ABC1K1 and ABC1K3, two of the six plastoglobule ABC1K kinases, physically interact, and the double mutant displays a conditional senescent-like phenotype. ABC1K1 and 3 regulate prenyl-lipid metabolism and phosphorylation targets likely include tocopherol cyclase, VTE1. Plastoglobules function as specialized thylakoid microdomains with local enrichment of selected proteins and metabolites.
Abstract
Plastoglobules (PGs) are plastid lipid-protein particles. This study examines the function of PG-localized kinases ABC1K1 and ABC1K3 in Arabidopsis thaliana. Several lines of evidence suggested that ABC1K1 and ABC1K3 form a protein complex. Null mutants for both genes (abc1k1 and abc1k3) and the double mutant (k1 k3) displayed rapid chlorosis upon high light stress. Also, k1 k3 showed a slower, but irreversible, senescence-like phenotype during moderate light stress that was phenocopied by drought and nitrogen limitation, but not cold stress. This senescence-like phenotype involved degradation of the photosystem II core and upregulation of chlorophyll degradation. The senescence-like phenotype was independent of the EXECUTER pathway that mediates genetically controlled cell death from the chloroplast and correlated with increased levels of the singlet oxygen–derived carotenoid β-cyclocitral, a retrograde plastid signal. Total PG volume increased during light stress in wild type and k1 k3 plants, but with different size distributions. Isolated PGs from k1 k3 showed a modified prenyl-lipid composition, suggesting reduced activity of PG-localized tocopherol cyclase (VTE1), and was consistent with loss of carotenoid cleavage dioxygenase 4. Plastid jasmonate biosynthesis enzymes were recruited to the k1 k3 PGs but not wild-type PGs, while pheophytinase, which is involved in chlorophyll degradation, was induced in k1 k3 and not wild-type plants and was localized to PGs. Thus, the ABC1K1/3 complex contributes to PG function in prenyl-lipid metabolism, stress response, and thylakoid remodeling.
INTRODUCTION
Plastoglobules (PGs) are plastid-localized lipid-protein particles with dynamic morphologies, and they are responsive to changing developmental states and environmental conditions (Bréhélin et al., 2007). In chloroplasts, PGs are formed by blebbing of the outer leaflet of the thylakoid membrane and remain attached to the thylakoid membrane system, providing a conduit for metabolite exchange between the two structures (Austin et al., 2006). Purification of PGs revealed a small proteome of ∼30 strongly enriched proteins (Vidi et al., 2006; Ytterberg et al., 2006; Lundquist et al., 2012a), as well as various prenyl-lipids, especially plastoquinone-9 and α-tocopherol, and nonpolar lipids, such as triacylglycerols (TAGs) and fatty acid phytyl esters (Gaude et al., 2007; Zbierzak et al., 2009; Eugeni Piller et al., 2012; Lippold et al., 2012). Although the lipid and protein composition of PGs is distinct from the associated thylakoids, their reversible shrinking and swelling indicates rapid and reversible exchange of components between the two structures. However, strong (but unknown) selection mechanisms must be in place to maintain the specialized PG composition.
Among the PG-enriched proteins, four have demonstrated enzymatic activity: Tocopherol cyclase (VTE1), which catalyzes the cyclization of quinone and is necessary for tocopherol and plastochromanol synthesis (Porfirova et al., 2002); an NAD(P)H dehydrogenase C1 (NDC1) that was shown to reduce a plastoquinone analog in vitro (Eugeni Piller et al., 2011); and phytyl esterases 1 and 2 (PES1/2 or DGAT3/4), which catalyze esterification of free phytol and fatty acid and display TAG synthase activity (Lippold et al., 2012). None of the PG proteins have predicted transmembrane domains, consistent with the monolayer nature of the PG, but the mechanism for a selective protein association with the PG is unclear. Metabolic enzymes acting on substrates deposited in the PG likely insert into the hydrophobic face, permitting access to lipid substrates, similar as has been suggested for Arabidopsis thaliana allene oxide synthase (AOS) based on x-ray crystallography (Lee et al., 2008).
A comprehensive functional model of the PG was constructed based on coexpression analysis using the PG proteins as baits, their hypothesized protein functions, the metabolite composition of the PG, and previous experimental studies (Lundquist et al., 2012a). Importantly, the coexpression network implicated four specific functions for the PG: (1) senescence, (2) plastid biogenesis, (3) prenyl-lipid metabolism, and (4) redox/photosynthetic regulation. Furthermore, we suggested that the PG is essentially a microdomain within the thylakoid membrane and likely serves as a platform to recruit proteins and metabolites into spatial proximity, to facilitate metabolic channeling or signal transduction, comparable to lipid rafts of plasma membranes. This study provides further support for such a microdomain or lipid raft–like model.
Among the enzymes of the PG are six proteins of the ancient ABC1 atypical kinase (ABC1K) family (Lundquist et al., 2012a, 2012b). A fundamental regulatory role for these PG proteins is suggested by their central positions as hubs in three of the four coexpression modules (Lundquist et al., 2012a). The first identified member of the ABC1K family, ABC1 or COQ8, is a nuclear-encoded yeast protein necessary for ubiquinone (coenzyme Q) synthesis in the mitochondria (Bousquet et al., 1991). The name derives from the molecular phenotype of the null mutant, which lacks activity of the bc1 complex due to the loss of ubiquinone. Several bacterial homologs were also found to be necessary for ubiquinone synthesis, specifically at the first monooxygenase step (Macinga et al., 1998; Poon et al., 2000). ABC1K protein sequences have sequence motifs typical of a kinase and are thought to function as regulators of ubiquinone synthesis via protein phosphorylation (Poon et al., 2000). Remarkable functional conservation has been demonstrated by complementation of the yeast abc1 mutant with homologs from Arabidopsis (At-ABC1K13) and human (Hs-ABC1K13) (Cardazzo et al., 1998; Xie et al., 2011).
The proliferation of the ABC1K family in photosynthetic organisms suggests a corresponding expansion in regulatory targets, likely including other enzymes of quinone/prenyl-lipid metabolism (Lundquist et al., 2012b). Consistent with this notion, PGs are rich reservoirs of various quinones and contain multiple enzymes involved in their metabolism (e.g., VTE1 and NDC1). Additionally, an Arabidopsis insertion mutant of the inner envelope-localized ABC1K8 revealed constitutively high accumulation of reactive oxygen species (ROS) and oxidative stress–associated genes, as well as sensitivity to cadmium stress (Jasinski et al., 2008). Although investigation of the PG-localized ABC1K1 using RNA interference (RNAi) suggested an impairment in chlorophyll degradation, the RNAi strategy used the full-length coding sequence of the gene, thus targeting many of the ABC1K transcripts and confounding the conclusions (Yang et al., 2012). Kinase activity of any of the ABC1Ks remains to be directly demonstrated, though indirect evidence from point mutants in the ABC1K domain of yeast, human, and bacterial ABC1K proteins strongly supports the putative kinase activity (Lagier-Tourenne et al., 2008; Mollet et al., 2008; Xie et al., 2011).
Here, we show that PG-localized ABC1K1 (AT4G31390) and ABC1K3 (AT1G79600) are involved in adaptation of Arabidopsis to increased light intensities during growth. Single or double null mutants of ABC1K1 and ABC1K3 showed rapid photobleaching and necrosis upon transition to a 10-fold increase in light intensity (from 120 to 1000 µmol photons m−2 s−1), while the double mutant (k1 k3) also showed enhanced sensitivity, demonstrating senescent-like degreening when transferred to only fivefold higher light intensity (from 120 to 520 µmol photons m−2 s−1). Elevated levels of β-cyclocitral were found in k1 k3 under fivefold light stress, implicating singlet oxygen (1O2) retrograde signaling in the degreening phenotype. Comparative proteomics and metabolite profiling of isolated PGs and other leaf fractions of wild-type and k1 k3 plants after light stress support a critical, regulatory role for the PG and ABC1Ks in photoacclimation and prenyl-lipid metabolism and likely during other adverse conditions that require rapid thylakoid remodeling.
RESULTS
Isolation of PG-Localized ABC1K Mutants
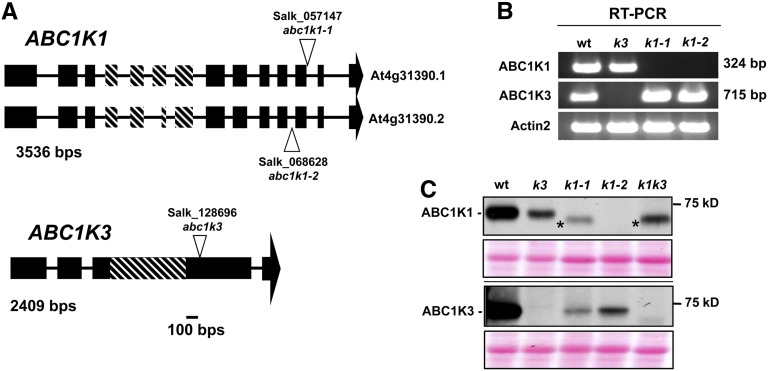
Previously, we reported that ABC1K1 and ABC1K3 positioned centrally within our PG gene coexpression network and together created a hub for the plastidic prenyl-lipid/protease functional module, suggesting a central regulatory role for the proteins (Lundquist et al., 2012a). Thus, we selected these two proteins with possible related or overlapping functions as the entry point to study PG function. We obtained two SALK T-DNA insertion mutants in ABC1K1 (k1-1 and k1-2) and one SALK T-DNA insertion mutant in ABC1K3 (k3) and confirmed their insertion sites by sequencing genomic DNA (Figure 1A). RT-PCR from mature leaf tissue showed that no full-length transcripts accumulated in the mutants, although truncated transcripts upstream of the insertion site were detected (Figure 1B; see Supplemental Figure 1 online). Immunoblotting with specific antibodies generated against the C-terminal regions of ABC1K1 and ABC1K3 (residues 578 to 682 and 556 to 711, respectively) did not detect full-length protein in the leaf tissue (Figure 1C). Low levels of a smaller protein product were observed by immunoblotting of total leaf extract in k1-1 (but not in the k1-2 allele), and this may represent truncated ABC1K1 protein; however, no peptides of ABC1K1 (or ABC1K3) were detected by mass spectrometry (MS) analysis of various proteome preparations (see further below). Interestingly, ABC1K3 accumulation levels were strongly reduced in both k1 alleles, and ABC1K1 level was also strongly reduced in the k3 allele, suggesting a mutual stabilization between these two proteins (Figure 1C). The mutual destabilization of ABC1K1 and ABC1K3 in k1-1, k1-2, and k3 showed that the single mutants were not independent and that specific functions for each kinase can therefore not easily be determined by single mutant analysis. Moreover, ABC1K1 and ABC1K3 tightly clustered together in the RNA coexpression analysis, suggesting that they may have similar or overlapping functions. Therefore, to test for functional redundancy or other genetic interactions, we created a k1-1 × k3 double mutant (hereafter named k1 k3) for phenotypic analysis.
Figure 1.
Identification of Null Mutant Lines for ABC1K1 (abc1k1-1 and abc1k1-2) and ABC1K3 (abc1k3).
(A) The structure of the ABC1K1 and ABC1K3 loci and the position of the T-DNA insertions. Exons are indicated by black boxes, introns are indicated by lines, and the 5′ and 3′ untranslated regions are not included. The diagonal hash marks indicate the location of the ABC1K domain (Lundquist et al., 2012b).
(B) RT-PCR amplification (35 cycles) of ABC1K1, ABC1K3, or ACTIN2 cDNA show that k1 and k3 mutants do not accumulate transcripts containing the C-terminal portions for ABC1K1 and ABC1K3, respectively. For information about primer location and additional RT-PCR analysis, see Supplemental Figure 1 online. wt, the wild type.
(C) Immunoblot with anti-ABC1K1 and ABC1K3 antisera of unstressed mature leaf tissue protein extracts. Thirty micrograms of protein was loaded per lane. The asterisk identifies a nonspecific product or a truncated ABC1K1 protein product. A Ponceau stain showing the region around RBCL is included as a loading control under each immunoblot.
ABC1K1 and ABC1K3 Physically Interact
Because the immunoblot suggested a mutual stabilization, we tested whether ABC1K3 could be immunoprecipitated with ABC1K1. Thylakoids with attached PGs were isolated from wild-type leaves as well as from k1 k3 leaves as a negative control. These thylakoids were incubated with four different reversible free amine cross-linkers [formaldehyde, disuccinimdyl tartrate (DST), 3,3′-dithiobis(sulfosuccinimidyl propionate) (DTSSP), and ethyleneglycolbis(sulfosuccinimidyl succinate) (Sulfo-EGS)], which differed in spacer length (2.0 to 16.1 Å). Following extensive optimization, we selected the best concentration for each. After cross-linking, the samples were solubilized in SDS, dialyzed and immunoprecipitated with affinity purified anti-ABC1K1 serum, followed by reversal of the cross-link and SDS-PAGE (see Supplemental Figure 2 online). The gel was stained with MS-compatible silver stain, and each lane was cut in slices, digested in the gel, and analyzed by nano-liquid chromatography–tandem MS (MS/MS) using an LTQ-Orbitrap mass spectrometer. The number of matched MS/MS spectra (spectral counts, SPC), adjusted for shared peptides between proteins (adjusted SPC or adjSPC), was used to determine whether proteins were enriched compared with the immunoprecipitation of the k1 k3 mutant (see Supplemental Data Set 1 online). ABC1K1 was identified with the highest number of adjSPC (489 adjSPC across the four cross-link experiments), indicating that the immunoprecipitation was successful. Among the proteins identified with at least 40 adjSPC, no protein was enriched compared with the negative control, with the exception of ABC1K3, which was identified (59 adjSPC) with two of the cross-linkers (formaldehyde and Sulfo-EGS) but not in the k1 k3 mutant (see Supplemental Data Set 1 online). Given that ABC1K3 is not very abundant (abundance rank 250 in 5-d light-stressed leaf; see further below) compared with the other identified proteins (ribulose biphosphate carboxylase oxygenase large subunit [RBCL] and proteins from photosystem II [PSII] and ATP synthase), the presence of ABC1K3 in the immunoprecipitation strongly suggests that it interacts with ABC1K1. This is consistent with the mutual destabilization in abc1k1 and abc1k3 and the coexpression of abc1k1 and abc1k3 in the RNA coexpression analysis. We then performed a reciprocal experiment with the most effective cross-linker (formaldehyde) using purified anti-ABC1K3 serum instead of anti-ABC1K1 serum. MS/MS analysis showed high levels of ABC1K3, indicating that the affinity purification for the bait was successful and ABC1K1 was strongly enriched, further supporting that ABC1K1 and ABC1K3 physically interact (see Supplemental Data Set 2 online).
Loss of ABC1K1 and ABC1K3 Caused a Conditional Light Stress Phenotype
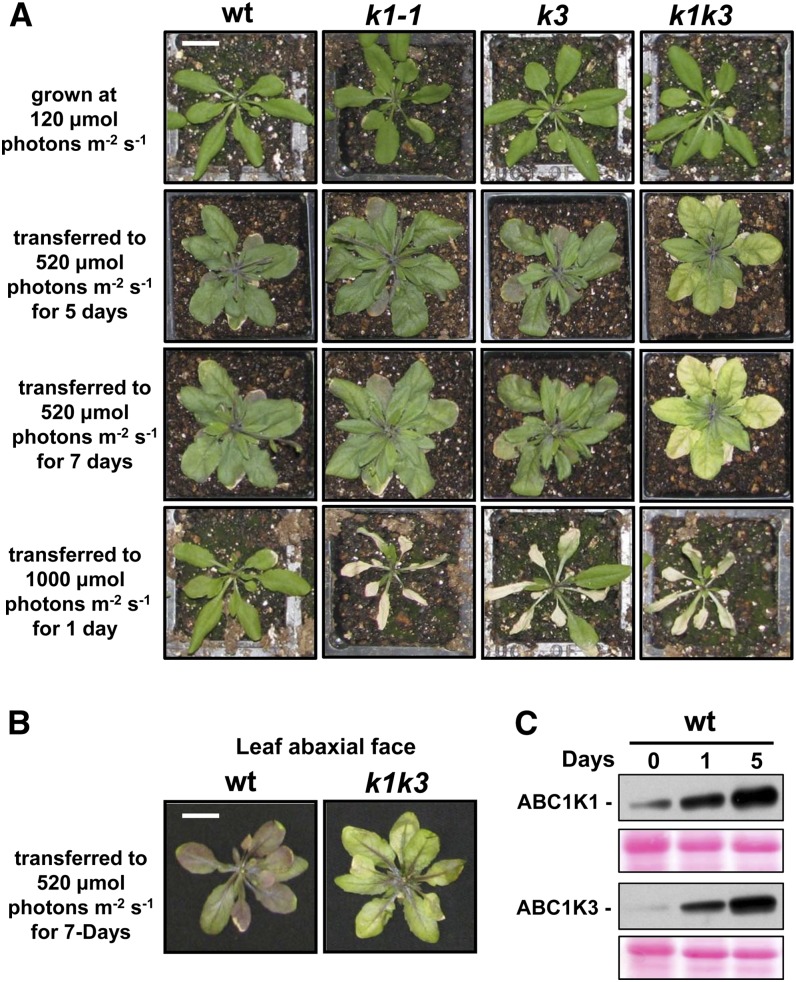
The single and double mutants did not display any visual phenotypes when grown under standard conditions in climate-controlled chambers (Figure 2A; see Supplemental Figure 3 online for a comparison of both k1 alleles). Given the expected role of PGs under suboptimal abiotic conditions, we tested various abiotic stresses. First, we tested a moderate light stress treatment by transferring plants from ∼120 µmol photons m−2 s−1 to ∼520 µmol photons m−2 s−1 (5× light stress) and followed the plants for several weeks. The wild-type and all single mutants (k1-1, k1-2, and k3) responded similarly to the increased light intensity by rapid leaf expansion and accumulation of anthocyanins (Figure 2; see Supplemental Figure 3 online). By contrast, k1 k3 demonstrated a senescence-like phenotype in which they began to visibly degreen after about 3 d and failed to accumulate anthocyanins outside of the vasculature (Figure 2A). This lack of high levels of anthocyanins is best observed at the leaf abaxial side (Figure 2B). Younger leaf tissue in k1 k3 appeared unaffected by the 5× light stress treatment, as has been reported for other oxidative/light stress treatments in other mutants (Havaux et al., 2000; Giacomelli et al., 2007). This synergistic effect in k1 k3 indicates (at least partially) nonredundant contributions of ABC1K1 and ABC1K3 to the light stress response. When wild-type, single, and double mutants grown at ∼120 µmol photons m−2 s−1 were transferred to 1000 µmol photons m−2 s−1 (10× light stress), the single and double mutants, but not the wild type, displayed a clear phenotype, showing rapid leaf bleaching and necrosis of the older leaves within 1 d (Figure 2A; see Supplemental Figure 3 online). Consistent with a role in light stress adaptation, ABC1K1 and ABC1K3 protein accumulation strongly increased more than fourfold and 19-fold, respectively, in response to the 5× light stress (Figure 2C).
Figure 2.
abc1k1-1 and abc1k3 Demonstrate a Conditional and Synergistic Light Stress Phenotype.
(A) Mature, 3-week-old plants grown under a 120 µmol photons m−2 s−1 light regime were transferred to either 520 or 1000 µmol photons m−2 s−1 and photographed at the described time points. wt, the wild type. Bar = 2 cm.
(B) The abaxial face of leaves of the wild type and k1 k3 treated with (5×) light stress treatment for 7 d demonstrate the lack of anthocyanin accumulation outside of the vasculature in k1 k3. Bar = 2 cm.
(C) Immunoblot with anti-ABC1K1 and anti-ABC1K3 antisera of mature leaf tissue protein extracts harvested after 0, 1, or 5 d of light stress. Twenty micrograms of protein from each sample was separated by SDS-PAGE and immunoblotted with the indicated antibody. A Ponceau stain showing the region around the RBCL is included as a loading control.
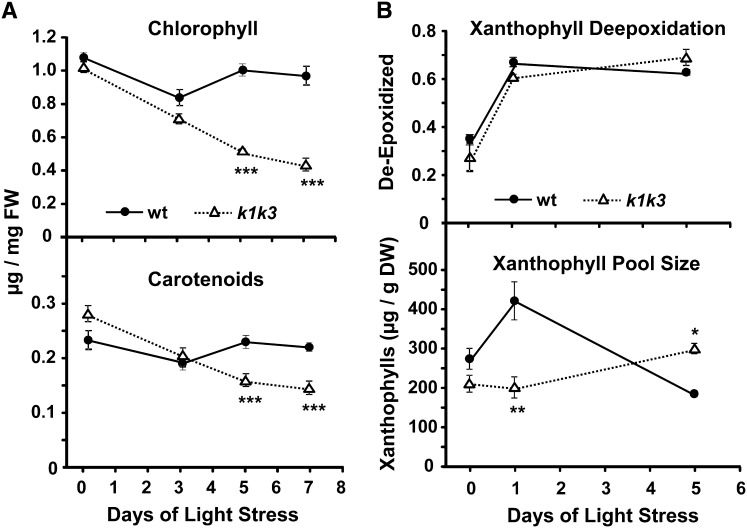
Measurement of extracted chlorophylls and carotenoids by absorption spectroscopy revealed a steady depletion of the photosynthetic pigments in k1 k3 over the course of the light stress treatment, decreasing to 50% (chlorophyll) and 66% (carotenoids) of prestress tissue after 7 d of light stress (Figure 3A). Quantification of xanthophyll pigments from leaf tissue by HPLC showed that the total xanthophyll pool size increased after the first day of stress in the wild type but not in k1 k3 (Figure 3B). However, the deepoxidation state of the xanthophylls was the same for the wild type and k1 k3, indicating that the xanthophyll cycle (i.e., conversion of violaxanthin into zeaxanthin) was not impaired in k1 k3.
Figure 3.
Accumulation of Photosynthetic Pigments Is Altered in k1 k3 under Light Stress.
(A) Chlorophyll (top panel) and carotenoid (bottom panel) levels were measured in mature leaf tissue by absorption spectroscopy prior to transfer to 5× light stress and after 3, 5, and 7 d of light stress. Error bars indicate the sd of three biological replicates. Significant genotypic effects were identified by Student’s t test and are indicated with one, two, or three asterisks (corresponding to P value < 0.05, P value < 0.01, or P value < 0.001, respectively). FW, fresh weight; wt, the wild type.
(B) Induction of the xanthophyll cycle and xanthophyll pool size in response to 5× light stress was measured using HPLC with a photodiode array detector. Xanthophyll deepoxidation (top panel) is measured as (Z+0.5A)/(Z+A+V), where Z is zeaxanthin, A is antheraxanthin, and V is violaxanthin. The xanthophyll pool size (bottom panel) is the total of Z, A, and V on a dry weight basis. Error bars indicate the sd of three biological replicates. Significant genotypic effects were identified by Student’s t test and are indicated with one, two, or three asterisks (corresponding to P value < 0.05, P value < 0.01, or P value < 0.001, respectively). DW, dry weight.
A comparable senescence-like degreening with a lack of anthocyanins has been demonstrated in a PG-localized FBN1a/1b/4 RNAi knockdown line under an ∼7× high light stress (120 µmol photons m−2 s−1 transferred to 850 µmol photons m−2 s−1) combined with cold stress (23 to 15°C) (Youssef et al., 2010). Surprisingly, this phenotype could be complemented by exogenous application of methyl jasmonate. By contrast, a similar methyl jasmonate treatment of k1 k3 failed to prevent or delay the degreening (see Supplemental Figure 4 online). Remarkably, the degreening phenotype of k1 k3 was also induced in mature leaves during drought stress or growth on limited nitrogen (at low light intensities) but not by applying cold treatment (see Supplemental Figure 5 online). The rate and extent of degreening under nitrogen-limiting conditions increased with lower levels of N concentration in the medium. This indicates that the conditional k1 k3 phenotype is not a strict light stress effect.
To determine if the degreening was reversible, wild-type and k1 k3 plants were subjected to 5× light stress for either 3, 5, or 7 d and then returned to prestress intensity. Mature leaves that had initiated degreening failed to regreen, instead continuing to degreen and gradually became white and eventually necrotic, albeit at a reduced rate than under the elevated light intensities (see Supplemental Figure 6 online). This suggests that the light stress treatment initiates an irreversible senescence-like response in mature leaves within 3 d, the kinetics of which are dependent on the prevailing light intensity. We note that the younger leaves did not bleach or degreen under these varying light conditions.
Since envelope-located ABC1K8 (OSA1) was reported to be important in defense against Cd, hydrogen peroxide (H2O2), and high light (Jasinski et al., 2008), we also evaluated the effect of Cd and H2O2 on root growth for the wild type, k1, k3, and k1 k3. However, all genotypes showed similar levels of reduced root length, indicating that k1 and k3 do not increase tolerance to Cd or H2O2 tolerance (data not shown).
To further investigate molecular functions of ABC1K1 and ABC1K3, we chose to study the wild type and k1 k3 under 5× light stress, as it provided viable tissue with a clear phenotype. Analysis of single mutants after short intervals of exposure to the 10× light stress prior to tissue death could also have been helpful, but the rapid induction of extensive cellular damage likely results in more pleiotropic phenotypes. Furthermore, the reciprocal effects of the single mutants on ABC1K1 and ABC1K3 protein abundance and the direct interaction between these kinases make it less likely that their functions are independent and it would be difficult to distinguish their individual roles. Therefore, the remainder of the experiments compares responses in wild-type versus k1 k3 plants before and after the 5× light stress.
Oxidative Stress in k1 k3
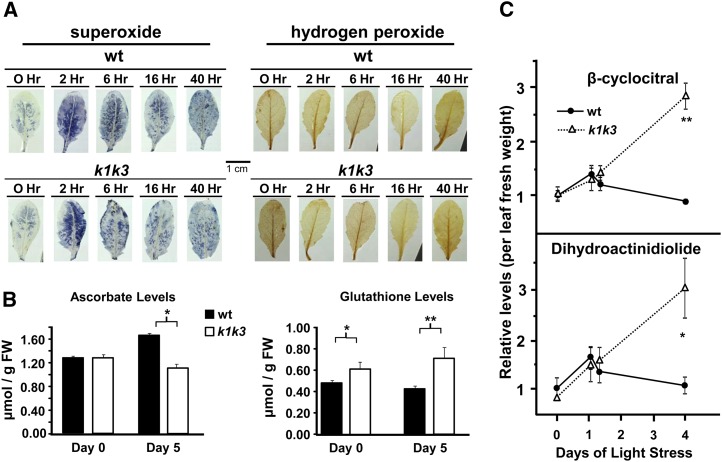
Oxidative stress in the form of ROS may act as the irreversible trigger for the degreening in k1 k3. ROS accumulation in the leaf was therefore studied by tissue stains specific for H2O2 and superoxide (O2−) immediately prior to the light transition and at 2, 6, and 16 h and 2 d after light transition (Figure 4A). A burst of accumulation of O2− throughout the leaf, 2 h after light transition, which tapered over the remainder of the time course, was observed in both genotypes. Prior to light transition, H2O2 accumulated mostly in the leaf vasculature and in small patches along the leaf margins in both genotypes. The light transition did not lead to enhanced accumulation in either genotype, and H2O2 accumulation in the vasculature in both genotypes decreased considerably at the 16-h and 2-d time points (Figure 4A).
Figure 4.
Analysis of Soluble ROS Defense Metabolites and Oxidative Stress Products.
From three biological replicates, significant genotypic effects were identified by Student’s t test and are indicated with one, two, or three asterisks (corresponding to P value < 0.05, P value < 0.01, or P value < 0.001, respectively).
(A) Cell stains for O2− and H2O2 in response to 5× light stress. Leaves were stained either with nitroblue tetrazolium, which develops a blue precipitate in the presence of superoxide (left-hand panels), or with diaminobenzidine, which develops a brown precipitate in the presence of hydrogen peroxide (right-hand panels). wt, the wild type.
(B) Total levels of ascorbate and glutathione equivalents were measured by absorption spectroscopy as described (Luwe et al., 1993) before and after 5× light stress. Error bars indicate the sd of four biological replicates. Levels of oxidized product are <5% in all pools (data not shown). FW, fresh weight.
(C) Elevated accumulation of β-carotene oxidation products was found in k1 k3 in rosette tissue in response to the 5× light stress treatment. Oxidation products were collected from tissue samples 10 h into the photoperiod on 0, 1, and 3 d of 5× light stress, as well as 5 h into the photoperiod of the first day of light stress. Data points are normalized to fresh weight. Error bars indicate the sd of three biological replicates.
The water-soluble ascorbate-glutathione cycle plays an important role in ROS scavenging and therefore changes in the ascorbate or glutathione levels may indicate changes in ROS levels (Foyer and Noctor, 2011). Total amounts and oxidation state of ascorbate and glutathione were measured from total leaf samples using absorption spectroscopy (Figure 4B). Prior to light stress, ascorbate levels were indistinguishable between the two genotypes, and glutathione was ∼20% higher in k1 k3. In response to the 5× light stress, ascorbate levels increased (∼30%) in the wild-type tissue, whereas glutathione levels increased (∼15%) in k1 k3 tissue. The oxidation states of ascorbate and glutathione were not significantly different between the genotypes. Thus, the investments in soluble ROS defense shifted somewhat from ascorbate- to glutathione-based defense in the case of k1 k3, in particular after light stress. The lower ascorbate levels are likely related to the reduced anthocyanin levels (Page et al., 2012).
Light Stress Induces 1O2 Retrograde Signaling via β-Cylcocitral in k1 k3 Plants
1O2 is a ROS produced as a byproduct of normal photosynthetic activity but is difficult to measure in vivo due to its short lifetime. Recently, oxidation products of β-carotene have been shown to be produced by reaction with 1O2; one of the resulting oxidation products, β-cyclocitral, subsequently mediated a 1O2 retrograde signal from the plastid (Ramel et al., 2012). Measurement of β-cyclocitral and the related β-carotene oxidation product, dihydroactinidiolide, prior to and subsequent to 5× light stress, revealed a small increase of both compounds during the first day of stress in wild-type and k1 k3 plants (Figure 4C). In the wild type, levels of β-cyclocitral and dihydroactinidiolide returned to prestress levels by the third day of light stress. By contrast, levels of both compounds continued to rise in k1 k3 reaching approximately threefold greater levels than the wild type (Figure 4C). These observations indicate that the inability for k1 k3 to adapt to elevated light intensity leads to excessive 1O2 production in the thylakoid and elevated production of the retrograde signaling molecule β-cyclocitral.
Elevated levels of 1O2 produced under excess excitation energy may activate an EXECUTER1 (EX1)- and EX2-dependent retrograde signaling pathway leading to programmed cell death (op den Camp et al., 2003). Therefore, we tested whether the light stress sensitivity in k1 k3 depended on EX1 and EX2. We generated the quadruple mutant ex1 ex2 k1 k3 and compared its phenotype with the wild type and both parents under both 5× and 10× light stress (Figures 5A and 5B). The ex1 ex2 mutant behaved like the wild type, and the quadruple mutant behaved like k1 k3 (Figures 5A and 5B), indicating that the light stress phenotype of k1 k3 is not dependent on the EX pathway. This EX-independent response is consistent with a β-cyclocitral–mediated retrograde signal because β-cyclocitral–induced changes in gene expression and photosynthetic adaptation were not affected by the ex1 mutation (Ramel et al., 2012).
Figure 5.
The Conditional Phenotype of k1 k3 Is Unaffected by Lesions in the EX1 and EX2 Loci Involved in Singlet Oxygen Retrograde Signaling.
Wild-type (wt) plants were grown up alongside ex1 ex2, k1 k3, and the corresponding quadruple mutant, ex1 ex2 k1 k3. All four genotypes grew and developed at the same rate. When plants were transferred to 520 (A) or 1000 (B) μmol photons m−2 s−1; ex1 ex2 did not display a conditional light stress phenotype, whereas the quadruple mutant displayed the same degreening or necrosis as k1 k3. Bars = 2 cm.
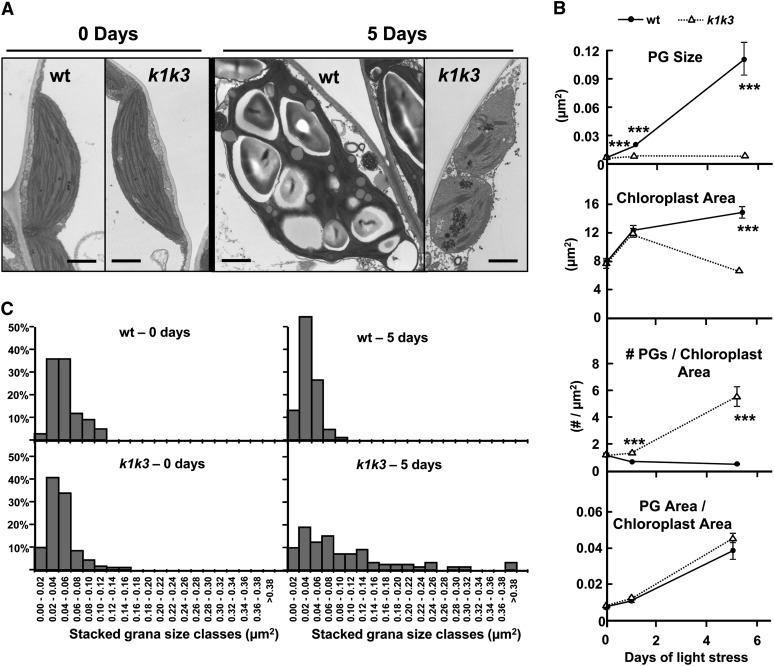
The Wild Type and k1 k3 Demonstrate Differential Changes in Plastid Ultrastructure under Light Stress
The impact of loss of ABC1K1 and ABC1K3 on the ultrastructure of chloroplasts was determined in the wild type and k1 k3 grown under standard growth and in response to 5× light stress. Prior to stress, the ultrastructures of the PG and chloroplasts were indistinguishable between the genotypes (Figure 6A). In response to the light stress, the plastid and PG ultrastructure was altered in both genotypes, albeit in different ways. However, the integrity of the chloroplasts was maintained, indicating that the degradation of photosynthetic pigments observed for k1 k3 was not due to a global dismantling of the photosynthetic organelles (Figure 6A).
Figure 6.
Plastid Ultrastructure in the Wild Type and k1 k3 Responds Differently to 5× Light Stress.
(A) Representative TEM micrographs after 0 and 5 d illustrate the genotype-specific responses to 5× light stress in the chloroplast ultrastructure. Mature leaf tissue was harvested 2 h after the onset of the photoperiod after 0, 1, and 5 d of 5× light stress. All micrographs are to the same scale. wt, the wild type. Bar = 1 µm.
(B) Graphs of four parameters measured from TEM micrographs. Each data point represents the mean measurement from 24 micrographs of three different plants. Error bars represent the se. The asterisks indicate P < 0.001 (Student’s t test) between the two genotypes at a given time point.
(C) Histogram demonstrating the granal hyperstacking in k1 k3 after 0 and 5 d of 5× light stress. At least 80 granal stacks from three individuals were measured for each genotype at each time point.
PG size in the wild type increased after 1 d of stress and increased further after 5 d of stress (Figures 6A and 6B). Concomitant with the increased size, the interior of many PGs in the wild type stained lighter with a solid black periphery. In fact, the staining was reminiscent of oil bodies derived from the endoplasmic reticulum (White et al., 2006; Huang et al., 2009). The chloroplast area also increased after 1 and 5 d of light stress, causing a subtle decrease in the number of PGs per chloroplast area in the wild type (Figure 6B). The PG size in k1 k3 remained remarkably consistent across the 5 d of light stress treatment, but the number of PGs per chloroplast area increased almost sixfold (Figure 6B), along with the induction of granal hyperstacking (Figure 6C). The PGs in k1 k3 stained solid black even after 5 d of light stress, lacking any light gray staining as observed in the wild-type light-stressed PGs. The chloroplasts of k1 k3 also swelled transiently after 1 d of light stress, similar to the wild type, but reverted back to near prestress levels after 5 d of stress. The two genotypes increased their ratio of PG area per chloroplast area in parallel, albeit using different methods; while the wild type increased the size of its PGs, k1 k3 increased its number of PGs (Figure 6B). Thus, after light stress, k1 k3 PGs had a higher volume:surface area ratio than the wild type, with implications for the accumulation of amphiphilic versus hydrophobic lipids (see further below and Discussion).
Comparative Quantitative Proteomics of the Wild Type and k1 k3
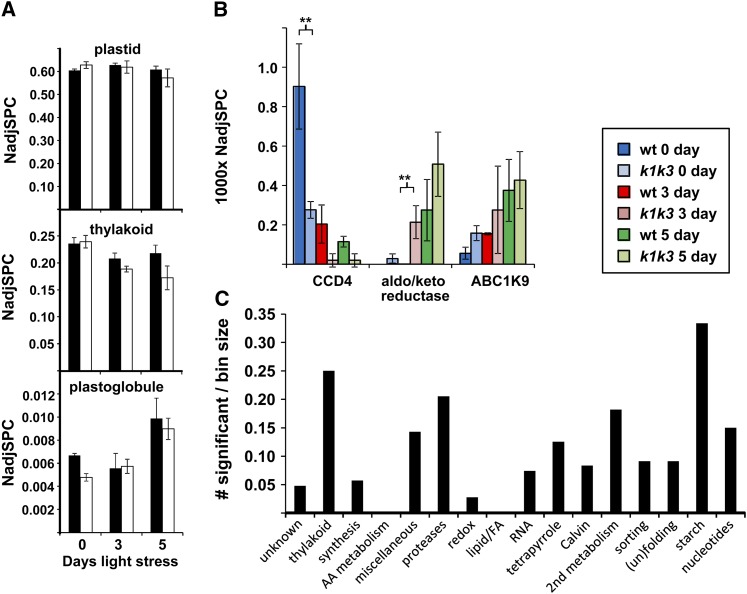
To further determine the molecular phenotype of wild-type and k1 k3 leaf tissue before and during light stress, we conducted a quantitative proteome analysis on total leaf rosette protein samples collected from the wild type and k1 k3 after 0, 3, and 5 d of 5× light stress. The proteomics workflow is summarized in Supplemental Figure 7 online. High correlation coefficients (0.932 to 0.994) between the three replicates of each genotype-time point were observed, indicating good reproducibility (see Supplemental Data Set 3 online). In total, 2274 proteins or protein groups were quantified and annotated based on the Plant Proteome Database and Huang et al. (2013) (see Supplemental Data Set 4 online); 810 identified proteins (or protein groups) were chloroplast localized, allowing for a meaningful analysis of the chloroplast protein responses. ABC1K1 and ABC1K3 were identified in two and three of the three time points of the wild type but not in k1 k3.
During the light stress treatment, the total plastid protein mass did not change in the wild type and decreased only 7% in k1 k3 (Figure 7A). After 5 d of light stress, the thylakoid proteome mass decreased by 7 and 28% in the wild type and k1 k3, respectively, whereas the PG proteome mass increased in both genotypes (Figure 7A). The accelerated loss of thylakoid protein mass in k1 k3 is consistent with the loss of chlorophyll in k1 k3 (Figure 3A). We identified 25 out of the 30 PG core proteins, as defined by Lundquist et al. (2012a). With the exception of FBN7b, all FBN core proteins (FBN1a,1b, 2, 4, 7a, and 8) increased 1.5-fold to threefold during light stress without significant genotypic differences, thus explaining most of the increase in PG protein mass (see Supplemental Figure 7 online). ABC1K9, the most abundant PG kinase and an uncharacterized aldo/keto reductase (AT1G06690) increased upon light stress in both genotypes, whereas carotenoid cleavage enzyme CCD4 in k1 k3 was already reduced prior to light stress compared with the wild type and was further downregulated after 3 and 5 d of light stress (Figure 7B). As will be shown below, the reduction in CCD4 is compatible with changes in PG carotenoid content.
Figure 7.
Response of the Chloroplast Proteome to 5× Light Stress in the Wild Type and k1 k3.
(A) Changes in protein mass investments (as determined by NadjSPC) in chloroplasts, PGs, and thylakoids in the wild type and k1 k3. Black bars, the wild type; white bars, k1 k3.
(B) The response of specific PG-localized proteins in the wild type (wt) and k1 k3 before and after transition to increased light intensity. CCD4, AT4G19170; aldo/keto reductase, AT1G06690; ABC1K9, AT5G05200. From three biological replicates, significant genotypic effects were identified by GLEE (P < 0.01; marked with two asterisks). Error bars in (A) and (B) indicate sd.
(C) Enrichment of specific plastid functions for significant (P < 0.01; GLEE) genotypic effects in individual proteins in k1 k3 before and after light stress. Only plastid functions with at least 20 proteins were considered.
We then compared the chloroplast protein mass investments across specific functions between the wild type and k1 k3. The most effected functions were the photosynthetic electron transport chain, proteolysis, secondary metabolism (mostly isoprenoid), and starch metabolism (Figure 7C). Here, we highlight only the most relevant genotypic effects; a more complete analysis is provided in Supplemental Figure 8 online. Quantitative information for all proteins can be found in Supplemental Data Set 4 online.
Effect on the Photosynthetic Apparatus
After 5 d of light stress, in particular the PSII core and the NDH complexes were both 50% reduced in k1 k3 compared with the wild type (see Supplemental Figure 9A online). In both genotypes, the total protein mass of Calvin cycle proteins increased by ∼30% after 3 d of light stress but returned to normal levels after 5 d of light stress (see Supplemental Data Set 4 online). Another significant genotypic change in the thylakoid proteome of k1 k3 was the strong reduction of the abundant calcium-sensing receptor phosphoprotein (CaS) in k1 k3 leaves (∼15 to 30% of the wild type) under stress (see Supplemental Figure 9B online). The function of CaS is unclear but is localized at the thylakoid membrane where it binds Ca2+ and is phosphorylated in a STATE TRANSITION KINASE8 (STN8)–dependent manner (Vainonen et al., 2008; Reiland et al., 2011; Rokka et al., 2011).
Effect on Plastid Proteases
We identified 39 proteases and peptidases in the plastid, localized in the thylakoid lumen (DegP1/5/8 and Plsp1), thylakoid membrane (FtsH1/2/5/8, SPPA, and EGY2), envelope (FtsH11 and RIP) and stroma, including the Clp system, Prep1, and various types of amino-peptidases (see Supplemental Data Set 4 online). The data indicate that Prep1 and the Clp protease system are likely involved in the accelerated turnover in k1 k3 during light stress, which is consistent with the tight coexpression of several CLP genes with ABC1K1 and ABC1K3. By contrast, thylakoid FtsH1/2/5/8 and DegP1/5/8 were downregulated during light stress in both genotypes (see Supplemental Figure 8C online).
Effect on Secondary Metabolism
Given the role of the PG in storage of isoprenoids, we carefully evaluated plastid enzymes in isoprenoid metabolism for genotypic effects. The three most abundant enzymes were involved in (1) the methylerythritol phosphate pathway (4-hydroxy-3-methylbutyl diphosphate synthase), (2) geranylgeranyl reductase, or (3) tocopherol and plastoquinone biosynthesis (VTE3) and did not show genotypic effects. But the next most abundant enzyme, CCD4, localized in PGs, was significantly threefold downregulated in k1 k3 already prior to light stress and was further downregulated after 3 and 5 d of light stress (Figure 7B). Furthermore, the total protein mass investment in the MEP or carotenoid pathways did not show any genotypic effects. Thus, within plastid isoprenoid metabolism, we only detected a strong effect on the abundance of PG-localized CCD4.
Effect on Tetrapyrrole Synthesis and Degradation
We identified most of the tetrapyrrole biosynthetic pathway that consistently decreased after 5 d of light stress in both genotypes and after 3 d of light stress in k1 k3 (see Supplemental Figure 9D online). We identified two enzymes involved in tetrapyrrole degradation, namely, thylakoid pheophorbide a oxygenase (PAO/ACD1) and red chlorophyll catabolite reductase (RCCR/ACD2) dually localized in plastids and mitochondria (see Supplemental Data Set 4 online). RCCR did not show genotypic or light stress effects; this lack of induction is consistent with a previous study showing that it was not regulated during leaf development and senescence (Pruzinská et al., 2005). PAO cleaves the photosensitizer pheophorbide, and induction of PAO is associated with leaf senescence (Pruzinská et al., 2005). PAO was never detected in the wild type nor in k1 k3 prior to light stress but was induced after 3 and 5 d of light stress in k1 k3, consistent with the net loss of chlorophyll.
Redox Control and ROS Defense Systems
Interestingly, total protein mass investment in ROS sensing, scavenging, and signaling was unchanged in k1 k3 (see Supplemental Figure 9E online), which is in agreement with the ROS staining for H2O2 and O2− (Figure 4) and provides further evidence that the degreening phenotype is a genetically controlled response to the elevated light intensity, rather than uncontrolled oxidative damage.
Comparison of the Isolated PG Protein Composition in the Wild Type and k1 k3
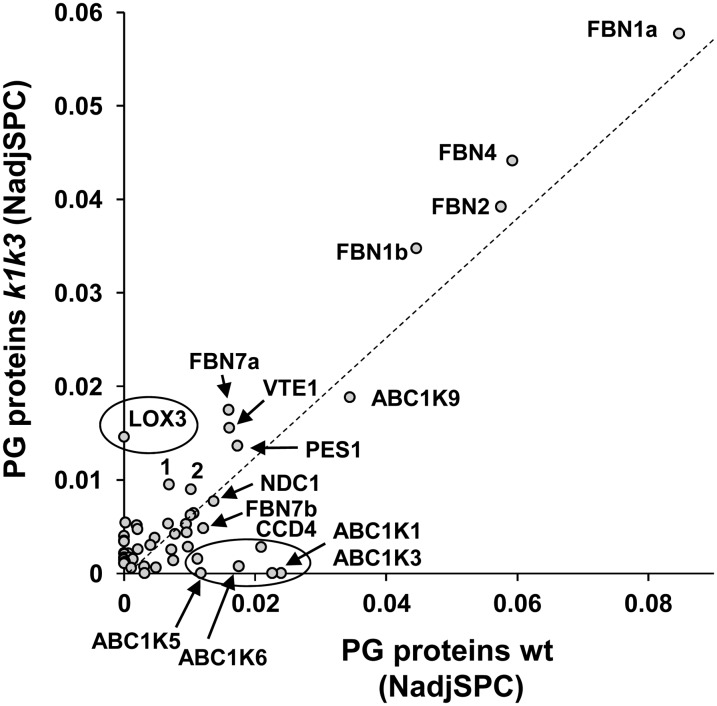
To determine the impact on the PG proteome specifically, we isolated PGs from the wild type and k1 k3 after 5 d of light stress and analyzed them by quantitative proteomics (in three biological replicates) using a similar workflow as for the total leaf proteomes. To exclude low abundant contaminants and assign proteins truly enriched in the PGs of the wild type and k1 k3, we applied a minimal abundance threshold of normalized adjSPC (NadjSPC) > 0.001 (corresponding to ∼0.1% of the extracted protein mass) and required a >15-fold enrichment (based on NadjSPC) compared with the respective leaf proteome from 5 d of light stress. This identified a total of 52 proteins, with 36 and 39 PG-enriched proteins in the wild type and k1 k3, respectively; 23 proteins were enriched in both genotypes (see Supplemental Data Set 5 and Supplemental Figure 10 online). Figure 8 shows the cross-correlation between protein abundance of these 52 proteins in the wild type and k1 k3 and points out which proteins were missing in k1 k3 PGs. Similar to the wild type, the most abundant proteins in k1 k3 were the four fibrillins FBN1a/1b/2/4, followed by ABC1K9, FBN7a, VTE1, and PES1. AB1K1 and ABC1K3 were not detected in k1 k3, indicating that even if perhaps truncated ABC1K1/3 proteins accumulate, they are either not in the PGs or at levels undetectable by our MS analysis. Interestingly, ABC1K5 was also not detected in k1 k3 PGs, and ABC1K6 was strongly downregulated in k1 k3; this suggests that ABC1K5 and ABC1K6 both require stabilization by ABC1K1 and ABC1K3. By contrast, the abundance of ABC1K9 was unchanged in k1 k3, whereas CCD4 was downregulated in k1 k3 PGs. These results for the core PG proteome are highly consistent with the observations for PG proteins from the total leaf proteome analysis presented above. In the next sections, we show that several additional proteins were highly enriched in k1 k3 PGs; these proteins were either not previously detected in isolated PGs from the wild type or were found in PGs but not sufficiently partitioned to the PG to be considered core PG proteins.
Figure 8.
Comparison of Protein Abundance in Isolated PGs from the Wild Type and k1 k3 after 5 d of Light Stress.
Abundance of proteins enriched in the wild type (wt) and/or k1 k3 PGs were plotted. The dashed line indicates the best fit. Those proteins deviating from this line (in particular those circled) are over- or underrepresented in k1 k3 PGs. 1, Unknown1 (AT4G13200); 2, SOUL protein (AT3G10130). Details and accession numbers can be found in Supplemental Data Set 3 online. The same plot with sd (n = 3) is shown in Supplemental Figure 9 online.
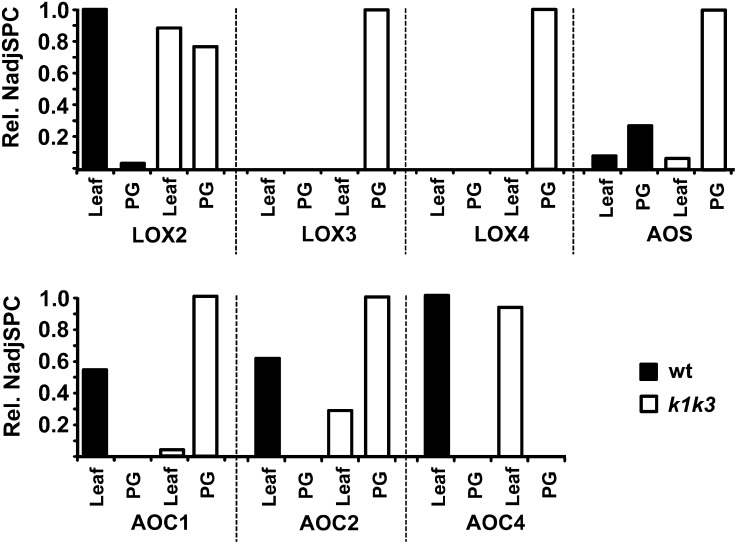
Recruitment of the Jasmonate Biosynthetic Pathway to PGs in k1 k3
A striking enrichment of lipoxygenase 3 (LOX3; AT1G17420) was found in k1 k3 PGs but was not detected in PGs from the wild type (Figure 8; see Supplemental Data Set 5 online). A second, but less abundant, lipoxygenase (LOX4; AT1G72520) was also found in PGs of k1 k3 but not in wild-type PGs. Plastid-localized LOX proteins are involved in the synthesis of the plant hormone jasmonate (JA), and the strong enrichment of LOX proteins in k1 k3 PGs suggested an increased lipid remobilization for JA production. Therefore, we reviewed the abundance of the chloroplast enzymes of the JA biosynthetic pathway in the leaves and PGs of the wild type and k1 k3 (Figure 9). The first step of the JA pathway consists of oxidation of the 18:3 or 16:3 polyunsaturated fatty acids (PUFAs) by the activity of LOX (LOX2/3/4; reviewed in Wasternack and Kombrink, 2010). In chloroplasts, these PUFAs are derived from the thylakoid membrane bilayer through cleavage of a glycerol lipid by the lipases DAD and DGL (Hyun et al., 2008; Wasternack and Kombrink, 2010) and perhaps others (Seo et al., 2009). Oxidized PUFAs are subsequently converted in two steps by AOS and allene oxide cyclase (AOC) into oxophytodienoic acid (OPDA). OPDA is transported from the plastid to the peroxisome where it is reduced by OPDA reductase, followed by three rounds of β-oxidation to generate JA (Gfeller et al., 2010b). Whereas LOX2 did not change in abundance at the whole-leaf level, it clearly associated much more strongly with k1 k3 PGs (Figure 9). Similarly, abundance of AOS in total leaf extracts was not altered in k1 k3 yet was strongly increased in PGs of k1 k3 (Figure 9). Protein accumulation patterns of the other enzymes that were identified in leaf or PG preparations, including the lower abundant LOX proteins and several isoforms of AOC, also indicated recruitment to the k1 k3 PGs.
Figure 9.
Distribution of Plastid-Localized JA Biosynthesis Enzymes in 5-d Light-Stressed Leaf or Isolated PG from the Wild Type and k1 k3.
NadjSPC are normalized within each protein, illustrating the protein’s relative depletion/enrichment in each total leaf or PG preparation. An additional AOC (AOC3) is putatively involved in JA biosynthesis but was not identified in leaf or PG preparations. The plastid-localized steps of the JA biosynthesis pathway begin with peroxidation of linolenic acid by 13-lipoxygenase (13-LOX), which may act on the free or esterified form of the fatty acid. The peroxidized fatty acid is subsequently epoxidized by AOS and cyclized by AOC to form OPDA, which is exported to the peroxisome for subsequent processing by OPDA reductase and three rounds of β-oxidation to produce the JA phytohormone. The relative NadjSPC of LOX2 is set to 0.0129; of LOX3, 0.0147; of LOX4, 0.0040; of AOS, 0.0657; of AOC1, 0.0012; of AOC2, 0.0012, of AOC4, 0.0001; and of OPR, 0.0001. wt, the wild type.
Localization of a Key Enzyme in Chlorophyll Degradation
We identified pheophytin pheophorbide hydrolase (pheophytinase) (PPH; AT5G13800) in each of the three PG preps in k1 k3 (with excellent 51% protein sequence coverage; 49 adjSPC) but not in wild-type PGs, nor in total leaf extracts from the wild type or k1 k3 (see Supplemental Data Set 5 online). PPH dephytylates the Mg-free chlorophyll pigment, pheophytin (phein), yielding pheophorbide and is an important component of the chlorophyll breakdown machinery of senescent leaves, upstream of PAO (Schelbert et al., 2009). The high confidence identification of PPH in isolated PGs shows that PPH is likely to be a bona fide PG protein, similar to PES1,2, which is involved in esterification of free phytol immediately downstream of PPH and to the putative upstream Mg2+-dechelatase (AT5G17450) that we identified recently (Lundquist et al., 2012a).
Metabolite Profiling of the Wild Type and k1 k3
To assess the accumulation of PG lipids, extracts of total lipid from wild-type and k1 k3 PGs were analyzed by thin layer chromatography (TLC; see Supplemental Figure 11 online). Surprisingly, neither genotype accumulated detectable levels of TAGs. Although TAGs have been shown to accumulate in PGs (Gaude et al., 2007), little is known about their accumulation patterns under stress. PGs in k1 k3 accumulated a much higher amount of lipid migrating near the solvent front and therefore must have a strongly nonpolar character. Significantly higher amounts of carotenoid (carotene and/or xanthophyll) migrating with the β-carotene standard were found in k1 k3 PGs, consistent with the prenyl-lipid-specific profiling described below. Several lipids migrating below the 16:0-18:0 fatty acyl standard were also elevated in the k1 k3 PGs. These compounds did not migrate with any of the molecular standards and may represent alternative acyl ester compounds such as carotenoid esters, fatty acid methyl esters, or fatty acid phytyl esters that have also been shown to accumulate in the PG (Lippold et al., 2012). Lower amounts of contaminating chlorophyll were found in k1 k3 PGs, consistent with the depleted chlorophyll levels in k1 k3 leaves.
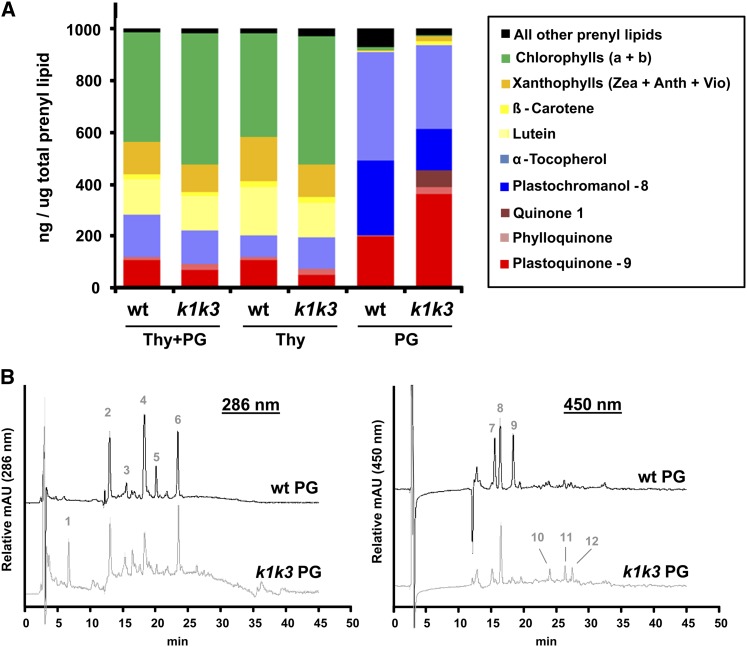
Several clues led us to hypothesize a function for ABC1K1 and ABC1K3 in prenyl-lipid metabolism (Lundquist et al., 2012b). We used an established method with C30 reverse-phase HPLC and photodiode array detector (Fraser et al., 2000) to characterize and quantify the prenyl-lipid profile of isolated PGs, thylakoids, and total leaves from the wild type and k1 k3 (Table 1; Figure 10; see Supplemental Data Set 6 online). Three additional metabolites (PQ-9ox, plastochromanol-8 [PC-8], and an unknown quinone, eluting at 24.5, 19.6, and 6.0 min, respectively) could not be identified with certainty because they were absent in the results of Fraser et al. (2000). Instead, we characterized them by HPLC-MS (see Supplemental Figures 12 to 14 online). The only plastoquinone identified by Fraser et al. (2000), eluting at 5.6 min, is likely PQ-9red, judging by its early elution on the C30 column and λmax of 275 nm (Redfearn and Whittaker, 1962; Martinis et al.). HPLC-MS was used to characterize the three additional metabolites, using the same elution gradient and a single quadrupole mass spectrometer with ammonium acetate (NH4+CH3COO−) added postcolumn to induce ionization. The metabolite eluting at 24.5 min was confirmed as PQ-9ox based on its absorption spectrum and ionized masses (see Supplemental Figure 12 online). The abundant tocochromanol, with a retention time of 19.6 min, was confirmed as PC-8 based on absorption spectrum and masses (see Supplemental Figure 13 online). Its redox state could not be determined from the molecular mass alone, because the oxidized and reduced PC-8 have the same molecular mass. Extracted ion chromatograms identified the other reduction state of PC-8 (PC-8ox), eluting 3.4 min later at 23.0 min, with much lower abundance (see Supplemental Figure 13 online).
Table 1. Quantitative Metabolite Profiles of Thylakoid and PG Prenyl Lipids in ng/μg Prenyl-Lipids.
| The Wild Type |
k1 k3 |
|||||
|---|---|---|---|---|---|---|
| Prenyl-Lipid Metabolite | Thy and PGa | Thya | PGa | Thy and PGa | Thya | PGa |
| Plastoquinone-9b | 104.1 ± 24.4 | 104.1 ± 16.8 | 193.8 ± 49.7 | 64.5 ± 28.6 | 46.7 ± 27.0 | 359.4 ± 53.7 |
| Phylloquinone | 11.1 ± 4.2 | 12.0 ± 6.6 | 4.9 ± 1.9 | 23.0 ± 2.5 | 24.1 ± 3.2 | 24.8 ± 7.9 |
| Quinone 1c,d | n.d. | n.d. | n.d. | n.d. | n.d. | 67.1 ± 70.5 |
| PC-8b | n.d. | n.d. | 289.4 ± 48.1 | n.d. | n.d. | 160.4 ± 76.6 |
| α-Tocopherol | 162.6 ± 42.6 | 82.6 ± 24.4 | 421.6 ± 75.4 | 131.4 ± 15.0 | 119.2 ± 59.9 | 324.3 ± 117.5 |
| γ-Tocopherol | n.d. | n.d. | n.d. | 10.4 ± 7.1 | 19.7 ± 19.1 | n.d. |
| Phytoenee | 0.4 ± 0.1 | 0.2 ± 0.3 | 0.9 ± 0.1 | n.d. | n.d. | 0.2 ± 0.1 |
| Lutein | 138.2 ± 7.0 | 186.4 ± 29.0 | 0.3 ± 0.2 | 132.2 ± 15.8 | 135.4 ± 19.4 | 4.4 ± 01.5 |
| Violaxanthin | 28.6 ± 4.2 | 38.8 ± 5.5 | 1.1 ± 0.7 | 15.1 ± 2.4 | 16.4 ± 4.3 | 3.4 ± 2.4 |
| Antheraxanthin | 16.3 ± 1.9 | 21.8 ± 2.6 | n.d. | 15.5 ± 3.1 | 17.6 ± 2.5 | n.d. |
| Zeaxanthin | 80.6 ± 28.4 | 111.1 ± 37.9 | 3.7 ± 0.8 | 74.3 ± 28.1 | 94.4 ± 12.7 | 14.2 ± 5.3 |
| Neoxanthin | 0.4 ± 0.1 | 0.3 ± 0.3 | n.d. | 0.1 ± 0.3 | 0.4 ± 0.1 | n.d. |
| α-Carotene | 4.0 ± 1.4 | 5.1 ± 1.7 | n.d. | 2.3 ± 0.3 | 2.4 ± 0.4 | n.d. |
| β-Carotene | 20.1 ± 7.0 | 24.4 ± 14.6 | 1.2 ± 0.6 | 16.0 ± 6.3 | 20.8 ± 8.5 | 10.5 ± 6.7 |
| Other prenyl-lipidsf | 421.8 ± 98.9 | 367.4 ± 23.9 | 83.2 ± 22.0 | 324.6 ± 15.3 | 330.0 ± 7.2 | 30.6 ± 16.1 |
n.d., metabolite not detected.
Lipids were extracted from thylakoid samples before and after sonication, which included both thylakoids and attached PGs or thylakoids from which most PGs were removed. PG are isolated PGs.
Oxidized and reduced forms.
Unknown compounds were tentatively assigned to a compound class based upon their absorption spectrum.
The molecular mass of quinone 1 was determined by MS to be 746.6 D.
Cis- and trans-forms.
Other prenyl-lipids include chlorophyll a and b, two unknown tocopherol compounds, eight unknown carotenoid compounds, and seven unknown quinone compounds, which were assigned to a compound class based on their absorption spectrum.
Figure 10.
Metabolite Profiling of Thylakoids and PGs.
(A) The abundance of selected prenyl-lipids normalized to the total pool of prenyl lipids were plotted from presonicated thylakoid preparations, containing thylakoid and bound PGs (Thy+PG) as well as subsequent fractions of thylakoid and isolated PGs. wt, the wild type.
(B) HPLC chromatographic traces of wild-type and k1 k3 PGs. Separation of prenyl-lipids was done on a reverse-phase C30 column using a methyl t-butyl ether mobile phase. k1 k3 is offset from the wild type along the y axis. Identification of metabolite peaks was based on retention time and absorption spectrum, established by Fraser et al. (2000) with the exception of PC-8, PQ-9, and Quinone 1, identified by HPLC-MS analysis (see Supplemental Figure 12 online). Abundant peaks are indicated by number as follows: (1) quinone 1 (molecular mass = 746.6 D), (2) α-tocopherol, (3) phylloquinone, (4) plastochromanol-8 (reduced), (5) 15-cis-phytoene, (6) PQ-9 (oxidized), (7) chlorophyll b, (8) zeaxanthin, (9) chlorophyll a, (10) unidentified carotenoid, (11) β-carotene, and (12) 9-cis-β-carotene. mAU, milli Absorbance Units.
The majority of the PG prenyl-lipid pool in both genotypes was composed of just three compounds: PQ-9, α-tocopherol, and PC-8. They made up 90% (w/w) of the wild-type PG pool and 85% of the k1 k3 PG pool (Table 1, Figure 10). Strikingly, the ratio of these compounds was altered in k1 k3; the relative amount of PQ-9 increased 1.9-fold and the amount of PC-8 and α-tocopherol decreased 1.8- and 1.3-fold, respectively. This pattern is consistent with a reduced activity of the PG-localized tocopherol cyclase (VTE1), which catalyzes cyclization of quinone substrates such as PQ-9 to tocochromanol products (Porfirova et al., 2002; Zbierzak et al., 2009). The protein level of VTE1 was unchanged in the isolated PGs (Figure 9), indicating altered regulation of activity, rather than degradation of VTE1 (see Discussion). An unknown quinone, eluting at 6.0 min, accumulated to ∼7% (w/w) of the pool size in the k1 k3 PG but was below the limit of detection in wild-type PGs as well as thylakoid preparations of both genotypes (Table 1). The mass spectrum associated with this chromatographic peak was consistent with a molecular mass of 746.6 (see Supplemental Figure 14 online). While the absorption maximum of this molecule at ∼264 nm is consistent with a quinone molecule and its mass was conclusively measured, no known products or intermediates of tocochromanol/quinone metabolism fit this mass. This compound may be a quinone biosynthesis intermediate, indicating impairment in a tocochromanol/quinone metabolic pathway or a prenyl-lipid oxidation product accumulating as a result of ROS scavenging. Its molecular mass and retention time eliminates it as a known VTE1 substrate, though it is indicative of a solanyl-based prenyl-lipid derivative. Additional experimentation will be necessary to identify this metabolite accumulating in k1 k3.
Parallel metabolite analyses of the thylakoid preparations, before and after stripping of the PGs, provided a comparison of metabolite distribution between thylakoids and PGs (Table 1, Figure 10; see Supplemental Data Set 6 online). Apart from chlorophyll a and b, the most abundant prenyl-lipids of the thylakoid preps were α-tocopherol, lutein, and plastoquinone (PQ-9), comprising ∼16, 13, and 10% of the prenyl-lipid fraction in the wild type, respectively (Figure 10). As an important structural pigment of the abundant light-harvesting complexes, it is not surprising that lutein was found to be the most prevalent carotenoid in the thylakoid but it was essentially absent in PGs. PQ-9 in the thylakoid was reduced to about half the level of the wild type in k1 k3 and was strongly redistributed toward the PG. Despite low relative abundance, phylloquinone increased about twofold in k1 k3 and also appeared to redistribute toward the PG. Thylakoid α-tocopherol levels appeared to be slightly increased in k1 k3 due to redistribution from the PG to the thylakoid preps. Similarly, a small proportion of γ-tocopherol was found in k1 k3 thylakoid preps but was absent from wild-type thylakoid preps. Surprisingly, no detectable PC-8 accumulated in the thylakoid preps of either genotype despite its abundance in the PG. In the wild type, nearly all carotenoids were found to distribute to the thylakoid fraction. However, a conspicuous accumulation of β-carotene and zeaxanthin was found in k1 k3 PGs, and these may be released from PSII and light-harvesting complex during disassembly and directed to the PG for storage or degradation (Table 2).
Table 2. Ratio of Selected Carotenoids Normalized to Chlorophyll b in PG Preps.
| Carotenoid | Wild Type | k1 k3 |
|---|---|---|
| Zeaxanthin | 1.2 | 8.6 |
| β-Carotene | 0.4 | 6.3 |
| Lutein | 0.1 | 2.6 |
| Violaxanthin | 0.4 | 2.0 |
| Phytoene | 0.3 | 0.1 |
Finally, we also determined the prenyl-lipid composition at the total leaf level before light stress and after 1 and 5 d of 5× light stress (see Supplemental Data Set 6 online). This showed that PQ-9 remains constant in the wild type, but it starts out lower in k1 k3 and then increases to the wild type level by day 5.
In summary, a modified abundance of quinone and tocochromanol compounds in the k1 k3 PG, including the appearance of an unidentified putative quinone compound, indicates reduced activity of PG-localized VTE1. The redistribution of several prenyl-lipids between the thylakoid and PG in k1 k3, notably the movement of plastoquinone and phylloquinone toward the PG and α-tocopherol toward the thylakoid, may reflect PG’s role in detoxification of ROS and redox signaling through shuttling of these redox active compounds or storage of excess metabolites released during turnover of the thylakoid membrane system.
DISCUSSION
Recent studies have suggested that chloroplast PGs are dynamic monolayer particles with specialized metabolite and protein content that play an active role in thylakoid formation, remodeling, and breakdown, rather than merely serving a passive storage function as long believed (reviewed in Bréhélin et al., 2007; Besagni and Kessler, 2013). The PG-localized ABC1K family likely plays a regulatory role in PG function, in particular through control of prenyl-lipid metabolism. This study provides new insight into the role of two of the six known PG-localized ABC1K kinases and shows that PGs function as highly specialized thylakoid microdomains that recruit and concentrate specific sets of proteins and metabolites. Absence of ABC1K1 or ABC1K3 results in a conditional high light stress phenotype in Arabidopsis manifested by photobleaching and rapid leaf necrosis. The corresponding k1 k3 double mutant showed a conditional enhanced light sensitivity, displaying loss of chlorophyll at lower light stress intensities. This degreening, senescence-like phenotype develops over several days and is irreversible, ultimately resulting in complete loss of pigment and subsequent necrosis. This degreening can also be induced by drought or by nitrogen limitation but not by cold treatment, suggesting that the degreening phenotype relates to disassembly and turnover of the thylakoid membrane and its content, rather than light stress per se. Through quantitative profiling of proteins and metabolites of leaves or chloroplast subfractions, as well as transmission electron microscopy (TEM) and ROS staining, we described a molecular phenotype of k1 k3 in response to moderate light stress that includes the following differences compared with light-stressed wild-type plants: (1) loss of PSII and other thylakoid complexes and hyperstacking of thylakoids; (2) lack of starch accumulation, without significant loss of Calvin cycle enzymes; (3) increased number of PGs but with unchanged size; (4) loss of chlorophyll and strong induction of PPH in PGs; (5) shift of PQ-9 from thylakoid to PG; (6) change in PG metabolites, in particular the accumulation of a novel quinone compound and several carotenoids; (7) recruitment of the JA biosynthesis enzymes to PG, but no general upregulation of the chloroplast enzymes; and (8) accumulation of β-cyclocitral likely functioning as a retrograde signal. Additionally, our results provide indirect evidence that targets for ABC1K1/3 phosphorylation may include PG-localized VTE1 and CCD4 both involved in prenyl-lipid metabolism and possibly ABC1K5 and ABC1K6. Finally, we provide support for the hypothesis that the PG is a microdomain within the thylakoid membrane and serves as a platform to recruit proteins and metabolites into spatial proximity, to facilitate metabolic channeling or signal transduction, comparable to lipid rafts of plasma membranes. This represents a paradigm shift in our view of the PG, which was long thought to be just a passive storage space for lipophilic metabolites.
Phenotypic Similarity between k1 k3 and a Mutant Impaired in Starch Biosynthesis and the Triose Phosphate/Phosphate Translocator
Results from the total leaf proteome analysis demonstrated significantly reduced accumulation of the photosystem I, PSII, and cytochrome b6f complexes under light stress. In particular, the PSII core complex was strongly downregulated, consistent with the degreening phenotype. A significant reduction in the NDH complex was also apparent after 5 d of light stress. Moreover, TEM analysis showed thylakoid hyperstacking in k1 k3 after light stress, indicative of an altered thylakoid composition, and shows striking resemblance to the hyperstacking in an Arabidopsis double mutant impaired in starch biosynthesis and the triose phosphate/phosphate translocator (adg1-1/tpt-1) (Häusler et al., 2009). The mutant has diminished utilization of photoassimilates, strongly diminished photosynthetic electron transport capacity and chlorophyll contents, increased PG content, and appears to have a more reduced redox state. The similarity between the two phenotypes (lack of starch, hyperstacking, loss of chlorophyll, etc.) suggest that k1 k3 also suffers a defect in carbon metabolism that cannot be attributed to loss of Calvin cycle protein abundance indicating crosstalk between the thylakoid and Calvin cycle metabolism. Such crosstalk was also evident in the Arabidopsis CEF1 mutant that was isolated in a forward genetics screen for cyclic electron flow mutants, which surprisingly mapped to chloroplast fru-1,6-bisphosphatase (Livingston et al., 2010).
A Retrograde Signaling Trigger
Conditional light stress phenotypes have been observed in a wide variety of chloroplast mutants and are typically the result of either direct large scale oxidative damage through production of ROS or genetically controlled degradation indirectly mediated by retrograde signaling to the nucleus. Such chloroplast mutants can be divided into different classes: for instance, those that accumulate metabolic intermediates that act as photosensitizers (e.g., chlorophyll biosynthesis intermediates; Meskauskiene et al., 2001); those that are defective in ROS defense systems (e.g., the ascorbate defense system; Giacomelli et al., 2006, 2007; Kangasjärvi et al., 2008); or mutants defective in regulation, expression, or assembly of the photosynthetic apparatus (Dall’Osto et al., 2007). The molecular analysis of the k1 k3 mutant under moderate light stress strongly suggests that a genetically controlled retrograde pathway, rather than direct oxidative damage, causes the relatively slow degreening phenotype under 5× light stress. It has been suggested that at least two independent pathways for 1O2 retrograde signaling exist, mediated either by β-cyclocitral (Ramel et al., 2012) or the EX proteins (Lee et al., 2007; Baruah et al., 2009), dictated by the source of 1O2 (see discussion in Woodson and Chory, 2012). β-Cyclocitral is a product of 1O2, and β-carotene and has recently been shown to act as a mediator of 1O2-specific retrograde signaling (Ramel et al., 2012). Our measurements of β-carotene oxidation products points to 1O2 retrograde signaling mediated by accumulation of β-cyclocitral, which drives the degreening process in k1 k3 and is consistent with multiple observations: (1) Increased production of H2O2 or O2− could not be detected in k1 k3, and ROS scavenging proteins were not increased in k1 k3 leaves; (2) the degreening was irreversible after only 3 d under the 5× light stress (when degreening was still not visible to the naked eye), indicating a committed genetically controlled pathway had been initiated; (3) the persistent, gradual decrease in chlorophyll and carotenoid is consistent with a systematic, controlled degradation of chlorophylls and carotenoids; and (4) the degreening phenotype was independent of the EX pathway, similar to β-cyclocitral–mediated retrograde signaling (Ramel et al., 2012). The source(s) of 1O2 production remains to be determined.
PG Morphology
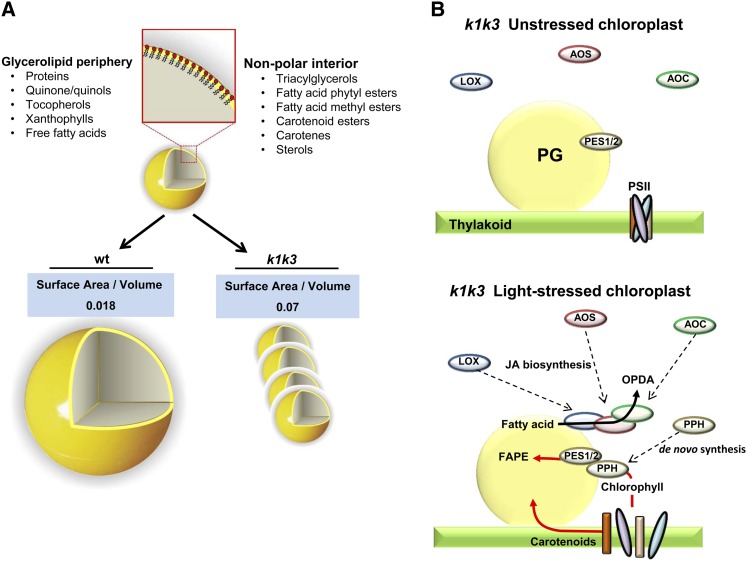
Upon the fivefold increase in light intensity, both k1 k3 and the wild type increased the total amount of PG mass per chloroplast, either by increasing the number (in k1 k3) or by increasing the size (in the wild type). Furthermore, the PG-localized FBNs increased their abundance approximately two- to threefold without significant genotypic effects, and this persistent increase explains most of the increase in PG protein mass in both genotypes. Several other PG proteins did not change their abundance during light stress in both genotypes. Together, this suggests that FBNs are needed to expand total PG mass but that they don’t control PG diameter. Consistently, overexpression of a pepper (Capsicum annuum) FBN homolog, CDSP34, in tobacco (Nicotiana tabacum) led to a five- to sevenfold higher number of PGs in chloroplasts, whereas the diameter did not change or slightly decreased (Rey et al., 2000).
The morphology of the PG has important implications for the accumulation of lipophilic metabolites. The single monolayer periphery and hydrophobic interior require that amphiphilic compounds, such as prenyl-lipids, accumulate at the periphery of the PG, while hydrophobic compounds, such as fatty acid phytyl esters (FAPEs) and TAG, deposit in the PG interior (Figure 11A). Thus, the ratio of amphiphilic-to-hydrophobic metabolites at the PG will be reflected in the surface area/volume ratio of the PG and, thus, the diameter of the approximately spherical PGs. An increasing diameter achieves a lower surface area/volume ratio, which would be desirable when hydrophobic compounds, such as FAPEs or TAGs, are preferentially accumulating at the PG. Conversely, a decreasing diameter will achieve a higher surface area/volume ratio, preferred when amphiphilic compounds, such as prenyl-lipids, are in higher abundance. From the TEM measurements of in situ PG dimensions, we estimated that the surface area/volume ratio is approximately 4 times higher in k1 k3 than in the wild type at 5 d of 5× light stress (Figure 11A), suggesting a higher ratio of amphiphilic lipids (e.g., prenyl-lipids) to hydrophobic lipids (e.g., TAGs or FAPEs) in k1 k3. Indeed, our prenyl-lipid profiling indicates that the amphiphilic PQ-9 pool in particular shifts to the PG from the thylakoid.
Figure 11.
Model for Recruitment to the PG under Light Stress and Its Influence on Surface Area/Volume.
(A) The three-dimensional cut-away representation of a PG illustrates the PG periphery composed of amphiphilic glycerolipids surrounding a nonpolar interior. The differential ultrastructures of the PG in response to 5× light stress in the wild type (wt) and k1 k3 genotypes are depicted.
(B) Recruitment of JA biosynthetic enzymes and photosystem metabolites to the PG occur simultaneous with increasing number of PGs in k1 k3 chloroplasts. Because the PPH protein has never been identified elsewhere in the plastid, we propose that it is synthesized de novo in response to the increased chlorophyll turnover of k1 k3 light-stressed leaves.
Several Steps of Chlorophyll Degradation and Recycling Occur at the PG
The degradation of chlorophyll must be carefully controlled as several of the degradation products are toxic (Mochizuki et al., 2010; Hörtensteiner and Kräutler, 2011). We observed strong evidence for localization of PPH to PGs, placing this key enzyme that is involved in cleavage of the phytol tail from the pheophorbide ring in the same compartment as the immediate downstream enzymes PES1 and 2 that are involved in detoxification of the phytol tail by esterification with fatty acid (Lippold et al., 2012). Indeed, the phytol tail, released during chlorophyll breakdown, has been shown to be deposited in the PG as FAPEs (Gaude et al., 2007). Moreover, a protein putatively involved in removing Mg2+ from the porphyrin ring (AT5G17450), immediately upstream of PPH, is also located in PGs (Lundquist et al., 2012a; see discussion in Besagni and Kessler, 2013). Therefore, the initial steps of chlorophyll degradation, including cleavage and storage of the phytol tail, appear to occur at the PG (Figure 11B). Interestingly, cleaved phytol can be recycled by phytol kinase (VTE5) for generation of tocopherol (Valentin et al., 2006), but VTE5 has not been localized to PGs, which is logical as it has several transmembrane domains making its localization incompatible with localization in the monolayer of the PG. However, it remains to be determined how VTE5 can act on the phytol. Breakdown of the soluble pheophorbide ring occurs outside the PG through the sequential action of PAO and RCCR; the leaf comparative proteome analysis showed that PAO is also strongly upregulated in k1 k3 following the light stress treatment, consistent with increased chlorophyll degradation rates.
Recruitment of the JA Biosynthetic Pathway to the PG Likely Facilitates Thylakoid Lipid Turnover
We observed strong evidence for recruitment of the three plastid JA biosynthetic enzymes (LOX, AOS, and AOC). The PG is a logical site for the initial steps of the JA biosynthetic pathway, which is initiated in the plastid from 16:3 and 18:3 PUFAs, which are found prominently in thylakoid membrane galactolipids. JA biosynthesis begins with enzymatic peroxidation of PUFAs catalyzed by 13-LOX enzymes (LOX2, 3, and 4). These PUFAs are derived from the thylakoid membrane bilayer through cleavage of a glycerol lipid by the lipases DAD and DGL (Hyun et al., 2008; Wasternack and Kombrink, 2010) and perhaps others (Seo et al., 2009). Oxidized PUFAs are subsequently converted in two steps by AOS and AOC into OPDA. OPDA is transported from the plastid to the peroxisome where it is reduced by OPDA reductase followed by three rounds of β-oxidation to generate JA (Gfeller et al., 2010b). In total leaf tissue, we did not observe elevated abundance of LOX, AOS, or AOC in k1 k3 relative to the wild type. However, all isoforms of these enzymes were enriched in the k1 k3 PGs, indicating recruitment of preexisting pools of these enzymes to the PG (Figures 9 and 11B).
JAs accumulate during natural and dark-induced senescence, but an increase in these compounds is not essential for the initiation or progression of these senescence processes (Zhang and Zhou, 2012). In fact, JA production during senescence has been suggested to be a consequence of increased thylakoid membrane turnover (Seltmann et al., 2010). We propose that the recruitment of JA biosynthetic enzymes to PGs in k1 k3 is a consequence of the degreening process, thereby facilitating the remobilization of thylakoid lipid and export out of the chloroplast. Production of JA involves several initial steps in the plastid, followed by export of the JA intermediate OPDA to the peroxisomes where JA is ultimately produced and formation of JA conjugates are needed for it to act as plant hormone (Fonseca et al., 2009; Gfeller et al., 2010a). It remains to be determined if the strong recruitment of plastid JA biosynthetic enzymes to the PG results in increased OPDA production and subsequent production of bioactive JA conjugates or if perhaps export of OPDA serves as a mechanism to simply transport fatty acids out of the chloroplast.
ABC1K1/3 Targets and Their Interactions
Phylogenetic analysis and limited amounts of experimental information suggest that the ancient function of the ABC1K family is the regulation of menaquinone metabolism (Lundquist et al., 2012a). Based on this, and the observation that ABC1K1 and ABC1K3 localize to the PG, which is rich in quinones and contains several enzymes of their metabolism, we hypothesized that ABC1K1 and ABC1K3 function in the regulation of plastidic quinone metabolism (Ytterberg et al., 2006; Bréhélin et al., 2007; Lundquist et al., 2012a). Consistently, we show here that loss of ABC1K1 and ABC1K3 alters the accumulation of prenyl-lipids, including quinones in the PG under light stress. A reduction of the abundant tocochromanols, α-tocopherol and PC-8, was found with a concomitant increase in PQ-9. This observation indicates reduced activity of the PG-localized VTE1 enzyme, responsible for cyclization of the quinones dimethylphytyl benzoquinone (DMPBQ) and PQ-9 to their tocochromanol products, α-tocopherol and PC-8, respectively. The dramatic accumulation to 7% of the PG prenyl-lipid pool of unknown quinone (746.6 D) in k1 k3, but not in the wild type, is interesting and shows that prenyl-lipid metabolism in the chloroplast is not fully understood. The possibility that the unknown quinone was the VTE1 substrate DMPBQ was considered; however, the molecular mass is inconsistent, and elution of DMPBQ from vte1 PGs did not match the elution time of the unknown quinone (data not shown). Importantly, the level of VTE1 enzyme was unchanged in k1 k3 PGs, further indicating an impaired activity of the VTE1 enzyme, rather than change in abundance. We suggest that ABC1K1 and/or ABC1K3 are/is responsible for activation of VTE1 under excess excitation energy, likely through phosphorylation. VTE1 was shown to be phosphorylated in a large-scale phosphoproteome analysis upon nitrogen starvation (Nakagami et al., 2010).
Carotenoids comprise a small fraction of the prenyl-lipid pool of wild-type light-stressed PGs, which is composed almost exclusively of phytoene. However, it was found that loss of ABC1K1 and ABC1K3 dramatically changed the carotenoid composition in the PG. Accumulation of β-carotene, and to a lesser extent the xanthophylls lutein, zeaxanthin, and violaxanthin, was apparent in PGs of k1 k3 (Table 2). Accumulation of β-carotene and the xanthophylls in the k1 k3 PGs supports the hypothesis that PGs are the site of deposition and degradation of carotenoids released from photosystems and light harvesting complexes of the thylakoid. There is one PG-localized enzyme that is predicted to catalyze carotenoid degradation, CCD4, which substrates are not known. Interestingly, CCD4 levels in k1 k3 PGs were reduced by more than threefold; whether accumulation of carotenoids and depletion of CCD4 at the k1 k3 PGs are directly related remains to be explored, but is an intriguing possibility. The reduced CCD4 level in k1 k3 compared with the wild type suggests a role for ABC1K1 or ABC1K3 in regulation of CCD4 stability.
Interestingly, a recent study in yeast showed that the kinase activity of the ABC1K (COX8) is essential for stabilization of components of the multisubunit complex involved in ubiquinone biosynthesis (Xie et al., 2012). Phosphorylation by ABC1K1 and/or ABC1K3 may similarly confer protein stability to CCD4. Coincidentally, phosphorylation of thylakoid STN7 has been shown to protect it from degradation (Willig et al., 2011). A connection between ABC1K1 and ABC1K3 in plastid protein degradation and prenyl-lipid metabolism was suggested by the mRNA coexpression patterns of ABC1K1 and ABC1K3. In addition to the reduced levels of the PG-localized CCD4 in k1 k3, three prenyl-lipid coexpressers of ABC1K1 and ABC1K3 (Lundquist et al., 2012a) were effected under light stress: Coexpressor geranylgeranyl diphosphate reductase was downregulated in k1 k3 after 3 d of stress, while zeaxanthin epoxidase and ζ-carotene desaturase were elevated in k1 k3 after 5 d of light stress. This warrants further investigation of their functional relationship to ABC1K1/3.
The synergistic light stress phenotype demonstrated in k1 k3 suggests distinct targets by the two kinases but does not rule out one or more common phosphorylation targets. It is likely that the targets need to be localized (at least transiently) to PGs to undergo phosphorylation, thereby limiting the potential target list. Various lines of evidence indicate that ABC1K1 and ABC1K3 interact, providing mutual stabilization. Furthermore, ABC1K5 and ABC1K6 are also destabilized in the absence of ABC1K1 and ABC1K3, suggesting a phosphorylation cascade or a complex involving all four kinases.
Conclusions
This work presented an investigation of Arabidopsis T-DNA insertion mutants affected in two PG-localized atypical protein kinases, ABC1K1 and ABC1K3. Evidence is presented that ABC1K1 and ABC1K3 are regulators of plastid metabolism, important for adaptation to excess excitation energy and crosstalk to the Calvin cycle. Targets of ABC1K1 and ABC1K3 remain elusive but likely are enzymes involved in prenyl-lipid metabolism, in particular PG-localized VTE1 and CCD4. Furthermore, ABC1K1 and ABC1K3 likely form a complex and also are required for the stability of ABC1K5 and ABC1K6, in turn affecting other PG functions and downstream effects on other processes in the chloroplast. The discovery of PPH in k1 k3 PGs further establishes the PG as the location for the initial steps of chlorophyll degradation. Our results provide compelling evidence that the PG recruits proteins and compounds into spatial proximity for control of metabolism and likely redox control, functioning as a microdomain facilitating concentration of metabolites and proteins to accommodate metabolic channeling and higher flux rates for specific reactions.
METHODS
Plant Materials and Plant Growth Conditions
The following SALK insertion mutants were acquired from the ABRC: abc1k1-1 (SALK 057147), abc1k1-2 (SALK 068628), and abc1k3-1 (SALK 128696). Plants were grown on Cornell Mix soil with a 16-h photoperiod of 120 µmol photons m−2 s−1 actinic light at a constant 60% relative humidity and 22°C ambient temperature. For light stress application, plants were transferred late in the vegetative growth stage to 520 µmol photons m−2 s−1 (5× light stress) or 1000 µmol photons m−2 s−1 (10× light stress) actinic light, while maintaining the same 16-h photoperiod, 60% relative humidity, and 22°C ambient temperature. For cold stress treatments, plants were transferred late in the vegetative growth stage to 4°C during the dark period, maintaining the same 16-h photoperiod with 120 µmol photons m−2 s−1 actinic light. For drought stress treatments, water was withheld from the soil for 1.5 weeks beginning during the vegetative growth stage. For nitrogen stress, plants were grown on Murashige and Skoog agar plates prepared from Murashige and Skoog basal salt micronutrient solution (Sigma-Aldrich) diluted to half strength. Non-nitrogen salts (MgSO4, CaCl2, and KH2PO4) were added back to half strength Murashige and Skoog concentration, while ammonium nitrate and potassium nitrate were added to one-half or one-eighth concentration. Sterilized seeds were sown on half nitrogen and one-eighth nitrogen plates and grown under a 16-h photoperiod at 120 µmol photons m−2 s−1.
RT-PCR Analysis
Total RNA was extracted from leaf tissue using the RNeasy plant mini kit (Qiagen) according to the manufacturer’s instructions. From total RNA, cDNA was synthesized by the Superscript III cDNA first-strand synthesis system (Invitrogen) according to the manufacturer’s instructions. cDNA samples were normalized by OD260, and PCR reactions were set up in the GoTaq Green Master Mix (Promega) using the appropriate primers pairs, as described below, and 1 μL of cDNA template. Reactions were run for 30 or 35 cycles with an annealing temperature of 56°C. The forward and reverse primers are as follows: K1 F:, 5′-CTGTTCTTAATCGGAGAGATGC-3′ and 5′-CCTCTTGATTCCAGCACTTTACTCC-3′; K1 M, 5′-CTTGACTGTTGGCGTAAGTGCTG-3′ and 5′-CATATACCCTCCTGCGTAAGC-3′; K1 B, 5′-ATCCAAATCCTGCACTTCGGG-3′ and 5′-CCAGTGTGGGAATATCTACTTCGT-3′; K3 F, 5′-GGTCAGTCTCTGGGTTTAACTC-3′ and 5′-CTTCATGTTCTTCTCTAAGTTGCCC-3′; K3 M, 5′-GCTAGAGACCATTTTCTCGTC-3′ and 5′-CTGACTCTCAATGGCAAGTTGC-3′; K3 B, 5′-CCCCTGATGTGGATGTTACTCCTA-3′ and 5′-TGCCTCTGGTTGTTGTAACACCT-3′; ACTIN2, 5′-CAAACGAGGGCTGGAACAAGACT-3′ and 5′-GCAACTGGGATGATATGGAAAAGA-3′.
Antibody Generation
The C termini of the ABC1K family members are highly divergent and thus provide specific sequence of adequate size for antigen production. The C termini of ABC1K1 (residues 578 to 682) and ABC1K3 (residues 556 to 771) were cloned and overexpressed in Escherichia coli with His6 tags and purified in high abundance on nickel-nitrilotriacetic acid resin. Antibodies were raised in rabbit against purified antigen. Aliquots of antisera from the final bleed were purified on an antigen column and used for immunoblotting and immunoprecipitation.
PG Isolation
PG isolations were performed as described (Lundquist et al., 2012a). All PG preparations were made from 5 d moderate light-stressed leaf tissue, collected in the morning of the sixth day of stress. The resulting floating pad of PGs was removed with a hypodermic disposable syringe, flash frozen in liquid N2, and stored at −80°C.
Quantification of Carotenoid Oxidation Products
Whole rosettes (∼500 mg) were harvested in 4 mL dichloromethane and homogenized with a Polytron Power Gen 125 homogenizer and sawtooth generator at full speed for 1 min. Following centrifugation for 3 min at 18,000g, the supernatant was extracted and evaporated to 500 μL under a gentle stream of nitrogen. Samples were filtered with a 0.45-μm syringe filter and stored under nitrogen at −80°C until use. Compounds were separated and identified by gas chromatography/MS using an Agilent 6890N GC equipped with split/splitless injector and a JandW Scientific DuraGuard DB-5ms (30 m × 0.25 mm × 0.25 um film with 10-m guard column) coupled to a JEOL-GCMate II mass spectrometer using electron impact ionization in selective ion monitoring mode. Samples of 2 μL were injected in splitless mode with an inlet purge wait time of 2 min. The following oven temperature program was used with helium as the carrier gas at a constant flow rate of 1.0 mL/min: from 50 to 160°C at a rate of 15°C/min, from 160 to 183°C at a rate of 3°C /min, and from 183 to 300°C at a rate of 23.4°C/min and held at 300°C for 5 min. Interface, injector, and ion source were kept at 280, 280, and 250°C, respectively. MS spectra were acquired with electron energy of 70 eV, preamp multiplication of ×100, and detector voltage of 300 V. Relative quantification of compounds was measured by the peak area of characteristic fragment ions normalized to the leaf fresh weight.
Leaf Tissue Stains for ROS
Two of the oldest leaves of three individuals from each genotype were harvested and submerged in 10 mL of diaminobenzidine staining solution (10 mM Tris-acetate, pH 4.0, and 0.05% [w/v] diaminobenzidine) or 10 mL of nitroblue tetrazolium staining solution (50 mM potassium phosphate, pH 7.8, 10 mM sodium azide, and 0.5% [w/v] nitroblue tetrazolium). Submerged tissue was vacuum infiltrated in a desiccator (twice for 10 min), breaking the vacuum between each event. Diaminobenzidine -infiltrated tissue was subsequently incubated in the dark at room temperature for 24 h. Nitroblue tetrazolium–infiltrated tissue was incubated under a benchtop lamp for 15 min on each side of the leaf, before terminating the reaction in warm 95% ethanol. Tissue from both treatments was destained in 95% ethanol at 45°C, changing the ethanol solution several times as needed. Destained tissue was stored in 60% glycerol. Images were collected using a Nikon d90 digital SLR equipped with a Tamron 90 mm f/2.8 macro lens. All images were white balanced and level-adjusted using Photoshop software (Adobe; v.11.0.2).
Ascorbate and Glutathione Measurements
Ascorbate and glutathione measurements were made spectrophotometrically as adapted from Luwe et al. (1993), essentially as described (Giacomelli et al., 2007).
TEM
Leaf tissue from three individuals of each genotype at each time point was harvested 1 h after the beginning of the photoperiod. Leaf margins and midribs were excluded, and the remaining leaf tissue was divided into 1 × 2-mm sections with a razor blade. Sections were processed as described (Lundquist et al., 2012a) using 2% glutaraldehyde, 2% paraformaldehyde, and subsequently 1% osmium tetroxide as fixative and 2% uranyl acetate as stain. Cured resin blocks were sectioned and imaged at Electron Microscopy Services, Colorado Springs, CO. Measurements of PG dimensions were made from cross sections using the ImageJ software program (National Institutes of Health).
PG Total Lipid Extraction and TLC
Lipids were extracted from PG preparations by chloroform/methanol extraction, essentially as described (Li-Beisson et al., 2010). Briefly, PG samples were normalized by OD600, lyophilized, and resuspended in 3 mL isopropanol. Samples were heated at 75°C for 15 min followed by addition of 1.5 mL chloroform and 0.6 mL water with 0.01% butylated hydroxytoluene. Samples were incubated at room temperature with shaking for 1 h. The organic phase was transferred to a new glass vial, and the aqueous phase was reextracted by adding 4 mL chloroform:methanol (2:1; v/v). Organic phases were pooled and washed with 1 M potassium chloride, separated by centrifugation for 2 min at 1700g, and the aqueous phase removed. The organic phase was washed by adding 2 mL water, centrifuging, and extracting the aqueous phase. The remaining organic phase was dried down under nitrogen gas and subjected immediately to TLC. For TLC, dried extracts were dissolved in 1 to 10 μL chloroform containing 0.01% butylated hydroxy-toluene and spotted on the concentrating zone of Silica Gel 60 analytical TLC plates (EMD Chemicals). Plates were developed in a mobile phase consisting of n-hexane:diethyl ether:acetic acid in a ratio of 90:10:1. Silica plates were stained with iodine vapor.
Total Leaf Protein Extraction
Protein was extracted from mature leaf tissue 3 h into the photoperiod. While working in the dark at 4°C, the mature leaf tissue was ground in liquid nitrogen and combined with 450 μL of protein extraction buffer (50 mM Tris, pH 8.0, 2% SDS, and protease inhibitor cocktail). Samples were vortexed for 25 s and then filtered through a 0.8-mL frit by centrifuging at 18,000g for 1 min. Protein concentrations of the supernatant were determined by the BCA assay kit (Pierce).
PG Sample Preparation and In-Gel Digestion
PG samples were normalized by OD600 and were lyophilized and solubilized in a modified Laemmli solubilization buffer (125 mM Tris-HCl, pH 6.8, 6% SDS, 10% β-mercaptoethanol, and 20% glycerol). Samples were shaken gently at 30°C for 15 min to ensure complete solubilization and subsequently heated at 80°C for 10 min. Samples were centrifuged to remove insoluble material, and proteins were separated by SDS-PAGE (6% acrylamide stacking and 12% separation). Each gel lane was cut in five slices, and proteins were digested with trypsin, as described by Friso et al. (2011).
Comparative Proteomics of Leaf Extracts from the Wild Type and k1 k3
Total leaf proteomes were extracted with SDS, separated by SDS-PAGE, and visualized by Coomassie Brilliant Blue staining (see Supplemental Figure 5A online). Each gel lane was cut in 12 slices for in-gel tryptic digestion, followed by peptide extraction and analysis by nano-liquid chromatography–electrospray ionization–MS/MS using an LTQ-Orbitrap mass spectrometer with a total of 216 liquid chromatography–MS/MS runs. The acquired MS data (1.67 million MS/MS spectra) were searched using MASCOT against the latest Arabidopsis thaliana gene models and filtered to minimize false-positive identification. Proteins were quantified based on the number of adjSPC. Proteins quantified mostly with adjSPC shared with homologs were grouped with these homologs for further quantitative analysis; 254 proteins were placed in 96 groups (see Supplemental Figure 5B online). For statistical analysis of quantitative protein comparison between the wild type and k1 k3 based on the spectral counting, we used as input NadjSPC and the GLEE software, which was developed in MATLAB version 7 (MathWorks) (A. Poliakov, L. Ponnala, P.D. Olinares, and K.J. van Wijk, unpublished data). GLEE is essentially an improved version of the PLGEM software (Pavelka et al., 2008) and benchmarked against QSpec (Choi et al., 2008). GLEE was run on a Windows platform with a cubic polynomial equation fitting and 1000 iterations for estimation of variation.
Proteome Analysis by Nano-Liquid Chromatography-LTQ-Orbitrap and Data Processing
Peptides prepared from in-gel digestion and in-solution digestion were analyzed by data-dependent MS/MS using an on-line LC-LTQ-Orbitrap (Thermo Electron) with dynamic exclusion, similar as described (Majeran et al., 2008). Peak lists (.mgf format) were generated using DTA supercharge (v1.19) software (http://msquant.sourceforge.net/) and searched with Mascot v2.2 (Matrix Science) against a combined database containing the Arabidopsis genome with protein-coding gene models and 187 sequences for known contaminants (e.g., keratin and trypsin) (a total of 33,013 entries) and concatenated with a decoy database where all the sequences were randomized. In total, this database contained 66,026 protein sequences. Off-line calibration for all precursors ions was done as described (Olinares et al., 2010). Each of the peak lists was searched using Mascot v2.2 (maximum P value of 0.01) for full tryptic peptides using a precursor ion tolerance set at ±6 ppm, fixed Cys carbamido-methylation and variable Met oxidation, protein N-terminal acetylation, Gln deamidation, and maximally one missed cleavage allowed. The maximum fragment ion tolerance (MS/MS) was 0.8 D. For semitryptic peptides, the search was performed with a precursor ion tolerance set at ±3 ppm, fixed Cys carbamido-methylation and variable Met oxidation, N-terminal acetylation, Gln deamidation, and maximally one missed cleavage allowed. Minimal ion score threshold was chosen such that a peptide false discovery rate below 1% was achieved. Using an in-house written filter, the search results were further filtered as follows: For identification with two or more peptides, the minimum ion score threshold was set to 30. For protein identification based on a single peptide, the minimum ion score threshold was set to 33, and the mass accuracy of the precursor ion was required to be within ±3 ppm. The peptide false discovery rate was calculated as 2 × (decoy hits)/(target + decoy hits) and below 1%. The false discovery rate of proteins identified with two or more peptides was zero. Peptides with less than seven amino acids were discarded.
Several Arabidopsis genes have more than one gene model, and in such cases, the protein form with the highest number of matched spectra was selected; if two gene models had the same number of matched spectra, the model with the lower digit was selected. For quantification, each protein accession was scored for total spectral counts, unique spectral counts (uniquely matching to an accession), and adjSPC. The latter assigns shared peptides to accessions in proportion to their relative abundance using unique spectral counts for each accession as a basis. The NadjSPC for each protein was calculated through division of adjSPC by the sum of all adjSPC values for the proteins from that gel lane. NadjSPC provides a relative protein abundance measure by mass.
Cross-Linking and Immunoprecipitation
Thylakoid preparations from the wild type and k1 k3 were prepared from 3-d light-stressed plants and diluted in Medium R (50 mM HEPES, pH 8.0, 5 mM MgCl2, and 170 µg/mL phenylmethylsulfonyl fluoride) to a chlorophyll concentration of 4 mg/mL. Aliquots were treated with formaldehyde, DST, DTSSP, and Sulfo-EGS at concentrations of 0.8% (w/v), 5 mM, 0.25 mM, and 0.75 mM, respectively. Reactions were incubated at 4°C for 2 h with gentle rotation and were subsequently quenched by applying SDS-PAGE solubilization buffer lacking reductant and incubating at room temperature for 1 h. Soluble material was dialyzed against immunoprecipitation buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.2% Nonidet P-40, and 170 µg/mL phenylmethylsulfonyl fluoride). Dialyzed samples were then incubated with anti-ABC1K1 Dynabeads (Invitrogen) for 2 h at 4°C with gentle rotation. Beads were washed with immunoprecipitation buffer, and bound protein was eluted with half-strength SDS-PAGE solubilization buffer at 70°C for 10 min. DST, DTSSP, and Sulfo-EGS cross-links were cleaved according to the manufacturer’s instructions, and the formaldehyde cross-link was cleaved by boiling for 30 min. The protein composition was analyzed by in-gel digestion and nanoHPLC-MS/MS according to the procedure described above.
Prenyl-Lipid Metabolite Extractions
Working under minimal light and at 4°C, PG preparations were normalized by OD600 and lyophilized. Prenyl-lipids were extracted from normalized, lyophilized tissue essentially as described (Fraser et al., 2000), except all solvent volumes were doubled. To lyophilized samples, 200 μL of methanol was added and was mixed for 10 min on a thermoshaker (Thermomixer R; Eppendorf) at 4°C. Two hundred microliters of 50 mM Tris-HCl, pH 7.5, + 1 M NaCl was added and shaken for another 10 min. Eight hundred microliters of chloroform was added, and the solution was incubated with gentle rocking for 15 min at 4°C. Sample was centrifuged at 3000g for 5 min to achieve thorough phase separation, and the hypophase was extracted by glass Pasteur pipette. Extraction was repeated once by adding an additional 800 μL of chloroform and again incubating for 15 min at 4°C and centrifuging. Extracted prenyl-lipids were dried down under a stream of nitrogen gas and stored at −20°C.
Prenyl-Lipid Metabolite Profiling by HPLC-Photodiode Array
Separation of the metabolite extracts by HPLC was accomplished according to the method described by Fraser et al. (2000). Chromatography was performed on a reverse-phase C30 column with 5-µm beads (250 × 4.6 mm; YMC) using an Agilent 1100 chromatography system with a flow rate of 1 mL/min that was connected to a photodiode array detector, monitoring continuously from 260 to 700 nm. Extractions were resuspended in 250 μL of ethyl acetate and passed through a 0.45-µm syringe filter. Sample injections (25 µL) were separated with mobile phases of (A) 100% methanol, (B) 80% methanol in water + 0.2% ammonium acetate, and (C) methyl t-butyl ether. A gradient elution was employed consisting of 95% A, 5% B isocratically for 12 min, a step to 80% A, 5% B, 15% C at 12 min, followed by a linear gradient to 30% A, 5% B, 65% C at 30 min, and finally a return to the initial conditions (95% A and 5% B) by 60 min. Peak identities were established by comparison of retention times with those established by Fraser et al. (2000) and the absorbance spectrum. Quantification was performed using the Chromeleon software package v6.80 (Waters) by measuring the area under the curve at each compound’s λmax. Absorbance response factors for λmax were calculated from calibration curves of a standard of each compound class: carotenoids (β-carotene), tocopherols (α-tocopherol), quinones (menadione), and chlorophyll (chlorophyll b).
Prenyl-Lipid Metabolite Identification by HPLC-MS
For MS analysis, metabolite extracts were chromatographically separated as above but connected in-line to a Waters Micromass ZMD 4000 single quadrupole mass spectrometer scanning alternately in (+) and (−) electrospray ionization modes, with a capillary voltage of 3500 V and cone voltages of 29 and 45 V for positive ion mode and negative ion mode, respectively. Postcolumn flow was split 75:25, producing a flow of 250 μL/min. Following the split, ammonium acetate (0.5% [w/v]) was injected using a syringe pump at a flow rate of 50 μL/min with a three-way adaptor.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT1G79600 (ABC1K3) and AT4G31390 (ABC1K1). SALK insertion mutants were acquired from the ABRC: abc1k1-1 (SALK 057147), abc1k1-2 (SALK 068628), and abc1k3-1 (SALK 128696).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Truncated Transcripts Accumulate in the abc1k1-1, abc1k1-2, and abc1k3 Mutants.
Supplemental Figure 2. SDS-PAGE Gel of IP with Anti-ABC1K1 Serum after Cross-Linking with Different Cross-Linkers of Thylakoids, Followed by Solubilization by SDS and Reversal of the Cross-Link.
Supplemental Figure 3. The Phenotypes of the Two ABC1K1 Alleles and the Wild Type under 5× Light Stress for 2-Week-Old Plants and 10× Light Stress for 3-Week Day-Old Plants.
Supplemental Figure 4. Treatment with methyl jasmonate Fails to Complement the 5× Light Stress Phenotype Found in k1 k3 Individuals.
Supplemental Figure 5. Responses of k1 k3 Plants to Various Environmental Stresses.
Supplemental Figure 6. Degreening in k1 k3 Continues to Progress Independent of Stress once Triggered.
Supplemental Figure 7. Comparative Proteomics of Total Leaf Samples.
Supplemental Figure 8. Light Stress Response (5× Light Stress) in the Wild Type and k1 k3 of the PG-Localized Fibrillins as Determined by Comparative Proteome Analysis of Total Leaf Proteins.
Supplemental Figure 9. Protein Abundance of Selected Proteins and Complexes in Wild-Type and k1 k3 Total Leaf Tissue.
Supplemental Figure 10. Comparison of Protein Abundance in PGs Isolated from the Wild Type and k1 k3.
Supplemental Figure 11. Thin Layer Chromatographic Separation of PGs Isolated from k1 k3 and the Wild Type after 5-d 5× Light Stress.
Supplemental Figure 12. Absorption Spectra and Electrospray Mass Spectra of Oxidized Plastoquinone-9.
Supplemental Figure 13. Absorption Spectra, Electrospray Mass Spectra, and Extracted Ion Chromatograms of Plastochromanol-8.
Supplemental Figure 14. Absorption Spectra and Electrospray Mass Spectra of an Unknown Quinone in k1 k3 PGs.
Supplemental Data Set 1. Proteins Identified by Immunoprecipitation with Anti-ABC1K1 Antiserum after Cross-Linking of Thylakoids from the Wild Type and k1 k3.
Supplemental Data Set 2. Proteins Identified by Immunoprecipitation with Anti-ABC1K3 Antiserum after Cross-Linking of Thylakoids from the Wild Type and k1 k3 with Formaldehyde.
Supplemental Data Set 3. Correlation Analysis between Replicates after Removal of Zero Pairs for the Comparative Proteome Analysis of Total Leaf Proteomes from the Wild Type and k1 k3 before Light Stress and after 3 and 5 d of Transfer to 5× Higher Light
Supplemental Data Set 4. Comparative Proteome Analysis of Total Leaf Proteomes from the Wild Type and k1 k3 before Light Stress and after 3 and 5 d of Transfer to 5× Higher Light Intensity.
Supplemental Data Set 5. Comparative Proteome Analysis of Isolated PGs from the Wild Type and k1 k3 after 5 d of Transfer to 5× Higher Light Intensity.
Supplemental Data Set 6. Metabolite Analysis of Total Leaves and Chloroplast Subfractions in the Wild Type and k1 k3.
Supplementary Material
Acknowledgments
We thank Richard Medville of Electron Microscopy Sciences (Colorado Springs, CO) for collection of TEM micrographs, Ivan Kerestzes of the Cornell NMR facility for assistance with the gas chromatography–MS analysis, and Greg Buda for assistance with photography of the leaf tissue stains. Funding was provided in part by a National Institutes of Health Chemical-Biological Interface training grant (5T32GM008500) awarded to P.K.L. Part of this work was carried out by using the resources of the Computational Biology Service Unit of Cornell University, which is partially funded by the Microsoft Corporation. This study was in part supported by funding to K.J.V.W. from the National Science Foundation (IOS-0701736 and IOS-0922560).
AUTHOR CONTRIBUTIONS
P.K.L. and K.J.V.W. devised most of the experiments and wrote the article. P.K.L. carried out most of the experiments. G.F., A.P., and E.R. carried out the MS analysis, and A.P. did the statistical analysis using GLEE. L.G. isolated the abc1k mutants, generated the k1 k3 double mutant, and carried out the initial light stress phenotyping. R.P.M. supported P.K.L. in the HPLC analysis of prenyl-lipids, and S.B.K. helped P.K.L. with liquid chromatography-MS–based metabolite analysis. M.A. carried out the TLC analysis of PG metabolites. L.P. carried out various types of correlation and statistical analysis of proteomics data and developed GLEE with A.P. Q.S. was in charge of the Plant Proteome Database and provided bioinformatics support for proteomics analysis. K.J.V.W. provided general oversight.
Glossary
- PG
plastoglobule
- TAG
triacylglycerol
- AOS
allene oxide synthase
- ROS
reactive oxygen species
- RNAi
RNA interference
- MS
mass spectrometry
- DST
disuccinimdyl tartrate
- DTSSP
3,3′-dithiobis(sulfosuccinimidyl propionate)
- MS/MS
tandem mass spectrometry
- adjSPC
adjusted for shared peptides between proteins
- PSII
photosystem II
- H2O2
hydrogen peroxide
- O2−
superoxide
- 1O2
singlet oxygen
- JA
jasmonate
- PUFA
polyunsaturated fatty acid
- AOC
allene oxide cyclase
- OPDA
oxophytodienoic acid
- PPH
pheophytin pheophorbide hydrolase (pheophytinase)
- TEM
transmission electron microscopy
- FAPE
fatty acid phytyl ester
- DMPBQ
dimethylphytyl benzoquinone
- TLC
thin layer chromatography
- NadjSPC
normalized adjSPC
- Sulfo-EGS
ethyleneglycolbis(sulfosuccinimidyl succinate)
References
- Austin J.R., II, Frost E., Vidi P.A., Kessler F., Staehelin L.A. (2006). Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18: 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah A., Simková K., Apel K., Laloi C. (2009). Arabidopsis mutants reveal multiple singlet oxygen signaling pathways involved in stress response and development. Plant Mol. Biol. 70: 547–563 [DOI] [PubMed] [Google Scholar]
- Besagni C., Kessler F. (2013). A mechanism implicating plastoglobules in thylakoid disassembly during senescence and nitrogen starvation. Planta 237: 463–470 [DOI] [PubMed] [Google Scholar]
- Bousquet I., Dujardin G., Slonimski P.P. (1991). ABC1, a novel yeast nuclear gene has a dual function in mitochondria: It suppresses a cytochrome b mRNA translation defect and is essential for the electron transfer in the bc 1 complex. EMBO J. 10: 2023–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréhélin C., Kessler F., van Wijk K.J. (2007). Plastoglobules: Versatile lipoprotein particles in plastids. Trends Plant Sci. 12: 260–266 [DOI] [PubMed] [Google Scholar]
- Cardazzo B., Hamel P., Sakamoto W., Wintz H., Dujardin G. (1998). Isolation of an Arabidopsis thaliana cDNA by complementation of a yeast abc1 deletion mutant deficient in complex III respiratory activity. Gene 221: 117–125 [DOI] [PubMed] [Google Scholar]
- Choi H., Fermin D., Nesvizhskii A.I. (2008). Significance analysis of spectral count data in label-free shotgun proteomics. Mol. Cell. Proteomics 7: 2373–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L., Fiore A., Cazzaniga S., Giuliano G., Bassi R. (2007). Different roles of alpha- and beta-branch xanthophylls in photosystem assembly and photoprotection. J. Biol. Chem. 282: 35056–35068 [DOI] [PubMed] [Google Scholar]
- Eugeni Piller L., Abraham M., Dörmann P., Kessler F., Besagni C. (2012). Plastid lipid droplets at the crossroads of prenylquinone metabolism. J. Exp. Bot. 63: 1609–1618 [DOI] [PubMed] [Google Scholar]
- Eugeni Piller L., Besagni C., Ksas B., Rumeau D., Bréhélin C., Glauser G., Kessler F., Havaux M. (2011). Chloroplast lipid droplet type II NAD(P)H quinone oxidoreductase is essential for prenylquinone metabolism and vitamin K1 accumulation. Proc. Natl. Acad. Sci. USA 108: 14354–14359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chico J.M., Solano R. (2009). The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2011). Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P.D., Pinto M.E.S., Holloway D.E., Bramley P.M. (2000). Technical advance: Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J. 24: 551–558 [DOI] [PubMed] [Google Scholar]
- Friso, G., Olinares, P.D.B., and van Wijk, K.J. (2011). The workflow for quantitative proteome analysis of chloroplast development and differentiation, chloroplast mutants, and protein interactions by spectral counting. In Chloroplast Research in Arabidopsis, R.P. Jarvis, ed (New York: Humana Press), pp. 265–282. [DOI] [PubMed] [Google Scholar]
- Gaude N., Bréhélin C., Tischendorf G., Kessler F., Dörmann P. (2007). Nitrogen deficiency in Arabidopsis affects galactolipid composition and gene expression and results in accumulation of fatty acid phytyl esters. Plant J. 49: 729–739 [DOI] [PubMed] [Google Scholar]
- Gfeller A., Dubugnon L., Liechti R., Farmer E.E. (2010b). Jasmonate biochemical pathway. Sci. Signal. 3: cm3. [DOI] [PubMed] [Google Scholar]
- Gfeller A., Liechti R., Farmer E.E. (2010a). Arabidopsis jasmonate signaling pathway. Sci. Signal. 3: cm4. [DOI] [PubMed] [Google Scholar]
- Giacomelli L., Masi A., Ripoll D.R., Lee M.J., van Wijk K.J. (2007). Arabidopsis thaliana deficient in two chloroplast ascorbate peroxidases shows accelerated light-induced necrosis when levels of cellular ascorbate are low. Plant Mol. Biol. 65: 627–644 [DOI] [PubMed] [Google Scholar]
- Giacomelli L., Rudella A., van Wijk K.J. (2006). High light response of the thylakoid proteome in Arabidopsis wild type and the ascorbate-deficient mutant vtc2-2. A comparative proteomics study. Plant Physiol. 141: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler R.E., Geimer S., Kunz H.H., Schmitz J., Dörmann P., Bell K., Hetfeld S., Guballa A., Flügge U.-I. (2009). Chlororespiration and grana hyperstacking: How an Arabidopsis double mutant can survive despite defects in starch biosynthesis and daily carbon export from chloroplasts. Plant Physiol. 149: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M., Bonfils J.P., Lütz C., Niyogi K.K. (2000). Photodamage of the photosynthetic apparatus and its dependence on the leaf developmental stage in the npq1 Arabidopsis mutant deficient in the xanthophyll cycle enzyme violaxanthin de-epoxidase. Plant Physiol. 124: 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S., Kräutler B. (2011). Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta 1807: 977–988 [DOI] [PubMed] [Google Scholar]
- Huang C.-Y., Chung C.-I., Lin Y.-C., Hsing Y.-I.C., Huang A.H.C. (2009). Oil bodies and oleosins in Physcomitrella possess characteristics representative of early trends in evolution. Plant Physiol. 150: 1192–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Friso G., Nishimura K., Qu X., Olinares P.D., Majeran W., Sun Q., van Wijk K.J. (2013). Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships; the concept of orthoproteomics. J. Proteome. Res. 12: 491–504 [DOI] [PubMed] [Google Scholar]
- Hyun Y., et al. (2008). Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Dev. Cell 14: 183–192 [DOI] [PubMed] [Google Scholar]
- Jasinski M., Sudre D., Schansker G., Schellenberg M., Constant S., Martinoia E., Bovet L. (2008). AtOSA1, a member of the Abc1-like family, as a new factor in cadmium and oxidative stress response. Plant Physiol. 147: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangasjärvi S., Lepistö A., Hännikäinen K., Piippo M., Luomala E.M., Aro E.M., Rintamäki E. (2008). Diverse roles for chloroplast stromal and thylakoid-bound ascorbate peroxidases in plant stress responses. Biochem. J. 412: 275–285 [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C., et al. (2008). ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am. J. Hum. Genet. 82: 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.-S., Nioche P., Hamberg M., Raman C.S. (2008). Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes. Nature 455: 363–368 [DOI] [PubMed] [Google Scholar]
- Lee K.P., Kim C., Landgraf F., Apel K. (2007). EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y., et al. (2010). Acyl-lipid metabolism. The Arabidopsis Book 8: e0133, /10.1199/tab.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippold F., vom Dorp K., Abraham M., Hölzl G., Wewer V., Yilmaz J.L., Lager I., Montandon C., Besagni C., Kessler F., Stymne S., Dörmann P. (2012). Fatty acid phytyl ester synthesis in chloroplasts of Arabidopsis. Plant Cell 24: 2001–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston A.K., Cruz J.A., Kohzuma K., Dhingra A., Kramer D.M. (2010). An Arabidopsis mutant with high cyclic electron flow around photosystem I (hcef) involving the NADPH dehydrogenase complex. Plant Cell 22: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist P.K., Poliakov A., Bhuiyan N.H., Zybailov B., Sun Q., van Wijk K.J. (2012a). The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol. 158: 1172–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist P.K., Davis J.I., van Wijk K.J. (2012b). ABC1K atypical kinases in plants: Filling the organellar kinase void. Trends Plant Sci. 17: 546–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luwe M., Takahama U., Heber U. (1993). Role of ascorbate in detoxifying ozone in the apoplast of spinach (Spinacia oleracea L.) leaves. Plant Physiol. 101: 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macinga D.R., Cook G.M., Poole R.K., Rather P.N. (1998). Identification and characterization of aarF, a locus required for production of ubiquinone in Providencia stuartii and Escherichia coli and for expression of 2′-N-acetyltransferase in P. stuartii. J. Bacteriol. 180: 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Zybailov B., Ytterberg A.J., Dunsmore J., Sun Q., van Wijk K.J. (2008). Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Mol. Cell. Proteomics 7: 1609–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinis J., Kessler F., Glauser G. (2011). A novel method for prenylquinone profiling in plant tissues by ultra-high pressure liquid chromatography-mass spectrometry. Plant Methods 7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene R., Nater M., Goslings D., Kessler F., op den Camp R., Apel K. (2001). FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Tanaka R., Grimm B., Masuda T., Moulin M., Smith A.G., Tanaka A., Terry M.J. (2010). The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 15: 488–498 [DOI] [PubMed] [Google Scholar]
- Mollet J., Delahodde A., Serre V., Chretien D., Schlemmer D., Lombes A., Boddaert N., Desguerre I., de Lonlay P., de Baulny H.O., Munnich A., Rötig A. (2008). CABC1 gene mutations cause ubiquinone deficiency with cerebellar ataxia and seizures. Am. J. Hum. Genet. 82: 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Sugiyama N., Mochida K., Daudi A., Yoshida Y., Toyoda T., Tomita M., Ishihama Y., Shirasu K. (2010). Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153: 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares P.D., Ponnala L., van Wijk K.J. (2010). Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol. Cell. Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- op den Camp, R.G., Przybyla, D., Ochsenbein, C., Laloi, C., Kim, C., Danon, A., Wagner, D., Hideg, E., Gobel, C., Feussner, I., Nater, M., and Apel, K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis Plant Cell 15: 2320–2332. [DOI] [PMC free article] [PubMed]
- Page M., Sultana N., Paszkiewicz K., Florance H., Smirnoff N. (2012). The influence of ascorbate on anthocyanin accumulation during high light acclimation in Arabidopsis thaliana: Further evidence for redox control of anthocyanin synthesis. Plant Cell Environ. 35: 388–404 [DOI] [PubMed] [Google Scholar]
- Pavelka N., Fournier M.L., Swanson S.K., Pelizzola M., Ricciardi-Castagnoli P., Florens L., Washburn M.P. (2008). Statistical similarities between transcriptomics and quantitative shotgun proteomics data. Mol. Cell. Proteomics 7: 631–644 [DOI] [PubMed] [Google Scholar]
- Poon W.W., Davis D.E., Ha H.T., Jonassen T., Rather P.N., Clarke C.F. (2000). Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J. Bacteriol. 182: 5139–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porfirova S., Bergmuller E., Tropf S., Lemke R., Dormann P. (2002). Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc. Natl. Acad. Sci. USA 99: 12495–12500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A., Tanner G., Aubry S., Anders I., Moser S., Müller T., Ongania K.H., Kräutler B., Youn J.Y., Liljegren S.J., Hörtensteiner S. (2005). Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol. 139: 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F., Birtic S., Ginies C., Soubigou-Taconnat L., Triantaphylidès C., Havaux M. (2012). Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfearn E.R., Friend J. (1962). Studies on plastoquinone\x{2014}1. Determination of the concentration and oxidation-reduction state of plastoquinone in isolated chloroplasts. Phytochemistry 1: 147–151 [Google Scholar]
- Reiland S., Finazzi G., Endler A., Willig A., Baerenfaller K., Grossmann J., Gerrits B., Rutishauser D., Gruissem W., Rochaix J.D., Baginsky S. (2011). Comparative phosphoproteome profiling reveals a function of the STN8 kinase in fine-tuning of cyclic electron flow (CEF). Proc. Natl. Acad. Sci. USA 108: 12955–12960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey P., Gillet B., Römer S., Eymery F., Massimino J., Peltier G., Kuntz M. (2000). Over-expression of a pepper plastid lipid-associated protein in tobacco leads to changes in plastid ultrastructure and plant development upon stress. Plant J. 21: 483–494 [DOI] [PubMed] [Google Scholar]
- Rokka A., Aro E.M., Vener A.V. (2011). Thylakoid phosphoproteins: Identification of phosphorylation sites. Methods Mol. Biol. 684: 171–186 [DOI] [PubMed] [Google Scholar]
- Schelbert S., Aubry S., Burla B., Agne B., Kessler F., Krupinska K., Hörtensteiner S. (2009). Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltmann M.A., Stingl N.E., Lautenschlaeger J.K., Krischke M., Mueller M.J., Berger S. (2010). Differential impact of lipoxygenase 2 and jasmonates on natural and stress-induced senescence in Arabidopsis. Plant Physiol. 152: 1940–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y.S., Kim E.Y., Kim J.H., Kim W.T. (2009). Enzymatic characterization of class I DAD1-like acylhydrolase members targeted to chloroplast in Arabidopsis. FEBS Lett. 583: 2301–2307 [DOI] [PubMed] [Google Scholar]
- Vainonen J.P., Sakuragi Y., Stael S., Tikkanen M., Allahverdiyeva Y., Paakkarinen V., Aro E., Suorsa M., Scheller H.V., Vener A.V., Aro E.-M. (2008). Light regulation of CaS, a novel phosphoprotein in the thylakoid membrane of Arabidopsis thaliana. FEBS J. 275: 1767–1777 [DOI] [PubMed] [Google Scholar]
- Valentin H.E., Lincoln K., Moshiri F., Jensen P.K., Qi Q., Venkatesh T.V., Karunanandaa B., Baszis S.R., Norris S.R., Savidge B., Gruys K.J., Last R.L. (2006). The Arabidopsis vitamin E pathway gene5-1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. Plant Cell 18: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidi P.A., Kanwischer M., Baginsky S., Austin J.R., Csucs G., Dörmann P., Kessler F., Bréhélin C. (2006). Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J. Biol. Chem. 281: 11225–11234 [DOI] [PubMed] [Google Scholar]
- Wasternack C., Kombrink E. (2010). Jasmonates: Structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem. Biol. 5: 63–77 [DOI] [PubMed] [Google Scholar]
- White D.A., Fisk I.D., Gray D.A. (2006). Characterisation of oat (Avena sativa L.) oil bodies and intrinsically associated E-vitamers. J. Cereal Sci. 43: 244–249 [Google Scholar]
- Willig A., Shapiguzov A., Goldschmidt-Clermont M., Rochaix J.-D. (2011). The phosphorylation status of the chloroplast protein kinase STN7 of Arabidopsis affects its turnover. Plant Physiol. 157: 2102–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J.D., Chory J. (2012). Organelle signaling: How stressed chloroplasts communicate with the nucleus. Curr. Biol. 22: R690–R692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L.X., Hsieh E.J., Watanabe S., Allan C.M., Chen J.Y., Tran U.C., Clarke C.F. (2011). Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim. Biophys. Acta 1811: 348–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L.X., Ozeir M., Tang J.Y., Chen J.Y., Jaquinod S.K., Fontecave M., Clarke C.F., Pierrel F. (2012). Overexpression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. J. Biol. Chem. 287: 23571–23581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., Zeng, X., Li, T., Liu, M., Zhang, S., Gao, S., Wang, Y., Peng, C., Li, L., and Yang, C. (2012). AtACDO1, an ABC1-like kinase gene, is involved in chlorophyll degradation and the response to photooxidative stress in Arabidopsis J. Exp. Bot. 63: 3959–3973 [DOI] [PubMed]
- Youssef A., Laizet Y., Block M.A., Maréchal E., Alcaraz J.-P., Larson T.R., Pontier D., Gaffé J., Kuntz M. (2010). Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress. Plant J. 61: 436–445 [DOI] [PubMed] [Google Scholar]
- Ytterberg A.J., Peltier J.B., van Wijk K.J. (2006). Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 140: 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbierzak A.M., Kanwischer M., Wille C., Vidi P.A., Giavalisco P., Lohmann A., Briesen I., Porfirova S., Bréhélin C., Kessler F., Dörmann P. (2009). Intersection of the tocopherol and plastoquinol metabolic pathways at the plastoglobule. Biochem. J. 425: 389–399 [DOI] [PubMed] [Google Scholar]
- Zhang, H., and Zhou, C. (October 25, 2012). Signal transduction in leaf senescence. Plant Mol. Biol., doi/10.1007/s11103-012-9980-4 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.