Abstract
This review aims to provide an overview of the technologies which make the nematode Caenorhabditis elegans an attractive genetic model system. We describe transgenesis techniques and forward and reverse genetic approaches to isolate mutants and clone genes. In addition, we discuss the new possibilities offered by genome engineering strategies and next generation genome analysis tools.
Introduction
The modern era of C. elegans research began less than fifty years ago when Sydney Brenner settled on the small nematode to tackle an ambitious goal laid out in his seminal paper published in 1974: “Some eight years ago, […] I decided that what was needed was an experimental organism which was suitable for genetical study and in which one could determine the complete structure of the nervous system. Drosophila, with about 105 neurons, is much too large, and, looking for a simpler organism, my choice eventually settled on the small nematode, Caenorhabditis elegans”.1 From the start, Brenner outlined a methodology that still prevails today: “One experimental approach to these problems is to investigate the effects of mutations on nervous systems. In principle, it should be possible to dissect the genetic specification of a nervous system in much the same way as was done for biosynthetic pathways in bacteria […]”.1 In the following years, Brenner and his colleagues completed a series of major projects that laid the foundations for C. elegans research: (1) they established the genetics of C. elegans and performed the first large scale mutant screens;1 (2) they described the complete and largely invariant lineage of all 959 somatic C. elegans cells (plus 131 cells that die by programmed cell death) using standard light microscopy;2, 3 (3) they achieved the complete reconstruction of the adult nervous system by using electron micrographs of serial sections.4
Over the years, a number of discoveries added considerable attractiveness to C. elegans as a biological model organism. One important advance was the generation of transgenic animals, achieved through an efficient transgenesis technique developed in 1991 by Mello et al.5 and still in use to this day. In 1994, GFP reporter technology introduced by Martin Chalfie, allowed the visualization of gene expression patterns, as well as protein localization in live animals.6 In 1998, the discovery of genetic interference by double-stranded RNA (RNAi)7 enabled alternative approaches for large scale genetic screens. In the same year, the completion of the C. elegans genome sequence8 provided the first comprehensive view over the complete genomic landscape of a metazoan organism. This review aims to describe the genetic toolbox available to C. elegans researchers by focusing on classical and novel techniques used for forward and reverse genetic analysis and recent developments in genome engineering by homologous recombination.
FORWARD GENETICS
Forward genetic screens in C. elegans have provided key mechanistic insights into deeply conserved developmental, homeostatic and pathogenic processes (see www.wormbook.org). We will describe here the different methods which can be used to generate and clone mutants from forward genetic screens. Previous monographs have discussed some of the more traditional methods in more detail (wormbook.org and 9, 10).
Random mutagenesis with chemical and physical mutagens
Random mutations can be generated by exposing worms to mutagenic agents such as EMS (Ethyl methanesulfonate), ENU (N-ethyl-N-nitrosourea) or trimethylpsoralen followed by ultraviolet light activation (TMP/UV).9 EMS is the most popular mutagen in C. elegans. It alkylates primarily guanine residues leading to mispairing during replication: alkylated G pairs to T instead of C. The resulting mutations are mainly G/C to A/T transitions. EMS also introduces deletions, yet at much lower frequency than point mutations. ENU is also a potent mutagen which generates transversions in addition to transitions. Mutagenesis by EMS and ENU was compared in a genetic suppressor screen of the rubber band phenotype induced by the unc-93(e1500) mutation.11 Both mutagens produced revertants at similar frequencies but the types of alleles and the number of alleles per known suppressor gene were different. In particular, 90% of EMS point mutants were G/C to A/T transitions, which are more likely to produce stop codons rather that missense mutations. In contrast, the rate of A/T to G/C transitions was greatly increased with ENU. This analysis may be partially biased by the A/T richness of unc-93 but has been largely confirmed by examining whole-genome sequencing data.12, 13 Interestingly, UV/TMP treatment appears to induce all types of transitions and transversions, with almost no bias.12 EMS is the most powerful mutagen causing 1.5 to 2 times more mutations than ENU and more than 3 times more variations than UV/TMP. Which mutagen to choose will therefore be determined by the type of desired mutations. EMS remains the best choice to obtain null alleles, however ENU generates the biggest diversity of missense alleles which can sometimes be more informative for functional studies. Gamma, UV and X-ray irradiation cause major chromosomal rearrangements and have been mostly used to generate duplications, inversions, deficiencies and translocations.9
Classical mutant mapping
Classically, mutants have been mapped to a genetic interval by a combination of the following techniques (extensively reviewed in “Genetic mapping and manipulation”, www.wormbook.org): (1) Two- or three-point mapping with genetic markers;14 (2) Mapping with deficiencies and duplications;15 (3) Single Nucleotide Polymorphism (SNP) mapping.16
Two- or three-point mapping with genetic markers relies on the measure of the genetic distance (i.e. recombination frequency) between phenotypic mutants (with known physical positions) and the gene to be mapped. This approach can be used to rapidly assign a given mutation to a chromosomal region, however it lacks resolutions due to the relatively small number of such “landmark” mutants. In addition, genetic interactions between the landmark mutant and the target gene can sometimes complicate mapping. Similarly, deficiencies and duplications are not always precisely defined at their extremities. Classical genetic mapping was therefore rapidly supplanted by the introduction of SNP mapping techniques.
SNP mapping in C. elegans is based on the sequence divergence between the reference strain N2 Bristol and the CB4856 Hawaiian strain.17 SNPs occur on average every 1,000 base pairs. Most SNPs are indeed single nucleotide changes, but small insertions or deletions are also common. SNPs which lead to restriction-fragment length polymorphisms can be easily detected by restriction digests while “silent” SNPs need to be sequenced. SNP mapping can rapidly assign mutants to a chromosome18 and narrow down the position of a mutation of interest to relatively few candidate ORFs. Note however that an analysis of the CB4856 Hawaiian strain by array comparative genome hybridization has revealed 141 deletions removing 1.54 Mb of DNA and 483 predicted genes from CB4856’s genome.19 In addition, complex genetic interactions20 and phenotypic variation of mutants in the Bristol or Hawaiian background can sometimes confuse mapping results. These and other limitations have encouraged the development of novel gene cloning techniques described in the next section.
New approaches for mapping mutants
Traditional mapping strategies can be time-consuming undertakings depending on the phenotypic complexity (e.g. tedious to score, variable phenotype, low penetrance) and genetic nature (e.g. suppressor or enhancer mutant) of the studied mutant. Two “next generation” technologies circumvent these problems.
Array Comparative Genome Hybridization
Array Comparative Genome Hybridization (aCGH) was first developed in C. elegans to accelerate the identification of “indel” (insertion/deletion) mutants for the C. elegans Knockout Consortium.19 It uses high density oligonucleotide microarrays (~380,000 probes/array) to detect sequence polymorphisms (SNP, indels) for the entire genome or specific regions of interest. The great sensitivity of aCGH allows the detection of heterozygous deletions as small as 141 bp. The resolution is directly related to the extent of the overlap between 50mer oligonucleotides on the array. In addition, aCGH can map a mutation to a small chromosomal interval (~200 – 400 kb) by analyzing the SNP distribution in homozygous mutants resulting from a single genetic cross of Hawaiian and N2 strains.21 In theory this approach should also be able to resolve polygenic traits.
Beyond SNP and deletion mapping, aCGH can directly detect homozygous single nucleotide mutations.22 To maximize the chances of detection, a maximum probe distance of less than 5 nucleotides is recommended. Given the probe capacity of the array, a mutation should therefore be mapped to a 1 Mb interval prior to aCGH, in order to achieve the highest probe density on both strands. Alternatively, multiple sub-arrays could be used to further increase coverage. For reference, aCGH has been successfully used to detect two single base lesions in the promoter of the cog-1 gene.23
Whole-Genome Sequencing
The most recent paradigm shift for C. elegans researchers is the development of whole-genome sequencing (WGS) methods. The first C. elegans mutant cloned by WGS (lsy-12(ot177)) was reported in 2008.24 lsy-12(ot177) was initially mapped by a single SNP mapping cross to a 4 Mb region of chromosome V containing ~1100 genes. Paired-end Illumina sequencing generated 3.1 Gb of mapped reads resulting in ~28-fold genome coverage. Analysis and filtering of these sequences produced a list of 80 candidate variants which were tested by Sanger sequencing. This list was further reduced to four candidates by focusing on exonic mutations and by comparing variants between the lsy-12(ot177) mutant strain and the N2 strain used for outcrossing. Only one of the four polymorphisms was a nonsense mutation and mapped to the lsy-12 gene. This demonstrated for the first time the feasibility of gene cloning by rough mapping and WGS.
The strengths of WGS are evident. (1) It is fast. Sequencing currently takes ~5 days and data analysis a few hours using optimized bioinformatic tools such as MAQgene25 or galign.26 (2) It is highly cost effective. Given the comparatively small size of the C. elegans genome (~100 Mb) and recent technology advances, a genome can be sequenced for less than US$1,000. This expense compares favorably with the time, personnel and reagent cost involved for classical mapping and is also expected to drop even much further. (3) It greatly simplifies cloning of mutants with subtle, low penetrance or difficult to score phenotypes. Multigenic, quantitative or behavioral genetic traits which are sensitive to genetic backgrounds and therefore hard to map by conventional approaches will be much more accessible with WGS. (4) In addition to point mutations, WGS is also able to detect deletions depending on sequencing depth.13 (5) Sequencing errors are negligible when sufficient coverage is reached, reducing the risks of false negatives. (6) As it is relatively easy to obtain sufficient amounts of DNA, the technology can also be used for analyzing lethal mutants. For example, a homozygous lethal mutant in the lsy-22 gene could be cloned by sequencing DNA extracted from a few hundred hand-picked, sterile worms.27
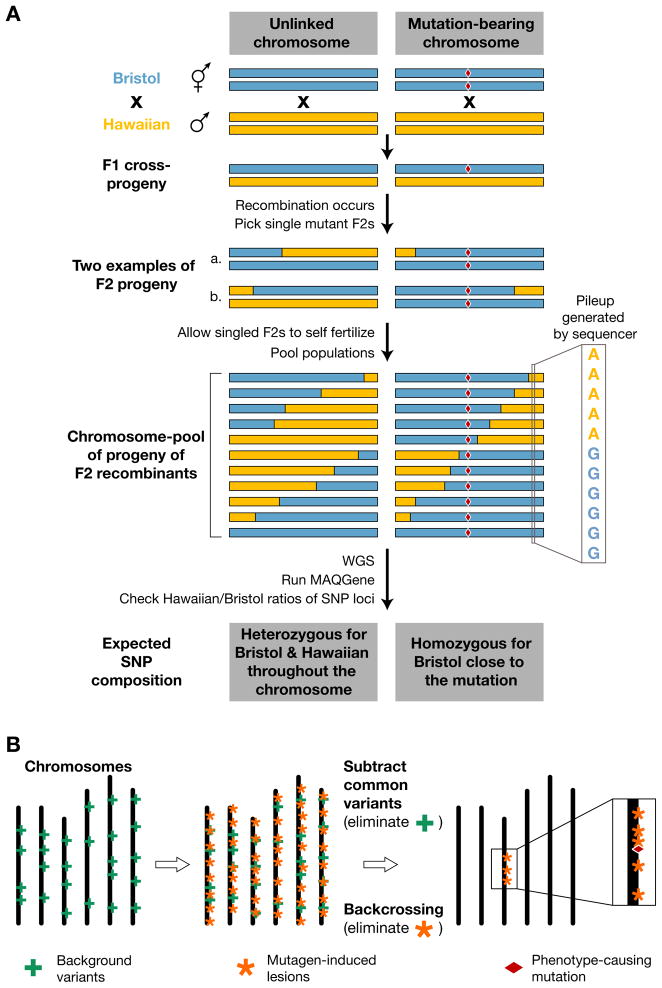
Chemical mutagenesis generates hundreds of sequence variants per genome.12, 13 Initially, genetic mapping information was therefore considered crucial, unless independent alleles of the same gene were available for WGS. A novel one-step strategy that combines SNP mapping and whole-genome sequencing has recently been described (see Fig. 1).28 Instead of focusing only on a few manually analyzed SNPs, this approach can theoretically take advantage of every SNP in the genome. The mutant strain isolated in an N2 Bristol background is first crossed with the polymorphic CB4856 Hawaiian strain and 20 to 50 mutant F2 recombinants are picked to single plates. These Bristol/Hawaiian recombinant lines are then pooled and analyzed by WGS. Chromosomal regions which are linked to the mutation are characterized by a strong increase in the Bristol to Hawaiian SNP ratio. In this proof-of-principle study, the SNP mapping information defined a ~2 Mb region, which only contained three protein-coding sequence variants.28
Figure 1. Mutant mapping by whole genome sequencing.
(A) WGS-SNP mapping. A homozygous mutant strain (mutation denoted by a red diamond) in a Bristol background is crossed with a SNP-containing Hawaiian strain. Mutant worms segregating from this cross are reselected in the F2 generation. These mutant lines carry chimeric chromosomes (containing Bristol and Hawaiian SNPs) generated by meiotic recombination. When DNA from multiple F2 recombinant lines is pooled and sequenced, then the relative number of Bristol vs. Hawaiian SNP at a given location in the genome will be a reflection of the relative distribution of recombinants in the pool. Therefore, only Bristol SNPs will be found close to the mutation of interest (adapted from Ref. 28).
(B) Mapping based on mutagen-induced DNA variation density across the genome. Mutagenized strains contain not only mutagen-induced damage (orange asterisks) but also carry background variants (green crosses) present in the mother strain before mutagenesis. By backcrossing a candidate mutant line to the strain used for the original mutagenesis, it is possible to eliminate most mutagen-induced lesions except for those which are closely linked to the phenotype-causing mutation (red diamond). In addition, background variation present in the unmutagenized strain can by identified by comparing independent backcrossed mutant lines. After filtering, the genomic region linked to the phenotype-causing mutation will be highlighted by a high-density cluster of mutagen-induced lesions (adapted from Ref. 29).
Another strategy takes advantage of the fact that phenotype-relevant EMS mutations should cluster around the mutation of interest after outcrossing (see Fig. 1).29 Rather than using the polymorphic Hawaiian strain, mutants retrieved from a genetic screen are backcrossed four to six times with the original strain used for mutagenesis. Their complete genome sequences are aligned to the N2 reference genome and variants which are common between at least two mutant strains (i.e. background mutations) are filtered out. Hotspots containing typical EMS mutations can then be detected by plotting sequence variants along chromosomes, highlighting the likely position of the mutant gene (see Fig. 1). Finally, analysis of individual variants provides a list of candidate genes which can be validated by other approaches such as phenotypic rescue, RNAi or analysis of additional alleles by Sanger sequencing.
Transposon-based insertional mutagenesis
Transposons are mobile DNA elements present in the genomes of all living organisms. While mobile elements are less efficient than chemical mutagens (see below), the resulting mutants can be more easily cloned because the transposon sequence serves as a molecular tag. This advantage will be less striking in the future, given the rapid development of whole-genome sequencing. However, transposons have unique and powerful downstream applications, i.e. generating custom deletions and engineering the genome.
Endogenous Transposons
Tc1 was the first transposable element identified in C. elegans and a founding member of the Tc1/mariner superfamily.30 These class II elements move throughout the host genome via a “cut-and-paste” mechanism in which a specific transposon-encoded transposase binds the two terminal inverted repeats of the transposon and catalyses its excision and reinsertion into a TA dinucleotide. In wild-type strains, Tc transposons are only mobile in somatic cells. However, “mutator” backgrounds can lift germline silencing31 and allow insertional mutagenesis using Tc elements. In 1985, long before any of the genome project tools were available, Iva Greenwald cloned the first C. elegans gene, lin-12/Notch, by using a transposon-tagging and chromosome walking approach.32 Subsequently, many genetic screens were performed with Tc1 and the localization of mutagenic Tc1 insertions was further simplified by the development of a “transposon-insertion display” technique.33–36
While Tc1 has been a useful tool for forward genetic screens, some limitations have hampered its routine use: (1) Transposition in mutator backgrounds is an uncontrolled process. Most mutator backgrounds derepress all classes of Tc1/mariner transposons and lead to an accumulation of transposon copies in the genome, making it harder to identify the mutagenic insertion. (2) Tc mutants are inherently unstable in mutator backgrounds. Spontaneous mobilization of a mutagenic insertion can sometimes lead to imprecise excision of the transposon leaving a DNA footprint which can remain mutagenic but is no longer linked to a transposon. (3) Tc1 transposons can be spliced out of pre-mRNAs.37 This results in a mature mRNA that either contains small insertions of Tc1 sequences and/or lacks gene sequences at the spliced junctions.
Given these limitations, a number of labs have attempted to mobilize heterologous transposons in the C. elegans germline (Himar, Sleeping Beauty, Minos). However, all of these transposons could only be mobilized in somatic cells so far. However, one publication reports heritable insertions of the medaka fish transposon Tol1.38 Although this system needs to be further characterized, it could serve as the basis for the a gene delivery system since, contrary to Tc1/mariner transposons, Tol1 can carry exogenous sequences and still be reasonably active.39 To date, the only efficient transposon mutagenesis strategy using a heterologous transposon is based on the mobilization of the Drosophila mauritiana element Mos1.40, 41
Mos1 Transposon Mutagenesis
Mos1 is a member of the Tc1/mariner transposon family. It only requires its transposase but no host cell factor for transposition. Therefore, it has been possible to mobilize Mos1 in vitro and in a number of evolutionarily distant species.42, 43 The Mos1 sequence is absent from the C. elegans genome making it a unique molecular feature which can be easily identified by standard molecular biology techniques (see Fig. 2).44 Mos1-containing strains can be outcrossed against N2 or any other N2 derived background, removing all unlinked copies. Mos1 transposons are only active when the Mos1 transposase is provided. Therefore, Mos1 insertions are stable in the genome once the source of transposase has been lost. Finally, a study of over 900 insertions demonstrated no strong insertional bias of Mos1 except for a site on chromosome I.45
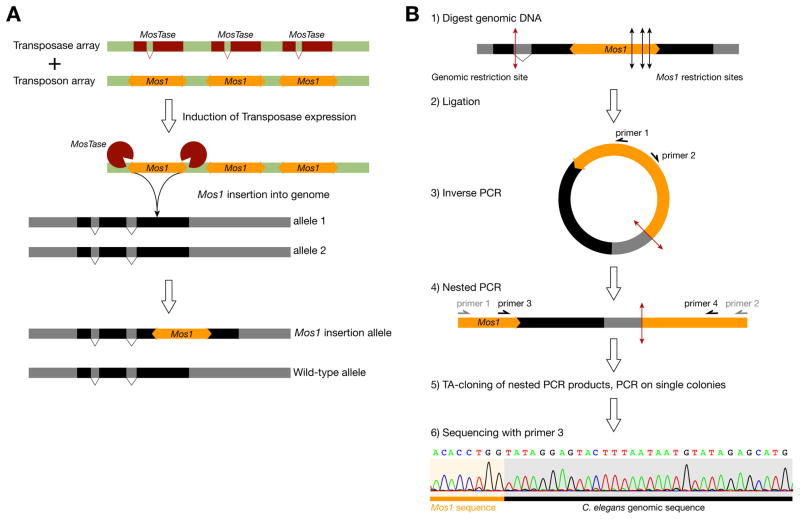
Figure 2. Mos1-based insertional mutagenesis.
(A) Mos1 can jump from an extrachromosomal transgene carrying multiple copies of the transposon into the genome, following the expression of the Mos1-specific transposase via a heat-shock inducible transgene. Mos1 insertions are generally random in the genome but always takes place at a TA dinucleotide.
(B) Mos1 insertions can be rapidly mapped with single nucleotide resolution by sequencing the flanking genomic sequences using an inverse PCR strategy (adapted from Ref. 44).
Mos1 mutagenesis is based on a binary system (see Fig. 2).40, 44 Two extrachromosomal transgenes are used: one that contains multiple copies of the Mos1 transposon (transposon array), and another that provides the transposase under the control of a heat-shock inducible promoter (transposase array). When the transposase activity is induced in doubly transgenic worms, Mos1 can jump from the “transposon array” into the genome. The extrachromosomal arrays are lost in subsequent generations, eliminating the risk for Mos1 re-excision. Mos1 is approximately 10 times less efficient than EMS as a mutagen under optimal conditions.46 This low efficiency has to be contrasted with the ease, speed and precision with which Mos1 can be located.44 Mos1 transposons in a given strain can be quickly mapped by inverse PCR and sequencing of the Mos1-flanking genomic regions, revealing their location with single base pair precision (see Fig. 2). On average, 2.5 insertions (range from 1 to 10) can be found per animal, making it easy to genetically associate a given insertion with a mutant phenotype. Mos1 has been used successfully in different forward genetic screens.47–50 In addition, Mos1 mutagenesis can also be used in C. briggsae, a related nematode species useful for comparative functional studies.51 Most importantly, each Mos1 insertion can be used as a starting point to engineer the genome, as described in detail later in this review.
Automating genetic screens
There are many clever ways to perform genetic screens in C. elegans52 but many of them still rely on a researcher sitting for innumerable hours behind a microscope sifting through mountains of worms. In contrast, “high throughput” screens make it possible to (1) approach genetic saturation, (2) use less efficient mutagens (e.g. transposons) and (3) increase the chances of finding rare and unusual alleles (e.g. temperature-sensitive, hyper-, anti- or neomorphic alleles). Recently, certain screens have been automated using “worm sorters” and microfluidic devices.
The COPAS (Complex Object Parametric Analyzer and Sorter) Biosort machine (Union Biometrica) can sort hundreds of worms per minute, based on fluorescence, optical density or length. It is used to distribute worms into multiwell plates, for example for high-throughput screening of chemical libraries53, 54 or to generate C. elegans libraries.55 But its most powerful feature is the ability to measure fluorescence profiles along the length of the animal at two different wavelength. This has allowed for example to analyse spatiotemporal promoter activity,56 identify mutants required for the execution of neuronal cell fates,57 or screen for genes involved in the innate immune response of C. elegans to pathogens.58
Although the COPAS sorter is able to distinguish fluorescence changes along the body axis, it is not adapted for high-resolution analysis. Microfluidic devices have now been developed that allow automated screening of live C. elegans.59, 60 Worms can be automatically loaded into “worm chips”, mechanically immobilized, imaged at high-resolution and sorted based on complex phenotypic criteria with high accuracy. However, these elaborate systems require more processing time, with throughput still being significantly smaller than the COPAS worm sorter. Alternatively, approaches combining high resolution confocal microscopy and image analysis software could be adapted for some screening approaches. By taking advantage of the reproducibility of the C. elegans cell lineage and anatomy, it was possible for example to automatically assess the expression profiles of ~100 genes in ~400 cells of L1 stage larvae61 and to analyse gene expression during embryonic development with single-cell resolution.62
REVERSE GENETICS
With the completion of the worm’s genome sequence over a decade ago,8 reverse genetic approaches have become increasingly popular. Instead of searching for genes implicated in a given process by phenotype (“forward genetics”), the goal of reverse genetics is to characterize the function of a selected gene by inactivating the expression of the gene either by RNAi or by isolating a loss-of-function allele.
Manipulating gene activity by RNAi
RNA interference (RNAi) was first discovered in C. elegans,7, 63 rapidly generalized to all other model systems from plants to vertebrates and recognized as a revolutionary approach to modify gene expression with potential medical applications. RNAi immediately became an attractive gene knock-down technology in C. elegans. With RNAi and the complete genome sequence, it was theoretically possible for the first time to easily and rapidly test the function of a large number of genes in a systematic fashion. This led to remarkable large scale screens for cytokinesis,64 essential genes and cell polarity,65 lifespan,66 fat regulation67 and gene function at the whole-genome level.68, 69 Hundreds of genes gained new functions through RNAi-based studies. However it also became clear that not all genes were affected by RNAi and affected genes often did not show phenotypes as strong as corresponding genetic null mutants. Intriguingly, RNAi seemed to be much more efficient in certain tissues, with neurons being often refractory to RNAi. This surprising result led researchers to investigate the basis for this tissue specificity and identify sensitized mutant backgrounds which display increased RNAi sensitivity.70–75
RNAi can be applied at any stage during development, which makes it easier to study essential genes or to study a gene’s function at different stages of development. Double-stranded RNA (dsRNA) can be efficiently delivered in worms by four protocols.76, 77 Worms can either be (1) injected with in vitro-transcribed dsRNA;7 (2) soaked in buffer containing concentrated in vitro-transcribed dsRNA;78 (3) fed bacteria expressing dsRNA from an engineered plasmid;79–81 (4) transformed with a vector expressing dsRNA in a given tissue.82–84 Injection of dsRNA into the gonad of young adult hermaphrodites, yields the greatest number of affected progeny. Surprising at first, injection into the intestine or body cavity also produces phenotypes in the progeny due to systemic RNAi.85 Younger larvae may also be injected when the effect of RNAi needs to be tested on the injected animal and not its progeny. Following injection, the effect of RNAi lasts several days, affecting the adult progeny of an injected worm. Recently, an approach has been developed which allows temperature-dependent induction of RNAi using the mec-8-dependent regulation of mec-2 intron 9 splicing.86 In addition, expressing the transmembrane protein SID-185 in neurons has been found to increase neuronal response to dsRNA delivered by feeding.87 Finally, clones for ~94% of all C. elegans genes are available from RNAi feeding libraries generated by the Ahringer and Vidal labs and can be procured from commercial suppliers (Source BioScience LifeSciences and Open Biosystems, respectively).
A few limitations of RNAi have to be kept in mind however: (1) A major disadvantage of RNAi compared to genetic mutation is the variable efficiency and potency of RNAi depending on the experimental conditions. (2) It is not possible to assess unambiguously the degree to which a gene’s function is knocked down by RNAi. This is a significant caveat when interpreting RNAi results. (3) As discussed previously, some tissue types are insensitive to RNAi in the wild-type context but sometimes also in sensitized backgrounds. (4) The efficiency of RNAi decreases rapidly when multiple genes are targeted at once, limiting genetic interactions tests based solely on RNAi. (5) A given RNAi clone can sometimes target more than one gene, based on sequence similarity.88 If no mutant allele is available to confirm an RNAi result, it is important to conduct RNAi with distinct non-overlapping dsRNA sequences derived from the target gene to avoid off-target effects.89
Inhibition of microRNA function by antisense oligoribonucleotides
MicroRNAs (miRNA) constitute a large number of specialized, non-protein coding regulatory genes.90 Their function can not be easily manipulated by conventional RNAi. When no genetic mutant is available for a miRNA of interest, its function can be inhibited by injecting antisense 2′-O-methyl oligoribonucleotides. Injection of these antisense compounds into larvae phenocopies let-7 loss-of-function defects.91 However, injection into adult hermaphrodite gonads does not result in efficient miRNA inhibition suggesting that antisense oligoribonucleotides are not transmitted to the progeny efficiently. This approach was recently optimized by cross-linking antisense 2′-O-methyl oligoribonucleotides to dextran, a soluble polysaccharide.92 These novel antisense conjugates can be conveniently injected into hermaphrodite gonads and their effects assessed in the progeny up to late larval stages. Conjugated oligoribonucleotides have been shown to efficiently inhibit miRNA function in hypodermis, vulva and neurons. Their effect is dose-dependent and highly specific. In addition, multiple miRNA can be targeted at once by mixing antisense reagents conjugated with different antisense oligoribonucleotides. This could facilitate the study of miRNA families and address potential functional redundancy.
Generating deletion mutants
Obtaining mutants by deletion library screening
Deletion libraries contain large numbers of worms organized in small pools that have been mutagenized with a deletion-inducing agent such as UV/TMP or EMS (see Fig. 3).93–95 When a deletion is of sufficient size, it can be detected by PCR with primers flanking the region of interest. Once a pool is found that reproducibly tests positive for a deletion, the worms in the pool are separated and rescreened until a single animal carrying the deletion is isolated. Two groups in the C. elegans community generate such mutants upon request: the Mitani Lab96 and the C. elegans gene knock-out consortium97 in Vancouver. So far, alleles for over 6000 genes have been generated by these two groups (Don Moerman, personal communication). Mutant strains are available from these consortia at a small fee and/or can be ordered from the CGC strain repository. In addition, individual labs can use published protocols to setup libraries to screen for gene deletions using EMS98 or TMP/UV mutagenesis.77, 99 Building and screening a deletion library is a fairly large-scale project which is best suited when multiple genes are targeted.
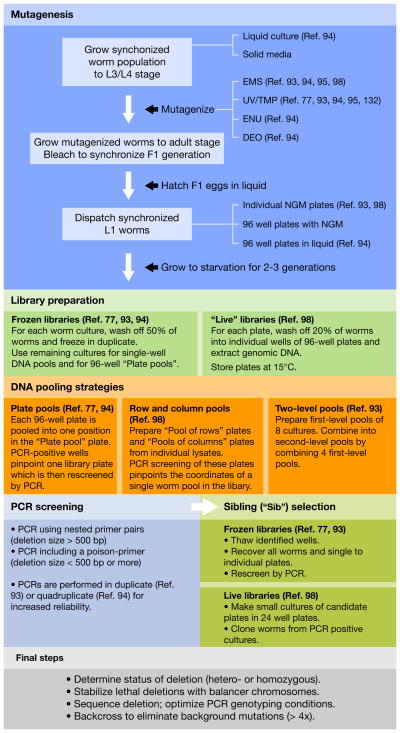
Figure 3. Generating deletion mutants by library screening.
Populations of randomly mutagenized worms can be established using different mutagens and different culture strategies. Mutant strains are either preserved in a frozen library or conserved on NGM plates in a “live” library. To optimize library screening, DNA samples extracted from all worm cultures can be pooled according to different pooling schemes. Deletions with sizes ranging from 500 bp to 1,500 bp are detected by nested PCR screening. Shorter deletions can be detected more easily by including a poison-primer. Once a candidate pool has been identified, the corresponding well is thawed from the frozen library and all recovered worms are cloned on individual plates for secondary rescreening. Similarly, worms from a “live” library are recultured in small pools and single worms are picked from PCR-positive pools. Once a single worm harboring a deletion has been isolated, a few steps should be undertaken before the strain can be used for functional studies.
Deletion mutants are very useful but a few issues need to be kept in mind: (1) worms retrieved from a library have been heavily mutagenized and usually carry many mutations in their genetic background, sometimes closely linked with the deletion of interest. (2) The size of the deletion is random and usually in the range of few hundred bases, which can be a problem for large genes and result in alleles which are not molecular nulls. (3) The endpoints of the deletion cannot be chosen, but only framed by the position of the screening primers. (4) Some deletions causing sterility or lethality can be difficult to recover and need to be stabilized using genetic balancers.99 (5) Some deletions can be complex rearrangements that are difficult to characterize.
Obtaining deletion mutants by transposon mobilization
Spontaneous Tc1 excision
Although Tc1 elements are fairly large DNA sequences which should disrupt gene function entirely, this is not always the case (see above). It is therefore important to derive a deletion mutant by remobilization of the transposon. In mutator backgrounds, deletions are generated at a low frequency after imprecise excision of the transposon. Most likely these events result from interrupted gene conversions with the sister strand.100 This can be used as an alternative to deletion library screening when a Tc1 transposon insertion is already available at an appropriate position in the gene. Such insertions can come from Tc1-based screens, specific PCR-based strategies targeted to a gene of interest,101 or from Tc1 mutant collections (CGMC, Lyon, France;36). This technique offers little to no control over the final structure of the deletion. In addition, PCR screening can be complicated by the frequent mobilization of somatic Tc1 insertions which can be confused with heritable germline events and require additional sib-selection steps. Finally, uncontrolled jumping of Tc1/mariner transposons in the mutator background is accompanied by increased morbidity of the progeny and the possibility of novel transposon insertions closely linked to the deletion site. To circumvent some of these limitations, a protocol has been developed to precisely engineer genomic deletions or gfp insertion alleles by transgene-instructed gene conversion.102 This strategy is based on an earlier protocol103 with a few optimizations: (1) two mutator backgrounds with increased Tc1 excision rates (mut-2 and mut-7 vs. mut-6) were used; (2) the total length of sequence homology was increased from 3 kb to 9 kb. These modifications increased the frequency of transgene-instructed gene conversion events significantly (4 × 10−4), however no further study has been published since this initial report.
Mos1 transposon remobilization
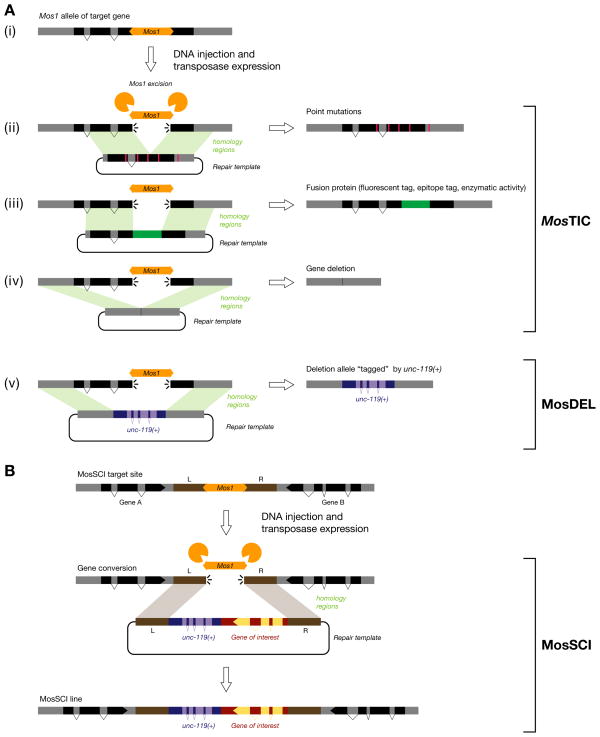
Generating deletion alleles through Mos1 transposon mobilization (MosDEL) is, in our opinion, the superior technique for this purpose (see Fig. 4). In order to put this technique in context with other Mos1-related techniques (MosTIC and MosSCI), we will discuss this approach in the “Genome engineering” section below.
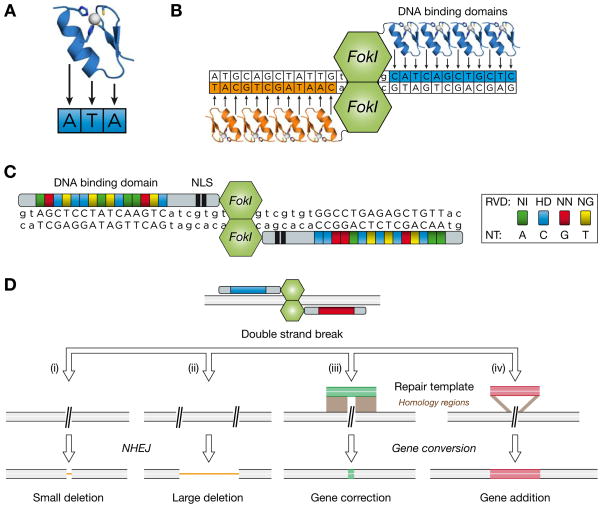
Figure 4. Genome editing with engineered nucleases.
(A) Structure of a Zinc Finger (ZF) DNA-binding module. Each individual ZF domain is optimized to recognize a three base pair sequence. Two cysteines and two histidines coordinate a zinc atom in each module.
(B) ZF nucleases (ZFN) are composed of at least four ZF domain repeats, linked to the FokI endonuclease domain. Specificity is increased by using obligatory dimers of two ZFN recognizing two sites which flank the targeted double-strand break site.
(C) Structure of TALE nucleases (TALEN). A central array of tandem repeats forms the specific DNA binding domain. Two nucleotides (repeat-variable di-residues, RVD) in each repeat determine the sequence preference of each module. Examples of four RVDs that recognize all four nucleotides (NT) are indicated. Two TALEN monomers bind to their target sites and allow the dimerization of the FokI endonuclease domain. TALENs also contain a nuclear localization signal (NLS).
(E) ZFNs and TALENs allow different types of genome editing strategies. A double strand-break (DSB) can be resolved by different pathways. (i) A single DSB can be repaired by non-homologous end-joining (NHEJ) and result in a small deletion of a few base pairs. Alternatively, (ii) two simultaneous DSB can generate a larger deletion allele. When a repair template with homology regions is provided, the DSB is resolved by gene conversion to (iii) insert a small sequence (as small as a single nucleotide, i.e. “gene correction”) or (iv) a larger transgene (“gene addition”).
Obtaining spontaneous deletions in the dog-1 mutant background
G4 DNA (guanine-quadruplex DNA) is a sequence motif that can fold into quadruplex structures in vitro. In vivo, such stable secondary structures can be problematic for DNA and RNA polymerases. Consistently, G4 DNA motifs have been identified as highly mutagenic sites in dog-1 mutant animals.104, 105 dog-1 is the C. elegans ortholog of the mammalian DNA helicase FANCJ, which is mutated in Fanconi anemia patients. There are 2907 sites matching the G4 DNA signature in the C. elegans genome. In a proof of principle study, candidate sites were chosen to isolate deletion mutants in 10 genes. In brief, 96 pools of dog-1(pk2247) mutant worms were grown to starvation. After DNA isolation, each pool was screened by PCR for deletions 5′ of various G4 DNA sites. This approach led to the isolation of 11 deletion alleles for the 10 targeted genes. This strategy could be generalized to the 1642 genes that are located 5′ of to the 2907 G4 DNA sites. Approximately three quarters of these genes had no available alleles at the time of the study.105
Generating targeted deletions by spontaneous gene conversion
Spontaneous recombination events between an engineered DNA template and a targeted genomic locus are extremely rare in C. elegans, most likely because injected DNA preferably forms extrachromosomal arrays and is therefore less available from homologous recombination. Early on, spontaneous gene conversion was obtained by injecting DNA into meiotic oocytes of individual worms which is technically challenging.106, 107 Another strategy has used microparticle bombardment to deliver a recombination template to a very large number of worms with little hands-on work.108, 109 The recombination template is composed of two homology arms which define the deletion end-points and flank a positive selection marker (unc-119(+)). In practice, this approach relies on generating hundreds of integrated lines in a unc-119(ed3) mutant background and selecting the rare homologous recombination events. When this strategy was used to disrupt the unc-54 and unc-22 genes, 1 and 3 homologous recombination events from 400 and 274 integrated lines were retrieved respectively.108 This strategy was recently refined by “tagging” the bombardment plasmid with a gfp reporter gene (located outside the homology arms) which makes it easy to distinguish recombination events from non-homologous integrations events110. In addition, FLP recombination sites were added upstream and downstream of unc-119(+) to create “clean” mutants by removing the cassette using tissue-specific FRT expression. Unfortunately, it was impossible to delete the unc-119 cassette in the germline and no further report using the technique has been published since.
Genome-scale identification of point mutations
Although chemical mutagens such as EMS can generate deletions (generally small and at a low frequency), they overwhelmingly induce point mutations in the genome. Two approaches have been devised to identify point mutants in individual genes throughout the entire genome.
TILLING: Targeting Induced Local Lesions in Genomes
TILLING aims to identify single nucleotide changes in target genes in a library of randomly mutagenized worms. PCR-amplified DNA from control and mutagenized worms is hybridized and digested with a single-strand-specific nuclease. Digested fragments are detected via the fluorescently-labeled primers used for amplification. The position of the mutation can be estimated by determining the size of the resulting DNA fragment on a denaturing polyacrylamide LI-COR gel. Using a library of 1,500 mutagenized worms, TILLING was successfully used to identified 71 mutations in 10 target genes. 3% are putative null alleles and 59% result in missense mutations. The remaining mutants are silent.111 TILLING appears not to have been used in C. elegans since this initial study.
Target-selected mutagenesis by high-throughput resequencing
Nonsense mutations can be identified from a library of mutagenized worms by cost-optimized high-throughput resequencing of target genes.112 First, a clonal library of 6,144 mutagenized F1 animals was generated and cryopreserved. Next, gene regions of interest were amplified by nested PCR and sequenced. Calculations based on a pilot screen suggest that knockout mutations for over 90% of C. elegans genes are present in this library. Furthermore, if every nonsynonymous amino acid change is taken into account, then every eighth amino of the proteome should be mutated. In order to reduce the screening complexity, the authors propose to only target genomic positions called NIMs (nonsense introducing mutations). Since 93% of EMS mutations are G to A transitions and based on the size of the ORFeome (~24 Mb), only 500,000 NIMs should be targeted in the genome. Theoretically, this could now be achieved by aCGH or even whole-genome sequencing.
Genome editing using sequence specific nucleases: ZFNs and TALENs
Genome editing using in vitro-engineered Zn finger nucleases (ZFNs)113 and TALE-based hybrid nucleases (TALENs)114, 115 has been recently demonstrated in C. elegans.116 These proteins combine a tailor-made DNA binding module with the endonuclease domain of the FokI enzyme (Fig. 4). Recognition of specific DNA sequences by a nuclease dimer is followed by DNA cleavage between the two DNA binding sites. Imprecise repair of this double-strand break, usually through error-prone non-homologous end-joining, results in small deletions or insertions of a few base pairs at the target locus, thereby likely disrupting the function of the targeted sequences, such as genes or cis-regulatory elements.116
Genome editing in C. elegans is a four step process. First, two complementary nucleases that recognize a specific site in the genome must be designed, constructed and validated. Next, 5′ and 3′ UTR sequences favorable for germline translation are added to maximize the expression of the enzymes in the C. elegans germline. This artificial ORF is then transcribed in vitro to obtain highly concentrated, 5′ capped, messenger RNA. Finally, RNA mixtures of the two complementary nucleases are directly injected into the gonad using the same procedure as for DNA germline transformation. Mutants alleles can then be selected in the F1 or F2 progeny of an injected animal. In the proof-of-principle study, mutant alleles were found in one percent of the total progeny of an injected animal (5% if an optimal 4-hour time window was considered).116 Whole genome analysis of one mutant strain revealed no off-target, ZFN-related lesions, confirming the very high specificity of this strategy. ZFN-based editing was also successfully applied to generate mutations in the related nematode species C. briggsae.116
Even though operating by similar overall principles, the TALE-based nucleases are more straight-forward to design, hence more cost-effective and will likely be the future method of choice. Different genome editing strategies which are used in other model systems113 could be adapted for C. elegans to allow for very precise genome engineering (Fig. 4D).
Engineering the genome: from single base changes to complex gene modification
The goal of genome engineering is to mutate a gene of interest or to modify a genomic locus by introducing in vitro-engineered sequences that, for example, contain affinity or reporter tags or alter the activity or localization of a gene. As mentioned previously, spontaneous gene conversion events are extremely rare in C. elegans. One efficient way to increase the rate of homologous recombination is to induce double-strand breaks (DSB) by reexcising a transposon. Two types of transposons have been successfully used in C. elegans: endogenous Tc1 and Drosophila Mos1.
Chromosomal double-strand breaks are catastrophic events which can be resolved using multiple cellular mechanisms.117 In brief, either DSBs are repaired either by using homology of DNA sequences flanking the breakpoint (gene conversion) or by religation of free DNA ends (non-homologous end-joining, NHEJ). If the break produces cohesive ends, NHEJ will restore the wild-type sequences. If the lesion is blunt, partial digestion and religation by NHEJ will introduce small footprints. When a homologous recombination pathway is used, 5′ to 3′ exonucleases digest DNA to generate single-stranded DNA regions which can carry out homology scanning. If this single stranded region anneals inappropriately within the same chromosome, DNA synthesis from this position and subsequent religation will generate a deletion around the DSB site. However this single stranded region can also anneal with a homologous donor on a different DNA fragment such as sister chromatids, homologous chromosomes or engineered homologous templates. In the later case, artificial sequences can then be incorporated into the chromosome at the DSB site via gene conversion, resulting in a modified genomic locus and engineered alleles.
Transgene-instructed genomic engineering upon Tc1 excision
Genome engineering in a template-dependent manner was first reported by Plasterk and Groenen.103 A strain containing a mutagenic Tc1 insertion in the unc-22 locus was micro-injected with a wild-type repair template containing silent sequence polymorphisms close to the Tc1 insertion site. These markers were flanked by 1.5 kb of homologous sequences to drive recombination. Phenotypic revertants of the Unc-22 phenotype were selected from the progeny of these transgenic animals and analysed. The observed reversion frequency was low (2 × 10−5) but three quarters of the revertants had incorporated sequence polymorphism up to ~200 bp from the DSB. This approach and an optimized protocol102 have however remained proof-of-principle experiments.
MosTIC: Mos1 excision induced Transgene Instructed gene Conversion
MosTIC is the first robust strategy that allows genome engineering in C. elegans in a targeted and controlled fashion (see Fig. 4).117 Mos1 excision is used to create a genomic double-strand break at the engineered locus. This DSB is then repaired by gene conversion using an in vitro-engineered template which is provided on an extrachromosomal transgene. A detailed protocol has been published recently.118 MosTIC is preceded by a Mos1 mutagenesis screen (see above) which provides the necessary transposon insertions. Alternatively, a large collection of Mos1 alleles has been generated and mapped by the NemaGENETAG consortium (searchable on WormBase).55, 119 For efficient MosTIC, Mos1 insertions should be chosen as close as possible to the target site. Indeed, the frequency of MosTIC events decreases in a bell shaped curve as a function of the distance from the Mos1 insertion site. Point mutations have been introduced up to 3 kb away from the insertion site but with a 20-fold lower efficiency than at the insertion site itself.117 A point mutation contained within a 1 kb-long region centered on the DSB will be transferred into the genome in 50% of MosTIC events.
The MosTIC repair template is composed of two homology arms of 1.5 kb which flank the target region and the Mos1 insertion site. The rate of MosTIC does not seem to increase when longer homology sequences are used117. This template is injected into the Mos1 carrying strain with one of two vectors used to trigger Mos1 transposase expression in the germline: (1) a heat-shock inducible construct using the Phsp-16.48 promoter (Phsp-16.48::MosTase); (2) a construct using the constitutive germline promoter Pglh-2 (Pglh-2::MosTase). In the first case, transgenic lines are established and amplified. Once sufficient transgenic animals are available, adult worms are heat-shocked and their progeny is screened for gene conversion events. MosTIC frequency generally ranges from 10−3 to 10−5 events per offspring of the heat-shocked animals. In the second case, gene conversion events can be detected in the F1 progeny of injected animals but more animals need to be injected since at best 1/10 will produce a MosTIC progeny.118
MosTIC events can be detected either by phenotypic reversion117 or by PCR screening.49 Note that not all phenotypic reversion events will be bona fide MosTIC. Indeed, DSBs repaired by NHEJ can sometimes regenerate functional alleles. In the case of PCR screening strategies, false positives can arise from PCR jumping.117 Therefore, PCR screening conditions should be optimized carefully beforehand.118 MosTIC has been used successfully to insert point mutations,117 fluorescent tags,49, 117 epitopes for commercial antibodies49 and affinity tags (unpublished). Such MosTIC alleles seem to replicate gene expression and protein localization with high accuracy and fidelity.49
MosDEL: Mos1-mediated deletion
The goal of MosDEL is to generate in vitro-designed deletions around a Mos1 insertion site in the genome (see Fig. 4).120 Based on available strains in WormBase (> 14,000), 99.4% of the 20,160 C. elegans genes are within 25 kb of at least one Mos1 insertion making them theoretically all targetable. Of these, 8,803 have no other genes between them and Mos1 so only the targeted gene would be removed. In addition, all Mos1 alleles obtained by insertional mutagenesis are usable for MosDEL.
The first step of MosDEL is to combine a Mos1 strain with the unc-119(ed3) mutant. These worms are then injected with a DNA mix containing (1) mCherry-expressing vectors (used to identify false positives due to extrachromosomal array formation), (2) the source of Mos1 transposase (Pglh-2::MosTase) and (3) the deletion template. The deletion template contains the C. briggsae unc-119(+) gene flanked by two homology arms which define the planed deletion end-points. When the Mos1-induced double-strand break is repaired using this transgene, the target region is deleted and Cbr-unc-119(+) is copied into the locus. This rescues the unc-119(ed3) mutation and results in a perfectly balanced deletion allele (see Fig. 4).
Approximately 100 P0 worms should be injected for MosDEL. In a proof-of-principle experiment aimed at deleting a 25 kb region close to the dpy-13 locus, injection of 81 animals resulted in 7 stable lines carrying a Cbr-unc-119(+) fragment (9%).120 Four of these seven lines were complete deletions of the 25 kb region (5%). No complete deletion of 35 or 50 kb could be recovered however. MosDEL currently appears as the most efficient tool to engineer precise gene deletions in the C. elegans genome.
Recent experiments show that antiobiotics neomycin121 and puromycin122 can be used as efficient selection markers in worms. Current efforts to facilitate MosDEL focus on using antibiotic markers (Christian Frøkjær-Jensen & Erik M. Jorgensen, pers. comm.) which will eliminate the need to carry an unc-119 mutation in the background.
EXPRESSING GENES IN C. ELEGANS
There are numerous reasons to introduce genes in C. elegans, including reporter gene analysis, transformation rescue of mutant phenotypes, manipulation of cell function, et cetera.
Classical C. elegans transgenesis
Transformation of C. elegans was first reported in the early 1980 in a study of the sex determination gene tra-3.123 Kimble et al. microinjected an amber codon-suppressing tRNA (sup-7(st5)) into the hermaphrodite gonad of tra-3 mutants carrying an amber allele which resulted in suppression of the Tra-3 phenotype in the progeny. Integrative107 and non-integrative124 DNA-mediated transformation was derived from this approach soon after.
DNA transformation by microinjection is a very fast, simple, inexpensive and efficient way to generate transgenic C. elegans strains.5, 10, 124 Injection requires little to no preparation and only few worms need to be injected to obtain transgenic lines. The C. elegans hermaphrodite germline is a syncytium containing hundreds of mitotically active oocytes at different stages of maturation and almost all nuclei share a common cytoplasm. Different DNA species can be co-injected (plasmids, PCR products, cosmids, fosmids, BACs, YACs) with selectable injection markers (fluorescent reporter genes, dominant or recessive phenotypic markers). This DNA mix will be processed by DNA end ligation and homologous recombination into large, heritable DNA concatemers called extrachromosomal arrays.5, 124 These episomes are recognized by the replication machinery and usually segregate like chromosomes during cytokinesis and meiosis. Detailed transgenesis protocols can be found here125 and here (see Jin in10).
The ease with which these transgenes can be generated has to be balanced against the limitations of this approach: (1) Classical transgenesis offers little to no control over expression levels. Extrachromosomal transgenes generally contain hundreds of copies of the transforming DNA and therefore almost always result in overexpression. To obtain a more physiological result, the concentration of a given DNA species can be reduced by diluting it with more carrier DNA. (2) The highly repetitive nature of extrachromosomal transgenes leads to silencing in the germline.126 This effect can be limited by generating “complex arrays” by co-injecting digested genomic DNA, preferably from a bacterial source. (3) Transgenes can rarely recreate the full genomic context and regulatory sequences necessary for accurate gene expression. Strikingly, the linear or circular nature of the injected DNA can influence the resulting expression patterns, with linear, vector-free DNA producing more accurate results.127 (4) Extrachromosomal arrays can evolve over time. In particular, their expressivity and stability can change rapidly between generations.124 It is therefore recommended to freeze transgenic lines as soon as possible and return to the original isolate if necessary. (5) Extrachromosomal arrays are semi-stable and not all progeny of a transgenic animal will inherit the transgene. Therefore they require manual maintenance unless a selectable co-injection marker is used. For example, transgenic animals can be generated in a pha-1(e2123ts) mutant background. These pha-1 mutants are viable at 15°C but require the wild-type pha-1 gene to survive at 25°C.128 Very recently, two groups have developed antibiotic selection systems that can be used in any genetic background. Neomycin (or G-418)121 and puromcyin122 are translation inhibitors which prevent the larval development of C. elegans. Ubiquitous expression of the corresponding resistance genes allows hands-off selection of transgenic animals and robust enrichment of large transgenic populations.
Stable integration of C. elegans transgenes
Integrative transgenesis was first achieved by injecting DNA into the nuclei of maturing oocytes.106, 107 Some integration events have also been reported when single-stranded oligonucleotides of 50 bases are co-injected with double-stranded DNA.5 However, these approaches are inefficient or technically challenging. Therefore, transgene integration is generally achieved by irradiating extrachromosomal lines with gamma or UV radiation (see Jin in 10 and 125). Transgene sequences are randomly integrated into the genome, most likely following erroneous repair of a chromosomal break. The resulting stable lines need to be extensively outcrossed to remove background mutations. Moreover, it is recommended to obtain independent integrated lines since some variability in expression can be observed, likely due to positional effects and the amount of integrated DNA.
Stable transformation by microparticle bombardment
Microparticle bombardment has been used successfully in C. elegans to generate single to low-copy integrated transformant.129, 130 Gold beads coated with DNA are projected at high velocity at the sample allowing penetration into cells. The small amount of DNA on each bead likely decreases the chances of extrachromosomal array formation, therefore favoring integration events. Bombardment lines show more reproducible gene expression than extrachromosomal lines, but some variability has been reported.131 This approach also allows expression of transgenes in the germline because the low copy number circumvents germline-silencing observed for multicopy extrachromosomal arrays.126 The low integration frequency (~ 5 × 10−5) requires a positive selection strategy (usually unc-119(ed3) rescue) and the use of large numbers of synchronized worms.129 Detailed instructions for transformation by microparticle bombardment can be found in132.
MosSCI: Generating single-copy transgenes by homologous recombination
The goal of MosSCI (Mos1-mediated single copy insertion) is to generate stable single-copy transgenic strains by transgene-instructed gene conversion (see Fig. 4).131 The MosSCI approach is based on MosTIC117 with some specific modifications: (1) MosSCI is performed at one of two genomic sites (on chromosome II and IV) which are located in the 3′ regions of two opposing genes. Therefore insertions there should minimally disrupt gene function. (2) MosSCI uses positive and negative selection strategies to identify bona fide gene conversion events. (3) A toolbox of vectors is available to rapidly generate MosSCI constructs (http://sites.google.com/site/jorgensenmossci/), in particular for germline expression.133
The first step for MosSCI is to build the recombination template. This vector contains two 1.5 kb long homology arms corresponding to one of the two genomic insertion site (see Fig. 4). These sequences flank the C. briggsae unc-119(+) gene used for positive selection and a multiple cloning site or two gateway att recombination sequences which are used to clone the transgene. Transgenes of 3 to 8 kb have been inserted into the genome using these vectors131 and unpublished results indicate that insert sizes can be as large as 17 kb (Frøkjær-Jensen and Jorgensen, personal communication). Transgene insertions generated by MosSCI should be designated by “Si” (following the unique laboratory identifier).
Two protocols can be used for MosSCI: (1) The direct insertion protocol: the MosSCI vector is co-injected with mCherry-expressing vectors and the Pglh-2::MosTase plasmid and recombination events are selected directly in the F2-F3 progeny of injected animals. This requires to inject at least 25–50 P0 animals, of which 2 to 5 will generate a MosSCI allele. On average 60% of these lines will contain complete, single-copy inserts. (2) The selection-based insertion protocol: the MosSCI vector is co-injected with the Phsp-16.48::MosTase construct, multiple vectors expressing mCherry in different tissues and a muscle-expressed gain-of-function mutant of the twk-18 potassium channel. MosSCI is triggered by heat-shocking these transgenic animals and mCherry and twk-18 are used as counterselection markers to eliminate animals which rescue the Unc-119 phenotype but are not true MosSCI lines. The direct insertion protocol is generally much faster (~10 days vs. one month) and requires less hands-on time. However, the selection-based insertion protocol can be scaled up significantly and displays higher success rates.
MosSCI recombination events are detected by the phenotypic rescue of unc-119(ed3). However, unc-119(ed3) mutant are fairly unhealthy and difficult to inject. Recently, an alternative strategy usable in any genetic background and based on neomycin resistance has been used successfully.121
Up-to-date information, detailed instructions, descriptions of reagents and strains can be found here (http://sites.google.com/site/jorgensenmossci/). Upcoming developments of this technique are: (1) vectors and strains for 5 new genomic sites; (2) an improved insertion method with a success rate above 50%; (3) a new, heat-shock inducible negative selection marker that simplifies the selection of insertion events; (4) in depth characterization of each site regarding it permissiveness for germline expression (Frøkjær-Jensen and Jorgensen, personal communication).
Spatial and temporal manipulation of gene expression
Cell-specific gene expression can be achieved easily in C. elegans using specific, heterologous promoter fragments and gene expression can be controlled temporally using broadly if not ubiquitously expressed heat-shock-induced promoters. One approach to achieve both temporal and spatial control has been to combine cell-specific rescue of a mutant defective in the heat-shock response (hsf-1(sy441)) with a transgene expressing a gene of interest under the control of a heat-shock-responsive promoter.134 Gene induction is therefore restricted to the hsf-1-expressing cells. This two component system was used to express gfp in the pharynx and neurons. It functions at all developmental stages and only requires a 30 min heat shock treatment. GFP expression was seen 1 h after heat shock and remained visible for 24 h. This system can also be used to study gene function as exemplified by the rescue of the dye filling defect of a mutant defective in neuron cilium formation and function.134
A second approach for controlled gene expression is based on the mec-8-dependent alternative splicing of mec-2.86 MEC-8 is an RNA binding protein that regulates the formation of the long mec-2a mRNA by promoting exon 9–10 over 9–9′ splicing. Hence, inclusion of the 9th mec-2 intron into another gene confers mec-8-dependent regulation. Using the temperature sensitive mec-8(ut218ts) allele, it is possible to control splicing and gene expression by shifting worms from the restrictive (25°C) to the permissive (15°C) temperature. This approach was used to activate yfp expression in neurons, hypodermis and intestine. However, although mec-8 is ubiquitously expressed early in development, it is restricted to some tissue types at later stages. Therefore gfp expression was seen in head and tail neurons but not in the ventral nerve cord, somewhat limiting this approach. A temperature sensitive mec-8(ut218ts) allele could be ectopically expressed in the targeted tissue to circumvent this limitation. By extension, this approach was used to selectively express genes in cells for which no specific promoters are known by using promoters with partially overlapping expression domains to express mec-8(ut218ts) and the targeted gene fused to the 9th mec-2 intron.86
Another approach is based on the use of FLP recombinases which allows for permanent induction of gene expression in any genetic background.110, 135, 136 The FLP enzyme catalyses the intra-molecular excision of DNA located between two FRT sites when they are oriented in the same direction (“FLP-out”). FLP-out activity in C. elegans was demonstrated by removing a cassette containing a fluorescent protein and a transcriptional termination sequence flanked by FRT sites.135, 136 Induction of FLP expression using a heat shock promoter resulted in total or partial loss of the FRT-flanked sequences and the “activation” of the downstream open reading frame. This system was used as a lineage tracing tool, to induce cell-specific RNAi, express a dominant negative transcription factor136 and to silence neurotransmission in a temporally and spatially controlled fashion using tetanus toxin.135 Both publications provide vector toolkits that can be used to adapt these technologies to specific biological questions. Recently, somatic FLP-mediated recombination has been demonstrated in the context of a genomic knock-in allele generated by biolistic transformation.110
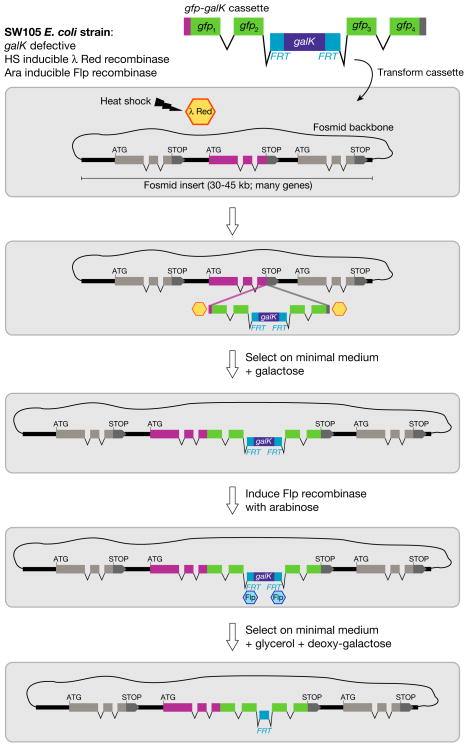
Building reporter gene constructs by fosmid recombineering
Determining a gene’s expression pattern is a necessary step in the characterisation of any C. elegans gene. This can be achieved by using transcriptional or translational gfp or LacZ reporters.6 Reporter gene strategies have been discussed in great detail in Ref. 137. High-throughput approaches were also made possible by the use of Gateway compatible vectors, in particular with the completion of the ORFeome138 and Promoterome projects.139 However, most of these reporters only contain a few kilobases of gene regulatory sequence, because of the technical limitations encountered when trying to manipulate much larger DNA sequences. Recently, three novel methods have been developed which use recombineering of fosmid clones or BACs to engineer more comprehensive reporter constructs (see Fig. 5).140–142
Figure 5. MosTIC, MosDEL and MosSCI strategies.
(A) The double-strand break induced by the excision of Mos1 (i) can be repaired by MosTIC using different repair templates to engineer point mutations (ii), fusion proteins (iii), or gene deletions (iv). In the case of MosDEL (v), a unc-119(+) mini-gene is used as a positive selection marker.
(B) The goal of MosSCI is to engineer single copy transgenes at a defined location in the genome. A double-strand break induced by Mos1 excision is repaired by homologous recombination with an in vitro-engineered template which contains the desired transgene in combination with the unc-119(+) mini-gene.
A fosmid library containing over 15,000 clones each carrying ~40 kb long genomic fragments is now available for C. elegans (Don Moerman, personal communication). Clones can be searched on WormBase or using a dedicated tool (http://elegans.bcgsc.ca/perl/fosmid/CloneSearch), and obtained through Source BioScience LifeSciences. Given the good genome coverage (~6x) of this library, it is usually possible to obtain a fosmid in which the gene of interest is at the center, flanked by large portions of genomic DNA, therefore including most of the likely regulatory sequences. The most recently developed recombineering protocol140 is fast (approximately one week), robust (owing to positive and negative selection steps) and requires little more than a freely available bacterial strain, minimal media, and a diverse set of cassettes including fluorescent proteins, affinity tags or bicistronic fluorescent reporters.140 Injection of fosmid as complex arrays (typically digested recombineered fosmid at 10–15 ng/μl, digested co-injection marker (2–7 ng/μl) and digested bacterial genomic DNA (120–150 ng/μl)) generates arrays that usually show excellent germline expression.140
Conclusion
C. elegans has not only been at the forefront as a model system to understand biological processes, but also at the forefront for establishing widely applicable technologies. GFP reporter technology and RNAi are already classic examples. A number of the techniques presented here will surely be further developed and significantly fine-tuned. For example, the recent introduction of antibiotic selection markers121, 122 offers much promise. Anticipated significant drops in the cost of whole genome sequencing will further popularize forward genetic screens but also provide reverse-generated (i.e. non-phenotype driven) allelic versions of many/most/all genes in the genome. The development of engineered Mos1 transposable elements, able to carry large cargo, will greatly simplify the generation of single copy transgenes and allow gene trapping strategies. Finally, efficient genome editing and engineering will open the door to a range of new experimental approaches. So far, only Mos1-based genome engineering was sufficiently efficient to be used routinely. The promise of ZFNs and TALENs lies in their ability to cause double-strand breaks with high efficiency and accuracy. Once this is combined with transgene-instructed repair, these nucleases and possibly other nucleases such as meganucleases143 will become the premier tools to mutate, modify and restore genes and gene function in C. elegans and related nematodes.
Figure 6. C. elegans fosmid recombineering.
Recombineering can be used to efficiently engineer a gene of interest in the context of a fosmid genomic clone. For example, a cassette containing a GFP tag and the FRT-galK-FRT selection module (FgF) can be inserted by λRed recombinase-mediated homologous recombination into any position of a gene. Recombination is carried out in the SW105 E. coli strain. Recombinant clones are selected on minimal medium with galactose. Excision of the FgF selection module is carried out by Flp recombinase (induced by addition of arabinose) and can be selected for in medium containing deoxy-galactose. Note that the FRT “scar” remaining after galK excision resides in an intron and therefore has no impact on protein coding sequences. Additional applications of fosmid recombineering include: sequence deletion, sequence replacement and fosmid extension. A comprehensive set of recombineering cassettes including different fluorescent reporters, affinity tags, nuclear localization signals, mutant FRT sites and gene replacement cassettes is available (adapted from Ref. 140).
Contributor Information
Thomas BOULIN, Email: boulin@biologie.ens.fr, Institut de Biologie de l’École Normale Supérieure, Biology Department, F-75005 Paris, France. Inserm, Unité 1024, F-75005 Paris, France. Centre National de la Recherche Scientifique, Unité Mixte de Recherche 8197, F-75005 Paris, France.
Oliver HOBERT, Columbia University Medical Center, HHMI, New York, USA.
References
- 1.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sulston JE. Neuronal cell lineages in the nematode Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol. 1983;48:443–452. doi: 10.1101/sqb.1983.048.01.049. [DOI] [PubMed] [Google Scholar]
- 3.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 4.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London B Biological Sciences. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 5.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 7.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 8.The-C elegans-Sequencing-Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 9.Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. Vol. 48. Academic Press; 1995. [Google Scholar]
- 10.Hope IA. C. elegans: a practical approach. Oxford: Oxford University Press; 1999. [Google Scholar]
- 11.De Stasio E, Lephoto C, Azuma L, Holst C, Stanislaus D, Uttam J. Characterization of revertants of unc-93(e1500) in Caenorhabditis elegans induced by N-ethyl-N-nitrosourea. Genetics. 1997;147:597–608. doi: 10.1093/genetics/147.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flibotte S, Edgley ML, Chaudhry I, Taylor J, Neil SE, Rogula A, Zapf R, Hirst M, Butterfield Y, Jones SJ, et al. Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics. 2010;185:431–441. doi: 10.1534/genetics.110.116616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarin S, Bertrand V, Bigelow H, Boyanov A, Doitsidou M, Poole RJ, Narula S, Hobert O. Analysis of multiple ethyl methanesulfonate-mutagenized Caenorhabditis elegans strains by whole-genome sequencing. Genetics. 2010;185:417–430. doi: 10.1534/genetics.110.116319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fay D. Genetic mapping and manipulation: chapter 1--Introduction and basics. WormBook. 2006:1–12. doi: 10.1895/wormbook.1.90.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fay D. Genetic mapping and manipulation: chapter 6--Mapping with deficiencies and duplications. WormBook. 2006:1–3. doi: 10.1895/wormbook.1.95.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fay D, Bender A. SNPs: introduction and two-point mapping. WormBook. 2008:1–10. doi: 10.1895/wormbook.1.93.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- 18.Davis MW, Hammarlund M, Harrach T, Hullett P, Olsen S, Jorgensen EM. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics. 2005;6:118. doi: 10.1186/1471-2164-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maydan JS, Flibotte S, Edgley ML, Lau J, Selzer RR, Richmond TA, Pofahl NJ, Thomas JH, Moerman DG. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Res. 2007;17:337–347. doi: 10.1101/gr.5690307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seidel HS, Rockman MV, Kruglyak L. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319:589–594. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flibotte S, Edgley ML, Maydan J, Taylor J, Zapf R, Waterston R, Moerman DG. Rapid high resolution single nucleotide polymorphism-comparative genome hybridization mapping in Caenorhabditis elegans. Genetics. 2009;181:33–37. doi: 10.1534/genetics.108.096487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maydan JS, Okada HM, Flibotte S, Edgley ML, Moerman DG. De Novo identification of single nucleotide mutations in Caenorhabditis elegans using array comparative genomic hybridization. Genetics. 2009;181:1673–1677. doi: 10.1534/genetics.108.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Meara MM, Bigelow H, Flibotte S, Etchberger JF, Moerman DG, Hobert O. Cis-regulatory mutations in the Caenorhabditis elegans homeobox gene locus cog-1 affect neuronal development. Genetics. 2009;181:1679–1686. doi: 10.1534/genetics.108.097832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarin S, Prabhu S, O’Meara MM, Pe’er I, Hobert O. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat Methods. 2008;5:865–867. doi: 10.1038/nmeth.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bigelow H, Doitsidou M, Sarin S, Hobert O. MAQGene: software to facilitate C. elegans mutant genome sequence analysis. Nat Methods. 2009;6:549. doi: 10.1038/nmeth.f.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaham S. galign: a tool for rapid genome polymorphism discovery. PLoS One. 2009;4:e7188. doi: 10.1371/journal.pone.0007188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flowers EB, Poole RJ, Tursun B, Bashllari E, Pe’er I, Hobert O. The Groucho ortholog UNC-37 interacts with the short Groucho-like protein LSY-22 to control developmental decisions in C. elegans. Development. 2010;137:1799–1805. doi: 10.1242/dev.046219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doitsidou M, Poole RJ, Sarin S, Bigelow H, Hobert O. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS One. 2010;5:e15435. doi: 10.1371/journal.pone.0015435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuryn S, Le Gras S, Jamet K, Jarriault S. A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics. 2010;186:427–430. doi: 10.1534/genetics.110.119230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bessereau JL. Transposons in C. elegans. WormBook. 2006:1–13. doi: 10.1895/wormbook.1.70.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori I, Moerman DG, Waterston RH. Analysis of a mutator activity necessary for germline transposition and excision of Tc1 transposable elements in Caenorhabditis elegans. Genetics. 1988;120:397–407. doi: 10.1093/genetics/120.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greenwald I. lin-12, a nematode homeotic gene, is homologous to a set of mammalian proteins that includes epidermal growth factor. Cell. 1985;43:583–590. doi: 10.1016/0092-8674(85)90230-2. [DOI] [PubMed] [Google Scholar]
- 33.Korswagen HC, Durbin RM, Smits MT, Plasterk RH. Transposon Tc1-derived, sequence-tagged sites in Caenorhabditis elegans as markers for gene mapping. Proc Natl Acad Sci U S A. 1996;93:14680–14685. doi: 10.1073/pnas.93.25.14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wicks SR, de Vries CJ, van Luenen HG, Plasterk RH. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev Biol. 2000;221:295–307. doi: 10.1006/dbio.2000.9686. [DOI] [PubMed] [Google Scholar]
- 35.Martin E, Laloux H, Couette G, Alvarez T, Bessou C, Hauser O, Sookhareea S, Labouesse M, Segalat L. Identification of 1088 new transposon insertions of Caenorhabditis elegans: a pilot study toward large-scale screens. Genetics. 2002;162:521–524. doi: 10.1093/genetics/162.1.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Linden AM, Plasterk RH. Shotgun cloning of transposon insertions in the genome of Caenorhabditis elegans. Comp Funct Genomics. 2004;5:225–229. doi: 10.1002/cfg.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rushforth AM, Anderson P. Splicing removes the Caenorhabditis elegans transposon Tc1 from most mutant pre-mRNAs. Mol Cell Biol. 1996;16:422–429. doi: 10.1128/mcb.16.1.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kodama K, Takagi S, Koga A. The Tol1 element of the medaka fish, a member of the hAT transposable element family, jumps in Caenorhabditis elegans. Heredity. 2008;101:222–227. doi: 10.1038/hdy.2008.47. [DOI] [PubMed] [Google Scholar]
- 39.Koga A, Shimada A, Kuroki T, Hori H, Kusumi J, Kyono-Hamaguchi Y, Hamaguchi S. The Tol1 transposable element of the medaka fish moves in human and mouse cells. J Hum Genet. 2007;52:628–635. doi: 10.1007/s10038-007-0161-2. [DOI] [PubMed] [Google Scholar]
- 40.Bessereau JL, Wright A, Williams DC, Schuske K, Davis MW, Jorgensen EM. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature. 2001;413:70–74. doi: 10.1038/35092567. [DOI] [PubMed] [Google Scholar]
- 41.Robert VJ, Bessereau JL. Manipulating the Caenorhabditis elegans genome using mariner transposons. Genetica. 2010;138:541–549. doi: 10.1007/s10709-009-9362-2. [DOI] [PubMed] [Google Scholar]
- 42.Coates CJ, Jasinskiene N, Miyashiro L, James AA. Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci U S A. 1998;95:3748–3751. doi: 10.1073/pnas.95.7.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gueiros-Filho FJ, Beverley SM. Trans-kingdom transposition of the Drosophila element mariner within the protozoan Leishmania. Science. 1997;276:1716–1719. doi: 10.1126/science.276.5319.1716. [DOI] [PubMed] [Google Scholar]
- 44.Boulin T, Bessereau JL. Mos1-mediated insertional mutagenesis in Caenorhabditis elegans. Nat Protoc. 2007;2:1276–1287. doi: 10.1038/nprot.2007.192. [DOI] [PubMed] [Google Scholar]
- 45.Granger L, Martin E, Segalat L. Mos as a tool for genome-wide insertional mutagenesis in Caenorhabditis elegans: results of a pilot study. Nucleic Acids Res. 2004;32:e117. doi: 10.1093/nar/gnh111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams DC, Boulin T, Ruaud AF, Jorgensen EM, Bessereau JL. Characterization of Mos1-mediated mutagenesis in Caenorhabditis elegans: a method for the rapid identification of mutated genes. Genetics. 2005;169:1779–1785. doi: 10.1534/genetics.104.038265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruaud AF, Bessereau JL. Activation of nicotinic receptors uncouples a developmental timer from the molting timer in C. elegans. Development. 2006;133:2211–2222. doi: 10.1242/dev.02392. [DOI] [PubMed] [Google Scholar]
- 48.Gally C, Eimer S, Richmond JE, Bessereau JL. A transmembrane protein required for acetylcholine receptor clustering in Caenorhabditis elegans. Nature. 2004;431:578–582. doi: 10.1038/nature02893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gendrel M, Rapti G, Richmond JE, Bessereau JL. A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature. 2009;461:992–996. doi: 10.1038/nature08430. [DOI] [PubMed] [Google Scholar]
- 50.Yook K, Hodgkin J. Mos1 mutagenesis reveals a diversity of mechanisms affecting response of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2007;175:681–697. doi: 10.1534/genetics.106.060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winter AD, Keskiaho K, Kukkola L, McCormack G, Felix MA, Myllyharju J, Page AP. Differences in collagen prolyl 4-hydroxylase assembly between two Caenorhabditis nematode species despite high amino acid sequence identity of the enzyme subunits. Matrix Biol. 2007;26:382–395. doi: 10.1016/j.matbio.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Jorgensen EM, Mango SE. The art and design of genetic screens: caenorhabditis elegans. Nat Rev Genet. 2002;3:356–369. doi: 10.1038/nrg794. [DOI] [PubMed] [Google Scholar]
- 53.Burns AR, Kwok TC, Howard A, Houston E, Johanson K, Chan A, Cutler SR, McCourt P, Roy PJ. High-throughput screening of small molecules for bioactivity and target identification in Caenorhabditis elegans. Nat Protoc. 2006;1:1906–1914. doi: 10.1038/nprot.2006.283. [DOI] [PubMed] [Google Scholar]
- 54.Kwok TC, Ricker N, Fraser R, Chan AW, Burns A, Stanley EF, McCourt P, Cutler SR, Roy PJ. A small-molecule screen in C. elegans yields a new calcium channel antagonist. Nature. 2006;441:91–95. doi: 10.1038/nature04657. [DOI] [PubMed] [Google Scholar]
- 55.Duverger Y, Belougne J, Scaglione S, Brandli D, Beclin C, Ewbank JJ. A semi-automated high-throughput approach to the generation of transposon insertion mutants in the nematode Caenorhabditis elegans. Nucleic Acids Res. 2007;35:e11. doi: 10.1093/nar/gkl1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, et al. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol. 2007;25:663–668. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- 57.Doitsidou M, Flames N, Lee AC, Boyanov A, Hobert O. Automated screening for mutants affecting dopaminergic-neuron specification in C. elegans. Nat Methods. 2008;5:869–872. doi: 10.1038/nmeth.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pujol N, Cypowyj S, Ziegler K, Millet A, Astrain A, Goncharov A, Jin Y, Chisholm AD, Ewbank JJ. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr Biol. 2008;18:481–489. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung K, Crane MM, Lu H. Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat Methods. 2008;5:637–643. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- 60.Gilleland CL, Rohde CB, Zeng F, Yanik MF. Microfluidic immobilization of physiologically active Caenorhabditis elegans. Nat Protoc. 2010;5:1888–1902. doi: 10.1038/nprot.2010.143. [DOI] [PubMed] [Google Scholar]
- 61.Liu X, Long F, Peng H, Aerni SJ, Jiang M, Sanchez-Blanco A, Murray JI, Preston E, Mericle B, Batzoglou S, et al. Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell. 2009;139:623–633. doi: 10.1016/j.cell.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murray JI, Bao Z, Boyle TJ, Boeck ME, Mericle BL, Nicholas TJ, Zhao Z, Sandel MJ, Waterston RH. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat Methods. 2008;5:703–709. doi: 10.1038/nmeth.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 64.Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 65.Zipperlen P, Fraser AG, Kamath RS, Martinez-Campos M, Ahringer J. Roles for 147 embryonic lethal genes on C. elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 2001;20:3984–3992. doi: 10.1093/emboj/20.15.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 67.Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 68.Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 69.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 70.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 71.Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 72.Wang D, Kennedy S, Conte D, Jr, Kim JK, Gabel HW, Kamath RS, Mello CC, Ruvkun G. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–597. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- 73.Schmitz C, Kinge P, Hutter H. Axon guidance genes identified in a large-scale RNAi screen using the RNAi-hypersensitive Caenorhabditis elegans strain nre-1(hd20) lin-15b(hd126) Proc Natl Acad Sci U S A. 2007;104:834–839. doi: 10.1073/pnas.0510527104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lehner B, Calixto A, Crombie C, Tischler J, Fortunato A, Chalfie M, Fraser AG. Loss of LIN-35, the Caenorhabditis elegans ortholog of the tumor suppressor p105Rb, results in enhanced RNA interference. Genome Biol. 2006;7:R4. doi: 10.1186/gb-2006-7-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhuang JJ, Hunter CP. Tissue specificity of Caenorhabditis elegans enhanced RNA interference mutants. Genetics. 2011;188:235–237. doi: 10.1534/genetics.111.127209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Timmons L. Delivery methods for RNA interference in C. elegans. Methods Mol Biol. 2006;351:119–125. doi: 10.1385/1-59745-151-7:119. [DOI] [PubMed] [Google Scholar]
- 77.Ahringer J, editor. Reverse genetics. WormBook. 2006 Available at: http://www.wormbook.org.
- 78.Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- 79.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 80.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 81.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Timmons L, Tabara H, Mello CC, Fire AZ. Inducible systemic RNA silencing in Caenorhabditis elegans. Mol Biol Cell. 2003;14:2972–2983. doi: 10.1091/mbc.E03-01-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tavernarakis N, Wang SL, Dorovkov M, Ryazanov A, Driscoll M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- 84.Esposito G, Di Schiavi E, Bergamasco C, Bazzicalupo P. Efficient and cell specific knock-down of gene function in targeted C. elegans neurons. Gene. 2007;395:170–176. doi: 10.1016/j.gene.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 85.Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 86.Calixto A, Ma C, Chalfie M. Conditional gene expression and RNAi using MEC-8-dependent splicing in C. elegans. Nat Methods. 2010;7:407–411. doi: 10.1038/nmeth.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C.elegans using SID-1. Nat Methods. 2010;7:554–559. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rual JF, Klitgord N, Achaz G. Novel insights into RNAi off-target effects using C.elegans paralogs. BMC Genomics. 2007;8:106. doi: 10.1186/1471-2164-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C.elegans germ cells into specific neuron types. Science. 2011;331:304–308. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 91.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zheng G, Ambros V, Li WH. Inhibiting miRNA in Caenorhabditis elegans using a potent and selective antisense reagent. Silence. 2010;1:9. doi: 10.1186/1758-907X-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jansen G, Hazendonk E, Thijssen KL, Plasterk RH. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat Genet. 1997;17:119–121. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- 94.Liu LX, Spoerke JM, Mulligan EL, Chen J, Reardon B, Westlund B, Sun L, Abel K, Armstrong B, Hardiman G, et al. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 1999;9:859–867. doi: 10.1101/gr.9.9.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gengyo-Ando K, Mitani S. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;269:64–69. doi: 10.1006/bbrc.2000.2260. [DOI] [PubMed] [Google Scholar]
- 96.Mitani S. Nematode, an experimental animal in the national BioResource project. Exp Anim. 2009;58:351–356. doi: 10.1538/expanim.58.351. [DOI] [PubMed] [Google Scholar]
- 97.Moerman DG, Barstead RJ. Towards a mutation in every gene in Caenorhabditis elegans. Brief Funct Genomic Proteomic. 2008;7:195–204. doi: 10.1093/bfgp/eln016. [DOI] [PubMed] [Google Scholar]
- 98.Lesa GM. Isolation of Caenorhabditis elegans gene knockouts by PCR screening of chemically mutagenized libraries. Nat Protoc. 2006;1:2231–2240. doi: 10.1038/nprot.2006.345. [DOI] [PubMed] [Google Scholar]
- 99.Barstead RJ, Moerman DG. C. elegans deletion mutant screening. Methods Mol Biol. 2006;351:51–58. doi: 10.1385/1-59745-151-7:51. [DOI] [PubMed] [Google Scholar]
- 100.Zwaal RR, Broeks A, van Meurs J, Groenen JT, Plasterk RH. Target-selected gene inactivation in Caenorhabditis elegans by using a frozen transposon insertion mutant bank. Proc Natl Acad Sci U S A. 1993;90:7431–7435. doi: 10.1073/pnas.90.16.7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rushforth AM, Saari B, Anderson P. Site-selected insertion of the transposon Tc1 into a Caenorhabditis elegans myosin light chain gene. Mol Cell Biol. 1993;13:902–910. doi: 10.1128/mcb.13.2.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barrett PL, Fleming JT, Gobel V. Targeted gene alteration in Caenorhabditis elegans by gene conversion. Nat Genet. 2004;36:1231–1237. doi: 10.1038/ng1459. [DOI] [PubMed] [Google Scholar]
- 103.Plasterk RH, Groenen JT. Targeted alterations of the Caenorhabditis elegans genome by transgene instructed DNA double strand break repair following Tc1 excision. EMBO J. 1992;11:287–290. doi: 10.1002/j.1460-2075.1992.tb05051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, Tijsterman M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol. 2008;18:900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 105.Pontier DB, Kruisselbrink E, Guryev V, Tijsterman M. Isolation of deletion alleles by G4 DNA-induced mutagenesis. Nat Methods. 2009;6:655–657. doi: 10.1038/nmeth.1362. [DOI] [PubMed] [Google Scholar]
- 106.Broverman S, MacMorris M, Blumenthal T. Alteration of Caenorhabditis elegans gene expression by targeted transformation. Proc Natl Acad Sci U S A. 1993;90:4359–4363. doi: 10.1073/pnas.90.10.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fire A. Integrative transformation of Caenorhabditis elegans. EMBO J. 1986;5:2673–2680. doi: 10.1002/j.1460-2075.1986.tb04550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Berezikov E, Bargmann CI, Plasterk RH. Homologous gene targeting in Caenorhabditis elegans by biolistic transformation. Nucleic Acids Res. 2004;32:e40. doi: 10.1093/nar/gnh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jantsch V, Pasierbek P, Mueller MM, Schweizer D, Jantsch M, Loidl J. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol Cell Biol. 2004;24:7998–8006. doi: 10.1128/MCB.24.18.7998-8006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vazquez-Manrique RP, Legg JC, Olofsson B, Ly S, Baylis HA. Improved gene targeting in C. elegans using counter-selection and Flp-mediated marker excision. Genomics. 2010;95:37–46. doi: 10.1016/j.ygeno.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 111.Gilchrist EJ, O’Neil NJ, Rose AM, Zetka MC, Haughn GW. TILLING is an effective reverse genetics technique for Caenorhabditis elegans. BMC Genomics. 2006;7:262. doi: 10.1186/1471-2164-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cuppen E, Gort E, Hazendonk E, Mudde J, van de Belt J, Nijman IJ, Guryev V, Plasterk RH. Efficient target-selected mutagenesis in Caenorhabditis elegans: toward a knockout for every gene. Genome Res. 2007;17:649–658. doi: 10.1101/gr.6080607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 114.Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci U S A. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 116.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted Genome Editing Across Species Using ZFNs and TALENs. Science. 2011 doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Robert V, Bessereau JL. Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J. 2007;26:170–183. doi: 10.1038/sj.emboj.7601463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Robert VJ, Katic I, Bessereau JL. Mos1 transposition as a tool to engineer the Caenorhabditis elegans genome by homologous recombination. Methods. 2009;49:263–269. doi: 10.1016/j.ymeth.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 119.Bazopoulou D, Tavernarakis N. The NemaGENETAG initiative: large scale transposon insertion gene-tagging in Caenorhabditis elegans. Genetica. 2009;137:39–46. doi: 10.1007/s10709-009-9361-3. [DOI] [PubMed] [Google Scholar]
- 120.Frokjaer-Jensen C, Davis MW, Hollopeter G, Taylor J, Harris TW, Nix P, Lofgren R, Prestgard-Duke M, Bastiani M, Moerman DG, et al. Targeted gene deletions in C. elegans using transposon excision. Nat Methods. 2010;7:451–453. doi: 10.1038/nmeth.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Giordano-Santini R, Milstein S, Svrzikapa N, Tu D, Johnsen R, Baillie D, Vidal M, Dupuy D. An antibiotic selection marker for nematode transgenesis. Nat Methods. 2010;7:721–723. doi: 10.1038/nmeth.1494. [DOI] [PubMed] [Google Scholar]
- 122.Semple JI, Garcia-Verdugo R, Lehner B. Rapid selection of transgenic C. elegans using antibiotic resistance. Nat Methods. 2010;7:725–727. doi: 10.1038/nmeth.1495. [DOI] [PubMed] [Google Scholar]
- 123.Kimble J, Hodgkin J, Smith T, Smith J. Suppression of an amber mutation by microinjection of suppressor tRNA in C. elegans. Nature. 1982;299:456–458. doi: 10.1038/299456a0. [DOI] [PubMed] [Google Scholar]
- 124.Stinchcomb DT, Shaw JE, Carr SH, Hirsh D. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol Cell Biol. 1985;5:3484–3496. doi: 10.1128/mcb.5.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Evans TC, editor. Transformation and microinjection. WormBook. 2006 Available at: http://www.wormbook.org.
- 126.Kelly WG, Fire A. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development. 1998;125:2451–2456. doi: 10.1242/dev.125.13.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Etchberger JF, Hobert O. Vector-free DNA constructs improve transgene expression in C. elegans. Nat Methods. 2008;5:3. doi: 10.1038/nmeth0108-3. [DOI] [PubMed] [Google Scholar]
- 128.Granato M, Schnabel H, Schnabel R. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 1994;22:1762–1763. doi: 10.1093/nar/22.9.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wilm T, Demel P, Koop HU, Schnabel H, Schnabel R. Ballistic transformation of Caenorhabditis elegans. Gene. 1999;229:31–35. doi: 10.1016/s0378-1119(99)00043-8. [DOI] [PubMed] [Google Scholar]
- 131.Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Strange K, editor. C. elegans: Methods and Applications. Vol. 351. Humana Press; 2006. [Google Scholar]
- 133.Zeiser E, Frokjaer-Jensen C, Jorgensen E, Ahringer J. MosSCI and Gateway Compatible Plasmid Toolkit for Constitutive and Inducible Expression of Transgenes in the C. elegans Germline. PLoS One. 2011;6:e20082. doi: 10.1371/journal.pone.0020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bacaj T, Shaham S. Temporal control of cell-specific transgene expression in Caenorhabditis elegans. Genetics. 2007;176:2651–2655. doi: 10.1534/genetics.107.074369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Davis MW, Morton JJ, Carroll D, Jorgensen EM. Gene activation using FLP recombinase in C. elegans. PLoS Genet. 2008;4:e1000028. doi: 10.1371/journal.pgen.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Voutev R, Hubbard EJ. A “FLP-Out” system for controlled gene expression in Caenorhabditis elegans. Genetics. 2008;180:103–119. doi: 10.1534/genetics.108.090274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Boulin T, Etchberger JF, Hobert O. Reporter gene fusions. WormBook. 2006 doi: 10.1895/wormbook.1.106.1. Available at: http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 138.Reboul J, Vaglio P, Rual JF, Lamesch P, Martinez M, Armstrong CM, Li S, Jacotot L, Bertin N, Janky R, et al. C. elegans ORFeome version 1. 1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- 139.Dupuy D, Li QR, Deplancke B, Boxem M, Hao T, Lamesch P, Sequerra R, Bosak S, Doucette-Stamm L, Hope IA, et al. A first version of the Caenorhabditis elegans Promoterome. Genome Res. 2004;14:2169–2175. doi: 10.1101/gr.2497604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tursun B, Cochella L, Carrera I, Hobert O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS One. 2009;4:e4625. doi: 10.1371/journal.pone.0004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sarov M, Schneider S, Pozniakovski A, Roguev A, Ernst S, Zhang Y, Hyman AA, Stewart AF. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat Methods. 2006;3:839–844. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- 142.Dolphin CT, Hope IA. Caenorhabditis elegans reporter fusion genes generated by seamless modification of large genomic DNA clones. Nucleic Acids Res. 2006;34:e72. doi: 10.1093/nar/gkl352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J Mol Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]