Abstract
Translation inhibition is a major but poorly understood mode of action of micro(mi)RNAs in plants and animals. In particular, the subcellular location where this process takes place is unknown. Here we show that the translation inhibition but not the mRNA cleavage activity of Arabidopsis miRNAs requires ALTERED MERISTEM PROGRAM1 (AMP1). AMP1 encodes an integral membrane protein associated with endoplasmic reticulum (ER) and ARGONAUTE1, the miRNA effector and a peripheral ER membrane protein. Large differences in polysome association of miRNA target RNAs are found between wild type and the amp1 mutant for membrane-bound but not total polysomes. This, together with AMP1-independent recruitment of miRNA target transcripts to membrane fractions, shows that miRNAs inhibit the translation of target RNAs on the ER. This study demonstrates that translation inhibition is an important activity of plant miRNAs, reveals the subcellular location of this activity, and uncovers a previously unknown function of the ER.
Keywords: AMP1, ER, membrane-bound polysome, microRNA, microsome, polysome, translation
Introduction
micro(mi)RNAs are ~21-nucleotide small RNAs that impact numerous biological processes in diverse eukaryotes. miRNAs repress target gene expression in two main manners, mRNA degradation and translation inhibition. mRNA degradation occurs through miRNA-guided transcript cleavage in plants and deadenylation followed by mRNA decay in animals (Fabian et al., 2010). Translation inhibition by miRNAs is poorly understood in plants and may entail multiple mechanisms in animals (Fabian et al., 2010; Fukaya and Tomari, 2012).
For animal miRNAs, the degrees of contribution by the two modes of action to target gene regulation have been controversial (Fabian et al., 2010). Recent studies that examined the translation inhibition and mRNA decay activities of zebrafish and Drosophila miRNAs with temporal resolution concluded that translation inhibition precedes target mRNA decay (Bazzini et al., 2012; Djuranovic et al., 2012). Therefore, translation inhibition is an integral and critical activity of animal miRNAs.
Unlike animal miRNAs, plant miRNAs display a high degree of sequence complementarity to their target mRNAs and were first found to guide target RNA cleavage through the endonucleolytic activity of ARGONAUTE1 (AGO1), the major miRNA effector (Baumberger and Baulcombe, 2005; Llave et al., 2002; Tang et al., 2003). This led to the assumption that RNA cleavage is the major mode of action of plant miRNAs (Jones-Rhoades et al., 2006). However, in cases where protein accumulation from miRNA target genes was examined, plant miRNAs were found to exert disproportionate effects on target gene expression at protein versus mRNA levels (Aukerman and Sakai, 2003; Chen, 2004; Gandikota et al., 2007). Furthermore, mutations in a number of genes, including AGO1 and AGO10 and the P-body component VARICOSE (VCS), compromise miRNA-mediated target repression at the protein but not the mRNA level (Brodersen et al., 2008; Yang et al., 2012). In addition, miRNAs and AGO1 are associated with polysomes (Lanet et al., 2009). These observations are usually interpreted to indicate that plant miRNAs inhibit target mRNA translation, but direct evidence showing that plant miRNAs inhibit protein synthesis from their target genes is lacking. In this study, through measurements of protein synthesis, we demonstrate that a plant miRNA indeed represses its target gene through translation inhibition.
Despite the widespread effects of plant miRNAs on target mRNA translation, the underlying mechanisms are completely unknown. Furthermore, the subcellular location where miRNA-mediated translation inhibition occurs has not been defined in eukaryotes. Here, we report that ALTERED MERISTEM PROGRAM1 (AMP1), a gene that impacts multiple developmental processes in Arabidopsis via an unknown mechanism (Chaudhury et al., 1993; Conway and Poethig, 1997; Helliwell et al., 2001; Hou et al., 1993; Jurgens et al., 1991; Mordhorst et al., 1998), and its paralog LIKE AMP1 (LAMP1), mediate the translation inhibition but not the RNA cleavage activity of plant miRNAs. Measurement of protein synthesis in vivo and examination of polysome profiles demonstrate that plant miRNAs inhibit target mRNA translation in an AMP1-dependent manner, and that AMP1 does not impact protein synthesis in general. AMP1 is an integral membrane protein associated with the rough ER and AGO1 is a peripheral membrane protein that co-localizes with ER. We show that miRNA target transcripts are associated with microsomes but are prevented from being recruited to membrane-bound polysomes by AMP1. This, together with the large differences in the effects of the amp1 mutation on the loading of miRNA target transcripts onto total versus membrane-bound polysomes, suggests that translation inhibition by plant miRNAs occurs on the ER. The presence of AMP1 homologs in animals implies that ER may be a common site of miRNA-mediated translation inhibition in eukaryotes.
Results
Mutations in AMP1 lead to pleiotropic developmental defects
This study began with the isolation of the amp1-30 allele in the gene ALTERED MERISTEM PROGRAM1 (AMP1) (see below) from an ethyl methanesulfonate mutagenesis screen. The genetic screen was conducted in a line in which a luciferase reporter gene undergoes transcriptional gene silencing through DNA methylation (Won et al., 2012) and was designed to identify players in DNA methylation. The line also contained a mutation in RNA DEPENDENT RNA POLYMERASE6 (RDR6) to prevent posttranscriptional transgene silencing (Dalmay et al., 2000; Mourrain et al., 2000). Therefore, the isolated mutant was actually a double mutant, which we refer to as rdr6-11 amp1-30 (or rdr6 amp1 for simplicity). rdr6 amp1 did not show altered reporter gene expression, but its morphological phenotypes were reminiscent of those of mutants defective in miRNA biogenesis or activity and prompted us to study this mutant. While the morphology of rdr6 plants is largely similar to wild type, pin-shaped cotyledons were present in 28% of rdr6 amp1 plants (Fig 1A, B; Table S1). The amp1 single mutant was later recovered and found to also exhibit radialized cotyledons (Table S1).
Figure 1.
The amp1-30 mutant exhibits pleiotropic developmental defects. (A) A 12-day-old rdr6-11 seedling with two cotyledons and two true leaves. (B) A 12-day-old rdr6-11 amp1-30 seedling with three pin-shaped cotyledons (marked by arrows) and four true leaves. Abnormal anthocyanin accumulation gives the seedling a purple color. (C) A phb-1d/+ plant. (D) A phb-1d/+amp1-30 plant with trumpet-shaped leaves. (E) An rdr6-11 embryo with a normal suspensor (arrow). (F) An rdr6-11 amp1-30 embryo showing over proliferation of the suspensor (arrow). (G) A cleared rdr6-11 cotyledon showing venation patterns similar to those in wild type. (H) A cleared rdr6-11 amp1-30 cotyledon showing incomplete venation. (I) A dissected rdr6-11 silique showing a full complement of seeds. (J) A dissected rdr6-11 amp1-30 silique with few seeds. Scale bars in (I–J), 3 mm. (K) Five-week-old wild type (left), amp1-30 (middle), and amp1-30 lamp1-1 (right) plants. (L) The AMP1 gene rescues the developmental defects of the amp1-30 mutant. Left, wild type; middle, amp1-30; right, amp1-30AMP1p::AMP1-GFP. See also Table S1 and Figure S1.
Defects in adaxial-abaxial polarity specification are known to result in radialized lateral organs (McConnell et al., 2001). The type III HD-Zip genes PHABULOSA (PHB), PHAVOLUTA (PHV) and REVOLUTA (REV), which are repressed by miR165/166 in the abaxial domain of organ primordia, specify adaxial identity, and failure to repress the genes leads to adaxialization and radialization of leaves (Emery et al., 2003; Mallory et al., 2004b). We thought that the phenotypes of amp1 could reflect defects in miR165/166-mediated repression of the HD-Zip genes. To test this genetically, we crossed amp1 with phb-1d/+, which harbors a miR165/166-resistant phb allele and displays mild leaf polarity defects (the phenotypes are mild because other HD-Zip genes are repressed by miR165/166) (McConnell et al., 2001) (Fig 1C). The amp1 phb-1d/+ plants had dramatic defects in leaf polarity – nearly all leaves became trumpet-shaped (Fig 1D). Therefore, amp1 is partially compromised in leaf polarity specification, and further studies revealed a defect in miR165/166-mediated repression of PHB, PHV, and REV (see below).
The amp1 mutant exhibited phenotypic similarities with mutants defective in miRNA biogenesis or function. Null mutants in the miRNA biogenesis gene DICERLIKE1 (DCL1) (Park et al., 2002; Reinhart et al., 2002) show over-proliferation of suspensor cells (Schwartz et al., 1994). The rdr6 amp1 mutant also showed such phenotypes (Fig 1E, F). Mutants in the P body gene VCS, which is required for miRNA-mediated translation inhibition (Brodersen et al., 2008), exhibit pointed cotyledons with incomplete veins and anthocyanin accumulation (Goeres et al., 2007). Both rdr6 amp1 and amp1 had incomplete veins (Fig 1G, H) and anthocyanin deposition in cotyledons (Fig 1B), although the phenotype was not fully penetrant (Table S1). rdr6 amp1 and amp1 plants were also reduced in fertility (Fig 1I, J) and stature (Fig 1K). These phenotypes raised the possibility of amp1 being compromised in miRNA biogenesis or activity.
AMP1 is required for miRNA-mediated target gene repression at the protein but not the mRNA level
To first evaluate whether AMP1 plays a role in miRNA biogenesis, we performed northern blotting to compare the steady-state levels of nine miRNAs between rdr6 and rdr6 amp1 (Fig S1A). No difference in miRNA accumulation was observed, which argued against a role of AMP1 in miRNA biogenesis.
Next, we examined the transcript levels of eight genes targeted by five different miRNAs using real-time RT-PCR in rdr6 amp1 and rdr6. Only CUC2 showed an increase in transcript abundance in rdr6 amp1 relative to rdr6 (Fig 2A). This argued against a general role of AMP1 in miRNA-guided transcript cleavage.
Figure 2.
AMP1 is dispensable for miRNA-mediated transcript cleavage, but is required for the activities of miRNAs in repressing the protein levels of target genes. *, P value < 0.05; **, P value < 0.01. (A) Steady-state levels of transcripts from eight miRNA target genes as determined by real-time RT-PCR in three independent experiments. UBQ5 was used as an internal control. (B–F) Western blots to determine the levels of various proteins. HSC70 served as the loading control. One biological replicated is shown; two others are in Fig S2. The numbers above the blots indicate relative protein levels as calculated from the three biological replicates. (B–C) CSD2 protein levels in various genotypes under Cu2+-limiting (miR398 present) and Cu2+-replete (miR398 absent) conditions. L50 and L70 are two independent AMP1p::AMP1-HA transgenic lines in the rdr6 amp1 background. (D) Levels of CSD2-HA or CSD2m-HA from rdr6 and rdr6 amp1 plants carrying a homozygous transgene at the same genomic location. L28 and L32 are two independent transgenic events for CSD2p::CSD2-HA; L4 and L5 are two independent transgenic events for CSD2p::CSD2m-HA. Plants were grown under Cu2+-limiting conditions. (E) Levels of PHB-MYC or PHBm-MYC from rdr6 and rdr6 amp1 plants carrying a homozygous transgene at the same genomic location. L52, L49, and L25 are three independent transgene insertion events for 35S::PHB-MYC; L2, L8, and L26 are three independent transgene insertion events for 35S::PHBm-MYC. (F) Levels of REV-MYC, CNA-MYC, and CUC1-MYC proteins. For each transgene, two to three independent transgene insertion events (as indicated by L followed by a number above the gel images) were evaluated. (G) Real-time RT-PCR to examine transcript levels from various transgenes in (D–F). Values were normalized to UBQ5. Standard deviations were calculated from three independent experiments. (H) Semi-quantitative 5′ RACE-PCR to detect the 3′ fragments generated through miRNA-guided cleavage of various target transcripts. UBQ5 is a loading control. (I) Mapping the position of miRNA-guided cleavage by cloning and sequencing the 5′ RACE products in (H). The miRNAs that target the transcripts are shown. The numbers above the sequences indicate the number of clones representing cleavage at the expected position (arrowhead) out of the total number of clones sequenced. See also Figure S2.
Finally, we determined whether the amp1 mutant was defective in miRNA-mediated target gene repression at the protein level. We first examined whether miR398-mediated repression of CSD2 (Sunkar et al., 2006) was compromised in rdr6 amp1. miR398 is strictly induced under Cu2+-limiting conditions and is not detectable under Cu2+-replete conditions (Sunkar et al., 2006). We found that CSD2 protein levels were higher in rdr6 amp1 than rdr6 when seedlings were grown in the absence of Cu2+ (Fig 2B; Fig S2A, B), but CSD2 mRNA levels were similar in the two genotypes (Fig 2A). In the presence of Cu2+, CSD2 protein levels were similar in the two genotypes (Fig 2B; Fig S2A, B). This, in conjunction with the unaltered levels of miR398 (Fig S1A), suggested that miR398’s activity to repress CSD2 at the protein level was compromised in rdr6amp1. The amp1 mutation was responsible for this molecular defect since the defect was rescued by AMP1p::AMP1-HA (Fig 2C; Fig S2A).
To conclusively demonstrate that the increase in CSD2 protein levels in rdr6 amp1 under Cu2+-limiting conditions reflected compromised miR398 activity, we generated CSD2p::CSD2-HA and CSD2p::CSD2m-HA transgenic lines in order to compare transgene expression between rdr6 and rdr6 amp1. In these lines, HA-tagged wild-type or miR398-resistant CSD2 was driven by the CSD2 promoter. Since plant transformation results in random transgene integration and transgene expression can be strongly influenced by transgene location, comparison of expression levels of independent transgenes in rdr6 versus rdr6 amp1 would not be meaningful. We obtained isogenic rdr6 and rdr6 amp1 lines in which each transgene was homozygous and inserted into a single genomic locus (see Supplemental Experimental Procedures) to enable the comparison of transgene expression between the two genotypes. We found that under Cu2+-limiting conditions (i.e. miR398 present), CSD2-HA levels were higher in rdr6 amp1 than rdr6, but CSD2m-HA levels were similar in the two genotypes (Fig 2D; Fig S2C). This demonstrated that miR398-mediated repression of CSD2 at the protein level was compromised by the amp1 mutation.
We next determined whether AMP1 also mediates the activities of other miRNAs. We generated isogenic rdr6 and rdr6 amp1 lines containing each of the following transgenes: 35S::PHB-MYC, 35S::CNA- MYC, and 35S::REV- MYC (all targeted by miR165/6 (Rhoades et al., 2002; Tang et al., 2003)), and 35S::CUC1- MYC (targeted by miR164 (Mallory et al., 2004a)). Note that the inclusion of the rdr6 mutation in these analyses was necessary to prevent transgene silencing. For each transgene, at least two independent transgenic events were evaluated. In all cases, an increase in transgene protein levels was found in rdr6 amp1 relative to rdr6 (Fig 2E, F; Fig S2 D–G). The miR165/6-resistant 35S::PHBm-MYC transgene was similarly expressed in rdr6 amp1 and rdr6 (Fig 2E; Fig S2D). The transcript levels from each of the transgenes were similar in rdr6 and rdr6 amp1 except for CUC1-MYC (Fig 2G).
To rule out an effect of amp1 on miRNA-mediated transcript cleavage, we performed semi-quantitative 5′ RACE RT-PCR to detect the 3′ cleavage products from five miRNA target genes in rdr6 and rdr6 amp1. No difference in the abundance of the 3′ cleavage products was detected (Fig 2H). In addition, the cleavage products were cloned and sequenced, which revealed that the precision of miRNA-mediated RNA cleavage was unaffected by the amp1 mutation (Fig 2I).
In conclusion, AMP1 is not required for miRNA-mediated cleavage of target mRNAs; instead, it mediates the activities of multiple miRNAs in reducing the protein levels of their target genes. The lack of an effect of the amp1 mutation on protein accumulation from miRNA-resistant transgenes indicates that AMP1 does not affect protein levels in general.
A plant miRNA inhibits the translation of its target mRNA in an AMP1-dependent manner
Although the disproportionate effects of plant miRNAs on target gene expression at protein versus mRNA levels have been attributed to their translation inhibition activities, plant miRNAs have not been definitively shown to inhibit protein synthesis. We took advantage of the amp1 mutant to determine whether plant miRNAs inhibit protein synthesis from their target mRNAs. To this end, we first determined the half-life of the CSD2-HA protein from a CSD2p::CSD2-HA transgenic line through pulse-chase experiments. We found the half-life to be at least 12 hours under Cu2+-limiting conditions (data not shown). The long half-life allowed us to ascertain protein synthesis rates from the transgene by incubating seedlings in 35S-Methionine media for short time periods (20 min and 40 min) followed by immunoprecipitation of CSD2-HA. The immunoprecipitated and radiolabeled CSD2-HA represented newly synthesized CSD2-HA, whereas western blots on the same samples displayed the steady-state levels of the protein (both labeled and unlabeled). As expected, in both rdr6 and rdr6 amp1, the amount of radiolabeled CSD2-HA increased over time, but the steady-state levels were the same at the two time points (Fig 3A, B; Fig S3A). The amount of newly synthesized CSD2-HA was higher at both time points in rdr6 amp1 than rdr6 under Cu2+-limiting conditions (Fig 3A, B; Fig S3A). Therefore, AMP1 was required for the inhibition of protein synthesis from CSD2-HA. To determine whether the translation inhibition by AMP1 required miR398, we performed the pulse labeling experiments in the presence of Cu2+. The amount of newly synthesized CSD2-HA was similar in rdr6 and rdr6 amp1 under Cu2+-replete conditions (Fig 3C, D; Fig S3B). Therefore, miR398 inhibits protein synthesis from the CSD2p::CSD2-HA transgene in an AMP1-dependent manner. Furthermore, the general profiles of labeled proteins were identical in rdr6 and rdr6 amp1 (Fig S3C), indicating that AMP1 does not impact protein synthesis in general.
Figure 3.
Measurement of protein synthesis from a CSD2p::CSD2-HA transgene in the rdr6 or rdr6 amp1 background. (A, C) Isogenic rdr6CSD2p::CSD2-HA and rdr6 amp1CSD2p::CSD2-HA seedlings (L32 in Fig. 2D) were labeled with 35S-Methionine for 20 or 40 min under Cu2+-limiting (miR398 present) or Cu2+-replete (miR398 absent) conditions. CSD2-HA was then immunoprecipitated, resolved by SDS-PAGE, and transferred to a membrane. The membrane was first subjected to western blotting to detect steady-state levels of CSD2-HA (bottom gel images) and then to autoradiography to detect labeled CSD2-HA (35S-CSD2-HA). Note that the steady-state CSD2-HA levels serve as a loading control. Under Cu2+-limiting conditions, the difference in steady-state CSD2-HA levels between rdr6 and rdr6 amp1 was already known (Fig 2D). By referencing Fig 2D, we conclude that the loading was comparable for rdr6 and rdr6 amp1. Under Cu2+-replete conditions, CSD2-HA steady-state levels should be nearly equal (Fig 2C). Therefore, loading was even for the four samples. (B, D) Quantification of 35S-CSD2-HA signals in (A, C). The rdr6 20 min sample was arbitrarily set to 1.0. The quantification was based on three independent replicates (this Figure and Figure S3). **, P value < 0.01. See also Figure S3.
AMP1 belongs to a conserved protein family in eukaryotes
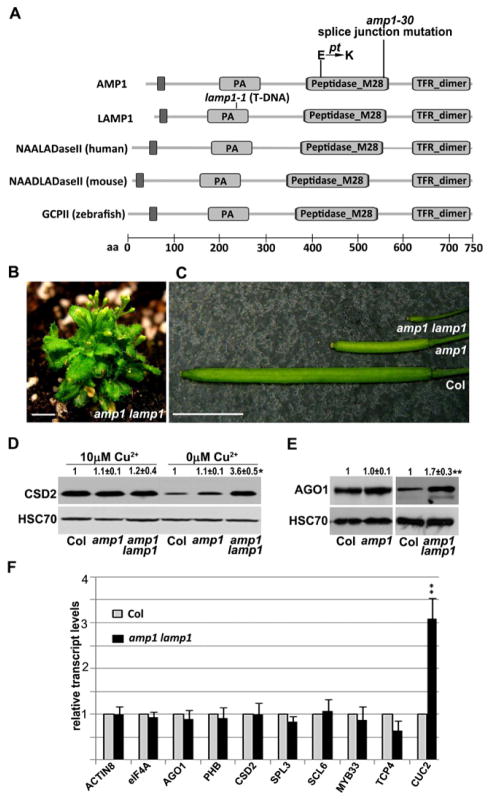
Using a map-based cloning strategy, we identified the molecular lesion of amp1-30 as a G-to-A change in the splice junction at the sixth intron in the AMP1 gene (Fig 4A). Two genomic clones AMP1p::AMP1-GFP and AMP1p::AMP1-HA both fully rescued the morphological defects (Fig 1L) as well as the molecular defects of rdr6 amp1 (i.e. the increase in CSD2 protein levels, Fig 2C; Fig S2A), demonstrating that AMP1 mediates the activities of miRNAs.
Figure 4.
AMP1 and its paralog LAMP1 have overlapping functions in mediating the translation inhibition activities of miRNAs. (A) Diagrams of AMP1, LAMP1, and three animal AMP1 homologs. The molecular lesions in two amp1 alleles are shown. PA, protease-associated domain; Peptidase_M28, a Pfam peptidase domain; TFR_dimer, a dimerization domain in transferrin receptor. pt is a previously described amp1 allele (Helliwell et al., 2001). (B) The amp1 lamp1 double mutant is extremely dwarfed. Scale bar, 6 mm. (C) Siliques from wild type (Col), amp1, and the amp1 lamp1 double mutant. Scale bar, 6 mm. (D, E) Western blots to determine CSD2 (D) and AGO1 (E) protein levels in wild type (Col), amp1, and amp1 lamp1. Note that the two AGO1 panels cannot be compared with each other due to differences in exposure time. (F) Real-time RT-PCR to determine the transcript levels of eight miRNA target genes and two non-targets (ACTIN8 and eIF4A) in wild type (Col) and amp1 lamp1. Values were normalized to UBQ5. Standard deviations were calculated from three independent experiments. In (D–F), P value < 0.05 (*) or 0.01 (**).
Homologs of plant AMP1 are easily recognized in animals (Fig 4A). These included six human genes, four encoding confirmed or putative glutamate carboxypeptidases or N-acetylated alpha-linked acidic dipeptidases (NAALADase) and two encoding the transferrin receptor. AMP1 and its homologs share a common organization of motifs/domains: an N-terminal transmembrane domain, a protease-associated (PA) domain, a peptidase domain, and a transferrin receptor dimerization domain (Fig 4A).
The presence of the peptidase domain in AMP1 raised the possibility that AMP1 acts to degrade proteins from miRNA target genes. Although this scenario would be consistent with AMP1’s role in repressing miRNA target gene expression at the protein level, it is not likely since we had shown that AMP1 inhibits protein synthesis (i.e. translation) from miRNA target genes (Fig 3). Nonetheless, we examined whether AMP1 affects the half-life of miRNA target proteins by performing pulse-chase experiments. As the long half-life of CSD2-HA made it unsuitable for this analysis, we focused on PHB-MYC with a much shorter half-life. The half-lives of PHB-MYC in rdr6PHB-MYC and rdr6 amp1PHB-MYC were not significantly different as measured from three independent experiments (Fig S3D, E), thus ruling out a role of AMP1 in degrading PHB-MYC.
An AMP1 paralog (At5g19740), LAMP1, showing 42% amino acid similarity to AMP1 is present in the Arabidopsis genome (Fig 4A). We identified a T-DNA insertion mutant, lamp1-1 and produced the amp1-30 lamp1-1 double mutant (in the wild-type RDR6 background; will be referred to as amp1 lamp1). The double mutant was extremely small (Fig 1K,4B) and sterile (Fig 4C), suggesting that the two genes have overlapping functions in development. The two genes also have overlapping molecular functions. miR398-mediated repression of CSD2 was more compromised in amp1lamp1 than in amp1 (Fig 4D, Fig S2H). The amp1 lamp1 double mutant, but not the amp1 single mutant, showed an increase in the protein but not the transcript levels from the AGO1 gene, which is targeted by miR168 (Vaucheret et al., 2004) (Fig 4E, F, Fig S2I). The amp1 lamp1 double mutant had normal levels of various miRNAs (Fig S1B) as well as most miRNA target mRNAs (Fig 4F), suggesting that both genes mediate the translation inhibition activities of miRNAs. Note that a significant increase in CSD2 protein levels was observed in rdr6 amp1 relative to rdr6 (Fig 2D) but not in amp1 relative to WT (Fig 4D). This suggests that the rdr6 mutation somehow enhances amp1’s defects in miRNA-mediated translation repression. We suspect that this reflects an indirect effect of RDR6, which enables the production of endogenous siRNAs (Peragine et al., 2004; Vazquez et al., 2004) that may share with miRNAs the translation repression machinery involving AMP1. The absence of endogenous siRNAs in rdr6 may allow miRNAs better access to the machinery to exert a stronger effect in translation repression.
AMP1 and AGO1 are localized to the ER
Given that both AMP1 and the miRNA effector AGO1 are required for miRNA-mediated translation repression (this study and (Brodersen et al., 2008)), we sought to determine whether the two proteins are associated or co-localize in vivo. Immunoprecipitation (IP) was conducted using anti-GFP antibodies with extracts from rdr6 and AMP1p::AMP1-GFP rdr6 amp1 plants. Western blotting using anti-AGO1 antibodies showed that AGO1 was enriched in the IP (Fig 5A). In the reciprocal experiment, in which AGO1 was immunoprecipitated from rdr6 and AMP1p::AMP1-HA rdr6 amp1 plants, AMP1-HA was found enriched in the IP (Fig 5B). This suggested that the two proteins are associated in vivo.
Figure 5.
AMP1 and AGO1 are associated in vivo, and both are localized to the ER. (A) Co-IP performed with an AMP1p::AMP1-GFP rdr6 amp1 line, in which the transgene fully rescues the amp1 phenotypes. WT, rdr6 plants without the transgene. IP was performed with anti-GFP antibodies, and the immunoprecipitates were subjected to western blotting with anti-GFP (left panel) or anti-AGO1 (right panel) antibodies. (B) Co-IP performed with an AMP1p::AMP1-HA rdr6 amp1 line, in which the transgene fully rescues the amp1 phenotypes. WT, rdr6 plants without the transgene. IP was conducted with anti-AGO1 antibodies and the immunoprecipitates were subjected to western blotting with anti-AGO1 (left panel) or anti-HA (right panel) antibodies. 1% of lysate was used to extract proteins as input in (A) and (B). (C–H) Co-localization of AMP1-GFP or YFP-AGO1 with ER-mCherry. AMP1-GFP (C–E) or YFP-AGO1 (F–H) was transiently co-expressed with ER-mCherry in N. benthamiana leaves. Fluorescence was observed separately (C and D; F and G), and the images were merged (E, H). (I) Quantification of co-localization between YFP-AGO1 or DCP1-CFP with ER-mCherry. Confocal z-stacks were collected from 30 and 23 cells for YFP-AGO1 and DCP1-CFP, respectively, and co-localization was quantified with the image analysis software IMARIS7.2. (J) AMP1-HA and AGO1 are present in a crude membrane fraction. Western blots were performed to detect various proteins in the total extract, the soluble fraction or the pellet after centrifugation at 100,000g. PEPC and HSC70 are a cytosolic and an ER luminal protein, respectively. (K) AMP1 is an integral membrane protein, whereas AGO1 is a peripheral membrane protein. Western blots were performed to detect the presence of AMP1-GFP and AGO1 in the soluble fraction (S) or the pellet (P) after membrane suspensions in buffers containing high salt or detergent concentrations or high pH were centrifuged at 100,000g. AMP1p::AMP1-GFP rdr6 amp1 plants were used for the assays. (L) A TEM image of a root tip section from an AMP1p::AMP1-HA rdr6 amp1 line immunolabeled with anti-HA antibodies. All immunogold particles are marked by red arrows. White arrowheads indicate ribosome particles on the surface of ER. Most ER tubules in the images are dotted with ribosomes and hence are rough ER. (M) A TEM image of a root tip section from an AMP1p::AMP1-HA rdr6 amp1 line immunolabeled for AMP1-HA (15nm gold particles; white arrows) and Calnexin (10nm gold particles; red arrows). See also Table S2 and Figure S4.
A previous study showed that GFP-AMP1 from a 35S::GFP-AMP1 transgenic line localized to the endoplasmic reticulum (ER) in Arabidopsis (Vidaurre et al., 2007), consistent with the presence of a transmembrane domain in AMP1. We sought to confirm the ER localization of AMP1 and determine whether AGO1 is also associated with ER. When transiently expressed in Nicotiana benthamiana, AMP1-GFP and ER-mCherry, an ER marker (Nelson et al., 2007), were found to co-localize in mesh-like patterns typical for the ER (Fig 5C–E). In leaves co-expressing YFP-AGO1 and ER-mCherry, YFP-AGO1 accumulated in many cytoplasmic granules that co-localized with ER-mCherry (Fig 5F–H, Fig S4A, B). Quantification of YFP-AGO1 signals revealed that most AGO1 granules were along the ER meshwork (Fig 5I). DECAPPING1 (DCP1), a P body marker, was also found in cytoplasmic granules (Fig S4C) (Xu et al., 2006), but the DCP1 foci did not co-localize with ER-mCherry (Fig 5I; Fig S4C).
To examine the membrane association of AMP1 and AGO1 in Arabidopsis, we separated extracts from an AMP1p::AMP1-HA rdr6 amp1 line into crude membrane and soluble fractions. AMP1-HA was exclusively in the membrane fraction (Fig 5J) and AGO1 was enriched in the membrane fraction (Brodersen et al., 2012) (Fig 5J). Under high salt or high pH conditions, AGO1 was displaced into the soluble fraction (Brodersen et al., 2012) (Fig 5K), whereas AMP1-HA remained in the membrane fraction (Fig 5K). Therefore, AMP1 is an integral membrane protein whereas AGO1 is a peripheral membrane protein. We further examined whether the membrane association of AGO1 was dependent on AMP1. Similar fractionation studies were conducted with rdr6 and rdr6 amp1. No difference in the distribution of AGO1 between soluble and membrane fractions was found in the two genotypes (Fig S4D), indicating that AMP1 is not required for the membrane association of AGO1.
Sucrose gradient-based membrane fractionation with a 35S::GFP-AMP1 line showed a shift of GFP-AMP1 to lighter fractions upon ribosome dissociation in the absence of Mg2+ (Vidaurre et al., 2007), suggesting that AMP1 is present on the rough ER. To confirm this finding with an AMP1 transgene driven by its own promoter, we fractionated membranes with the AMP1::AMP1-GFP rdr6 amp1 line and examined the distribution of AMP1-GFP and AGO1 along the sucrose gradient. In the presence of Mg2+, AMP1-GFP was found in dense and medium-dense fractions and AGO1 was in medium dense fractions (Fig S4E). In the presence of EDTA that chelates Mg2+, AMP1-GFP shifted to medium-dense and light fractions and AGO1 shifted to light fractions (Fig S4E). The Mg2+-induced shift towards heavier fractions for the two proteins suggest that both are associated with membranes bound by ribosomes or large protein complexes. The distribution of the two proteins in the presence of Mg2+ partially overlapped, consistent with the two proteins acting together.
To confirm the rough ER localization of AMP1, we performed immunogold labeling for AMP1-HA in an AMP1p::AMP1-HA rdr6amp1 line and wild type (a negative control). AMP1-HA signals in AMP1p::AMP1-HA rdr6amp1 were associated with tubular structures characteristic of ER (Fig 5L). Quantification of AMP1-HA signals in AMP1p::AMP1-HA rdr6 amp1 and wild type revealed that the labeling was specific (Table S2). Co-localization with the ER marker Calnexin confirmed the ER localization of AMP1-HA (Fig 5M). The ER with AMP1-HA signals was also associated with ribosomes (Fig 5L), suggesting that AMP1 localizes to the rough ER. However, as rough ER was the main form of ER in the root tip cells examined, we could not exclude an association of AMP1 with smooth ER.
In conclusion, AMP1 is an integral membrane protein localized on the rough ER. AGO1 is a peripheral ER membrane protein that is also possibly associated with the rough ER. The rough ER association of AGO1 is supported by the observed Mg2+-induced shift in sucrose gradients (Fig S4E) and the presence of AGO1 on polysomes (Lanet et al., 2009). Given the ER association of both proteins, the observed co-IP between AMP1 and AGO1 (Fig 5A, B) could be attributable to the two proteins being in close proximity on the same membrane rather than being in the same protein complex.
miRNA target transcripts are similarly associated with total polysomes in wild type and amp1 lamp1
To begin to determine how AMP1 mediates the translation inhibition activity of miRNAs, we examined the distribution of miRNA target transcripts along polysomes in wild type and the amp1 lamp1 double mutant. We first extracted total polysomes (TPs), which are composed of soluble as well as membrane- and cytoskeleton-associated polysomes, and resolved them on sucrose density gradients. The A254 absorption profiles of TPs from WT and amp1 lamp1 were largely similar (Fig S5A), suggesting that AMP1 does not affect translation in general. The TP gradient was collected into 17 fractions, and the amount of mRNAs in each fraction was determined by real-time RT-PCR. Five miRNA target transcripts, PHB, CSD2, CUC2, SCL6, and TCP4 (all targeted by different miRNAs), as well as the non-target UBQ5 RNA, were similarly distributed along the TP profiles in wild type and amp1 lamp1 (Fig S5B).
miRNA target transcripts show enhanced association with membrane-bound polysomes in amp1 lamp1
The requirement for ER-localized AMP1 for the translation inhibition activity of miRNAs suggests that miRNAs inhibit the translation of their target mRNAs on the ER. To test this hypothesis, we examined the distribution of miRNA target transcripts along membrane-bound polysomes (MBPs). We isolated microsomes through sucrose step gradient centrifugation (Fig 6A) using optimized conditions that allowed for the preservation of AMP1, AGO1 and intact ribosomal RNAs in the microsomes (Fig S5C, D). The quality of the microsome fraction was shown by the presence of the ER membrane protein SEC12, the absence of the soluble protein PEPC, and the type and quality of the rRNA species (Fig S5C, D). We first quantified the levels of various transcripts, including those targeted by miRNAs, in the microsome fraction. The proportion of microsome-associated RNA in total RNA for most tested genes - both miRNA targets as well as non-targets such as UBQ5, eIF4A and ACTIN8 - was similar in wild type and amp1 lamp1 (Fig 6B), suggesting that AMP1 is not required for the membrane association of these transcripts. AMP1 is also dispensable for the membrane association of the AGO1 protein. AGO1 was present in the microsome fraction in both wild type and amp1 lamp1 and was at a higher level in the latter (Fig 6C), consistent with its increased abundance in the double mutant (Fig 4E, Fig S2I).
Figure 6.
Profiles of membrane-bound polysomes (MBPs) from wild type (Col) and the amp1 lamp1 double mutant, and the distribution of various mRNAs along MBPs. (A) A diagram showing the schemes used for the isolation of total polysomes (TPs) and MBPs. (B) Quantification of various transcripts in the microsome fraction by real-time RT-PCR. Eight miRNA target mRNAs and three non-targets (UBQ5, ACTIN8, and eIF4A) were assayed. The amount of a transcript in the microsome fraction is expressed as a proportion of the total amount for the RNA in the cell. The error bars were calculated from three biological repeats. (C) AMP1 or LAMP1 is not required for the microsome association of AGO1. Western blots were performed to determine the levels of AGO1 in total extract and in the microsome fraction. SEC12 and HSC70 serve as an ER marker and a loading control, respectively. (D) A254 absorption profiles of MBPs. 17 sucrose gradient fractions were collected. (E) Distribution of mRNAs along the sucrose gradients in (D) as determined by real-time RT-PCR. AGO1, PHB, CSD2, SPL3, SCL6, and TCP4 are miRNA target transcripts. UBQ5, eIF4A, ACTIN8, At5g66770, At5g28770, and At2g01570 are non-target transcripts. The amount of a specific mRNA in each of the 17 fractions is expressed as the percentage of the total amount of the RNA in microsomes. Fractions with negligible amounts of transcripts are not shown. The error bars were calculated from three biological replicates. P values < 0.05 (*) or 0.01 (**). See also Figure S5.
We next extracted membrane-bound polysomes (MBPs) from the microsome fraction and subjected the MBPs to sucrose density gradient fractionation (Fig 6A). The A254 absorption profiles of MBPs (from either wild type or amp1 lamp1) differed from those of TPs in that the 80S peak was smaller (compare Fig 6D and Fig S5A). Similar observations were made for MBPs from other organisms (Birckbichler and Pryme, 1978; Noll and Burger, 1974). The MBP gradients were collected into 17 fractions, and the amount of mRNAs in each fraction was quantified by real-time RT-PCR. miRNA target transcripts, such as AGO1, PHB, CSD2, SPL3, SCL6, and TCP4, showed a significant increase in MBP association in the double mutant, whereas non-target transcripts such as UBQ5, eIF4A, ACTIN8, At2g01570, At5g28770, and At5g66770 were similarly distributed along MBP fractions in wild type and amp1 lamp1 (Fig 6E). Note that At2g01570, At5g28770, and At5g66770, which encode transcription factors, were included in the analysis because their transcript levels were more similar to those of the miRNA target transcripts than the abundant UBQ5, eIF4A, and ACTIN8 transcripts. The similar degree of microsome association of miRNA target transcripts in wild type and amp1 lamp1 (Fig 6B) contrasts with their preferential loading onto MBPs in amp1 lamp1 (Fig 6E) and indicates that miRNAs inhibit the translation of their target transcripts on membranes. The requirement for the ER protein AMP1 for this activity suggests that ER is the site of miRNA-mediated translation inhibition.
Discussion
ER as the site of miRNA-mediated translation repression
The subcellular location where miRNAs repress the translation of their target mRNAs has been unknown. In this study, we show that miRNA-mediated translation inhibition but not target RNA cleavage requires AMP1. Characterization of the subcellular localization of AMP1 and comparison of the distribution of miRNA target RNAs on TPs versus MBPs reveal that miRNA-mediated translation inhibition occurs on the ER. Given that homologs of AMP1 are present in animal genomes, it is possible that the connection between the ER and translation inhibition by miRNAs is conserved. Consistent with this, miRNA effectors, AGO1 in plants and Ago2 in mammals, are membrane-associated (Brodersen et al., 2012; Cikaluk et al., 1999).
It is of interest to integrate the knowledge of the ER as the site of translation inhibition by miRNAs with existing knowledge on the subcellular compartmentalization of miRNA biogenesis/action. This study, together with several studies in animals (Gibbings et al., 2012; Gibbings et al., 2009; Lee et al., 2009), establishes the endomembrane system as important sites where miRNA biogenesis/action or its regulation takes place. On the other hand, cytoplasmic granules such as P bodies have also been implicated in RNA silencing (Fabian et al., 2010). Genetic evidence supports a role of the P body component VCS in miRNA-mediated translational repression in Arabidopsis (Brodersen et al., 2008). In cultured human cells, the miRNA let-7 and its target transcript are both localized to cytoplasmic foci adjacent to P bodies (Pillai et al., 2005). How translation repression on the ER is mechanistically connected to P bodies is currently unknown. The P body component DCP1 does not co-localize with ER (Fig 5I, Fig S4C).
Translation on the ER is not restricted to mRNAs encoding membrane or secreted proteins
Our studies also ascribe a new function to the ER. ER is well known as the site of translation of mRNAs encoding membrane or secreted proteins. The classical view is that mRNAs encoding membrane or secreted proteins are recruited to the ER via the signal recognition particle in a translation-dependent manner. However, many studies report puzzling observations that a large portion of the yeast or mammalian transcriptome, including numerous transcripts encoding soluble proteins, is ER-associated or translated on membrane-bound polysomes (Kraut-Cohen and Gerst, 2010; Lerner et al., 2003; Reid and Nicchitta, 2012). Although the ER association of transcripts is likely to have multifaceted functions, we propose that one function is to allow miRNA-mediated inhibition of translation to occur. We envision that miRNA target transcripts are sequestered on the ER or in a compartment linked to the ER to prevent their translation. While the prevention of their translation requires AMP1, the membrane association of the transcripts as well as the AGO1 protein occurs in an AMP1-independent manner.
ER-cytosol partitioning of translation and miRNA activities
The similar loading of miRNA target transcripts onto TPs and the differential loading of the transcripts onto MBPs in wild type and amp1 lamp1 indicate that miRNA-mediated translation inhibition occurs on membranes. Given that a small portion (less than 15%) of the total transcript pool of a miRNA target gene is associated with microsomes (Fig 6B), how could the inhibition of this portion result in the observed differences in protein output between wild type and amp1 lamp1? We suspect that the observed levels of membrane-associated transcripts (Fig 6B) were an under-estimate because membrane breakage during the experimental procedure would reduce the yield of intact microsomes. However, it is still likely that a large fraction of the transcript pool is present in the cytosol and it remains an open question whether ER-associated translation inhibition is sufficient to contribute to the overall activities of miRNAs. One possibility is that translation in the cytosol is also inhibited by miRNAs, and the lack of differences in transcript distribution along TPs between wild type and amp1 lamp1 could be due to pseudo-polysomes (large RNA-protein complexes) induced by miRNAs, which were observed in Drosophila (Thermann and Hentze, 2007). However, under this scenario, translation inhibition in the cytosol also needs to be AMP1-dependent for it to account for the protein output differences between wild type and amp1 lamp1. An alternative hypothesis is that translation on the ER is much more efficient than that in the cytosol, such that inhibition of a small, ER-associated portion of the transcript pool is sufficient to account for the differences in protein output. In fact, an analysis of ER-cytosol partitioning of transcripts and their translation status as reflected by their ribosome association in HEK293 cells suggested that ER serves as the site of translation for a broad set of transcripts including those encoding soluble proteins (Reid and Nicchitta, 2012). Furthermore, the study suggested that translation is more efficient on the ER than in the cytosol in terms of ribosome density per transcript as well as the processivity of translation elongation (Reid and Nicchitta, 2012).
The translation inhibition activity of plant miRNAs is probably crucial for development
Plant miRNAs are key players in almost all developmental events and regulate target transcripts through two modes of action, cleavage and translation inhibition. Previous studies on a number of miRNAs such as miR156, miR172, and miR398 show that both modes of action act upon the same target transcripts (Aukerman and Sakai, 2003; Brodersen et al., 2008; Chen et al., 2004; Chen, 2004; Gandikota et al., 2007; Kasschau et al., 2003; Schwab et al., 2005; Sunkar et al., 2006). In this study, we extend similar observations to miR164 and miR165/6 and show that the two modes of action are genetically separable (Fig 2). This raises the question regarding the relative contribution of each mode of action to plant development. The cleavage activity is likely crucial for plant development, as a slicer-defective AGO1 transgene fails to rescue the developmental defects of an ago1 null mutant (Carbonell et al., 2012). How about the contribution of translation inhibition to plant development?
Mutations in AMP1 lead to pleiotropic developmental defects such that amp1 alleles were independently isolated in a number of genetic screens focusing on various aspects of plant development and the gene was given many names, such as AMP1 (Chaudhury et al., 1993), CONSTITUTIVE MORPHOGENESIS2 (Hou et al., 1993), HAUPTLING (Jurgens et al., 1991), MULTIFOLIA (Lee, 2009), and PRIMORDIA TIMING (Mordhorst et al., 1998), to reflect the phenotypes of interest. AMP1 is a homolog of human glutamate carboxypeptidase II, and an active site glutamate (E) in the human enzyme corresponds to E404 in AMP1, which is mutated to lysine (K) in the pt allele of AMP1 (Fig 4A) (Helliwell et al., 2001). This led to the hypothesis that AMP1 produces an as yet unidentified small molecule that impacts multiple aspects of plant development.
In this study, we uncover a role of AMP1 and LAMP1 in miRNA-mediated translation inhibition but not transcript cleavage. Given that plant miRNAs impact many aspects of plant development, it is not surprising that amp1 alleles were isolated in a number of genetic screens focusing on various aspects of plant development. The severe developmental defects of the amp1 lamp1 double mutant suggest that translation inhibition is an essential activity of plant miRNAs. However, it cannot be excluded that AMP1 and LAMP1 possess miRNA-independent functions and these functions contribute to the pleiotropic developmental defects of the amp1 lamp1 mutant.
Experimental Procedures
Measurement of protein synthesis and protein half-life by metabolic labeling and immunoprecipitation (IP)
For pulse-labeling-based measurement of protein synthesis, rdr6 CSD2-HA and rdr6 amp1 CSD2-HA seedlings were incubated in ATS liquid medium containing 800μCi TRAN 35S-LABEL (MP Biochemicals) for 20 and 40 min. IP was then performed with anti-HA affinity matrix (Roche) to precipitate both labeled (newly synthesized) and non-labeled (steady-state) CSD2-HA, which were subsequently detected by autoradiography and western blotting, respectively. Pulse-chase experiments were performed to measure the half-life of PHB-MYC. rdr6 PHB-MYC and rdr6 amp1 PHB-MYC seedlings were incubated in ATS liquid medium containing 1mCi TRAN 35S-LABEL for 5h. After labeling, plants were washed with ATS liquid medium and incubated in ATS liquid medium containing 1mM Methionine/Cysteine and 100 μg/mL cycloheximide for 0–3h. IP was performed with anti-c-myc affinity gel (Sigma).
Isolation and fractionation of membrane bound polysomes
To isolate membrane-bound polysomes (MBPs), 2g seedlings were ground in liquid nitrogen, and the powder was suspended in 7ml microsome extraction buffer (MEB, see Supplemental Experimental Procedures). The slurry was filtered with 2 layers of miraclothand centrifuged at 10,000g for 15 min at 4°C to remove debris. 100μl of the supernatant was saved as the total RNA input, and the rest was loaded onto the top of a 2.5ml 0.6M/2.5ml 1.7M sucrose step gradient and centrifuged at 140,000g in a Beckman SW41 rotor for 1 h at 4°C. The microsome fraction, which sediments to the 0.6M/1.7M layer interface, was carefully transferred into a new tube. The microsome fraction was diluted 10 times by MEB and centrifuged at 140,000g for 0.5 h to pellet the microsomes. The microsomes were dissolved in 8ml polysome extraction buffer (PEB; see Supplemental Experimental Procedures). Next, sucrose density gradient fractionation was conducted on the microsomes following the same procedure as for total polysomes (Mustroph et al., 2009).
See Supplemental Experimental Procedures for details on the above-described methods as well as additional methods and procedures.
Supplementary Material
Highlights.
miRNA-mediated translation inhibition but not translation in general requires AMP1
AMP1 is dispensable for miRNA-mediated target RNA cleavage
AMP1 is an integral membrane protein that is localized to the rough ER
AMP1 inhibits the loading of miRNA target transcripts onto membrane-bound polysomes
Acknowledgments
We are grateful to Dr. Julia Bailey-Serres for technical advice on polysome profiling. We thank Drs. Kathryn Barton, Nam-Hai Chua, Daniel Kleibenstein, Elliot Meyerowitz, and Zhiyong Wang for sharing plasmids, seeds, or antibodies. We thank Julia Bailey-Serres, Theresa Dinh, Elizabeth Luscher, Kestrel Rogers, and Yuanyuan Zhao for comments on the manuscript. The work was funded by National Institutes of Health (GM061146) and by Howard Hughes Medical Institute and Gordon and Betty Moore Foundation (through Grant GBMF3046) to X. Chen, by the Research Grants Council of Hong Kong (CUHK466610, CUHK466011 and CUHK2/CRF/11G) to L.J, and by National Science Foundation of China to X. Cao, B.M., and X. Chen. L.L. was supported by a fellowship from China Scholarship Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci U S A. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birckbichler PJ, Pryme IF. Fractionation of membrane-bound polysomes, free polysomes, and nuclei from tissue-cultured cells. Eur J Biochem. 1978;33:368–373. doi: 10.1111/j.1432-1033.1973.tb02692.x. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Schaller H, Khafif M, Schott G, Bendahmane A, Voinnet O. Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:1778–1783. doi: 10.1073/pnas.1112500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A, Fahlgren N, Garcia-Ruiz H, Gilbert KB, Montgomery TA, Nguyen T, Cuperus JT, Carrington JC. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell. 2012;24:3613–3629. doi: 10.1105/tpc.112.099945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury AM, Letham S, Craig S, Dennis ES. amp1: A mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precious flowering. Plant J. 1993;4:907–916. [Google Scholar]
- Chen J, Li WX, Xie D, Peng JR, Ding SW. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microrna in host gene expression. Plant Cell. 2004;16:1302–1313. doi: 10.1105/tpc.018986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cikaluk DE, Tahbaz N, Hendricks LC, DiMattia GE, Hansen D, Pilgrim D, Hobman TC. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol Biol Cell. 1999;10:3357–3372. doi: 10.1091/mbc.10.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway LJ, Poethig RS. Mutations of Arabidopsis thaliana that transform leaves into cotyledons. Proc Natl Acad Sci U S A. 1997;94:10209–10214. doi: 10.1073/pnas.94.19.10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Fukaya T, Tomari Y. MicroRNAs mediate gene silencing via multiple different pathways in Drosophila. Mol Cell. 2012;48:825–836. doi: 10.1016/j.molcel.2012.09.024. [DOI] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007;49:683–693. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- Goeres DC, Van Norman JM, Zhang W, Fauver NA, Spencer ML, Sieburth LE. Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell. 2007;19:1549–1564. doi: 10.1105/tpc.106.047621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Chin-Atkins AN, Wilson IW, Chapple R, Dennis ES, Chaudhury A. The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell. 2001;13:2115–2125. doi: 10.1105/TPC.010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Von Arnim AG, Deng XW. A new class of Arabidopsis constitutive photomorphogenic genes involved in regulating cotyledon development. Plant cell. 1993;5:329–339. doi: 10.1105/tpc.5.3.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Jurgens G, Mayer U, Ruiz R, Berleth T, Misera S. Genetic analysis of pattern formation in the Arabidopsis embryo. Development. 1991;1(suppl):27–38. [Google Scholar]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- Kraut-Cohen J, Gerst JE. Addressing mRNAs to the ER: cis sequences act up! Trends Biochem Sci. 2010;35:459–469. doi: 10.1016/j.tibs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Lanet E, Delannoy E, Sormani R, Floris M, Brodersen P, Crete P, Voinnet O, Robaglia C. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell. 2009;21:1762–1768. doi: 10.1105/tpc.108.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B-h. Ecotype-dependent genetic regulaton of bolting time in the Arabidopsis mutants with increased number of leaves. J Microbial Biotechnol. 2009;19:542–546. [PubMed] [Google Scholar]
- Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim do H, Cho IS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, Nicchitta CV. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. Rna. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B. MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol. 2004a;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 2004b;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Mordhorst AP, Voerman KJ, Hartog MV, Meijer EA, van Went J, Koornneef M, de Vries SC. Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics. 1998;149:549–563. doi: 10.1093/genetics/149.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. ArabidopsisSGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Mustroph A, Juntawong P, Bailey-Serres J. Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol. 2009;553:109–126. doi: 10.1007/978-1-60327-563-7_6. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Noll M, Burger MM. Membrane-bound and free polysomes in transformed and untransformed fibroblast cells. J Mol Bio. 1974;90:215–236. doi: 10.1016/0022-2836(74)90369-6. [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Reid DW, Nicchitta CV. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem. 2012;287:5518–5527. doi: 10.1074/jbc.M111.312280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Schwartz BW, Yeung EC, Meinke DW. Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development. 1994;120:3235–3245. doi: 10.1242/dev.120.11.3235. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Vidaurre DP, Ploense S, Krogan NT, Berleth T. AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development. 2007;134:2561–2567. doi: 10.1242/dev.006759. [DOI] [PubMed] [Google Scholar]
- Won SY, Li S, Zheng B, Zhao Y, Li D, Zhao X, Yi H, Gao L, Dinh TT, Chen X. Development of a luciferase-based reporter of transcriptional gene silencing that enables bidirectional mutant screening in Arabidopsis thaliana. Silence. 2012;3:6. doi: 10.1186/1758-907X-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang JY, Niu QW, Chua NH. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–3398. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wu G, Poethig RS. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:315–320. doi: 10.1073/pnas.1114673109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.