digested with BamHI-XmaI and cloned into pREP41X (Forsburg, 1993). pREP41X-eYFP and pREP42X-FtsZ2-1-eCFP were described previously, and transformation and growth of S. pombe strain MBY192 were performed as in previous reports (Yang et al., 2011; TerBush and Osteryoung, 2012).
Microscopy
For analyses of chloroplast phenotypes, 3- to 5-mm young expanding leaves from 2.5-week-old seedlings were fixed and analyzed as described previously (Pyke and Leech, 1991). Mesophyll cells were observed using differential interference contrast optics on a Leica DMI3000B inverted microscope and captured with a Leica DFC320 camera.
Tissue samples of plants for immunofluorescence labeling of FtsZ1 or FtsZ2 were collected from the same stage seedlings as described above, prepared, sectioned, and incubated with FtsZ1 and FtsZ2-1 antibodies as reported previously (Stokes et al., 2000; Vitha et al., 2001). The FtsZ1 and FtsZ2-1 primary antibodies were labeled by Alexa Fluor 488 goat anti-rabbit IgG (1:500) (Invitrogen). For observing ARC341-598-eYFP and FtsZ2-1-eCFP in Schizosaccharomyces pombe, 2 μL fresh cell culture was dropped on poly-l-Lys–treated slides. Alexa Fluor 488, chlorophyll, eCFP, and eYFP fluorescence signals were detected and images acquired using a Leica DMRA2 microscope equipped with appropriate filter sets and Q-Capture camera control software (Q-Imaging) as described (TerBush and Osteryoung, 2012). Image analysis and quantitative analyses of chloroplast number per cell were performed using ImageJ version 1.37 (National Institutes of Health).
Immunoblotting
Plant proteins were extracted from 2-week-old seedlings of different individuals and immunoblotting was performed as reported (Stokes et al., 2000). Comparison of FtsZ1 and FtsZ2-1 levels in individual lines was performed as described (McAndrew et al., 2001). For detecting ARC3-Myc and ARC31-598-eYFP fusion proteins, anti-GFP monoclonal antibody (Clontech), anti-c-Myc monoclonal antibody, and anti-mouse HRP secondary antibody (Pierce Biotechnology) were diluted 1:6000, 1:2000, and 1:10,000, respectively. ARC3 and MinD1 were detected using anti-ARC3 anti-MinD1 polyclonal antibodies (Shimada et al., 2004; Nakanishi et al., 2009) and anti-rabbit HRP secondary antibody diluted 1:3000, 1:5000, and 1:10,000, respectively. Polyvinylidene fluoride membranes were developed using a SuperSignal West Pico Chemiluminescent Substrate kit (Pierce Biotechnology).
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (TAIR) or GenBank/EMBL databases under the following names and accession numbers: ARC3 (At1g75010), FtsZ1 (At5g55280), FtsZ2-1 (At2g36250), FtsZ2-2 (At3g52750), MinD1 (ARC11, At5g24020), and MinE1 (ARC12, At1g69390). Germplasm identification numbers of alleles used in this work are arc3-2 (SALK_057144), arc11 (TAIR stock number CS281), arc3-1 arc11 (TAIR CS485), ftsZ1 (SALK_073878), ftsZ2-1 (GABI-Kat 596H04), ftsZ2-2 (SALK_134970), and arc12 (TAIR CS16472).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Quantitative Analysis of Chloroplast Number versus Mesophyll Cell Size in Young Expanding Leaves.
Supplemental Figure 2. Chloroplast Phenotype and FtsZ Morphology in Wild-Type Plants with Lower Accumulation of P35S-ARC3-Myc.
Supplemental Figure 3. Chloroplast Phenotypes and FtsZ Morphologies in the minD1-1 Single Mutant and the minD1-1 arc12 Double Mutant.
Supplemental Figure 4. FtsZ2-eCFP Morphology in Additional S. pombe Cells with Higher Accumulation of ARC341-598-eYFP.
Supplemental Figure 5. Localization of ARC31-598-eYFP in Wild-Type and ftsZ1 Plants.
Supplemental Figure 6. Yeast Two-Hybrid Assays between ARC3 and Truncated Variants of FtsZ2-1 and FtsZ2-2.
Supplemental Figure 7. Effect of ARC341-598 on E. coli FtsZ Assembly in S. pombe.
Supplemental Table 1. Primers Used in This Article Shown 5′ to 3′.
Supplementary Material
Acknowledgments
We thank Hiroshi Shimada for providing homozygous arc3-2 seeds and Arabidopsis ARC3 antibodies, Shin-ya Miyagishima for providing Arabidopsis MinD1 antibodies, Kevin A. Pyke for providing arc12 seeds, and Susan Forsburg, Mohan Balasubramanian, and Ramanujam Srinivasan for S. pombe strains and vectors used in this study. We also thank Yue Yang and Yi Liu for help in generating the arc3-2 arc12 double mutant and Yamato Yoshida for comments and discussion on this article. We thank the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through Grant DE-FG02-06ER15808 for funding all transgenic and genetic analyses performed in Arabidopsis and the National Science Foundation (Grant 1121943) for funding experiments in S. pombe.
AUTHOR CONTRIBUTIONS
M.Z. and K.W.O. designed research and wrote the article. M.Z., A.J.S., D.K.K.-K., and A.D.T. carried out experiments. M.Z., A.J.S., and D.K.K.-K. analyzed data.
Glossary
- Col-0
Columbia-0
- TAIR
The Arabidopsis Information Resource
- DIC
differential interference contrast
- Rubisco
ribulose-1,5-bis-phosphate carboxylase/oxygenase
References
- Aldridge C., Møller S.G. (2005). The plastid division protein AtMinD1 is a Ca2+-ATPase stimulated by AtMinE1. J. Biol. Chem. 280: 31673–31678 [DOI] [PubMed] [Google Scholar]
- Bi E.F., Lutkenhaus J. (1991). FtsZ ring structure associated with division in Escherichia coli. Nature 354: 161–164 [DOI] [PubMed] [Google Scholar]
- Bi E., Lutkenhaus J. (1993). Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175: 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colletti K.S., Tattersall E.A., Pyke K.A., Froelich J.E., Stokes K.D., Osteryoung K.W. (2000). A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 10: 507–516 [DOI] [PubMed] [Google Scholar]
- Dajkovic A., Lan G., Sun S.X., Wirtz D., Lutkenhaus J. (2008). MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 18: 235–244 [DOI] [PubMed] [Google Scholar]
- de Boer P.A.J. (2010). Advances in understanding E. coli cell fission. Curr. Opin. Microbiol. 13: 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A.J., Crossley R.E., Rothfield L.I. (1992). Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 174: 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kafafi S., Mukherjee S., El-Shami M., Putaux J.L., Block M.A., Pignot-Paintrand I., Lerbs-Mache S., Falconet D. (2005). The plastid division proteins, FtsZ1 and FtsZ2, differ in their biochemical properties and sub-plastidial localization. Biochem. J. 387: 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H.P., Taylor D.W., Taylor K.A., Bramhill D. (1996). Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. USA 93: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S.L. (1993). Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21: 2955–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X., Shih Y.L., Zhang Y., Rothfield L.I. (2001). The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98: 980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M.T., Hashimoto H., Kazama Y., Abe T., Yoshida S., Sato N., Itoh R.D. (2008). The assembly of the FtsZ ring at the mid-chloroplast division site depends on a balance between the activities of AtMinE1 and ARC11/AtMinD1. Plant Cell Physiol. 49: 345–361 [DOI] [PubMed] [Google Scholar]
- Fujiwara M.T., Nakamura A., Itoh R., Shimada Y., Yoshida S., Møller S.G. (2004). Chloroplast division site placement requires dimerization of the ARC11/AtMinD1 protein in Arabidopsis. J. Cell Sci. 117: 2399–2410 [DOI] [PubMed] [Google Scholar]
- Fujiwara M.T., Sekine K., Yamamoto Y.Y., Abe T., Sato N., Itoh R.D. (2009). Live imaging of chloroplast FtsZ1 filaments, rings, spirals, and motile dot structures in the AtMinE1 mutant and overexpressor of Arabidopsis thaliana. Plant Cell Physiol. 50: 1116–1126 [DOI] [PubMed] [Google Scholar]
- Gao H.B., Kadirjan-Kalbach D., Froehlich J.E., Osteryoung K.W. (2003). ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 100: 4328–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn J.M., Froehlich J.E., Osteryoung K.W. (2008). Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20: 2460–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn J.M., Miyagishima S.Y., Yoder D.W., Osteryoung K.W., Vitha S. (2007). Chloroplast division. Traffic 8: 451–461 [DOI] [PubMed] [Google Scholar]
- Glynn J.M., Yang Y., Vitha S., Schmitz A.J., Hemmes M., Miyagishima S.Y., Osteryoung K.W. (2009). PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. 59: 700–711 [DOI] [PubMed] [Google Scholar]
- Gray M.W. (1992). The endosymbiont hypothesis revisited. Int. Rev. Cytol. 141: 233–357 [DOI] [PubMed] [Google Scholar]
- Hu Z., Gogol E.P., Lutkenhaus J. (2002). Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl. Acad. Sci. USA 99: 6761–6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Lutkenhaus J. (1999). Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34: 82–90 [DOI] [PubMed] [Google Scholar]
- Hu Z., Lutkenhaus J. (2001). Topological regulation of cell division in E. coli. Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7: 1337–1343 [DOI] [PubMed] [Google Scholar]
- Hu Z., Lutkenhaus J. (2003). A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol. Microbiol. 47: 345–355 [DOI] [PubMed] [Google Scholar]
- Hu Z., Mukherjee A., Pichoff S., Lutkenhaus J. (1999). The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R., Fujiwara M., Nagata N., Yoshida S. (2001). A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol. 127: 1644–1655 [PMC free article] [PubMed] [Google Scholar]
- Leech R.M., Thomson W.W., Plattaloia K.A. (1981). Observations on the mechanism of chloroplast division in higher plants. New Phytol. 87: 1–9 [Google Scholar]
- Lutkenhaus J. (2007). Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76: 539–562 [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. (2008). Min oscillation in bacteria. Adv. Exp. Med. Biol. 641: 49–61 [DOI] [PubMed] [Google Scholar]
- Maple J., Aldridge C., Møller S.G. (2005). Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J. 43: 811–823 [DOI] [PubMed] [Google Scholar]
- Maple J., Chua N.H., Møller S.G. (2002). The topological specificity factor AtMinE1 is essential for correct plastid division site placement in Arabidopsis. Plant J. 31: 269–277 [DOI] [PubMed] [Google Scholar]
- Maple J., Møller S.G. (2007). Interdependency of formation and localisation of the Min complex controls symmetric plastid division. J. Cell Sci. 120: 3446–3456 [DOI] [PubMed] [Google Scholar]
- Maple J., Vojta L., Soll J., Møller S.G. (2007). ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep. 8: 293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. (2005). FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6: 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrison J.L., Rutherford S.M., Robertson E.J., Lister C., Dean C., Leech R.M. (1999). The distinctive roles of five different ARC genes in the chloroplast division process in Arabidopsis. Plant J. 18: 651–662 [DOI] [PubMed] [Google Scholar]
- Mazouni K., Domain F., Cassier-Chauvat C., Chauvat F. (2004). Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol. Microbiol. 52: 1145–1158 [DOI] [PubMed] [Google Scholar]
- McAndrew R.S., Froehlich J.E., Vitha S., Stokes K.D., Osteryoung K.W. (2001). Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 127: 1656–1666 [PMC free article] [PubMed] [Google Scholar]
- McAndrew R.S., Olson B.J., Kadirjan-Kalbach D.K., Chi-Ham C.L., Vitha S., Froehlich J.E., Osteryoung K.W. (2008). In vivo quantitative relationship between plastid division proteins FtsZ1 and FtsZ2 and identification of ARC6 and ARC3 in a native FtsZ complex. Biochem. J. 412: 367–378 [DOI] [PubMed] [Google Scholar]
- Meinhardt H., de Boer P.A.J. (2001). Pattern formation in Escherichia coli: A model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl. Acad. Sci. USA 98: 14202–14207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingorance J., Tadros M., Vicente M., González J.M., Rivas G., Vélez M. (2005). Visualization of single Escherichia coli FtsZ filament dynamics with atomic force microscopy. J. Biol. Chem. 280: 20909–20914 [DOI] [PubMed] [Google Scholar]
- Miyagishima S.Y. (2011). Mechanism of plastid division: From a bacterium to an organelle. Plant Physiol. 155: 1533–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.Y., Froehlich J.E., Osteryoung K.W. (2006). PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18: 2517–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., Suzuki K., Kabeya Y., Miyagishima S.Y. (2009). Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr. Biol. 19: 151–156 [DOI] [PubMed] [Google Scholar]
- Neff M.M., Neff J.D., Chory J., Pepper A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 14: 387–392 [DOI] [PubMed] [Google Scholar]
- Okazaki K., Kabeya Y., Miyagishima S.Y. (2010). The evolution of the regulatory mechanism of chloroplast division. Plant Signal. Behav. 5: 164–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson B.J.S.C., Wang Q.A., Osteryoung K.W. (2010). GTP-dependent heteropolymer formation and bundling of chloroplast FtsZ1 and FtsZ2. J. Biol. Chem. 285: 20634–20643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M., Anderson D.E., Erickson H.P. (2009). Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 28: 3476–3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., McAndrew R.S. (2001). The plastid division machine. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 315–333 [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Stokes K.D., Rutherford S.M., Percival A.L., Lee W.Y. (1998). Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., Vierling E. (1995). Conserved cell and organelle division. Nature 376: 473–474 [DOI] [PubMed] [Google Scholar]
- Park K.T., Wu W., Lovell S., Lutkenhaus J. (2012). Mechanism of the asymmetric activation of the MinD ATPase by MinE. Mol. Microbiol. 85: 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S., Lutkenhaus J. (2001). Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183: 6630–6635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plattaloia K.A., Thomson W.W. (1977). Chloroplast development in young sesame plants. New Phytol. 78: 599–605 [Google Scholar]
- Popp D., Iwasa M., Narita A., Erickson H.P., Maéda Y. (2009). FtsZ condensates: An in vitro electron microscopy study. Biopolymers 91: 340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A. (1999). Plastid division and development. Plant Cell 11: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. (1991). Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 96: 1193–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. (1992). Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Physiol. 99: 1005–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. (1994). A genetic analysis of chloroplast division and expansion in Arabidopsis thaliana. Plant Physiol. 104: 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M., de Boer P.A.J. (1999). Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland S.L., Fu X., Sayed M.A., Zhang Y., Cook W.R., Rothfield L.I. (2000). Membrane redistribution of the Escherichia coli MinD protein induced by MinE. J. Bacteriol. 182: 613–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A.J., Glynn J.M., Olson B.J.S.C., Stokes K.D., Osteryoung K.W. (2009). Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol. Plant 2: 1211–1222 [DOI] [PubMed] [Google Scholar]
- Shen B., Lutkenhaus J. (2009). The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD. Mol. Microbiol. 72: 410–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Lutkenhaus J. (2010). Examination of the interaction between FtsZ and MinCN in E. coli suggests how MinC disrupts Z rings. Mol. Microbiol. 75: 1285–1298 [DOI] [PubMed] [Google Scholar]
- Shimada H., Koizumi M., Kuroki K., Mochizuki M., Fujimoto H., Ohta H., Masuda T., Takamiya K. (2004). ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 45: 960–967 [DOI] [PubMed] [Google Scholar]
- Shiomi D., Margolin W. (2007). The C-terminal domain of MinC inhibits assembly of the Z ring in Escherichia coli. J. Bacteriol. 189: 236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.G., Johnson C.B., Vitha S., Holzenburg A. (2010). Plant FtsZ1 and FtsZ2 expressed in a eukaryotic host: GTPase activity and self-assembly. FEBS Lett. 584: 166–172 [DOI] [PubMed] [Google Scholar]
- Srinivasan R., Mishra M., Wu L.F., Yin Z.C., Balasubramanian M.K. (2008). The bacterial cell division protein FtsZ assembles into cytoplasmic rings in fission yeast. Genes Dev. 22: 1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes K.D., McAndrew R.S., Figueroa R., Vitha S., Osteryoung K.W. (2000). Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 124: 1668–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M.R., Amasino R.M., Young J.C., Krysan P.J., Austin-Phillips S. (2000). The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush A.D., Osteryoung K.W. (2012). Distinct functions of chloroplast FtsZ1 and FtsZ2 in Z-ring structure and remodeling. J. Cell Biol. 199: 623–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., Froehlich J.E., Koksharova O., Pyke K.A., van Erp H., Osteryoung K.W. (2003). ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15: 1918–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., McAndrew R.S., Osteryoung K.W. (2001). FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153: 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrens A.N., Jones M.D., Lechler R.I. (1997). Splicing by overlap extension by PCR using asymmetric amplification: An improved technique for the generation of hybrid proteins of immunological interest. Gene 186: 29–35 [DOI] [PubMed] [Google Scholar]
- Wilson M.E., Jensen G.S., Haswell E.S. (2011). Two mechanosensitive channel homologs influence division ring placement in Arabidopsis chloroplasts. Plant Cell 23: 2939–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Glynn J.M., Olson B.J.S.C., Schmitz A.J., Osteryoung K.W. (2008). Plastid division: Across time and space. Curr. Opin. Plant Biol. 11: 577–584 [DOI] [PubMed] [Google Scholar]
- Yang Y., Sage T.L., Liu Y., Ahmad T.R., Marshall W.F., Shiu S.H., Froehlich J.E., Imre K.M., Osteryoung K.W. (2011). CLUMPED CHLOROPLASTS 1 is required for plastid separation in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18530–18535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder D.W., Kadirjan-Kalbach D., Olson B.J.S.C., Miyagishima S.Y., Deblasio S.L., Hangarter R.P., Osteryoung K.W. (2007). Effects of mutations in Arabidopsis FtsZ1 on plastid division, FtsZ ring formation and positioning, and FtsZ filament morphology in vivo. Plant Cell Physiol. 48: 775–791 [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Miyagishima S.Y., Kuroiwa H., Kuroiwa T. (2012). The plastid-dividing machinery: Formation, constriction and fission. Curr. Opin. Plant Biol. 15: 714–721 [DOI] [PubMed] [Google Scholar]
- Zhang M., Hu Y., Jia J.J., Li D.P., Zhang R.J., Gao H.B., He Y.K. (2009). CDP1, a novel component of chloroplast division site positioning system in Arabidopsis. Cell Res. 19: 877–886 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.