This work examines the MAP kinase cascade encoded by MEKK1, MKK1/MKK2, and MPK4, finding that deletion of the MEKK1/2/3 tandem gene family produces a wild-type plant and that the autoimmune phenotypes of mutants in this pathway require MEKK2. The key determinant of autoimmunity was found to be the level of MEKK2 expression, which was shown to be regulated by MPK4 activity.
Abstract
An Arabidopsis thaliana mitogen-activated protein (MAP) kinase cascade composed of MEKK1, MKK1/MKK2, and MPK4 was previously described as a negative regulator of defense response. MEKK1 encodes a MAP kinase kinase kinase and is a member of a tandemly duplicated gene family with MEKK2 and MEKK3. Using T-DNA insertion lines, we isolated a novel deletion mutant disrupting this gene family and found it to be phenotypically wild-type, in contrast with the mekk1 dwarf phenotype. Follow-up genetic analyses indicated that MEKK2 is required for the mekk1, mkk1 mkk2, and mpk4 autoimmune phenotypes. We next analyzed a T-DNA insertion in the MEKK2 promoter region and found that although it does not reduce the basal expression of MEKK2, it does prevent the upregulation of MEKK2 that is observed in mpk4 plants. This mekk2 allele can rescue the mpk4 autoimmune phenotype in a dosage-dependent manner. We also found that expression of constitutively active MPK4 restored MEKK2 abundance to wild-type levels in mekk1 mutant plants. Finally, using mass spectrometry, we showed that MEKK2 protein levels mirror MEKK2 mRNA levels. Taken together, our results indicate that activated MPK4 is responsible for regulating MEKK2 RNA abundance. In turn, the abundance of MEKK2 appears to be under cellular surveillance such that a modest increase can trigger defense response activation.
INTRODUCTION
Mitogen-activated protein (MAP) kinase cascades play important roles in the signaling pathways that are activated when plants are exposed to pathogenic microorganisms (Colcombet and Hirt, 2008; Rodriguez et al., 2010; Rasmussen et al., 2012). Using biochemical and genetic approaches, a specific Arabidopsis thaliana MAP kinase cascade composed of MEKK1, MKK1/MKK2, and MPK4 has been shown to be involved in defense signaling (Petersen et al., 2000; Ichimura et al., 2006; Mészáros et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007; Gao et al., 2008; Qiu et al., 2008). MEKK1 is a MAP kinase kinase kinase, MKK1 and MKK2 are a redundant pair of MAP kinase kinases, and MPK4 is a MAP kinase. Exposure of Arabidopsis plants to bacterial pathogens, fungal pathogens, or elicitors such as the flagellin peptide flg22 results in the rapid activation of MPK4 via this cascade within a few minutes of treatment (Ichimura et al., 2006; Mészáros et al., 2006; Suarez-Rodriguez et al., 2007; Gao et al., 2008). Genetic analyses have suggested that this cascade serves as a negative regulator of defense response since plants homozygous for mekk1, mkk1 mkk2, or mpk4 all have similar phenotypes characterized by severe dwarfism coupled with the constitutive activation of defense response genes (Petersen et al., 2000; Ichimura et al., 2006; Mészáros et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007; Gao et al., 2008; Qiu et al., 2008). In addition to the autoimmune phenotype, mpk4 plants also display defects in root cell cytokinesis, pollen meiotic cytokinesis, and microtubule dynamics that are not shared with mekk1 or mkk1 mkk2 mutant plants, indicating that MPK4 has additional roles outside of defense signaling (Kosetsu et al., 2010; Takahashi et al., 2010; Beck et al., 2011; Zeng et al., 2011).
Pathogens can inject proteins called effectors into host cells to inhibit the ability of the host to mount an effective defense response. The bacterial effector HopAI1 has been shown to inactivate the kinase activities of the MAP kinases MPK3, MPK4, and MPK6 in Arabidopsis by dephosphorylating a conserved Thr in the activation loops of these proteins via a phosphothreonine lyase activity (Zhang et al., 2007; Zhang et al., 2012). In order to defend against bacterial effectors, plants have evolved resistance proteins (R proteins) that act as countermeasures against the effectors. Zhang et al. (2012) recently reported the identification of a nucleotide binding Leucine-rich repeat protein named suppressor of mkk1 mkk2 2 (SUMM2) that appears to be acting as an R protein downstream of the MEKK1, MKK1/MKK2, MPK4 pathway. In their model, MPK4 acts as a negative regulator of SUMM2. When HopAI1 is injected into a plant cell, MPK4 is inactivated. Inactivation of MPK4 relieves the negative regulation of SUMM2 and causes the activation of defense responses. The autoimmune and dwarf phenotypes of mekk1, mkk1 mkk2, and mpk4 plants were found to be rescued by the summ2 mutation, consistent with the hypothesis that SUMM2 is a positive regulator of defense response that acts downstream of this MAP kinase cascade (Zhang et al., 2012).
If the MEKK1, MKK1/MKK2, MPK4 pathway is the subject of R protein surveillance, then knocking out one of its components by mutation would be predicted to cause the activation of defense response, which is consistent with the phenotypes displayed by mekk1, mkk1 mkk2, and mpk4 plants. The apparent negative regulation of defense attributed to this MAP kinase cascade may therefore be a reflection of the fact that this pathway is guarded by an R protein, rather than an indication that the primary role of this cascade is to serve as a negative regulator of defense. Analysis of the pathogen response phenotypes of mekk1 summ2 and mkk1 mkk2 summ2 mutant plants indicated that this MAP kinase pathway acts as a positive regulator of basal defense against Hyaloperonospora arabidopsidis and Pseudomonas syringae, rather than as a negative regulator (Zhang et al., 2012). However, recent experiments involving a constitutively active form of MPK4 have suggested that MPK4 may instead act as a negative regulator of defense signaling since Arabidopsis plants expressing constitutively active MPK4 were found to be more susceptible to P. syringae than the wild type (Berriri et al., 2012). These conflicting results could be due to differences in the experimental approaches used to study this signaling pathway.
MEKK1 is located on chromosome IV as part of a tandemly duplicated gene family along with MEKK2 and MEKK3. In this study we describe a reverse-genetic analysis of the MEKK1/2/3 gene family in which we used a novel high-throughput genotyping system (Clark and Krysan, 2007; Su et al., 2011) to identify a deletion allele that disrupted all three members of this gene family. The method used to produce this deletion allele may be of general interest since ∼17% of the genes in Arabidopsis are members of tandemly duplicated arrays (Arabidopsis Genome Initiative, 2000), and there is currently no method available for efficiently deleting selected regions of the Arabidopsis genome.
Our reverse-genetic analysis of the MEKK1/2/3 gene family indicated that MEKK2 acts downstream of the MEKK1, MKK1/MKK2, MPK4 cascade as a positive regulator of defense response. Kong et al. (2012) recently arrived at the same conclusion using a forward-genetic approach. Their study involved screening for extragenic suppressors of mkk1 mkk2 and resulted in the identification of mutant alleles of MEKK2 that could rescue the mkk1 mkk2 autoimmune phenotype. They also determined that MEKK2 acts upstream of the SUMM2 nucleotide binding Leucine-rich repeat gene. Interestingly, flg22-induced cellular responses and resistance to virulent and avirulent strains of P. syringae were not affected by the mekk2 mutations (Kong et al., 2012).
Our study confirms this recently described role of MEKK2 and also provides significant new insights into the mechanism of MEKK2 regulation and function. In particular, we used a T-DNA line carrying an insertion in the promoter region of MEKK2 to show that a relatively modest upregulation of MEKK2 expression is responsible for the autoimmune phenotypes observed when the MEKK1, MKK1/MKK2, MPK4 pathway is disrupted. In addition, using a constitutively active version of MPK4, we demonstrate that activated MPK4 is responsible for keeping MEKK2 expression in check. Our data suggest that Arabidopsis has a mechanism for monitoring the level of MEKK2 such that an increase in MEKK2 protein abundance serves as a trigger for defense pathway activation. Our work provides an example of how the abundance of a protein kinase can potentiate the tipping point between the activation and the nonactivation of defense response.
RESULTS
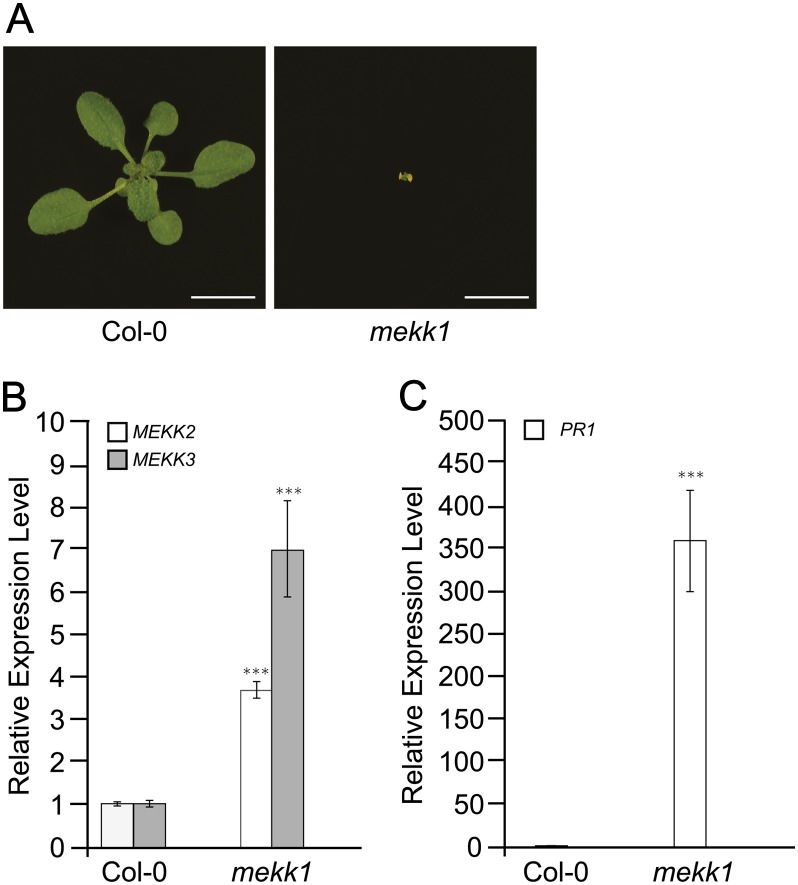
It has been previously shown that null mutants of the MAP kinase kinase kinase gene MEKK1 are severe dwarf plants that constitutively express defense response genes such as pathogenesis-related protein 1 (PR1) (Ichimura et al., 2006; Nakagami et al., 2006; Suarez-Rodriguez et al., 2007). The two closest homologs of MEKK1 in Arabidopsis are MEKK2 and MEKK3, and these three genes constitute a tandemly duplicated gene family located within a 25-kb region of chromosome four. Using quantitative RT-PCR (qRT-PCR), we determined that both MEKK2 and MEKK3 are upregulated in mekk1 mutant plants when compared with the wild type (Figure 1). The expression level of MEKK2 was found to be ∼3.5-fold higher in the mekk1 background than in the wild type, while MEKK3 was approximately sevenfold higher. The defense response marker gene PR1 is also strongly upregulated in mekk1 mutants (Figure 1). We were next interested in determining if this upregulation of MEKK2 and MEKK3 was a cause or a consequence of the mutant phenotypes displayed by mekk1 plants.
Figure 1.
MEKK2 and MEKK3 Are Upregulated in mekk1 Dwarf Plants.
(A) Wild-type (Col-0) and mekk1 mutant plants grown for 19 d on soil. Bars = 1 cm.
(B) and (C) mRNA levels for MEKK2, MEKK3, and PR1 are shown relative to the wild type (Col-0) as determined by qRT-PCR using the PP2A gene as an internal standard. Values are the mean of three independent biological replicates with error bars indicating the se. Asterisks indicate a significant difference from the Col-0 sample as determined by a t test with P < 0.001.
[See online article for color version of this figure.]
Isolation of a mekk1 mekk2 mekk3 Deletion Mutant
The most direct approach to determining if MEKK2 and/or MEKK3 play a role in causing the mekk1 mutant phenotype would be to generate mutant lines that inactivate various combinations of the MEKK1, MEKK2, and MEKK3 loci. Because these genes are members of a tandemly duplicated gene family, tight genetic linkage between the genes makes generating double and triple mutants technically challenging. Our initial strategy for addressing this challenge was to take advantage of the propensity of dissociation (Ds) transposons to hop to locations that are tightly linked to their genomic launch pads (Bancroft and Dean, 1993). Toward this end, T-DNA insertion alleles for MEKK1 and MEKK3 were obtained from the WiscDsLox collection (Woody et al., 2007). We initially sought to mobilize the Ds transposons present in these lines so that we could use localized Ds reinsertions to produce double mutants within the MEKK gene family. However, we determined that the Ds transposons present in these particular WiscDsLox lines were no longer able to transpose at a detectable rate.
Given the failure of the Ds transposon strategy described above, we were interested in determining if it would be practical to produce a mekk1 mekk3 double mutant line by simply screening a large F2 population derived from a mekk1(+/−) mekk3(+/−) parent to find rare recombinants. In the F1 generation, the mekk1 and mekk3 T-DNA lines would be in the trans configuration. In order to eventually produce homozygous mekk1 mekk3 plants, it would therefore be necessary to first identify an F2 individual in which a crossover event had brought the mekk1 and mekk3 mutant alleles into the cis configuration. Based on the published recombination rate within this region of chromosome IV, we expected crossovers to occur between the mekk1 and mekk3 T-DNA insertions at a rate of ∼1 in 4000 meioses (Drouaud et al., 2006).
Using the high-throughput Ice-Cap system developed in our lab for growing and genotyping Arabidopsis seedlings in 96-well plates (Krysan, 2004; Clark and Krysan, 2007; Su et al., 2011), we determined the genotypes of ∼2000 F2 progeny produced by a mekk1(+/−) mekk3(+/−) parent plant. One individual homozygous for the mekk3 allele and heterozygous for mekk1 was identified, indicating that recombination had occurred in one of the gametes that brought the mutant alleles into cis. This plant was allowed to self-pollinate, and homozygous mekk1 mekk3 plants were identified by PCR in the next generation.
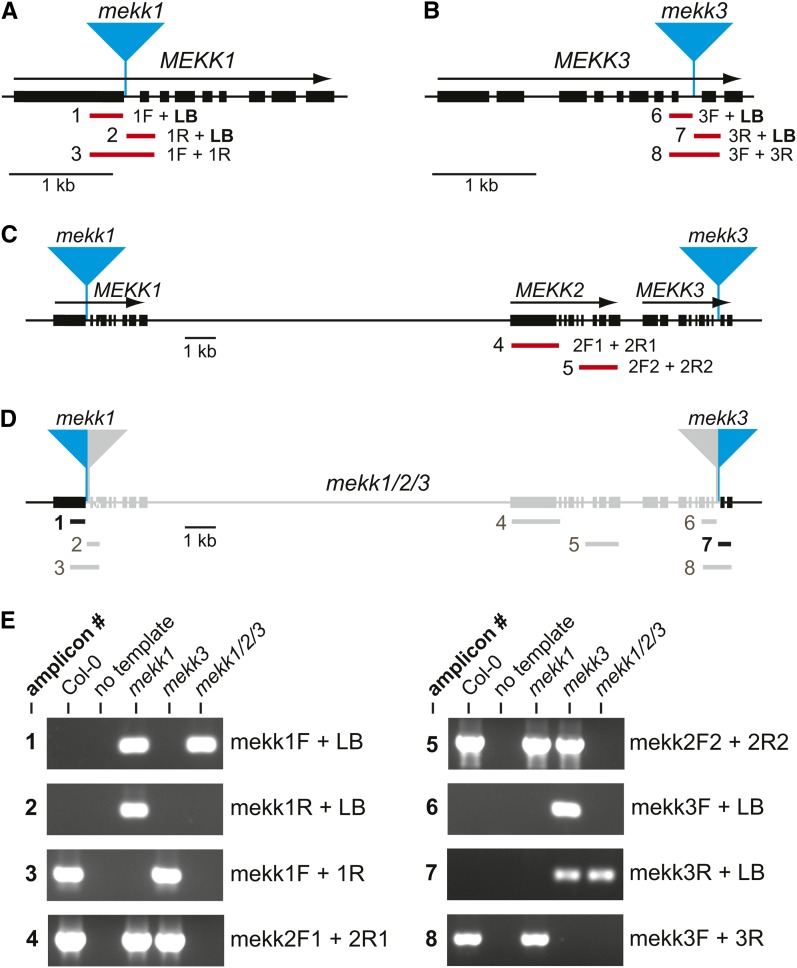
In order to more carefully analyze these mekk1 mekk3 plants, a number of PCR amplifications were performed. To begin, we determined that T-DNA left border sequences are present on both sides of the T-DNA insertions in the original mekk1 and mekk3 single-mutant lines. PCR targeting these T-DNA inserts can therefore be performed using either an upstream or downstream gene-specific primer plus a T-DNA left border primer. The presence of the left border at both the upstream and downstream junctions of a T-DNA insertion has been previously shown to be the most common arrangement of T-DNA borders in a survey of Arabidopsis T-DNA lines (Clark and Krysan, 2010).
PCR amplification targeting the upstream and downstream sides of the mekk1 T-DNA insert produced the expected products when an mekk1 single-mutant plant was tested (Figure 2). However, when mekk1 mekk3 plants were analyzed, only the upstream amplicon was detected (Figure 2). No product was produced by the primer pair targeting the downstream side of the mekk1 T-DNA insertion in the mekk1 mekk3 plants. The opposite result was seen with the MEKK3 locus. The upstream side of the mekk3 T-DNA was found to be missing in the mekk1 mekk3 line, while the downstream side was present. “Upstream” and “downstream” refer to the directions of transcription of the MEKK1, MEKK2, and MEKK3 genes, as shown in Figure 2.
Figure 2.
Structural Analysis of the mekk1/2/3 Deletion Allele.
(A) and (B) Locations of the mekk1 and mekk3 T-DNA insertions in the single mutant lines used to generate the mekk1/2/3 deletion. Thick black lines indicate exons, and arrows indicate direction of transcription. The numbered red lines indicate the locations of the PCR amplicons tested in (E). 1F, 1R, 3F, and 3R are gene-specific PCR primers. LB indicates a T-DNA left border–specific primer.
(C) Predicted structure of a hypothetical mekk1 mekk3 double mutant. 2F1, 2R1, 2F2, and 2R2 are MEKK2 gene-specific PCR primers.
(D) Predicted structure of the mekk1/2/3 deletion allele. Numbered black lines indicate PCR amplicons that were detected in mekk1/2/3 plants, while gray bars indicate those that were not. The region of the genome predicted to be missing in the mekk1/2/3 allele is indicated in gray.
(E) Agarose gel electrophoresis of PCR products. Numbers to the left of each gel refer to the amplicon numbers shown in (A) and (B). “Col-0” is wild-type Columbia, and “no template” is the negative control. The primer pairs used for each PCR amplification are indicated to the right of each gel image.
The unexpected PCR results described above suggested that the recombination event that brought the mekk1 and mekk3 T-DNA alleles into the cis configuration was not a simple crossover. We hypothesized that an unequal crossover may have occurred between the two copies of chromosome IV in the F1 parent plant that was directed by homology between the T-DNA vector sequences present at the mekk1 and mekk3 mutant loci, rather than by homology of the intervening Arabidopsis chromosomal DNA. The result of this hypothesized recombination event would be the deletion of all the chromosomal DNA between the mekk1 and mekk3 T-DNA insertions, including the T-DNA junction sequences on the downstream side of mekk1 and those on the upstream side of the mekk3 (see Supplemental Figure 1 online). This model also predicts that the MEKK2 gene would be missing in the mekk1 mekk3 line.
In order to test this model, we performed PCR using primers that targeted two regions of the MEKK2 locus. The expected products were produced when template from wild-type, mekk1 single mutant, or mekk3 single mutant plants was used. However, no PCR products were detected when the mekk1 mekk3 line was tested (Figure 2). This result was consistent with the hypothesis that the mekk1 mekk3 line carries a deletion of the MEKK2 locus. The precise mechanism by which this deletion was formed will be the focus of a separate study. Regardless of the mechanism, however, it is clear from the PCR genotyping results that these plants are triple-null mutants for MEKK1, MEKK2, and MEKK3. For brevity, we will refer to this deletion allele as the mekk1/2/3 allele.
The mekk1 Mutant Phenotype Is Rescued by mekk2
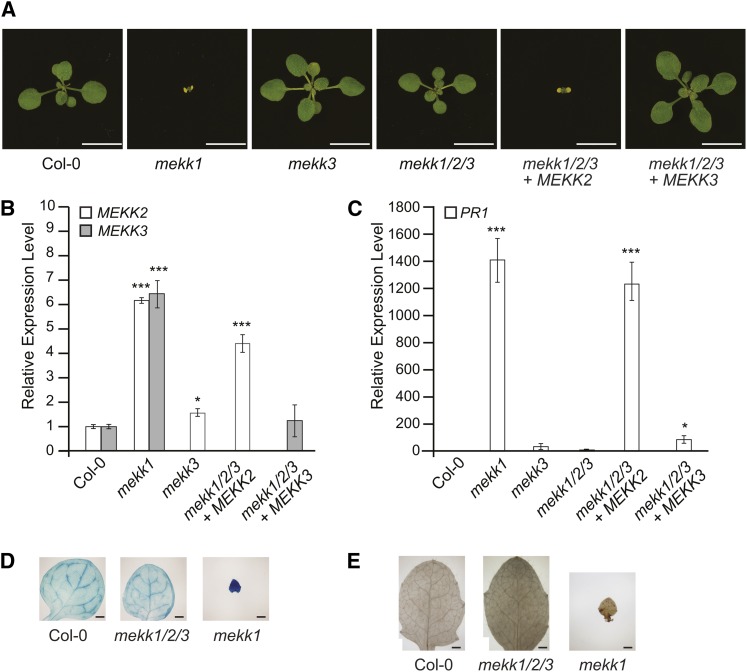
Plants homozygous for the mekk1/2/3 allele are wild-type in appearance, in stark contrast with the dwarf phenotype displayed by mekk1 single mutants (Figure 3). mekk1 mutants also show constitutive activation of defense response genes, such as PR1, as well as increased cell death and reactive oxygen species production in leaf tissue. Using qRT-PCR analysis to measure PR1 gene expression, trypan blue staining to visualize cell death, and 3,3-diaminobenzidine (DAB) staining to monitor reactive oxygen species production (Figure 3), we observed that all of these aspects of the mekk1 mutant phenotype were absent in mekk1/2/3 plants. To further document the extent of the phenotypic rescue, we also photographed the plants at various developmental stages (see Supplemental Figure 2 online).
Figure 3.
MEKK2 Is Responsible for the mekk1 Autoimmune Phenotype.
(A) Plants with the indicated genotypes grown for 16 d on soil. mekk1/2/3 + MEKK2 and mekk1/2/3 + MEKK3 indicate lines carrying a genomic rescue construct for the indicated gene. Bars = 1 cm.
(B) and (C) mRNA levels for MEKK2, MEKK3, and PR1 are shown relative to the wild type (Col-0) as determined by qRT-PCR using the PP2A gene as an internal standard. Values are the mean of three independent biological replicates with error bars indicating the se. Because the MEKK2 and MEKK3 loci are deleted in the mekk1/2/3 line, no PCR products are produced for those genes when mekk1/2/3 plants are analyzed. For this reason, no values are present on the graph for those samples. Three asterisks indicate a significant difference from the Col-0 sample as determined by a t test with P < 0.001, and one asterisk indicates P < 0.05.
(D) Trypan blue staining was used to visualize cell death in the leaves of 2-week-old plants. Bars = 0.5 mm.
(E) DAB staining was used to visualize hydrogen peroxide accumulation in the leaves of 2-week-old plants. Bars = 0.5 mm.
We next determined that deletion of MEKK2 is responsible for rescuing the mekk1 mutant phenotype in mekk1/2/3 plants. When a wild-type copy of MEKK3 under the control of its native promoter was stably introduced into the mekk1/2/3 background by Agrobacterium tumefaciens transformation, no change in phenotype was observed. By contrast, introducing MEKK2 into mekk1/2/3 plants led to the reappearance of the signature mekk1 dwarf phenotype (Figure 3). For this experiment, a T-DNA construct carrying MEKK2 under the control of its native promoter was first introduced into wild-type Columbia-0 (Col-0) plants by Agrobacterium transformation, and transgenic lines were selected that displayed a wild-type phenotype. The MEKK2 expression construct present in one of these wild-type lines was then moved into the mekk1/2/3 background by genetic crossing. Plants homozygous for the mekk1/2/3 allele that also carried the MEKK2 expression construct displayed the mekk1 dwarf phenotype (Figure 3). Taken together, the phenotypic and genetic analyses described above demonstrate that the mutant phenotypes displayed by mekk1 single mutant plants are dependent on the presence of the MEKK2 gene.
The mekk1/2/3 Allele Rescues mkk1 mkk2 Mutant Plants
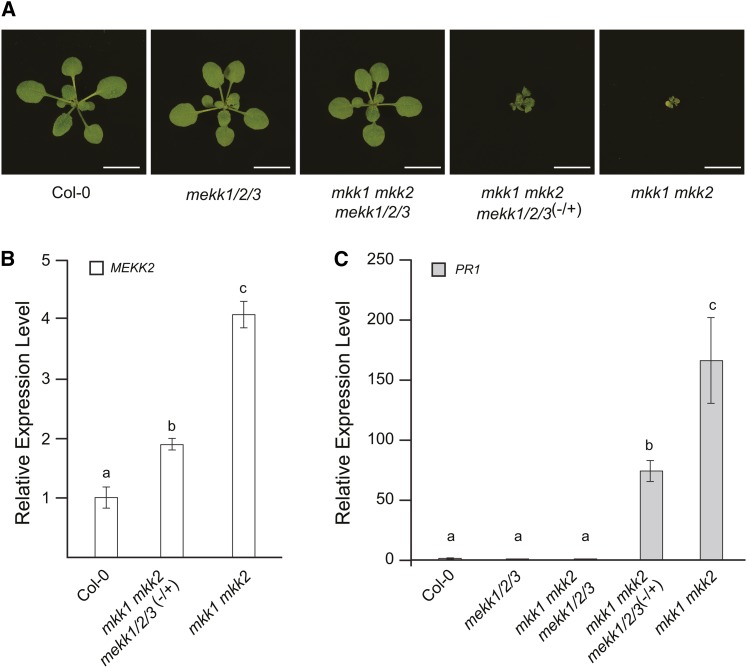
The functionally redundant MAP kinase kinases MKK1 and MKK2 have been previously shown to act downstream of MEKK1 (Mészáros et al., 2006; Gao et al., 2008; Qiu et al., 2008). Similar to mekk1 mutants, mkk1 mkk2 double mutants are dwarf plants that constitutively express pathogen response genes. Using genetic crosses, we produced plants carrying the mekk1/2/3 allele along with T-DNA insertion alleles of mkk1 and mkk2. Analysis of the resulting progeny confirmed that mkk1 mkk2 double mutants were dwarf plants that expressed the PR1 gene at a high level (Figure 4). By contrast, mekk1/2/3 mkk1 mkk2 plants were indistinguishable from the wild type. We also analyzed the phenotypes of mkk1 mkk2 plants that were heterozygous for the mekk1/2/3 allele. These plants showed an intermediate phenotype relative to mkk1 mkk2 and wild-type Col-0 in terms of plant size, MEKK2 expression level, and PR1 expression level (Figure 4). These results suggested that there may be a gene dosage effect related to the role of MEKK2 in causing the mutant phenotypes of mkk1 mkk2 plants.
Figure 4.
The mekk1/2/3 Deletion Rescues the mkk1 mkk2 Autoimmune Phenotype in a Dosage-Dependent Manner.
(A) Plants with the indicated genotypes grown for 17 d on soil. Bars = 1 cm.
(B) and (C) mRNA levels for MEKK2 and PR1 are shown relative to the wild type (Col-0) as determined by qRT-PCR using the PP2A gene as an internal standard. Values are the mean of three independent biological replicates with error bars indicating the se. Because the MEKK2 locus is deleted in the mekk1/2/3 line, no PCR products are produced for that gene when mekk1/2/3 plants are analyzed. For this reason, no value is shown on the MEKK2 expression graph for the mekk1/2/3 genotype. An analysis of variance (ANOVA) was performed on the gene expression data, followed by Tukey's post-hoc analysis (α = 0.05). Means not sharing the same letter are significantly different.
[See online article for color version of this figure.]
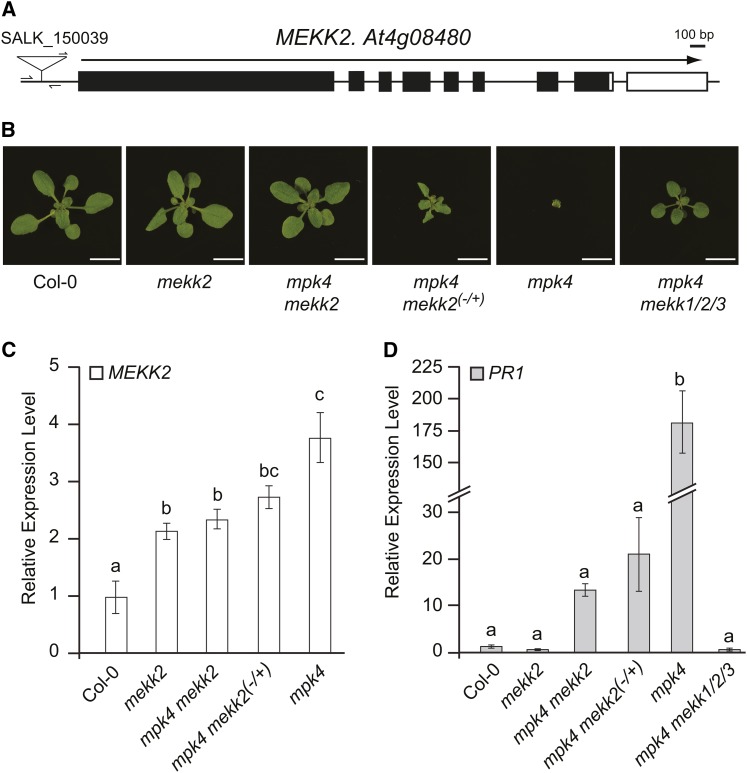
MEKK2 Expression Level Is a Critical Determinant of the mpk4 Mutant Phenotype
Mutation of MPK4 leads to the same autoimmune and dwarf plant phenotypes described above for mekk1 and mkk1 mkk2 plants (Petersen et al., 2000). Using genetic crosses, we produced mekk1/2/3 mpk4 plants and determined that the mekk1/2/3 allele was indeed able to rescue the mpk4 dwarf phenotype and restore PR1 expression to near wild-type levels (Figure 5). In order to more specifically test the role of MEKK2 in this process, we obtained a T-DNA line from the Salk collection (Alonso et al., 2003) carrying an insertion 385 bp upstream of the MEKK2 start codon (Figure 5). No T-DNA lines were available with insertions located inside of the coding region of the gene. As shown in Figure 5, the mekk2 T-DNA insertion allele is able to rescue the mpk4 dwarf plant phenotype and restore PR1 gene expression to near-normal levels in mekk2 mpk4 plants. We also observed a dosage effect of the mekk2 T-DNA allele on the mpk4 dwarf phenotype. Specifically, mekk2(-/+) mpk4 plants were intermediate in size when compared with mpk4 and the wild type, similar to the dosage effect described above for mekk1/2/3 and mkk1 mkk2.
Figure 5.
A T-DNA Insertion in the Promoter Region of MEKK2 Rescues the mpk4 Autoimmune Phenotype in a Dosage-Dependent Manner.
(A) Structure of the MEKK2 gene. Black boxes indicate exons, and the white box indicates the 3′-untranslated region. The white triangle indicates the location of the T-DNA insertion allele used in our study. Bar = 100 bp.
(B) Plants with the indicated genotypes grown for 17 d on soil. Bars = 1 cm.
(C) and (D) mRNA levels for MEKK2 and PR1 are shown relative to the wild type (Col-0) as determined by qRT-PCR using the PP2A gene as an internal standard. Values are the mean of three independent biological replicates with error bars indicating the se. Because the MEKK2 locus is deleted in the mekk1/2/3 line, no PCR products are produced for that gene when mekk1/2/3 plants are analyzed. For this reason, no value is shown on the MEKK2 expression graph for the mekk1/2/3 genotype. ANOVA was performed on the gene expression data followed by Tukey's post-hoc analysis (α = 0.05). Means not sharing the same letter are significantly different.
[See online article for color version of this figure.]
We next measured MEKK2 expression using qRT-PCR and determined that the mekk2 T-DNA insertion does not reduce the expression level of MEKK2 (Figure 5). In fact, mekk2 plants express MEKK2 at a level approximately twofold higher than the wild type. This surprising result raised the question of how this mekk2 allele is able to rescue the mpk4 mutant phenotype. Analysis of MEKK2 expression levels in the remaining genotypes in this experiment provided an explanation. Most importantly, we found that the level of MEKK2 expression in mekk2 mpk4 plants is only 2.4-fold higher than the wild type, compared with the 3.8-fold upregulation that is seen in mpk4 dwarf plants (Figure 5). Although the difference in MEKK2 expression level observed between mpk4 and mekk2 mpk4 plants is relatively modest, we consistently observed the same pattern in multiple independent experiments: mekk2 mpk4 plants always express MEKK2 at a lower level than mpk4 dwarf plants. The mekk2 T-DNA insertion appears to interfere with the upregulation of MEKK2 that occurs in mpk4 plants. The fact that this mekk2 allele can rescue the mpk4 dwarf phenotype suggests that a modest increase in MEKK2 expression level is a critical determinant of the mutant phenotype displayed by mpk4 plants. Analysis of PR1 gene expression in these lines indicated that rescue of the dwarf plant phenotype correlated with reduced PR1 expression (Figure 5). Although the difference was not found to be statistically significant, we did observe that PR1 expression levels were moderately higher in mekk2 mpk4 plants when compared with wild-type controls. This elevated level of PR1 expression could be due to the fact that these plants are not MEKK2 null mutants. The level of MEKK2 protein present in this genotype may be sufficient to cause a mild activation of defense response when compared with the wild type.
We were next interested in determining if MEKK2 is upregulated in wild-type plants in response to exposure to pathogens or microbial elicitors. We surveyed the publicly available Affymetrix gene chip data using the University of Toronto’s eFP Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). None of the pathogen or elicitor treatments caused a substantial increase in MEKK2 expression (see Supplemental Figures 4 and 5 online).
mpk4 Cytokinesis Defects Are Not Rescued by mekk1/2/3 or mekk2
In addition to the autoimmune and dwarf phenotypes, mpk4 mutants also display defects in cytokinesis that are particularly apparent in root cells (Kosetsu et al., 2010; Beck et al., 2011). These cytokinesis defects are not present in mekk1 and mkk1 mkk2 mutant lines, suggesting that MPK4 has additional functional roles outside of the MEKK1, MKK1/MKK2, MPK4 defense signaling pathway. We found that the mekk2 and mekk1/2/3 alleles were not able to rescue the cytokinesis defects caused by the mpk4 mutation. The root cytokinesis defects caused by the mpk4 mutation can be assayed by growing seedlings on vertically oriented agar plates. When grown in this manner, the roots of mpk4 seedlings are substantially shorter than the wild type and appear kinked (Kosetsu et al., 2010; Beck et al., 2011). These mpk4 root phenotypes were not rescued by the mekk2 allele (see Supplemental Figure 2 online). The mekk1/2/3 allele also did not rescue these mpk4 root phenotypes (data not shown).
Overexpression of MEKK2 Recapitulates the mekk1, mkk1/2, and mpk4 Mutant Phenotypes
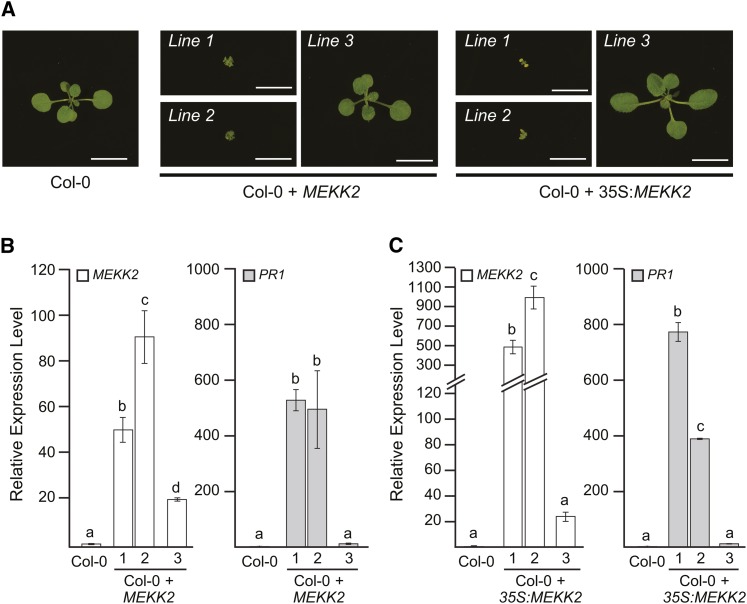
The data presented above indicated that MEKK2 expression level is a key determinant of the autoimmune and dwarf phenotypes displayed by mekk1, mkk1 mkk2, and mpk4 mutant plants. Based on this model, we would predict that overexpression of MEKK2 in a wild-type genetic background would cause a dwarf phenotype and PR1 overexpression. To test this model, we screened a number of transgenic lines that had been transformed with a T-DNA vector carrying the MEKK2 gene under the control of its native promoter. Twenty-five primary transformants carrying the MEKK2 expression construct were identified on agar plates using hygromycin selection. Resistant seedlings were then transferred to soil and grown for 3 weeks. We observed plants with wild-type and dwarf phenotypes in the T1 generation in approximately equal numbers. Representative dwarf and wild-type plants carrying the MEKK2 expression construct are shown in Figure 6. Gene expression analysis indicated that plant phenotype became more severe with increasing MEKK2 expression level: Lines expressing high levels of MEKK2 were dwarf and those with lower MEKK2 levels were wild-type in appearance. PR1 expression also increased with more severe dwarf stature and higher MEKK2 expression level (Figure 6).
Figure 6.
Overexpression of MEKK2 Causes an Autoimmune Phenotype.
(A) The wild type (Col-0), three independent transgenic lines expressing MEKK2 from its native promoter (Col-0 + MEKK2), and three independent transgenic lines expressing MEKK2 via the cauliflower mosaic virus 35S promoter (Col-0 + 35S:MEKK2) grown on soil for 17 d. All transgenic lines are primary transformants in the wild-type (Col-0) genetic background. Bars = 1 cm.
(B) and (C) mRNA levels for MEKK2 and PR1 are shown relative to the wild type (Col-0) as determined by qRT-PCR using the PP2A gene as an internal standard. Values are the mean of three independent biological replicates with error bars indicating the se. ANOVA was performed on the gene expression data followed by Tukey's post-hoc analysis (α = 0.05). Means not sharing the same letter are significantly different.
[See online article for color version of this figure.]
We next performed a similar experiment using a T-DNA construct in which the MEKK2 coding sequence was placed under the transcriptional control of the cauliflower mosaic virus 35S promoter. Once again, we found that plants expressing high levels of MEKK2 were dwarfs, while those expressing lower levels of MEKK2 appeared wild-type (Figure 6). PR1 expression also increased with increasing MEKK2 expression and decreasing plant size in these lines. Taken together, these transgenic experiments indicate that overexpression of MEKK2 is able to phenocopy the loss of MEKK1, MKK1/MKK2, or MPK4.
Genetic Background Potentiates the Effect of MEKK2 Overexpression
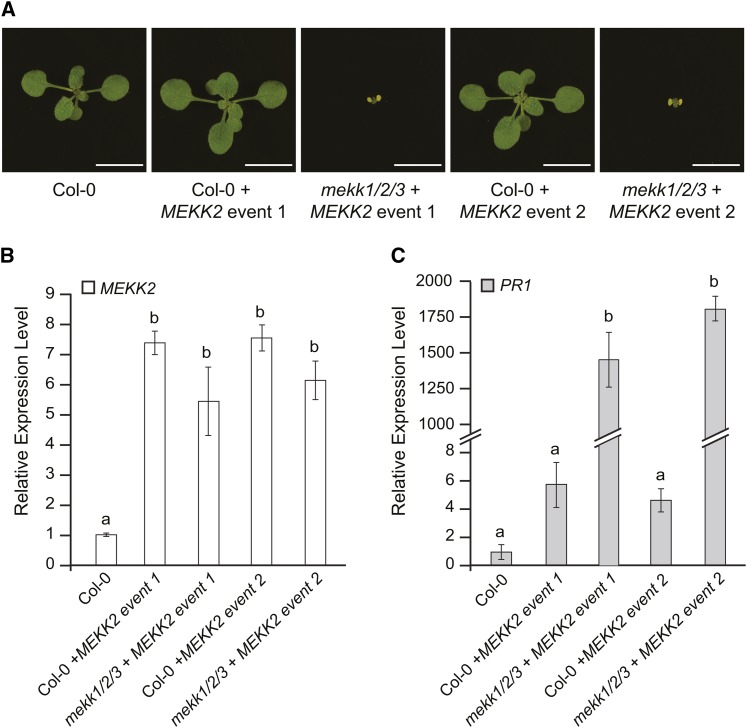
mekk1 dwarf plants express MEKK2 at a level approximately fourfold higher than that of wild-type plants. In the overexpression experiments shown in Figure 6, we saw a clear relationship between MEKK2 expression level and plant phenotype. If we consider the absolute level of MEKK2 expression observed in those experiments, however, we see that the transgenic plants showing a wild-type phenotype express MEKK2 at a level that is up to 20-fold higher than wild-type Col-0 control plants. If MEKK2 expression level was the sole determinant of the dwarf phenotype, then one would expect plants with MEKK2 levels 20-fold higher than the wild type to be dwarfed since MEKK2 expression is only elevated fourfold in mekk1 dwarfs. We hypothesized that genetic background might affect the sensitivity of the different genotypes to MEKK2 overexpression. To test this hypothesis, we took two phenotypically normal PMEKK2:MEKK2 overexpression lines produced in the Col-0 background and crossed them with plants carrying the mekk1/2/3 allele.
When these two independent transgenic events were moved from the Col-0 background into the mekk1/2/3 genetic background, the characteristic dwarf and autoimmune phenotypes appeared (Figure 7). Plants carrying the same transgenic events in the original wild-type background were phenotypically normal. qRT-PCR analysis indicated that MEKK2 was expressed at similar levels in both genetic backgrounds for both of these transgenic events. For all of the samples, the level of MEKK2 expression was in the range of fivefold to 10-fold above the wild-type control. In each case, moving the transgenic event into the mekk1/2/3 genetic background led to a slight decrease in the MEKK2 expression level. This phenomenon could be due to the absence of the native MEKK2 locus in the mekk1/2/3 plants, which would reduce the total number of copies of the MEKK2 expression unit present in those plants. The results of this experiment indicate that mekk1/2/3 plants are more sensitive to the level of MEKK2 overexpression than the wild type, with similar levels of MEKK2 expression causing a dwarf phenotype in the mekk1/2/3 background but a wild-type phenotype in the Col-0 background.
Figure 7.
Genetic Background Potentiates the Effect of MEKK2 Overexpression.
(A) The wild type (Col-0) and plants derived from two independent transgenic events carrying the MEKK2 native promoter expression construct grown for 17 d on soil. The independent transgenic events, labeled “event 1” and “event 2,” were originally produced in the wild-type Col-0 background. Each independent event was then transferred into the mekk1/2/3 genetic background by genetic crossing. Bars = 1 cm.
(B) and (C) mRNA levels for MEKK2 and PR1 are shown relative to the wild type (Col-0) as determined by qRT-PCR using the PP2A gene as an internal standard. Values are the mean of three independent biological replicates with error bars indicating the se. ANOVA was performed on the gene expression data followed by Tukey's post-hoc analysis (α = 0.05). Means not sharing the same letter are significantly different.
[See online article for color version of this figure.]
MEKK2 Protein Level Correlates with MEKK2 mRNA Level
The experiments described above indicate that the level of MEKK2 mRNA is a key determinant of the autoimmune phenotypes displayed by mekk1, mkk1 mkk2, and mpk4 mutant plants. We next wanted to determine if the relationship observed between MEKK2 expression and plant phenotype could be extended to the protein level. For this experiment, we used selective reaction monitoring (SRM) mass spectrometry to perform quantitative analysis of MEKK2 protein level in wild-type, mekk2, mpk4, mekk2 mpk4, and mekk1/2/3 plants. The SRM method makes use of stable isotope-labeled peptide standards to quantitate protein abundance. Because the publically available proteomic databases contained very limited data regarding MEKK2, candidate MEKK2 peptides for SRM analysis were chosen using in silico predictions. For this analysis, only fully tryptic peptides with no missed cleavages were considered, and sequences containing residues prone to chemical modification during sample handling were also avoided (i.e., Cys and Met). All of the peptides chosen for analysis were unique to the MEKK2 protein based on the current annotation of the Arabidopsis genome.
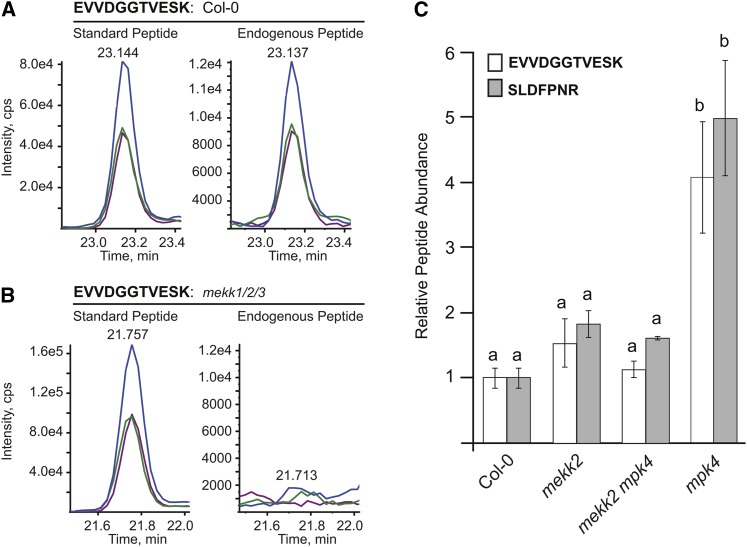
We produced a list of 11 candidate peptides, obtained heavy-labeled synthetic versions of each, and performed a pilot experiment to determine if any of the corresponding MEKK2 endogenous peptides could be detected in Arabidopsis total-protein extracts. For this analysis, digested protein extracts were spiked with heavy-labeled MEKK2 peptide standards and prefractionated offline using strong cation exchange (SCX) chromatography. SCX fractions were then analyzed independently using online nanoflow reverse-phase chromatography coupled to an AB SCIEX QTRAP 5500 triple quadrupole mass spectrometer. From this analysis, we found that two of the MEKK2 peptides, corresponding to amino acids 150 to 156 and 326 to 336, were reproducibly detectable in the wild-type, mekk2, mpk4, and mekk2 mpk4 samples. As a negative control, we also analyzed mekk1/2/3 protein extracts and found, as expected, that both of these endogenous peptides were absent, consistent with the facts that the MEKK2 gene is deleted in this genotype and these peptides are unique to MEKK2. The extracted ion chromatograms for the standard and endogenous peptides displayed the same transition patterns, indicating no interference from isobaric peptides (Figure 8; see Supplemental Figure 6 online). Additionally, tandem mass spectrometry was used to confirm the peptide identities (see Supplemental Figure 7 online).
Figure 8.
MEKK2 Protein Abundance as Determined by SRM Mass Spectrometry.
(A) Extracted ion chromatograms (XICs) for MEKK2 peptide EVVDGGTVESK. Three parent-ion to fragment-ion transitions were monitored for this peptide: y7 is represented by the purple trace, y8 by blue, and y9 by green. Only the strongest transition, y8, was used for quantification. The left panel displays XIC for the isotope-labeled peptide standard, while the right trace shows XIC for the endogenous peptide extracted from wild-type plants. The standard and endogenous peptides assayed in the same run have the same retention times and patterns of relative fragment ion intensity.
(B) XIC for protein extracted from mekk1/2/3 plants as a negative control. No endogenous MEKK2 peptide was observed.
(C) The abundance of the indicated MEKK2 peptides is presented relative to their levels in wild-type Col-0. For each genotype, the abundance of the peptide was first determined relative to the isotope-labeled internal standard peptide. These values were then normalized to the average value obtained for wild-type Col-0. Three independent biological replicates were measured for each genotype, and each biological replicate was analyzed using two to four injection replicates. XICs for the SLDFPNR peptide are presented in Supplemental Figure 6 online. ANOVA was performed separately for each peptide followed by Tukey's post-hoc analysis (α = 0.05). Means not sharing the same letter are significantly different.
To quantify the relative abundance of MEKK2 protein, we measured the peak areas of the extracted ion chromatograms for each endogenous peptide as well as the corresponding heavy-labeled standard present in each sample. Three independent biological replicates were performed for each genotype, and peptide abundance was normalized to the average value observed for the wild type (Figure 8). The level of MEKK2 protein predicted from peptide abundance is four- to fivefold higher in mpk4 dwarf plants when compared with the wild type, whereas the phenotypically wild-type mekk2 mpk4 plants have an MEKK2 protein level that is not significantly different from the wild type. These results demonstrate that MEKK2 protein levels reflect MEKK2 mRNA levels and that the autoimmune phenotype correlates with elevated MEKK2 protein abundance.
Expression of Constitutively Active MPK4 Restores Normal MEKK2 Expression in mekk1 Mutant Plants
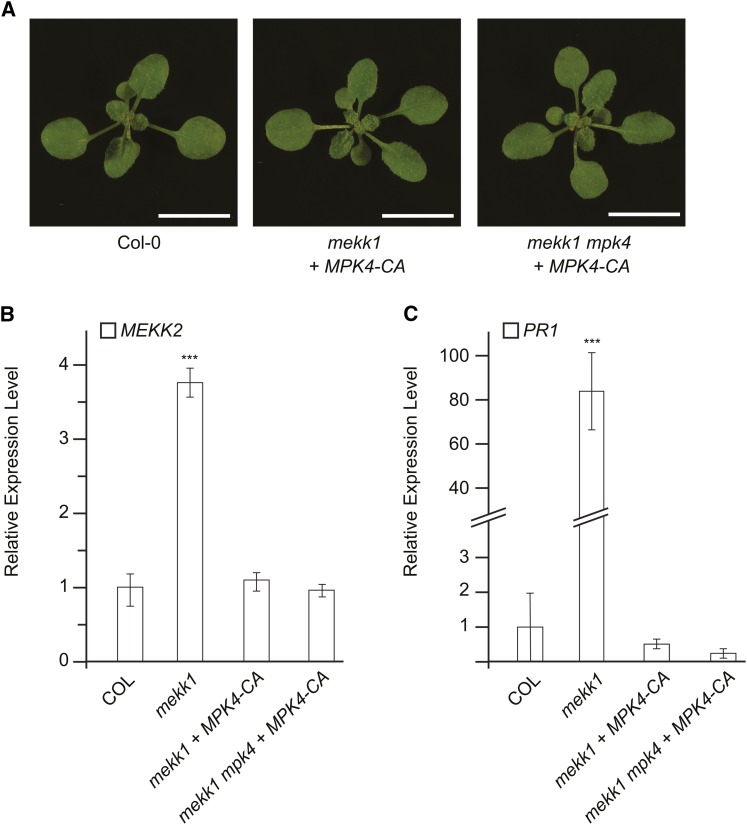
Taken together, the results presented above demonstrate that the autoimmune phenotype seen in mekk1, mkk1 mkk2, and mpk4 plants is caused by increased MEKK2 abundance. In order to further explore the connection between this MAP kinase cascade and MEKK2 regulation, we made use of a recently described constitutively active mutant form of MPK4 (Berriri et al., 2012). It has been previously shown that expression of MPK4D198G E202A in mekk1 mutant plants is able to largely rescue the dwarf and autoimmune phenotypes of that genetic background (Berriri et al., 2012). We obtained transgenic lines expressing MPK4D198G E202A in both the mekk1 and mekk1 mpk4 mutant backgrounds and measured the gene expression levels of MEKK2 and PR1 (Figure 9). In both genetic backgrounds, expression of MPK4D198G E202A was able to restore MEKK2 expression to the normal level seen in wild-type plants. This result demonstrates that activated MPK4 negatively regulates the abundance of MEKK2 RNA. It has been reported that the constitutive kinase activity of MPK4D198G E202A is ∼10% that of fully activated MPK4 (Berriri et al., 2012). This level of MPK4 kinase activity is apparently sufficient to maintain the level of MEKK2 inhibition seen in wild-type plants.
Figure 9.
Constitutively Active MPK4 Reduces the Level of MEKK2 Expression in the mekk1 and mekk1 mpk4 Mutant Backgrounds.
(A) Plants with the indicated genotypes grown for 17 d on soil. “+ MPK4-CA” indicates plants carrying a transgene expressing a constitutively active variant of MPK4 due to its D198G and E202A mutations. Bars = 1 cm.
(B) and (C) RNA levels for MEKK2 and PR1 are shown relative to the wild type (Col-0) as determined by qRT-PCR using the PP2A gene as an internal standard. Values are the mean of three independent biological replicates with error bars indicating the se. Asterisks indicate a significant difference from the Col-0 sample as determined by a t test with P < 0.001.
[See online article for color version of this figure.]
DISCUSSION
The reverse-genetic analysis of MAP kinase signaling that we report in this study began with the identification of a deletion mutation affecting the tandemly duplicated genes MEKK1, MEKK2, and MEKK3. This mekk1/2/3 allele was isolated by screening ∼2000 progeny of a selfed mekk1(+/−) mekk3(+/−) trans-heterozygote to search for rare recombinants that brought the mekk1 and mekk3 T-DNA alleles into the cis configuration. The apparent genotype of the individual that we isolated from this screen was mekk1(+/−) mekk3(−/−). Upon further PCR analysis, however, we determined that this line appeared to be carrying a deletion that encompassed the entire MEKK1, MEKK2, MEKK3 gene family. The first clue that a deletion was present in this line came from our observation that the T-DNA border junctions present on the inside flanks of the mekk1 and mekk3 T-DNA loci were absent in mekk1/2/3 homozygous plants. Follow-up analysis indicated that the MEKK2 gene was also absent in these plants. The exact structures of the T-DNA insertions present in the original mekk1 and mekk3 single mutant lines is not known. Based on PCR analysis, however, it appears that there are at least two copies of the WiscDsLox vector present at each insertion in an inverted-repeat orientation. In a recent survey of T-DNA lines from the Salk collection, it was found that 40 out of the 64 lines analyzed had this apparent inverted-repeat structure (Clark and Krysan, 2010). We hypothesized that the deletion present in the mekk1/2/3 line arose by an unequal crossover event between the T-DNA vector sequences present in the mekk1 and mekk3 alleles. At this point, we do not have direct proof of this hypothetical mechanism, but it seems to provide the most straightforward explanation of how a deletion could arise in lines carrying two closely spaced T-DNA insertions.
One prediction of this model is that the meiotic event that produced the mekk1/2/3 deletion should also have yielded a duplication of the MEKK1/2/3 locus, which would have segregated into another gamete. The design of the PCR screen that we used to isolate the mekk1/2/3 deletion would not have detected this hypothetical MEKK1/2/3 duplication. We are in the process of further exploring the mechanism by which the mekk1/2/3 deletion was formed, which will include screening for this predicted duplication event. For the purposes of studying the MEKK1/2/3 gene family, however, the mechanism by which the mekk1/2/3 deletion was formed is of little importance. The relevant fact is that we were able to obtain a mutant line that was homozygous for null mutants of MEKK1, MEKK2, and MEKK3.
A straightforward strategy for quickly producing targeted deletions in the Arabidopsis genome is currently not available. However, there is a substantial need for such a technology in order to facilitate the reverse-genetic analysis of tandemly duplicated genes. It remains to be seen if the mechanism by which the mekk1/2/3 deletion was produced can be used as a general strategy for producing targeted deletions in Arabidopsis. We recovered one deletion event out of 2000 progeny screened in our experiments. With a single observation, it is impossible to calculate the rate with which such a deletion would be expected to occur. We are currently investigating the rate with which such T-DNA directed deletions arise in Arabidopsis. If these deletions do occur with a reasonable frequency, then one could imagine a simple strategy for deleting targeted regions of the genome whereby a pair of T-DNA insertions flanking a region of interest would first be chosen. These T-DNA insertions would then be combined into a single plant by genetic crossing to produce a trans-heterozygote, and a number of progeny would then be screened by PCR to identify the desired deletion. The Ice-Cap method for high-throughput seedling growth and genotyping developed in our laboratory makes it practical for one scientist to easily genotype a thousand seedlings in a single day (Krysan, 2004; Clark and Krysan, 2007; Su et al., 2011). If desired deletions could be identified by simply genotyping a few thousand F2 progeny, then the Ice-Cap method would make T-DNA directed deletion a viable strategy for producing targeted deletions throughout the Arabidopsis genome.
Our genetic analysis of the MEKK1, MKK1/2, MPK4 signaling pathway revealed that MEKK2 acts downstream of MPK4 and functions as a positive regulator of defense response. A similar conclusion regarding the position of MEKK2 in this signaling pathway was recently reported by Kong et al. (2012). In addition to confirming this recent work, our study also provides significant new insight into the mechanism of MEKK2 function. Specifically, we were able to pinpoint MEKK2 expression level as the key determinant of the autoimmune phenotypes that arise when this MAP kinase cascade is disrupted. In addition, we have shown that activated MPK4 is responsible for inhibiting MEKK2 gene expression. Initially, we observed using qRT-PCR that MEKK2 expression is upregulated approximately four- to fivefold in mekk1, mkk1 mkk2, and mpk4 dwarf plants. However, it was not clear from this data alone if MEKK2 upregulation was a cause or a consequence of the mutant phenotypes. The mekk2 allele that we characterized in our study provided us with a valuable tool for addressing this question because it carried a T-DNA insertion in the promoter region of the MEKK2 gene.
Plants homozygous for the mekk2 promoter insertion allele express MEKK2 at a level approximately twofold higher than the wild type. These plants have a normal phenotype, indicating that mild upregulation of MEKK2 does not noticeably affect the plants. Surprisingly, the mekk2 allele is able to largely rescue the mpk4 dwarf and autoimmune phenotypes, despite the fact that it does not suppress the basal expression level of the MEKK2 gene. The reason that this mekk2 allele can rescue the mpk4 phenotype appears to be due to its ability to prevent the upregulation of MEKK2 that normally occurs in the mpk4 mutant background. The mekk2 T-DNA insertion must disrupt the MEKK2 promoter region in a manner that results in this diminished upregulation.
By bringing MEKK2 expression back down to near wild-type levels, the mekk2 allele is able to rescue the mpk4 mutant phenotype. The gene-dosage effect of mekk2 on mpk4 rescue that we observed provided further evidence that a modest change in MEKK2 expression level is responsible for the mpk4 phenotype. In particular, mpk4 plants that are heterozygous for mekk2 display a partial-rescue phenotype, suggesting that one wild-type copy of MEKK2 does not provide enough MEKK2 expression to cause the full mpk4 dwarf phenotype. We also observed that overexpression of MEKK2 in the wild-type Col-0 background produced the characteristic dwarf and autoimmune phenotypes, whether the overexpression was driven by the MEKK2 native promoter or the cauliflower mosaic virus 35S promoter.
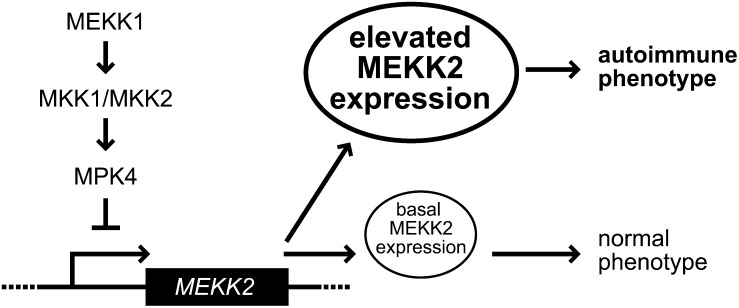
To further characterize the connection between the MEKK1-MKK1/2-MPK4 cascade and MEKK2 regulation, we made use of a recently described constitutively active version of MPK4. This modified protein has two amino acid substitutions, D198G and E202A, that increase its basal kinase activity to a level ∼10% that of activated wild-type MPK4 (Berriri et al., 2012). Expression of MPK4D198G E202A was shown to largely rescue the dwarf and autoimmune phenotypes of mekk1 plants (Berriri et al., 2012). Using these transgenic lines, we observed that MEKK2 levels are restored to normal in mekk1 plants expressing MPK4D198G E202A. This result demonstrated that activated MPK4 is responsible for moderating MEKK2 expression. A basal level of active MPK4 must be present in wild-type plants. When MEKK1 is knocked out, the wild-type MPK4 present in those plants is apparently not sufficiently activated to keep MEKK2 expression in check. Since we have shown that a T-DNA insertion in the promoter of MEKK2 interferes with MEKK2 upregulation, we hypothesize that activated MPK4 is required for the inhibition of MEKK2 transcription in wild-type plants (Figure 10).
Figure 10.
Model for the Role of MEKK2 Regulation in Autoimmunity.
MPK4 acts downstream of MEKK1 and MKK1/2 as a negative regulator of MEKK2 RNA abundance. Activated MPK4 is required for the repression of MEKK2 expression. In a wild-type plant, MEKK2 is expressed at a basal level. When the MEKK1-MKK1/2-MPK4 signaling pathway is disrupted, MEKK2 abundance increases, which in turn triggers the autoimmune response.
It has been established that mRNA and protein levels do not always correlate with each other for a given gene in a given population of cells (Greenbaum et al., 2003). For this reason, we used SRM, a highly sensitive mass spectrometry–based method, to directly quantitate the level of MEKK2 protein in our samples. Heavy isotope–labeled peptide standards were synthesized that corresponded to tryptic peptides predicted to be unique to MEKK2 in Arabidopsis based on an in silico analysis. By spiking known amounts of these labeled peptides into our samples, we were able to obtain quantitative measures of the abundance of MEKK2 protein in plant total protein extracts from the various genotypes. Because MEKK2 is a relatively low-abundance protein, high-performance liquid chromatography fractionation of the total protein extracts using SCX chromatography was implemented as an orthogonal separation technique to improve the sensitivity of detection. The high selectivity of SRM data acquisition allowed us to reproducibly identify and quantitate MEKK2, a protein that has otherwise been largely undetected in proteomic data sets. The results obtained from this analysis demonstrated that the relative levels of MEKK2 protein present in the various genotypes agreed with the levels of mRNA determined by RT-PCR. mpk4 dwarf plants had the highest MEKK2 protein level, and the rescued mekk2 mpk4 plants had MEKK2 level similar to that of mekk2 and the wild type.
The experiments described above all indicate that MEKK2 upregulation is responsible for the dwarf and autoimmune phenotypes observed when the MEKK1, MKK1/MKK2, MPK4 pathway is disrupted by mutation. When MEKK2 protein abundance exceeds a certain limit, the defense response is activated. In the mpk4 mutant background, the critical level of MEKK2 expression appears to be somewhere between two- and fourfold higher than the expression level seen in the wild type. For comparison, we observed that the critical level of MEKK2 expression in the wild-type background was substantially greater, with the tipping point between the wild-type and autoimmune phenotypes occurring when MEKK2 expression was 20- to 40-fold above the wild type. These results suggest that the mpk4 mutant background is sensitized to the effects of MEKK2 overexpression. This conclusion was further supported by our finding that approximately fivefold overexpression of MEKK2 in the mekk1/2/3 background caused a dwarf phenotype, but the same level of overexpression driven by the same transgenic event in the wild-type background resulted in plants with a normal phenotype.
The recent identification of SUMM2 as a potential R protein acting to guard the MEKK1, MKK1/MKK2, MPK4 pathway suggests that the apparent negative regulatory role of MEKK1, MKK1/MKK2, and MPK4 in defense signaling may be due to the fact that this pathway is the subject of R gene surveillance (Zhang et al., 2012). This model is consistent with the observations that the HopAI1 effector molecule can target this MAP kinase cascade (Zhang et al., 2007; Zhang et al., 2012). If SUMM2 is acting to guard the MEKK1, MKK1/MKK2, MPK4 pathway, our findings indicate that MEKK2 protein level must play a critical role in the signaling mechanism that leads to SUMM2-mediated defense activation. An important next step in the analysis of MEKK2 function will be to determine the molecular mechanisms by which an increase in MEKK2 protein abundance can trigger SUMM2–mediated defense response. It has also been shown that MPK4 interacts with MEKK2 in vivo and phosphorylates it in vitro (Kong et al., 2012). These results suggest that there is a direct physical and biochemical connection between the MEKK1, MKK1/MKK2, MPK4 pathway and MEKK2, which may be related to the differences in sensitivity to MEKK2 overexpression that we observed in the different genetic backgrounds.
Our work has demonstrated that activated MPK4 is responsible for inhibiting the expression of MEKK2. When this negative regulation is disrupted, an autoimmune response is mounted. These findings provide an example of how a modest increase in the expression level of a protein kinase can switch a plant from a normal state to an autoimmune state. Future work will be needed to integrate this new information regarding MEKK2 protein dynamics within the broader context of defense signaling.
METHODS
Plant Material and Genotyping
Arabidopsis thaliana plants were grown in soil under continuous light at 18 to 22°C. mekk1 (At4g08500; WiscDsLox339H07), mekk2 (At4g08480; SALK_150039), mekk3 (At4g08470; WiscDsLox472D11), mkk1 (At4g26070; SALK_027645), mkk2(At4g29810; SAIL_511_H01), and mpk4 (At4g01370, SALK_056245) T-DNA lines, all in the Col-0 ecotype, were obtained from the ABRC (http://abrc.osu.edu/). Constitutively active MPK4D198G E202A transgenic lines were obtained from Jean Colcombet (Berriri et al., 2012). Plants carrying multiple T-DNA insertions were generated by genetic crossing and PCR-based genotyping. PCR primers used for genotyping are listed in Supplemental Table 1 online.
Gene Expression Analysis
qRT-PCR was used to measure mRNA levels. Total RNA was isolated from the rosettes of soil-grown plants using the RNeasy Plant Mini Kit (Qiagen). The SuperScript III first-strand cDNA synthesis system (Invitrogen) was used to produce cDNA from these RNA samples using a poly-T primer to prime cDNA synthesis. Quantitative PCR was then performed on these cDNA samples using the CFX96 detection system (Bio-Rad Laboratories). The SYBR/FAM emission/excitation filter set was used to detect fluorescence from the EvaGreen dye (Biotium) present in the reactions. All PCR amplifications were run using the following cycling parameters: 95°C for 10 s, 58°C for 30 s, and 72°C for 30 s, for 50 cycles. Melt curve analysis was used to verify the identity of the PCR products produced in these reactions. The total volume of each PCR was 20 μL and contained 0.2 μM of each primer, 1× EvaGreen nucleotide binding dye, 0.2 mM each deoxynucleotide triphosphate (Promega), 75 mM Tris-HCl, pH 9, 20 mM (NH4)2SO4, 3 mM MgCl2, 0.01% (v/v) Tween 20, and Taq polymerase. The oligonucleotide primers used for qRT-PCR analysis are listed in Supplemental Table 2 online. Phosphatase PP2A (At1g13320) was used as the reference gene for normalization of the qRT-PCR data (Czechowski et al., 2005). For each qRT-PCR experiment, three independent biological replicates were performed with separate RNA extractions, cDNA syntheses, and qRT-PCR amplifications for each replicate. In addition, each biological replicate was analyzed using two independent qRT-PCR amplifications. For each sample, the expression level relative to wild-type Columbia was determined by first normalizing to the internal standard provided by the PP2A gene and then normalizing the data for a given gene to the average value obtained for wild-type Columbia for that experiment.
Plasmid Construction and Agrobacterium tumefaciens–Mediated Transformation
Genomic expression clones of MEKK2 and MEKK3 were constructed by PCR amplifying the two loci from wild-type Arabidopsis (ecotype Col-0) genomic DNA template using the following primers. MEKK2 forward, 5′-AAAACTGCAGAAACTAATCTAAAAATTCCGCAAAGAGATG-3′, and reverse, 5′-AAAAGAGCTCAGTTTTCAACGGGGCAAATATTGTA-3′; MEKK3 forward, 5′-TTTTAAGCTTATTTTATTAGATTGCCTCCATAGGTG-3′, and reverse, 5′-AAAAGAGCTCTTTTCAAATAGGGGTAAAATGGTCA-3′. These primers added unique recognition sites for the restriction enzymes PstI and SacI to the MEKK2 amplicon and HindIII and SacI to the MEKK3 amplicon. These restriction sites were used to ligate the PCR products into the T-DNA binary vector pMDC32 (Curtis and Grossniklaus, 2003). Digestion of the vector with these restriction enzymes removed the 35S promoter present in pMDC32 during the cloning process, and the resulting plasmids carried the MEKK2 or MEKK3 genomic clone under the control of their native promoters plus the hygromycin resistance cassette as a plant selectable marker. The MEKK2 clone included 1210 bp of genomic sequence upstream of the MEKK2 start codon and 792 bp of downstream from the stop codon. The MEKK3 clone had 4590 bp upstream of the start codon and 831 bp downstream from the stop codon. The MEKK2 and MEKK3 genomic clones used for this study were fully sequenced to obtain error-free clones.
The plasmid expressing the MEKK2 coding sequence under the control of the cauliflower mosaic virus 35S promoter was constructed by PCR amplifying the MEKK2 coding region from wild-type Arabidopsis (ecotype Col-0) genomic DNA template using the following primers: forward, 5′- AAAAGGCGCGCCATGAAGAAGTCGTCGGATAAGTCAC-3′, and reverse, 5′- AAAATTAATTAAAGTTTTCAACGGGGCAAATATTGTA-3′. These primers added unique recognition sites for the restriction enzymes AscI and PacI to the amplicon and allowed it to be cloned into pMDC32 such that the MEKK2 coding sequence was under the transcriptional control of the 35S promoter.
The T-DNA vectors described above were introduced into Agrobacterium and used to transform Arabidopsis plants using a modified floral dip procedure (Clough and Bent, 1998). Primary transformants were selected by growth on 0.7% agar (w/v) plates containing 0.5× Murashige and Skoog basal salt mixture with 2 mg/mL hygromycin. Plants were transferred to soil after 7 to 10 d of growth on the agar plates.
Histological Analysis
Trypan blue staining to visualize cell death was performed as previously described (Weigel and Glazebrook, 2002). DAB staining to visualize hydrogen peroxide accumulation in leaf tissue was performed as previously described (Nakagami et al., 2006).
SRM Mass Spectrometry
Protein was extracted from the supernatant of homogenized 3-week-old soil-grown seedlings using a previously described methanol/chloroform/water method (Wessel and Flügge, 1984). Protein concentration was measured using a BCA assay kit (Thermo Scientific Pierce). Two milligrams of protein was reduced with 5 mM DTT (45 min, 50°C) and alkylated using 15 mM iodoacetamide (45 min, room temperature) before undergoing protease digestion. Samples were first digested with LysC (Wako Chemicals) in a 4 M urea solution at a 1:50 enzyme-to-protein ratio for 3 h at 37°C. Samples were then diluted to 1.2 M urea using 50 mM NH4HCO3 and digested with trypsin (Promega) at a 1:100 enzyme-to-protein ratio overnight at 37°C.
Samples were acidified using 0.5% formic acid (FA) to stop enzymatic digestion, and isotopically labeled peptides standards, synthesized by Sigma-Aldrich PEPscreen crude synthesis platform, were spiked in at known dilutions. Samples were desalted using C-18 solid phase extraction columns (Waters). SCX sample fractionation was performed using a polysulfoethyl aspartamide column (4.6 × 200 mm; PolyLC) on a Waters Alliance 2795 HPLC. The gradient used for separation consisted of 0 to 40% buffer B (5 mM KH2PO4, 350 mM KCl, and 30% acetonitrile (ACN), pH 2.65) over 50 min at a flow rate of 1 mL/min. The gradient was followed by an 80% buffer B wash and reequilibration with buffer A (5mM KH2PO4 and 30% ACN, pH 2.65). Blanks were run between samples to ensure no carryover. Sample fractions were frozen, lyophilized, and desalted using solid phase extraction.
Peptide quantitation was performed using an Eksigent NanoLC-Ultra 2D system with cHiPLC nanoflex microfluidic C18 column (75 μm, 120 Å) coupled to an AB SCIEX 5500 QTRAP mass spectrometer. SRM method development was performed using peptide standards and MRMPilot software (AB SCIEX). For each peptide, three parent ion-to-fragment ion transitions were monitored with a dwell time of 115 ms. Analytical separation was performed using a linear gradient of 2 to 35% buffer B (ACN and 0.1% FA) (Honeywell Burdick and Jackson) over 70 min at a flow rate of 300 nL/min. Gradient was followed by a 90% buffer B wash and reequilibration with buffer A (0.1% FA) (Honeywell Burdick and Jackson). Peak areas were integrated using the automatic MQ4 function in MultiQuant software (AB SCIEX). Peptide quantitation was achieved by comparing the area ratio of endogenous peptides to peptide standards within each run. For each sample, peptides were quantified from two to four injection replicates for three independent biological replicates. Tandem mass spectrometry data acquisition was achieved using IDA methods in Analyst software (AB SCIEX). Three EPI scan were summed to generate tandem mass spectrometry spectra.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MEKK1, AT4G08500; MEKK2, AT4G08480; MEKK3, AT4G08470; MKK1, AT4G26070; MKK2, AT4G29810; MPK4, AT4G01370; PR1, AT2G14610; and PP2A, AT1G13320.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Proposed Model for the Formation of the mekk1/2/3 Deletion.
Supplemental Figure 2. Time-Course Comparison of mekk1/2/3 and Col-0 Growth.
Supplemental Figure 3. mekk2 Does Not Rescue the Root Cell Cytokinesis Defects Present in mpk4 Plants.
Supplemental Figure 4. Expression of MEKK2 in Response to Pseudomonas syringae Exposure.
Supplemental Figure 5. Expression of MEKK2 in Response to Elicitor Exposure.
Supplemental Figure 6. Quantitation of the MEKK2 Peptide SLDFPNR by Selective Reaction Monitoring Mass Spectrometry.
Supplemental Figure 7. Tandem Mass Spectrometry Analysis to Confirm the Identities of the MEKK2 Peptides.
Supplemental Table 1. PCR Primers Used for Genotyping.
Supplemental Table 2. PCR Primers Used for qRT-PCR.
Supplementary Material
Acknowledgments
We thank Jean Colcombet for the generous gift of the transgenic lines expressing constitutively active MPK4. This work was supported by the National Science Foundation MCB Award 0447750 to P.K. and MCB Award 0929395 to M.R.S.
AUTHOR CONTRIBUTIONS
S.-H.S., S.M.B., and P.K. conceived the project. S,-H.S., S.M.B., N.Z., K.S., M.R.S., and P.K. designed the research. S.-H.S., S.M.B., N.Z., K.S., and P.K performed the research. S.-H.S., S.M.B., N.Z., K.S., and P.K analyzed the data. S.-H.S., K.S., and P.K. wrote the article.
Glossary
- MAP
mitogen-activated protein
- qRT-PCR
quantitative RT-PCR
- DAB
3,3-diaminobenzidine
- Col-0
Columbia-0
- SRM
selective reaction monitoring
- SCX
strong cation exchange
- FA
formic acid
- ACN
acetonitrile
- ANOVA
analysis of variance
- XIC
extracted ion chromatogram
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bancroft I., Dean C. (1993). Transposition pattern of the maize element Ds in Arabidopsis thaliana. Genetics 134: 1221–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Komis G., Ziemann A., Menzel D., Samaj J. (2011). Mitogen-activated protein kinase 4 is involved in the regulation of mitotic and cytokinetic microtubule transitions in Arabidopsis thaliana. New Phytol. 189: 1069–1083 [DOI] [PubMed] [Google Scholar]
- Berriri S., Garcia A.V., Dit Frey N.F., Rozhon W., Pateyron S., Leonhardt N., Montillet J.L., Leung J., Hirt H., Colcombet J. (2012). Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell 24: 4281–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K.A., Krysan P.J. (2007). Protocol: An improved high-throughput method for generating tissue samples in 96-well format for plant genotyping (Ice-Cap 2.0). Plant Methods 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K.A., Krysan P.J. (2010). Chromosomal translocations are a common phenomenon in Arabidopsis thaliana T-DNA insertion lines. Plant J. 64: 990–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colcombet J., Hirt H. (2008). Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 413: 217–226 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud J., Camilleri C., Bourguignon P.Y., Canaguier A., Bérard A., Vezon D., Giancola S., Brunel D., Colot V., Prum B., Quesneville H., Mézard C. (2006). Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots”. Genome Res. 16: 106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18: 1190–1198 [DOI] [PubMed] [Google Scholar]
- Greenbaum D., Colangelo C., Williams K., Gerstein M. (2003). Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K., Casais C., Peck S.C., Shinozaki K., Shirasu K. (2006). MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 281: 36969–36976 [DOI] [PubMed] [Google Scholar]
- Kong Q., Qu N., Gao M., Zhang Z., Ding X., Yang F., Li Y., Dong O.X., Chen S., Li X., Zhang Y. (2012). The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24: 2225–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosetsu K., Matsunaga S., Nakagami H., Colcombet J., Sasabe M., Soyano T., Takahashi Y., Hirt H., Machida Y. (2010). The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell 22: 3778–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan P. (2004). Ice-cap. A high-throughput method for capturing plant tissue samples for genotype analysis. Plant Physiol. 135: 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mészáros T., Helfer A., Hatzimasoura E., Magyar Z., Serazetdinova L., Rios G., Bardóczy V., Teige M., Koncz C., Peck S., Bögre L. (2006). The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 48: 485–498 [DOI] [PubMed] [Google Scholar]
- Nakagami H., Soukupová H., Schikora A., Zárský V., Hirt H. (2006). A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 281: 38697–38704 [DOI] [PubMed] [Google Scholar]
- Petersen M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Qiu J.L., Zhou L., Yun B.W., Nielsen H.B., Fiil B.K., Petersen K., Mackinlay J., Loake G.J., Mundy J., Morris P.C. (2008). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M.W., Roux M., Petersen M., Mundy J. (2012). MAP kinase cascades in Arabidopsis innate immunity. Front. Plant Sci. 3: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Su, S.H., Clark, K.A., Gibbs, N.M., Bush, S.M., and Krysan, P.J. (2011). Ice-Cap: A method for growing Arabidopsis and tomato plants in 96-well plates for high-throughput genotyping. J. Vis. Exp. 57: 3280. [DOI] [PMC free article] [PubMed]
- Suarez-Rodriguez M.C., Adams-Phillips L., Liu Y., Wang H., Su S.H., Jester P.J., Zhang S., Bent A.F., Krysan P.J. (2007). MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Soyano T., Kosetsu K., Sasabe M., Machida Y. (2010). HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol. 51: 1766–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Wessel D., Flügge U.I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Woody S.T., Austin-Phillips S., Amasino R.M., Krysan P.J. (2007). The WiscDsLox T-DNA collection: An Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant Res. 120: 157–165 [DOI] [PubMed] [Google Scholar]
- Zeng Q., Chen J.G., Ellis B.E. (2011). AtMPK4 is required for male-specific meiotic cytokinesis in Arabidopsis. Plant J. 67: 895–906 [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y., Ba H., Zhou J., Zhang Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.