This work identifies dioxobilin-type catabolites to represent more than 90% of the chlorophyll breakdown products in Arabidopsis. Their formation from formyloxobilin-type precursors is catalyzed by CYP89A9 in an unusual oxidative deformylation reaction. Our findings reveal a transformation in the complex pathway of chlorophyll breakdown that may also occur in other plant species.
Abstract
Nonfluorescent chlorophyll catabolites (NCCs) were described as products of chlorophyll breakdown in Arabidopsis thaliana. NCCs are formyloxobilin-type catabolites derived from chlorophyll by oxygenolytic opening of the chlorin macrocycle. These linear tetrapyrroles are generated from their fluorescent chlorophyll catabolite (FCC) precursors by a nonenzymatic isomerization inside the vacuole of senescing cells. Here, we identified a group of distinct dioxobilin-type chlorophyll catabolites (DCCs) as the major breakdown products in wild-type Arabidopsis, representing more than 90% of the chlorophyll of green leaves. The molecular constitution of the most abundant nonfluorescent DCC (NDCC), At-NDCC-1, was determined. We further identified cytochrome P450 monooxygenase CYP89A9 as being responsible for NDCC accumulation in wild-type Arabidopsis; cyp89a9 mutants that are deficient in CYP89A9 function were devoid of NDCCs but accumulated proportionally higher amounts of NCCs. CYP89A9 localized outside the chloroplasts, implying that FCCs occurring in the cytosol might be its natural substrate. Using recombinant CYP89A9, we confirm FCC specificity and show that fluorescent DCCs are the products of the CYP89A9 reaction. Fluorescent DCCs, formed by this enzyme, isomerize to the respective NDCCs in weakly acidic medium, as found in vacuoles. We conclude that CYP89A9 is involved in the formation of dioxobilin-type catabolites of chlorophyll in Arabidopsis.
INTRODUCTION
Senescence, the ultimate phase of leaf development in higher plants, is phenotypically defined by leaf yellowing, which results from the quantitative loss of chlorophyll. In many plant species, chlorophyll has been shown to be broken down in a multistep pathway to a group of structurally similar colorless linear tetrapyrroles, termed nonfluorescent chlorophyll catabolites (NCCs) (Kräutler and Matile, 1999; Moser et al., 2009; Hörtensteiner and Kräutler, 2011). NCCs are chlorophyll a-derived formyloxobilin-type linear tetrapyrroles where the chlorin macrocycle is oxygenolytically opened in the so-called northern meso-position. Thereby, the C5-carbon atom bridging pyrrole rings A and B is retained as formyl group attached to ring B (Kräutler et al., 1991) (Figure 1). This is in contrast with heme degradation by heme oxygenase, where the corresponding carbon atom is lost as CO, and biliverdin (a dioxobilin) is formed as degradation product (Unno et al., 2007).
Figure 1.
Structural Outline of Chlorophyll Breakdown with Representative Key Compounds.
In Arabidopsis, chlorophylls are degraded by a known pathway that is commonly present in other higher plant species. Thereby, a pFCC (the C1 epimer of epi-pFCC) that is formed inside senescing chloroplasts is ultimately converted to five NCCs (with different groups R1 to R3, for example, At-NCC-1, R1 = CH3, R2 = O-β-glucopyranosyl, and R3 = H), which are derived from isomerization of corresponding FCCs in the vacuole. Relevant catabolite intermediates are depicted along with the enzymes catalyzing their formation. Alternatively, as shown in this work, FCCs are converted by CYP89A9 to FDCCs and the latter isomerize into corresponding NDCCs. As an example, conversion of pFCC to pFDCC and further to pNDCC is depicted. Pyrrole rings (A and B) and relevant atoms are labeled in the structure of pFCC. For abbreviations, see text.
The pathway of chlorophyll breakdown can be divided into two parts: early reactions that take place within senescing chloroplasts and that result in the formation of a colorless primary fluorescent chlorophyll catabolite (pFCC) (Mühlecker et al., 1997). The reactions catalyzing chlorophyll-to-pFCC conversion are commonly present in land plants and therefore represent the core part of the pathway. The intermediates of breakdown upstream of pFCC are considered potent phototoxins, and metabolic channeling was proposed to occur among these early reactions to minimize the risk of phototoxicity (Kräutler and Matile, 1999; Sakuraba et al., 2012). The key reaction of the pathway is catalyzed by pheophorbide a oxygenase (PAO), a plant-specific Rieske-type monooxygenase, which catalyzes the already mentioned opening of the macrocycle of pheophorbide a, the Mg- and phytol-free intermediate of breakdown (Hörtensteiner et al., 1998; Pruzinská et al., 2003) (Figure 1). The product of the oxygenation reaction, red chlorophyll catabolite (Kräutler et al., 1997), is reduced to pFCC by red chlorophyll catabolite reductase in a stereo-specific manner. Thus, depending on the plant species as the source of red chlorophyll catabolite reductase one of two stereoisomers (i.e., pFCC and epi-pFCC) is formed (Mühlecker et al., 1997, 2000; Pruzinská et al., 2007).
The second part of the pathway of chlorophyll breakdown is characterized by largely species-specific modification at different peripheral positions within pFCC, giving rise to modified FCCs (Pruzinská et al., 2005; Moser et al., 2012) and hypermodified FCCs (Moser et al., 2008b; Hörtensteiner and Kräutler, 2011). The reactions catalyzing these modifications occur in the cytosol, implying that pFCC is exported from the chloroplast. After import of modified FCCs into the vacuole, spontaneous nonenzymatic isomerization to respective NCCs occurs because of the acidic pH of the vacuolar sap (Oberhuber et al., 2003). Thus, it is commonly accepted now that peripheral modifications present in NCCs (Figure 1) reflect modification reactions that occurred at the level of FCCs. In Arabidopsis thaliana, for example, five NCCs (At-NCCs) were identified that differ from each other at the C71, C82, and/or O134 position (R1, R2, and R3 in Figure 1) (Pruzinská et al., 2005; Müller et al., 2006). The occurrence of different NCCs in a given plant species has been explained by the possibility that the corresponding variety of modified FCCs are imported to the vacuole; however, to date, the affinities of the vacuolar transporters for such different FCCs have not been investigated.
As NCCs were found to accumulate in various senescent leaves, they were considered as the final breakdown products of chlorophyll, but the question whether they might be further metabolized in leaves was apparent (Ulrich et al., 2011). In senescent leaves of Cercidiphyllum japonicum, for example, a single NCC was estimated to represent more than 90% of the original chlorophyll content (Curty and Engel, 1996), but, for example, the major NCC of spinach (Spinacia oleracea) accounted for only 20% of the degraded chlorophyll (Berghold et al., 2002). In several cases, additional presumed breakdown products of chlorophyll were identified in senescent leaves (e.g., yellow- and pink-colored tetrapyrrolic catabolites) derived from the main NCC in C. japonicum (Moser et al., 2008a; Ulrich et al., 2011). In addition, two isomeric nonfluorescent dioxobilin-type chlorophyll catabolites (NDCCs) were identified in barley (Hordeum vulgare). These were thought to be formed by unspecific oxidation events from Hv-NCC-1 (Losey and Engel, 2001), the major NCC in barley (Kräutler et al., 1991). Recently, a structurally similar NDCC was identified in naturally senescent leaves of Norway maple (Acer platanoides), representing ∼50% of the chlorophyll present in green leaves, while NCCs were not detected (Müller et al., 2011). The molecular constitution of the maple NDCC was elucidated by spectroscopic means. These newer studies showed that NDCCs, rather than NCCs, are the major degradation products of chlorophyll in some senescent leaves, and they suggested that (N)DCCs were formed by still enigmatic, catabolic enzyme activities, rather than by nonphysiological degradation events.
Here, we show that in Arabidopsis NDCCs, not NCCs, are the major breakdown products of chlorophyll. We further identified cytochrome P450 monooxygenase CYP89A9 as being responsible for NDCC accumulation during Arabidopsis leaf senescence. Using a combination of genetic, biochemical, and metabolic approaches, CYP89A9 was shown to catalyze a previously unknown deformylation reaction of fluorescent chlorophyll catabolite (FCC)-type chlorophyll breakdown intermediates. These findings reveal a novel basic transformation in the complex pathway of chlorophyll breakdown that may not only be relevant in Arabidopsis, but might also occur in other plant species (Müller et al., 2011).
RESULTS
NDCCs Are the Major Chlorophyll Catabolites in Arabidopsis.
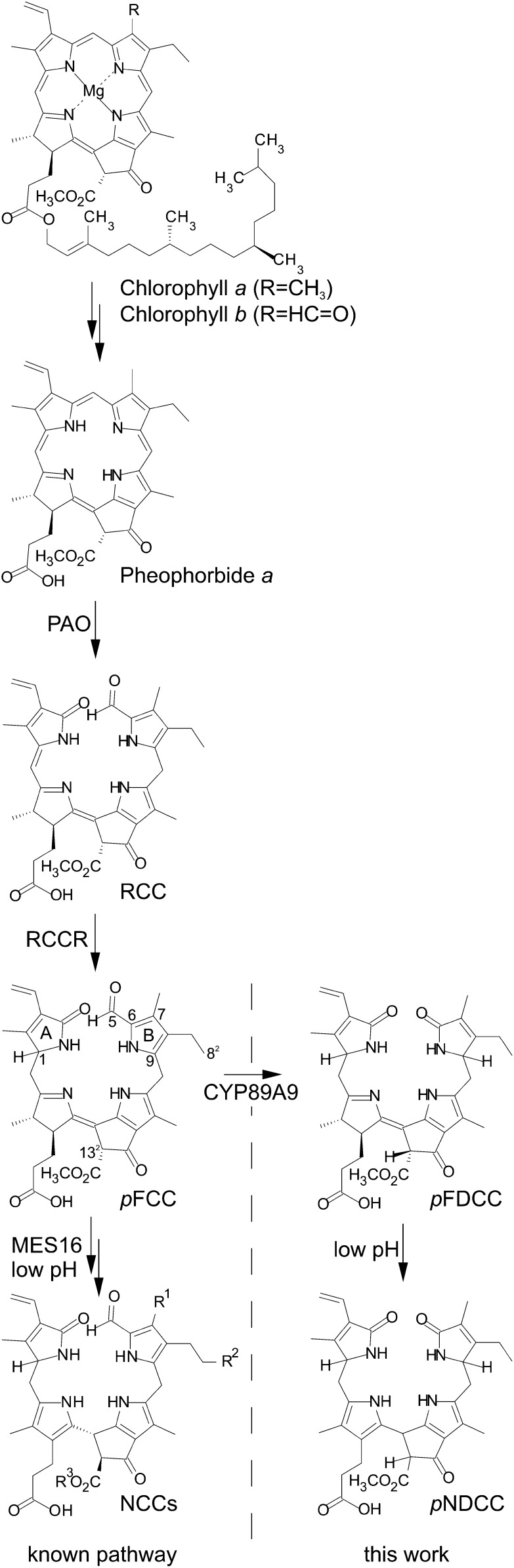
NCCs have been identified as breakdown products of chlorophyll that accumulate during leaf senescence in many angiosperms, including annual species and deciduous trees. However, in some cases, NDCCs were reported to represent products of chlorophyll breakdown. To explore the possibility that, besides NCCs (Pruzinská et al., 2005), NDCCs might also accumulate in the model plant Arabidopsis, we used HPLC to reanalyze extracts of senescent leaves. Leaf senescence was induced by dark treatment of detached leaves, a procedure that is well known to efficiently induce chlorophyll breakdown (Pruzinská et al., 2005; van der Graaff et al., 2006; Schelbert et al., 2009). Like other NCCs, At-NCCs exhibit a characteristic absorption maximum at around 315 nm and equally strong absorption at 254 nm (Figure 2A). When comparing Arabidopsis leaf extracts with detection at these two wavelengths, several fractions were identified that strongly absorbed at 254 nm but only weekly at 315 nm (Figure 2C). At least five of these fractions showed absorption spectra (Figure 2B; see Supplemental Figure 1A online) that were described for the Norway maple NDCC (Müller et al., 2011), indicating that they could represent related NDCCs (At-NDCCs) in Arabidopsis.
Figure 2.
NDCCs Are the Major Catabolites in Arabidopsis.
(A) UV absorption spectrum of At-NCC-1.
(B) UV absorption spectra of At-NDCC-1.
(C) Colorless catabolites of dark-incubated (8 d) leaves of Col-0 were separated by HPLC. A254 and A320 are shown. For clarity, only the relevant part of the HPLC traces is shown; inset, sums of NDCCs and NCCs accumulating during dark-induced senescence. Values are means of three replicates, and error bars represent sd.
(D) A 600-MHz 1H-NMR spectrum of At-NDCC-1 (in CD3OD, 283 K) and constitutional formula of At-NDCC-1. NMR signals indicating hydrogen atoms at C9 and C1 are labeled.
The most polar, major NDCC from Arabidopsis (At-NDCC-1) was thoroughly analyzed by spectroscopy methods (for detailed spectroscopic data, see Methods). At-NDCC-1 was revealed to represent a nonfluorescent dioxobilane-type catabolite (Figure 2D, inset) as follows. The molecular formula of At-NDCC-1 was determined as C33H38N4O8 by mass spectrometry (MS), by which the quasimolecular ion [C33H38N4O8+H]+ was observed at a mass-to-charge ratio (m/z) = 619.3. In the 1H-NMR spectra of At-NDCC-1 (in CD3OD, 10°C) (Figure 2D), signals of all 30 exchange-inert carbon-bound hydrogen atoms were observed. A singlet near 9 ppm was absent, which is a characteristic of the formyl hydrogen atom of NCCs (Kräutler et al., 1991). Instead, a multiplet at δ = 4.34 ppm and a double doublet at δ = 4.11 ppm indicated hydrogen atoms at positions C9 and C1, respectively, as is typical for NDCCs (Müller et al., 2011) (Figure 2D).
Both NDCC and NCC abundances increased during the course of leaf senescence; however, the amounts of NDCCs exceeded NCCs by a factor of about 10 (inset in Figure 2C). In addition, after 8 d of dark-induced senescence, NDCCs accounted for more than 75% of the degraded chlorophyll, demonstrating them to represent the by far most abundant type of chlorophyll catabolite in Arabidopsis.
CYP89A9 Is Required for NDCC Accumulation in Arabidopsis.
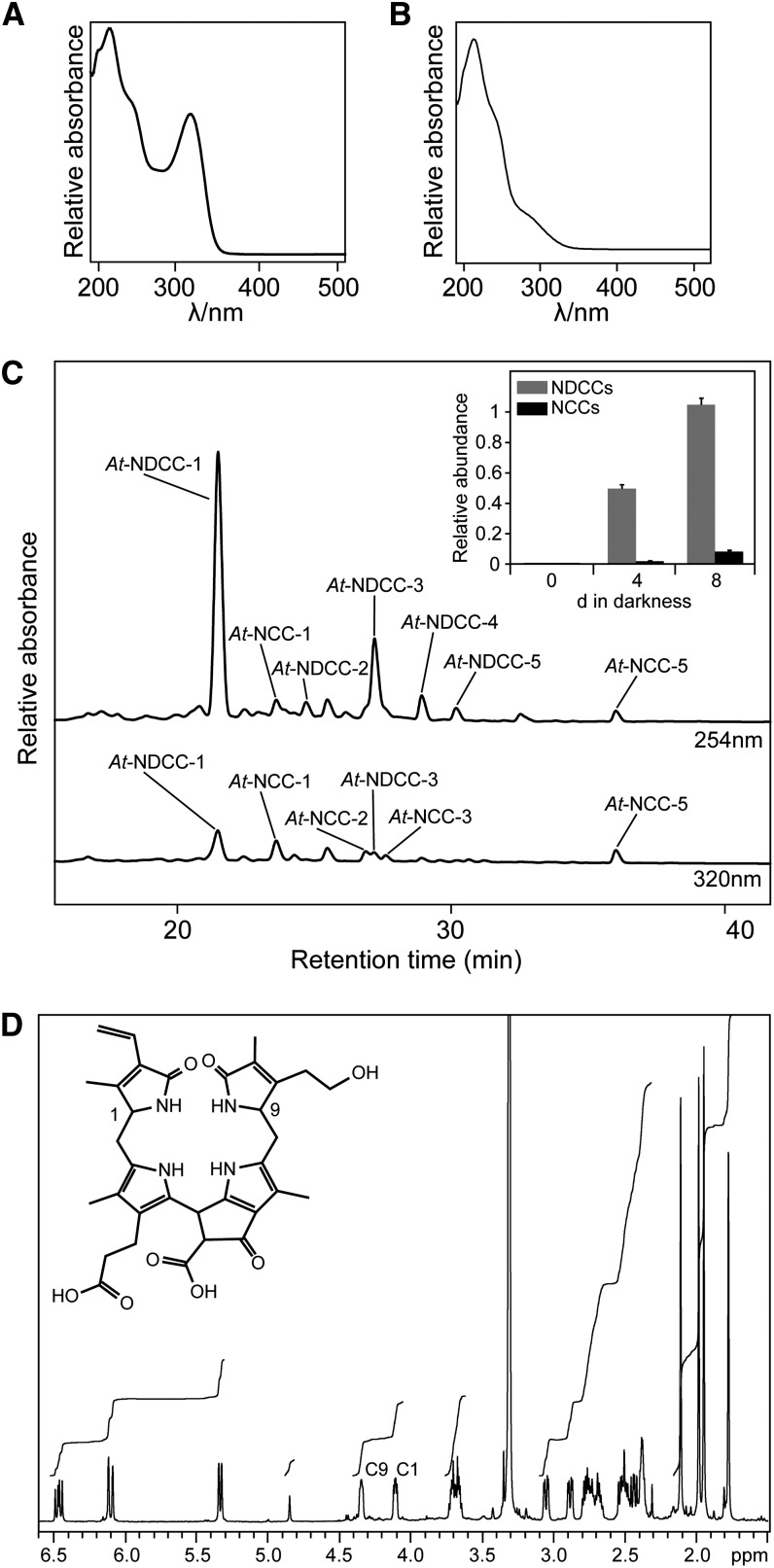
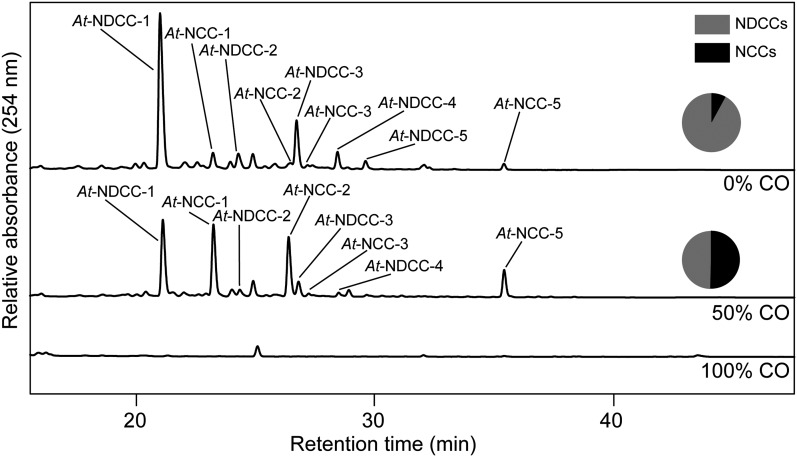
The identification of NDCCs as the major chlorophyll catabolites in Arabidopsis called for elucidating the mechanism of their formation. Two possible pathways were addressed: (1) formation from a chlorin-type substrate, such as chlorophyll or pheophorbide, whose macrocycle could be opened by a heme oxygenase-like reaction (i.e., under the loss of the C5-carbon atom); or (2) oxidative deformylation of the C5-formyl group present in FCCs and NCCs (Figure 1). We excluded the first possibility because pao1, an Arabidopsis PAO mutant, retains large quantities of chlorophyll during leaf senescence (Pruzinská et al., 2005). If NDCC formation would involve an activity for chlorin ring cleavage that is distinct from PAO, degreening should not be blocked in pao1. Alternatively, PAO itself could exhibit a heme oxygenase-like activity; however, in vitro activity studies clearly demonstrated a specificity of PAO toward FCC formation (Hörtensteiner et al., 1995). To identify likely candidates for C5-deformylation, we tested whether cytochrome P450 monooxygenases (P450s) could be involved. P450s have been shown to catalyze different types of reactions (Schuler et al., 2006; Mizutani and Ohta, 2010; Bak et al., 2011), including deformylation of C-bound formyl groups (Halkier, 1996; Guengerich, 2001). We exploited the ability of CO to effectively inhibit P450s (Schuler, 1996); when senescence was induced in detached leaves in an atmosphere composed of a 1:1 ratio (v/v) of CO and ambient air, the NDCC-to-NCC ratio was shifted to higher amounts of NCCs when compared with incubation at ambient air (Figure 3). This supported the idea that P450s could catalyze NDCC formation. Based on the fact that several genes encoding chlorophyll catabolic enzymes are highly expressed in senescent rosette leaves, cauline leaves, and petals and/or sepals (Zimmermann et al., 2004; Pruzinská et al., 2005), we reasoned that the genes for P450s involved in NDCC formation could be similarly induced. Using the Genes Search Tool of the Genevestigator platform (Zimmermann et al., 2004) and the above-mentioned tissues as targets, we found two P450 genes, CYP89A9 and CYP71B19, among the first hundred genes that are similarly expressed (see Supplemental Table 1 online). As expected, this gene list also contained PAO, STAY-GREEN, NONYELLOW COLORING1, and PHEOPHYTINASE, four senescence-regulated chlorophyll catabolic genes. To analyze the potential role of the two P450 candidate genes, we characterized chlorophyll breakdown in T-DNA insertion mutants (see Supplemental Figure 2A online) that fail to express the respective full-length gene transcripts (see Supplemental Figure 2B online). These lines did not show any obvious visible phenotype during normal development and high light treatment, and upon senescence induction, chlorophyll was degraded like in the wild type. During senescence, cyp71b19 mutants accumulated wild-type patterns of catabolites (see Supplemental Figure 2C online) and were not considered further. By contrast, both investigated cyp89a9 mutants did not accumulate NDCCs but had more than 10-fold increased levels of NCCs (Figure 4; see Supplemental Figure 3A online). In cyp89a9-1, NCCs accumulated up to 1.1 μmol per g fresh weight, which corresponded to more than 90% of degraded chlorophyll. UV/Vis (see Supplemental Figure 1B online) and MS analysis of the NCCs of cyp89a9-1 (for MS data, see Methods) confirmed them to be identical to the NCCs found in the Columbia-0 (Col-0) wild type (Pruzinská et al., 2005). The cyp89a9-2 mutation is in the Landsberg erecta (Ler) background, and Ler has been shown to be a natural METHYLESTERASE16 (MES16) mutant (Christ et al., 2012). MES16 is responsible for demethylation of the C132-carboxymethyl group of FCCs. Because of this, the NDCC/NCC catabolite patterns differed between Col-0 and Ler (Figure 4B; see Supplemental Figure 3A online). In addition, O134-desmethyl FCCs that accumulate in mes16 mutants (hence, also in Ler) were shown to isomerize more slowly to the corresponding NCCs (i.e., these FCCs accumulate and, as a consequence, the leaves will fluoresce) (Christ et al., 2012). To confirm that the catabolite pattern seen in cyp89a9-2 is due to the absence of both CYP89A9 and MES16, we crossed cyp89a9-1 with mes16-1 (a MES16 mutant in Col-0 background). In this line, chlorophyll catabolite patterns were identical to cyp89a9-2 (see Supplemental Figure 3 online).
Figure 3.
NDCC Formation Is Inhibited by CO.
Detached wild-type leaves were incubated in the dark for 5 d in glass containers containing 0, 50, and 100% (v/v) CO mixed with ambient air. Colorless catabolites were analyzed by HPLC. HPLC traces at A254 are shown. Pie graphs depict the relative amounts of NDCCS and NCCs. In 100% CO atmosphere, chlorophyll degradation was inhibited and no colorless catabolites were detected. For more details, see Methods.
Figure 4.
Colorless Catabolites Accumulating in Dark-Incubated (8 d) Leaves of cyp89a9 Mutants.
(A) Colorless catabolites of Col-0 and cyp89a9-1 were separated by HPLC. For clarity, only the relevant part of the A254 traces is shown. The inset shows chlorophyll degradation of Col-0 and cyp89a9-1 during dark-induced senescence.
(B) NDCC and NCC abundance in Col-0, cyp89a9-1, Ler, and cyp89a9-2. Values are means of three replicates, and error bars represent sd.
CYP89A9 Is a Fluorescent Chlorophyll Catabolite Deformylase
The NDCC deficiency of cyp89a9 mutants indicated that CYP89A9 may catalyze their formation in wild-type plants. To analyze this hypothesis, we tested in vitro activity of recombinant CYP89A9 expressed in Sf9 insect cells. FCCs were considered as likely substrates for CYP89A9 because when expressed as a fusion with green fluorescent protein in Arabidopsis mesophyll protoplasts, CYP89A9 localized outside chloroplasts and, as shown for most extraplastidial P450 enzymes (Schuler et al., 2006; Bassard et al., 2012), most likely to the endoplasmic reticulum (ER) (Figure 5). P450 activity requires a P450 reductase; therefore, ARABIDOPSIS THALIANA P450 OXIDOREDUCTASE1 (ATR1), one of the two Arabidopsis P450 reductases (Jensen and Møller, 2010), was expressed in Sf9 cells as well. Using epi-pFCC (Mühlecker et al., 2000) as substrate, four fractions of fluorescent compounds were obtained in a time-dependent manner in the presence of CYP89A9 (Figure 6A). They exhibited identical spectra with a prominent absorption maximum at around 360 nm (Figure 6B; see Supplemental Figure 1C online). MS analysis (for MS data, see Methods) demonstrated these compounds to be isomers of a constitutionally uniform fluorescent dioxobilin-type chlorophyll catabolite (FDCC) (Figure 6B), which we termed epi-pFDCCs because they derived from epi-pFCC. They likely differ in the configuration at C132 and C9 (Figure 6B); however, this was not further investigated. To test the substrate specificity of CYP89A9, assays were performed also with mes16-FCC-2 (pFCC with an additional hydroxyl group at C82) and the corresponding NCC, mes16-NCC-2 (Christ et al., 2012) (Figure 6C). Only the FCC, but not the isomeric NCC, was converted by CYP89A9, supporting the specificity of CYP89A9 for FCCs. Under acidic conditions, the epi-pFDCCs isomerized into NDCCs (Figure 6D; see Supplemental Figure 1 online), indicating that in vivo formation of NDCCs from corresponding FDCCs might occur after their import into the vacuole.
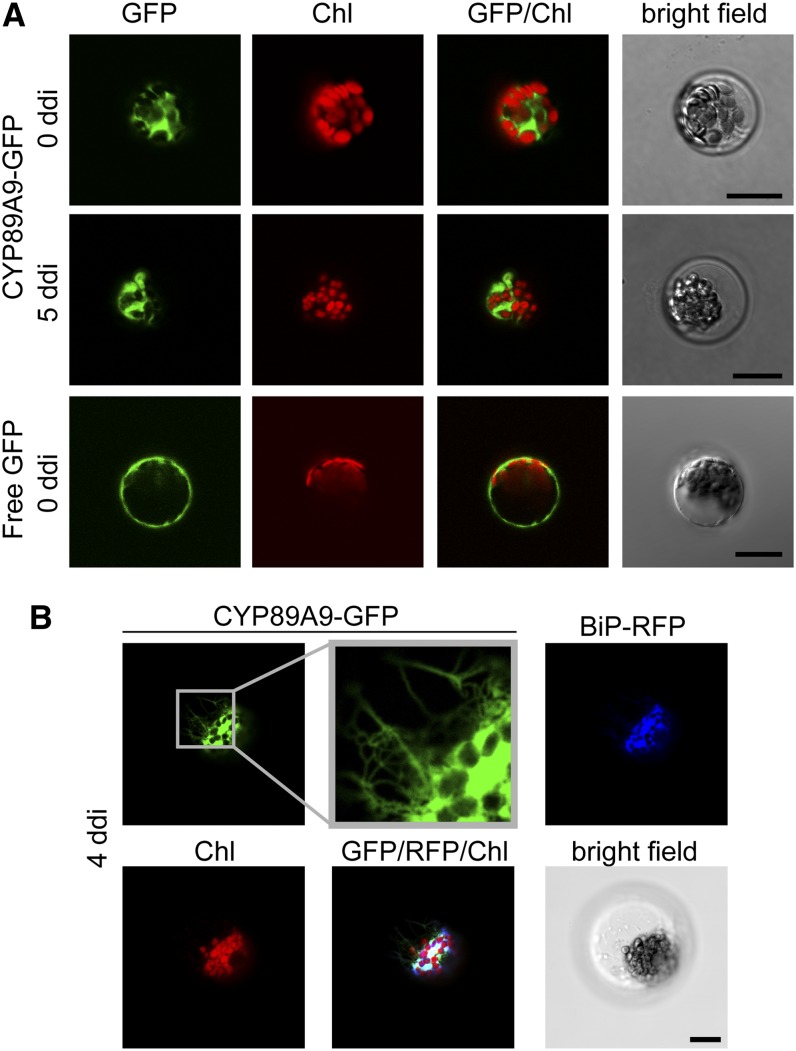
Figure 5.
CYP89A9-GFP Likely Localizes to the ER in Arabidopsis Mesophyll Protoplasts.
(A) Transient expression of CYP89A9-GFP and free GFP in protoplasts isolated from green (0 d of dark incubation [ddi]) or senescent (5 ddi) leaves.
(B) Colocalization of CYP89A9-GFP with BiP-RFP in protoplasts isolated from senescent (4 ddi) leaves. GFP, RFP, and chlorophyll autofluorescence was examined by confocal laser scanning microscopy. Note the ER-like membrane network visible in the GFP zoom.
Bars = 20 μm.
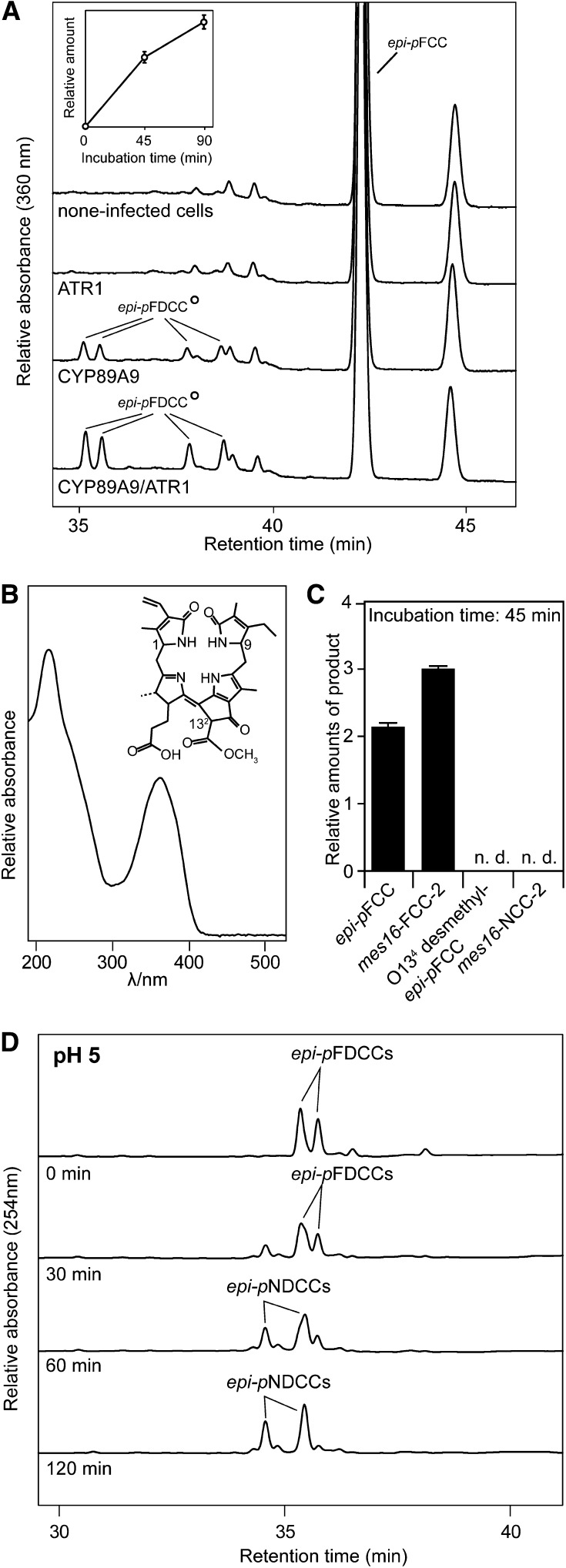
Figure 6.
Analysis of Recombinant CYP89A9.
(A) HPLC analysis of assays employing Sf9 microsomes expressing CYP89A9 and ATR1, and epi-pFCC as substrate. HPLC traces at A360 after 90 min of incubation at 28°C are shown. For clarity, only the relevant part of the HPLC traces is shown. The inset shows the relative amounts of formed epi-pFDCCs. Values are means of three replicates. Error bars indicate sd.
(B) UV absorption spectrum of epi-pFDCCs produced with recombinant CYP89A9 and chemical constitution of (epi-)pFDCCs. Relevant stereocenters are labeled.
(C) Substrate specificity of CYP89A9. Values are means of three replicates. Error bars indicate sd.
(D) FDCC-to-NDCC isomerization assays. The two most polar epi-pFDCCs produced with recombinant CYP89A9 were incubated at pH 5.0 for up to 120 min and analyzed by HPLC. A254 is shown.
FCC Deformylation Precedes FCC Demethylation
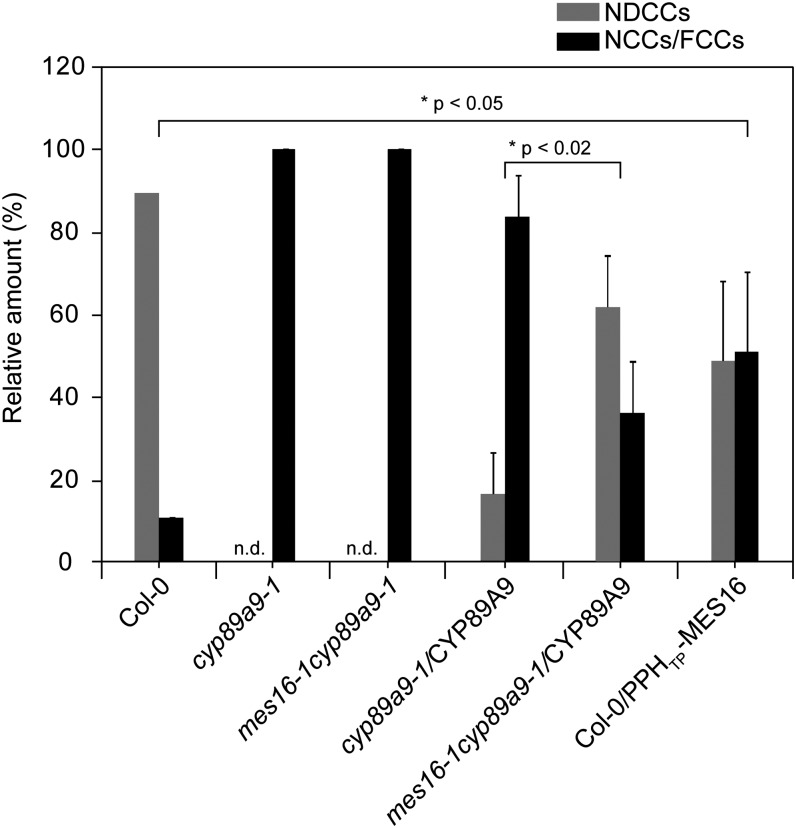
During our analysis of CYP89A9 substrate specificity in vitro, it turned out that the enzyme did not convert O134-demethylated epi-pFCC to the respective FDCC (Figure 6C). This indicated that, although both CYP89A9 and MES16 act in the cytosol and therefore could compete for the same substrate in vivo, deformylation by CYP89A9 might precede catabolite demethylation by MES16. To investigate this in more detail, we used a line (Col-0/PPHTP-MES16) in which MES16 was targeted to the chloroplast (Christ et al., 2012) (i.e., demethylation was allowed to occur before FCC export to the cytosol). In this line, the NDCC-to-NCC ratio was significantly shifted toward more NCCs compared with Col-0 (Figure 7; see Supplemental Figure 4 online). This supports the idea that in Col-0 wild-type plants, CYP89A9 precedes MES16 action to a large extent leading to the formation of NDCCs, while only a small fraction of catabolites may be first demethylated, thus ending in the formation of NCCs. Further evidence for this was obtained by CYP89A9 complementation tests in cyp89a9 mutants. When analyzing the relative amounts of NDCCs and NCCs as a measure of complementation, absence of MES16 in the mes16-1 cyp89a9-1 double mutant resulted in more efficient complementation compared with (the MES16-containing line) cyp89a9-1 (Figure 7).
Figure 7.
FCC Deformylation Precedes FCC Demethylation.
Colorless catabolites accumulating in dark-incubated (7 d) leaves of Col-0, cyp89a9-1, mes16-1 cyp89a9-1, cyp89a9-1/CYP89A9, mes16-1cyp89a9-1/CYP89A9, and Col-0/PPHTP-MES16 were separated by HPLC. Relative amounts are means of n replicates, and error bars represent se. Col-0, cyp89a9-1, and cyp89a9-1 mes16-1, n = 3; cyp89a9-1/CYP89A9 and Col-0/PPHTP-MES16, n = 6; cyp89a9-1mes16-1/CYP89A9, n = 5. For the transgenic lines, replicates correspond to independent transformation events. The significance of differences between Col-0 and Col-0/PPHTP-MES16 and between cyp89a9-1/CYP89A9 and mes16-1cyp89a9-1/CYP89A9 was determined using a one-tailed t test with Welch correction. n.d., not detected.
DISCUSSION
NCCs have been known for more than 20 years to represent colorless products of chlorophyll breakdown during leaf senescence (Kräutler et al., 1991). They are seemingly end products of a multistep pathway (Hörtensteiner and Kräutler, 2011) that involves an acid-catalyzed isomerization of intermediary FCCs inside the vacuoles of senescing cells (Oberhuber et al., 2003). Both FCCs and NCCs are linear tetrapyrroles that can be classified as formyloxobilin-type tetrapyrroles because the C5-carbon, bridging pyrrole rings A and B in chlorophyll, is present as formyl group. Here, we show that, in Arabidopsis, NCCs are only minor products of chlorophyll breakdown and up to 90% of chlorophyll is metabolized to a different type of catabolites, termed here as dioxobilin-type chlorophyll catabolite (DCCs). Compared with NCCs, NDCCs miss the C5-carbon (Figure 1). In a few reports, NDCCs were observed as presumed chlorophyll degradation products (Losey and Engel, 2001; Djapic and Pavlovic, 2008; Djapic et al., 2009; Müller et al., 2011), but their means of formation remained unclear. Here, we show that Arabidopsis CYP89A9 specifically deformylates FCCs to the corresponding FDCCs and thus defines the biochemical basis for the generation of DCCs (i.e., FDCC and NDCC) (Figure 1). In Arabidopsis, DCCs and NCCs account for almost all chlorophyll that is present in green leaves, indicating that these catabolites are not metabolically recycled or further degraded for use as carbon or nitrogen source. This is similar to other plant species, such as Norway maple, C. japonicum, and oilseed rape (Brassica napus), where DCCs/NCCs were calculated to represent the major fraction of degraded chlorophyll (Mühlecker and Kräutler, 1996; Kräutler and Matile, 1999; Müller et al., 2011).
The identification of NDCCs as major degradation products of chlorophyll in Arabidopsis is intriguing because structurally they are closely related to dioxobilins, which are products of heme degradation. Hence, regarding the structures of the main catabolites, chlorophyll breakdown in Arabidopsis resembles heme degradation (Müller et al., 2011). Mechanistically, however, DCC formation totally differs from the latter. Heme oxygenase uses the iron-containing porphyrin, heme, as substrate for oxygenolytic ring opening and extrusion of the meso-carbon as CO. Thereby, the dioxobilin biliverdin is directly formed (Unno et al., 2007). By contrast, during chlorophyll breakdown, PAO catalyzes chlorin ring opening of the metal-free breakdown intermediate pheophorbide a (Pruzinská et al., 2003) (Figure 1). Thereby, the C5-carbon is retained in FCCs and NCCs. The C5-carbon is only subsequently removed by oxidative deformylation of FCCs through the here discovered P450 monooxygenase, yielding dioxobilin-type FDCCs. In analogy to FCC-to-NCC isomerization (Oberhuber et al., 2003), FDCCs isomerize to NDCCs under acidic conditions (Figure 6).
The key role of CYP89A9 in DCC formation is supported by the fact that cyp89a9 mutants did not accumulate NDCCs but contained proportionally higher quantities of NCCs (Figure 4). This also implied that NDCCs and NCCs derive from a common source of precursors. Interestingly, although CYP89A9 specifically reacted with FCCs, it was not equally active toward different tested FCCs (Figure 6). Thus, O134-desmethyl epi-pFCC was not deformylated, despite the fact that almost all NCCs (Pruzinská et al., 2005) and At-NDCC-1, structurally characterized here (Figure 2), contain a free C132 carboxyl group. This indicates that CYP89A9 activity precedes demethylation by MES16 in vivo. In agreement with this idea is the finding that wild-type plants in which MES16 was targeted to the chloroplast and consequently demethylated FCCs before export from the organelle and exposure to CYP89A9, exhibited a cyp89a9 mutant–like phenotype (i.e., abundance of NCCs was substantially increased) (Figure 7). There is increasing evidence in the literature that chloroplasts and the ER are associated through defined membrane contact sites (Andersson et al., 2007; Tan et al., 2011). Such contact sites have been shown to have an important function for lipid transfer between chloroplast and the ER and might involve protein–protein interaction between the partaking membranes. It is reasonable to assume that with regard to chlorophyll breakdown, pFCC released from senescing chloroplasts might directly be transferred to CYP89A9, which most likely localizes to the ER (Figure 5). Thus, NCCs found in the wild type might be the products of FCCs that escaped CYP89A9 and were demethylated first.
Cytochrome P450 monooxygenases are encoded by one of the largest gene families in plants with 244 members in Arabidopsis (Bak et al., 2011). Based on phylogenetic relationships, CYP clans have been defined. CYP89A9 belongs to the highly divergent CYP71 clan, representing almost 50% of all known plant CYPs (Nelson and Werck-Reichhart, 2011). The CYP89 family first appeared in the Magnoliids (i.e., in early angiosperms) and shares a common ancestor with the most closely related CYP77 family. CYP71 clan members possess a great diversity of functions, including metabolism of amino acid derivatives, isoprenoids, alkaloids, fatty acids, and hormones and are often involved in plant–insect interactions (Mizutani and Ohta, 2010; Nelson and Werck-Reichhart, 2011; Schuler, 2011). Our work extends the substrate spectrum of CYPs to linear tetrapyrroles. It is tempting to speculate that CYP89 orthologs catalyze catabolite deformylation also in other plant species. Indeed, besides the Brassicales (Arabidopsis), CYP89 genes are present in several plant taxa (Nelson and Werck-Reichhart, 2011) containing species that were shown to produce NDCCs, such as Rosales (Norway maple; Müller et al., 2011) and Liliopsida (barley; Losey and Engel, 2001), but not Saxifragales (Parrotia persica; Djapic and Pavlovic, 2008). Within Arabidopsis, CYP89A9 is at the base of phylogeny of the seven-member CYP89 family (Bak et al., 2011), indicating that its recruitment for chlorophyll catabolism occurred rather early in evolution. Functions for the six CYP89A9 paralogs remain unknown. Based on the fact that cyp89a9 mutants are devoid of any DCCs (Figure 4), we assume that none of these paralogs is involved in FCC deformylation. However, CYP89A2 is within a wider network of genes that are coregulated with CYP89A9 (Obayashi et al., 2009) and clusters with CYP89A5, CYP89A6, CYP89A5, and two UDP-glucosyltransferase genes on chromosome 1 of Arabidopsis. Structures of At-NCCs (Pruzinská et al., 2005) indicate catabolite modifications at C82 that may require additional P450 and glucosyltransferase activities, but it remains to be shown whether CYP89A9 paralogs are involved. Furthermore, many P450s are coregulated with genes related to chloroplast function, such as photosynthesis and phytohormone metabolism (Ehlting et al., 2008). It can be speculated that additional P450s with so far unknown function may be involved in chlorophyll metabolism. This may be the case also in other plant species; for example, P450 enzymes could be responsible for dihydroxylation of the C3 vinyl group as found in NCCs from barley, spinach, and maize (Zea mays) (Hörtensteiner and Kräutler, 2011).
Chemically, the conversion of FCCs to FDCCs is an oxidative deformylation. Several P450s are known that deformylate their substrates, thereby forming alkenes as products (Roberts et al., 1991; Guengerich, 2001). However, the proposed reaction of CYP89A9, producing a γ-lactam upon deformylation of an α-formyl-pyrrole unit, differs substantially from the previously described deformylations. In contrast with heme degradation, the by-product of the CYP89A9 reaction most probably is formate, which in plants can be metabolized either by formate dehydrogenase to CO2 and water (Li et al., 2002) or enter C1-metabolic pathways via conjugation to tetrahydrofolate (Chen et al., 1997). A result of the observed deformylation process is the new formation of a stereocenter at C9 of DCCs (Figure 2D). It is most likely generated in a multistep mechanism, thus allowing for the formation of two stereoisomers at C9 (Figure 6B). This explains the occurrence in in vitro CYP89A9 assays of several isomeric FDCCs (Figure 6A). C9-epimeric NDCCs were identified in senescent barley leaves (Losey and Engel, 2001). By contrast, Norway maple accumulates a single, stereochemically uniform NDCC (Müller et al., 2011), indicating that depending on the plant species and/or conditions, DCC formation might or might not be stereoselective.
More than 20 years ago, the first NCC was identified in senescent barley leaves and characterized as a formyloxobilane-type tetrapyrrole (Kräutler et al., 1991). Since then, NCCs have been isolated from many different plant species (Kräutler, 2008; Hörtensteiner and Kräutler, 2011). By contrast, Norway maple was observed to accumulate one NDCC exclusively, but NCCs were absent (Müller et al., 2011), while in Arabidopsis, as shown here, both forms of catabolites occur simultaneously. Considering that P450-dependent reactions consume energy and absence of CYP89A9 activity in respective mutants does not cause an obvious phenotype that may indicate an important function for the plant, one may ask the question, why (for example) does Arabidopsis specifically recruited CYP89A9 for (ultimately) NDCC formation, while other species can degrade chlorophyll without such activity? Modifications of pFCC that occur at different peripheral positions increase its polarity. This is seen in reversed-phase HPLC, where all catabolites downstream of pFCC elute before pFCC. At-NDCC-1 is the most polar of all NDCCs and NCCs in Arabidopsis (Figure 2B). Thus, it could be argued that CYP89A9, like other pFCC-modifying activities (Pruzinská et al., 2005), helps to increase the polarity of catabolites, required for storage inside the vacuolar sap (Matile et al., 1988). In addition, it has been shown that chlorophyll is broken down in order to detoxify this potentially phototoxic pigment when the photosynthetic machinery is dismantled during senescence (Hörtensteiner, 2004). Thus, a key feature of chlorophyll breakdown is to abolish light absorption. In this respect, CYP89A9 activity is responsible for the loss of absorption around 320 nm, characteristic for the NCCs, and leaves behind colorless pigments that also display low-light absorption in the UV-A and UV-B ranges.
In summary, we identified NDCCs as the major chlorophyll catabolites in wild-type Arabidopsis, while NCCs represent only a minor fraction. NDCCs are the products of acid-catalyzed isomerization from FDCCs. FDCCs are biosynthesized from FCCs by CYP89A9, a P450 enzyme of Arabidopsis, in an unprecedented oxidative deformylation reaction. Catabolite deformylation does not seem to occur in all plant species, but it remains to be shown how widely distributed NDCCs are as novel final degradation products of chlorophyll.
METHODS
Plant Material and Senescence Induction
Arabidopsis thaliana ecotype Col-0 was used as the wild type. Ecotype Ler was also analyzed in experiments including cyp89a9-2 (Ler background). Ler is a natural mutant in the MES16 gene (Christ et al., 2012). T-DNA insertion lines were from the following collections: SALK lines (Alonso et al., 2003), AT3G26170-2 (cyp71b19-2), SALK_149952; SAIL lines (Sessions et al., 2002), AT3G26170-1 (cyp71b19-1), SAIL_1165_B02; John Innes Centre suppressor mutator lines (Tissier et al., 1999), AT3G03470-1 (cyp89a9-1), SM_3_39636; John Innes Centre gene trap lines (Sundaresan et al., 1995), AT3G03470-2 (cyp89a9-2), GT_5_22744. SALK, SAIL, SM, and GT lines were obtained from the European Arabidopsis Center, Nottingham, UK. Homozygous plants were identified by PCR using T-DNA–, transposon-, and gene-specific primers as listed in Supplemental Table 2 online. Likewise, a homozygous mes16-1 cyp89a9-1 double mutant was identified by PCR.
Plants were grown on soil either in 8-h-light/16-h-dark (short day) or 12-h-light/12-h-dark (12:12) photoperiods under fluorescent light of 60 to 120 µmol photons m−2 s−1 at 22°C or at 16 h light/8 h dark (long day) in a greenhouse with fluence rates of 100 to 200 µmol photons m−2 s−1 at 22°C. For senescence induction, leaves from 8-week-old (short day) or 5-week-old (12:12 and long day) plants were excised and incubated in permanent darkness on wet filter paper for up to 8 d at ambient temperature. For dark incubation in CO atmosphere, leaves from short-day-grown Col-0 plants were excised and incubated for 5 d in glass containers containing 0, 50, and 100% (v/v) CO mixed with ambient air.
Biocomputational Methods
Search for coexpressed genes in senescent rosette leaves, cauline leaves, petals, and sepals was performed with the Gene Search Tool Anatomy of the Genevestigator platform (Zimmermann et al., 2004) using a limit of 100 genes.
Green Fluorescent Protein Fusion Production and Confocal Microscopy
CYP89A9 was amplified by PCR with the Expand high-fidelity PCR system (Roche Applied Science) from clone U67778 (ABRC) using the primers listed in Supplemental Table 2 online, introducing XmaI-NheI restriction sites at the ends. The PCR fragment was then cloned into the pGEM-T-easy vector (Promega) and after XmaI-NheI digest subcloned into pUC18-spGFP6 (Meyer et al., 2006), thereby producing a fusion of CYP89A9 with the N terminus of green fluorescent protein (GFP). Arabidopsis mesophyll protoplasts were isolated from 6-week-old short-day-grown Col-0 plants according to published procedures (Endler et al., 2006; Schelbert et al., 2009). Cell numbers were quantified with a Neubauer chamber and adjusted to a density of 2 to 3 × 106 protoplasts mL−1. CYP89A9-GFP was cotransformed with the ER marker BiP-RFP (for red fluorescent protein) (Min et al., 2007) in protoplasts isolated from green or dark-induced senescent leaves by 20% polyethylene glycol transformation (Meyer et al., 2006). Free GFP expressed from empty pUC18-spGFP6 was used as a control for cytosolic localization. Transformed cells were incubated for 24 h in the dark at room temperature before laser scanning confocal microscopy (Leica TCS SP5; Leica Microsystems). GFP and RFP fluorescence were imaged at an excitation wavelength of 488 and 561 nm, respectively. The detection channel windows were set as follow: 495 to 530 nm (GFP), 593 to 619 nm (RFP), and 643 to 730 nm (chlorophyll autofluorescence).
RNA Isolation and RT-PCR
RNA was isolated using the RNeasy plant kit (Qiagen) from green and dark-incubated leaves of plants grown in the short-day photoperiod. After DNA digestion with RQ1 DNase (Promega), first-strand cDNA was synthesized using M-MLV reverse transcriptase (Promega). PCR was performed with nonsaturating numbers of amplification cycles as shown in Supplemental Figure 2 online using gene-specific primers as listed in Supplemental Table 2 online.
epi-pFCC, O134-Desmethyl epi-pFCC, mes16-FCC-2, and NCC Preparation
The preparation of epi-pFCC (from Capsicum annuum fruit extracts) is described elsewhere (Christ et al., 2012). O134-desmethyl epi-pFCC was produced from epi-pFCC with MES16 (Christ et al., 2012). mes16-FCC-2 and mes16-NCC-2 were extracted from dark-incubated leaves (5 d) of mes16-1cyp89a9-1 grown in 12:12 photoperiod in 50 mM phosphate buffer, pH 7/methanol (1:3, v/v), concentrated using C18-SepPak cartridges (Waters) and purified by HPLC.
Heterologous Expression of CYP89A9 and ATR1, and Activity Determination
ATR1 and CYP89A9 were amplified by PCR (Expand high-fidelity PCR system) from clone pda02355 obtained from the RIKEN resource (Seki et al., 2002) and clone U67778 (ABRC), respectively, using the primers listed in Supplemental Table 2 online and then cloned into pFastbac1 (Invitrogen) via EcoRI. CYP89A9 and ATR1 were also cloned in pFastbac DUAL (Invitrogen) using KpnI and EcoRI restriction sites, respectively, allowing simultaneous expression of the two genes with one bacmid. After sequencing, the constructs were used for the preparation of recombinant bacmid DNAs by transformation of Escherichia coli strain DH10Bac (Invitrogen). Insect cell transfection and recombinant protein expression were conducted as outlined in the bac-to-bac manual (Invitrogen), using Sf9 cells grown in monolayer cultures. For expression of the recombinant proteins, Sf9 cells were maintained in BD BaculoGold TNM-FH insect medium (BD Biosciences) supplemented with 200 mM 5-aminolevulinic acid and 200 mM ferrous sulfate to increase the low heme synthetic capacity of the insect cells (Saito et al., 2004). For preparation of microsomal fractions, infected cells were concentrated by centrifugation and resuspended in lysis buffer consisting of 20 mM potassium phosphate, pH 7.3, 20% (v/v) glycerol, 1 mM EDTA, and 1 mM DTT complemented with a protease inhibitor cocktail (Complete; Roche Applied Science). The cells were homogenized on ice with a glass-Teflon homogenizer, and cell debris was removed by centrifugation at 1000g for 10 min. The supernatant was further centrifuged at 100,000g for 1 h. The pellet (microsomal fraction) was homogenized in lysis buffer with a syringe through a 0.5-mm-wide needle, frozen in liquid nitrogen, and stored at −80°C until use.
Standard assays with CYP89A9 and ATR1 microsomes (total volume 100 µL) consisted of 40 µg of microsome proteins, 10 µM of substrate, and 0.1 mM NADPH. After incubation at 28°C in darkness for different periods of time as indicated in Figure 6, the reactions were stopped by the addition of methanol to a final concentration of 50%. After centrifugation (2 min at 16,000g), samples were analyzed by HPLC as described below. Microsomes produced with bacmids generated from pFastbac1 constructs were used for standard assays. Noninfected cells or cells infected only with ATR1 bacmid were used as controls. For scaling up the standard assay to produce higher amounts of products obtained when using epi-pFCC as substrate, microsomes generated with the pFastbac DUAL construct were used. epi-pFDCCs were purified by HPLC prior to MS analysis (see below).
Analysis of Chlorophyll and Chlorophyll Catabolites
Quantification of Chlorophyll
Pigments were extracted from liquid nitrogen–homogenized tissue during 2 h at −20°C in 10% (v/v) 0.2 M Tris-HCl, pH 8.0, in acetone, precooled to –20°C (5 mL g−1 fresh weight). After centrifugation twice (4 min, 16,000g, 4°C), supernatants were analyzed by spectrophotometry (Strain et al., 1971).
Colorless Chlorophyll Catabolites
Plant material was ground in liquid nitrogen, and colorless catabolites were extracted with 3 volumes (w/v) of 50 mM phosphate buffer (P-buffer), pH 7/methanol (1:3, v/v), and analyzed by HPLC as described (Christ et al., 2012).
Peak detection was performed with sequential monitoring using a PA-100 photodiode array detector (200 to 700 nm; ThermoFisher Scientific) and a RF2000 fluorescence detector (excitation at 320 nm, emission at 450 nm; ThermoFisher Scientific). Chlorophyll catabolites were identified by their absorption (FCCs, FDCCs, NCCs, and NDCCs) and fluorescence (FDCCs and FCCs) properties. Relative amounts of FCCs, FDCCs, NCCs, and NDCCs were determined by peak areas at 254 nm using the approximation that absorption at this wavelength is similar in the different types of catabolites. In addition, NCCs were quantified at 320 nm (Oberhuber et al., 2001).
cyp89a9-1 and mes16-1cyp89a-1 Complementation
CYP89A9 was amplified by PCR as described above and cloned via EcoRI-HindIII into pHannibal (Wesley et al., 2001). The pHannibal cassette containing cauliflower mosaic virus 35S promoter, the CYP89A9 open reading frame, and an OCS terminator was excised with NotI and introduced into NotI-restricted pGreen0179 (Hellens et al., 2000). Arabidopsis cyp89a9-1 and mes16-1cyp89a9-1 plants were transformed by the floral dip method (Clough and Bent, 1998). Transformants were selected on hygromycin, and resistant T2 plants were used for further analysis.
In Vitro FDCC-to-NDCC Isomerization
The isomerization assays consisted of 11.2 µM of purified epi-pFDCCs produced with recombinant CYP89A9 (see above) and 80 mM phosphate buffer, pH 5. Aliquots were taken after incubation times as indicated in Figure 6D, and isomerization was stopped by adding Tris-HCl, pH 8.0, to a concentration of 350 mM. Methanol was added to a final concentration of 35% (v/v) prior to analysis by HPLC as described above.
MS and NMR
Spectroscopy
For spectroscopic analyses, the following instruments and settings were employed: UV/VIS spectra: Hitachi U-3000 spectrophotometer; λmax (nm) (εrel). CD spectra: JASCO J715; λmin/max (nm) (Δε). 1H- and 13C-NMR: Bruker UltraShield 600 MHz Avance II+ [δ (C1HD2COD) = 3.31 ppm, δ(13CD3OD) = 49.0 ppm] (Gottlieb et al., 1997). MS: Finnigan MAT 95, electrospray ionization (ESI) source, positive ion mode, 1.4-kV spray voltage (At-DNCC-1); Finnigan LCQ classic, ESI source, positive ion mode, spray voltage 4.25 kV, solvent methanol/water (10 mM NH4OAc) 1:1 (v/v); m/z (percentage of intensity, type of ion).
HPLC
The HPLC used for MS analysis was a Hewlett Packard series 1100 HPLC system with an online degasser, an Agilent quaternary pump, a diode array detector (DAD) and a fluorescence detector (FLD).
Analytical HPLC
For analytical HPLC the system was equipped with an injection loop (200 μL; Rheodyne valve), and a Phenomenex hyperclone ODS (5 μm; 250 × 4.6 mm i.d.) column (at room temperature) connected to a Phenomenex ODS (4 × 3 mm i.d.) pre-column. The column was eluted at room temperature at a flow rate of 0.5 ml min−1 with solvent A (MeOH) and solvent B (P-buffer, pH 7) using the following solvent composition (A/B, v/v; as a function of time): 0 to 5 min, 20/80; 5 to 55 min, 20/80 to 70/30; 55 to 60 min, 70/30 to 100/0; 60 to 70 min, 100/0; 70 to 75 min, 100/0 to 80/20. For the separation of the four products of CYP89A9/ATR1 assays, solvent A (MeOH) and solvent B (10 mM ammonium acetate buffer) were used with the following solvent composition (A/B, v/v; as a function of time): 0 to 5 min, 30/70; 5 to 55 min, 30/70 to 65/35; 55 to 70 min, 65/35 to 75/25; 70 to 80 min, 75/25 to 100/0; 80 to 90 min, 100/0; 90 to 95 min, 100/0 to 30/70.
Preparative HPLC
For preparative HPLC, a Phenomenex HyperClone ODS (5 μm; 250 × 21.2 mm i.d.) column protected with a Phenomenex ODS (10 mm × 5 mm i.d.) pre-column was used. The column was eluted at room temperature with a flow rate of 5 ml min−1 with solvent A (MeOH) and solvent B (P-buffer, pH 7) with the following solvent composition (A/B, v/v; as a function of time): 0 to 5 min, 25/75; 5 to 123 min, 25/75 to 38/62; 123 to 200 min, 38/62 to 64/36; 200 to 210 min, 64/36 to 100/0; 210 to 230 min, 100/0.
Spectroscopy Characterization of At-NDCC-1
Senescent leaves (16.5 g) of Arabidopsis were ground in a mortar in liquid nitrogen, mixed with sea sand, and extracted with 16 mL of methanol. The obtained slurry was filtrated through a Buchner funnel, and the extraction with methanol was repeated seven times. The collected green extract (160 mL) was diluted with 30 mL P-buffer, pH 7.0, and washed twice with 160 mL n-hexane. The solution was reduced under vacuum to 70 mL, diluted with 600 mL P-buffer, pH 7.0, and filtrated. The mixture was loaded on a Sep-Pak Vac 20cc (5 g) C18 cartridge (Waters), washed with water (50 mL), and eluted with methanol (15 mL). The solvent was removed under reduced pressure on a rotary evaporator. The crude product was dissolved in 800 μL methanol/P-buffer (50:50, v/v), diluted with 500 μL P-buffer, and centrifuged for 5 min at 13,000 rpm. The clear brown solution was injected into the preparative HPLC. The At-NDCC-1–containing fraction was collected, diluted with 4 volumes of water, and applied to a Sep-Pak classic C18 cartridge. The At-NDCC-1 was obtained salt-free by washing with 20 mL of water and eluting with 5 mL of methanol. The solvents were removed in vacuum to give 0.770 mg of At-NDCC-1.
The following spectroscopic data were obtained: UV/Vis (methanol, 6.9 × 10−4 M) λmax (εrel): 288 sh (0.29), 239 sh (1.00), 221 (1.29); CD (methanol, 6.9 × 10−4 M) λmin/max (Δε): 311 (5.3), 284 (−26.4), 250 (7.1), 231 (17.1). 1H-NMR (600 MHz, CD3OD, 283 K): δ [ppm] = 1.78 [s, CH3 (71)], 1.95 [s, CH3 (181)], 1.99 [s, CH3 (21)], 2.11 [s, CH3 (121)], 2.38 [m, CH2 (172)], 2.45 [dd, J = 14.4/5.1 Hz, CHA (20)], 2.50 [m, CHA (81)], 2.54 [m, CHA (10)], 2.69 [m, CHA (171)], 2.75 [m, CHB (171)], 2.78 [m, CHB (81)], 2,89 [dd, J= 14.4/9.4 Hz, CHB (20)], 3.05 [dd, J= 14.6/4.9 Hz, CHB (10)], 3.66 [m, CHA (82)], 3.70 [m, CHB (82)], 4.11 [dd, J= 9.3/5.1 Hz, CH (1)], 4.34 [m, J ca 5 Hz, CH (9)], 4.84 [s, CH (15)], 5.33 [dd, J = 11.7/1.94 Hz, CHA (32)], 6.09 [dd, J = 17.7/1.91 Hz, CHB (32)], 6.46 [dd, J = 17.6/11.7 Hz, CH (31)]; 13C-NMR (150 MHz, CD3OD, 283 K, 13C-signal assignments from HSQC- and HMBC-experiments): δ (ppm) = 8.2 (71), 9.1 (181), 9.3 (121), 12.2 (21), 21.6 (171), 30.0 (10), 30.6 (20), 30.7 (81), 38.1 (15), 39.1 (172), 60.3 (9), 60.9 (82), 61.5 (1), 71.8 (132), 112.4 (12), 115.5 (18), 118.6 (32), 119.9 (17), 123.4 (19), 125.7 (16), 126.7 (31), 128.5 (3), 130.6 (7), 133.6 (11), 156.0 (8), 162.0 (14), 174.9 (4), 176.6 (6), 176.7 (133), and 181.0 (173). ESI-MS: m/z (%) = 657.3 (33, [M+K]+); 633.3 (10); 621.71 (6), 620.3 (35), 619.3 (100, C33H39N4O8, [M+H]+); 613.3 (53, [M-CO2+K]+); 601.3 (31, [M-H2O+H]+); 575.3 (15, [M-CO2+H]+); 496.2 (26, [M-C7H9NO (ring A)+H]+; 478.2 (6, [M-ring A-H2O+H]+); and 452.2 (32, [M-CO2-ring A+H]+).
MS Data of NCCs from cyp89a9-1
For MS analysis, five fractions containing NCCs from senescent leaf extracts of cyp89a9-1 were isolated by HPLC, analyzed by ESI ionization, and tentatively identified with the known five At-NCCs (Pruzinská et al., 2005; Müller et al., 2006).
ESI-MS
The following spectroscopic data were obtained: At-NCC-1: m/z (%) = 831.0 (39, [M+K]+); 815.1 (52, [M+Na]+); 795.1 (15), 794.1 (49), 793.2 (100, C40H49N4O13, [M+H]+); 787.3 (4, [M-CO2+K]+); 779.1 (6, [M-2H2O+Na]+); 775.2 (14, [M-H2O+H]+); 771.3 (17, [M-CO2+Na]+); 749.1 (17, [M-CO2+H]+); 670.1 (5, [M-C7H9NO (ring A)+H]+); 631.2 (8, [M-C6H10O5 (Glc)+H]+); 626.2 (6, [M-CO2-ring A+H]+); 613.2 (4, [M-H2O-Glc+H]+); 587.1 (4, [M-CO2-Glc+H]+); 479.2 (8). At-NCC-2: m/z (%) = 669.0 (17, [M+K]+); 653.0 (15, [M+Na]+); 633.1 (10), 632.2 (39), 631.1 (100, C34H39N4O8, [M+H]+); 613.1 (29, [M-H2O+H]+); 609.2 (6, [M-CO2+Na]+); 587.1 (14, [M-CO2+H]+); 508.0 (4, [M-ring A+H]+); 490.0 (4, [M-H2O-ring A+H]+); 464.1 (8, [M-CO2-ring A+H]+). At-NCC-3: m/z (%) = 669.0 (61, [M+K]+); 653.1 (58, [M+Na]+); 633.1 (15), 632.0 (36), 631.0 (100, C34H39N4O8, [M+H]+); 613.1 (94, [M-H2O+H]+); 609.2 (26, [M-CO2+Na]+); 595.2 (45, [M-2H2O+H]+); 585.1 (13); 569.2 (54, [M-H2O-CO2+H]+); 508.0 (7, [M-ring A+H]+); 490.1 (9, [M-H2O-ring A+H]+); 472.0 (10, [M-2H2O-ring A+H]+); 464.1 (11, [M-CO2-ring A+H]+); 446.1 (11, [M-H2O-CO2-ring A+H]+). At-NCC-4: m/z (%) = 845.1 (30, [M+K]+); 829.2 (55, [M+Na]+); 809.2 (24), 808.1 (44), 807.1 (100, C41H51N4O13, [M+H]+); 793.3 (20); 775.2 (28, [M-MeOH+H]+); 684.1 (12, [M-ring A+H]+); 652.1 (4, [M-MeOH-ring A+H]+); 645.1 (11, [M-Glc+H]+); 613.1 (7, [M-MeOH-Gluc+H]+). At-NCC-5: m/z (%) = 653.0 (49, [M+K]+); 637.1 (46, [M+Na]+); 617.0 (12), 616.1 (36), 615.0 (100, C34H39N4O7, [M+H]+); 597.2 (39, [M-H2O+H]+); 593.3 (54, [M-CO2+Na]+); 585.1 (13); 571.0 (20, [M-CO2+H]+); 492.00 (6, [M-ring A+H]+); 474.1 (9, [M-H2O-ring A+H]+); and 448.0 (15, [M-CO2-ring A+H]+).
MS Data of epi-pFDCCs from CYP89A9/ATR1 Assays with epi-pFCC as Substrate
For MS analysis, the four products of the CYP89A9/ATR1 assay with epi-pFCC as substrate were isolated by HPLC and analyzed by ESI ionization.
ESI-MS
The following spectroscopic data were obtained: epi-pFDCC-1: m/z (%) = 677.1 (13, [M+K+Na-H]+); 655.2 (19, [M+K]+); 639.3 (24, [M+Na]+); 619.2 (9), 618.1 (33), 617.1 (100, C34H41N4O7 [M+H]+); 607.2 (6, [M-MeOH+Na]+); 585.1 (19, [M-MeOH+H]+; 559.2 (11); 519.2 (13); 492.1 (31, [M- C7H11NO (ring B)+H]+); 460.0 (5, [M-ring B-MeOH+H]+); epi-pFDCC-2: m/z (%) = 677.1 (9, [M+K+Na-H]+); 655.1 (16, [M+K]+); 639.1 (24, [M+Na]+); 619.1 (8), 618.1 (39), 617.1 (100, [M+H]+); 585.1 (14, [M-MeOH+H]+); 519.2 (4); 492.1 (13, [M-ring B+H]+); 460.0 (4, [M-ring B-MeOH+H]+); epi-pFDCC-3: m/z (%) = 677.1 (4, [M+K+Na-H]+); 655.1 (14, [M+K]+); 639.2 (16, [M+Na]+); 619.1 (9), 618.1 (39), 617.1 (100, [M+H]+); 585.1 (8, [M-MeOH+H]+); 492.1 (9, [M-ring B+H]+); epi-pFDCC-4: m/z (%) = 677.1 (4, [M+K+Na-H]+); 655.1 (10, [M+K]+); 639.1 (15, [M+Na]+); 618.1 (9), 618.1 (39), 617.1 (100, [M+H]+); 585.1 (8, [M-MeOH+H]+); and 492.1 (9, [M-ring B+H]+).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACT2, At3g18780; ATR1, At4g24520; BiP, At5g42020; CYP71B19, At3g26170; CYP89A9, At3g03470; MES16, At4g16690; and PPH, At5g13800.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. UV/Vis Spectra of Chlorophyll Catabolites Identified in This Study.
Supplemental Figure 2. Analysis of Gene Expression in cyp89a9 and cyp71b19 Mutants and Colorless Catabolites of cyp71b19 Mutants.
Supplemental Figure 3. Analysis of the cyp89a9-2 Mutant in the Ler Background, a Natural mes16 Mutant.
Supplemental Figure 4. Colorless Catabolites Accumulating in Dark-Incubated (8 d) Leaves of Col-0, cyp89a9-1, and Col-0/PPHTP-MES16.
Supplemental Table 1. Search for Coexpressed Genes in Senescent Leaves, Cauline Leaves, Petals, and Sepals.
Supplemental Table 2. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Science Foundation (Grant 31003A_132603) and the National Center of Competence in Research Plant Survival, a research program of the Swiss National Science Foundation (to S.H.) and by the Austrian National Science Foundation (Fonds zur Förderung der wissenschaftlichen Forschung Projects P-19596 and L-472 to B.K.).
AUTHOR CONTRIBUTIONS
B.K. and S.H. designed the research. B.C., I.S., S.M., N.B., A.E., and T.M. performed research. B.C., I.S., T.M., B.K., and S.H. analyzed data. B.C., T.M., B.K., and S.H. wrote the article.
Glossary
- NCC
nonfluorescent chlorophyll catabolite
- pFCC
primary fluorescent chlorophyll catabolite
- PAO
pheophorbide a oxygenase
- NDCC
nonfluorescent dioxobilin-type chlorophyll catabolites
- MS
mass spectrometry
- m/z
mass-to-charge ratio
- Col-0
Columbia-0
- Ler
Landsberg erecta
- FDCC
fluorescent dioxobilin-type chlorophyll catabolite
- DCC
dioxobilin-type chlorophyll catabolites
- FCC
fluorescent chlorophyll catabolite
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- RFP
red fluorescent protein
- ESI
electrospray ionization
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Andersson M.X., Goksör M., Sandelius A.S. (2007). Optical manipulation reveals strong attracting forces at membrane contact sites between endoplasmic reticulum and chloroplasts. J. Biol. Chem. 282: 1170–1174 [DOI] [PubMed] [Google Scholar]
- Bak S., Beisson F., Bishop G., Hamberger B., Höfer R., Paquette S., Werck-Reichhart D. (2011). Cytochromes p450. The Arabidopsis Book 9: e0144, /10.1199/tab.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassard J.E., Mutterer J., Duval F., Werck-Reichhart D. (2012). A novel method for monitoring the localization of cytochromes P450 and other endoplasmic reticulum membrane associated proteins: A tool for investigating the formation of metabolons. FEBS J. 279: 1576–1583 [DOI] [PubMed] [Google Scholar]
- Berghold J., Breuker K., Oberhuber M., Hörtensteiner S., Kräutler B. (2002). Chlorophyll breakdown in spinach: On the structure of five nonfluorescent chlorophyll catabolites. Photosynth. Res. 74: 109–119 [DOI] [PubMed] [Google Scholar]
- Chen L.F., Chan S.Y., Cossins E.A. (1997). Distribution of folate derivatives and enzymes for synthesis of 10-formyltetrahydrofolate in cytosolic and mitochondrial fractions of pea leaves. Plant Physiol. 115: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ B., Schelbert S., Aubry S., Süssenbacher I., Müller T., Kräutler B., Hörtensteiner S. (2012). MES16, a member of the methylesterase protein family, specifically demethylates fluorescent chlorophyll catabolites during chlorophyll breakdown in Arabidopsis. Plant Physiol. 158: 628–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curty C., Engel N. (1996). Detection, isolation and structure elucidation of a chlorophyll a catabolite from autumnal senescent leaves of Cercidiphyllum japonicum. Phytochemistry 42: 1531–1536 [Google Scholar]
- Djapic N., Pavlovic M. (2008). Chlorophyll catabolite from Parrotia persica autumnal leaves. Rev. Chim. 59: 878–882 [Google Scholar]
- Djapic N., Pavlovic M., Arsovski S., Vujic G. (2009). Chlorophyll biodegradation product from Hamamelis virginiana autumnal leaves. Rev. Chim. 60: 398–402 [Google Scholar]
- Ehlting J., Sauveplane V., Olry A., Ginglinger J.F., Provart N.J., Werck-Reichhart D. (2008). An extensive (co-)expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol. 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A., Meyer S., Schelbert S., Schneider T., Weschke W., Peters S.W., Keller F., Baginsky S., Martinoia E., Schmidt U.G. (2006). Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol. 141: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb H.E., Kotlyar V., Nudelman A. (1997). NMR chemical shifts of common laboratory solvents as trace impurities. J. Org. Chem. 62: 7512–7515 [DOI] [PubMed] [Google Scholar]
- Guengerich F.P. (2001). Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem. Res. Toxicol. 14: 611–650 [DOI] [PubMed] [Google Scholar]
- Halkier B.A. (1996). Catalytic reactivities and structure/function relationships of cytochrome P450 enzymes. Phytochemistry 43: 1–21 [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2004). The loss of green color during chlorophyll degradation—A prerequisite to prevent cell death? Planta 219: 191–194 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Kräutler B. (2011). Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta 1807: 977–988 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Vicentini F., Matile P. (1995). Chlorophyll breakdown in senescent cotyledons of rape, Brassica napus L.: Enzymatic cleavage of phaeophorbide a in vitro. New Phytol. 129: 237–246 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Wüthrich K.L., Matile P., Ongania K.-H., Kräutler B. (1998). The key step in chlorophyll breakdown in higher plants. Cleavage of pheophorbide a macrocycle by a monooxygenase. J. Biol. Chem. 273: 15335–15339 [DOI] [PubMed] [Google Scholar]
- Jensen K., Møller B.L. (2010). Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry 71: 132–141 [DOI] [PubMed] [Google Scholar]
- Kräutler, B. (2008). Chlorophyll catabolites. In Progress in the Chemistry of Organic Natural Products, W. Herz, H. Falk, G.W. Kirny, R.E. Moore, and C. Tann, eds (Vienna, Austria: Springer), pp. 1–43. [DOI] [PubMed] [Google Scholar]
- Kräutler B., Matile P. (1999). Solving the riddle of chlorophyll breakdown. Acc. Chem. Res. 32: 35–43 [Google Scholar]
- Kräutler B., Jaun B., Bortlik K.-H., Schellenberg M., Matile P. (1991). On the enigma of chlorophyll degradation: The constitution of a secoporphinoid catabolite. Angew. Chem. Int. Ed. Engl. 30: 1315–1318 [Google Scholar]
- Kräutler B., Mühlecker W., Anderl M., Gerlach B. (1997). Breakdown of chlorophyll: partial synthesis of a putative intermediary catabolite. Helv. Chim. Acta 80: 1355–1362 [Google Scholar]
- Li R., Moore M., Bonham-Smith P.C., King J. (2002). Overexpression of formate dehydrogenase in Arabidopsis thaliana resulted in plants tolerant to high concentrations of formate. J. Plant Physiol. 159: 1069–1076 [Google Scholar]
- Losey F.G., Engel N. (2001). Isolation and characterization of a urobilinogenoidic chlorophyll catabolite from Hordeum vulgare L. J. Biol. Chem. 276: 8643–8647 [DOI] [PubMed] [Google Scholar]
- Matile P., Ginsburg S., Schellenberg M., Thomas H. (1988). Catabolites of chlorophyll in senescing barley leaves are localized in the vacuoles of mesophyll cells. Proc. Natl. Acad. Sci. USA 85: 9529–9532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A., Eskandari S., Grallath S., Rentsch D. (2006). AtGAT1, a high affinity transporter for γ-aminobutyric acid in Arabidopsis thaliana. J. Biol. Chem. 281: 7197–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min M.K., Kim S.J., Miao Y., Shin J., Jiang L., Hwang I. (2007). Overexpression of Arabidopsis AGD7 causes relocation of Golgi-localized proteins to the endoplasmic reticulum and inhibits protein trafficking in plant cells. Plant Physiol. 143: 1601–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M., Ohta D. (2010). Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 61: 291–315 [DOI] [PubMed] [Google Scholar]
- Moser S., Müller T., Ebert M.O., Jockusch S., Turro N.J., Kräutler B. (2008b). Blue luminescence of ripening bananas. Angew. Chem. Int. Ed. Engl. 47: 8954–8957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser S., Müller T., Holzinger A., Lütz C., Kräutler B. (2012). Structures of chlorophyll catabolites in bananas (Musa acuminata) reveal a split path of chlorophyll breakdown in a ripening fruit. Chemistry 18: 10873–10885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser S., Müller T., Oberhuber M., Kräutler B. (2009). Chlorophyll catabolites - Chemical and structural footprints of a fascinating biological phenomenon. Eur. J. Org. Chem. 2009: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser S., Ulrich M., Müller T., Kräutler B. (2008a). A yellow chlorophyll catabolite is a pigment of the fall colours. Photochem. Photobiol. Sci. 7: 1577–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlecker W., Kräutler B. (1996). Breakdown of chlorophyll: Constitution of nonfluorescing chlorophyll-catabolites from senescent cotyledons of the dicot rape. Plant Physiol. Biochem. 34: 61–75 [Google Scholar]
- Mühlecker W., Kräutler B., Moser D., Matile P., Hörtensteiner S. (2000). Breakdown of chlorophyll: A fluorescent chlorophyll catabolite from sweet pepper (Capsicum annuum). Helv. Chim. Acta 83: 278–286 [Google Scholar]
- Mühlecker W., Ongania K.-H., Kräutler B., Matile P., Hörtensteiner S. (1997). Tracking down chlorophyll breakdown in plants: Elucidation of the constitution of a 'fluorescent' chlorophyll catabolite. Angew. Chem. Int. Ed. Engl. 36: 401–404 [Google Scholar]
- Müller T., Moser S., Ongania K.-H., Pružinská A., Hörtensteiner S., Kräutler B. (2006). A divergent path of chlorophyll breakdown in the model plant Arabidopsis thaliana. ChemBioChem 7: 40–42 [DOI] [PubMed] [Google Scholar]
- Müller T., Rafelsberger M., Vergeiner C., Kräutler B. (2011). A dioxobilane as product of a divergent path of chlorophyll breakdown in Norway maple. Angew. Chem. Int. Ed. Engl. 50: 10724–10727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D., Werck-Reichhart D. (2011). A P450-centric view of plant evolution. Plant J. 66: 194–211 [DOI] [PubMed] [Google Scholar]
- Oberhuber M., Berghold J., Breuker K., Hörtensteiner S., Kräutler B. (2003). Breakdown of chlorophyll: A nonenzymatic reaction accounts for the formation of the colorless “nonfluorescent” chlorophyll catabolites. Proc. Natl. Acad. Sci. USA 100: 6910–6915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhuber M., Berghold J., Mühlecker W., Hörtensteiner S., Kräutler B. (2001). Chlorophyll breakdown - on a nonfluorescent chlorophyll catabolite from spinach. Helv. Chim. Acta 84: 2615–2627 [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37 (Database issue): D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A., Anders I., Aubry S., Schenk N., Tapernoux-Lüthi E., Müller T., Kräutler B., Hörtensteiner S. (2007). In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19: 369–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A., Tanner G., Anders I., Roca M., Hörtensteiner S. (2003). Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 100: 15259–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A., Tanner G., Aubry S., Anders I., Moser S., Müller T., Ongania K.-H., Kräutler B., Youn J.-Y., Liljegren S.J., Hörtensteiner S. (2005). Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol. 139: 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E.S., Vaz A.D.N., Coon M.J. (1991). Catalysis by cytochrome P-450 of an oxidative reaction in xenobiotic aldehyde metabolism: Deformylation with olefin formation. Proc. Natl. Acad. Sci. USA 88: 8963–8966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Hirai N., Matsumoto C., Ohigashi H., Ohta D., Sakata K., Mizutani M. (2004). Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134: 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., Schelbert S., Park S.-Y., Han S.-H., Lee B.-D., Andrès C.B., Kessler F., Hörtensteiner S., Paek N.-C. (2012). STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 24: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelbert S., Aubry S., Burla B., Agne B., Kessler F., Krupinska K., Hörtensteiner S. (2009). Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M.A. (1996). Plant cytochrome P450 monooxygenases. Crit. Rev. Plant Sci. 15: 235–284 [Google Scholar]
- Schuler M.A. (2011). P450s in plant-insect interactions. Biochim. Biophys. Acta 1814: 36–45 [DOI] [PubMed] [Google Scholar]
- Schuler M.A., Duan H., Bilgin M., Ali S. (2006). Arabidopsis cytochrome P450s through the looking glass: A window on plant biochemistry. Phytochem. Rev. 5: 205–237 [Google Scholar]
- Seki M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strain H.H., Cope B.T., Svec W.A. (1971). Analytical procedures for the isolation, identification, estimation and investigation of the chlorophylls. Methods Enzymol. 23: 452–476 [Google Scholar]
- Sundaresan V., Springer P., Volpe T., Haward S., Jones J.D., Dean C., Ma H., Martienssen R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9: 1797–1810 [DOI] [PubMed] [Google Scholar]
- Tan X.L., Wang Q.Y., Tian B.X., Zhang H.A., Lu D.L., Zhou J. (2011). A Brassica napus lipase locates at the membrane contact sites involved in chloroplast development. PLoS ONE 6: e26831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A.F., Marillonnet S., Klimyuk V., Patel K., Torres M.A., Murphy G., Jones J.D. (1999). Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich M., Moser S., Müller T., Kräutler B. (2011). How the colourless ‘nonfluorescent’ chlorophyll catabolites rust. Chemistry 17: 2330–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno M., Matsui T., Ikeda-Saito M. (2007). Structure and catalytic mechanism of heme oxygenase. Nat. Prod. Rep. 24: 553–570 [DOI] [PubMed] [Google Scholar]
- van der Graaff E., Schwacke R., Schneider A., Desimone M., Flügge U.I., Kunze R. (2006). Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.