Abstract
Herpes simplex virus type 1 (HSV-1) immediate-early (IE) regulatory protein ICP0 is required for efficient progression of infected cells into productive lytic infection, especially in low-multiplicity infections of limited-passage human fibroblasts. We have used single-cell-based assays that allow detailed analysis of the ICP0-null phenotype in low-multiplicity infections of restrictive cell types. The major conclusions are as follows: (i) there is a threshold input multiplicity above which the mutant virus replicates normally; (ii) individual cells infected below the threshold multiplicity have a high probability of establishing a nonproductive infection; (iii) such nonproductively infected cells have a high probability of expressing IE products at 6 h postinfection; (iv) even at 24 h postinfection, IE protein-positive nonproductively infected human fibroblast cells exceed the number of cells that lead to plaque formation by up to 2 orders of magnitude; (v) expression of individual IE proteins in a proportion of the nonproductively infected cells is incompletely coordinated; (vi) the nonproductive cells can also express early gene products at low frequencies and in a stochastic manner; and (vii) significant numbers of human fibroblast cells infected at low multiplicity by an ICP0-deficient virus are lost through cell death. We propose that in the absence of ICP0 expression, HSV-1 infected human fibroblasts can undergo a great variety of fates, including quiescence, stalled infection at a variety of different stages, cell death, and, for a minor population, initiation of formation of a plaque.
Herpes simplex virus type 1 (HSV-1) immediate-early (IE) regulatory protein ICP0 stimulates the expression of all temporal classes of HSV-1 genes and many heterologous genes in transfection reporter assays (reviewed in references 14 and 28). The abilities of ICP0 to stimulate initiation of lytic infection and induce reactivation of quiescent viral genomes have led to the suggestion that it plays a key role in regulating the balance between lytic and latent HSV-1 infection. The function of the protein itself has been extensively studied, and an increasingly detailed picture of its interactions with cellular proteins and its biochemical functions is emerging (3, 4, 15, 16, 18, 20, 23, 25-27, 36, 37, 50). One of the key functions of ICP0 is the ubiquitin E3 ligase activity conferred by the RING finger domain in the N-terminal third of the protein (4). This activity leads to the proteasome-dependent degradation of several cellular proteins, resulting in the disruption of centromeres and discrete nuclear structures known as promyelocytic leukemia protein (PML) nuclear bodies or ND10 and probably in the modulation of other, as-yet-incompletely explored pathways (for example, the inhibition of an induction of interferon-responsive gene expression triggered by the incoming virus particle) (11, 35, 39).
Soon after ICP0 was identified as an activator of gene expression, ICP0-null mutant viruses were constructed (42, 48). It was found that ICP0 was not essential for virus replication in cultured cells but that the mutant viruses grew poorly if the multiplicity of infection (MOI) was low. This phenotype is particularly marked in limited-passage human fibroblast cells, while in BHK and Vero cells the phenotype is less severe, and in U2OS cells ICP0-deficient HSV-1 mutants grow as well as the wild-type virus (51). A thorough and elegant study has described the properties of ICP0-deficient viruses in Vero cells in terms of their plaque-forming efficiency and their ability to express the major viral transcriptional transactivator protein ICP4 in individual cells (7). An intriguing finding was that the number of Vero cells that were infected by the ICP0 mutant viruses, as judged by expression of ICP4, far exceeded the number of PFU. In contrast, in human fibroblasts, viruses that are defective in ICP0 and other important viral regulatory proteins become repressed and can be maintained in a quiescent state in which little or no expression from the viral genome can be detected (38, 40, 43). Whether simple ICP0-null mutant viruses behave in an identical fashion in human fibroblasts has not been formally investigated.
We have examined the fates of individual human fibroblast cells infected at low MOIs with wild-type HSV-1 strain 17 and ICP0-null mutant dl1403. We found that even in this most restrictive cell type, the number of cells that express viral proteins following low-MOI infection by dl1403 greatly exceeds the number of cells that initiate the formation of a plaque. Viral proteins could be detected in many of these nonproductively infected cells for extended periods and, in the case of ICP4, in amounts similar to those in productively infected cells. The nonproductively infected cells could be assigned to four classes: quiescent, with no detectable viral protein expression; stalled and expressing an apparently incomplete set of IE proteins; stalled at the IE stage; or stalled at a stage at which expression of some early proteins has occurred but DNA replication has not been initiated. Our data emphasize the complications concerning the analysis of the properties of ICP0 mutant viruses and suggest that single-cell assays at low MOIs provide a more realistic description of the ICP0-defective phenotype than methods that utilize whole-population approaches.
MATERIALS AND METHODS
Cells and viruses.
BHK cells were propagated in Glasgow modified Eagle's medium containing 100 U of penicillin per ml and 100 μg of streptomycin per ml and supplemented with 10% newborn calf serum and 10% tryptose phosphate broth. Vero and human osteosarcoma U2OS cells were grown in Glasgow modified Eagle's medium or Dulbecco's modified Eagle's medium, respectively, supplemented with 10% fetal calf serum and antibiotics as described above. Human fetal foreskin fibroblast cells (HFFF-2; European Collection of Cell Cultures) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 1% glutamine, and antibiotics as described above.
HSV-1 strain 17 syn+ was the wild-type strain used in these studies. The mutant viruses were derived from strain 17 and were as follows: ICP0-null mutant dl1403 (48); virus FXE, which expresses ICP0 with its RING finger domain deleted (13); vEG-dl110, which expresses enhanced green fluorescent protein (EGFP) from the IE-1 transcription unit in place of ICP0 (29); and virus in1411, which has an insertion mutation that inactivates expression of ICP4 (41). Viral stocks were prepared from BHK cells infected at a low MOI or from the complementing BHK M49 cell line in the case of in1411. When the cells exhibited extensive cytopathic effect, the medium was harvested, clarified by low-speed centrifugation, and stored at 4°C. Examination of viral stocks prepared by this procedure by electron microscopy showed a high proportion of complete particles with filled capsids and very small amounts of cellular debris. The PFU titers of these stocks were stable for several weeks. Particle counts of the virus stocks were determined by mixing virus samples with a preparation of 250-nm-diameter latex beads (Agar Scientific) of known concentration in a solution of negative stain, adsorbing the mixture onto carbon-coated electron microscope grids, and then counting the relative numbers of virus particles and beads. Virus titers were determined in a variety of cell types as described in Results.
Construction and preliminary analysis of virus dl0C4.
Virus dl0C4, a derivative of ICP0-null mutant virus dl1403 that expresses ICP4 linked to enhanced cyan fluorescent protein (ECFP) from otherwise normal diploid IE3 genes, was constructed as follows. Plasmid pIE3-ECFP-ICP4 (19) was transfected into BHK cells with infectious dl1403 DNA. Plaque isolates expressing the autofluorescent ICP4 protein were purified until homogenous viral populations giving 100% fluorescent plaques and no trace of the nonfusion protein on Western blots had been isolated. One such isolate was selected for the preparation of a working stock of dl0C4. Analysis of the expression of ECFP-linked ICP4 and typical early proteins during productive infection by dl0C4 was performed by Western blotting in a manner analogous to the characterization of virus vECFP-ICP4 (19). Except for the production of the ECFP-ICP4 fusion protein, no differences could be detected between the growth and gene expression characteristics of dl0C4 and dl1403 (data not shown).
Antibodies.
The following antibodies were used: anti-ICP4 monoclonal antibody (MAb) 58S (46); anti-ICP4 MAb 10176 and rabbit polyclonal serum r74 (12); anti-ICP0 MAb 11060 (17); anti-ICP8 rabbit polyclonal serum r515 (30); MAb 7381, which also recognizes ICP8 and was a kind gift from Howard Marsden; anti-ICP6 rabbit polyclonal serum r76 (10); anti-UL42 MAb Z1F11 (45); anti-ICP27 MAb H1113 (1), which was purchased from the Goodwin Institute for Cancer Research; and anti-VP5 MAb DM165 (31), which was a kind gift from Frazer Rixon. MAb 3E6 recognizes native forms of EGFP, ECFP, and enhanced yellow fluorescent protein (Qbiogene). Horseradish peroxidase-conjugated sheep anti-mouse and goat anti-rabbit secondary antibodies were purchased from Sigma and were used at dilutions of 1/1,000 and 1/50,000, respectively. Secondary antibodies used for immunofluorescence were fluorescein isothiocyanate-conjugated sheep anti-mouse immunoglobulin G (IgG) (Sigma), Alexa 488-conjugated goat anti-rabbit and goat anti-mouse IgGs (Molecular Probes), and Cy3-conjugated goat anti-mouse and Cy3-conjugated goat anti-rabbit IgGs (Amersham).
Western blots.
Whole-cell extracts of transfected cells were prepared by washing cell monolayers with phosphate-buffered saline (PBS) and then lysing the cells in an appropriate volume of sodium dodecyl sulfate-gel boiling mix. After boiling, the proteins were separated by electrophoresis on sodium dodecyl sulfate-7.5% polyacrylamide gels and electrophoretically transferred to nitrocellulose filters. The filters were blocked overnight in PBS containing 0.1% Tween 20 and 5% dry milk and then incubated with primary antibodies for 2 h in the same buffer. After extensive washing, the filters were incubated with horseradish peroxidase-conjugated secondary antibody in PBS-0.1% Tween 20-2% dry milk for 1 h and then washed again extensively before detection of bound antibody by enhanced chemiluminescence (NEN) and exposure to film. ICP0 and ICP4 were detected by using MAbs 11060 and 10176, respectively. The use of an antibody mixture to detect proteins ICP4, UL39, ICP8, and UL42 was as described previously (22).
Confocal microscopy.
Cells on glass coverslips were washed with PBS, fixed with formaldehyde (5% [vol/vol] in PBS containing 2% sucrose), and then permeabilized with 0.5% NP-40 in PBS with 10% sucrose. The coverslips were incubated with for 1 h at room temperature with primary antibodies diluted in PBS containing 1% newborn calf serum and then washed several times before treatment with secondary antibodies in the same manner. The coverslips were then mounted with Citifluor AF1 and examined with a Zeiss LSM 510 confocal microscope and the 488- and 543-nm excitation laser lines. The data from the channels were collected sequentially by using the appropriate band pass filters built into the instrument. Data were collected with fourfold averaging at a resolution of 1,024 by 1,024 pixels, using optical slices of between 0.5 and 1 μm. The microscope was a Zeiss Axioplan with a ×63 oil immersion objective lens (numerical aperture, 1.4). Data sets were processed by using the LSM 510 software and then exported for processing with Adobe Photoshop. Protein expression in individual cells was compared semiquantitatively by using the Palette option in the LSM 510 tools to ensure that the detectors were not saturated during image acquisition on control samples. Images of other samples were captured by using exactly the same acquisition variables as for the relevant control. Average pixel intensities across an infected cell nucleus were compared with those of control infected and uninfected cells by using the Profile option in the LSM 510 tools.
FACS analysis.
Cells in 35-mm-diameter dishes were infected with dl1403 or HSV-1 strain 17+ at the appropriate MOI and then harvested for fluorescence-activated cell sorter (FACS) analysis at various times thereafter. The cells were washed and suspended in cell dissociation buffer (Sigma), and then total cell numbers in the sample were determined in a cell counting chamber. The cells were fixed with 5% (vol/vol) formaldehyde in PBS and then permeabilized with 0.1% saponin in PBS containing 2% calf serum. The cells were washed and resuspended in 100 μl of PBS-2% calf serum containing antibodies to detect the following proteins: ICP4, MAb 58S hybridoma supernatant at 1/100; ICP0, MAb 11060 ascites at 1/1,000; and ECFP or EGFP, MAb 3E6 at 1/1,000. After incubation at room temperature for 1 h, the cells were washed twice and then incubated with fluorescein isothiocyanate-conjugated sheep anti-mouse IgG. After a further incubation for 1 h, then cells were washed twice and then resuspended in 0.5 ml of PBS-2% calf serum. The samples were analyzed on a Becton Dickinson FACSCalibur machine in comparison with mock-infected control samples prepared in parallel. Positive cells were gated by comparison with the uninfected controls stained with the appropriate antibody. The PFU titers of the virus stocks used for FACS analysis were determined with the appropriate cell types at the same time in each experiment.
Apoptosis assays.
HFFF-2 cells were either mock infected or infected with dl1403 or HSV-1 strain 17 at an MOI of 1 U2OS PFU per cell, and then 12 h later the detached cells were harvested by centrifugation and mixed with the adherent cells that had been released and suspended in cell dissociation buffer. The cells were pelleted by centrifugation and washed twice with 1 ml of PBS. The cells were stained for apoptotic markers by using the annexin V-phycoerythrin apoptosis detection kit (BD PharMingen) according to the manufacturer's instructions. The cells were subsequently analyzed by flow cytometry (FACSCalibur; Becton Dickinson), using dual-channel detection of apoptotic (annexin V positive), necrotic (7-amino actinomycin D positive), and apoptotic and necrotic cell populations with the reagents provided in the kit. The scatter plots were gated according to the mock-infected control to determine the percentages of cells in these populations.
RESULTS
Rationale of the study.
While the understanding of the biochemical and other activities of ICP0 has increased rapidly over the past few years, the consequences of these activities for the biological properties of HSV-1 have remained less clear. An underlying problem is that the defect of ICP0-deficient viruses is exhibited only at low MOIs in certain cell types, while many assays of virus behavior of necessity utilize high-MOI conditions to establish an infection. We set out to study the properties of viruses that are deficient only in ICP0 under conditions in which the phenotype is exhibited: low-multiplicity infection of restrictive cell types. It was first necessary to establish as accurately as possible the basic characteristics of infection of permissive and restrictive cell types by an ICP0-deficient virus and, in particular, to distinguish quantitatively between “low” and “high” MOIs. Therefore, the first sections below concern a partial reiteration of previously published material in order to establish a robust baseline from which the later sections in Results can expand.
Comparison of particle numbers, infected-cell-forming units, and PFU of wild-type and ICP0-null mutant HSV-1.
The number of virus particles in typical stocks of HSV-1 strain 17 and ICP0-null mutant dl1403 were determined by electron microscopy, and then their PFU and ICP4-positive cell fluorescence-forming units (FFU) titers were determined in Vero, U2OS, and HFFF-2 cells (Table 1). The results were consistent with those of previous studies (7, 47, 51): stocks of strain 17 and dl1403 had similar PFU titers in U2OS cells, but dl1403 formed plaques about 20- and 1,000-fold less efficiently on Vero and HFFF-2 cells, respectively, than on U2OS cells. Note that because strain 17 also formed plaques less efficiently on HFFF-2 cells than on U2OS cells, the defect of dl1403 in HFFF-2 cells compared to strain 17 is less than the apparent 1,000-fold deficiency (Table 1). The PFU titers of strain 17 varied by up to fourfold between the different cell types, with HFFF-2 cells giving the lowest titers and therefore the highest particle-to-PFU ratio (Table 2). This difference might be explained by cell type variations in the efficiency of virus adsorption, penetration, trafficking within the cytoplasm, or even initiation of IE gene expression. As in previous studies, the PFU titer of dl1403 in HFFF-2 cells was nonlinear, with the lowest dilution giving fewer plaques than expected from the previous plate in the 10-fold dilution series; the given titers were calculated from the plate giving the most plaques.
TABLE 1.
Particle, PFU, and ICP4 FFU titers of typical stocks of HSV-1 strain 17 and dl1403 on different cell typesa
| Virus | Particles | Vero cells
|
U2OS cells
|
HFFF cells
|
|||
|---|---|---|---|---|---|---|---|
| FFU | PFU | FFU | PFU | FFU | HFFF PFU | ||
| Strain 17 | 1.0 × 1010 | 4.8 × 108 | 2.0 × 108 | 5.4 × 108 | 9.1 × 107 | 4.8 × 107 | 4.8 × 107 |
| dl1403 | 2.9 × 109 | 2.6 × 108 | 6.2 × 106 | 2.3 × 108 | 9.2 × 107 | 9.3 × 106 | 1.1 × 105 |
All values are in units per milliliter. ICP4 FFU titers were determined 6 h after infection. Repeat determinations showed that the experimental variation was less than twofold for PFU and FFU determinations but up to fivefold for particle determinations.
TABLE 2.
Average particle-to-PFU, particle-to-ICP4 FFU, and ICP4 FFU-to-PFU ratios of stocks of HSV-1 strain 17 and mutant viruses on different cell typesa
| Virus | Vero cells
|
U2OS cells
|
HFFF-2 cells
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Particles/PFU | FFU/PFU | Particles/FFU | Particles/PFU | FFU/PFU | Particles/FFU | Particles/PFU | FFU/PFU | Particles/FFU | |
| Strain 17 | 32 | 2.1 | 16 | 58 | 3.7 | 23 | 157 | 0.9 | 148 |
| dl1403 | 459 | 33 | 16 | 32 | 2.0 | 28 | 22,400 | 75 | 605 |
| FXE | 210 | 19 | 11 | 74 | 5.6 | 12 | 27,000 | 86 | 320 |
ICP4 FFU titers were determined 6 h after infection. The data show averages of the ratios of five stocks of strain 17, four stocks of dl1403, and one stock of FXE. The degree of variation in these ratios between individual stocks was according to the variability described in Table 1, footnote a.
Table 2 shows averages of particle-to-PFU, particle-to-ICP4 FFU, and ICP4 FFU-to-PFU ratios in the three cell types determined from independent stocks of strain 17 and dl1403. Consistent with previous data (7), the ICP4 FFU titer of dl1403 in Vero cells was around 25-fold greater than the PFU titer (Table 1), although the particle-to-ICP4 FFU ratio of dl1403 in Vero cells was typically similar to that of strain 17 (Table 2). Therefore, the plaque-forming defect of dl1403 in Vero cells is principally due to a reduction in the probability that ICP4-positive cells will initiate the formation of a plaque. However, dl1403 was as efficient at forming plaques from cells expressing ICP4 as strain 17 in U2OS cells (Table 2), indicating that stocks of dl1403 do not contain an increased proportion of intrinsically defective particles or genomes. The ICP4 FFU/PFU ratios of strain 17 in Vero cells and of both strain 17 and dl1403 in U2OS cells were reproducibly greater than unity. This indicates that even under permissive conditions, not all potentially infected cells give rise to visible plaques, so the number of at least partially biologically active particles in the stocks of both strain 17 and dl1403 is greater than the PFU titers would imply.
Some surprising results emerged from parallel studies with HFFF-2 cells. Expression from marker genes incorporated into the genomes of multiply defective viruses that, in addition to a defect in ICP0, fail to express active VP16, ICP4, and/or other viral IE proteins rapidly becomes undetectable in human fibroblasts (reviewed in reference 38). However, Table 2 shows that HFFF-2 cells infected with dl1403 and expressing ICP4 were readily detected by FACS analysis at 6 h after infection. Although the ICP4 FFU titer left a considerable number of potentially infectious particles unaccounted for, it is clear that a significant part of the dl1403 phenotype in HFFF-2 cells is a failure to produce plaques from cells that express ICP4. The levels of ICP4 expressed in this population of dl1403-infected cells were similar to those in strain 17-infected cells at the same time point (see below).
The majority of the dl1403-infected HFFF-2 cells that are expressing ICP4 at 6 h after infection retain detectable ICP4 for extended periods. HFFF-2 cells were infected with dl1403 at a multiplicity of 1 U2OS PFU per HFFF-2 cell (which is expected to give around 10% ICP4-positive cells) and then maintained in medium containing acycloguanosine to inhibit DNA replication and subsequent plaque development. FACS analysis of such cultures at 6, 24, and 48 h after infection showed that at 24 h the number of ICP4-positive cells had not greatly decreased from that at 6 h, and even after 48 h the number of ICP4-positive cells was between 40 and 80% of that at 6 h.
Estimation of a threshold above which ICP0-negative viruses replicate normally in HFFF-2 cells.
The replication of ICP0-negative viruses in Vero or BHK cells is multiplicity dependent, in that poor infection by mutant virus at low MOIs can be overcome at higher multiplicities (7, 9, 48). We set out to estimate an approximate numerical value of the boundary between high- and low-MOI dl1403 infection in HFFF-2 cells. U2OS and HFFF-2 cells were infected with HSV-1 strain 17 and dl1403 at MOIs of 5, 1, and 0.2 U2OS PFU per cell in both cell types, and then total virus yields were determined 16 h later. Productive virus infection spreads rapidly in HFFF-2 cells, so that at 16 h after infection with strain 17 at low MOI, plaques or small groups of cells with a spreading infection could be seen by immunofluorescence staining (data not shown). The infectious cycle proceeds very rapidly in secondarily infected cells because they receive very large doses of virus particles. This could explain why the yields of strain 17 in HFFF-2 cells were slightly greater and less affected by input multiplicity than those in U2OS cells (Fig. 1).
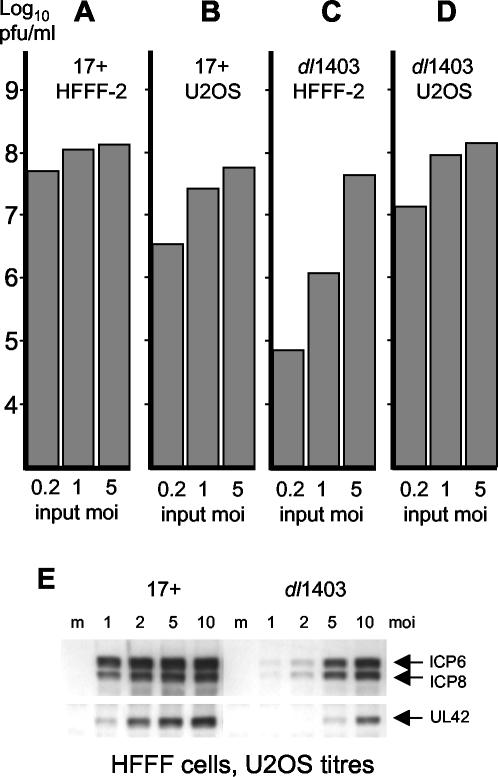
FIG. 1.
Growth properties of HSV-1 strain 17 (17+) and dl1403 in HFFF-2 and U2OS cells. (A to D) Yields of strain 17 and dl1403 (both titrated in U2OS cells) in HFFF-2 and U2OS cells 16 h after infection at the stated multiplicities, based on U2OS cell titers. (E) Western blot analysis of selected HSV-1 strain 17 and dl1403 protein expression in HFFF-2 cells at various input MOIs. The samples were harvested at 6 h postinfection. The relevant details are marked. Lane m, mock-infected cells.
As expected, the yields of dl1403 and strain 17 in U2OS cells were similar at all MOIs (Fig. 1). In HFFF-2 cells, however, dl1403 infection at an MOI of 5 U2OS PFU per cell (equivalent to about 0.005 HFFF-2 PFU per cell [Table 1]) gave only a fourfold reduction in yield from that of strain 17 at the same multiplicity (Fig. 1A and C). Because of the secondary spread noted above, this likely underestimates the real dl1403 defect, but it indicates that an MOI of 5 U2OS PFU per HFFF-2 cell is not far below the multiplicity at which dl1403 will infect even this restrictive cell type fully efficiently. In contrast, at MOIs of 1 and 0.2 U2OS PFU per cell, the yields of dl1403 in HFFF-2 cells were 110- and almost 700-fold reduced, respectively, from those of strain 17 at the same multiplicities. These data illustrate the multiplicity dependence of the dl1403 defect and reveal that there is a broad MOI threshold above which the virus replicates normally. This threshold is surprisingly low, and this may explain why dl1403 titers in HFFF-2 cells are nonlinear (see above); as more potentially infectious particles are applied, suddenly a large number or even most of the cells become productively infected. An important qualification to this concept of threshold is that it is not a tight figure, partly because cells in a population will receive a number of particles that varies according to the Poisson distribution and partly because even at very low MOIs (such as in a plaque assay), some cells receiving only a single particle will commit to lytic infection. Operationally, the threshold concept suggests that above a certain MOI most HFFF-2 cells infected with dl1403 become productively infected and produce yields of virus equivalent to those in a wild-type virus infection at the same MOI; the infection has become ICP0 independent. In order to assess the contribution of ICP0 to productive virus infection in any particular cell type, the input MOI must be safely below the MOI threshold determined for that cell type.
Western blotting experiments indicated that dl1403 gene expression in HFFF-2 cells at a U2OS MOI of 1 was barely detectable compared to that of strain 17 at the same multiplicity at 6 h postinfection (Fig. 1E). However, at multiplicities of 5 and 10 U2OS PFU per HFFF-2 cell, the levels of early gene expression (ICP6, ICP8, and UL42) in dl1403-infected cultures at 6 h were substantially increased (Fig. 1E) and were close to those in the strain 17 infection at an MOI of 2, reflecting an increase in the number of cells that had become productively infected. Taken together, the results in Fig. 1 suggest that the threshold level of dl1403 infection in HFFF-2 cells, above which infected cells become committed to lytic infection, must be close to 10 U2OS PFU per HFFF-2 cell, that is, about 0.01 HFFF-2 PFU per HFFF-2 cell. This conclusion is consistent with the observation that the majority of HFFF-2 cells infected with dl1403 at 10 U2OS PFU per cell develop replication compartments, and at this multiplicity strain 17- and dl1403-infected samples are indistinguishable in this respect (data not shown).
Similar virus yield experiments with Vero cells gave a value for the threshold of approximately 1 U2OS PFU (0.05 Vero PFU) per Vero cell (data not shown), a result consistent with previous studies (5, 9). These threshold values are also consistent with analyses of dl1403 viral gene expression in Vero, U2OS, and HFL cells at different input multiplicities (7, 9, 13).
A major phenotype of ICP0-null mutant virus dl1403 is failure to progress beyond the IE phase of infection.
It follows from the discussion above that the phenotype of ICP0-deficient viruses may best be studied with infections that are safely below the relevant threshold multiplicity. Furthermore, in that a significant part of the defect of ICP0-null mutant viruses in both Vero and HFFF-2 cells is the failure of cells that express ICP4 to initiate plaque formation, population-based methods such as those measuring bulk protein or mRNA levels will obscure the cell-to-cell variation that appears to be a hallmark of the ICP0 phenotype. Therefore, we have taken two approaches to determine the fate of individual cells in low-multiplicity dl1403 infections, i.e., FACS and confocal microscopy analysis. We note that these approaches detect antibody-reactive species in single cells and do not take account of current transcriptional activity or relative protein or mRNA stabilities. The FACS analysis experiments were conducted at multiplicities that gave 5 to 10% positive cells to ensure data reliability. This equates to the formation of 250 to 500 dl1403 plaques on a 35-mm-diameter plate of HFFF-2 cells, i.e., higher than the multiplicities used in a plaque assay or in the fluorescence microscopy approach.
HFFF-2 cells on coverslips were infected with dl1403 at an MOI of 0.1 U2OS PFU (which is expected to give about 10 plaques and 750 to 1,000 ICP4-positive cells). The cells were overlaid with medium containing human serum to eliminate secondary infections and then costained 24 h later to detect IE proteins ICP4 and ICP27 or ICP4 and ICP8 (as an example of a typical early gene product). Coverslips of HFFF-2 cells were infected with strain 17 in parallel at a U2OS cell multiplicity of 0.001 (which is expected to give around 100 plaques). Images of the strain 17 plaques (not shown) were captured by confocal microscopy, taking care to avoid saturation of the detectors so that the digital image intensities were directly proportional to the relative levels of protein expression. These settings were stored and reused to analyze plaques in the dl1403-infected samples, so that direct comparison of the relative viral protein expression levels in strain 17 and dl1403 plaques was possible.
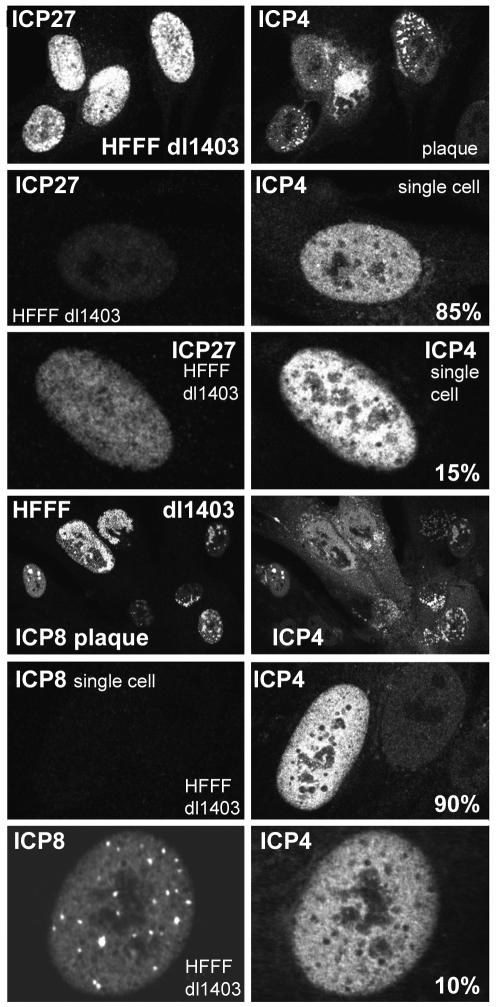
As expected, viral protein expression as estimated by this method was similar in strain 17 and dl1403 plaque cells (data not shown). However, as predicted by the PFU and FFU titers of dl1403 in HFFF-2 cells (Tables 1 and 2), the dl1403-infected coverslips contained a few plaques and many isolated, individual infected cells that contained ICP4 in a mainly diffuse distribution without any indication of replication compartment formation. Cells of this type could persist in this state for at least a further 24 h, and therefore they are referred to below as being stalled or nonproductively infected. The stalled cells were examined under the same image capture conditions as used for the plaques (Fig. 2). Quantitation of the fluorescence signal intensities (see Materials and Methods) indicated that ICP4 was present in these cells at levels of 25 to 100% of those in productively infected cells in plaques, but the levels of ICP27 varied more extensively. The majority (60%) of ICP4-positive stalled cells contained ICP27 at levels below 5% of those in plaque cells, 25% contained medium levels (5 to 50% of average plaque cell intensity), and a minority (15%) contained levels of ICP27 that were between 50 and 100% of those in plaque cells. On the other hand, the great majority (90%) of the ICP4-positive stalled cells contained ICP8 either at levels that were undetectable (even after increasing the detection efficiency of the image capture conditions) or at levels less than 5% of those in plaque cells. A minority (10%) of the nonproductively infected cells contained readily detectable ICP8, but in these cells neither ICP4 nor ICP8 was recruited into replication compartments characteristic of productively infected cells. (Although the example shown in Fig. 2 contains punctate ICP8 foci, these are not replication compartments because they have not recruited ICP4. Most ICP4- and ICP8-positive cells had mainly diffuse distributions of both proteins [see, for example, the distribution of ICP8 in Fig. 4].) These results show that the dl1403-infected HFFF-2 cells that contain ICP4 but have not initiated the formation of a plaque can be stalled at a variety of different stages: cells that contain an incomplete complement of IE proteins, cells that contain IE but not early proteins, and cells that contain at least some early proteins but have failed to initiate DNA replication (based on the absence of distributions of ICP4 and ICP8 that are characteristic of replication compartments). Although only 10% of the ICP4-positive singly infected cells expressed ICP8, since ICP4-positive cells exceeded cells committed to form plaques by about 75-fold (Table 2), the number of these ICP8-positive but apparently nonproductive infected cells still exceeded the number of plaque-committed cells by a factor of about 7.
FIG. 2.
Immunofluorescence analysis of HFFF-2 cells nonproductively infected with dl1403 at low multiplicity at 24 h after infection. The fluorescence intensity of isolated cells stained for ICP4 and ICP27 or for ICP4 and ICP8 was compared with that of cells in the small number of plaques on the coverslip. The images in the six upper panels were captured under identical scanning conditions and have been printed without alteration of brightness or contrast. The same is true for the six lower panels. Further details on the relative amounts of ICP27 and ICP8 expressed in nonproductively infected cells are given in the text.
FIG. 4.
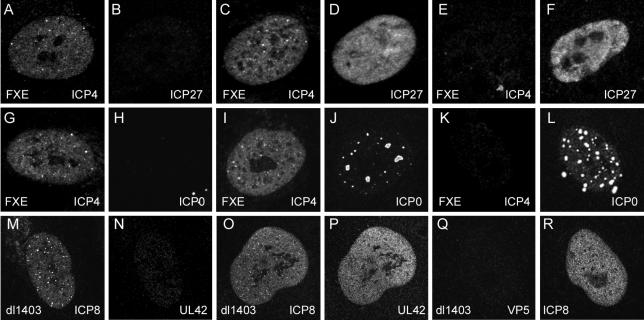
Stochastic expression of IE proteins in ICP0 mutant FXE-infected HFFF-2 cells and of early proteins in dl1403-infected HFFF-2 cells. In all cases the cells were fixed 24 h after infection. Each pair of panels (A and B, C and D, etc.) shows a single nonproductively infected cell stained for the indicated pairs of viral proteins. Panels A to L show FXE-infected cells, with all images captured under very similar scanning conditions. Panels M to R show dl1403-infected cells, also captured under very similar scanning conditions. The panels have been reproduced without further modification of brightness or contrast.
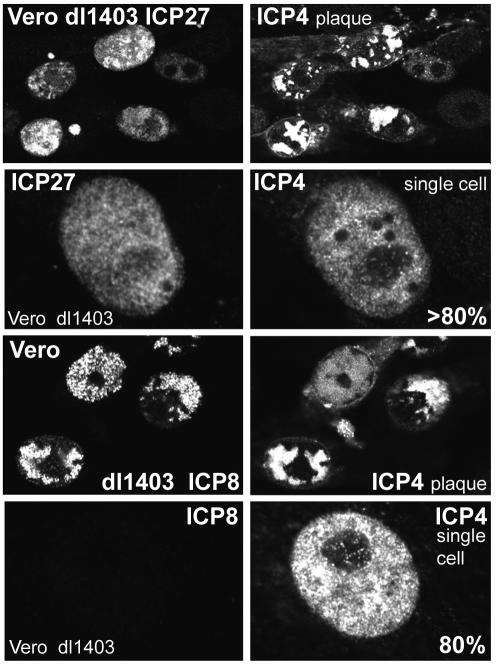
Similar experiments were conducted with Vero cells and dl1403 at an input multiplicity of 0.02 U2OS PFU per cell (Fig. 3). Again, a mixture of plaques and stalled or nonproductively infected cells was observed. As in HFFF-2 cells, the levels of ICP4 expression in productively and nonproductively infected Vero cells were similar. In contrast to the variable expression of ICP27 in nonproductively infected HFFF-2 cells, more than 80% of isolated Vero cells that contained ICP4 also contained ICP27 at levels comparable to those in cells in a plaque. The number of ICP4-positive cells that contained ICP8 was also greater in Vero cells than in HFFF-2 cells (data not shown), although 80% of stalled Vero cells had very low or undetectable levels of ICP8 (Fig. 3). Taken with the titers of particles, PFU, and FFU in Vero cells, these data indicate that the principal phenotype of dl1403 in Vero cells is that infection becomes stalled at the IE phase. Since the number of these stalled cells is close to the number of potential dl1403 infectious units as determined for U2OS cells (Tables 1 and 2), it follows that there is little evidence for any completely repressed or quiescent dl1403 genomes after low-multiplicity infection of Vero cells.
FIG. 3.
Immunofluorescence analysis of Vero cells nonproductively infected with dl1403 at low multiplicity at 24 h after infection. The fluorescence intensity of isolated cells stained for ICP4 and ICP27 or for ICP4 and ICP8 was compared with that of cells in the small number of plaques on the coverslip. The images in the four upper panels were captured under identical scanning conditions and have been printed without alteration of brightness or contrast. The same is true for the four lower panels. Further details on the relative amounts of ICP27 and ICP8 expressed in nonproductively infected cells are given in the text.
The differences between HFFF-2 and Vero cells in the relative proportions of ICP4-positive cells that also express ICP27 and/or ICP8 emphasizes that these variables are cell type dependent. For example, the proportion of ICP4-positive dl1403-infected NT2 cells that are ICP8 positive 24 h after a low-MOI infection is much lower than that seen in HFFF-2 cells (data not shown).
Complementation and reactivation of ICP0-deficient viruses.
Table 1 shows that the titer of ICP4-positive cells was about 10-fold greater in U2OS cells than in HFFF-2 cells infected with dl1403. Although there were many more ICP4-positive HFFF-2 cells than cells that became committed to plaque formation, there were large numbers of HFFF-2 cells that should have encountered potentially infectious dl1403 particles (on the basis of U2OS cell titers) that could not be accounted for by either PFU or ICP4 FFU titers. Therefore, we tested whether an ICP0-null mutant virus could be complemented to infect HFFF-2 cells as efficiently as HSV-1 strain 17. Virus dl0C4 (see Materials and Methods) contains the ICP0 deletion mutation of dl1403 and the ECFP-linked ICP4 fusion protein of vECFP-ICP4 (19). The ECFP fusion moiety has no detectable effect on ICP4 functionality (19), and virus dl0C4 behaved in a manner very similar to that of dl1403 in all assays tested, including analysis by Western blotting, growth curves, and titration of PFU and FFU in Vero, HFFF-2, and U2OS cells (data not shown). The ECFP fusion allows the determination of the number of cells that express ICP4 from the ICP0-deficient virus in populations in which ICP4 is also being expressed from the complementing virus. HFFF-2 cells were infected with dl0C4 in the absence or the presence of HSV-1 strain 17, and then the number of cells expressing ECFP-linked ICP4 was measured by FACS analysis with autofluorescent protein-specific antibody 3E6 (ECFP is not efficiently detected by autofluorescence by this method). At the same time, the ICP4 FFU titer of dl0C4 in U2OS cells and the proportion of HFFF-2 cells infected by the complementing virus were determined by FACS. The results showed that the intrinsic ICP4 FFU titer of dl0C4 in HFFF-2 cells (3.7 × 106 FFU/ml) could be increased to 4.4 × 107 FFU/ml in the presence of complementing virus, a value close to the ICP4 FFU titer of that stock of dl0C4 in U2OS cells (4.7 × 107 FFU/ml). Therefore, a high percentage of the HFFF-2 cells that receive dl0C4 virus particles capable of expressing ICP4 in U2OS cells can be detected in a coinfection complementation experiment.
The ICP4 FFU, PFU, and complementation data indicate that viruses that lack only functional ICP0 produce at least three classes of infected HFFF-2 cells: those committed to plaque formation, those that contain ICP4 in stalled infections for at least 24 to 48 h, and those that do not express ICP4 but can nonetheless be complemented by a coinfecting ICP0-positive virus. We tested whether the population of ICP4-negative infected cells could be detected by later superinfection with a virus that expresses ICP0. In this type of experiment, cells are initially infected with the ICP0-null mutant at a very low multiplicity, and then, after a period of 24 h or more in which stalled, nonproductive, or quiescent infections are established, the cells are superinfected with an ICP4-defective virus that expresses ICP0. Although this is different from the situation of reactivation of latent virus in animal models, the term reactivation has commonly been applied to such experiments with cultured cells because they are distinct from the complementation coinfection experiments described above. HFFF-2 cells were infected with an input of dl1403 that was expected to give no plaques on the plate, and then the following day the plates were infected with ICP4-defective virus in1411 (41) at an input adjusted to infect about 50% of the cells. The total potential PFU titer of the sample of dl1403 in U2OS cells was determined in parallel, and the proportion of HFFF-2 cells actually infected by in1411 was calculated by FACS analysis of ICP0-expressing cells (to provide a correction factor to allow for cells initially receiving dl1403 particles but not subsequently infected by in1411). The results showed that the intrinsic titer of the stock of dl1403 on HFFF-2 cells (3.7 × 104 PFU/ml) was increased to 2.9 × 107 PFU/ml, which is close to the U2OS titer of the stock used (1.9 × 107 PFU/ml) and considerably greater than the ICP4 FFU titer of the stock (2.6 × 106 FFU/ml on HFFF-2 cells). These data demonstrate that a high proportion of the potentially infectious dl1403 particles in low-multiplicity, nonproductive HFFF-2 cells infections can be accounted for by later reactivation. Therefore, ICP4-negative nonproductively infected HFFF-2 cells must be present, and it is likely that both ICP4-positive and ICP4-negative cells can be reactivated to initiate plaque formation. However, the ICP0 mutant viruses in even the ICP4-negative infected HFFF-2 cells are not initially completely transcriptionally quiescent (see below).
Evidence for partially uncoordinated viral gene expression in HFFF-2 cells infected with ICP0-deficient mutants.
Given that HFFF-2 cells nonproductively infected with dl1403 could contain ICP4 without simultaneously containing ICP27 (Fig. 2), we asked whether other viral proteins could be expressed in a similarly uncoordinated manner. Virus FXE expresses a form of ICP0 that lacks the crucial RING finger domain (13), thus eliminating most functions of the protein and rendering the virus almost as disabled in HFFF-2 cells as the complete deletion mutant dl1403 (Table 2). HFFF-2 cells were infected with FXE at low multiplicity, and then the sample was examined 24 h later for the appearance of plaques and nonproductively infected cells by staining for ICP4 in combination with either ICP0 or ICP27. Image capture conditions were standardized as described for the data in Fig. 2 and 3 to allow a quantitative comparison of the relative levels of protein expression in productively and nonproductively infected cells. The nonproductively infected cells were easily recognized by being isolated and having readily detectable levels of ICP4 but no replication compartments. Images of several such cells were collected with similar data collection settings; typical examples are shown in Fig. 4.
As with the comparable dl1403 results (Fig. 2), examples of HFFF-2 cells nonproductively infected with FXE and containing ICP4 but little ICP27 were common (Fig. 4A and B), while cells containing both proteins (Fig. 4C and D) or containing ICP27 and little ICP4 (Fig. 4E and F) were also present. Surprisingly, large numbers of cells that contained high levels of mutant ICP0 but no detectable ICP4 (Fig. 4K and L) or containing mutant ICP0 but no ICP27 (data not shown) were readily detectable. These findings could not be explained by one of the antibodies in the combination detecting its cognate protein with greater intrinsic efficiency than the other, since similar results were obtained with alternative antibody combinations (data not shown). Most ICP4-containing nonproductively infected cells were also ICP0 positive (Fig. 4I and J), although there were rare examples of ICP4-positive cells that contained very low levels of ICP0 (Fig. 4G and H). These results suggest that the normal coordinate regulation of IE genes has been partially lost in the nonproductively infected cells, at least at the level of detectable protein.
The partially uncoordinated presence of IE proteins in the absence of functional ICP0 raises the question whether those ICP4-positive dl1403-infected HFFF-2 cells that also express ICP8 (Fig. 2) have similar uncoordinated expression of early gene products. HFFF-2 cells were infected with dl1403 under conditions expected to give a nonproductively infected cell population, and then ICP8 and UL42 (another typical early gene product) were detected by simultaneous antibody staining 24 h later. Preliminary experiments indicated that, like the case with ICP8 (Fig. 2), a low percentage of ICP4-positive nonproductively infected cells could express amounts of UL42 that were equivalent to those in productively infected cells at the edges of developing plaques (data not shown). Although most stalled infected cells that contained UL42 also contained ICP8 (Fig. 4O and P), there were many cells that contained ICP8 but not UL42 (Fig. 4M and N). Thus, it appears that early proteins may be expressed in an apparently stochastic manner in nonproductively infected cells. This could explain why the nonproductively infected cells that contain ICP4 and ICP8 have failed to produce replication compartments; they may have failed to express one of the other proteins that are essential for this process.
Double staining for ICP8 and VP5, a late protein that requires DNA replication for efficient expression, indicated that the ICP8-positive nonproductively infected cells (characterized by a lack of replication compartment formation) did not express VP5 above background levels at 24 h after infection (Fig. 4Q and R). Therefore, although gene expression within the IE and early protein classes is at least partially uncoordinated in individual HFFF-2 cells nonproductively infected with ICP0 mutant viruses, the requirement for DNA replication for efficient late gene expression has been retained. The partially uncoordinated expression of viral proteins within a particular temporal class described here is quite distinct from differences in the relative levels of individual early or late products in populations of cells infected with viruses with lesions in other regulatory proteins, such as ICP4 or ICP27. The examples in this paper illustrate that the relative levels of expression of proteins in the same kinetic class can vary greatly in individual cells, illustrating the stochastic nature of the phenomenon.
A class of HFFF-2 cells infected by ICP0-deficient viruses express products from the IE1 but not the IE3 gene loci.
The fluorescence results with virus FXE indicated the presence of an excess population of cells that contained the mutant ICP0 protein but not ICP4. To determine if the IE1 gene expression activity was higher in ICP0-null mutant low-multiplicity infection of HFFF-2 cells, we studied virus vEG-dl110, which contains the EGFP open reading frame in place of that of ICP0 in an otherwise normal IE1 transcription unit (29). In combination with the ICP4-null mutant in1411, this virus allows the parallel determination of all of the variables studied above: IE1 and ICP4 expression by FACS. complementation of IE1 and ICP4 FFU titers by coinfection with an ICP0-positive virus, and reactivation of nonproductively infected cells to initiate plaque formation. The results were compared with the FFU and PFU titers of vEG-dl110 in U2OS cells and hence the number of potentially infectious units.
Table 3 shows that strain 17 and vEG-dl110 had similar PFU titers in U2OS cells and that their IE1 and ICP4 FFU titers were similar to each other and about threefold greater than the PFU titers of both viruses in this cell type. These data are consistent with those in Table 2. In HFFF-2 cells infected at low multiplicity, the ICP4 FFU titer of strain 17 at 6 h was surprisingly but reproducibly less than the ICP0 FFU titer, but at 24 h after infection in the presence of acycloguanosine the ICP4 and ICP0 FFU titers were identical. This indicates that at low multiplicity, there appears to be a slight lag in expression of ICP4 even in strain 17 infections of HFFF-2 cells. Although the ICP4 FFU titer at 6 h was similar to the PFU titer (see also Table 2), at 24 h the FFU titers had increased to a figure close to the PFU titer of strain 17 in U2OS cells. These data suggest that there must be a population of wild-type strain 17-infected HFFF-2 cells that do not progress to plaque formation, a conclusion that is consistent with the increased particle-to-PFU ratio of strain 17 in HFFF-2 cells compared to Vero and U2OS cells (Table 2). In the case of vEG-dl110 in HFFF-2 cells, the IE1 FFU titer at 6 h was significantly greater than the ICP4 FFU titer, and indeed it was close to the titer of virus that could be reactivated by later superinfection. This suggests that most cells that establish quiescent or stalled infections can be initially detected by IE1 gene expression activity. At 24 h after infection, the ICP4 FFU titer had decreased slightly from that at 6 h, but the IE1 FFU titer had dropped markedly. When both IE gene products were detected in the same 24 h sample simultaneously, the titer was greater than either the IE1 or ICP4 FFU titer alone. This observation can be explained by the cells that express either IE1 products or ICP4 but not both (Fig. 4).
TABLE 3.
Titers of IE1- and ICP4-positive cells, complemented FFU titers, and reactivated PFU titers of vEG-dl110, in comparison with PFU titers in HFFF-2 and U2OS cells, and relevant comparative titers of HSV-1 strain 17a
| Parameter | U2OS cell titers
|
HFFF-2 cell titers
|
||
|---|---|---|---|---|
| Strain 17 | vEG-dl110 | Strain 17 | vEG-dl110 | |
| PFU | 1.5 × 108 | 2.2 × 108 | 6.7 × 107 | 1.5 × 105 |
| 6-h FFU | ||||
| ICP4 | 4.8 × 108 | 5.8 × 108 | 5.5 × 107 | 4.0 × 107 |
| IE1 | 5.2 × 108 | 6.0 × 108 | 1.0 × 108 | 1.8 × 108 |
| IE1 + ICP4 | 6.6 × 108 | 7.0 × 108 | 1.0 × 108 | 1.9 × 108 |
| 24-h FFU | ||||
| ICP4 | NDb | ND | 1.9 × 108 | 3.2 × 107 |
| IE1 | ND | ND | 1.9 × 108 | 5.2 × 107 |
| IE1 + ICP4 | ND | ND | 1.9 × 108 | 6.0 × 107 |
| 6-h FFU + in1411 | ||||
| ICP4 | ND | ND | ND | 3.4 × 108 |
| IE1 | ND | ND | ND | 3.3 × 108 |
| Reactivated PFU | ND | ND | ND | 2.0 × 108 |
IE3 FFU titers were determined by ICP4 staining for all three viruses. IE1 FFU were determined by staining for EGFP in vEG-dl110 infections. IE1-plus-ICP4 titers reflect cell numbers detected by staining simultaneously for ICP4 and EGFP. Further details are given in the text. The experiment was repeated, and similar results were obtained.
ND, not determined.
Consistent with the complementation and reactivation data for viruses dl0C4 and dl1403 presented above, both IE1 and ICP4 FFU titers of vEG-dl110 could be complemented by coinfection with in1411 to values close to those at 6 h in U2OS cells, and about half of these could be reactivated to plaque formation by superinfection with in1411, to give a titer very close to the PFU titer of vEG-dl110 in U2OS cells. These data begin to give a quantitative description of all of the possible fates of HFFF-2 cells that encounter a potentially infectious ICP0-null mutant virus particle.
It is important to note that the issue of possible differential mRNA and protein stability complicates the interpretation of these data. It is possible that the hybrid IE1 mRNA or the EGFP protein is less stable than the IE3 mRNA or the ICP4 protein; these methods do not allow conclusions to be drawn about the actual transcriptional status of the IE genes in the stalled infected cells. Indeed, stalled cells retain readily detectable amounts of ICP4 for many hours even in the presence of cycloheximide (data not shown), so the presence of this and other viral proteins in nonproductively infected cells cannot be assumed to indicate ongoing transcription.
A proportion of HFFF-2 cells infected at low multiplicity with dl1403 undergo cell death.
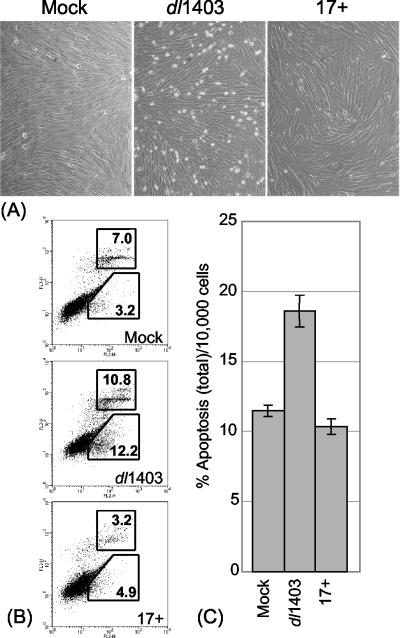
During the course of these studies it was observed that HFFF-2 cultures infected at low MOIs with dl1403, but not those infected with strain 17, contained many rounded cells at 12 h after infection (Fig. 5A). FACS detection of cells positive for cell surface annexin V (a commonly used marker for apoptotic cell death) and for 7-AAD staining (a marker for necrosis) showed that the proportion of annexin V-positive cells was not increased in the strain 17-infected samples but that the apoptotic and necrotic-apoptotic populations were significantly increased in the dl1403-infected samples (Fig. 5). The average increase of about 6% above background of dead or dying cells equates to about half the number that would express ICP4 under these infection conditions. We refer to these cells as apoptotic for simplicity and because cell surface annexin V staining is an accepted apoptotic marker, but with the proviso that it was not possible to test for other apoptotic markers such as caspase cleavage because of the low proportion of cells undergoing these events. The window for observation of these apoptotic cells was quite narrow; they did not appear at earlier times, but at later times they became detached from the cell sheet and were lost. Apoptotic cells were not observed at higher dl1403 MOIs (U2OS MOI of 10), probably because at this multiplicity most dl1403-infected HFFF-2 cells become committed to lytic infection (see above). Such cells will not be susceptible to apoptosis because of the battery of mechanisms that the virus employs to inactivate host apoptotic pathways (reviewed in reference 2).
FIG. 5.
Induction of necrosis and apoptosis in a proportion of HFFF-2 cells infected at a low MOI with dl1403. (A) Low-magnification phase-contrast images of cells 12 h after mock infection (left) or infection with dl1403 (center) or strain 17 (17+) (right), both at a U2OS MOI of 1. (B) FACS analysis of cells after mock infection (top) or infection with dl1403 (middle) or strain 17 (bottom), both at an MOI of 1. The cells were simultaneously stained for apoptotic and necrotic cells by detection of annexin V (horizontal axis) and 7-AAD (vertical axis). The upper and lower boxes show the gates selected to quantify apoptotic-necrotic and apoptotic cells, respectively, and the numbers show the percentages of the cells within the gated areas. (C) Analysis of total apoptotic and apoptotic-necrotic cells in similar dl1403 and strain 17 infections; results are averages from three independent experiments, and the error bars show standard errors of the means.
Although these apoptotic and necrotic cells were reproducibly observed by examination of infected plates and by FACS, it is noteworthy that the nonproductively infected cells analyzed by immunofluorescence had normal morphologies (Fig. 2 to 4) and were stably maintained for at least 24 h. Thus, the extended presence of IE proteins in cells harboring stalled dl1403 infections does not generally trigger apoptosis over this time period. Since the deletion in dl1403 also affects the latency-associated transcript (LAT) region and LATs have been implicated in protection from apoptosis (reviewed in reference 24), we tested whether virus FXE (which has only a very small deletion in the minor LAT region) also induced an apoptotic population. The results showed that FXE was similar to dl1403 in this regard (data not shown), indicating that it is the lack of functional ICP0 rather than a modification of the LATs that is responsible for the effect.
Summary of the fates of HFFF-2 cells infected at low multiplicity by an ICP0-negative virus.
Table 4 shows a summary of the predicted fates of 5 × 105 U2OS or HFFF-2 cells infected with 50,000 particles of typical stocks of strain 17 and an ICP0-negative virus. The predictions have been abstracted from the data in Tables 1, 2, and 3; the relevant sections in Results, and replicate experiments. Unavoidable experimental variability (on the order of twofold) limits the precision of these predictions, so the data in Table 4 should be taken as a reflection of the relative frequencies of the various fates rather than as definitive numerical values. The titers of IE1-positive cells for the mutant virus have been taken from results with vEG-dl110.
TABLE 4.
Estimation of the relative frequencies of fates of cells receiving single potentially infectious ICP0-null mutant virus particlesa
| Cell type and class | Value for:
|
|
|---|---|---|
| Strain 17 | Δ1CP0 | |
| U2OS cells | ||
| IE1-positive cells at 6 h | 5,000 | 5,000 |
| ICP4-positive cells at 6 h | 5,000 | 5,000 |
| PFU | 1,600 | 1,600 |
| HFFF-2 cells | ||
| IE1-positive cells at 6 h | 1,000 | 1,200 |
| ICP4-positive cells at 6 h | 550 | 400 |
| Complemented ICP4-positive cells at 6 h | NDc | 2,700 |
| IE1-positive cells at 24 h | 1,900 | 450 |
| ICP4-positive cells at 24 h | 1,900 | 300 |
| Cells that can be reactivated to PFU | ND | 1,600 |
| IE-positive stalled cells | ND | 600 |
| IE-negative quiescent cellsb | ND | 1,000 |
| Apoptotic cells at 12 h | <20 | 200 |
| PFU | 750 | 2 |
The numbers represent an interpretation of the data in Tables 1, 2, and 3 and give approximate predictions for a theoretical infection with 50,000 particles of an average viral stock on a 35-mm-diameter plate of 5 × 105 U2OS or HFFF-2 cells (MOI, 0.1 particle per cell). Variability in the original experimental data limits the precision of the predictions, which are intended to give a reasonable, rounded overview of the frequency of the possible outcomes.
The dl1403 IE-negative quiescent cell titer has been assumed to be the difference between the number of cells that can be reactivated to PFU and those that are IE positive and nonproductively infected at 24 h. The number of apoptotic cells was determined from infections at an MOI of 1 U2OS PFU per HFFF-2 cell. Apoptotic cells in strain 17 infections were not higher than those in the mock sample at this multiplicity.
ND, not determined.
The critical features of this summary are as follows: (i) strain 17 and an ICP0-negative virus have average particle-to-PFU ratios of around 30 in U2OS cells, giving in this theoretical example around 1,600 cells that would be committed to plaque formation; (ii) both viruses produce about threefold more IE1-positive than plaque-committed U2OS cells at 6 h postinfection, and most or all of the IE1-positive cells are also expressing ICP4; (iii) this dose of strain 17 in HFFF-2 cells gives far fewer IE1-positive cells than in U2OS cells at 6 h; (iv) the number of ICP4-expressing cells in HFFF-2 cells infected with strain 17 lags behind the IE1-positive cell titer at 6 h but is equivalent at 24 h; (v) the numbers of IE1- and ICP4-positive HFFF-2 cells in ICP0 mutant virus infections at 6 h are broadly similar to those in a strain 17 infection; (vi) the number of ICP4-positive HFFF-2 cells at 6 h in ICP0 mutant infections can be further increased by coinfection with a complementing virus; (vii) by 24 h postinfection, the number of IE1 (EGFP)-positive HFFF-2 cells in ICP0 mutant infections has dropped substantially; (viii) superinfection of such cells at 24 h with an ICP0-positive virus results in a PFU titer similar to that in U2OS cells and greater than the number of total IE-positive cells at 24 h, reflecting the presence of truly quiescent virus at this time point; (ix) low-multiplicity dl1403 infection results in a number of cells becoming apoptotic that is about half that of the ICP4-positive cells, but the latter have normal morphologies and most can be retained for at least a further 24 h; (x) the number of dl1403-infected HFFF-2 cells that become committed to plaque formation is around 700-fold less than those that express IE products at 6 h; and (xi) the differential presence of IE and early proteins in such cells is not consistent with the established coordinate regulation within particular kinetic classes observed in high-multiplicity productive infections.
DISCUSSION
This paper presents a thorough analysis of the consequences of deletion of the gene encoding ICP0 for HSV-1 gene expression and infection in limited-passage human fibroblast cells. These studies have built on much previously published work, particularly from the laboratory of P. Schaffer, who noted that ICP0-deficient viruses produce many more ICP4-expressing infected Vero cells than would be expected on the basis of PFU titers (7). We have found that a similar situation exists in human fibroblasts, cells that are considerably more restrictive for ICP0 mutant virus replication than Vero cells. Furthermore, we found that all of the HFFF-2 cells that have received dl1403 particles and can be later detected by reactivation can be detected by IE gene expression activity at 6 h. It is clear that viral gene expression is not immediately repressed in HFFF-2 cells infected with ICP0-negative viruses at low multiplicity and that IE protein-positive cells can persist without progression to lytic infection. The ICP0 phenotype in many of these cells is characterized by a failure to progress beyond the IE phase, which is what is observed when ICP0 function is inhibited by the proteasome inhibitor MG132 (18).
The conclusion that the predominant phenotype of ICP0-deficient viruses, particularly in the least permissive cell types, is for all viral gene expression to become repressed has been built up from a number of studies using a variety of mutant viruses. At first sight, this conclusion conflicts in part with the analysis presented in this paper. However, in only one previous study has gene expression in individual cells infected with a simple ICP0-deficent virus been examined in any detail, with results entirely consistent with those presented here (7). An important consideration is that studies of viral gene expression in quiescently infected cultured cells have generally used multiply defective viruses that fail to express ICP0 and one or more other significant or essential regulators of viral gene expression, such as VP16, ICP4, or ICP27. The lack of these proteins in addition to ICP0 would have substantial consequences for gene expression from the defective viral genome and could explain why even genes driven by strong constitutive promoters become repressed within the first 24 h after infection. On the other hand, simple ICP0-deficient viruses are VP16 positive, and this would be able to stimulate authentic IE gene expression at early times of infection, even in the absence of ICP0.
It is likely that an initial burst of VP16-stimulated IE gene transcription produces sufficient mRNA to explain the IE protein levels that we observe and that IE mRNA and protein stability enables the persistence of the IE protein-positive cells for extended periods. Therefore, repression of the viral genome undoubtedly occurs in simple ICP0-negative virus infections, but this process is insufficiently rapid to preclude synthesis of significant levels of IE proteins in many cells. The stochastic nature of protein expression within the IE and early classes in these cells suggests that the repression mechanism does not develop uniformly over the entire genome but that selected regions may escape for times sufficient to allow readily detected protein expression. Unfortunately, it is not possible to determine for how long active viral transcription persists in individual cells infected with dl1403 at low multiplicity. However, it is clear that ICP4 can persist for extended periods and that its presence is insufficient to activate the bulk of early gene transcription.
The data in this paper emphasize the probabilistic nature of the ICP0-defective phenotype. This probability aspect is accompanied by a threshold effect, such that cells that would otherwise support only a stalled or quiescent infection become productively infected provided that enough viral genomes are present. The value of this threshold is difficult to estimate precisely, but Fig. 1 suggests that it is close to 10 U2OS PFU per HFFF-2 cell (equivalent to about 300 particles, or 20 to 30 ICP4 FFU as measured in U2OS cells). An intriguing possible explanation for this threshold effect is provided by the apparent loss of precise coordinate regulation of IE and early proteins in nonproductively infected cells (Fig. 2 to 4). This apparently stochastic viral protein expression in the absence of ICP0 means that even if a cell is expressing both ICP4 and ICP8, it may not develop a productive infection because not all essential early genes will be expressed. If a cell contains several such genomes that are expressing IE and early proteins in an incompletely coordinated fashion, then at some point a full complement of viral proteins will be present and productive infection may ensue. An alternative but not mutually exclusive possibility, and an explanation put forward by many groups, is that the multiplicity dependence of ICP0-negative mutants is due to titration of the cellular repression mechanism by an increased viral genome load.
A consequence of the threshold effect is that investigations of treatments that might inhibit the activity of ICP0 are likely to be informative only if wild-type virus is used well below the threshold multiplicity in the relevant cell type. For example, in Vero cells the full effect of the lack of ICP0 activity may be seen only if the multiplicity is reduced to 0.1 U2OS PFU per cell (data not shown). Even at this multiplicity, complete inhibition of ICP0 activity would not reduce the number of Vero cells expressing ICP4 and would be expected to reduce the number of ICP8-positive cells and PFU by about 5- and 20-fold, respectively (Table 1 and Fig. 3). Thus, the available window to determine whether a treatment inhibits ICP0 function is quite narrow.
In addition to the threshold effect that determines the multiplicity at which most cells in a population become productively infected, in very-low-multiplicity dl1403 infections there is a small proportion of HFFF-2 cells that develop a productive infection despite the inoculum being far below the threshold multiplicity (a 35-mm-diameter plate giving 100 plaques will have been infected at an average multiplicity of about 5 particles per cell, whereas the apparent threshold is about 300 particles per cell). Some feature of the individual cell must determine whether the outcome is plaque production, a stalled or quiescent infection, or apoptosis. The nature of this variable is far from clear. It may be pertinent that cells released from growth arrest exhibit a transiently increased ability to support ICP0-null mutant plaque formation (6), and cyclin-dependent kinase activity is required for viral infection (44). However, several reports link ICP0 with resistance to the effects of interferon and the possible role of PML and ND10 in the interferon response (8, 11, 21, 32-35, 39, 49). It is possible that an interferon response in HFFF-2 cells infected at low multiplicity with dl1403 could lead to shut-down of viral gene expression or even apoptosis and that some cells escape this response. More recently, the presence of ICP0 has been found to inhibit circularization of parental HSV-1 genomes, and this too could contribute to the fate of cells in low-multiplicity infections (23).
Finally, it is possible that single-cell analysis of low-multiplicity infections could be useful in the study of other nonessential viral regulatory genes, or even of mutations in essential genes that retain partial function, whose phenotypes may become masked in high-multiplicity whole-population experimental approaches.
Acknowledgments
This work was supported by the Medical Research Council.
We are grateful for the constructive suggestions and comments on the manuscript of Duncan McGeoch, Chris Preston, and Nigel Stow. Susan Graham and Frazer Rixon made available some of the antibodies used in this work.
REFERENCES
- 1.Ackermann, M., D. K. Braun, L. Pereira, and B. Roizman. 1984. Characterization of herpes simplex virus 1 alpha proteins 0, 4, and 27 with monoclonal antibodies. J. Virol. 52:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubert, M., and J. A. Blaho. 2001. Modulation of apoptosis during herpes simplex virus infection in human cells. Microbes Infect. 3:859-866. [DOI] [PubMed] [Google Scholar]
- 3.Boutell, C., and R. Everett. 2003. The HSV-1 regulatory protein ICP0 interacts with and ubiquitinates p53. J. Biol. Chem. 278:36596-36602. [DOI] [PubMed] [Google Scholar]
- 4.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, W., T. L. Astor, L. M. Liptak, C. Cho, D. M. Coen, and P. A. Schaffer. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J. Virol. 67:7501-7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, W., and P. A. Schaffer. 1991. A cellular function can enhance gene expression and plating efficiency of a mutant defective in the gene for ICP0, a transactivating protein of herpes simplex virus type 1. J. Virol. 65:4078-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, W., and P. A. Schaffer. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77:7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., and S. Silverstein. 1992. Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J. Virol. 66:2916-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conner, J., J. Murray, A. Cross, J. B. Clements, and H. S. Marsden. 1995. Intracellular localisation of herpes simplex virus type 1 ribonucleotide reductase subunits during infection of cultured cells. Virology 213:615-623. [DOI] [PubMed] [Google Scholar]
- 11.Eidson, K. M., W. E. Hobbs, B. J. Manning, P. Carlson, and N. A. DeLuca. 2002. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J. Virol. 76:2180-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R., A. Cross, J. Tyler, and A. Orr. 1993. An epitope within the DNA-binding domain of the herpes simplex virus immediate early protein Vmw175 is conserved in the varicella-zoster virus gene 62 protein. J. Gen. Virol. 74:1955-1958. [DOI] [PubMed] [Google Scholar]
- 13.Everett, R. D. 1989. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J. Gen. Virol. 70:1185-1202. [DOI] [PubMed] [Google Scholar]
- 14.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 15.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D., A. Orr, and M. Elliott. 1991. High level expression and purification of herpes simplex virus type 1 immediate early polypeptide Vmw110. Nucleic Acids Res. 19:6155-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., A. Orr, and C. M. Preston. 1998. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 17:7161-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett, R. D., G. Sourvinos, and A. Orr. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 77:3680-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu, H., and B. Roizman. 2003. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc. Natl. Acad. Sci. USA 100:8963-8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harle, P., B. Sainz, Jr., D. J. Carr, and W. P. Halford. 2002. The immediate-early protein, ICP0, is essential for the resistance of herpes simplex virus to interferon-alpha/beta. Virology 293:295-304. [DOI] [PubMed] [Google Scholar]
- 22.Hsu, W. L., and R. D. Everett. 2001. Human neuron-committed teratocarcinoma nt2 cell line has abnormal nd10 structures and is poorly infected by herpes simplex virus type 1. J. Virol. 75:3819-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson, S. A., and N. A. DeLuca. 2003. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc. Natl. Acad. Sci. USA 100:7871-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 16:79-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 alpha regulatory protein ICP0 with elongation factor 1delta: ICP0 affects translational machinery. J. Virol. 71:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawaguchi, Y., M. Tanaka, A. Yokoymama, G. Matsuda, K. Kato, H. Kagawa, K. Hirai, and B. Roizman. 2001. Herpes simplex virus 1 alpha regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc. Natl. Acad. Sci. USA 98:1877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knipe, D. M., P. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus. 2001. Fields virology, 4th ed. Lippincott-Williams and Wilkins, Philadelphia, Pa.
- 29.Lomonte, P., and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G1 into S phase of the cell cycle. J. Virol. 73:9456-9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsden, H. S., A. M. Cross, G. J. Francis, A. H. Patel, K. MacEachran, M. Murphy, G. McVey, D. Haydon, A. Abbotts, and N. D. Stow. 1996. The herpes simplex virus type 1 UL8 protein influences the intracellular localization of the UL52 but not the ICP8 or POL replication proteins in virus-infected cells. J. Gen. Virol. 77:2241-2249. [DOI] [PubMed] [Google Scholar]
- 31.McClelland, D. A., J. D. Aitken, D. Bhella, D. McNab, J. Mitchell, S. M. Kelly, N. C. Price, and F. J. Rixon. 2002. pH reduction as a trigger for dissociation of herpes simplex virus type 1 scaffolds. J. Virol. 76:7407-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mossman, K. L., and J. R. Smiley. 2002. Herpes simplex virus ICP0 and ICP34.5 counteract distinct interferon-induced barriers to virus replication. J. Virol. 76:1995-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholl, M. J., L. H. Robinson, and C. M. Preston. 2000. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J. Gen. Virol. 81:2215-2218. [DOI] [PubMed] [Google Scholar]
- 36.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preston, C. M. 2000. Repression of viral transcription during herpes simplex virus latency. J. Gen. Virol. 81:1-19. [DOI] [PubMed] [Google Scholar]
- 39.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preston, C. M., and M. J. Nicholl. 1997. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J. Virol. 71:7807-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell, J., E. C. Stow, N. D. Stow, and C. M. Preston. 1987. Abnormal forms of the herpes simplex virus immediate early polypeptide Vmw175 induce the cellular stress response. J. Gen. Virol. 68:2397-2406. [DOI] [PubMed] [Google Scholar]
- 42.Sacks, W. R., and P. A. Schaffer. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61:829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schang, L. M., A. Rosenberg, and P. A. Schaffer. 1999. Transcription of herpes simplex virus immediate-early and early genes is inhibited by roscovitine, an inhibitor specific for cellular cyclin-dependent kinases. J. Virol. 73:2161-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schenk, P., and H. Ludwig. 1988. The 65 K DNA binding protein appears early in HSV-1 replication. Arch. Virol. 102:119-123. [DOI] [PubMed] [Google Scholar]
- 46.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stow, E. C., and N. D. Stow. 1989. Complementation of a herpes simplex virus type 1 Vmw110 deletion mutant by human cytomegalovirus. J. Gen. Virol. 70:695-704. [DOI] [PubMed] [Google Scholar]
- 48.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67:2571-2585. [DOI] [PubMed] [Google Scholar]
- 49.Taylor, J. L., D. Unverrich, W. J. O'Brien, and K. W. Wilcox. 2000. Interferon coordinately inhibits the disruption of PML-positive ND10 and immediate-early gene expression by herpes simplex virus. J. Interferon Cytokine Res. 20:805-815. [DOI] [PubMed] [Google Scholar]
- 50.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao, F., and P. A. Schaffer. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69:6249-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]